Abstract
Protein glycosylation (e.g., N-linked glycosylation) is known to play an essential role in both cellular functions and secretory pathways; however, our knowledge of in vivo N-glycosylated sites is very limited for the majority of fungal organisms including Aspergillus niger. Herein, we present the first extensive mapping of N-glycosylated sites in A. niger by applying an optimized solid phase glycopeptide enrichment protocol using hydrazide-modified magnetic beads. The enrichment protocol was initially optimized using both mouse blood plasma and A. niger secretome samples, and it was demonstrated that the protein-level enrichment protocol offered superior performance over the peptide-level protocol. The optimized protocol was then applied to profile N-glycosylated sites from both the secretome and whole cell lysates of A. niger. A total of 847 N-glycosylated sites from 330 N-glycoproteins (156 proteins from the secretome and 279 proteins from whole cells) were confidently identified by LC–MS/MS. The identified N-glycoproteins in the whole cell lysate were primarily localized in the plasma membrane, endoplasmic reticulum, Golgi apparatus, lysosome, and storage vacuoles, supporting the important role of N-glycosylation in the secretory pathways. In addition, these glycoproteins are involved in many biological processes including gene regulation, signal transduction, protein folding and assembly, protein modification, and carbohydrate metabolism. The extensive coverage of N-glycosylated sites and the observation of partial glycan occupancy on specific sites in a number of enzymes provide important initial information for functional studies of N-linked glycosylation and their biotechnological applications in A. niger.
Keywords: filamentous fungi, Aspergillus niger, N-linked glycosylation, N-glycosylated site, hydrazide chemistry, secretome
Graphical Abstract
INTRODUCTION
Filamentous fungi, such as Aspergillus niger, are well-known for their industrial applications. They are used to produce a diverse variety of products ranging from human therapeutics to specialty chemicals.1–6 Popularly known as an “industrial workhorse”, A. niger is capable of secreting large amounts of native proteins such as hydrolytic enzymes (e.g., glucoamylase) into the growth medium. Many of those hydrolytic enzymes are widely used for biomass conversion into ethanol and other renewable chemicals and advanced biofuel precursors through fermentation processes. Similar to other microbial expression systems, A. niger has also been utilized as an expression host for heterologous proteins of industrial/pharmaceutical significance.7 Some filamentous fungi are also known as pathogens in both humans8,9 and plants.10 Because of their importance, an increasing number of fungal genomes have recently been sequenced,11–15 laying the foundation for functional genomics studies, for example, protein post-translational modifications (PTMs), in filamentous fungi.
Protein glycosylation is one of the most common and structurally diverse PTMs. N- and O-linked protein glycosylation represent two of the most common types of glycosylation that occur on asparagine (N) and serine/threonine (S/T) residues, respectively. N-linked glycosylation has been implicated in a variety of biochemical and cellular processes, including protein secretion, stability and translocation, maintenance of cell structure, receptor ligand interactions and cell signaling, cell cell recognition, pathogen infection, and host defense.16–18 Despite their importance, few studies have been performed on the glycoproteome in fungal systems. Therefore, the full range of biochemical and cellular functions of N-glycosylation in these systems is still waiting to be determined.16,19,20 In this work, we present the first global profiling of N-glycosylated sites in A. niger for gaining initial functional insight on the N-glycosylated proteins.
With the recent advances in proteomic technologies, LC–MS/ MS has become the key tool for analyzing PTMs, including N-glycosylation.21,22 Similar to other PTMs, specific enrichment of the N-glycopeptides is essential in order to comprehensively characterize the often low abundance glycopeptides. A number of enrichment methods, including lectin affinity,23–26 hydrophilic interaction,27,28 and solid phase extraction using hydrazide chemistry,29–33 have been developed and applied for characterizing both N-glycoproteins and N-glycosylated sites.
For this study, we adapted the solid phase enrichment of glycopeptide (SPEG) approach originally developed by Zhang et al.34 using hydrazide chemistry, where chemically oxidized glycans are captured onto hydrazide-modified magnetic beads and N-glycopeptides are released using the deglycosylating enzyme PNGase F.29,35 The enrichment protocols were first assessed with both mouse plasma and A. niger secretome by comparing the peptide-level and protein-level enrichments. The validated protocol was subsequently applied for N-glycopeptide enrichment in both the secretome and the whole cell lysate samples of A. niger to gain an extensive coverage of N-glycosylated sites in the proteome. A total of 847 unique N-glycosylated sites from 330 N-linked glycoproteins from both the secretome and the whole cells were confidently identified in A. niger. Complex glycosylation patterns were observed in the aspects of multiple glycosylation sites per protein as well as partial glycan occupancies on specific sites. The extensive coverage of N-linked glycoproteins and their glycosylated sites provides detailed information regarding cellular localization, molecular function, and biological processes for which the identified glycoproteins are potentially involved.
EXPERIMENTAL SECTION
Materials
Mouse blood plasma (protein concentration of 42 mg/mL) was purchased from Equitech Bio, Inc. (Kerrville, TX). Hydrazide-modified magnetic beads with 1 and 5 μm diameters were obtained from Bioclone, Inc. (San Diego, CA). The magnetic bead separator was from Invitrogen (Carlsbad, CA). The glycerol free PNGase F was from New England Biolabs, Inc. (Ipswich, MA). All other reagents were from Sigma Aldrich (St. Louis, MO) unless otherwise noted.
Strain, Media, and Culture Methods
A. niger (NRRL3122), obtained from the American Type Culture Collection (Rockville, MD), was grown on potato dextrose agar (PDA) plates at 30 °C for culture maintenance and spore production. Cultures were incubated for 4 days and the spores were harvested by washing with sterile 0.8% Tween 80 (polyoxyethylenesorbitan monooleate). Spores were enumerated with a hemacytometer. Aliquots of the resulting spore suspension (1 109 spores/mL) were used to inoculate baffled-flask liquid cultures. For preparation of both the secretome and whole cell lysate, 2 106 spores/mL were inoculated and grown in 200 mL of modified minimal medium in 1 L baffled flasks in an Innova 4330 refrigerated incubator shaker (New Brunswick Scientific, NJ) at 30 °C and 200 rpm. The modified minimal medium per liter contained 18.2 g of D-sorbitol, 6 g of NaNO3, 0.52 g of KCl, 0.52 g of MgSO4(H2O)7, 1.52 g of KH2PO4, 200 μL of 1 M CaCl2, 1 g of yeast extract, and 2 mL of Tween 80 (100%). The yeast extract is the water-soluble portion of autolyzed yeast by self-digestion of the cellular constituents of yeast cells using its own enzymes, in which all proteins are broken down to small peptides (<1 kDa) and amino acids. The medium pH was adjusted to 6.5 with NaOH before autoclaving. The medium per liter also contained 50 mg of Na4EDTA, 0.512 mg of ZnSO4(H2O)7, 1.4 mg of MnCl2(H2O)4, 0.4 mg of CoCl2-(H2O)6, 0.4 mg of CuSO4(H2O)5, 0.034 mg of Na2MoO4-(H2O)2, 1.9 mg of H3BO3, 1 mg of FeSO4(H2O)7, 1 mg of biotin, 1 mg of pyridoxine, 1 mg of thiamine, 1 mg of riboflavin, 1 mg of PABA (p-aminobenzoic acid), and 1 mg of nicotinic acid. The spores were grown for the first 15 h in modified minimal medium. Then, the cultures were grown another 2 h before harvesting following the addition of 10 mM of D-xylose for the induction of glycosyl hydrolase production or water as the noninduction condition.
Protein Extraction
To prepare the secretome sample, the mycelia were separated from the culture medium by filtration through two layers of sterile miracloth (Calbiochem, San Diego, CA) and the supernatant was collected separately. The mycelia were washed 3 times with ice-cold 50 mM phosphate buffer (pH 7.0), immediately frozen in liquid nitrogen for 10 min, and stored at –80 °C until further use. The filtered medium (supernatant) was centrifuged at 15 000g at 4 °C for 10 min to remove any cell debris. The proteins in the supernatant were concentrated to ~40-fold at 4 °C and 4000g in a swinging bucket rotor with an Amicon ultra-15 centrifugal filter (MWCO 10 kDa, Millipore, Billerica, MA). Residual yeast extract (molecular weight typically <1 kDa) not consumed in cell growth was also removed by this filtering process. The concentrated secreted proteins were precipitated overnight at –20 °C using 5 vol of acetone. After centrifugation at 4 °C and 15 000g for 15 min, the precipitated proteins were washed twice with cold 80% acetone (–20 °C) and solubilized in 8 M urea buffer for 2 h at room temperature. The sample was centrifuged again at 15 000g to remove insoluble materials and the protein concentration was measured using the BCA (bicinchoninic acid) assay (Pierce, Rockford, IL).
To extract proteins from whole cells, frozen mycelia were ground into a fine powder using a chilled mortar and pestle, and then resuspended in the extraction buffer containing 100 mM Tris-HCl, 2% sodium dodecyl sulfate (SDS), 1% dithiothreitol (DTT), 10% glycerol, and 0.5% protease inhibitor cocktail (P8215, Sigma, St. Louis, MO). After incubating at room temperature for 1 h with intermittent vortexing, insoluble material was removed by centrifugation (15 000g for 15 min). The sub-sequent preparation of the total proteins was the same as the preparation for the secretome sample described above.
