Abstract
Preclinical imaging is critical in the development of translational strategies to detect diseases and monitor response to therapy. The National Cancer Institute Co-Clinical Imaging Resource Program was launched, in part, to develop best practices in preclinical imaging. In this context, the objective of this work was to develop a 1-hour, multiparametric magnetic resonance image-acquisition pipeline with triple-negative breast cancer patient-derived xenografts (PDXs). The 1-hour, image-acquisition pipeline includes T1- and T2-weighted scans, quantitative T1, T2, and apparent diffusion coefficient (ADC) parameter maps, and dynamic contrast-enhanced (DCE) time-course images. Quality-control measures used phantoms. The triple-negative breast cancer PDXs used for this study averaged 174 ± 73 μL in volume, with region of interest–averaged T1, T2, and ADC values of 1.9 ± 0.2 seconds, 62 ± 3 milliseconds, and 0.71 ± 0.06 μm2/ms (mean ± SD), respectively. Specific focus was on assessing the within-subject test–retest coefficient-of-variation (CVWS) for each of the magnetic resonance imaging metrics. Determination of PDX volume via manually drawn regions of interest is highly robust, with ∼1% CVWS. Determination of T2 is also robust with a ∼3% CVWS. Measurements of T1 and ADC are less robust with CVWS values in the 6%–11% range. Preliminary DCE test–retest time-course determinations, as quantified by area under the curve and Ktrans from 2-compartment exchange (extended Tofts) modeling, suggest that DCE is the least robust protocol, with ∼30%–40% CVWS.
Keywords: MRI, TNBC, PDX, Test–retest, multiparametric MRI, T1, T2, DCE, ADC, small animal, preclinical, mouse
Introduction
Triple-negative breast cancer (TNBC) is an aggressive tumor characterized by poor outcomes and higher relapse rates compared with other subtypes of breast cancer. Pathologic complete response often serves as an important endpoint in the treatment of TNBC following neoadjuvant chemotherapy. It is critical to identify patients who will respond to neoadjuvant chemotherapy and, thus, avoid the use of ineffective treatments in nonresponding patients. Toward that end, advanced quantitative imaging (QI) strategies have been developed and evaluated for predicting or assessing response to therapy in breast cancer. Although significant progress has been made in advancing such QI approaches, preclinical imaging remains a critical component in the translational pipeline of validating advanced QI methods for applications in drug discovery and assessment of response to therapy.
It is well appreciated that established tumor cell lines fail to fully recapitulate the microstructural/environmental, cellular, molecular, genetic and epigenetic properties, including abnormal vasculature with higher blood-vessel permeability, found in clinical TNBC tumors (1). To that end, patient-derived tumor xenografts (PDXs) are considered to provide more faithful tumor models than traditional orthotopic implantation of established tumor cell lines. The use of PDXs also ushers in new paradigms involving coclinical trials in which QI applied to PDXs can be implemented in the corresponding patient in a clinical setting and vice versa. However, there are challenges in developing optimal quantitative pipelines to assess response to therapy in a preclinical setting. Most preclinical (small-animal) magnetic resonance imaging (MRI) studies of cancer models involve tumor-cell implantation into the brain or leg (thigh) of the subject [see, for example (2–5)], which can be readily secured in the small-animal bed/cradle of the scanner, thus minimizing artifacts caused by respiratory motion. However, in light of the significance of the tumor microenvironment in tumor growth and pathogenesis following cell implantation, it is relevant that TNBC PDXs be implanted into mammary fat pads, which are more susceptible to respiratory motion–related imaging artifacts.
