Abstract
Background
The small ubiquitin-like modifier 1 (SUMO1) and small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3) are long noncoding RNAs (lncRNAs). The prognostic significance of SUMO1 and SUMO1P3 expression in non-small cell lung cancer (NSCLC) remains unclear. This study aimed to use clinical, genetic, and survival data from the Cancer Genome Atlas (TCGA), to analyze the prognostic significance of SUMO1 and SUMO1P3 expression in the two main subtypes of NSCLC, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).
Material/Methods
Data were acquired from TCGA and in silico survival analysis was performed. SUMO1 and SUMO1P3 expression were compared between patients with LUAD and LUSC. Patient outcome was assessed as complete remission (CR), partial remission (PR), stable disease (SD), and progressive disease (PD). Recurrence-free survival (RFS) was defined as the survival time from primary surgery to the time of locoregional or distant recurrence of lung cancer.
Results
SUMO1P3 was significantly increased in LUSC and LUAD tissues compared with adjacent normal lung tissue and was significantly co-expressed with SUMO1. SUMO1P3 expression was significantly increased in patients with LUAD but not LUSC with reduced RFS after primary or follow-up treatment. Although patients with LUAD who had high SUMO1 or SUMO1P3 expression had reduced RFS compared with low expression groups, univariate and multivariate analysis showed that only SUMO1P3 expression was independently associated reduced RFS (HR, 1.418; 95% CI, 1.041–1.930; p=0.027).
Conclusions
SUMO1P3 expression was an independent indicator of reduced RFS in patients with LUAD.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; Prognosis; SUMO-1 Protein
Background
Worldwide, lung cancer is a common malignancy that has a high annual mortality. In the US in 2018, it was estimated that there were 234,030 new cases of lung cancer and 154,050 deaths due to lung cancer [1]. About 85% of cases of lung cancer are non-small cell lung carcinoma (NSCLC) include the two main histological subtypes of lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). These two subtypes of NSCLC vary significantly in their molecular biology and clinical features [2–4]. The high frequency of epidermal growth factor (EGFR) gene mutations and ALK-EML4 translocations have provided new targeted therapy for patients with LUAD, which are less effective for patients with LUSC [5]. In terms of survival, patients with LUAD have reduced overall survival (OS) compared with patients with LUSC [6]. The molecular differences between the two subtypes of NSCLC indicate that they may have different prognostic biomarkers.
Long noncoding RNAs (lncRNAs) contain more than 200 nucleotides and have no protein encoding capability. Small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3) is a lncRNA that is upregulated in several cancers and exerts its oncogenic effects via multiple pathways [7–11]. For example, SUMO1P3 enhances the progression of breast cancer by negatively regulating the expression of miR-320a [10], and promotes abnormal growth and invasion of pancreatic cancer cells by facilitating the epithelial-mesenchymal (EMT) transition [11]. Also, SUMO1P3 expression may have a role as a prognostic marker in cancer. In bladder cancer, the expression of SUMO1P3 is associated with advanced TNM stage and increased histological grade [7]. The expression of SUMO1P3 shows has been shown to have a prognostic role in predicting overall survival (OS) in patients with pancreatic cancer [11].
Pseudogenes can be functionally linked with protein-coding genes [12,13]. A recent study showed that the small ubiquitin-like modifier 1 (SUMO1) gene mediated a post-translational modification process called SUMOylation, which was also upregulated in NSCLC [14]. SUMO1 dysregulation might be associated with chemosensitivity of NSCLC cells [15]. Also, SUMO1 overexpression can promote changes in the cell cycle and the progression and invasion of NSCLC cells by enhancing NF-κB expression [14]. However, the expression profiles of SUMO1P3 and the prognostic value of SUMO1P3 and SUMO1 in NSCLC remain to be characterized.
Therefore, this study aimed to use clinical, genetic, and survival data from the Cancer Genome Atlas (TCGA), to analyze the prognostic significance of SUMO1 and SUMO1P3 expression in the two main subtypes of NSCLC, LUAD and LUSC. Patient outcome was assessed as complete remission (CR), partial remission (PR), stable disease (SD), progressive disease (PD), overall survival (OS), and recurrence-free survival (RFS).
Material and Methods
Data acquisition and analysis
Three levels of data included clinical, genetic, and survival data were obtained from the Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) databases and were obtained using the University of California Santa Cruz (UCSC) Xena Functional Genomics Explorer (https://xenabrowser.net/) [16]. The data from cases of primary lung cancer tissue and the adjacent normal lung tissue were extracted for analysis. Data from patients who received neoadjuvant chemotherapy for LUSC (N=9) and LUAD (N=3) were excluded. Cases without RNA-Seq data were also excluded for LUSC (N=3) and LUAD (N=6). After the screening, 494 cases of primary LUSC and 511 cases of primary LUAD were included for analysis.