Protein Digestion for Global Analysis
The secretome proteins were denatured in 8 M urea for 1 h and reduced with 10 mM DTT for an additional 30 min at 37 °C. The free thiols were then alkylated with 40 mM iodoacteamide at 37 °C in the dark for 1 h. The proteins were diluted 10-fold with 50 mM NH4HCO3 buffer (pH 7.8) and digested at 37 °C for 3 h with sequencing grade modified porcine trypsin (Promega, Madison, WI) at a 1:50 trypsin to protein ratio (w/w). The digested samples were loaded on a 1-mL SPE C18 column (Supelco, Bellefonte, PA) and washed with 4 mL of 0.1% trifluoroacetic acid (TFA)/5% acetonitrile (ACN). Peptides were eluted from the SPE column with 1 mL of 0.1% TFA/ 80% ACN and lyophilized.
Protein-Level N-Glycopeptide Enrichment
The modified protein-level enrichment protocol was similar to those previously described30,31 in which hydrazide-modified magnetic beads were used instead of the original agarose bead. Protein samples were diluted with coupling buffer (100 mM sodium acetate, 150 mM NaCl, pH 5.5) to a final concentration of 1–5 mg/mL and then oxidized with 15 mM sodium periodate (NaIO4) at room temperature in the dark for 1 h. Eight molar urea was added to solubilize whole cell proteins during oxidation. The excess NaIO4 was removed after oxidation by performing buffer exchange with coupling buffer containing 0.2% CHAPS using an Amicon ultra-4 centrifugal filter (MWCO 30 kDa, Millipore, Billerica, MA). The concentrated proteins were then incubated overnight at room temperature with an appropriate amount (10 mg beads for up to 2 mg proteins) of hydrazide-modified magnetic beads prewashed twice with 1 mL of coupling buffer. Unbound proteins in the supernatant were removed using the magnetic bead separator. The beads were then washed 3 times with 2 mL of coupling buffer and 6 times with 2 mL of urea solution (8 M urea, 0.4 M NH4HCO3, pH 8.3) to remove nonspecifically bound proteins.
Glycoproteins bound on the beads were denatured and reduced by 8 M urea and 10 mM DTT for 1 h at 37 °C, and alkylated with 40 mM iodoacteamide for 1 h in the dark at 37 °C. The glycoproteins were then washed with 8 M urea and 50 mM NH4HCO3 three times each. The glycoproteins on the beads were then digested overnight at 37 °C with trypsin with a 1:100 enzyme to protein mass ratio. Nonglycopeptides were removed by washing 6 times with each of the following four solutions in the order listed: (1) 80% ACN and 0.1% TFA, (2) 8 M urea in 0.4 M NH4HCO3 with 0.1% SDS, (3) dimethyformamide (DMF), and (4) 0.1 M NH4HCO3 (pH 7.8). N-glycopeptides were released by incubating the beads with 3 μL of PNGase F (500 units/ μL) in 1 mL of 0.1 M NH4HCO3 buffer in a thermomixer (Cat # 022670107, Eppendorf, Westbury, NY) at 37 °C and 900 rpm for 5 h. Released glycopeptides were collected from the supernatant and the magnetic beads were then washed twice with 200 μL of 80% ACN and 0.1% TFA. The wash solutions were combined with the supernatant collected from the PNGase F incubation and dried down to a final volume of 100 μL using a Speed-vac concentrator.
Peptide-Level N-Glycopeptide Enrichment
The modified peptide-level enrichment protocol was similar to the one previously described.35 Digested peptides were oxidized with 15 mM NaIO4 for 1 h in the dark at room temperature. After oxidation, the excess NaIO4 was removed by SPE C-18 cleanup. Oxidized peptides eluted in 80% ACN and 0.1% TFA were incubated overnight with an appropriate amount (10 mg beads for up to 2 mg proteins) of prewashed hydrazide magnetic beads in the thermomixer at room temperature. Nonglycopeptides in the supernatant were removed by using the magnetic separator. Nonspecifically bound peptides were removed by washing 6 times with the same four solutions in the same order as described in the previous section. Glycopeptides on the beads were released and collected with the same protocol as described in the protein-level enrichment.
LC–MS/MS Analysis
Enriched glycopeptide samples were analyzed using a fully automated, custom-built, four-column capillary LC system coupled online to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) via an electrospray ionization (ESI) interface manufactured in-house. The 75 μm (inner diameter) × 65 cm fused silica capillary reversed phase column (Polymicro Technologies, Phoenix, AZ) was prepared using the slurry-packing procedure with 3 μm diameter Jupiter C18 bonded particles (Phenomenex, Torrence, CA) at 8000 psi. The mobile phases consisted of solution A (0.1% formic acid in water) and solution B (0.1% formic acid in ACN). An exponential gradient starting with 100% of mobile phase A to 60% of mobile phase B over the course of 100 min was employed with a flow-rate of ~500 nL/min over the separation column. The instrument was operated in data-dependent mode with an m/z range of 400–2000 with a resolution of 100K for full MS scans. The 10 most abundant ions from the MS analysis were selected for MS/MS analysis using a normalized collision energy setting of 35% and a dynamic exclusion duration of 1 min. The temperature of the heated capillary and the ESI voltage were 200 °C and 2.2 kV, respectively.
Data Analysis
LC–MS/MS raw data were converted into “.dta” files using Extract_MSn (version 3.0) from Bioworks Cluster 3.2 (Thermo Scientific), and the SEQUEST algorithm (version 27, revision 12) was used to independently search all the MS/MS spectra against the DOE Joint Genome Institute (JGI) A. niger v3.0 protein database that had 11 200 entries. The false discovery rate (FDR) was estimated based on the decoy-database searching methodology.36,37 The following dynamic modifications were included during the SEQUEST analysis: carbamidomethylation of cysteine, oxidation of methionine, and a PNGase F-catalyzed conversion of asparagine to aspartic acid at the site of glycan attachment (a mass increase of 0.9848 Da). MS Generating-Function (MSGF) scores were generated for each identified spectrum as described previously by computing rigorous p-values (spectral probabilities).38 The following filtering criteria were used to control the FDR to be <0.01%: (1) MSGF scores were less than 1 × 10−8 and 1 × 10−10 for fully and partially tryptic peptides, respectively, and mass measurement error less than 5 ppm; (2) for cases where nonmonoisotopic peaks were picked for fragmentation with mass measurement errors of 1 Da or more, an MSGF score less than 1 × 10−11 for both fully and partially tryptic peptides was used. The presence of at least one N-X-S/T motif, where X is any amino acids except proline, was required for all final N-glycopeptide identifications. For all glycopeptides identified with mass measurement errors greater than 1 Da, manual inspection of the raw MS spectra was performed and peptide identifications were confirmed when the large errors were found to be due to reporting the wrong monoisotopic peaks and the actual mass errors were within 5 ppm. The glycosylated sites were assigned based on the motif information within peptides. For peptides containing multiple N-X-S/T motifs, the sites of modification were also manually inspected based on MS/MS fragment ions. Uniprot and gene ontology (GO) definitions, and other terms extracted from text-based annotation files downloaded from the JGI Web site (http://genome.jgi-psf.org/) were used to annotate the identified proteins.
RESULTS
Analytical Strategy
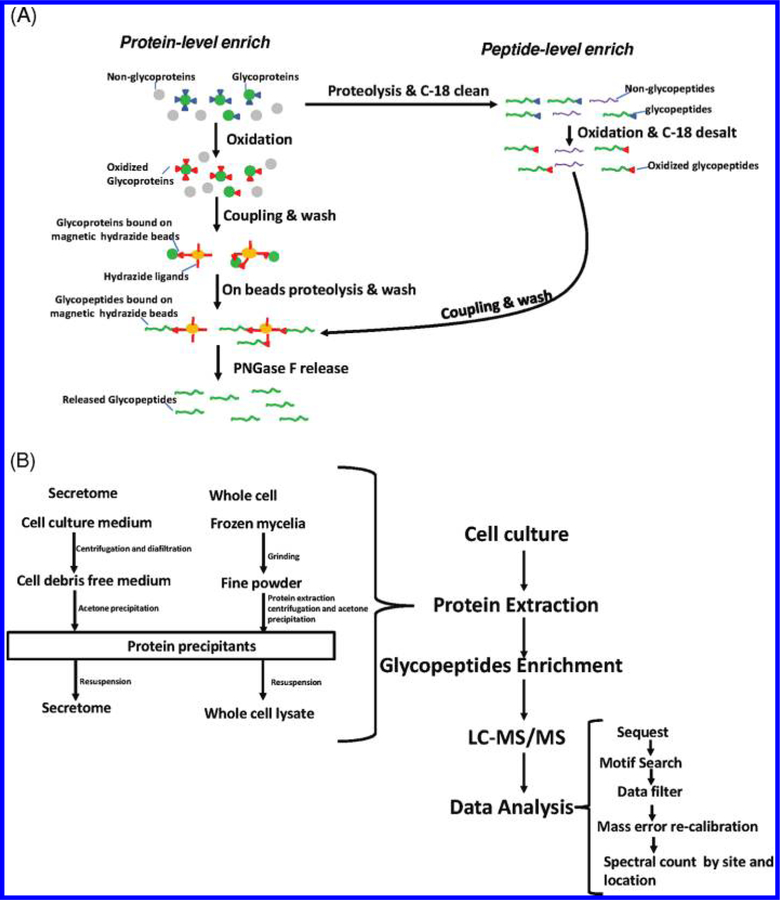
Figure 1 shows a schematic illustration of the overall enrichment and analysis of N-glycopeptides using hydrazide chemistry. Both protein- and peptide-level enrichment protocols have been applied in proteomic studies within different organisms including mammalian samples30,31,34,35 and zebrafish.39 In this method, the cis-diols of glycans are oxidized to aldehydes using NaIO4. The oxidized glycans are captured by hydrazide-modified magnetic beads through formation of covalent hydrazone bonds between the oxidized aldehyde and hydrazide groups on the beads. The N-glycopeptides are finally released from magnetic beads with PNGase F. The enzyme converts the glycan attached asparagine residues within the glycopeptides into aspartic acid, resulting in a mass increment of 0.9848 Da for each N-glycosylated site. Recently, Berven et al. performed an assessment of glycopeptide capture protocols and observed differences in performance between the protein-level and peptide-level captures using hydrazide-modified magnetic beads.31 Building upon their observations, we performed further evaluation of the enrichment protocols as shown in Figure 1A using mouse plasma and A. niger secretome samples in order to determine the optimal protocol for mapping the N-glycosylated sites in A. niger. Given the importance of N-glycosylation in cellular regulation40–42 and protein secretion,20 we analyzed the N-linked glycoproteins in both the secretome and whole cell lysate of A. niger. Figure 1B illustrates the workflow for profiling N-glycosylated sites with glycan occupancy in the A. niger secretome and the whole cell lysates. Briefly, the extracted proteins from either the secretome or the whole cell lysate were enriched by hydrazide magnetic beads, and the final enriched deglycosylated peptides were analyzed by LC–MS/MS. The confidence of the identified peptides was based on mass measurement accuracy (<5 ppm) and MSGF probability scores.
