Summary
Derivation of functional skeletal muscle stem cells from pluripotent cells without genetic modification has proven elusive. Here, we show that teratomas formed in adult skeletal muscle differentiate in vivo to produce large numbers of α7-Integrin+ VCAM-1+ myogenic progenitors. When FACS-purified and transplanted into diseased muscles, teratoma-derived myogenic progenitors demonstrate very high engraftment potential. As few as 40,000 cells can reconstitute ~80% of the tibialis anterior muscle volume. Newly generated fibers are innervated, express adult myosins, and ameliorate dystrophy-related force deficit and fatigability. Teratoma-derived myogenic progenitors also contribute quiescent PAX7+ muscle stem cells, enabling long-term maintenance of regenerated muscle and allowing muscle regeneration in response to subsequent injuries. Transcriptional profiling reveals that teratoma-derived myogenic progenitors undergo an embryonic to adult maturation when they contribute to the stem cell compartment of regenerated muscle. Thus, teratomas are a rich and accessible source of potent transplantable skeletal muscle stem cells.
Keywords: muscle stem cells, satellite cells, pluripotent stem cells, teratoma, myogenesis
eTOC blurb
Kyba and colleagues show that functional skeletal muscle stem cells can be produced from mouse pluripotent stem cells without genetic modification through teratoma formation. As few as 40,000 teratoma-derived cells can regenerate 80% of total muscle volume, improve force generation, and mature into functional muscle stem cells in vivo.
Introduction
PAX7+ satellite cells are responsible for muscle maintenance and regeneration after injury throughout life (Gunther et al., 2013; Seale et al., 2000; von Maltzahn et al., 2013). They are rare, making up 1–2% of the mononuclear fraction of skeletal muscle (Bosnakovski et al., 2008), itself a small fraction of total muscle mass, which is comprised mainly of multinucleated muscle fibers. Satellite cells associate intimately with fibers, residing under the fiber basal lamina (Mauro, 1961), and cannot be isolated without destroying muscle tissue, therefore only relatively small biopsies are feasible for human transplantation. Although freshly isolated satellite cells have tremendous regenerative potential (Arpke et al., 2013; Collins et al., 2005; Hall et al., 2010; Sacco et al., 2008), it is insufficient to enable a meaningful therapy from a small biopsy. In culture, satellite cells activate and convert into myoblasts, whose transplantation potential is limited (Gussoni et al., 1992; Mendell et al., 1995; Montarras et al., 2005; Sacco et al., 2008). Embryonic stem (ES) cells and induced pluripotent stem (iPS) cells have unlimited expansion potential, theoretically enabling large numbers of derivative cells for transplantation, however skeletal myogenesis does not arise spontaneously from pluripotent cells in vitro and although progress is being made (Chal et al., 2015; Shelton et al., 2014), cells capable of generating functional force-producing muscle after transplantation have only been derived through genetic modification of pluripotent cells to overexpress PAX3 (Darabi et al., 2008; Filareto et al., 2013) or PAX7 (Darabi et al., 2012).
The skeletal muscle lineage derives from a complex morphogenetic pathway, somitogenesis, involving precisely-timed mesenchymal condensation, patterning by neural tube and notochord, and delamination of myogenic progenitors. In vitro methods have not yet approached this complexity of morphogenesis, however teratomas derived from pluripotent stem cells implanted into live hosts are capable of producing highly complex mature tissues: hair follicles, glands, and other structures. Also, it has been reported that transplantable hematopoietic stem cells arise within teratomas in both the mouse (Suzuki et al., 2013), and the human system (Amabile et al., 2013). We therefore investigated teratomas for signs of skeletal myogenic progenitor formation, evaluated the nature of these progenitors, and investigated their in vivo muscle formation, force generation, and stem cell compartment engraftment potential.
Results
α7-Integrin+ VCAM-1+ teratoma cells are skeletal muscle progenitors
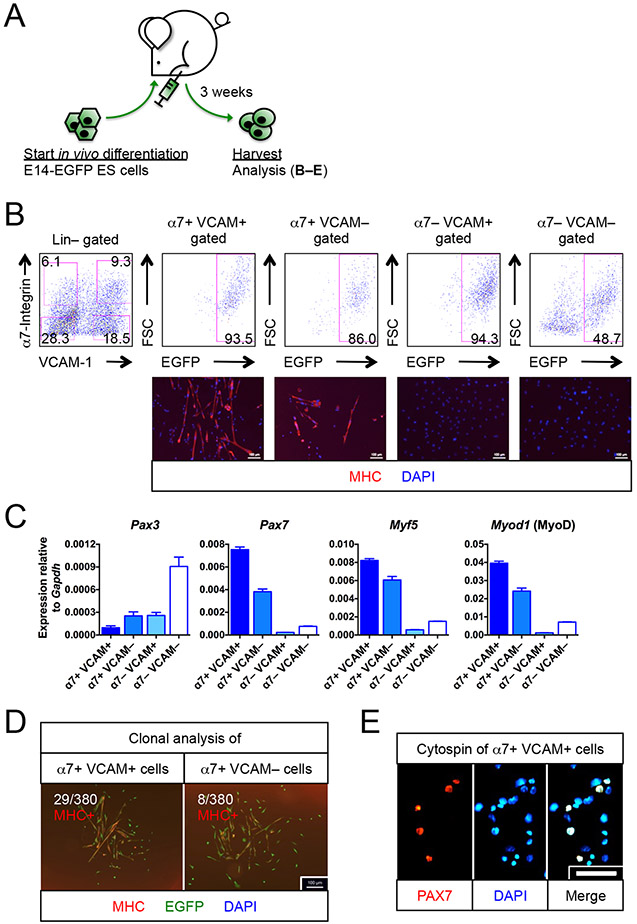
To maximize access of teratoma-derived cells to a pro-myogenic environment, we implanted EGFP+ murine ES cells (E14-EGFP ES cells) (Ismailoglu et al., 2008) into injured, irradiated tibialis anterior (TA) muscles of NSG-mdx4Cv mice. These animals are both immune- and dystrophin-deficient and therefore allow not only facile engraftment, but unequivocal assignment of donor identity (DYSTROPHIN+) to regenerated muscle tissue (Arpke et al., 2013). Prior to implantation, hind limbs were irradiated to impair host satellite cells, and TA muscles were injected with cardiotoxin to kill host fibers and to stimulate myogenesis. Using flow cytometry on three week teratomas (Figure 1A), we evaluated the population of cells negative for the hematopoietic and endothelial markers CD45 and CD31 (Lin−) with antibodies to the satellite cell markers α7-integrin and VCAM-1 (hereafter referred to as α7 and VCAM respectively) (Blanco-Bose et al., 2001; Chan et al., 2013; Fukada et al., 2007; Jesse et al., 1998; Seale et al., 2004). The α7+ VCAM+ population was abundant, forming about 10% of the total Lin− fraction, and the majority of α7+ VCAM+ cells were also EGFP+, i.e., donor-derived (Figures 1B and S1A). Teratomas also contained host-derived hematopoietic, endothelial, and other cells, demonstrating that the teratoma vigorously interacts with its host, with potential effects on differentiation (Figure S1B). We found minimal expression of other satellite cell markers on Lin− cells, such as CD34 or CXCR4 (Figure S1C). While α7+ VCAM+ cells were prominent at 3 weeks and beyond, their emergence could first be detected at 2 weeks post-ES cell implant (Figures S1D-E).
Figure 1. Myogenic progenitors are found in teratomas.
(A)Schematic of generating myogenic progenitors from EGFP-labeled E14 (E14-EGFP) ES cells in vivo.
(B) E14-EGFP ES cell-derived myogenic progenitors. FACS profiling (top row) of 3 week-old teratomas revealed the presence of α7+ VCAM+ and α7+ VCAM− putative myogenic progenitors. Immunostaining (bottom row) confirmed their myogenic identity (MHC+) (n=6 biological replicates). The other 2 fractions, α7− VCAM+ and α7− VCAM−, had minimal myogenic potentials (n=4 biological replicates). Scale bar represents 100 μm.
(C) Quantitative RT-PCR for markers for muscle stem cells (Pax3, Pax7), activated myogenic progenitor cells (Myf5) and myogenic-committed cells (Myod1) (n=6, from 2 biological replicates). Note that Pax3 is also a marker of neuroectoderm derivatives.
(D) Clonal analysis showing that single α7+ VCAM+ or α7+ VCAM− cells were capable of forming MHC+ myogenic colonies with differentiated myoblasts and multi-nuclei myotubes. Ratio indicates number of colonies developed per number of single cells seeded (n=5 biological replicates). Scale bar represents 100 μm.
(E) Cytospins of α7+ VCAM+ cells showing that 30% of which expressed PAX7+, a muscle stem cell transcription factor (n=4 biological replicates). Scale bar represents 100 μm.
α7, α7-integrin. VCAM, VCAM-1. ES cells, embryonic stem cells. Lin, lineage cocktail comprising antibodies against CD45 (hematopoietic) and CD31 (endothelial). MHC, myosin heavy chain. Mean ± SEM is shown in (C). See also Figure S1.
We sorted cells based on α7 and VCAM expression, and found that the α7+ fractions contained mRNA for myogenic transcription factors (Figure 1C). Interestingly, although both α7+ fractions express the embryonic myogenic factor Pax3, its expression is more abundant in the double negative fraction, indicating that most Pax3 expression in teratomas probably comes from neuroectodermal cells. Both α7+ fractions generated differentiated myotubes in vitro, however at a single cell level, myogenic clones were more frequent in the α7+ VCAM+ population (Figure 1D). We evaluated PAX7 protein expression by immunostaining cytospins of Lin− α7+ VCAM+ cells. This revealed the population to be heterogeneous, with about 30% of cells expressing PAX7 (Figure 1E). To determine the proliferative capacity of these cells, we subjected them to long-term ex vivo culture. We found that teratoma-derived α7+ VCAM+ cells could be exponentially expanded up to at least 12 passages, with the expanded cells retaining robust myogenic potential (Figure S1F). This is notable, as myoblasts from adults are reported to lose myogenic potential with extended passage (Penton et al., 2016).