Recognizing the challenges of preclinical imaging using realistic models of cancer, the National Cancer Institute recently launched the Co-Clinical Imaging Resource Program to develop best practices in preclinical imaging and support clinical trials. In this context, the objective of this work was to develop and implement a 1-hour, multiparametric MRI acquisition pipeline with orthotopic TNBC PDXs. Tumor cells were implanted into the fourth (inguinal/abdominal) mammary fat pad of NSGTM mice and characterized by a pipeline of preclinical MRI experiments that included T1-weighted (T1W) and T2-weighted (T2W) scans, quantitative T1, T2, and apparent diffusion coefficient (ADC) parametric maps, and a dynamic contrast-enhanced (DCE) protocol.
Conceptually, the project progressed through 3 stages: (1) the precision and accuracy of the MRI acquisition pipeline's protocols were assessed via phantoms with known MR properties; (2) artifacts from respiratory motion were suppressed by paying careful attention to mouse handling/placement within the scanner's cradle/bed, and, finally (3) the in vivo test–retest reproducibility (precision) of MRI metrics was determined with orthotopic TNBC PDXs.
Methods
Generation of TNBC PDX
TNBC PDX.
Tumors were generated in collaboration with the Human and Mouse-Linked Evaluation of Tumors (HAMLET) core of the Washington University School of Medicine's Institute of Clinical and Translational Sciences (http://digitalcommons.wustl.edu/hamlet/). Details regarding animals, surgeries, and tumor xenografts may be found in the report by Li et al. (6). All animal experiments were conducted in compliance with the Guidelines for the Care and Use of Research Animals established by Washington University's Institutional Animal Care and Use Committee.
Experiments used NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NOD/SCID) female mice obtained from The Jackson Laboratory (Bar Harbor, ME). Mouse mammary fat pads were humanized as previously described (7). Tumor samples from patients with breast cancer were placed in ice-chilled DMEM/F12 medium, necrotic tissue and fat were removed, and the samples were cut into 1- × 1-mm2 pieces. Tissue fragments were engrafted into the mammary glands. When xenograft tumors reached 1.5 cm in diameter, they were harvested and dissociated into a single-cell suspension.
Implantation of breast cancer cells used an inverted Y-shaped incision along the thoracic–inguinal region to expose the mammary glands. Two-to-4 million tumor cells mixed with Matrigel in a volume of 30 μL were injected into the fourth inguinal mammary fat pad. The skin was gathered, and the incision closed with wound clips. Following engraftment, tumor growth was monitored by means of calipers.
MRI Scanner and RF Coil Configuration
All experiments were performed on an Agilent/Varian (Santa Clara, CA) DirectDrive 4.7-T small-animal MR scanner built around an Oxford Instruments (Abingdon, UK) horizontal superconducting magnet, with a gradient/shim coil assembly having a 12-cm inner-bore diameter and providing 60 G/cm with 200-μs rise time. All MR images, including phantoms, were acquired using the same laboratory-built actively decoupled transmit (volume, 7.5-cm inner diameter) and receive (surface, 2.5-cm inner diameter) coil pair (8).
Homogeneous Aqueous Phantoms
Phantoms composed of homogeneous aqueous solutions, doped with various amounts of manganese chloride (9) to span the T1 and T2 values encountered in vivo, were used to set pulse sequence parameters and quantify performance under ideal conditions.
Six-Compartment Variable Relaxation Phantom.
A 6-compartment phantom was used to bridge the nominal range of in vivo T1 and T2 values expected during the DCE time course. This phantom was then used to set the flip angle (FA) in the high temporal resolution (3 seconds) DCE protocol. Solutions of 6 different MnCl2 concentrations (12.5, 25, 50, 100, 200, and 400 μM) were prepared in deionized water using manganese chloride tetrahydrate (MilliporeSigma, Burlington, MA). Further, 0.3 mL of each solution was transferred into separate 5-mm glass NMR tubes, and the 6 tubes were packed together as a bundle.
The top of the bundle was placed just within the plane of the surface coil, and data were collected from image slices perpendicular to the bundle axis. Thus, the MRI slice plane and surface-coil plane were parallel and orthogonal to the axial orientation of the 6-tube phantom, and all 6 tubes—as defined by the slice plane cutting through the tubes—were at the same fixed distance from the surface coil. The offset distance from the surface coil to the slice plane used for the 6-tube phantom was typical of the distance from the surface coil through the center of the PDXs used in this study.
