Abstract
Background
Aurora kinase B (AURKB) is an important carcinogenic factor in various tumors, while its role in clear cell renal cell carcinoma (ccRCC) still remains unclear. This study aimed to investigate its prognostic value and mechanism of action in ccRCC.
Methods
Gene expression profiles and clinical data of ccRCC patients were downloaded from The Cancer Genome Atlas database. R software was utilized to analyze the expression and prognostic role of AURKB in ccRCC. Gene set enrichment analysis (GSEA) was used to analyze AURKB related signaling pathways in ccRCC.
Results
AURKB was expressed at higher levels in ccRCC tissues than normal kidney tissues. Increased AURKB expression in ccRCC correlated with high histological grade, pathological stage, T stage, N stage and distant metastasis (M stage). Kaplan-Meier survival analysis suggested that high AURKB expression patients had a worse prognosis than patients with low AURKB expression levels. Multivariate Cox analysis showed that AURKB expression is a prognostic factor of ccRCC. GSEA indicated that genes involved in autoimmune thyroid disease, intestinal immune network for IgA production, antigen processing and presentation, cytokine-cytokine receptor interaction, asthma, etc., were differentially enriched in the AURKB high expression phenotype.
Conclusions
AURKB is a promising biomarker for predicting prognosis of ccRCC patients and a potential therapeutic target. In addition, AURKB might regulate progression of ccRCC through modulating intestinal immune network for IgA production and cytokine-cytokine receptor interaction, etc. signaling pathways. However, more research is necessary to validate the findings.
Keywords: ccRCC, AURKB, TCGA database, Prognosis, GESA
Introduction
Renal cell carcinoma (RCC) is one of the most common malignant tumors of the urinary system, and clear cell renal cell carcinoma (ccRCC) is the most common pathological subtype (Srigley et al., 2013). Morbidity and mortality of ccRCC are increasing year by year, while the mechanism of ccRCC development still remain unclear (Dutcher, 2013). Hence, biomarkers that can be used to diagnose, treat and predict prognosis of ccRCC, are urgently needed.
Aurora kinase B (AURKB), located on human chromosome 17p13.1, encodes a member of the aurora kinase subfamily of serine/threonine kinases. Previous researches have reported that aberrant AURKB expression is related to tumorigenesis and progression of tumors (Zhu et al., 2019). Single nucleotide polymorphisms (SNPs) of AURKB were associated with occurrence of gastric cancer (GC). rs2289590 in AURKB might contribute to susceptibility for the development of gastric cancer (Mesic et al., 2017). In thyroid cancer, Sorrentino et al. (2005) found that AURKB was not detected in normal thyroid tissue, but it was overexpressed in thyroid carcinoma. Further experiments indicated that silencing AURKB can obviously inhibit the growth of thyroid carcinoma cells. Hence, they thought that AURKB was an important protein in the progression of thyroid carcinomas and a promising candidate for targeted treatment. Besides, AURKB also plays an important role in non-neoplastic disease. In asthenozoospermia, over-expression of AURKB might be associated with development of asthenozoospermia. Over-expression of AURKB can decrease glycolytic activities, conferring to the occurrence and progression of asthenozoospermia (Zhou et al., 2018). Generally, the several researches have suggested the important role of AURKB in tumors and non-neoplastic disease. However, few studies about the relationship between AURKB and ccRCC have been reported so far and the role of AURKB in ccRCC remains elusive.
In this work, we attempted to reveal the significance of AURKB expression in ccRCC and the mechanisms related to ccRCC progression. We compared AURKB mRNA expression between tumor tissues and normal tissues. We then analyzed the relationship between AURKB mRNA expression and clinical parameters of ccRCC and correlated them with patients’overall survival (OS) and disease-free survival (DFS). Results indicated that patients with high AURKB expression have poorer prognosis than patients with low AURKB expression. In addition, to further understand the AURKB-related biological pathways involved in ccRCC, Gene set enrichment analysis (GSEA) was performed. Results showed that twenty-one genes were evidently enriched in patients with high AURKB expression, including intestinal immune network for IgA production, cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, cell cycle and cell adhesion molecules (CAMs), etc.
Material and Methods
Database
Gene expression profiles of ccRCC patients and clinical data of patients such as age, gender, pathological stage, histological grade, survival, and outcome were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). In addition, Drug sensitivity data of ccRCC cell lines were obtained from genomics of drug sensitivity in cancer (GDSC) database (https://portals.broadinstitute.org/ccle/about). We then utilized R software (R Core Team, 2018) to process all data.
Firstly, we extracted clinical data of ccRCC patients and data of gene expression profiles. We then obtained clinical data of 530 patients who possessed complete OS information and a gene expression matrix document. Secondly, we obtained expression of AURKB data from the gene expression matrix document and analyzed the relationship between expression of AURKB and clinical parameters including age, gender, histological grade, pathological stage, T stage, N stage, and M stage. Thirdly, the ccRCC patients be divided into two groups based on median value of AURKB expression (high AURKB expression group and low AURKB expression group) and analyzed their overall survival (OS) and disease-free survival (DFS). Fourthly, we utilized some clinical parameters, that correlated with prognosis of ccRCC, and AURKB to construct a prognostic model. Finally, we analyzed that the difference between sensitivity of AURKB targeted drug and other targeted drugs for ccRCC.
Gene set enrichment analysis
Gene expression profiles of ccRCC patients were divided into two groups (high expression group and low expression group) according to the median value of expression of AURKB. GSEA was utilized to detect potential mechanisms underlying the effect of AURKB expression on ccRCC prognosis. Gene set permutations were performed 1,000 times for each analysis. Gene sets with a p-value <0.05 and false discovery rate (FDR) <0.05 were regarded as significantly enriched.