Figure 1.
(A) A schematic overview of the protein- and peptide-level enrichment strategies using hydrazide modified magnetic beads. For protein-level enrichment, glycans are first oxidized by NaIO4. The oxidized glycoproteins are directly captured by hydrazide beads followed by on-beads digestion. Following washing, the glycopeptides were released by PNGase F. For peptide-level enrichment, the proteins are initially digested with trypsin and subjected to oxidation. The oxidized glycopeptides are then captured by beads. The captured peptides are then subjected to washing and releasing steps same as protein-level enrichment. (B) A flowchart for profiling N-glycosylated sites in the A. niger secretome and the whole cells. Details on the secretome and whole cell lysate preparation, subsequent enrichment, and data analysis are provided in the Experimental Section.
Assessment of the Protein-Level and Peptide-Level Enrichment Protocols
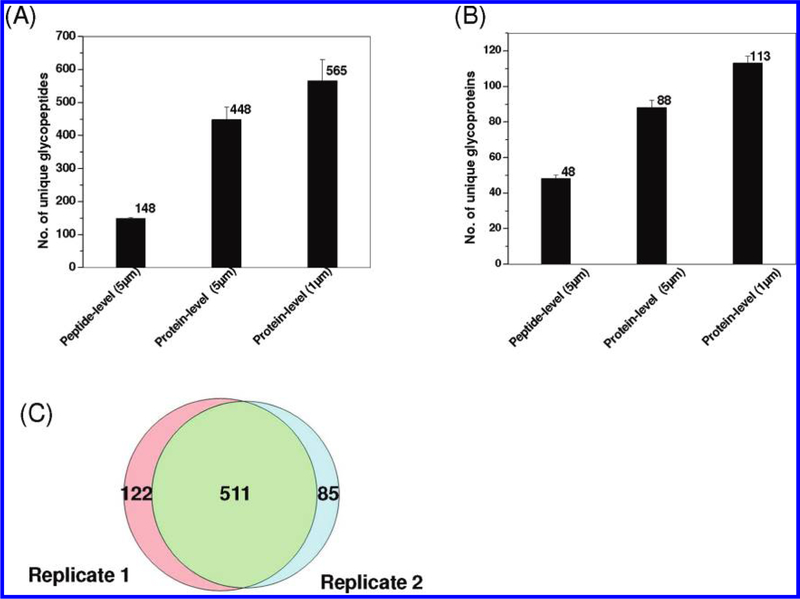
Mouse plasma was initially used to evaluate the performance of the protein- and peptide-level enrichment protocols using magnetic beads. A total of 50 μL of mouse plasma (~2 mg total proteins) was used for each experiment with ~10 mg hydrazide-modified magnetic beads for enrichment. All experiments were performed in duplicate with each enriched sample analyzed twice by LC–MS/MS to assess the overall performance of the protocols. Figure 2A,B shows the number of unique glycopeptides and glycoproteins identified from mouse plasma using both protein-and peptide-level enrichments with two different sizes of magnetic beads. While similar reproducibility was observed for all three separate experiments, the protein-level enrichment protocol resulted in nearly 3 times more unique glycopeptides than that with the peptide-level enrichment protocol when the same 5 μm beads were used. Additionally, 1 μm hydrazide magnetic beads seemed to be advantageous in providing 25% more glycopeptide identifications than the 5 μm beads. Enrichment specificity was also estimated based on the percentage of glycopeptides among all peptides identified passing the same filtering criteria. The peptide-level enrichment specificity was nearly 90%. The enrichment specificity was increased from 71% with 5 μm beads to 84% with 1 μm beads for protein-level enrichment. The better results observed for 1 μm beads are presumably due to the higher density of the functional groups (hydrazide) on 1 μm beads (200 μm ol/g beads) than that on 5 μm beads (180 μm ol/g beads), which leads to less nonspecific binding. Compared with magnetic beads based enrichment work reported by Berven et al., which identified 141 glycopeptides and 61 unique glycoproteins,31 the number of glycopeptide and glycoprotein identifications was significantly higher (565 unique glycopeptides and 113 unique glycoproteins) in our study, presumably due to better optimized washing conditions before and after on-bead proteolysis, in addition to performance differences on the LC–MS platforms used.
Figure 2.
Summary of protocol optimization results using mouse plasma. Comparisons of unique glycopeptides (A) and glycoproteins (B) identified using different enrichment conditions and the overlap of unique glycopeptides between two processing replicates (C) using the best condition (protein-level enrichment with 1 μm magnetic beads). Both 5 and 1 μm hydrazide-modified magnetic beads were applied along with the protein-level and peptide-level enrichment protocols. Each bar indicates the average number of identifications along standard deviation from two processing replicates for the given protocol. The protein-level enrichment with 1 μm magnetic beads offered the most unique N-glycopeptide and glycoprotein identifications.
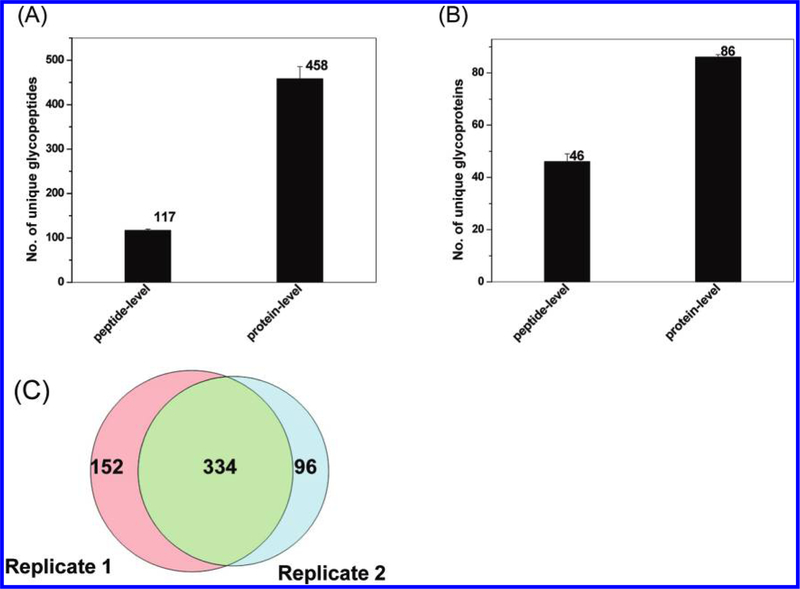
The enrichment protocols were further evaluated using A. niger secretome as shown in Figure 3. Glycopeptides from 1 mg of the secretome were enriched using 10 mg of the 1 μm hydrazide magnetic beads. The results were consistent with those observed in mouse plasma. A total of 458 unique glycopeptides were identified with 70% specificity for the protein-level enrichment, whereas only 117 unique glycopeptides were identified with nearly ~99% enrichment specificity for the peptide-level enrichment. While the peptide-level enrichment resulted in a higher specificity of glycopeptide capture consistent with the previous observation,31 the protein-level capture consistently resulted in much more glycopeptide and glycoprotein identifications with relatively high specificity. Figures 2C and 3C show the overlap of unique glycopeptides between two independently enriched glycopeptide samples (processing replicates) from mouse plasma and A. niger secretome, respectively, based on the protein-level enrichment protocol using 1 μm magnetic beads. The degree of overlap for peptide identification was ~75%, which suggests relatively good reproducibility in the overall process. On the basis of these results, the protein-level SPEG protocol with 1 μm magnetic beads offered the best performance for profiling N-linked glycopeptides.
Figure 3.
Summary of protocol optimization results using A. niger secretome. Comparisons of unique glycopeptides (A) and glycoproteins (B) identified using the peptide-level and protein-level enrichment protocols and the overlap of unique glycopeptides between two process replicates using protein-level enrichment (C). The 1 μm magnetic beads were used for the experiments. Each bar indicates the average number of identifications along standard deviation from two processing replicates for the given protocol.