The following data were extracted from the included cases, including RNA-Seq data of gene expression, gender, age at diagnosis, history of tobacco smoking, tumor stage, grade, primary outcome following treatment, follow-up outcome, the presence of residual tumor, and canonical mutation in KRAS/EGFR/ALK, recurrence status, recurrence-free survival (RFS) in days, and overall survival (OS) in days. The longest follow-up was approximately 20 years. Primary treatment and follow-up outcome following treatment were defined as complete remission (CR), partial remission (PR), stable disease (SD), and progressive disease (PD). The CR, PR, and SD cases were all considered to be without disease progression. OS was defined as the survival time from the date of primary surgery to the time of death. Recurrence-free survival (RFS) was defined as the survival time from the date of primary surgery to the time of locoregional or distant recurrence of lung cancer. The differences in SUMO1 and SUMO1P3 expression between patients with controlled disease, CR, PR, and SD, and uncontrolled PD were compared.
Statistical analysis
Data integration and statistical analysis were performed using GraphPad Prism version 7.04 (GraphPad Software, La Jolla, CA, USA) and SPSS version 25.0 software (IBM, Chicago, IL, USA). One-way analysis of variance (ANOVA) with post hoc Tukey’s multiple comparisons and Welch’s unequal variance t-test were performed for multiple group comparison and comparison between two groups, respectively. Kaplan-Meier survival curves were used, and the Youden index compared SUMO1 and SUMO1P3 expression with receiver operating characteristic (ROC) analysis for prognostic significance for recurrence or mortality. The log-rank test was performed to compare the difference between the survival curves. The independent prognostic value of SUMO1 and SUMO1P3 expression was evaluated using results from univariate and multivariate Cox regression models. SUMO1 and SUMO1P3 expression were treated as continuous variables in the model. A P-value <0.05 was considered to be statistically significant.
Results
Both SUMO1 and SUMO1P3 were significantly upregulated in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) tumor tissues compared with adjacent normal tissues
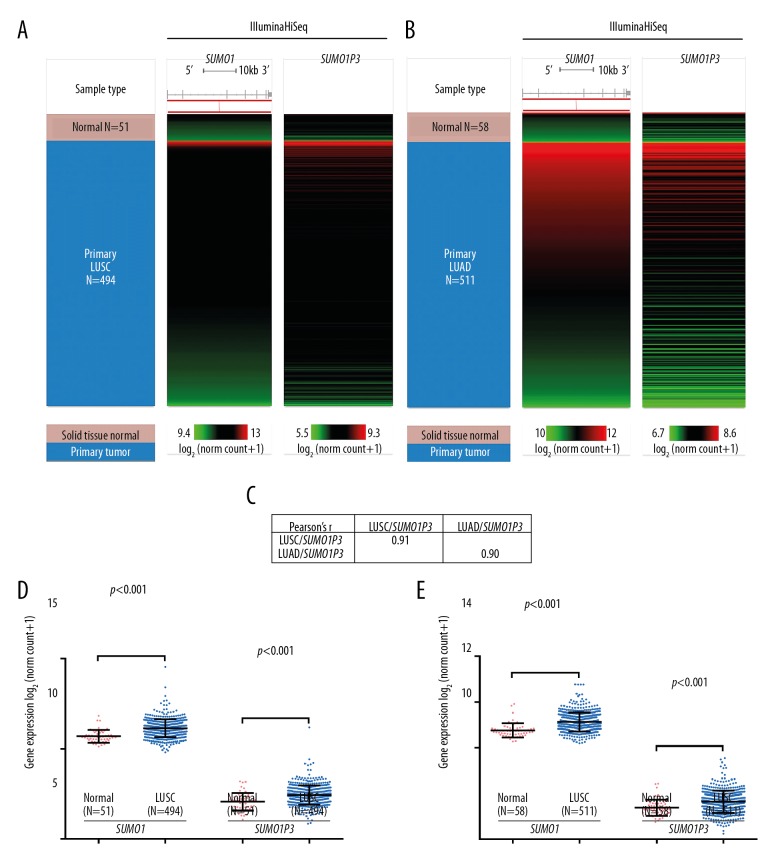
In this study, patient outcome was assessed as complete remission (CR), partial remission (PR), stable disease (SD), progressive disease (PD), and overall survival (OS). Recurrence-free survival (RFS) was defined as the survival time from the date of primary surgery to the time of locoregional or distant recurrence of lung cancer. Although the upregulation of SUMO1 in lung cancer has previously been reported [14], the expression profile of its pseudogene SUMO1P3 has not been characterized. Using RNA-Seq data from around 500 tumor samples, we compared the expression of SUMO1P3 in both LUSC and LUAD. SUMO1P3 showed a similar expression profile as SUMO1 in both LUSC and LUAD (Figure 1A, 1B). SUMO1P3 was strongly co-expressed with SUMO1 in LUSC (Pearson’s r=0.91) and LUAD (Pearson’s r=0.90) tissues (Figure 1C). Similar to SUMO1, SUMO1P3 expression was significantly increased in both LUSC and LUAD tissues compared with their adjacent normal tissues (p<0.001) (Figure 1D, 1E).