Statistical analysis
All statistical analyses were performed through R software and p <0.05 was regarded as statistically significant. The relationship between expression levels of AURKB and clinical parameters was analyzed via the Wilcoxon signed-rank test, Kruskal-Wallis test and logistic regression. The correlation between expression levels of AURKB, and patients’OS and DFS were analyzed using the Kaplan–Meier method. Univariate Cox analysis was used to select possible prognostic factors, and multivariate Cox analysis was utilized to verify the correlations between AURKB mRNA expression and survival along with other clinical features. A receiver operating characteristic (ROC) curve was used to evaluate the accuracy of models that predicted prognosis using the survival ROC package. An area under the curve (AUC) value of 0.75 or bigger was deemed an excellent predictive value, and values of 0.6 or larger were regarded as acceptable for survival predictions. The chi-square test be used to compare difference between sensitivity of target drugs in ccRCC cell lines.
Results
Clinical parameters of patients
The clinical data of 530 ccRCC patients were obtained from the TCGA database, and included age, gender, histological grade, pathological stage, survival, and outcome, etc. (Table 1).
Table 1. Clinical data of the ccRCC patients.
| Clinical parameters | Variable | Total (530) | Percentages (%) |
|---|---|---|---|
| Age | <60 | 245 | 46% |
| ≥60 | 285 | 54% | |
| Gender | Female | 186 | 35.56% |
| Male | 344 | 64.44% | |
| Histological grade | G1 | 14 | 2.64% |
| G2 | 227 | 42.83% | |
| G3 | 206 | 38.86% | |
| G4 | 75 | 14.15% | |
| GX | 5 | 0.94% | |
| Unknow | 3 | 0.56% | |
| Pathological stage | Stage I | 265 | 50.00% |
| Stage II | 57 | 10.75% | |
| Stage III | 123 | 23.20% | |
| Stage IV | 82 | 15.47% | |
| Unknow | 3 | 0.56% | |
| T stage | T1 | 271 | 51.21% |
| T2 | 69 | 12.84% | |
| T3 | 179 | 33.89% | |
| T4 | 11 | 2.04% | |
| M stage | M0 | 420 | 79.24% |
| M1 | 78 | 14.71% | |
| Mx | 30 | 5.66% | |
| Unknow | 2 | 0.37% | |
| N stage | N0 | 239 | 45.09% |
| N1 | 16 | 3.01% | |
| NX | 275 | 51.88% | |
| Disease free status | Recurred/Progressed | 125 | 23.58% |
| Disease free | 308 | 58.11% | |
| Unknow | 97 | 18.30% | |
| Survival status | Death | 166 | 31.32% |
| Alive | 364 | 68.68% |
High AURKB expression in ccRCC
Expression levels of AURKB in 539 ccRCC and 72 normal kidney tissues were compared via Wilcoxon signed-rank test, and the results showed that AURKB was highly expressed in ccRCC compared to normal kidney tissues (p < 0.05) (Fig. 1A). We further analyzed the expression of AURKB in 72 pairs of ccRCC tissues and matched non-cancerous adjacent tissues using Wilcoxon singed-rank test, and found that AURKB was significantly overexpressed in ccRCC tissues (p < 0.05) (Fig. 1B). These results suggested that AURKB may be a carcinogenic gene in ccRCC.
Figure 1. AURKB was significantly overexpressed in ccRCC compared to normal or adjacent normal tissues.
(A) AURKB was significantly upregulated in cancer tissues compared to normal tissues (p < 0.05). (B) AURKB was expressed at higher levels in ccRCC (p < 0.05) compared to 72 pairs of non-cancerous adjacent tissues.
Correlations between AURKB expression and clinical parameters in ccRCC patients
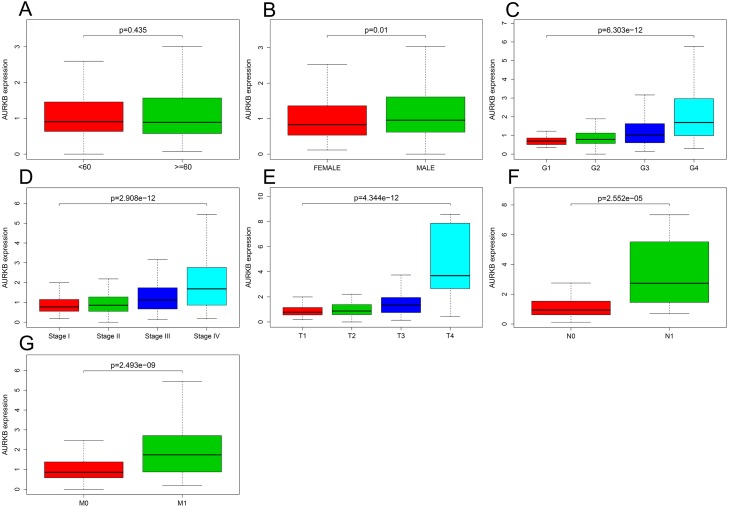
The relationship between AURKB expression and patients’ clinical parameters was analyzed by R software. Results indicated that with the increase of AURKB expression, these clinical parameters (histological grade, pathological stage, T stage, N stage and M stage) also elevated (all p <0.05). Furthermore, expression of AURKB in male was higher than female (p <0.05) (Fig. 2).
Figure 2. Association of AURKB expression with clinical parameters.
(A) Age (p > 0.05); (B) gender (p < 0.05); (C) histological grade (p < 0.05); (D) pathological stage (p < 0.05); (E) T stage (p < 0.05); (F) N stage (p < 0.05); (G) M stage (p < 0.05).