N-Glycosylation Sites with Glycan Occupancy in the Secretome and Whole Cells of A. niger
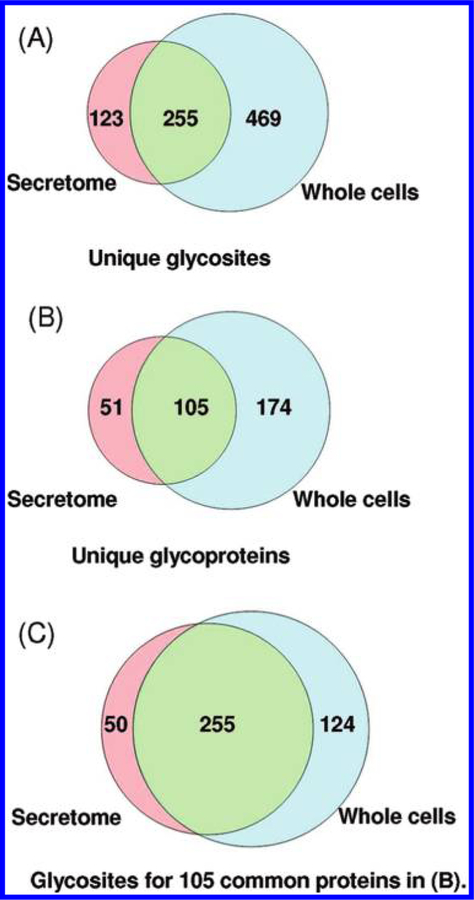
The protein-level SPEG protocol validated above was then applied to profile N-glycosylated sites in the secretome and the whole cell lysate of A. niger. Samples collected from noninduction (sorbitol) and glycosyl hydrolase induction (D-xylose) growth conditions were subjected to analyses. It should be noted that we did not attempt to provide a quantitative comparative investigation on the glycoproteome due to the limitation on the quantitative aspect. While the protocol appears to be qualitatively reproducible (Figures 2 and 3), we recognize that stable isotope labeling needs to be incorporated for quantitative comparison43 because the reproducibility of peptide identifications is significantly associated with instrument sensitivity of each MS analysis, especially when analyzing very small enriched samples. Both the secretome and the whole cell lysate samples from the two conditions were subjected to glycopeptide enrichment in duplicate to generate 8 final enriched peptide samples for LC–MS/MS analysis. Following stringent filtering (see details in Experimental Sections), a total of 7617 spectra were confidently identified as N-glyco-peptides from the combined data with the consensus N-X-S/T motif, corresponding to 1554 unique glycopeptides (Supplementary Table 1) and 847 N-glycosylated sites (Supplementary Table 2) in 330 N-glycoproteins (Supplementary Table 3). Figure 4 shows the overlap of the identified N-glycosylated sites and N-glycoproteins between the secretome and whole cells. In this study, 724 and 378 N-glycosylated sites (Figure 4A) that corresponded to 279 and 156 N-glycoproteins (Figure 4B) were identified in the whole cells and the secretome, respectively. Among them, 105 N-glycoproteins were common to both compartments, while the remaining N-glycoproteins were only detected in the whole cells or the secretome. Selected enzymes and proteins involved in various key biological processes were listed in Table 1, which included a number of glycoside hydro-lases, peptidases and proteases, oxidases, phosphatases and phospholipases, and other miscellaneous proteins. These proteins included many secretory proteins44 reported previously such as glucoamylase, 6-hydroxy-D-nicotine oxidase, aspartic acid protease, and GPI-anchored protein. Several proteins that were not previously reported in the secretome44,45 were also identified as glycoproteins, including acid trehalase and α-L-fucosidase.
Figure 4.
Overlap of glycosites and glycoproteins between the secretome and whole cells of A. niger. (A) The number of unique N-glycosites; (B) the number of unique N-glycoproteins; (C) the number of unique glycosites for 105 common proteins identified in (B). These results are the combined data obtained from all MS runs from both xylose and sorbitol growth conditions.
Table 1.
Selected Enzymes Identified As Glycoproteins in A. niger Secretome and Whole Cells
| reference ID | protein description |
no. of N-glycosylated sites |
spectral count (secretome) |
spectral count (whole cells) |
site IDs* |
|---|---|---|---|---|---|
| Glycoside Hydrolases | |||||
| 43342 | Alpha-glucosidase 2 | 3 | 3 | 10 | 279, 376, 655 |
| 52061 | Acid trehalase | 14 | 35 | 84 | 48, 138, 240, 310, 325, 621, 647, 670, 808, 832, 946, 969, 1005, 1047 |
| 53033 | 1,3-Beta-glucanosyltransferase gel3 | 5 | 32 | 151 | 38, 56,150, 168, 413 |
| 53702 | Alpha-L-fucosidase 2 | 4 | 3 | 6 | 258, 280, 659, 703 |
| 55270 | Beta-1,3-endoglucosidase | 3 | 21 | 14 | 55, 375, 598 |
| 56782 | Beta-glucosidase 1 | 9 | 15 | 33 | 211, 252, 322, 387, 442, 523, 542, 690, 712 |
| 138876 | Beta-mannosidase | 5 | 8 | 16 | 79, 483, 665, 673, 793 |
| 140567 | Alpha-amylase A | 3 | 83 | 6 | 45, 198, 218 |
| 194447 | Endoglucanase/exoglucanase | 2 | 13 | 1 | 113, 131 |
| 196122 | Probable glycosidase crf2 | 2 | 81 | 7 | 61, 266 |
| 205517 | Alpha-1,2-mannosidase | 3 | 13 | 4 | 97, 117, 184 |
| 205670 | Thermostable beta-glucosidase | 13 | 56 | 269 | 124, 148, 242, 251, 357, 390, 413, 444, 573, 576, 665, 696, 718 |
| 207264 | Alpha-galactosidase | 3 | 52 | 0 | 156, 232, 282 |
| 208214 | Beta-1,3-glucosidase | 1 | 19 | 2 | 272 |
| 213597 | Glucoamylase | 3 | 783 | 83 | 195, 206, 419 |
| Proteases | |||||
| 55493 | Tripeptidyl-peptidase 1 | 9 | 255 | 193 | 44, 95, 202, 230, 257, 363, 440, 525, 611 |
| 55665 | Tripeptidyl-peptidase 1 | 7 | 133 | 100 | 103, 128, 212, 230, 451, 462, 569 |
| 56689 | Serine protease | 8 | 28 | 41 | 55, 65, 74, 143, 260, 277, 377, 455 |
| 191956 | Aspartic acid protease | 5 | 14 | 31 | 64, 108, 123, 145, 295 |
| 211797 | Acid protease | 4 | 13 | 41 | 136, 143, 247, 163 |
| Oxidases | |||||
| 198099 | Chloroperoxidase | 3 | 20 | 28 | 131, 225, 240 |
| 45804 | 6-hydroxy-D-nicotine oxidase | 5 | 40 | 21 | 43, 57, 324, 361, 374 |
| 38973 | Mitomycin radical oxidase | 5 | 28 | 29 | 121, 261, 304, 448, 472 |
| 131352 | Laccase (copper-containing oxidase) | 6 | 37 | 24 | 23, 199, 316, 325, 413, 421 |
| Phosphatases and Phospholipases | |||||
| 57215 | Phosphate-repressible acid phosphatase | 4 | 18 | 11 | 110, 161, 295, 578 |
| 56950 | Lysophospholipase | 13 | 8 | 97 | 51, 70, 80, 94, 110, 179, 233, 250, 296, 364, 474, 526, 554 |
| 210730 | Mono- and diacylgerol lipase | 2 | 13 | 0 | 59, 150 |
| Miscellaneous Proteins | |||||
| 51794 | Ribonuclease | 2 | 45 | 16 | 97, 106 |
| 52629 | Glycolipid-anchored surface protein | 7 | 26 | 59 | 219, 255, 311,341, 359, 385,429 |
| 55604 | Aldose 1-epimerase | 6 | 16 | 44 | 88, 132, 149, 191, 290, 380 |
| 36604 | Glutamyl-tRNA(Gln) amidotransferase subunit A |
8 | 36 | 36 | 23, 46, 209, 301, 321, 393, 416, 465 |
| 50333 | 3-phytase | 3 | 4 | 14 | 162, 378, 387 |
Site ID indicates the amino acid residue site numbered starting from the N-terminus of protein sequence in the FASTA database.
More glycosylated sites were identified for the proteins from the whole cells than those from the secretome in the 105 common glycoproteins (Figure 4C). Fifty sites were only observed in secretory glycoproteins from the secretome and 124 sites only in the whole cell glycoproteins besides the 255 sites observed in glycoproteins from both the secretome and the whole cells. Figure 5 further illustrates the distribution of glycoproteins based on the number of glycosylated sites observed per protein. Among the N-glycoproteins identified in the secretome, 43% (67 out of 156) glycoproteins were identified with only one glycosylated site. The N-glycoproteins identified in the whole cells also had similar distribution, where approximately 5% of them were heavily glycosylated with more than 8 glycosylated sites. The results suggest that glycosylation patterns of N-glycoproteins may be different between secreted glycoproteins and the glycoproteins expressed from the same genes inside the cells.
Figure 5.