The teratomas described above were formed in muscle that was irradiated and cardiotoxin-injured to enhance myogenesis and to minimize host contribution to the α7+ VCAM+ compartment. To evaluate the necessity of irradiation and injury, we formed teratomas in hosts pretreated with in various ways, and evaluated the development of myogenic progenitors (Figure S1G-H). EGFP+ α7+ VCAM+ cells were found in all scenarios, and they had comparable potentials in forming myotubes in vitro and fibers after transplantation (below). Nevertheless, the irradiated and injured hosts showed much greater enrichment of EGFP+ cells within the α7+ VCAM+ fraction, and thus teratomas were formed in irradiated, injured hosts for all following experiments.
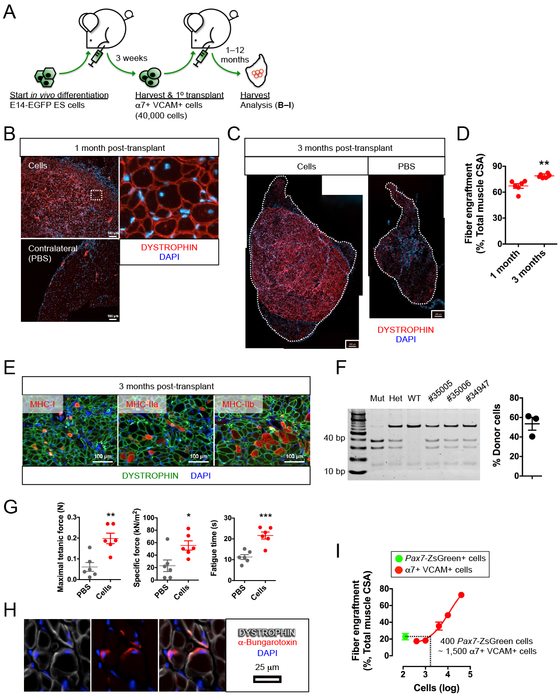
Primary myogenic cells from teratomas have extremely high in vivo regenerative potential
To test the in vivo regenerative potential of these α7+ VCAM+ cells, we performed transplants 40,000 EGFP+ Lin− α7+ VCAM+ cells into TA muscles of new NSG-mdx4Cv recipients (Figure 2A). Prior studies transplanting myoblasts (Partridge et al., 1989) or PAX3/7-modified pluripotent cells (Darabi et al., 2008) have used one or more orders of magnitude more cells. We began with this relatively low number in order to transplant a number of PAX7+ cells close to the number of endogenous PAX7+ cells in a single TA muscle (Brack et al., 2005). One month post-transplant, we observed muscle regeneration on a scale that significantly surpasses previous reports. The engraftment of DYSTROPHIN+ fibers was pervasive (Figures 2B-C). At one month post-transplant, this accounted for approximately 2/3 of the TA muscle, and at three months, about 80% of total muscle cross-sectional area (Figures 2C-D). These newly formed DYSTROPHIN+ fibers were of varied phenotype, consisting of both slow (MHC-I) and fast (MHC-IIa, MHC-IIb) twitch fibers (Figure 2E and Figure S2A).
Figure 2. ES cell-derived myogenic progenitors reconstitute functional fibers.
(A) Schematic of functional evaluation of E14-EGFP ES cell-derived α7+ VCAM+ myogenic progenitors.
(B–F) ES cell-derived α7+ VCAM+ myogenic progenitors engrafted and differentiated into functional muscle fibers.
(B) α7+ VCAM+ myogenic progenitors engrafted and differentiated into DYSTROPHIN+ muscle fibers (top left) (n=18 biological replicates). Areas depicted by the white dotted rectangle are magnified to show individual fibers (top right). Minimal DYSTROPHIN+ revertant fibers were observed in the contralateral muscle with PBS injection (bottom left). Scale bar represents 100 μm.
(C) Engraftment (DYSTROPHIN+ fibers) at 3 months comparing injected (left) to contralateral PBS-injected (right) (n=6 biological replicates). The whole TA is outlined. Scale bar represents 200 μm.
(D) Quantitation of fiber engraftment (DYSTROPHIN+ fibers) at 1 month and 3 months (n=6 biological replicates). **p<0.01 versus 1 month.
(E) Engrafted DYSTROPHIN+ muscle fibers consist of slow (MHC-I) and fast (MHC-IIa, MHC-IIb) twitch fibers (n=6 biological replicates). Scale bar represents 100 μm.
(F) Contribution of donor nuclei in transplanted muscle. SCID PCR of total genomic DNA from transplanted muscles (left) and quantification (right). The first 3 control lanes represent: Mut, PrkdcSCID/SCID; Het, Prkdc+/SCID; WT: Prkdc+/+. The 3 right lanes represent transplanted muscles.
(G) Ex vivo physiological assessment revealed functional improvement at 3 months after α7+ VCAM+ cells transplantation (n=6 biological replicates). *p<0.05, **p<0.01, ***p<0.001 versus PBS (vehicle).
(H) Cross section showing pre-synaptic staining with α-bungarotoxin in DYSTROPHIN+ fibers (n=3 biological replicates). Scale bar represents 25 μm.
(I) Comparison of Pax7-ZsGreen satellite cells and α7+ VCAM+ teratoma cells: 400 Pax7-ZsGreen cells are equivalent to 1,500 α7+ VCAM+ cells for fiber engraftment (n=4–5 biological replicates). Note that the raw data for generating the dose-response relationship is shown in Figure S2F.
α7, α7-integrin. VCAM, VCAM-1. CSA, cross-sectional area. ES cells, embryonic stem cells. Lin, lineage cocktail comprising antibodies against CD45 (hematopoietic) and CD31 (endothelial). MHC, myosin heavy chain. Mean ± SEM is shown in (D), (F), (G) and (I). See also Figure S2.
Since the recipient mice were irradiated to prevent the contribution of endogenous cells for regeneration, we infer that the regenerated DYSTROPHIN+ fibers predominately derive from donor cells. Nevertheless, we wished to rigorously evaluate the donor:host ratio using a direct approach. Because the NSG-mdx4Cv recipient mice are homozygous for a single nucleotide polymorphism in the Prkdc gene (the SCID mutation), we amplified this region from total genomic DNA from transplanted TA muscles and evaluated frequency of donor vs. host DNA sequence (Figure 2F). This revealed the average donor contribution to be over 50% of the total genomic DNA content of the transplanted TA. It is notable that the TA muscle contains many host-derived non-myogenic cell types such as fibroblasts, endothelial cells and hematopoietic cells, all of which contribute to the host component of this measurement, so this value is consistent with the donor fibers being almost exclusively derived from donor nuclei.
To ensure that differentiation via teratoma had not led to undesirable changes in the cells being transplanted, we evaluated karyotypes of α7+ VCAM+ cells. This revealed that teratoma-derived cells did not acquire any numerical and structural chromosomal abnormality, and they retained a normal karyotype after serial passages (Figure S2B). We further evaluated the status of the regenerated muscle for signs of abnormally maintained proliferation and differentiation. Embryonic MHC, a marker of recently-generated fibers, was present in only a negligible portion of DYSTROPHIN+ fibers at 1 month post-transplant and its expression was completely absent after 3 months (Figure S2C), indicating that the newly formed fibers were maturing and that new fibers were not being generated. To determine whether an abnormal population of proliferating mononuclear cells was present, we administered a 3-day pulse of 5-ethynyl-2’-deoxyuridine (EdU), at 3 months post-transplant and harvested the TA muscles 5 weeks later. We did not observe any EdU incorporation nor any embryonic MHC positivity in DYSTROPHIN+ fibers (Figure S2D), indicating that the engrafted cells became quiescent over time. Neither did we observe any secondary teratomas forming after transplantation, in well over 100 transplantations of teratoma-derived α7+ VCAM+ cells, 12 of which were evaluated at one year. Taken together, this data suggests that teratoma formation per se does not induce carcinogenicity.
To test the impact of irradiation, which eliminates host competition, and cardiotoxin injury, which initiates widespread regeneration, we tested transplants of teratoma-derived α7+ VCAM+ cells without irradiation and injury. Although engraftment was lower in these cases, α7+ VCAM+ cells were capable of forming large numbers of new fibers in irradiated-only muscles, injured-only muscles, and also non-irradiated uninjured muscles (Figure S2E).
To determine whether the newly-formed muscle tissue was functional, we measured its force-generation capability (Figure 2G). The maximal tetanic force generated by the α7+ VCAM+ cell-regenerated muscles was approximately 3 times greater than that of the sham contralateral controls, indicating that the new muscle fibers are functional, i.e., capable of generating force when stimulated. The engrafted muscles also showed greatly increased specific force (maximal force normalized to the size of the muscle). This improved quality of force metric reflects the fact that the newly engrafted muscle is DYSTROPHIN+ and thus less diseased, while the disease process was ongoing in the contralateral leg and possibly amplified due to the impairment of host satellite cells by the irradiation. In addition, the time to fatigue after repetitive contractions was much increased in the cell-transplanted TAs vs. contralateral controls, further reflecting the DYSTROPHIN+ nature of the newly-formed muscle.
To further demonstrate the functionality of these newly formed fibers, we tested whether they were innervated and thereby integrated into the recipient environment. Staining with the presynaptic marker α-bungarotoxin revealed its close proximity to DYSTROPHIN+ fibers, suggesting the presence of neuromuscular junctions in these fibers (Figure 2H).