Figure 1.
Both SUMO1 and SUMO1P3 were significantly upregulated in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) tissue compared with adjacent normal lung tissue. (A, B) Heatmap showing the expression profile of SUMO1 and SUMO1P3 in tissues from patients with lung squamous cell carcinoma (LUSC) (n=494) and lung adenocarcinoma (LUAD) (n=511) compared with adjacent normal tissues (n=51 and n=58, respectively). (C) Pearson’s r-value showing the correlation between SUMO1 and SUMO1P3 in LUSC (n=494) and LUAD (n=511) tissues. (D, E) Plot chart comparing the expression of SUMO1 and SUMO1P3 in LUSC (n=494) and LUAD (n=511) tissues compared with their respective adjacent normal tissues (n=51 and n=58, respectively).
Patients with LUAD with PD after primary treatment or follow-up treatment showed significantly increased expression of SUMO1P3
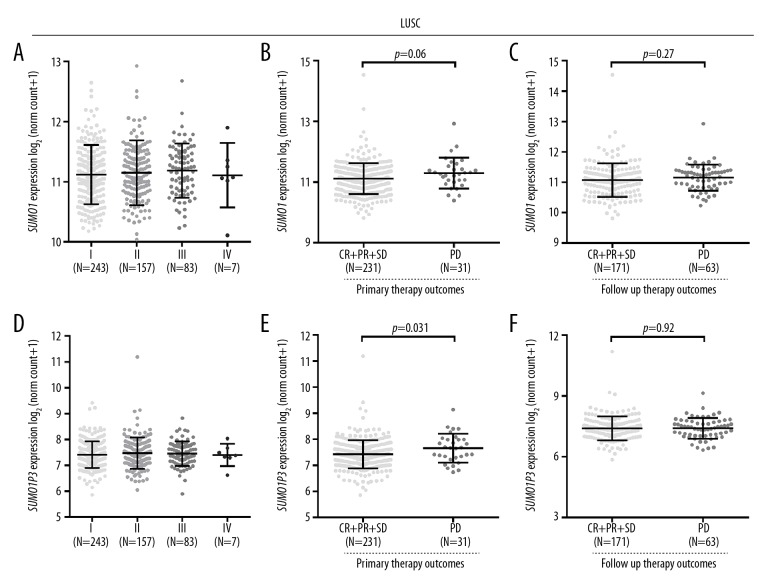
In patients with LUSC, no significant differences in SUMO1 or SUMO1P3 expression were observed in tumors with different stage (Figure 2A, 2D). As SUMO1 expression was previously reported to be associated with chemosensitivity of lung cancer, SUMO1 and SUMO1P3 expression were analyzed for patients with different outcome following treatment. In patients with LUSC, no significant differences were observed in SUMO1 expression between the PD and CR, PR, and SD groups after either primary therapy or follow-up therapy (Figure 2B, 2C). SUMO1P3 expression was significantly increased in patients with LUSC with PD after primary therapy, compared with patients with CR, PR, and SD (Figure 2E). However, this difference was not observed in follow-up therapy (Figure 2F).
Figure 2.
SUMO1 and SUMO1P3 expression in patients with lung squamous cell carcinoma (LUSC) in different stages or with different therapeutic responses. (A, D) SUMO1 (A) and SUMO1P3 (D) expression in patients with LUSC at different stages (B, C, E, F) SUMO1 (B, C) and SUMO1P3 (E, F) expression in patients with LUSC with different therapeutic responses after primary therapy (B, E) or after follow-up therapy (C, F).
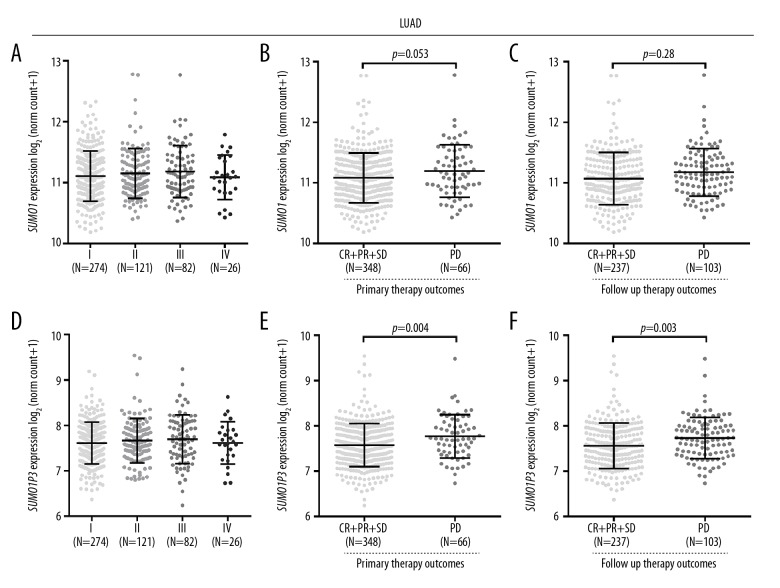
Then, we analyzed the associations in patients with LUAD. No significant differences were found in SUMO1 or SUMO1P3 expression for patients in different stages of lung cancer (Figure 3A, 3D). SUMO1 expression was significantly increased in patients with PD after follow-up therapy, compared with that in patients with CR, PR, and SD (Figure 3C). This difference was not observed after primary therapy (Figure 3B). SUMO1P3 expression was significantly increased in patients with PD after primary therapy or follow-up therapy, compared with that in patients with CR, PR, and SD (Figure 3E, 3F).