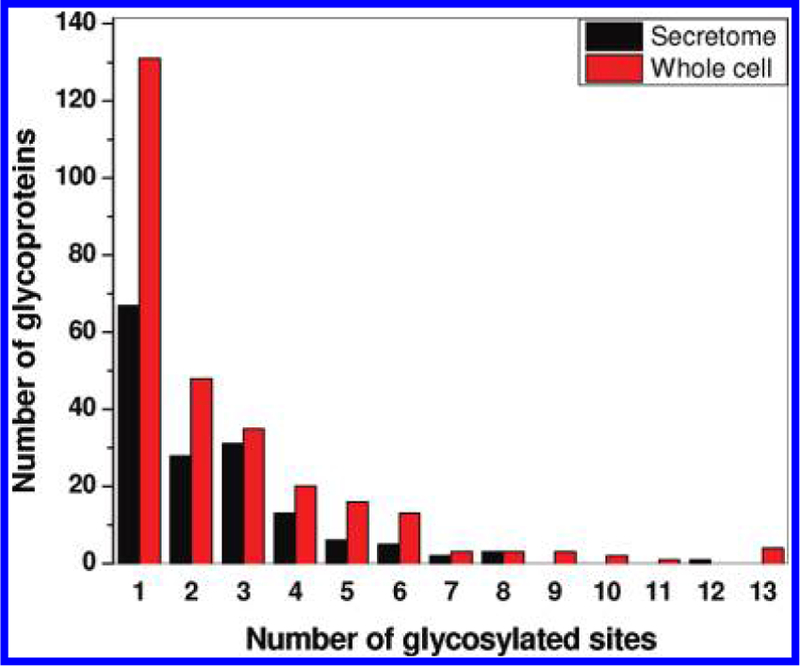
Distribution of glycoproteins identified in both the secretome and whole cells based on the number of glycosylated sites per protein. While ~40% glycoproteins were identified with only one N-glycosite, a majority of glycoproteins have multiple glycosites.
Partial Glycan Occupancy Observed on Specific N-Glycosylation Sites
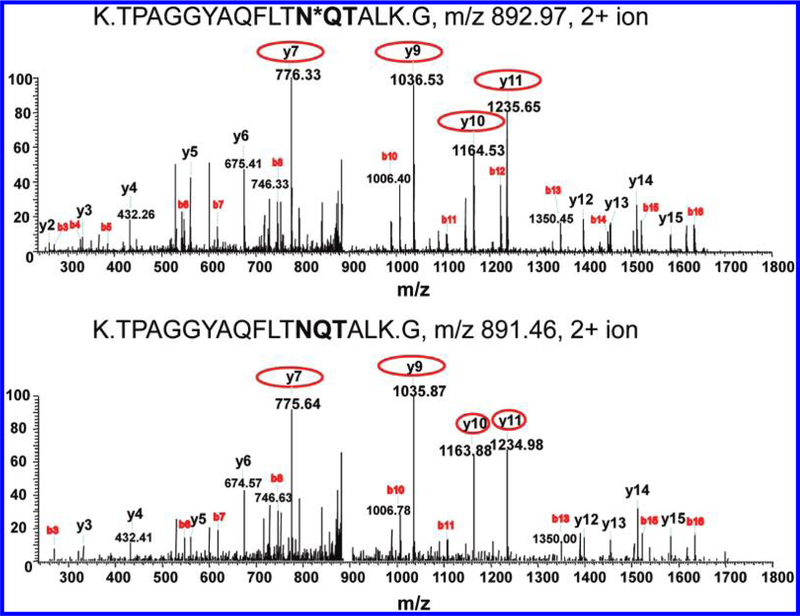
By comparing the peptide identifications from the global digests and enriched N-glycopeptide samples, we observed the evidence that some glycosylation sites were only partially glycosylated because the same peptide was observed in enriched samples as a glycosylated form as well as in global digests as a nonglycosylated form. Figure 6 shows a representative pair of MS/MS spectra for peptide K.TPAGGYAQFLTNQTALK.G from protein 36604, a putative amidase, in both glycosylated and nonglycosylated forms. The MS/MS spectra of 2+ ions with m/ z= 892.97 and m/z= 891.46 resulted in the confident identification of the fully tryptic, formerly N-glycosylated peptide (post-cleavage by PNGase F) in enriched sample (top) and nonglycosylated version in the global digest (bottom). The ~1 Da increase in the serial y-ions clearly indicates the modification on the specific site with NQT motif. Table 2 listed 18 N-glycosylation sites with partial glycan occupancy from 16 proteins observed in the secretome, which included hydrolytic enzymes (glucosidase, protease, and peptidase) and several other enzymes as well as some unknown proteins.
Figure 6.
MS/MS spectra indicative of partial glycosylation. Asterisk on N*QT indicates the N-glycosylated site with the consensus motif in bold. (A) Formerly N-glycosylated form, in which the asparagine was converted to aspartic acid at the site of glycan attachment due to PNGase F cleavage, leading to a mass increase of 0.9848 Da; (B) nonmodified form of the peptide originating from protein 36604 (glutaminyl-tRNA synthase). The N-glycosylated site is indicated by an asterisk, and the consensus motif appears in bold. The y-fragment ions highlighted by circles in the top panel show ~1 Da increase compared to those ions in the bottom panel, indicative of the modification.
Table 2.
List of Partially Glycosylated Sites Observed in the A. niger Secretome
| reference ID |
protein description |
peptide sequence |
site residue |
spectral count (unmodified) |
spectral count (glycosylated) |
|---|---|---|---|---|---|
| 170223 | Endo-1,3(4)-beta-glucanase 1 | K.KGIEN*VTTPIETNK.F | 227 | 1 | 2 |
| 176050 | Unknown protein | R.QSAN*FTDTESQK.V | 95 | 2 | 19 |
| 179912 | Cholinesterase | R.GFEHN*DTNVFLGIPFAETTAGENR.W | 42 | 3 | 2 |
| 204301 | Glycolipid-anchored surface protein 5 | K.N*FTGYGLPLFLSEYGCNTNK.R | 251 | 4 | 3 |
| 205670 | Thermostable beta-glucosidase B | K.HYAGYDIENWHN*HSR.L | 242 | 1 | 4 |
| 205670 | Thermostable beta-glucosidase B | K.N*NTNVSALLWGGYPGQSGGFALR.D | 573 | 1 | 2 |
| 208214 | Glucan 1,3-beta-glucosidase | K.GTDVFYFEAFDEVWKPN*STGDNGQSMDEK.H | 272 | 1 | 19 |
| 211032 | Tripeptidyl-peptidase 1 | K.LN*NSDLPQVISTSYGEDEQTIPVPYAR.T | 346 | 4 | 4 |
| 36048 | Unknown protein | T.N*ATTPSTFGIMSAR.S | 26 | 12 | 31 |
| 36604 | Cytokinin dehydrogenase 2 | K.TPAGGYAQFLTN*QTALK.G | 321 | 5 | 9 |
| 37529 | Unknown protein | K.SPGATTYDLCLYDVCGSGN*GSGR.V | 121 | 2 | 11 |
| 37529 | Unknown protein | R.LFTPGCDPN*TTSFDLSIFHR.Q | 63 | 3 | 1 |
| 38973 | Mitomycin radical oxidase | K.AHPDATISGAALEFTIAN*ITSDLFYEAVER.F | 304 | 2 | 12 |
| 42759 | Unknown protein | K.IGN*YSFQNVVGVGHSAGSTLTQAITTQYPK.D | 195 | 4 | 7 |
| 45021 | Pectin lyase | K.IGSN*TSIIGK.D | 95 | 2 | 2 |
| 52703 | Serine protease | K.LQFDN*SSR.S | 162 | 1 | 11 |
| 54860 | Unknown protein | K.VLTNGTGNYCASAQEDN*ATLEAMVR.A | 269 | 2 | 6 |
| 56689 | Serine protease | K.SAFGGSATN*LSNQDFVSSLSYGLDSFQSR.N | 277 | 7 | 1 |
Functional Categories of N-Glycoproteins in A. niger
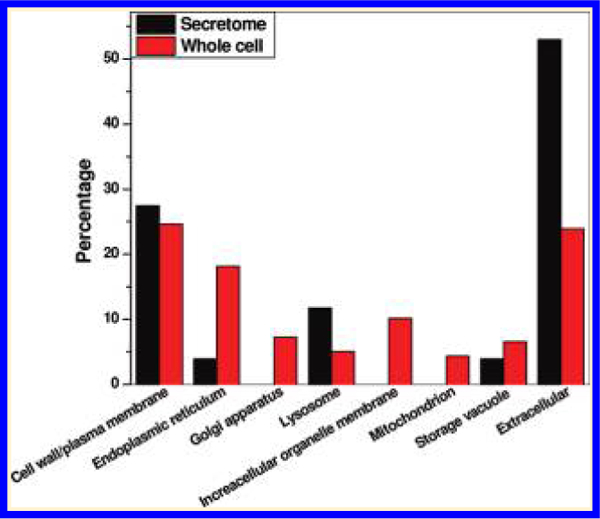
To provide an overall picture of the general functions of these identified N-linked glycoproteins, functional analyses were performed based on the Gene Ontology (GO) information in terms of cellular component, molecular function, and biological process. Figure 7A shows the distributions of glycoproteins in different cellular components. As expected, the secreted proteins were mainly annotated as in the extracellular, cell surface or plasma membrane, lysosome, and storage vacuole compartments. In addition to these components, the intracellular glycoproteins were localized in endoplasmic reticulum (ER), Golgi apparatus, the membranes of other organelles, and mitochondria.
Figure 7.
Distribution of glycoproteins identified in both the secretome and whole cells based on functional categories. (A) cellular component; (B) molecular function; and (C) biological process. The Y-axis indicates the percentage of total glycoproteins identified either in the secretome or in the whole cells that belongs to a given functional category.