The substantial muscle regenerative potential of these α7+ VCAM+ myogenic cells encouraged a direct comparison with bona fide satellite cells. We used the Pax7-ZsGreen reporter mouse (Bosnakovski et al., 2008) to isolate adult satellite cells by FACS, and compared fiber formation of these to fiber formation of various numbers of teratoma-derived α7+ VCAM+ cells, establishing the dose-response relationship for fiber engraftment (Figure 2I and S2F). Remarkably, 1,500 teratoma-derived α7+ VCAM+ cells are functionally equivalent to 400 freshly isolated Pax7-ZsGreen+ cells.
Previously published data with transplantation from in vitro differentiated pluripotent cells have shown only small clusters of at most 1-200 fibers. Using the same injury/irradiation protocol in the same host strain, we directly compared teratomas differentiation to in vitro differentiation. We adopted a monolayer differentiation protocol (Chal et al., 2013) and detected α7+ VCAM+ cells with myogenic potential after 4 weeks of culture (Figure S2G). We subsequently transplanted 40,000 α7+ VCAM+ sorted cells and harvested the transplanted TA muscles 3 months later. This comparison supported the published data in showing that cells generated via in vitro differentiation give much poorer engraftment than cells derived via teratoma formation (Figure S2G).
To demonstrate that the development of myogenic progenitors was not restricted to the 129P2/Ola background from which our ES cells derive, we repeated the experiments using another ES cell line with a C57BL/6N background (C57BL/6N-PRX-B6N #1). Similarly, α7+ VCAM+ cells were found in 3-week-old C57BL/6N teratomas, and upon transplantation into NSG-mdx4Cv recipients, formed DYSTROPHIN+ muscle fibers (Figure S2H).
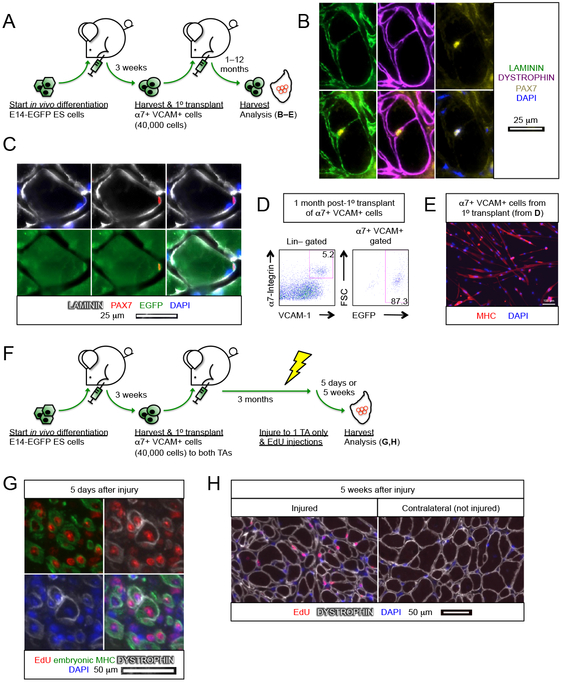
A subpopulation of cells engrafts as functional muscle stem cells
Long-term maintenance of new skeletal muscle is ultimately dependent on the ability of the transplanted cells to contribute to the skeletal muscle stem cell pool. Immunostaining revealed PAX7+ EGFP+ cells associated with DYSTROPHIN+ fibers under the basal lamina (Figure 3A-C). We did not observe teratoma-derived cells in the skeletal muscle interstitium nor any circulating teratoma-derived myogenic cells in the peripheral blood by FACS (data not shown). We then analyzed a cohort of recipients for contribution to the stem cell compartment by digesting the transplanted TAs into individual cells, and testing for donor-derived (EGFP+) cells in the mononuclear fraction. Transplanted muscles had an abundant α7+ VCAM+ population within the Lin− fraction, and the great majority of these cells were donor derived, i.e., EGFP+ (Figure 3D). Re-isolated donor-derived cells differentiated into multinucleated myotubes in vitro, indicating that they are indeed myogenic progenitors (Figure 3E).
Figure 3. ES cell-derived myogenic progenitors reconstitute the muscle stem cell compartment.
(A) Schematic of evaluating the contribution of E14-EGFP ES cell-derived α7+ VCAM+ myogenic progenitors in the muscle stem cell compartment.
(B) PAX7+ cell associated with a DYSTROPHIN+ fiber under the basal lamina (n=6 biological replicates). Scale bar represents 25 μm.
(C) Donor derived PAX7+ EGFP+ muscle stem cell under the basal lamina (n=6 biological replicates). Scale bar represents 25 μm.
(D) FACS analysis of transplanted muscles revealed that the majority of α7+ VCAM+ muscle stem cells are also EGFP+, i.e., donor derived (n=7 biological replicates).
(E) Re-harvested α7+ VCAM+ EGFP+ cells (from C) differentiated into multi-nuclei MHC+ myotubes upon culture (n=7 biological replicates). Scale bar represents 100 μm.
(F) Schematic of evaluating the regenerative potency of transplanted E14-EGFP ES cell-derived α7+ VCAM+ myogenic progenitors upon subsequent injury.
(G,H) Transplanted α7+ VCAM+ cells regenerate upon subsequent injury. α7+ VCAM+ cells were transplanted into both left and right TA muscles and 3 months later only the left TA muscles were re-injured. EdU was administered 2–4 days after re-injury. Muscles were then harvested (G) 5 days or (H) 5 weeks after injury.
(G) The presence of EdU+ embryonic MHC+ fibers 5 days after re-injury indicates regeneration. Some EdU+ embryonic MHC+ fibers start to re-express DYSTROPHIN (n=4 biological replicates). Scale bar represents 50 μm.
(H) EdU+ DYSTROPHIN+ fibers in the injured muscle 5 weeks post-injury. Note that in the contralateral muscle that was not re-injured after the primary transplant, only EdU− DYSTROPHIN+ fibers were present (n=4 biological replicates). Scale bar represents 50 μm.
α7, α7-integrin. VCAM, VCAM-1. ES cells, embryonic stem cells. Lin, lineage cocktail comprising antibodies against CD45 (hematopoietic) and CD31 (endothelial). MHC, myosin heavy chain.
To rigorously characterize the regenerative potential of the engrafted mononuclear cell population, we tested the regenerated muscle by re-injury (Figure 3F). Three months after performing primary transplants of teratoma-derived α7+ VCAM+ cells into both TAs, we re-injured one of the muscles and administered EdU for 3 days. TA muscles were then harvested 5 days or 5 weeks after the re-injury. In the 5-day post-injury cohort, EdU+ embryonic MHC+ cells were readily observed, indicating robust regeneration after injury (Figure 3G). In the 5-week post-injury cohort, EdU+ DYSTROPHIN+ fibers were now found in the re-injured TA muscle whereas the DYSTROPHIN+ muscle fibers of the contralateral TA were EdU−. This demonstrates that the engrafted, quiescent, mononuclear fraction is capable of proliferating in response to re-injury, and of generating a secondary regenerate (Figure 3H).
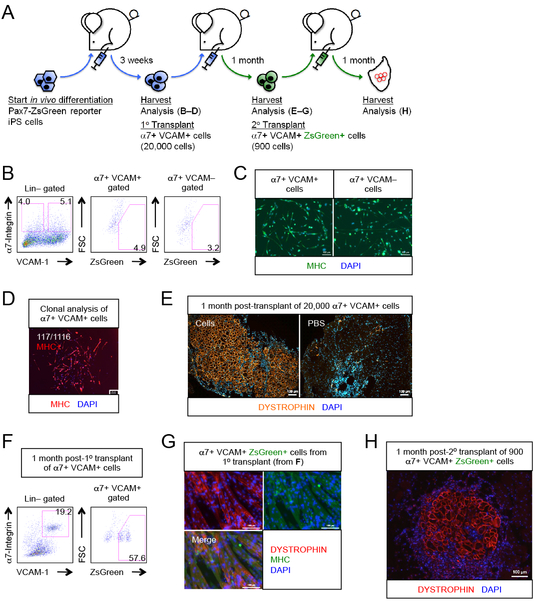
We next performed a series of studies using iPS cells that we generated from the Pax7-ZsGreen reporter mouse, in which quiescent satellite cells express the ZsGreen reporter (Bosnakovski et al., 2008). In the initial teratomas, we found surprisingly that the Pax7-ZsGreen reporter was not expressed in a significant fraction of the α7+ VCAM+ population (Figures 4A-B). Nevertheless, like their ES cell-derived cognates, the iPS cell-derived α7+ VCAM+ cells could differentiate into MHC-expressing multinucleated myotubes in vitro, both in bulk and clonally (Figures 4C-D). When transplanted into NSG-mdx4Cv recipients, the iPS cell-derived α7+ VCAM+ cells formed abundant myofibers (Figure 4E). Multiple independent Pax7-ZsGreen iPS cell clones gave similar results (Figure S3A). Most importantly, in these transplanted muscles we now found a large population of Pax7-ZsGreen+ α7+ VCAM+ cells (Figures 4F and S3B), thus although the Pax7-ZsGreen reporter is not expressed in the α7+ VCAM+ cells of the teratoma, it comes on in the α7+ VCAM+ cells of the newly regenerated muscle. To confirm that the donor-derived mononuclear Pax7-ZsGreen cells in the regenerated muscle were indeed skeletal muscle stem cells, we isolated them and tested their myogenic differentiation potential, both in vitro by colony assays, and in vivo by secondary transplantation. They produced MHC+ DYSTROPHIN+ myotubes in vitro (Figure 4G) and myofibers in NSG-mdx4Cv hosts (Figure 4H). This demonstrates that in addition to regenerating muscle fibers and rebuilding damaged muscle, teratoma-derived myogenic progenitors populate this new muscle with mature PAX7+ α7+ VCAM+ definitive skeletal muscle stem cells.