Logistic regression analysis shown that increased AURKB expression in ccRCC was obviously correlated with gender (OR = 1.49 for Female vs. Male, p = 0.029), histological grade (OR = 2.44 for G1/G2 vs. G3/G4, p = 6.97E-07), pathological stage (OR = 1.17 for stage I vs. stage III, p = 0.000; OR = 3.62 for stage I vs. stage IV, p = 2.42683E-06 ), TNM stage (OR = 2.84 for T1/T2 vs. T3/T4, p = 3.12E−08; OR = 4.75 for N0 vs. N1, p = 0.017; OR = 2.99, for M0 vs. M1, p = 4.8776E-05) (Table 2). These results indicated that ccRCC with increased AURKB expression is prone to progress to a more advanced stage, lymph node metastasis distant metastasis.
Table 2. AURKB expression correlated with clinical parameters (logistic regression).
| Clinical parameters | Total (N) | Odds ratio in AURKB expression | p-Value |
|---|---|---|---|
| Age | |||
| <60 vs. ≥60 | 530 | 0.92(0.65–1.30) | 0.6631356 |
| Gender | |||
| Female vs. Male | 530 | 1.49(1.04–2.13) | 0.02927836 |
| Histological grade | |||
| G1/G2 vs. G3/G4 | 523 | 2.44(1.72–3.48) | 6.97E−07 |
| Pathological stage | |||
| Stage I vs. Stage II | 326 | 1.17(0.65–2.08) | 0.5907123 |
| Stage I vs. Stage III | 394 | 2.26(1.46–3.52) | 0.000242585 |
| Stage I vs. Stage IV | 352 | 3.62(2.14–6.28) | 2.42683E−06 |
| T stage | |||
| T1/T2 vs. T3/T4 | 530 | 2.84(1.97–4.14) | 3.12E−08 |
| Lymph nodes (N stage) | |||
| N0 vs. N1 | 255 | 4.75(1.48–21.10) | 0.01705434 |
| Distant metastasis (M stage) | |||
| M0 vs. M1 | 498 | 2.99(1.78–5.17) | 4.8776E−05 |
Prognostic role of AURKB expression in ccRCC Patients
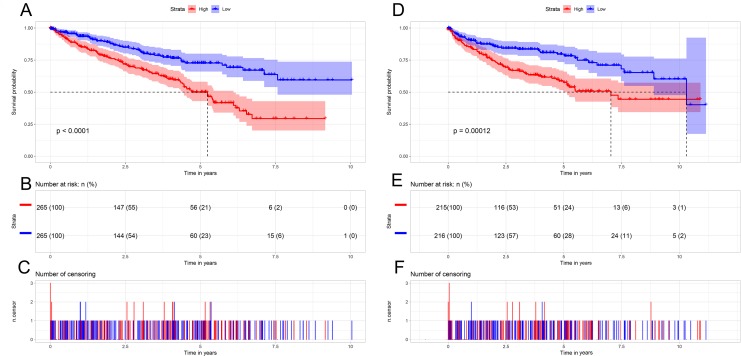
To further understand the prognostic role of AURKB expression in ccRCC, all ccRCC patients were categorized according to the median AURKB expression value (high AURKB expression group and low AURKB expression group). Patients who lacked complete clinical data were excluded from the analysis. Kaplan–Meier survival analysis indicated that the high AURKB expression group had worse prognosis compared with the low AURKB expression group (p <0.05) (Fig. 3). The univariate analysis indicated that high AURKB expression was associated with poorer OS and DFS (p <0.05). Other clinical parameters, such as pathological stage and histological grade, also correlated with worse OS and DFS (p <0.05) (Table 3).
Figure 3. Kaplan-Meier survival curves of patients with ccRCC based on AURKB expression levels.
(A) The Kaplan-Meier curves, (B) number at risk, and (C) number of censoring of OS in ccRCC. (D) The Kaplan-Meier curves, (E) number at risk, and (F) number of censoring of DFS in ccRCC. High expression of AURKB was correlated with poorer OS and DFS in ccRCC patients.
Table 3. Univariate and multivariate cox regression analyses for OS and DFS in ccRCC patients.
| Variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Overall survival | ||||
| Age | 1.03(1.01–1.04) | 0.00000229 | 1.03(1.02–1.05) | 0.00000107 |
| Gender | 0.93(0.67–1.28) | 0.662936583 | ||
| Histological grade | 2.29(1.85–2.83) | 1.94E−14 | 1.40(1.10–1.79) | 0.005937457 |
| Pathological stage | 1.88(1.64–2.16) | 4.67E−20 | 1.60(1.03–2.48) | 0.034897661 |
| T stage | 1.94(1.63–2.29) | 1.5E−14 | 0.87(0.58–1.31) | 0.528085268 |
| M stage | 4.28(3.10–5.90) | 7.45E−19 | 1.45(0.74–2.84) | 0.27403342 |
| AURKB | 1.13(1.09–1.16) | 2.42E−13 | 1.09(1.05–1.14) | 0.00000276 |
| Disease free survival | ||||
| Age | 1.00(0.99–1.02) | 0.20947696 | ||
| Gender | 1.35(0.91–2.01) | 0.132201537 | ||
| Histological grade | 2.97(2.29–3.83) | 8.11792E−17 | 1.73(1.33–2.25) | 4.49123E-05 |
| Pathological stage | 2.65(2.22–3.17) | 5.19396E−27 | 2.33(1.30–4.18) | 0.004223284 |
| T stage | 2.5(2.02–3.08) | 1.92212E−17 | 0.79(0.46–1.33) | 0.384923805 |
| M stage | 8.52(5.87–12.37) | 1.95879E−29 | 1.47(0.63–3.43) | 0.361781114 |
| AURKB | 1.13(1.09–1.18) | 3.0124E−09 | 1.07(1.01–1.13) | 0.019973352 |
To confirm the prognostic value of AURKB expression, multivariate analysis was performed. The results showed that age, histological grade, pathological stage and AURKB expression were independently associated with OS (p <0.05), and histological grade, pathological stage and AURKB were independently correlated with DFS (p <0.05). Overall, these results suggest that AURKB is an independent prognostic factor of ccRCC (Fig. 4 and Table 3).