Figure 7B shows the distribution of molecular functions for proteins identified in both secretome and whole cells. Peptidase, glucosidase, hydrolase, and binding were the major molecular functions of N-glycoproteins observed in both the secretome and the whole cells. Glycoproteins involved in a number of molecular functions were observed either uniquely in the cells or with significantly higher percentage in the cells than in the secretome. These categories included transferase, synthase, oxidase, nuclease, lipase, and several other categories (Figure 7B). The results indicate that many glycoproteins involved in these categories are not secreted by the cells. Figure 7C displays the distribution of glycoproteins in terms of biological process. These glycoproteins were involved in different metabolic processes, proteolysis, and catabolic processes in both the secretome and the whole cell lysate, which were consistent with the known functions of A. niger as a producer of hydrolytic enzymes. In addition to these biological processes common to the secretome and the whole cells, the data also revealed glycoproteins that were involved in a number of intracellular biological processes such as biosynthetic process, protein folding, protein modification, regulation, and response to oxidative stress. Table 3 lists some selected proteins involved in different intracellular biological processes.
Table 3.
Selected Glycoproteins Involved in Different Intracellular Biological Processes
| reference ID |
protein description |
no. of N-glycosylated sites |
spectral count (whole cells) |
site IDs | biological process and cellular component |
|---|---|---|---|---|---|
| 54378 | Cell wall alpha-1,3-glucan synthase | 13 | 82 | 1040, 825, 84, 602, 591, 220, 858, 869, 653, 378, 1022, 810, 832 | Polysaccharide biosynthetic process (cell wall) |
| 180332 | Cell wall synthesis protein | 1 | 8 | 313 | Cell wall biogenesis (cell wall) |
| 42808 | Mucin | 6 | 31 | 54, 456, 468, 532, 627, 692 | Protein complex assembly (extracellular or plasma membrane) |
| 49729 | DNAJ protein homologue | 1 | 9 | 311 | Protein folding (membrane) |
| 124206 | Uncharacterized J domain-containing protein | 1 | 7 | 42 | Protein folding (ER) |
| 212533 | Palmitoyl-protein thioesterase | 3 | 13 | 96, 231, 225 | Protein modification process (Golgi or lysosome) |
| 54756 | Vacuolar protein sorting/targeting protein PEP1 | 2 | 11 | 326, 963 | Protein targeting (Golgi) |
| 206219 | C-8 sterol isomerase | 1 | 3 | 49 | Steroid biosynthetic process (ER) |
| 52489 | Protein glycosyltransferase | 1 | 18 | 317 | Amino acid glycosylation (ER) |
| 209072 | Protein mannosyltransferase | 2 | 4 | 394, 413 | Amino acid glycosylation (ER) |
| 209577 | Glycoprotein glucosyltransferase | 2 | 15 | 57, 325 | Amino acid glycosylation (ER) |
| 53826 | Probable mannosyltransferase | 1 | 5 | 72 | Amino acid glycosylation (membrane) |
| 52517 | Putative serine carboxypeptidase | 6 | 69 | 159, 185, 317, 409, 500, 586 | Proteolysis (storage vacuole) |
| 43917 | Putative serine carboxypeptidase | 3 | 13 | 48, 128, 180 | Proteolysis (storage vacuole) |
DISCUSSION
The significance of filamentous fungi in biomass degradation and biofuel production, as well as other industrial applications, has been well recognized due to their ability in producing important enzymes and other products.5,46–52 It has been known that N-glycosylation exerts its effects in different biological functions53–55 and heterologous protein production.2 The well-known role of N-glycosylation is in the protein secretory pathway.20 Our work represents the first proteomics effort for profiling the in vivo N-glycosylated sites in a filamentous fungus, A. niger, though enzymes involved in the glycosylation pathway have been globally predicted previously with the genome sequence database.56 By applying magnetic bead-based hydrazide chemistry and high mass accuracy LC–MS/MS, N-glycopeptides along with specific glycosylated sites were identified with high confidence. The identified 847 N-glycosylated sites in 330 glycoproteins from the secretome and the whole cells represent an important resource for using A. niger to gain better understanding of N-glycosylation in cellular functions, secretory pathways, and protein production.
In this study, we demonstrated that the magnetic bead-based hydrazide chemistry offers a robust strategy for enriching N-linked glycopeptides from either the secretome or the whole cell lysate of the filamentous fungi. While this current work was only a qualitative profiling effort, we anticipate that comparative studies can be performed to study the functional changes of N-linked glycosylation in different filamentous strains with or without genetic alterations, at different growth and developmental stages, or different growth conditions by coupling with stable isotope labeling methods.43 The better performance of the protein-level enrichment protocol might be due to a better capture efficiency to the beads since glycoproteins often carry multiple glycosylated sites per protein, and once the glycan on one site is captured, the glycans on other glycosylated sites on the same protein may also be captured on the bead with a better efficiency. In addition, peptide-level enrichment involves multiple steps of SPE cleanup, which leads to significant sample loss. This method should also be applicable to many other organisms for profiling N-glycosylated sites since most glycan structures are oxidizable, with the exception being rare abnormal complex glycans that have a hindered cis-diol group. Furthermore, the captured glycopeptides are released by PNGase F, which has been reported to be effective for glycan cleavage in many organisms.29,35 However, bacterial organisms such as Campylobacter jejuni have been reported with N-glycans resistant to PNGase F due to the special structure of linking sugar.27 Plant or insect glycans that have α−1,3-fucosylated asparagine-bound GlcNAc cannot be cleaved by PNGase F, but can be cleaved by PNGase A.40
The broad coverage of N-glycosylated sites from both the secretome and the whole cell lysate provides an overview of cellular functions of N-glycoproteins. The intracellular N-glycoproteins were observed to be mainly distributed in ER, Golgi apparatus, lysosome, and storage vacuoles, which are all known to play a role along protein secretory pathways. Our identifications of N-glycosylation in a number of transferases, glycosidases, and proteins known to be involved in protein folding provide further support of the role of N-linked glycosylation in protein post-translational modifications. The role of N-glycosylation is also reflected by the identification of specific glycoproteins in various intracellular regulatory enzymes such as cell wall α−1,3-glucan synthase, cell elongation protein, transferase, phosphatase, triacylglycerol lipase, lysophospholipase, oxidase, reductase, and nuclease, all of which have a role in cellular regulation (selected glycoproteins listed in Table 1). The role of N-glycosylation in signal transduction pathways and gene regulation was supported by the identification of N-glycosylation in the transcriptional regulator, zinc-binding protein, Gβ protein, and RING finger proteins. Specific intracellular glycoproteins in ER and Golgi apparatus (see Table 3 and Supplemental Table 3) involved in protein folding and protein modification,57–59 two critical processes in secretory pathways, were also identified. These data support the importance of N-glycosylation for the functions of intracellular organelles such as ER and Golgi in protein quality control and trafficking. The identified N-glycosylated sites from specific proteins involved in different biological processes or molecular functions provide a basis for future systematic studies of the cellular growth and development under different culture environments and biofuels and protein production in filamentous fungi.
Interestingly, many glycoproteins contain multiple N-glycosylated sites, in agreement with one of the proposed N-linked glycan functions: promotion of protein folding and stabilization.57–59 One example is protein 205670, a thermally stable β-glucosidase, which was found to be heavily glycosylated in both the secretome (12 sites) and the whole cell (11 sites). N-linked glycans are also known to serve a structural role in the cell wall.60,61 Some cell wall proteins were, in fact, found to be heavily glycosylated. For example, proteins 54378 and 56240 are cell wall synthesis protein and serine-rich adhesion protein, respectively.62 Both proteins had 13 N-glycosylated sites and were only present in the whole cell lysate. Another observation was that the same glycoprotein exhibits different glycosylation patterns between the whole cell lysate and the secretome (Figures 4C and 5). Secreted glycoproteins are known to be possible proteolytic fragments of the precursor proteins expressed in the cells.63,64 The pattern of glycosylated sites may also be different between the precursor proteins in the cells and the mature secreted proteins. Just as an example, protein ID 56950, a lysophospholipase protein, was observed with 13 glycosylated sites in whole cells, while only 4 sites were identified in the secretome.
Additionally, we observed evidence of partial glycan occupancy for a number of glycosylation sites. The reason for partial glycosylation and its function in biological processes is not clear. Partial glycosylation in those enzymes suggests that their function/activities may be regulated by the extent of glycosylation. Functions of partial glycosylation in the selected proteins of mammalian systems were previously investigated.24,65,66 For example, the function of voltage-gated potassium channel, Kv3.1 glycoprotein, in neuroblastoma cells is regulated by the extent of glycosylation at the sites N220Q and N229Q.67 Picard et al.66 observed that the partial glycosylation of antithrombin III asperagine-135 is caused by the serine in the third position of its N-linked glycosylation consensus sequence, which is responsible for production of the β-antithrombine III isoform. Such partial glycosylation may constitute a new paradigm for post-translational regulation of gene/protein functions. The observation of multiple glycosylation sites per protein as well as the evidence of partial glycosylation suggests that glycosylation patterns are potentially dynamic and therefore the complexity of protein N-glycosylation may have much greater significance than anticipated. Understanding the endogenous and exogenous factors governing this process and its roles in protein production and enzymatic functions will benefit applications aimed at industrial production of proteins and chemicals from fungi.
In summary, our data from protocol optimization show that the protein-level enrichment protocol using 1 μm beads provides improved coverage of N-linked glycopeptides, and a basis for future studies on the role of N-linked glycosylation in, for example, cellular regulation in order to achieve improved industrial production. The method is broadly applicable for global profiling of N-glycosylated sites in different organisms. In the application to A. niger, the coverage of 847 glycosylated sites from 330 glycoproteins provided important insights into the functions of N-linked glycoproteins involved in this industrially important fungal model organism. The data also revealed complex glycosylation patterns in terms of multiple glycosylated sites per protein as well as patterns of partial glycosylation for different glycosylation sites. The biological implications of partial and complex glycosylation patterns warrant further investigations.