Figure 4. Myogenic progenitors derived from teratomas are muscle stem cells.
(A) Schematic of transplantation experiment using Pax7-ZsGreen reporter iPS cells to test whether PAX7+ muscle stem cells are generated.
(B) α7+ VCAM+ and α7+ VCAM− myogenic progenitors were found in teratomas generated from Pax7-ZsGreen iPS cells, but ZsGreen was barely expressed in both fractions (n=18 biological replicates).
(C) iPS teratoma-derived α7+ VCAM+ and α7+ VCAM− myogenic progenitors differentiated into MHC+ myogenic derivatives upon culture (n=3 biological replicates). Scale bar represents 100 μm.
(D) Clonal analysis showing that single Pax7-ZsGreen reporter iPS cell-derived α7+ VCAM+ cells were capable of forming MHC+ myogenic colonies with differentiated myoblasts and multi-nucleated myotubes (n=12 biological replicates). Ratio indicates number of colonies developed per number of single cells seeded. Scale bar represents 100 μm.
(E) iPS teratoma-derived α7+ VCAM+ myogenic progenitors differentiated into muscle fibers upon transplantation (n=12 biological replicates). Scale bar represents 100 μm.
(F) FACS analysis of transplanted muscles revealed that a significant fraction of the α7+ VCAM+ population is ZsGreen+, i.e., donor derived muscle stem cells (n=10 biological replicates). Compare to 4B, above.
(G) In vitro culture of α7+ VCAM+ ZsGreen+ muscle stem cells (from F) produced MHC+ DYSTROPHIN+ muscle fibers (n=10 biological replicates). Scale bar represents 100 μm.
(H) α7+ VCAM+ ZsGreen+ muscle stem cells engrafted into the muscle fiber compartment upon transplantation (n=3 biological replicates). Scale bar represents 100 μm.
α7, α7-integrin. VCAM, VCAM-1. ES cells, embryonic stem cells. Lin, lineage cocktail comprising antibodies against CD45 (hematopoietic) and CD31 (endothelial). MHC, myosin heavy chain. See also Figure S3.
Notably, the Pax7-ZsGreen reporter was expressed in many more α7+ VCAM+ cells in regenerated muscle than in teratomas, suggesting a maturation from an embryonic progenitor (Bober et al., 1994; Goulding et al., 1994) into a PAX7+ quiescent adult satellite cell (Kuang et al., 2006) only after removal from the teratoma and transplantation into adult muscle.
α7-Integrin+ VCAM-1+ teratoma cells mature after transplantation
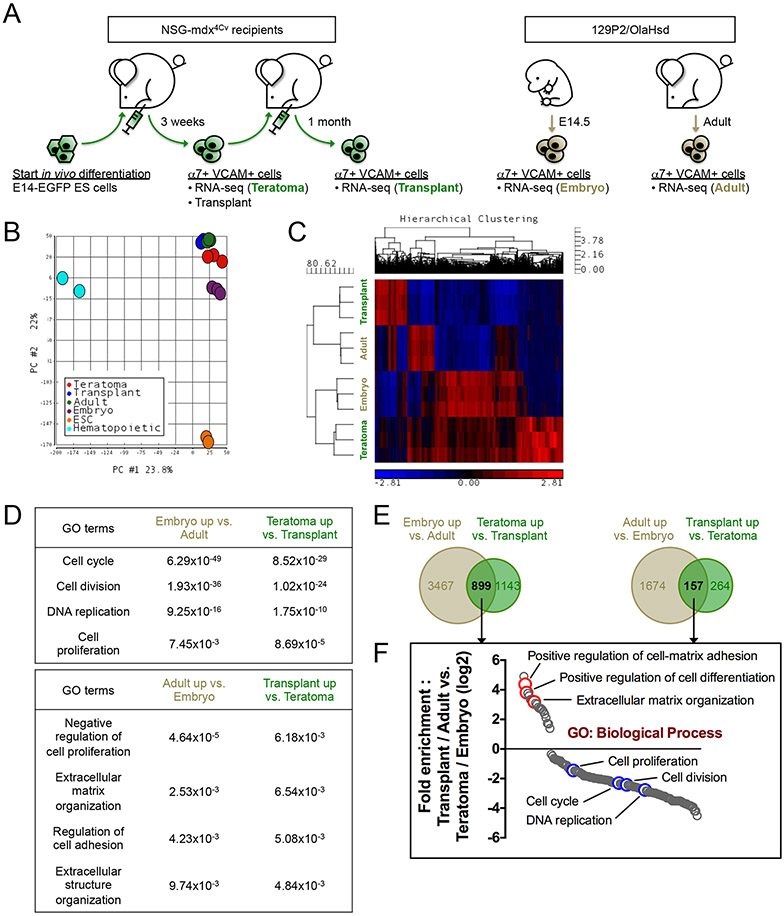
To better understand the nature of teratoma-derived α7+ VCAM+ myogenic progenitors before and after transplantation, we performed RNA-seq on α7+ VCAM+ cells isolated from teratomas, their mononuclear progeny after transplantation and repopulation of the new muscle satellite cell compartment, and for comparison from α7+ VCAM+ myogenic progenitors of isogenic (129P2/OlaHsd) E14.5 embryos and hind limbs of 8-week-old 129P2 mice (Teratoma, Transplant, Embryo, and Adult, respectively; Figure 5A). It is important to emphasize that all four populations are very similar in nature; they were all highly purified on α7+ and VCAM+ and they are all myogenic. To illustrate this point, we performed Principal Component Analysis (PCA) including non-myogenic cell types such as undifferentiated ES cells and hematopoietic progenitors (datasets from ENCODE, www.encodeproject.org), and indeed all 4 α7+ VCAM+ populations cluster very close together (Figure 5B and Figure S4A). Hierarchical clustering of differential genes revealed that the transcriptome of the teratoma α7+ VCAM+ population was most closely related to that of the embryonic α7+ VCAM+ population, while the α7+ VCAM+ cells in transplanted regenerated muscle were most closely related to the adult satellite cell population (Figure 5C and Table S1-2). Indeed, Gene Ontology (GO) analysis showed similar enrichment between the Teratoma-Transplant comparison and the Embryo-Adult comparison (Figure 5D). Notably, genes that are upregulated in both the Teratoma and Embryo samples were related to cell division and DNA replication, reminiscent of an embryonic myoblast signature of rapid growth and proliferation (Figure 5E-F). On the other hand, both Transplant and Adult α7+ VCAM+ cells are enriched for genes pertaining to extracellular matrix regulation, likely involved in satellite cell niche signaling and quiescence (Figure 5E-F).
Figure 5. Teratoma-derived myogenic cells mature into muscle stem cells after transplantation.
(A) Schematic of samples used for transcriptome analysis. α7+ VCAM+ myogenic cells were isolated from E14-ES cell teratomas (Teratoma), transplanted TA muscles (Transplant), E14.5 embryos (Embryo) and 8-week-old adult hindlimbs (Adult) for RNA-seq (n=3 biological replicates).
(B) Principal Component Analysis (PCA). The transcriptomes of the 4 α7+ VCAM+ myogenic populations (Teratoma, Transplant, Embryo and Adult) are very similar to each other, in comparison with those of ES cells and hematopoietic progenitors. The ES cells and the hematopoietic progenitors RNA-seq datasets were obtained from ENCODE (encodeproject.org).
(C) Hierarchical clustering of differentially expressed genes demonstrates a transcriptome similarity between Transplant myogenic cells and Adult satellite cells, and between Teratoma cells and Embryo progenitors.
(D) Gene Ontology (GO) Biological Process terms denoting genes enriched in the Embryo-Adult comparison (left column) and the Teratoma-Transplant comparison (right column). p-values of the GO terms are indicated.
(E) Venn diagrams showing differential genes commonly upregulated by Teratoma / Embryo progenitors and Transplant / Adult cells. The number of differential genes is indicated.
(F) Fold enrichment analysis of genes obtained from (E) based on GO Biological Process.
α7, α7-integrin. VCAM, VCAM-1. ES cells / ESC, embryonic stem cells. GO, Gene Ontology. See also Table S1-S4.
To gain insight into why teratoma-derived skeletal myogenic progenitors have such high engraftment efficiency, we performed GO analysis on the Teratoma-Embryo comparison and the Transplant-Adult comparison (Figure S4B-C and Table S3-4). Interestingly, Teratoma α7+ VCAM+ cells are enriched for genes related to immune response and cell migration, suggesting that teratoma cells may have an enhanced ability to evade the host’s remaining immune system (the NSG mice still have functional neutrophils and monocytes) and to migrate to distant areas of the muscle, away from the site of injection. In contrast, comparing to the Adult sample, genes enriched in the Transplant sample are related to muscle development, perhaps indicating that maturation is an ongoing process. Taken together, our data support that teratoma-derived α7+ VCAM+ cells are embryonic in nature, but after transplantation into adult muscle undergo an in vivo maturation into quiescent muscle stem cells.
Discussion
Here we describe a simple and efficient method for generating skeletal myogenic progenitors from pluripotent stem cells. Via teratoma formation within the TA muscle, α7+ VCAM+ cells arise robustly within three weeks, and more importantly, on a per cell basis, these cells have remarkable in vivo regenerative potential, developing into muscle fibers with similar efficiency to freshly isolated satellite cells, and capable of regenerating 80% or more of the TA muscle fibers, and over 50% of the total genomic DNA of the post-transplant TA muscle. With conventional tissue culture, it has proven difficult to efficiently derive myogenic progenitors from pluripotent stem cells, and to date, the only functional force-generating repopulating cells reported have been derived through genetic modification to overexpress PAX3 or PAX7 (Darabi et al., 2012; Darabi et al., 2008).