Figure 4. Forest plots for multivariate cox regression analyses.
(A) Age, histological grade, pathological stage and AURKB are independently correlated with OS; (B) Histological grade, pathological stage and AURKB are independently associated with DFS.
Prognostic models of AURKB expression and new nomograms
As the expression of AURKB plays an important role in OS and DFS in ccRCC, we attempted to explore whether it can be used to create better prognostic models. Two new nomograms were constructed to predict OS and DFS, 3 and 5 years after surgery (Figs. 5A and 5C). A ROC curve was used to estimate the accuracy of the two models, and the results indicated that the two models were able to accurately predict OS and DFS at 3 and 5 years after surgery (the areas under the ROC curve were 0.792 (3-year OS), 0.748 (5-year OS), 0.851 (3-year DFS) and 0.837 (5-year DFS)) (Figs. 5B and 5D).
Figure 5. Nomogram and ROC plots for the prediction of outcome in patients with ccRCC.
Nomogram for the prediction of OS (A) and PFS (B) at 3 and 5 years after surgery. ROC curve evaluated the accuracy of the two models (C and D), and the areas under the ROC curve were 0.792 (3-year OS), 0.748 (5-year OS), 0.851 (3-year DFS) and 0.837 (5-year DFS), and indicated a good accuracy.
Sensitivity analysis of AURKB targeted drug
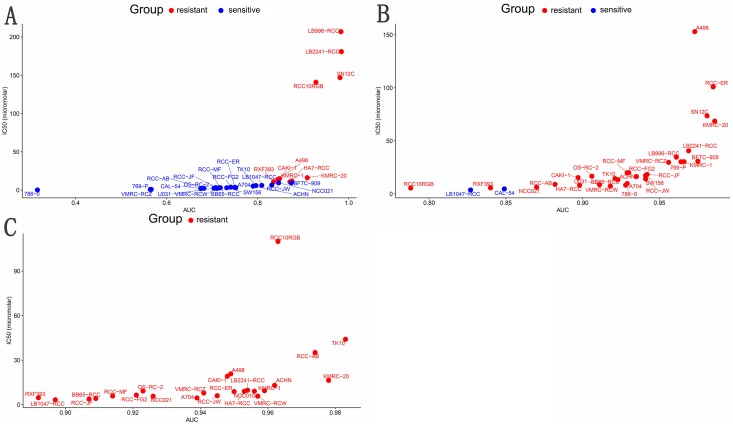
Cabozantinib (targets: VEGFR, MET, RET, KIT, FLT1, FLT3, FLT4, TIE2, AXL) and Axitinib (targets: PDGFR, KIT, VEGFR) are common target drug for ccRCC. Genentech Cpd 10 (targets: AURKA, AURKB) also is a target drug. To further validate AURKB might become a potential treatment target in ccRCC, drug sensitivity analysis of ccRCC cell lines was performed. Results showed that 2 ccRCC cell lines (n = 31) were sensitive to Cabozantinib, 21 ccRCC cell lines (n = 31) were sensitive to Genentech Cpd 10, and none ccRCC cell lines (n = 24) are sensitive to Axitinib (Table 4 and Fig. 6). These results suggested that ccRCC cell lines were more sensitive to Genentech Cpd 10 than Cabozantinib and Axitinib (p < 0.05), and AURKB probably become a promising target to treat ccRCC.
Table 4. Drugs sensitivity analysis of ccRCC cell lines.
| Genentech Cpd 10 vs. Cabozantinib | Genentech Cpd 10 vs. Axitinib | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | Total (N) | Sensitive | Resistent | χ2 | p-value | Drug | Total (N) | Sensitive | Resistent | χ2 | p-Value |
| Genentech Cpd 10 | 31 | 21 | 10 | 22.395 | 2.22E−06 | Genentech Cpd 10 | 31 | 21 | 10 | 23.508 | 1.244E-06 |
| Cabozantinib | 31 | 2 | 29 | Axitinib | 24 | 0 | 24 | ||||
Figure 6. Drugs sensitivity analysis of ccRCC cell ines.
(A) Genentech Cpd 10, (B) Cabozantinib, (C) Axitinib. Those cell lines whose IC50 value greater than max screening concentration was regarded as resistant to target drugs. Blue represents sensitive ccRCC cell lines to target drugs; red indicates resistant ccRCC cell lines to target drugs; IC50, natural log half maximal inhibitory concentration; AUC, Area under the dose-response curve.
Identification of AURKB related signaling pathways
GSEA was used to screen signaling pathways involved in ccRCC between low and high AURKB expression data set. GSEA indicated significant differences (FDR < 0.05, NOM p-value < 0.05) in enrichment of MSigDB Collection (c2.cp.v6.2.symbols.gmt).