Figure 3.
SUMO1 and SUMO1P3 expression in patients with lung adenocarcinoma (LUAD) in different stages or with different therapeutic responses. (A, D) SUMO1 (A) and SUMO1P3 (D) expression in patients with LUAD in different stages. (B, C, E, F) SUMO1 (B, C) and SUMO1P3 (E, F) expression in patients with LUAD with different therapeutic responses after primary therapy (B, E) or after follow-up therapy (C, F).
Patients with LUAD with poor survival outcomes had significantly increased SUMO1P3 expression
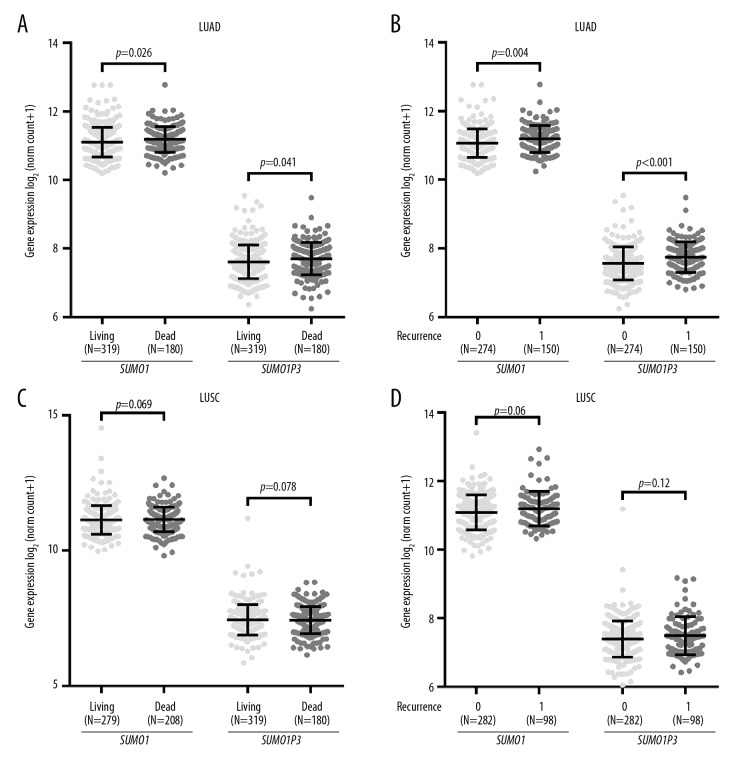
Since we found that SUMO1 and SUMO1P3 upregulation was associated with PD in both patients with LUSC and LUAD, we further examined patients with different survival indicators. In patients with LUAD, the patients with the poor survival indicators or death or tumor recurrence had significantly increased SUMO1 and SUMO1P3 expression, compared with the patients with favorable survival indicators, who were alive or without recurrence (Figure 4A, 4B). These trends were not observed in patients with LUSC (Figure 4C, 4D).
Figure 4.
SUMO1 and SUMO1P3 expression in patients with different survival indicators. (A–D) Comparison of SUMO1 and SUMO1P3 expression in patients with lung adenocarcinoma (LUAD) (A, B) and lung squamous cell carcinoma (LUSC) (C, D) with different survival indicators. Recurrence, 0: no recurrence; 1: with recurrence.
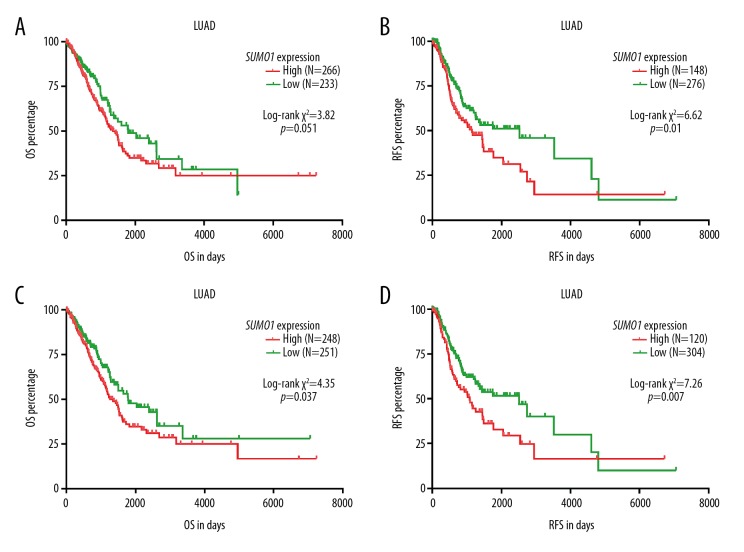
To further explore the association between the upregulation of SUMO1 and SUMO1P3 and survival outcomes in patients with LUAD, Kaplan-Meier survival curves were generated and compared to determine the survival differences. The patient group with high SUMO1 expression had a significantly reduced RFS (Figure 5B). No significant difference was observed in terms of OS (Figure 5A). In contrast, the patient group with high SUMO1P3 expression had a significantly shorter OS (p=0.037) and RFS (p=0.007) compared with the group with low SUMO1P3 expression (Figure 5C, 5D).