Another somatic stem cell type that has proven difficult to derive from unmodified pluripotent stem cells is the hematopoietic stem cell (HSC). Although pluripotent cells differentiate efficiently into blood progenitors, these progenitors do not have the capacity to engraft adults long-term, unless genetically modified with self-renewal factors (Kyba et al., 2002; Perlingeiro et al., 2001). It was recently reported that transplantable blood progenitors can be identified within teratomas (Suzuki et al., 2013), although in some cases with low engraftment potential (Amabile et al., 2013). It is remarkable that the skeletal muscle stem cells isolated from teratomas function equivalently to if not better than their definitive adult cognates in transplantation assays. When diluted to determine their fiber-generation potential on a per cell basis, they showed about 30% the activity of freshly isolated satellite cells, however this is about four orders of magnitude higher than other non-satellite cell muscle regenerative cells described to date. Because we see no evidence of unwanted cell types one month post-transplant, the moderate difference in efficiency of engraftment teratoma-derived α7+ VCAM+ cells is probably due to a cell-intrinsic difference in capacity, however we cannot rule out that some fraction of the sorted cells fail to engraft. However, comparing maximum engraftment potential at higher cell numbers, the fiber generation potential of satellite cells becomes non-linear earlier, such that satellite cells peak at restoring around one third of the TA muscle (Arpke et al., 2013), while with teratoma-derived α7+ VCAM+ cells, we find a mean of 70% of the muscle restored at one month, and 80% at 3 months. This difference is probably due to the non-adult character of the teratoma-derived cells, a result supported by the gene expression analysis. The adult TA muscle, together with its stem cells, derives entirely from a small embryonic founder population, thus the embryonic progenitors must have greater muscle generation potential than their adult derivatives; however it is also worth considering that their embryonic character may endow them with altered migratory potential, thus allowing them to contribute to fibers over a greater range from the site of injection. In fact, GO term analysis from our RNA-seq data supported the notion that teratoma-derived cells might present different immune and migration responses, thereby allowing superior regeneration potential. It is reasonable to assume that stem cells of other lineages could be isolated with equal effectiveness from teratomas, and given the results described here with muscle, and above with blood, this idea merits further investigation.
The fact that cells are spontaneously differentiated within and isolated from teratomas raises the question of whether they might themselves be teratomagenic. However, unlike spontaneous teratomas in adults, those from pluripotent cells are not carcinogenic; it is only the pluripotent status of their founder cells that presents a risk of overgrowth. It is important to point out that cells derived from pluripotent cells differentiated via conventional in vitro methods present no less risk. In fact, previous transplantation studies with pluripotent cells modified to overexpress PAX3 and differentiated in vitro found that teratomas developed in some recipients if non-mesodermal cells were not eliminated (Darabi et al., 2008). This problem was solved by FACS sorting the progenitors of interest away from the bulk of differentiating cells. Teratomas have even arisen when porcine fetal tissue was grafted into adult recipients. In these studies, isolated liver, pancreas and lung, taken from early developmental stages, but well after pluripotent cells are thought no longer to be present, generated teratomas when implanted under the kidney capsule (Eventov-Friedman et al., 2005). Therefore, regardless of whether differentiation is performed in vitro, in vivo, or from fetal donors, cell purification must be equally rigorous to address the risks of teratomagenesis in the recipient, and teratoma differentiation is no more risky than in vitro differentiation in this regard. In the current study, no teratomas were observed in any mice transplanted with teratoma-derived α7+ VCAM+ cells, followed for over 12 months. In addition, we found no evidence of abnormally maintained proliferation, as measured by EdU incorporation, in muscle reconstituted from teratoma-derived α7+ VCAM+ cells. The engrafted mononuclear fraction stays quiescent as long as the muscle is not injured. Upon re-injury however, these cells readily incorporate EdU, proliferate, and generate new fibers, as would be expected from normal quiescent skeletal muscle stem cells.
Differentiation via teratoma provides several advantages: it is technically simple, inexpensive, and capable of producing large quantities of skeletal muscle stem cells. However, its most striking feature is the scale of contribution to tissue regeneration after transplantation. The greatest engraftment previously documented in studies of monolayer in vitro differentiated pluripotent cells has been of less than 200 fibers, and this is in the case of transplanting on the order of 1 million cells. In contrast, myogenic progenitors derived from teratomas produce thousands of fibers, contributing to approximately 80% of the regenerated TA muscle fibers and over 50% of the total genomic DNA content of the recipient TA muscle, but with orders of magnitude fewer cells transplanted. This level of engraftment is necessary for meaningful force generation and therefore provides a benchmark by which future methods can be compared. In future, this approach may be useful for the development of novel disease models in which pluripotent cells derived from genetic myopathies are used to reconstitute a functional TA muscle in mouse, thereby allowing the disease process to be studied in vivo. It will be important to determine the extent to which human teratomas differentiate similarly to mouse, and whether they also produce skeletal muscle progenitors with high-level engraftment potential. Although no secondary teratomas were ever observed in this study, caution would demand a highly methodical evaluation of safety before considering the use of in vivo differentiation via teratoma to obtain human skeletal muscle stem cells for clinical use. In addition, scalability and efficiency in the human system will need to be addressed robustly. However, taken together, differentiation via teratoma represents an interesting and accessible means of generating cells for skeletal muscle regeneration.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Kyba (kyba@umn.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
ES and iPS cells culture
E14-EGFP male mouse ES cells (Ismailoglu et al., 2008), C57BL/6N-PRX-B6N #1 male mouse ES cells (The Jackson Laboratory #012448, Bar Harbor, ME, via Mouse Genetics Laboratory, University of Minnesota) and Pax7-ZsGreen reporter mouse iPS cells were cultured on irradiated mouse embryonic fibroblasts (MEFs) in maintenance medium consisting of: Knock-Out Dulbecco’s Minimum Essential Medium (KO-DMEM) (Life Technologies #10829-018, Grand Island, NY), 15% ES cells-qualified fetal bovine serum (ES-FBS) (Gemini Bio-Products #100-119, West Sacramento, CA), 1% non-essential amino acids (NEAA) (Life Technologies #11140-050), 1% penicillin/streptomycin (P/S) (Life Technologies #15140-122), 2 mM Glutamax (Life Technologies #SCR006), 0.1 mM β-mercaptoethanol (Sigma #M3148, St. Louis, MO) and 500 U/ml leukemia inhibitory factor (Millipore #ESG1107, Temecula, CA), at 37 °C in 5% CO2. On the day of transplantation for generating teratomas, ES and iPS cells were trypsinized and plated on a tissue culture flask for 45-60 min to remove MEFs.
Irradiated MEFs generation
MEFs were harvested from E14.5 embryos. After removing the head and the internal organs, the remaining parts were minced into little pieces and digested with 0.25% trypsin-EDTA (Life Technologies #25200-072) for 20 min at 37 °C. Cells from 3-4 embryos were pooled and cultured in a T75 flask in DMEM (HyClone #SH30081.01, Logan, UT) with 10% FBS (PEAK serum #PS-FBS, Wellington, CO) and 2 mM Glutamax at 37 °C in 5% CO2. Cells were passaged at 1:4-1:5 when confluent. By passage 4, cells were irradiated with 5000 cGy using a RS 2000 Biological Research Irradiator (Rad Source Technologies, Suwanee, GA) to generate irradiated MEFs.
Pax7-ZsGreen reporter iPS cells generation
For iPS cell generation, fibroblasts cultures were established by trypsin digestion of tail-tip biopsies taken from 3 week-old Pax7-ZsGreen male mice (Bosnakovski et al., 2008). Tail tip fibroblasts were seeded on irradiated MEFs in DMEM with 10% FBS and infected with Oct4 (Addgene #13366), Sox2 (Addgene #13367) and Klf4 (Addgene #13370) retroviruses (gifts from Shinya Yamanaka via Addgene, Cambridge, MA) (Takahashi and Yamanaka, 2006). Twenty-four hours after transduction, medium was switched to ES cell maintenance medium, and 2–3 weeks later, iPS colonies were individually isolated (Filareto et al., 2013).
Animals
Housing, husbandry and all procedures involving animals used in this study were performed in compliance with the protocol (#1408-31770A, #1708-35046A) approved by the University of Minnesota Institutional Animal Care and Use Committee and under institutional assurances of AAALAC accreditation (#000552, as of Nov 2015), USDA research facility registration (USDA No. 41-R-0005), and PHS Animal Welfare Assurance approval (A3456-01). Mice were group housed (up to 4 animals per cage for males and 5 for females) on a 12:12 hr light-dark cycle, with free access to food and water in individually ventilated specific pathogen free (SPF) cages. All mice used were healthy and were not involved in any previous procedures nor drug treatment unless indicated otherwise. NSG-mdx4Cv mice were generated by crossing NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice and B6Ros.Cg-Dmdmdx-4Cv/J (mdx4Cv) mice, as previously reported (Arpke et al., 2013). For teratoma formation and cells transplantation, 3–4 month-old homozygous NSG-mdx4Cv mice from both sexes were randomly allocated to experimental groups. Our preliminary results do not suggest any sex influence to the study. Pax7-ZsGreen mice, from which Pax7-ZsGreen reporter iPS cells derive, were generated by pronuclear injection of a mouse Pax7 locus-containing BAC with exon 1 of Pax7 coding sequence replaced by a ZsGreen coding sequence, as previously reported (Bosnakovski et al., 2008). Pax7-ZsGreen mice used were heterozygous in a C57BL/6 × 129P2/OlaHsd hybrid background. Wildtype 129P2/OlaHsd mice were obtained from Envigo (Indianapolis, IN).