The temperature of the air surrounding the phantom was regulated (37°C) using a temperature control system (SA Instruments, Stony Brook, NY); 1 hour was allowed for temperature equilibration. The pulse-sequence parameters were those used subsequently for in vivo DCE-MRI of PDX-bearing mice with the exception that only a single slice was interrogated for a single measurement at each of 18 FAs from 2° to 45°.
In a separate set of experiments, each tube was examined individually and in quintuplicate at 37°C using a small solenoid radiofrequency (RF) coil and standard MR spectroscopy relaxation measurement to determine T1 and T2. Parameters varied for each sample: repetition time (TR) > 5 times the expected T1, TI, and echo time (TE) arrayed to cover 3–5 expected decay e-foldings. Nonlinear least-squares, monoexponential modeling was used to estimate T1 and T2 values and the r1 and r2 relaxivities of MnCl2.
Single-Compartment Phantom.
A 5-mL spherical plastic phantom was filled with 150-μM MnCl2 solution to evaluate the inherent measurement accuracy and precision of the MRI protocols used herein. This concentration was chosen, based on the measured relaxivities (r1 and r2) of Mn2+ determined herein, to broadly mimic that of in vivo PDX. The temperature of air surrounding the phantom was controlled (37°C), as described earlier. The pulse-sequence parameters were those used subsequently for in vivo scanning of PDX-bearing mice (ie, same slice thickness, multiple slices through the phantom in a field of view (FOV) approximating the typical PDX location). Analysis used a 1-mL region of interest (ROI) manually drawn in a region immediately below the surface coil, anticipating and mimicking PDX versus surface-coil positioning. The signal-to-noise ratio (SNR) was nominally in the same range encountered herein with PDX in vivo. T1, T2, and ADC measurements were repeated in quintuplicate, with the phantom removed from and repositioned within the scanner after each measurement set.
In Vivo MRI of PDX-Bearing Mice
Mice were maintained on a temperature-controlled warm-water pad and anesthetized with isoflurane/O2 (1.2%–1.5% isoflurane) throughout the experiment. Within the scanner, the mice were positioned on their sides in a 2.2-cm-inner-diameter half-cylinder cradle to allow the tumor, facing up, to be located immediately under the surface coil. Tape and a modified ridged plastic panel (10) were placed judiciously to isolate the diaphragm and restrict respiratory motion to the chest area, away from the tumor and lower abdomen. Figure 1 shows a sketch of the experimental setup. Figure 2 compares representative DCE time-course data (single-slice tumor ROI averaged) in the absence and presence of respiratory motion constraints. Respiratory rate and body temperature (rectal probe) were monitored, and core temperature was regulated (37°C) with a small-animal physiologic monitoring and control unit (SA Instruments), which was also used for respiratory gating.
Figure 1.
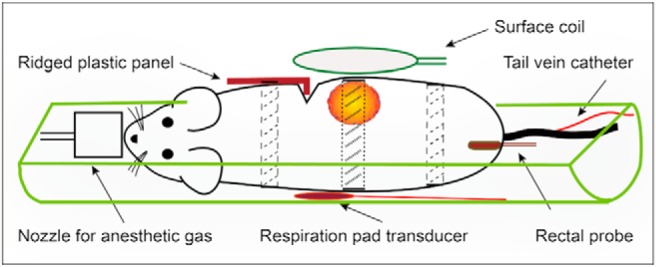
Sketch of animal positioning: Each patient-derived xenograft (PDX)-bearing mouse was placed on its side in a half-cylinder cradle. Respiratory motion–induced artifacts were minimized with (i) a ridged plastic panel (10) and (ii) judicious use of tape at locations above and below the tumor (dashed lines) and applied very lightly over the tumor (dotted lines). The tape over the mouse body also secured the ridged plastic panel in place. The surface-coil receiver was placed immediately above the tumor (8).