Supplementary Material
ACKNOWLEDGMENT
Portions of this work were supported by DOE Early Career Research Award and National Institutes of Health grant RR18522. Upstream cell culture was conducted in Fungal Biotechnology Laboratory, which was supported by the DOE Office of the Biomass Program. Proteomics experiments were performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) Office of Biological and Environmental Research national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus. PNNL is multiprogram national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO 1830.
ABBREVIATIONS:
- PTM
post-translational modification
- SPEG
solid phase enrichment of glycopeptides
- MSGF
MS generating function
- GO
gene ontology
- ER
endoplasmic reticulum
Footnotes
Supporting Information
A complete list of identified glycopeptides, glycosylated sites, and glycoproteins along with corresponding spectral count information is available in a Microsoft Excel worksheet. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Nevalainen KM; Te’o VS; Bergquist PL Heterologous protein expression in filamentous fungi. Trends Biotechnol 2005, 23 (9), 468–474. [DOI] [PubMed] [Google Scholar]
- (2).Gerngross TU Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol 2004, 22 (11), 1409–4414. [DOI] [PubMed] [Google Scholar]
- (3).Punt PJ; van Biezen N; Conesa A; Albers A; Mangnus J; van den Hondel C Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol 2002, 20 (5), 200–206. [DOI] [PubMed] [Google Scholar]
- (4).Schuster E; Dunn-Coleman N; Frisvad JC; Van Dijck PW On the safety of Aspergillus niger—A review. Appl. Microbiol. Biotechnol 2002, 59 (4–5), 426–435. [DOI] [PubMed] [Google Scholar]
- (5).Magnuson JK; Lasure LL Organic acid production by filamentous fungi. Adv. Fungal Biotechnol. Ind., Agric., Med 2004, 307–340.
- (6).Sauer M; Porro D; Mattanovich D; Branduardi P Microbial production of organic acids: Expanding the markets. Trends Biotechnol 2008, 26 (2), 100–108. [DOI] [PubMed] [Google Scholar]
- (7).Devchand M; Gwynne DI Expression of heterologous proteins in Aspergillus. J. Biotechnol 1991, 17 (1), 3–9. [DOI] [PubMed] [Google Scholar]
- (8).Segal BH; Barnhart LA; Anderson VL; Walsh TJ; Malech HL; Holland SM Posaconazole as salvage therapy in patients with chronic granulomatous disease and invasive filamentous fungal infection. Clin. Infect. Dis 2005, 40 (11), 1684. [DOI] [PubMed] [Google Scholar]
- (9).Cutler SJ; Fooks AR; van der Poel WHM Public health threat of new, reemerging, and neglected zoonoses in the industrialized world. Emerging Infect. Dis 2010, 16 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Maor R; Shirasu K The arms race continues: battle strategies between plants and fungal pathogens. Curr. Opin. Microbiol 2005, 8 (4), 399–404. [DOI] [PubMed] [Google Scholar]
- (11).Galagan JE; Calvo SE; Cuomo C; Ma LJ; Wortman JR; Batzoglou S; Lee SI; Basturkmen M; Spevak CC; Clutterbuck J; Kapitonov V; Jurka J; Scazzocchio C; Farman M; Butler J; Purcell S; Harris S; Braus GH; Draht O; Busch S; D’Enfert C; Bouchier C; Goldman GH; Bell-Pedersen D; Griffiths-Jones S; Doonan JH; Yu J; Vienken K; Pain A; Freitag M; Selker EU; Archer DB; Penalva MA; Oakley BR; Momany M; Tanaka T; Kumagai T; Asai K; Machida M; Nierman WC; Denning DW; Caddick M; Hynes M; Paoletti M; Fischer R; Miller B; Dyer P; Sachs MS; Osmani SA; Birren BW Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438 (7071), 1105–1115. [DOI] [PubMed] [Google Scholar]
- (12).Machida M; Asai K; Sano M; Tanaka T; Kumagai T; Terai G; Kusumoto K; Arima T; Akita O; Kashiwagi Y; Abe K; Gomi K; Horiuchi H; Kitamoto K; Kobayashi T; Takeuchi M; Denning DW; Galagan JE; Nierman WC; Yu J; Archer DB; Bennett JW; Bhatnagar D; Cleveland TE; Fedorova ND; Gotoh O; Horikawa H; Hosoyama A; Ichinomiya M; Igarashi R; Iwashita K; Juvvadi PR; Kato M; Kato Y; Kin T; Kokubun A; Maeda H; Maeyama N; Maruyama J; Nagasaki H; Nakajima T; Oda K; Okada K; Paulsen I; Sakamoto K; Sawano T; Takahashi M; Takase K; Terabayashi Y; Wortman JR; Yamada O; Yamagata Y; Anazawa H; Hata Y; Koide Y; Komori T; Koyama Y; Minetoki T; Suharnan S; Tanaka A; Isono K; Kuhara S; Ogasawara N; Kikuchi H Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438 (7071), 1157–1161. [DOI] [PubMed] [Google Scholar]
- (13).Dean RA; Talbot NJ; Ebbole DJ; Farman ML; Mitchell TK; Orbach MJ; Thon M; Kulkarni R; Xu JR; Pan H; Read ND; Lee YH; Carbone I; Brown D; Oh YY; Donofrio N; Jeong JS; Soanes DM; Djonovic S; Kolomiets E; Rehmeyer C; Li W; Harding M; Kim S; Lebrun MH; Bohnert H; Coughlan S; Butler J; Calvo S; Ma LJ; Nicol R; Purcell S; Nusbaum C; Galagan JE; Birren BW The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434 (7036), 980–986. [DOI] [PubMed] [Google Scholar]
- (14).Galagan JE; Calvo SE; Borkovich KA; Selker EU; Read ND; Jaffe D; FitzHugh W; Ma LJ; Smirnov S; Purcell S; Rehman B; Elkins T; Engels R; Wang S; Nielsen CB; Butler J; Endrizzi M; Qui D; Ianakiev P; Bell-Pedersen D; Nelson MA; Werner-Washburne M; Selitrennikoff CP; Kinsey JA; Braun EL; Zelter A; Schulte U; Kothe GO; Jedd G; Mewes W; Staben C; Marcotte E; Greenberg D; Roy A; Foley K; Naylor J; Stange-Thomann N; Barrett R; Gnerre S; Kamal M; Kamvysselis M; Mauceli E; Bielke C; Rudd S; Frishman D; Krystofova S; Rasmussen C; Metzenberg RL; Perkins DD; Kroken S; Cogoni C; Macino G; Catcheside D; Li W; Pratt RJ; Osmani SA; DeSouza CP; Glass L; Orbach MJ; Berglund JA; Voelker R; Yarden O; Plamann M; Seiler S; Dunlap J; Radford A; Aramayo R; Natvig DO; Alex LA; Mannhaupt G; Ebbole DJ; Freitag M; Paulsen I; Sachs MS; Lander ES; Nusbaum C; Birren B The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422 (6934), 859–868. [DOI] [PubMed] [Google Scholar]
- (15).Katinka MD; Duprat S; Cornillot E; Metenier G; Thomarat F; Prensier G; Barbe V; Peyretaillade E; Brottier P; Wincker P; Delbac F; El Alaoui H; Peyret P; Saurin W; Gouy M; Weissenbach J; Vivares CP Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414 (6862), 450–453. [DOI] [PubMed] [Google Scholar]
- (16).De Groot PW; Ram AF; Klis FM Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol 2005, 42 (8), 657–675. [DOI] [PubMed] [Google Scholar]
- (17).Helenius A; Aebi M Intracellular functions of N-linked glycans. Science 2001, 291 (5512), 2364–2369. [DOI] [PubMed] [Google Scholar]
- (18).Meynial-Salles I; Combes D In vitro glycosylation of proteins: an enzymatic approach. J. Biotechnol 1996, 46 (1), 1–14. [DOI] [PubMed] [Google Scholar]
- (19).Mechref Y; Novotny MV Structural investigations of glycoconjugates at high sensitivity. Chem. Rev 2002, 102 (2), 321–369. [DOI] [PubMed] [Google Scholar]
- (20).Roth J Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem. Rev 2002, 102 (2), 285–303. [DOI] [PubMed] [Google Scholar]
- (21).Witze ES; Old WM; Resing KA; Ahn NG Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 2007, 4 (10), 798–806. [DOI] [PubMed] [Google Scholar]
- (22).Pan S; Chen R; Aebersold R; Brentnall TA Mass spectrometry based glycoproteomics—From a proteomics perspective. Mol. Cell. Proteomics 2011, 10 (1), R110 003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zielinska DF; Gnad F; Wisniewski JR; Mann M Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141 (5), 897–907. [DOI] [PubMed] [Google Scholar]
- (24).Drake PM; Schilling B; Niles RK; Braten M; Johansen E; Liu H; Lerch M; Sorensen DJ; Li B; Allen S; Hall SC; Witkowska HE; Regnier FE; Gibson BW; Fisher SJ A lectin affinity workflow targeting glycosite-specific, cancer-related carbohydrate structures in trypsin-digested human plasma. Anal. Biochem 2011, 408 (1), 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Zeng Z; Hincapie M; Pitteri SJ; Hanash S; Schalkwijk J; Hogan JM; Wang H; Hancock WS; Proteomics A Platform combining depletion, multi-lectin affinity chromatography (M-LAC), and isoelectric focusing to study the breast cancer proteome. Anal. Chem 2011, 83 (12), 4845–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kullolli M; Hancock WS; Hincapie M Automated platform for fractionation of human plasma glycoproteome in clinical proteomics. Anal. Chem 2010, 82 (1), 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Scott NE; Parker BL; Connolly AM; Paulech J; Edwards AV; Crossett B; Falconer L; Kolarich D; Djordjevic SP; Hojrup P; Packer NH; Larsen MR; Cordwell SJ Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 2011, 10 (2), M000031–MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Calvano CD; Zambonin CG; Jensen ON Assessment of lectin and HILIC based enrichment protocols for characterization of serum glycoproteins by mass spectrometry. J. Proteomics 2008, 71 (3), 304–317. [DOI] [PubMed] [Google Scholar]