MODEL DETAILS
Cell transplantation and harvest
Recipient NSG-mdx4Cv mice (3–4 month-old) were anesthetized with ketamine (150 mg/kg, i.p., Akorn NDC:59399-114-10, Lake Forest, IL) and xylazine (10 mg/kg, i.p., Akorn NDC:59399-111-50), and both hindlimbs were irradiated with 1200 cGy, two days prior to intramuscular injection of cells. A lead shield permitted exposure only to the hindlimbs. One day prior to transplantation, cardiotoxin (10 μM in 15 μl, Sigma #C9759) was injected into both TA muscles of each mouse to induce muscle injury. For teratoma induction, 250,000 ES cells or 1,000,000 iPS cells were resuspended in 10 μl sterile PBS (HyClone #SH30256.01) and injected using a Hamilton syringe (Hamilton, Reno, NV). For transplantation of myogenic progenitors, sorted α7+ VCAM+ cells (40,000, 20,000 and 20,000 cells from E14 ES cell-, C57BL/6N ES cells- and iPS cell-derived teratomas respectively, and 900 cells from transplanted TAs) were injected into the left TA while PBS was injected into the right TA. TAs were harvested at 3–4 weeks (for teratomas) or 1–12 months (for myogenic progenitors) after transplantation and were analyzed by FACS or immunohistochemistry.
Cardiotoxin re-injury and EdU administration
Transplanted TA muscles were re-injured with cardiotoxin (10 μM in 15 μl) injections 3 months after the primary transplant. Two days later, EdU (5-ethynyl-2’-deoxyuridine) (Invitrogen #A10044, Carlsbad, CA) was administered intraperitoneally (0.1 mg/20 g body weight) twice a day for 3 days. Muscles were harvested 5 days or 5 weeks after injury.
Functional evaluation on isolated muscles
Mice were anesthetized with Avertin (250 mg/kg, i.p., Sigma #T48402) and TA muscles were isolated and connected to a force transducer (model #FT03, Grass Instrument, West Warwick, RI) in the Radnoti 4 Chamber Tissue-Organ Bath apparatus (ADInstruments, Colorado Springs, CO). Isolated tissues were bathed in Ringer solution (120.5 mM NaCl (Fisher Bioreagents #BP358-212, Pittsburgh, PA), 20.4 mM NaHCO3 (Sigma #S7277), 10 mM glucose (Sigma #G7021), 4.8 mM KCl (Fisher Bioreagents #BP366-500), 1.6 mM CaCl2 (Sigma #21097), 1.2 mM MgSO4 (Sigma #M7506), 1.2 mM NaH2PO4 (Sigma #S8282), 1.0 mM sodium pyruvate (Sigma #P5280), adjusted to pH 7.4) at 25 °C with 95% O2/5% CO2 perfusion. A pair of platinum electrodes was placed longitudinally on either side of the muscles for electrical stimulation using square wave pulses at 25 V, 0.2 ms duration and 150 Hz. Muscles were maintained at optimum length (L0), which was empirically determined to generate the maximal tetanic force (F0) upon stimulation. Fatigue time was defined as the time required for force to drop to 30% of F0 after 1-min pulse of stimulation. Total muscle cross-sectional area (CSA) was calculated by dividing muscle mass by the product of muscle length and muscle density (1.06 mg/mm3). Specific force (sF0) was subsequently calculated by normalizing F0 to CSA. Data acquisition was performed in a PowerLab 8/30 using the LabChart software (both ADInstruments).
FACS
Dissociated cells were incubated with antibodies (APC anti-α7-Integrin, AbLab #67-0010-05, Vancouver, Canada; APC anti-β1-Integrin, eBioscience #17-0291-82; RRID:AB_1210793, San Diego, CA; PE-Cy7 anti-CD31, BD Biosciences Cat#561410; RRID:AB_10612003, San Jose, CA; Biotin anti-CD34, eBioscience Cat#13-0341-81; RRID:AB_466424; PE-Cy7 anti-CD45, BD Biosciences Cat#552848; RRID:AB_394489; Biotin anti-CXCR4, eBioscience Cat#13-9991-82; RRID:AB_10609202; Biotin anti-VCAM-1, BD Biosciences Cat#553331; RRID:AB_10053328; PE streptavidin, BD Biosciences Cat#554061; all at 0.5 μl per 1 million cells) on ice for 30 min. Propidium iodide (PI) (1 μg/ml, Sigma #P4170) was added to differentiate between live and dead cells. Only live cells (PI−) were counted. FACS analysis and cell sorting were performed in a BD FACSAriaII (BD Biosciences, San Diego, CA) using the FACSDiva software (BD Biosciences). Single-cell precision was used for sorting single cells into 96-well plate for clonal analysis, and 4-way purity precision was used for bulk sort. Data were analyzed using FlowJo (FLOWJO LLC, Ashland, OR). Further information on antibodies used is listed in Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC α7-Integrin | AbLab | Cat#67-0010-05 |
| Alexa Fluor 555 α-Bungarotoxin | Invitrogen | Cat#B35451; RRID:AB_2617152 |
| APC β1-Integrin (CD29) | eBioscience | Cat#17-0291-82; RRID:AB_1210793 |
| PE-Cy7 CD31 (PECAM) | BD Biosciences | Cat#561410; RRID:AB_10612003 |
| Biotin CD34 | eBioscience | Cat#13-0341-81; RRID:AB_466424 |
| PE-Cy7 CD45 | BD Biosciences | Cat#552848; RRID:AB_394489 |
| Biotin CXCR4 | eBioscience | Cat#13-9991-82; RRID:AB_10609202 |
| DYSTROPHIN | Abcam | Cat#ab15277; RRID:AB_301813 |
| EGFP | Abcam | Cat#ab13970; RRID:AB_300798 |
| Embryonic myosin heavy chain | Developmental Studies Hybridoma Bank | Cat#F1.652; RRID:AB_528358 |
| Laminin | Sigma | Cat#L9393; RRID:AB_477163 |
| MHC-I | Developmental Studies Hybridoma Bank | Cat#BA-D5; RRID:AB_2235587 |
| MHC-IIa | Developmental Studies Hybridoma Bank | Cat#SC-71; RRID:AB_2147165 |
| MHC-IIb | Developmental Studies Hybridoma Bank | Cat#BF-F3; RRID:AB_2266724 |
| MYOD1 | Santa Cruz Biotechnology | Cat#sc-304; RRID:AB_631992 |
| PAX7 | Developmental Studies Hybridoma Bank | Cat#PAX7; RRID:AB_528428 |
| Sarcomeric MHC | Developmental Studies Hybridoma Bank | Cat#MF-20; RRID:AB_2147781 |
| Biotin VCAM-1 | BD Biosciences | Cat#553331; RRID:AB_394787 |
| Streptavidin-PE | BD Biosciences | Cat#554061; RRID:AB_10053328 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| KO-DMEM | Life Technologies | Cat#10829-018 |
| DMEM, high glucose | HyClone | Cat#SH30081.01 |
| DMEM/F12 | Cellgro | Cat#15-090-CV |
| FBS | PEAK serum | Cat#PS-FBS |
| ES cell-qualified FBS | Gemini Bio-Products | Cat#100-119 |
| Horse serum | HyClone | Cat#SH30074.03 |
| Non-essential amino acids | Life Technologies | Cat#11140-050 |
| Penicillin/streptomycin | Life Technologies | Cat#15140-122 |
| Glutamax | Millipore | Cat#SCR006 |
| β-Mercaptoethanol | Sigma | Cat#M3148 |
| Leukemia inhibitory factor | Millipore | Cat#ESG1107 |
| Chick embryo extract | US Biological | Cat#C3999 |
| Insulin-transferrin-selenium | Life Technologies | Cat#41400045 |
| Basic FGF | R&D Systems | Cat#233-FB/CF |
| Propidium iodide | Sigma | Cat#P4170 |
| Paraformaldehyde | Sigma | Cat#P6148 |
| Triton X-100 | Sigma | Cat#X100 |
| Bovine serum albumin | Fisher Bioreagents | Cat#BP1605-100 |
| PBS | HyClone | Cat#SH30256.01 |
| Gelatin | Sigma | Cat#G2500 |
| 0.25% Trypsin-EDTA | Life Technologies | Cat#25200-072 |
| DAPI | Life Technologies | Cat#D3571 |
| OCT solution | Scigen | Cat#4586 |
| 2-Methylbutane | Sigma | Cat#320404 |
| Acetone | Sigma | Cat#179124 |
| DAKO Retrieval Solution | Agilent | Cat#S169984-2 |
| Immu-Mount | Thermo Scientific | Cat#9990402 |
| Cardiotoxin | Sigma-Aldrich | Cat#C9759 |
| Ketamine (VetaKet) | Akorn | NDC:59399-114-10 |
| Xylazine (AnaSed) | Akorn | NDC:59399-111-50 |
| Avertin (tribromoethanol) | Sigma | Cat#T48402 |
| EdU | Invitrogen | Cat#A10044 |
| NaCl | Fisher Bioreagents | Cat#BP358-212 |
| NaHCO3 | Sigma | Cat#S7277 |
| Glucose | Sigma | Cat#G7021 |
| KCl | Fisher Bioreagents | Cat#BP366-500 |
| CaCl2 | Sigma | Cat#21097 |
| MgSO4 | Sigma | Cat#M7506 |
| NaH2PO4 | Sigma | Cat#S8282 |
| Sodium pyruvate | Sigma | Cat#P5280 |
| GoTaq Flexi DNA polymerase | Promega | Cat#M8298 |
| Premix Ex Taq (probe qPCR) master mix | Clontech | Cat#RR39WR |
| AluI | New England BioLabs | Cat#R0137S |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | Cat#74106 |
| Verso cDNA Synthesis Kit | Thermo Scientific | Cat#AB1453A |
| SMARTer Stranded Total RNA-Seq Kit – Pico Input Mammalian Kit | Clontech | Cat#634411 |
| Click-iT EdU Alexa Fluor 555 Imaging Kit | Invitrogen | Cat#C10338 |
| GeneJET Genomic DNA Purification Kit | Thermo Scientific | Cat#K0721 |
| Deposited Data | ||
| RNA-seq | This paper | GEO:GSE92892 |