Figure 2.
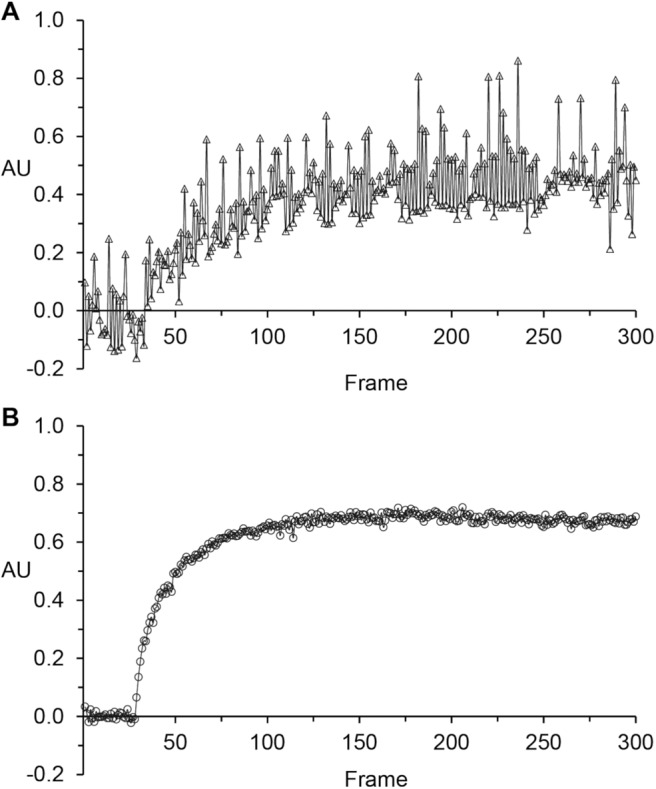
Representative examples comparing a dynamic contrast-enhanced (DCE) time course (2D slice, tumor region of interest [ROI] averaged) in the absence (A) and presence (B) of good motion control.
For all protocols except DCE, the initial “test” portion of the protocol was performed in the morning of a given day, and the second “retest” portion of the protocol was performed in the afternoon of the same day (n = 13). Typically, the interval between test and retest portions of the protocol was 3–4 h, during which time, each mouse was returned to its cage and allowed access to food and water ad libitum. For the test–retest DCE protocol, the initial “test” portion of the protocol was performed in the afternoon of a given day following the retest T1-, T2-, and ADC-map data acquisitions, and the second “retest” portion of the DCE protocol was performed the following morning, allowing time for washout and excretion of contrast agent (n = 4). Typically, the interval between the test and retest portions of the DCE protocol was 16–20 h, during which time, each mouse was returned to its cage and allowed access to food and water ad libitum. In aggregate, these test–retest scanning sessions were conducted over a 4-month period.
Setting up the mouse in the scanner and acquiring scout, T1W, and T2W images took ∼15 minutes. During this time, the animal's core temperature stabilized at the desired 37°C target. A schematic of the overall image acquisition pipeline including DCE is shown in Figure 3, and pulse sequence parameters are given below and summarized in Table 1.
Figure 3.

Magnetic resonance imaging (MRI) Data Acquisition Pipeline. The quantitative multiparameter protocol (tumor volume, T1- T2-, and apparent diffusion coefficient [ADC]-maps, and DCE) was designed to take ∼1 hour per subject. Pulse sequence parameters are listed in the text and Table 1. The DCE protocol was only performed on an n = 4 subset of the n = 13 test–retest cohort.
Table 1.