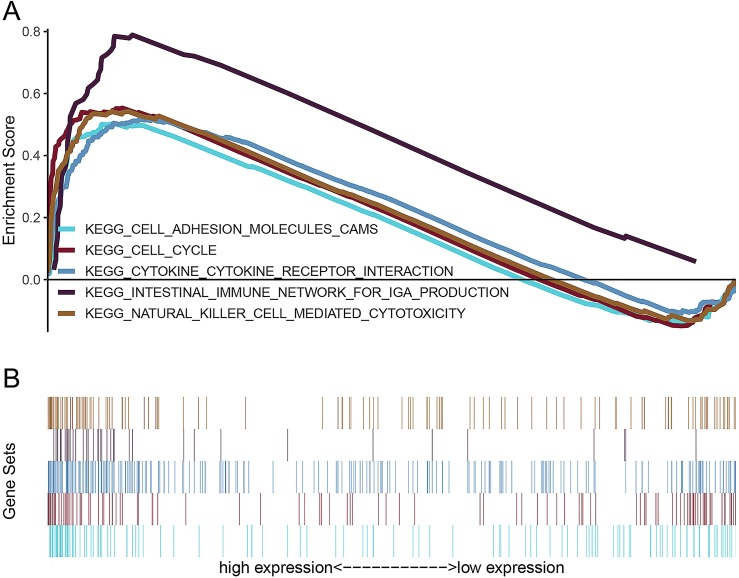
Twenty-one signaling pathways involved in autoimmune thyroid disease, intestinal immune network for IgA production, antigen processing and presentation, cytokine-cytokine receptor interaction, asthma, type I diabetes mellitus, primary immunodeficiency, graft versus host disease, allograft rejection, base excision repair, homologous recombination, natural killer cell mediated cytotoxicity, cytosolic DNA sensing pathway, viral myocarditis, hematopoietic cell lineage, DNA replication, systemic lupus erythematosus, leishmania infection, cell cycle, cell adhesion molecules (CAMs) and proteasome were differentially enriched in the AURKB high expression phenotype (Table 5). Five signaling pathways that may be closely connected to the progression of ccRCC tumors are shown in Fig. 7.
Table 5. Gene sets enriched in the high AURKB expression phenotype.
| Gene set name | NES | NOM p-value | FDR q-value |
|---|---|---|---|
| KEGG_AUTOIMMUNE_THYROID_DISEASE | 2.3979816 | 0 | 0.00331709 |
| KEGG_INTESTINAL_IMMUNE_NETWORK_FOR_IGA_PRODUCTION | 2.3240876 | 0 | 0.004554952 |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION | 2.2065759 | 0.007889546 | 0.012016102 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 2.1696072 | 0.001915709 | 0.013810257 |
| KEGG_ASTHMA | 2.1645305 | 0 | 0.011436771 |
| KEGG_TYPE_I_DIABETES_MELLITUS | 2.139911 | 0 | 0.012462393 |
| KEGG_PRIMARY_IMMUNODEFICIENCY | 2.0994904 | 0 | 0.014886827 |
| KEGG_GRAFT_VERSUS_HOST_DISEASE | 2.0993276 | 0 | 0.013095205 |
| KEGG_ALLOGRAFT_REJECTION | 2.0911534 | 0 | 0.012559181 |
| KEGG_BASE_EXCISION_REPAIR | 2.047824 | 0.001901141 | 0.016551593 |
| KEGG_HOMOLOGOUS_RECOMBINATION | 2.047304 | 0.003913894 | 0.015096117 |
| KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 2.0085354 | 0.015655577 | 0.019628806 |
| KEGG_CYTOSOLIC_DNA_SENSING_PATHWAY | 2.00772 | 0 | 0.018204125 |
| KEGG_VIRAL_MYOCARDITIS | 1.993407 | 0.009940358 | 0.019238696 |
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | 1.9526478 | 0.005825243 | 0.024869524 |
| KEGG_DNA_REPLICATION | 1.9470055 | 0.013461539 | 0.024576483 |
| KEGG_SYSTEMIC_LUPUS_ERYTHEMATOSUS | 1.9417096 | 0.013944224 | 0.02391057 |
| KEGG_LEISHMANIA_INFECTION | 1.9209716 | 0.027559055 | 0.026700974 |
| KEGG_CELL_CYCLE | 1.8969755 | 0.025590552 | 0.030597683 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 1.8593365 | 0.03206413 | 0.037772134 |
| KEGG_PROTEASOME | 1.8296527 | 0.028248588 | 0.044589847 |
Notes.
- NES
- normalized enrichment score
- NOM
- nominal
- FDR
- false discovery rate
Gene sets with NOM p-value <0.05 and FDR q-value <0.05 were regarded as significantly enriched.
Figure 7. Enrichment plots from gene set enrichment analysis (GSEA).
(A) Enrichment sore, (B) gene sets. Intestinal immune network for IgA production, cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, cell cycle, and cell adhesion molecules (CAMs) might be highly correlated with progression of ccRCC.
Discussion
Many studies have suggested that AURKB plays a vital role in tumorigenesis and tumor progression (Zhu et al., 2019; Mesic et al., 2017; Kotian et al., 2017). AURKB has been shown to be involved in the development of breast cancer, and its expression has been associated with breast cancer prognosis (Liao et al., 2018; Naorem, Muthaiyan & Venkatesan, 2019). In non-small cell lung cancer (NSCLC), AURKB has been shown to be overexpressed and correlated with poorer prognosis of patients. Its overexpression was shown to significantly promote proliferation of NSCLC cells via inhibiting the p53-related pathway. In addition, expression of AURKB has also been associated with drug resistance in NSCLC. Overexpression of AURKB increased drug resistance in NSCLC cells, whereas AURKB knockdown re-sensitized NSCLC cells to chemotherapeutic drugs (Yu et al., 2018). In colorectal cancer (CRC), AURKB has also been shown to act as an important oncogenic factor, be involved in the development of CRC, and promoted drug resistance and progression of CRC though regulation of the Wnt signaling pathway and the p53-related pathway (Subramaniyan, Kumar & Mathan, 2017; Nair et al., 2009; Wu et al., 2011; Pohl et al., 2011). All these studies have suggested that AURKB promotes carcinogenesis and is associated with drug resistance.