| ES-Bruce4 RNA-seq | www.encodeproject.org | GEO:GSE93453 |
| Hematopoietic multipotent progenitor cell RNA-seq | www.encodeproject.org | GEO:GSE90209 |
| Experimental Models: Cell Lines | ||
| Mouse embryonic fibroblasts | This paper | N/A |
| E14-EGFP mouse ES cells | Ismailoglu et al., 2008 | N/A |
| C57BL/6N-PRX-B6N #1 mouse ES cells | The Jackson Laboratory (via Mouse Genetics Laboratory, University of Minnesota) | Stock#012448 |
| Pax7-ZsGreen mouse iPS cells | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| NSG-mdx4Cv mice | Arpke et al., 2013 | N/A |
| Pax7-ZsGreen mice | Bosnakovski et al., 2008 | N/A |
| 129P2/OlaHsd mice | Envigo | N/A |
| Oligonucleotides | ||
| Taqman qPCR assay: Gapdh | Applied Biosystems | Mm99999915_g1 |
| Taqman qPCR assay: Myf5 | Applied Biosystems | Mm00435125_m1 |
| Taqman qPCR assay: Myod1 | Applied Biosystems | Mm00440387_m1 |
| Taqman qPCR assay: Pax3 | Applied Biosystems | Mm00435491_m1 |
| Taqman qPCR assay: Pax7 | Applied Biosystems | Mm00834079_m1 |
| SCID PCR forward primer | GGA AAA GAA TTG GTA TCC AC | N/A |
| SCID PCR reverse primer | AGT TAT AAC AGC TGG GTT GGC | N/A |
| Recombinant DNA | ||
| pMXs-Oct3/4 | Addgene | Plasmid#13366 |
| pMXs-Sox2 | Addgene | Plasmid#13367 |
| pMXs-Klf4 | Addgene | Plasmid#13370 |
| Software and Algorithms | ||
| ImageJ (v2.0.0-rc-65/1.52a) | NIH | https://imagej.nih.gov/ij/ |
| FlowJo (v7.6.3) | FLOWJO LLC | https://www.flowjo.com/ |
| Prism (v6.07) | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| ZEN (v2.3 pro) | Zeiss | https://www.zeiss.com/ |
| LabChart (v6.1.1) | ADInstruments | https://www.adinstruments.com/products/labchart/ |
| FACSDiva (v6.1.3) | BD Biosciences | http://www.bdbiosciences.com/ |
Myogenic differentiation of cultured cells
To access in vitro myogenic potential of various cell fractions or single cells, FACS-sorted cells were cultured in myogenic medium: DMEM/F12 (Cellgro #15-090-CV, Manassas, VA), 20% FBS, 10% horse serum (HyClone #SH30074.03), 10 ng/ml basic FGF (R&D Systems #233-FB/CF, Minneapolis, MN), 1% P/S, 2 mM Glutamax and 0.5% chick embryo extract (US Biological #C3999, Salem, MA). After 8 days in culture, cells were analyzed for MHC positivity by immunostaining. For long-term expansion experiments, cells were cultured in myogenic expansion medium: DMEM/F12, 20% FBS, 10 ng/ml basic FGF, 1% P/S, 2 mM Glutamax and 0.1 mM β-mercaptoethanol, and passaged every 7 days. To assess myotube formation, cells were switched to myogenic differentiation medium: high-glucose DMEM, 2% horse serum, 1% insulin-transferrin-selenium (Life Technologies #41400045) and 1% P/S for 3 days, and followed by immunostaining.
Gene expression analysis
Total RNA was extracted using RNeasy Mini Kit (Qiagen #74106, Valencia, CA), and subsequent genomic DNA removal and reverse transcription (RT) were performed using Verso cDNA Synthesis Kit (Thermo Scientific #AB1453A, Pittsburgh, PA). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed in triplicates using Taqman probes (Applied Biosystems, Carlsbad, CA) and Premix Ex Taq (probe qPCR) master mix (Clontech #RR39WR, Mountain View, CA). Expression of individual genes was subsequently analyzed by the ΔCt method in relative to the expression of the housekeeping gene Gapdh in a QuantStudio 6 Flex Real-Time PCR System using QuantStudio Real-Time PCR Software (both Applied Biosystems).
SCID PCR
Genomic DNA was extracted using the GeneJet Genomic DNA Purification kit (Thermo Scientific #K0721). DNA was amplified by GoTaq Flexi DNA polymerase (Promega #M8298, Madison, WI) using the following primers: forward: GGA AAA GAA TTG GTA TCC AC, reverse: AGT TAT AAC AGC TGG GTT GGC. PCR product was then digested by AluI (New England BioLabs #R0137S, Ipswich, MA) and analyzed in an 8% polyacrylamide gel.
Immunostaining on sorted cells
Cells were fixed with 4% paraformaldehyde (PFA) (Sigma #P6148) for 60 min, permeabilized with 0.3% Triton X-100 (Sigma #X100) for 30 min, and blocked with 3% bovine serum albumin (BSA) (Fisher Bioreagents #BP1605-100) for 1 hr, all at room temperature. Primary antibodies (anti-DYSTROPHIN at 1:250, Abcam Cat#ab15277; RRID:AB_301813, Cambridge, UK; anti-MHC at 1:20, Developmental Studies Hybridoma Bank (DSHB) Cat#MF-20; RRID:AB_2147781, Iowa City, IA; anti-MYOD1 at 1:100, Santa Cruz Biotechnology Cat#sc-304; RRID:AB_631992, Dallas, TX; and anti-PAX7 at 1:100, DSHB Cat#PAX7; RRID:AB_528428) were incubated overnight at 4 °C followed by Alexa Fluor-448, −555 or −647 conjugated secondary antibodies (Life Technologies) for 60 min at room temperature. Cells were counterstained with 4’,6-diamidino-2-phenylindole (DAPI) (Life Technologies #D3571). For cytospins, dissociated cells were spun onto coverslips via cytology funnels (Biomedical Polymers, Gardner, MA), and were air dried for at least 1 hr before subsequent fixation and immunostaining as described above. Images were acquired with a Zeiss Axio Imager M1 upright microscope with an AxioCam HRc camera using the ZEN software (Zeiss). Further information on antibodies used is listed in Key Resources Table.
Immunostaining on muscle sections
TA muscles were harvested and embedded with optimal cutting temperature (OCT) solution (Scigen #4586, Gardena, CA) and snap frozen with liquid nitrogen-cooled 2-methylbutane (Sigma #320404). For EGFP staining, TA muscles were prefixed with 4% PFA overnight at 4 °C following by a 20%/30% sucrose gradient treatment before embedding. Tissues were sectioned at 10 μm with a Leica CM3050 S cryostat (Leica Microsystems, Buffalo Grove, IL). For DYSTROPHIN staining, sections were fixed with ice-cold acetone (Sigma #179124) for 5 min. For PAX7 staining, sections were fixed with 4% PFA for 10 min, following by antigen retrieval with DAKO Retrieval Solution (Agilent #S169984-2, Santa Clara, CA) at 95 °C for 20 min. Subsequent immunostaining procedures were identical to those of sorted cells as described above, except that coverslips were mounted with Immu-Mount (Thermo Scientific #9990402) before imaging. Primary antibodies used are Alexa Fluor 555 anti-α-bungarotoxin (1:100, Invitrogen Cat#B35451; RRID:AB_2617152), anti-DYSTROPHIN (1:250, Abcam Cat#ab15277; RRID:AB_301813), anti-EGFP (1:500, Abcam Cat#ab13970; RRID:AB_300798), anti-embryonic MHC (1:20, DSHB Cat#F1.652; RRID:AB_528358), anti-laminin (1:500, Sigma Cat#L9393; RRID:AB_477163), anti-MHC-I (1:100, DSHB Cat#BA-D5; RRID:AB_2235587), anti-MHC-IIa (1:100, DSHB Cat#SC-71; RRID:AB_2147165), anti-MHC-IIb (1:100, DSHB Cat#BF-F3; RRID:AB_2266724), and anti-PAX7 (1:10, DSHB Cat#PAX7; RRID:AB_528428). Further information on antibodies used is listed in Key Resources Table.
Fiber counting and area measurement
Fiber counting and cross section area measurement were performed using ImageJ with the colocalization plugin (NIH). DYSTROPHIN or laminin staining was used to define the cross section area of muscle fibers.
RNA-seq
α7+ VCAM+ cells were FACS-sorted from E14 ES cell teratomas, transplanted TA muscles, and from E14.5 embryos and 8-week-old adult hind limbs from 129P2/OlaHsd mice (Envigo, Indianapolis, IN). Total RNA was extracted with in-column genomic DNA removal using RNeasy Mini Kit, and of which 10 ng was used for sequencing libraries creation using SMARTer Stranded Total RNA-Seq Kit – Pico Input Mammalian Kit (Clontech #634411). Paired-end 50 base-pair sequencing was performed using an Illumina HiSeq2500 (Illumina, San Diego, CA), producing 4–8 million raw reads per sample.
QUANTIFICATION AND STATISTICAL ANALYSIS
Software
FACS data acquisition were performed in FACSDiva v6.1.3 (BD) and analyzed in FlowJo v7.6.3 (FLOWJO LLC). Quantitative PCR data acquisition were performed in QuantStudio Real-Time PCR Software v1.3 (Applied Biosystems). Force measurements were acquired using LabChart v6.1.1 (ADInstruments). Immunostaining data were acquired using ZEN v2.3 pro (Zeiss). Fiber counting and measurements were performed with ImageJ v2.0.0-rc-65/1.52a (NIH).