Figure 5.
Kaplan-Meier curves of overall survival (OS) and recurrence-free survival (RFS) in patients with lung adenocarcinoma (LUAD). (A–D) Kaplan-Meier curves of overall survival (OS) (A, C) and recurrence-free survival (RFS) (B, D) in patients with lung adenocarcinoma (LUAD). Patients were separated into two groups according to the Youden Index of SUMO1 (A, B) and SUMO1P3 (C, D) expression in receiver operating characteristic (ROC) analysis for mortality (A, C) or recurrence (B, D).
SUMO1P3 expression was an independent predictor of shorter RFS in patients with LUAD
The clinical parameters of patients with LUAD included in survival analysis are summarized in Table 1. To examine the prognostic significance of SUMO1 and SUMO1P3 expression in patients with LUAD, we further conducted univariate and multivariate analysis based on the Cox proportional hazard model. In univariate analysis, advanced T status, N status, and the presence of residual lung tumors were risk factors of unfavorable OS and RFS (Tables 2, Table 3). However, by treating SUMO1 and SUMO1P3 expression as continuous variables, we found that they were not risk factors for reduced OS (Table 2). However, SUMO1P3 expression was a risk factor for reduced RFS (Table 3). Multivariate analysis showed that SUMO1P3 was an independent prognostic factor for RFS (HR, 1.418; 95% CI, 1.041–1.930; p=0.027) (Table 3). We further compared the expression of SUMO1P3 between lung tumor tissue and adjacent normal tissue and the low SUMO1P3 expression group in the RFS analysis. The results showed that there was no significant difference between these two groups (Figure 6).
Table 1.
Summary of the clinical parameters of LUAD patients included in survival analysis.
| Parameters | LUAD patients included in survival analysis (N=499) |
|---|---|
| Age (y, mean ±SD) | 65.33±9.92 |
| Gender | |
| Male | 228 |
| Female | 271 |
| Pathologic T | |
| T1/T2 | 433 |
| T3/T4 | 63 |
| No data | 3 |
| Pathologic N | |
| N0 | 323 |
| N1/N2/N3 | 165 |
| No data | 11 |
| Pathologic stages | |
| I/II | 387 |
| III/IV | 104 |
| No data | 8 |
| Smoking history | |
| 1 | 72 |
| 2/3/4/5 | 413 |
| No data | 14 |
| Residual tumors | |
| No | 334 |
| Yes | 16 |
| No data | 149 |
| Mutation in KRAS/EGFR/ALK | |
| No | 92 |
| Yes | 125 |
| No data | 282 |
| OS data available | 499 |
| RFS data available | 424 |
Smoking history: 1 – lifelong non-smoker; 2 – current smoker; 3 – current reformed smoker (for >15 yrs); 4 – current reformed smoker (for ≤15 yrs); 5 – current reformed smoker (duration not specified).
Table 2.
Univariate analysis of OS in patients with LUAD.
| Parameters | Univariate analysis | |||
|---|---|---|---|---|
| p | HR | 95% CI (lower/upper) | ||
| Age (continuous) | 0.307 | 1.008 | 0.993 | 1.024 |
| Gender | ||||
| Male (N=228) | 1.000 | |||
| Female (N=271) | 0.843 | 0.971 | 0.724 | 1.302 |
| Smoking history | ||||
| 1 (N=72) | 1.000 | |||
| 2/3/4/5 (N=413) | 0.596 | 0.894 | 0.592 | 1.351 |
| Pathological T stages | ||||
| T3/T4 (N=63) | 1.000 | |||
| T1/T2 (N=433) | <0.001 | 0.423 | 0.289 | 0.620 |
| Pathological N stages | ||||
| N1/N2/N3 (N=165) | 1.000 | |||
| N0 (N=323) | <0.001 | 0.391 | 0.290 | 0.527 |
| Pathological stages | ||||
| III/IV (N=104) | 1.000 | |||
| I/II (N=387) | <0.001 | 0.385 | 0.281 | 0.527 |
| Residual tumors | ||||
| Yes (N=16) | 1.000 | |||
| No (N=334) | <0.001 | 0.248 | 0.139 | 0.443 |
| Mutation in KRAS/EGFR/ALK | ||||
| Yes (N=92) | 1.000 | |||
| No (N=125) | 0.570 | 1.134 | 0.734 | 1.753 |
| SUMO1 expression | 0.273 | 1.204 | 0.864 | 1.676 |
| SUMO1P3 expression | 0.285 | 1.170 | 0.877 | 1.560 |
Smoking history: 1 – lifelong non-smoker; 2 – current smoker; 3 – current reformed smoker (for >15 yrs); 4 – Current reformed smoker (for ≤15 yrs); 5 – current reformed smoker (duration not specified); NX – regional lymph nodes cannot be assessed; RX – the presence of residual tumor cannot be assessed.