In this work, we sought to identify the role of AURKB expression in ccRCC progression, particularly, its role as a prognostic factor in ccRCC. Moreover, we also attempted to screen AURKB-related signaling pathways in ccRCC to contribute to the understanding the potential mechanism involved in the regulation of ccRCC development by AURKB.
Firstly, we compared that expression of AURKB in ccRCC and normal tissues. The results showed that AURKB was overexpressed in ccRCC tissues compared to normal tissues, and its expression was associated with pathological stage, histological grade, T stage, M stage, and N stage.
Secondly, Kaplan–Meier survival analysis showed that compared to the low AURKB expression group, the high AURKB expression group of patients had poorer OS and DFS. Moreover, some variables were also associated with the prognosis of ccRCC patients, including pathological stage, histological grade, T stage, and M stage. In addition, multivariate analysis confirmed that AURKB expression was a prognostic factor. Another, the drug sensitivity analysis of ccRCC cell lines suggested that various cell lines were sensitive to Genentech Cpd 10, and AURKB might be a promising target to treat ccRCC.
Finally, we constructed prognostic models of AURKB expression, and the area under the curve (AUC) values proved that the new prognostic models can accurately predict OS and PFS. Furthermore, AURKB related signaling pathways in ccRCC were analyzed by GSEA, and results suggested that intestinal immune network for IgA production, cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, cell cycle and cell adhesion molecules (CAMs), correlate with progression of ccRCC. It has been shown that intestinal immune network for IgA production play a pivotal role in tumor progression (Liang et al., 2018). Yang et al. (2018) have found that activation of intestinal immune network for IgA production signaling pathway promoted malignant behavior of tumor cells. Additionally, cytokine-cytokine receptor interaction was a significant immune signaling pathway, as it can modulate interaction of cytokines, thereby regulating occurrence and progression of cancers (Tumino et al., 2019; Nagarsheth, Wicha & Zou, 2017). Natural killer cell mediated cytotoxicity plays a vital role in modulating tumor microenvironment; their activation was closely related to the progression of tumors and prognosis of cancer patients (Malmberg et al., 2017; Chan, Wucherpfennig & De Andrade, 2019; Bassani et al., 2019). The cell cycle controls the progression of tumors (Roy et al., 2017), and its activation can significantly promote proliferation of tumor cells, thus accelerating tumor growth (Eifler & Vertegaal, 2015; Mast et al., 2019). Increasing amount of evidence has indicated that alterations in the adhesion properties of neoplastic cells play an important role in the development and progression of tumors, and cell adhesion molecules (CAMs) are involved in the adhesion of neoplastic cells, and participate in metastasis, migration and invasion of tumors (De Méndez & Bosch, 2011; Xin, Dong & Guo, 2015; Okegawa et al., 2004). All these results suggest that AURKB promotes oncogenesis and progression of ccRCC through regulating multiple signaling pathways.
At present, many studies have already indicated that AURKB is a promising therapeutic target in various cancers, such as non-small cell lung cancer (NSCLC) (Bertran-Alamillo et al., 2019), gastric cancer (GC) (He et al., 2019), leukemia (He et al., 2016), prostate cancer (PC) (Addepalli et al., 2010), and breast cancer (Han et al., 2017). Additionally, in the present study, we found that AURKB is a promising biomarker in the treatment of ccRCC and a predictor of prognosis.
Inevitably, our study also has several limitations. Firstly, the data we analyzed in the present study were extracted from several public databases, which had not been verified. Secondly, the mechanisms by which AURKB regulates the occurrence and progression of ccRCC need further exploration. Finally, our study found that ccRCC cell lines were more sensitive to AURKB-targeting drug (Genentech Cpd 10) than the conventional targeted drugs (Cabozantinib and Axitinib). However, more studies are necessary to identify whether AURKB could be used as a target for ccRCC treatment.
Conclusions
In summary, our study suggests that AURKB is over-expressed in ccRCC, and it is a valuable prognostic factor for predicting OS and DFS of ccRCC patients. AURKB can promote development of ccRCC via various signaling pathways including intestinal immune network for IgA production, cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, cell cycle and cell adhesion molecules (CAMs). In addition, AURKB might be a promising therapeutic target for ccRCC. However, more research is required to verify the findings of this study.
Supplemental Information
Funding Statement
The study was supported by the Natural Science Foundation of Hainan Province (No. 819MS136). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Bangbei Wan conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Yuan Huang conceived and designed the experiments, performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Bo Liu and Likui Lu performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables.
Cai Lv conceived and designed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files.