- (29).Zhang H; Li XJ; Martin DB; Aebersold R Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 2003, 21 (6), 660–666. [DOI] [PubMed] [Google Scholar]
- (30).Liu T; Qian WJ; Gritsenko MA; Camp DG II; Monroe ME; Moore RJ; Smith RD Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res 2005, 4 (6), 2070–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Berven FS; Ahmad R; Clauser KR; Carr SA Optimizing performance of glycopeptide capture for plasma proteomics. J. Proteome Res 2010, 9 (4), 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stahl-Zeng J; Lange V; Ossola R; Eckhardt K; Krek W; Aebersold R; Domon B High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 2007, 6 (10), 1809–1817. [DOI] [PubMed] [Google Scholar]
- (33).Sun B; Ranish JA; Utleg AG; White JT; Yan X; Lin B; Hood L Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol. Cell. Proteomics 2007, 6 (1), 141–149. [DOI] [PubMed] [Google Scholar]
- (34).Zhang H; Li X.-j.; Martin DB; Aerbersold R Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol 2003, 21 (6), 660–665. [DOI] [PubMed] [Google Scholar]
- (35).Tian Y; Zhou Y; Elliott S; Aebersold R; Zhang H Solid-phase extraction of N-linked glycopeptides. Nat. Protoc 2007, 2 (2), 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Elias JE; Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4 (3), 207–214. [DOI] [PubMed] [Google Scholar]
- (37).Qian WJ; Liu T; Monroe ME; Strittmatter EF; Jacobs JM; Kangas LJ; Petritis K; Camp DG; Smith RD Probability-Based Evaluation of Peptide and Protein Identifications from Tandem Mass Spectrometry and SEQUEST Analysis: The Human Proteome. J. Proteome Res 2005, 4, 53–62. [DOI] [PubMed] [Google Scholar]
- (38).Kim S; Gupta N; Pevzner PA Spectral probabilities and generating functions of tandem mass spectra: A strike against decoy databases. J. Proteome Res 2008, 7 (8), 3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Baycin-Hizal D; Tian Y; Akan I; Jacobson E; Clark D; Wu A; Jampol R; Palter K; Betenbaugh M; Zhang H GlycoFish: a database of zebrafish N-linked glycoproteins identified using SPEG method coupled with LC/MS. Anal. Chem 2011, 83 (13), 5296–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kukuruzinska MA; Lennon K Growth-related coordinate regulation of the early N-glycosylation genes in yeast. Glycobiology 1994, 4 (4), 437–443. [DOI] [PubMed] [Google Scholar]
- (41).Lennon K; Bird A; Kukuruzinska MA Deregulation of the first N-glycosylation gene, ALG7, perturbs the expression of G1 cyclins and cell cycle arrest in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun 1997, 237 (3), 562–565. [DOI] [PubMed] [Google Scholar]
- (42).Motteram J; Lovegrove A; Pirie E; Marsh J; Devonshire J; van de Meene A; Hammond-Kosack K; Rudd JJ Aberrant protein N-glycosylation impacts upon infection-related growth transitions of the haploid plant-pathogenic fungus Mycosphaerella graminicola. Mol. Microbiol 2011, 81 (2), 415–33. [DOI] [PubMed] [Google Scholar]
- (43).Tian Y; Bova GS; Zhang H Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal. Chem 2011, 83 (18), 7013–7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tsang A; Butler G; Powlowski J; Panisko EA; Baker SE Analytical and computational approaches to define the Aspergillus niger secretome. Fungal Genet. Biol 2009, 46 (Suppl. 1), S153–S160. [DOI] [PubMed] [Google Scholar]
- (45).Lu X; Sun J; Nimtz M; Wissing J; Zeng AP; Rinas U The intra- and extracellular proteome of Aspergillus niger growing on defined medium with xylose or maltose as carbon substrate. Microb. Cell Fact 2010, 9, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Sánchez C Lignocellulosic residues: biodegradation and bio-conversion by fungi. Biotechnol. Adv 2009, 27 (2), 185–194. [DOI] [PubMed] [Google Scholar]
- (47).Singh P; Sulaiman O; Hashim R; Rupani P; Peng LC Biopulping of lignocellulosic material using different fungal species: A review. Rev. Environ. Sci. Biotechnol 2010, 9 (2), 141–151. [Google Scholar]
- (48).Karaffa L; Kubicek CP Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl. Microbiol. Biotechnol 2003, 61 (3), 189–196. [DOI] [PubMed] [Google Scholar]
- (49).Hoffmeister D; Keller NP Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep 2007, 24 (2), 393–416. [DOI] [PubMed] [Google Scholar]
- (50).Archer D; Connerton I; MacKenzie D Filamentous fungi for production of food additives and processing aids. Food Biotechnol 2008, 99–147. [DOI] [PubMed]
- (51).Yang B; Dai Z; Ding SY; Wyman CE Enzymatic hydrolysis of cellulosic biomass. Biofuels 2011, 2 (4), 421–450. [Google Scholar]
- (52).Manzoni M; Rollini M Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Appl. Microbiol. Biotechnol 2002, 58 (5), 555–564. [DOI] [PubMed] [Google Scholar]
- (53).Haltiwanger RS; Lowe JB Role of glycosylation in development. Annu. Rev. Biochem 2004, 73 (1), 491–537. [DOI] [PubMed] [Google Scholar]
- (54).Cabral CM; Liu Y; Sifers RN Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem. Sci 2001, 26 (10), 619–624. [DOI] [PubMed] [Google Scholar]
- (55).Helenius A Intracellular functions of N-linked glycans. Science 2001, 291 (5512), 2364. [DOI] [PubMed] [Google Scholar]
- (56).Deshpande N; Wilkins MR; Packer N; Nevalainen H Protein glycosylation pathways in filamentous fungi. Glycobiology 2008, 18 (8), 626–637. [DOI] [PubMed] [Google Scholar]
- (57).Varki A Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 1993, 3 (2), 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Imperiali B; Ottesen JJ Uniquely folded mini-protein motifs. J. Pept. Res 1999, 54 (3), 177–184. [DOI] [PubMed] [Google Scholar]
- (59).Kern G; Kern D; Jaenicke R; Seckler R Kinetics of folding and association of differently glycosylated variants of invertase from Saccharomyces cerevisiae. Protein Sci 1993, 2 (11), 1862–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Schaffer C; Kahlig H; Christian R; Schulz G; Zayni S; Messner P The diacetamidodideoxyuronic-acid-containing glycan chain of Bacillus stearothermophilus NRS 2004/3a represents the secondary cell-wall polymer of wild-type B. stearothermophilus strains. Microbiology 1999, 145 (Pt 7), 1575–1583. [DOI] [PubMed] [Google Scholar]
- (61).Schaffer C; Messner P The structure of secondary cell wall polymers: how Gram-positive bacteria stick their cell walls together. Microbiology 2005, 151 (Pt 3), 643–651. [DOI] [PubMed] [Google Scholar]
- (62).Siboo IR; Chambers HF; Sullam PM Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets. Infect. Immun 2005, 73 (4), 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Shapiro RI; Wen D; Levesque M; Hronowski X; Gill A; Garber EA; Galdes A; Strauch KL; Taylor FR Expression of Sonic hedgehog-Fc fusion protein in Pichia pastoris. Identification and control of post-translational, chemical, and proteolytic modifications. Protein Expression Purif 2003, 29 (2), 272–283. [DOI] [PubMed] [Google Scholar]
- (64).Doucet A; Butler GS; Rodriguez D; Prudova A; Overall CM Metadegradomics: Toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol. Cell. Proteomics 2008, 7 (10), 1925–1951. [DOI] [PubMed] [Google Scholar]
- (65).Nicolaes GAF; Villoutreix BO; Dahlbäck B Partial glycosylation of Asn2181 in human factor V as a cause of molecular and functional heterogeneity. Modulation of glycosylation efficiency by mutagenesis of the consensus sequence for N-linked glycosylation. Biochemistry 1999, 38 (41), 13584–13591. [DOI] [PubMed] [Google Scholar]
- (66).Picard V; Ersdal-Badju E; Bock SC Partial glycosylation of antithrombin III asparagine-135 is caused by the serine in the third position of its N-glycosylation consensus sequence and is responsible for production of the β-antithrombin III isoform with enhanced heparin affinity. Biochemistry 1995, 34 (26), 8433–8440. [DOI] [PubMed] [Google Scholar]
- (67).Hall MK; Cartwright TA; Fleming CM; Schwalbe RA Importance of glycosylation on function of a potassium channel in neuroblastoma cells. PLoS One 2011, 6 (4), e19317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.