Table 3.
Univariate and multivariate analysis of RFS in LUAD.
| Parameters | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| p | HR | 95% CI (lower/upper) | p | HR | 95% CI (lower/upper) | |||
| Age (Continuous) | 0.356 | 1.008 | 0.991 | 1.025 | ||||
| Gender | ||||||||
| Male (N=191) | 1.000 | |||||||
| Female (N=233) | 0.516 | 1.114 | 0.805 | 1.541 | ||||
| Smoking history | ||||||||
| 2/3/4/5 (N=345) | 1.000 | |||||||
| 1 (N=65) | 0.454 | 1.199 | 0.746 | 1.926 | ||||
| Pathological T stages | ||||||||
| T3/T4 (N=46) | 1.000 | |||||||
| T1/T2 (N=375) | 0.002 | 0.468 | 0.291 | 0.754 | 0.018 | 0.546 | 0.330 | 0.903 |
| Pathological N stages | ||||||||
| N1/N2/N3 (N=133) | 1.000 | |||||||
| N0 (N=279) | 0.005 | 0.622 | 0.448 | 0.863 | 0.062 | 0.685 | 0.460 | 1.020 |
| Pathological stages | ||||||||
| III/IV (N=80) | 1.000 | |||||||
| I/II (N=337) | 0.009 | 0.599 | 0.407 | 0.882 | 0.519 | 0.849 | 0.517 | 1.396 |
| Residual tumors | ||||||||
| Yes (N=12) | 1.000 | |||||||
| No (N=273) | 0.000 | 0.262 | 0.126 | 0.543 | ||||
| Mutation in KRAS/EGFR/ALK | ||||||||
| Yes (N=77) | 1.000 | |||||||
| No (N=108) | 0.787 | 1.063 | 0.684 | 1.653 | ||||
| SUMO1 expression | 0.095 | 1.342 | 0.950 | 1.898 | ||||
| SUMO1P3 expression | 0.016 | 1.451 | 1.073 | 1.962 | 0.027 | 1.418 | 1.041 | 1.930 |
Smoking history: 1 – lifelong non-smoker; 2 – current smoker; 3 – current reformed smoker (for >15 yrs); 4 – Current reformed smoker (for ≤15 yrs); 5 – Current reformed smoker (duration not specified); NX – regional lymph nodes cannot be assessed; RX – the presence of residual tumor cannot be assessed.
Figure 6.
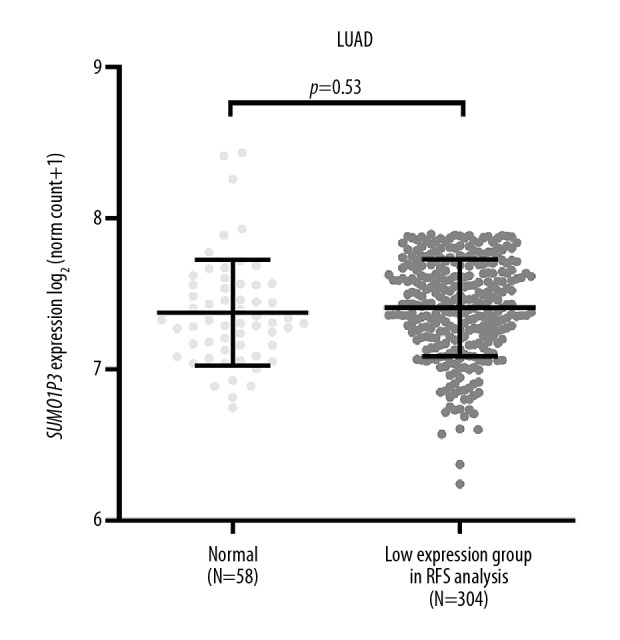
Comparison of SUMO1P3 expression between adjacent normal tissue and the low expression group in recurrence-free survival (RFS) analysis.