References
- Addepalli et al. (2010).Addepalli MK, Ray KB, Kumar B, Ramnath RL, Chile S, Rao H. RNAi-mediated knockdown of AURKB and EGFR shows enhanced therapeutic efficacy in prostate tumor regression. Gene Therapy. 2010;17(3):352–359. doi: 10.1038/gt.2009.155. [DOI] [PubMed] [Google Scholar]
- Bassani et al. (2019).Bassani B, Baci D, Gallazzi M, Poggi A, Bruno A, Mortara L. Natural killer cells as key players of tumor progression and angiogenesis: old and novel tools to divert their pro-tumor activities into potent anti-tumor effects. Cancer. 2019;11(4):461. doi: 10.3390/cancers11040461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertran-Alamillo et al. (2019).Bertran-Alamillo J, Cattan V, Schoumacher M, Codony-Servat J, Giménez-Capitán A, Cantero F, Burbridge M, Rodríguez S, Teixidó C, Roman R, Castellví J, García-Román S, Codony-Servat C, Viteri S, Cardona AF, Karachaliou N, Rosell R, Molina-Vila MA. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nature Communications. 2019;10(1):1812. doi: 10.1038/s41467-019-09734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Wucherpfennig & de Andrade (2019).Chan LL, Wucherpfennig KW, de Andrade LF. Visualization and quantification of NK cell-mediated cytotoxicity over extended time periods by image cytometry. Journal of Immunological Methods. 2019;469:47–51. doi: 10.1016/j.jim.2019.04.001. [DOI] [PubMed] [Google Scholar]
- de Méndez & Bosch (2011).de Méndez MT, Bosch AL. Abnormal immunoexpression of cell adhesion molecules (CAMs) in cervical cancer. International Journal of Surgical Pathology. 2011;19(6):733–742. doi: 10.1177/1066896909343435. [DOI] [PubMed] [Google Scholar]
- Dutcher (2013).Dutcher JP. Recent developments in the treatment of renal cell carcinoma. Therapeutic Advances in Urology. 2013;5(6):338–353. doi: 10.1177/1756287213505672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler & Vertegaal (2015).Eifler K, Vertegaal ACO. SUM Oylation-mediated regulation of cell cycle progression and cancer. Trends in Biochemical Sciences. 2015;40(12):779–793. doi: 10.1016/j.tibs.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han et al. (2017).Han EH, Min JY, Yoo SA, Park SJ, Choe YJ, Yun HS, Lee ZW, Jin SW, Kim HG, Jeong HG, Kim HK, Kim ND, Chung YH. A small-molecule inhibitor targeting the AURKC-IκBα interaction decreases transformed growth of MDA-MB-231 breast cancer cells. Oncotarget. 2017;8(41):69691–69708. doi: 10.18632/oncotarget.18883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2019).He J, Qi Z, Zhang X, Yang Y, Liu F, Zhao G, Wang Z. Aurora kinase B inhibitor barasertib (AZD1152) inhibits glucose metabolism in gastric cancer cells. Anti-Cancer Drugs. 2019;30(1):19–26. doi: 10.1097/CAD.0000000000000684. [DOI] [PubMed] [Google Scholar]
- He et al. (2016).He SJ, Shu LP, Zhou ZW, Yang T, Duan W, Zhang X, He ZX, Zhou SF. Inhibition of Aurora kinases induces apoptosis and autophagy via AURKB/p70S6K/RPL15 axis in human leukemia cells. Cancer Letters. 2016;382(2):215–230. doi: 10.1016/j.canlet.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Kotian et al. (2017).Kotian S, Zhang L, Boufraqech M, Gaskins K, Gara SK, Quezado M, Nilubol N, Kebebew E. Dual inhibition of HDAC and tyrosine kinase signaling pathways with CUDC-907 inhibits thyroid cancer growth and metastases. Clinical Cancer Research. 2017;23(17):5044–5054. doi: 10.1158/1078-0432.CCR-17-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang et al. (2018).Liang L, Zeng JH, Qin XG, Chen JQ, Luo DZ, Chen G. Distinguishable prognostic signatures of left- and right-sided colon cancer: a study based on sequencing data. Cellular Physiology and Biochemistry. 2018;48(2):475–490. doi: 10.1159/000491778. [DOI] [PubMed] [Google Scholar]
- Liao et al. (2018).Liao Y, Liao Y, Li J, Li J, Fan Y, Xu B. Polymorphisms in AURKA and AURKB are associated with the survival of triple-negative breast cancer patients treated with taxane-based adjuvant chemotherapy. Cancer Management and Research. 2018;10:3801–3808. doi: 10.2147/CMAR.S174735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg et al. (2017).Malmberg KJ, Carlsten M, Björklund A, Sohlberg E, Bryceson YT, Ljunggren HG. Natural killer cell-mediated immunosurveillance of human cancer. Seminars in Immunology. 2017;31:20–29. doi: 10.1016/j.smim.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Mast et al. (2019).Mast JM, Tse D, Shee K, Lakshmi Kuppusamy M, Kmiec MM, Kálai T, Kuppusamy P. Diarylidenylpiperidones, H-4073 and HO-3867, induce G2/M cell-cycle arrest, apoptosis and inhibit STAT3 phosphorylation in human pancreatic cancer cells. Cell Biochemistry and Biophysics. 2019;77(2):109–119. doi: 10.1007/s12013-019-00873-6. [DOI] [PubMed] [Google Scholar]
- Mesic et al. (2017).Mesic A, Markocic E, Rogar M, Juvan R, Hudler P, Komel R. Single nucleotide polymorphisms rs911160 in AURKA and rs2289590 in AURKB mitotic checkpoint genes contribute to gastric cancer susceptibility. Environmental and Molecular Mutagenesis. 2017;58(9):701–711. doi: 10.1002/em.22129. [DOI] [PubMed] [Google Scholar]