RNA-seq analysis
RNA-seq reads were processed in Galaxy via Minnesota Supercomputing Institute, University of Minnesota (Afgan et al., 2016) with TopHat (Galaxy v2.1.0) (Kim et al., 2013) and Cufflinks (Galaxy v2.2.1.0) (Trapnell et al., 2010) for transcriptome mapping and alignment against the Mus musculus genome (mm10). Undifferentiated mouse ES cell RNA-seq dataset and mouse hematopoietic multipotent progenitor cell dataset were obtained from GEO:GSE93453 and GEO:GSE90209 respectively via ENCODE (www.encodeproject.org). Further analysis including hierarchy clustering and differential genes determination was performed using Partek Genomic Suite v6.0 using default parameters (Partek, St. Louis, MI). Significant difference was set with a false discovery rate-adjusted p-value <0.05 and an absolute fold change >2.5. Gene Ontology (GO) analysis was performed using DAVID Bioinformatics Resources (Huang da et al., 2009a, b). RNA-seq datasets can be accessed on GEO (GSE92892).
Statistical analysis
Data are expressed as mean ± SEM. Graphs and statistics are prepared with Prism v6.07 (GraphPad Software, La Jolla, CA). Dose-response relationship is modeled using a log-linear regression model with variable slope. The number of replicates for individual experiments is indicated in the corresponding figure legend. Statistical significance is determined by Student’s t-test for comparison between two treatment groups, or one-way analysis of variance (ANOVA) with Tukey post-hoc test for comparison among three or more treatment groups. Statistical significance is set as p<0.05.
DATA AND SOFTWARE AVAILABILITY
The RNA-seq datasets have been deposited in the GEO under ID code GSE92892.
Supplementary Material
Table S1, related to Figure 5. Differential genes between Transplant and Teratoma samples in RNA-seq analysis
Table S2, related to Figure 5. Differential genes between Adult and Embryo samples in RNA-seq analysis
Table S3, related to Figure 5. Differential genes between Teratoma and Embryo samples in RNA-seq analysis
Table S4, related to Figure 5. Differential genes between Transplant and Adult samples in RNA-seq analysis
Highlights.
Teratomas are rich in α7-Integrin+ VCAM-1+ myogenic progenitors.
40,000 teratoma-derived α7+ VCAM+ cells reconstitute 80% of TA muscle fiber volume.
New fibers generate force and ameliorate dystrophin-related force deficiency.
Teratoma-derived myogenic progenitors mature into PAX7+ muscle stem cells in vivo.
Acknowledgements
The authors would like to thank Jinjoo Kang, Olivia Recht and Cara-Lin Lonetree for their help in genotyping and animal husbandry. The monoclonal antibodies to PAX7, embryonic MHC, MHC, MHC-I, MHC-IIa and MHC-IIb were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. Plasmids pMXs-Oct3/4, pMXs-Sox2 and pMXs-Klf4 were gifts from Shinya Yamanaka via Addgene. The study was supported by the National Institute for Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants R01 NS083549 (to M.K.) and R01 AR055299 (to R.C.R.P.), Regenerative Medicine Minnesota discovery science grant RMM 102516 001 (to S.S.K.C.), and by the Greg Marzolf Jr. Foundation.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44, W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile G, Welner RS, Nombela-Arrieta C, D'Alise AM, Di Ruscio A, Ebralidze AK, Kraytsberg Y, Ye M, Kocher O, Neuberg DS, et al. (2013). In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood 121, 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpke RW, Darabi R, Mader TL, Zhang Y, Toyama A, Lonetree CL, Nash N, Lowe DA, Perlingeiro RC, and Kyba M (2013). A new immuno-, dystrophin-deficient model, the NSG-mdx(4Cv) mouse, provides evidence for functional improvement following allogeneic satellite cell transplantation. Stem Cells 31, 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Bose WE, Yao CC, Kramer RH, and Blau HM (2001). Purification of mouse primary myoblasts based on alpha 7 integrin expression. Exp Cell Res 265, 212–220. [DOI] [PubMed] [Google Scholar]
- Bober E, Franz T, Arnold HH, Gruss P, and Tremblay P (1994). Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 120, 603–612. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Li W, Thet S, Cleaver O, Perlingeiro RC, and Kyba M (2008). Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells 26, 3194–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Bildsoe H, and Hughes SM (2005). Evidence that satellite cell decrement contributes to preferential decline in nuclear number from large fibres during murine age-related muscle atrophy. J Cell Sci 118, 4813–4821. [DOI] [PubMed] [Google Scholar]
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 33, 962–969. [DOI] [PubMed] [Google Scholar]
- Chan SS, Shi X, Toyama A, Arpke RW, Dandapat A, Iacovino M, Kang J, Le G, Hagen HR, Garry DJ, et al. (2013). Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell 12, 587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, and Morgan JE (2005). Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122, 289–301. [DOI] [PubMed] [Google Scholar]
- Darabi R, Arpke RW, Irion S, Dimos JT, Grskovic M, Kyba M, and Perlingeiro RC (2012). Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 10, 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, Kyba M, and Perlingeiro RC (2008). Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med 14, 134–143. [DOI] [PubMed] [Google Scholar]
- Eventov-Friedman S, Katchman H, Shezen E, Aronovich A, Tchorsh D, Dekel B, Freud E, and Reisner Y (2005). Embryonic pig liver, pancreas, and lung as a source for transplantation: optimal organogenesis without teratoma depends on distinct time windows. Proc Natl Acad Sci U S A 102, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filareto A, Parker S, Darabi R, Borges L, Iacovino M, Schaaf T, Mayerhofer T, Chamberlain JS, Ervasti JM, McIvor RS, et al. (2013). An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat Commun 4, 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, and Takeda S (2007). Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448–2459. [DOI] [PubMed] [Google Scholar]
- Goulding M, Lumsden A, and Paquette AJ (1994). Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120, 957–971. [DOI] [PubMed] [Google Scholar]
- Gunther S, Kim J, Kostin S, Lepper C, Fan CM, and Braun T (2013). Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell 13, 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussoni E, Pavlath GK, Lanctot AM, Sharma KR, Miller RG, Steinman L, and Blau HM (1992). Normal dystrophin transcripts detected in Duchenne muscular dystrophy patients after myoblast transplantation. Nature 356, 435–438. [DOI] [PubMed] [Google Scholar]
- Hall JK, Banks GB, Chamberlain JS, and Olwin BB (2010). Prevention of muscle aging by myofiber-associated satellite cell transplantation. Science Transl Med 2, 57ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Ismailoglu I, Yeamans G, Daley GQ, Perlingeiro RC, and Kyba M (2008). Mesodermal patterning activity of SCL. Exp Hematol 36, 1593–1603. [DOI] [PubMed] [Google Scholar]
- Jesse TL, LaChance R, Iademarco MF, and Dean DC (1998). Interferon regulatory factor-2 is a transcriptional activator in muscle where It regulates expression of vascular cell adhesion molecule-1. J Cell Biol 140, 1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, and Rudnicki MA (2006). Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol 172, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, and Daley GQ (2002). HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29–37. [DOI] [PubMed] [Google Scholar]
- Mauro A (1961). Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol 9, 493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JR, Kissel JT, Amato AA, King W, Signore L, Prior TW, Sahenk Z, Benson S, McAndrew PE, Rice R, et al. (1995). Myoblast transfer in the treatment of Duchenne's muscular dystrophy. N Engl J Med 333, 832–838. [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, and Buckingham M (2005). Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064–2067. [DOI] [PubMed] [Google Scholar]
- Partridge TA, Morgan JE, Coulton GR, Hoffman EP, and Kunkel LM (1989). Conversion of mdx myofibres from dystrophin-negative to -positive by injection of normal myoblasts. Nature 337, 176–179. [DOI] [PubMed] [Google Scholar]
- Penton CM, Badarinarayana V, Prisco J, Powers E, Pincus M, Allen RE, and August PR (2016). Laminin 521 maintains differentiation potential of mouse and human satellite cell-derived myoblasts during long-term culture expansion. Skelet Muscle 6, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlingeiro RC, Kyba M, and Daley GQ (2001). Clonal analysis of differentiating embryonic stem cells reveals a hematopoietic progenitor with primitive erythroid and adult lymphoid-myeloid potential. Development 128, 4597–4604. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, and Blau HM (2008). Self-renewal and expansion of single transplanted muscle stem cells. Nature 456, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Ishibashi J, Holterman C, and Rudnicki MA (2004). Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev Biol 275, 287–300. [DOI] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, and Rudnicki MA (2000). Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786. [DOI] [PubMed] [Google Scholar]
- Shelton M, Metz J, Liu J, Carpenedo RL, Demers SP, Stanford WL, and Skerjanc IS (2014). Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Reports 3, 516–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Yamazaki S, Yamaguchi T, Okabe M, Masaki H, Takaki S, Otsu M, and Nakauchi H (2013). Generation of engraftable hematopoietic stem cells from induced pluripotent stem cells by way of teratoma formation. Mol Ther 21, 1424–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, and Rudnicki MA (2013). Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A 110, 16474–16479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 5. Differential genes between Transplant and Teratoma samples in RNA-seq analysis
Table S2, related to Figure 5. Differential genes between Adult and Embryo samples in RNA-seq analysis
Table S3, related to Figure 5. Differential genes between Teratoma and Embryo samples in RNA-seq analysis
Table S4, related to Figure 5. Differential genes between Transplant and Adult samples in RNA-seq analysis
Data Availability Statement
The RNA-seq datasets have been deposited in the GEO under ID code GSE92892.