Discussion
In this study, patient outcome was assessed as complete remission (CR), partial remission (PR), stable disease (SD), progressive disease (PD), and overall survival (OS). Recurrence-free survival (RFS) was defined as the survival time from the date of primary surgery to the time of locoregional or distant recurrence of lung cancer. The findings showed that SUMO1P3 expression was significantly increased in both lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) tissues compared with adjacent normal tissues. In patients with LUAD, SUMO1P3 expression was significantly increased in the PD group after primary therapy or follow-up therapy, compared with the CR, PR, and SD group. However, these trends were not confirmed in cases of LUSC. Also, although patients with LUAD with high SUMO1 or SUMO1P3 expression tended to have worse survival compared with the low expression group, the results of univariate and multivariate analysis indicated that only SUMO1P3 expression had independent prognostic value in terms of RFS (HR, 1.418; 95% CI, 1.041–1.930; p=0.027). Further studies are required to determine whether SUMO1P3 expression has the potential to be a specific prognostic biomarker in patients with LUAD.
Recent studies have shown the oncogenic properties of SUMO1P3 in several human malignancies. SUMO1P3 has been shown to act as a miR-320a sponge and participate in the regulation of cell proliferation, migration, and invasion of breast cancer cells [10]. Knockdown of SUMO1P3 has been shown to inhibit the expression of cyclin D1, vimentin, and VEGF-A, resulting in restored E-cadherin expression in xenograft colon tumor tissues [17]. These molecular mechanisms can partly explain the correlation between increased SUMO1P3 expression and poor prognosis in malignancy.
In mammals, four SUMO encoding genes, SUMO1-4 have been identified, among which only SUMO1, SUMO2, and SUMO3 have been extensively studied. SUMO2 and SUMO3 share 99% similarity with identical amino acid sequences, while SUMO1 only has around 50% similarity with SUMO2 and SUMO3 [18]. Based on their sequence similarity, their expression levels, and their response to stress, SUMO1, SUMO2, and SUMO3 are categorized into subfamilies [19,20]. SUMOylation directly results in subsequent changes in substrate protein localization, stability, interactions or function to influence a series of cellular processes, including the regulation of the transcriptional process, recycling of proteins, and cellular apoptosis [18]. Previous studies have shown that dysregulated SUMO1 expression is associated with chemoresistance of some tumors, including testicular germ cell tumors [21], osteosarcoma [22], and NSCLC [15]. The findings in the present study showed that patients with LUAD with PD after follow-up therapy had increased expression of SUMO1, which supported that SUMO1 might have a role in modulating therapeutic responses.
Pseudogenes have previously been regarded as nonfunctional, but increasing studies have shown that although they have lost their protein-coding ability, pseudogenes exert a wide range of regulatory functions. For example, the PTEN pseudogene (PTENP1) is targeted by PTEN-targeting microRNAs to maintain the protein expression of PTEN [12]. The long noncoding RNA (lncRNA) pituitary tumor-transforming 3, pseudogene (PTTG3P) can promote hepatocellular carcinoma cell growth and metastasis by enhancing the expression of PTTG1 and activating PI3K/AKT signaling [23]. lncRNA RAS suppressor protein 1 pseudogene 2 (RSU1P2) is involved in the tumorigenesis of cervical cancer by acting as a competing endogenous RNA (ceRNA) against let-7a [24]. These findings indicate that the pseudogene might exert regulatory effects on the expression of the corresponding gene. Correlation analysis has shown that SUMO1P3 was strongly co-expressed with SUMO1 in both LUAD and LUSC, suggesting that there might be interactions between these two molecules as potential therapeutic targets [25–27]. However, it is necessary to validate the prognostic value of SUMO1P3 with independent studies and to study whether it could be detected using tumor byproducts in the blood, such as cell-free DNA or circulating tumor cells. These future studies might determine the future clinical potential of SUMO1P3 as a diagnostic or prognostic cancer biomarker.
Conclusions
SUMO1P3 expression was an independent indicator of reduced recurrence-free survival (RFS) in patients with LUAD.