- Nagarsheth, Wicha & Zou (2017).Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nature Reviews Immunology. 2017;17(9):559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair et al. (2009).Nair JS, Ho AL, Tse AN, Coward J, Cheema H, Ambrosini G, Keen N, Schwartz GK. Aurora B kinase regulates the postmitotic endoreduplication checkpoint via phosphorylation of the retinoblastoma protein at serine 780. Molecular Biology of the Cell. 2009;20(8):2218–2228. doi: 10.1091/mbc.E08-08-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naorem, Muthaiyan & Venkatesan (2019).Naorem LD, Muthaiyan M, Venkatesan A. Integrated network analysis and machine learning approach for the identification of key genes of triple-negative breast cancer. Journal of Cellular Biochemistry. 2019;120(4):6154–6167. doi: 10.1002/jcb.27903. [DOI] [PubMed] [Google Scholar]
- Okegawa et al. (2004).Okegawa T, Pong RC, Li Y, Hsieh JT. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochimica Polonica. 2004;51(2):445–457. [PubMed] [Google Scholar]
- Pohl et al. (2011).Pohl A, Azuma M, Zhang W, Yang D, Ning Y, Winder T, Danenberg K, Lenz HJ. Pharmacogenetic profiling of Aurora kinase B is associated with overall survival in metastatic colorectal cancer. The Pharmacogenomics Journal. 2011;11(2):93–99. doi: 10.1038/tpj.2010.18. [DOI] [PubMed] [Google Scholar]
- R Core Team (2018).R Core Team . Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- Roy et al. (2017).Roy D, Sheng GY, Herve S, Carvalho E, Mahanty A, Yuan S, Sun L. Interplay between cancer cell cycle and metabolism: challenges, targets and therapeutic opportunities. Biomedicine and Pharmacotherapy. 2017;89:288–296. doi: 10.1016/j.biopha.2017.01.019. [DOI] [PubMed] [Google Scholar]
- Sorrentino et al. (2005).Sorrentino R, Libertini S, Pallante PL, Troncone G, Palombini L, Bavetsias V, Spalletti-Cernia D, Laccetti P, Linardopoulos S, Chieffi P, Fusco A, Portella G. Aurora B overexpression associates with the thyroid carcinoma undifferentiated phenotype and is required for thyroid carcinoma cell proliferation. Journal of Clinical Endocrinology and Metabolism. 2005;90(2):928–935. doi: 10.1210/jc.2004-1518. [DOI] [PubMed] [Google Scholar]
- Srigley et al. (2013).Srigley JR, Delahunt B, Eble JN, Egevad L, Epstein JI, Grignon D, Hes O, Moch H, Montironi R, Tickoo SK, Zhou M, Argani P. The international society of urological pathology (ISUP) vancouver classification of renal neoplasia. American Journal of Surgical Pathology. 2013;37(10):1469–1489. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- Subramaniyan, Kumar & Mathan (2017).Subramaniyan B, Kumar V, Mathan G. Effect of sodium salt of Butrin, a novel compound isolated from Butea monosperma flowers on suppressing the expression of SIRT1 and Aurora B kinase-mediated apoptosis in colorectal cancer cells. Biomedicine and Pharmacotherapy. 2017;90:402–413. doi: 10.1016/j.biopha.2017.03.086. [DOI] [PubMed] [Google Scholar]
- Tumino et al. (2019).Tumino N, Martini S, Munari E, Scordamaglia F, Besi F, Mariotti FR, Bogina G, Mingari MC2, Vacca P, Moretta L. Presence of innate lymphoid cells in pleural effusions of primary and metastatic tumors: functional analysis and expression of PD-1 receptor. International Journal of Cancer. 2019 doi: 10.1002/ijc.32262. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2011).Wu L, Ma CA, Zhao Y, Jain A. Aurora B interacts with NIR-p53, leading to p53 phosphorylation in its DNA-binding domain and subsequent functional suppression. Journal of Biological Chemistry. 2011;286(3):2236–2244. doi: 10.1074/jbc.M110.174755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Dong & Guo (2015).Xin M, Dong XW, Guo XL. Role of the interaction between galectin-3 and cell adhesion molecules in cancer metastasis. Biomedicine and Pharmacotherapy. 2015;69:179–185. doi: 10.1016/j.biopha.2014.11.024. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2018).Yang Z, Tao Y, Xu X, Cai F, Yu Y, Ma L. Bufalin inhibits cell proliferation and migration of hepatocellular carcinoma cells via APOBEC3F induced intestinal immune network for IgA production signaling pathway. Biochemical and Biophysical Research Communications. 2018;503(3):2124–2131. doi: 10.1016/j.bbrc.2018.07.169. [DOI] [PubMed] [Google Scholar]
- Yu et al. (2018).Yu J, Zhou J, Xu F, Bai W, Zhang W. High expression of Aurora-B is correlated with poor prognosis and drug resistance in non-small cell lung cancer. International Journal of Biological Markers. 2018;33(2):215–221. doi: 10.1177/1724600817753098. [DOI] [PubMed] [Google Scholar]
- Zhou et al. (2018).Zhou R, Zhang Y, Du G, Han L, Zheng S, Liang J, Huang X, Qin Y, Wu W, Chen M, Wu D, Song L, Fu G, Lv S, Xia Y, Lu C, Wang X. Down-regulated let-7b-5p represses glycolysis metabolism by targeting AURKB in asthenozoospermia. Gene. 2018;663:83–87. doi: 10.1016/j.gene.2018.04.022. [DOI] [PubMed] [Google Scholar]
- Zhu et al. (2019).Zhu Q, Ding L, Zi Z, Gao S, Wang C, Wang Y, Zhu C, Yuan Z, Wei F, Cai Q. Viral-mediated AURKB cleavage promotes cell segregation and tumorigenesis. Cell Reports. 2019;27(5):1633–1636. doi: 10.1016/j.celrep.2019.04.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplemental Files.