Footnotes
Source of support: This study was supported by the National Science Foundation of China (No. 81802801 and 81773375), the National High Technology Research and Development Program of China (No. 2015AA020904), and the Support Program for Science and Technology of Sichuan Province, China (2017SZ0202)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Pinto JA, Vallejos CS, Raez LE, et al. Gender and outcomes in non-small cell lung cancer: An old prognostic variable comes back for targeted therapy and immunotherapy? ESMO Open. 2018;3(3):e000344. doi: 10.1136/esmoopen-2018-000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegfried JM, Lin Y, Diergaarde B, et al. Expression of PAM50 genes in lung cancer: evidence that interactions between hormone receptors and HER2/HER3 contribute to poor outcome. Neoplasia. 2015;17(11):817–25. doi: 10.1016/j.neo.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L, Yao Y, Wang Y, et al. Preparation and anti-cancer evaluation of promiximab-MMAE, an anti-CD56 antibody drug conjugate, in small cell lung cancer cell line xenograft models. J Drug Target. 2018;26(10):905–12. doi: 10.1080/1061186X.2018.1450413. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Moreno P, Brambilla E, Thomas R, Soria JC. Squamous cell carcinoma of the lung: Molecular subtypes and therapeutic opportunities. Clin Cancer Res. 2012;18(9):2443–51. doi: 10.1158/1078-0432.CCR-11-2370. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Nagai K, Yoshida J, et al. Conventional clinicopathologic prognostic factors in surgically resected nonsmall cell lung carcinoma. A comparison of prognostic factors for each pathologic TNM stage based on multivariate analyses. Cancer. 1999;86(10):1976–84. doi: 10.1002/(sici)1097-0142(19991115)86:10<1976::aid-cncr14>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Zhan Y, Liu Y, Wang C, et al. Increased expression of SUMO1P3 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Oncotarget. 2016;7(13):16038–48. doi: 10.18632/oncotarget.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mei D, Song H, Wang K, et al. Upregulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30(4):709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, He P, Xie X, Sun C. Knockdown of SUMO1P3 represses tumor growth and invasion and enhances radiosensitivity in hepatocellular carcinoma. Mol Cell Biochem. 2019;450(1–2):125–34. doi: 10.1007/s11010-018-3379-8. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Song Z, Feng C, et al. The long noncoding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am J Transl Res. 2017;9(12):5594–602. [PMC free article] [PubMed] [Google Scholar]
- 11.Tian C, Jin Y, Shi S. Long non-coding RNA SUMO1P3 may promote cell proliferation, migration, and invasion of pancreatic cancer via EMT signaling pathway. Oncol Lett. 2018;16(5):6109–15. doi: 10.3892/ol.2018.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumor biology. Nature. 2010;465(7301):1033–38. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi H, Arao T, Togashi Y, et al. The OCT4 pseudogene POU5F1B is amplified and promotes an aggressive phenotype in gastric cancer. Oncogene. 2015;34(2):199–208. doi: 10.1038/onc.2013.547. [DOI] [PubMed] [Google Scholar]
- 14.Ke C, Zhu K, Sun Y, et al. SUMO1 promotes the proliferation and invasion of non-small cell lung cancer cells by regulating NF-kappaB. Thorac Cancer. 2019;10(1):33–40. doi: 10.1111/1759-7714.12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han JY, Lee GK, Yoo SY, et al. Association of SUMO1 and UBC9 genotypes with tumor response in non-small-cell lung cancer treated with irinotecan-based chemotherapy. Pharmacogenomics J. 2010;10(2):86–93. doi: 10.1038/tpj.2009.46. [DOI] [PubMed] [Google Scholar]
- 16.Goldman M, Craft B, Kamath A, et al. The UCSC Xena system for cancer genomics data visualization and interpretation [abstract] Cancer Res. 2017;77(13 Suppl) Abstract 2584. [Google Scholar]
- 17.Zhang LM, Wang P, Liu XM, Zhang YJ. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res. 2017;9(12):5461–72. [PMC free article] [PubMed] [Google Scholar]
- 18.Daniel JA, Cooper BH, Palvimo JJ, et al. Analysis of SUMO1-conjugation at synapses. Elife. 2017;6 doi: 10.7554/eLife.26338. pii: e26338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayak A, Muller S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014;15(7):422. doi: 10.1186/s13059-014-0422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreou AM, Tavernarakis N. SUMOylation and cell signalling. Biotechnol J. 2009;4(12):1740–52. doi: 10.1002/biot.200900219. [DOI] [PubMed] [Google Scholar]
- 21.Wu YC, Ling TY, Lu SH, et al. Chemotherapeutic sensitivity of testicular germ cell tumors under hypoxic conditions is negatively regulated by SENP1-controlled sumoylation of OCT4. Cancer Res. 2012;72(19):4963–73. doi: 10.1158/0008-5472.CAN-12-0673. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D, Yu K, Yang Z, et al. Silencing Ubc9 expression suppresses osteosarcoma tumorigenesis and enhances chemosensitivity to HSV-TK/GCV by regulating connexin 43 SUMOylation. Int J Oncol. 2018;53(3):1323–31. doi: 10.3892/ijo.2018.4448. [DOI] [PubMed] [Google Scholar]
- 23.Huang JL, Cao SW, Ou QS, et al. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer. 2018;17(1):93. doi: 10.1186/s12943-018-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Guo X, Que S, et al. LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget. 2017;8(27):43768–81. doi: 10.18632/oncotarget.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai Q, Wang Y, Wang R, et al. Design, synthesis and biological evaluation of a novel tubulin inhibitor 7a3 targeting the colchicine binding site. Eur J Med Chem. 2018;156:162–79. doi: 10.1016/j.ejmech.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Wang Y, Wu Y, et al. Therapeutic potential of an anti-HER2 single chain antibody-DM1 conjugates for the treatment of HER2-positive cancer. Signal Transduct Target Ther. 2017;2:17015. doi: 10.1038/sigtrans.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y, Yu L, Su X, et al. Synthesis, characterization and targeting chemotherapy for ovarian cancer of trastuzumab-SN-38 conjugates. J Control Release. 2015;220(Pt A):5–17. doi: 10.1016/j.jconrel.2015.09.058. [DOI] [PubMed] [Google Scholar]