Pulse Sequences and Parameters
| MRI | VnmrJ-4.2A: Pulse Sequence | 2D Slice Matrix Size | Averages | TR (ms) | TE (ms) | ETL (Echo Train Length) | Flip Angle (FA) |
|---|---|---|---|---|---|---|---|
| T1W | Gradient Echo Multislice | 128 × 128 | 4 | 100 | 2 | – | 30° |
| T2W | Fast Spin Echo Multislice | 128 × 128 | 4 | 2000 | 52 | 4 | – |
| T2W with Fat Sat | Fast Spin Echo Multislice | 128 × 128 | 4 | 2000 | 52 | 4 | – |
| T2-Map | Multiecho Multislice | 64 × 64 | 1 | 6000 | 16 × 16 | – | – |
| ADC | Spin Echo Multislice | 64 × 64 | 2 | 1000 | 23 | – | – |
| T1-Map | Gradient Echo Multislice VFA | 64 × 64 | 8 | 50 | 1.6 | – | 5°, 10°, 15°, 30°, 50° |
| DCE | Gradient Echo Multislice | 64 × 64 | 1 | 47 | 1.4 | – | 35° |
T1W and T2W images were acquired at identical resolution: matrix size = 128 × 128, FOV = 25.6 × 25.6 mm2, slice thickness = 1 mm, number of transaxial slices = 15. Following T1W and T2W anatomic scanning, quantitative T1-, T2-, ADC-maps and a DCE time series of images were acquired using a reduced matrix size, 64 × 64.
T2W and T1W Images.
T1W images were collected with a 2D gradient-echo multislice sequence: TR = 100 milliseconds, TE = 2 milliseconds, averages = 4; FA = 30°, data-acquisition time = 52 seconds. T2W images with and without fat saturation were collected with a 2D fast-spin-echo multislice sequence: echo train length (ETL) = 4, TR = 2 seconds, effective TE = 52 milliseconds, averages = 4, 90° fat saturation RF pulse = 6 milliseconds, data-collection time = 4 minutes 32 seconds each.
T1 and T2 Maps.
T1 mapping used a variable flip angle, 2D gradient-echo pulse sequence: FA = 5°, 10°, 15°, 30°, and 50°; TR = 50 milliseconds; TE = 1.6 milliseconds; averages = 8; data-collection time = 2 minutes 8 seconds. T2 mapping used a multispin-echo, multislice pulse sequence: TR = 6 seconds, TE = 16 milliseconds, ΔTE = 16 milliseconds, echoes = 16, average = 1, data-acquisition time = 7 minutes 12 seconds.
ADC Maps.
ADC mapping used a respiratory-gated, spin-echo, multislice sequence: TR = 1.0 seconds, TE = 23 milliseconds, averages = 2, δ = 3 milliseconds, Δ = 15 milliseconds, b-value = 850 s/mm2, orthogonal b-vector directions = 3, and b = 0. Total ADC-map data-acquisition time, which is dependent on respiratory rate, was ∼12 minutes.
DCE MRI.
DCE time-series data (preliminary, 4 test–retest pairs) were collected with a gradient-echo multislice pulse sequence: TR = 47 milliseconds, TE = 1.4 milliseconds, FA 35°, temporal resolution = 3 seconds, repetitions = 300, total scan time = 15 minutes. At 1.5 minutes after the start of the DCE experiment, a 100-μL bolus of 50% (V/V) Dotarem® (Guerbet LLC USA, Princeton, NJ) in saline, ∼1.25 mmole/kg body weight, was manually administrated over 3 seconds via a tail-vein catheter.
Data Analysis
Analysis Strategies and Software.
The multislice T2W images from each mouse were used for tumor volume measurement/segmentation with ITK-SNAP (11). ROIs were manually drawn around each tumor, downsampled to 64 × 64, transferred to the lower-resolution parameter maps and DCE images, and then refined, as appropriate, using the ROI boundaries evident from the different contrasts present in the T1, T2, and ADC maps or DCE time course.
T1, T2, and ADC parametric maps and uncertainties were derived with the Bayesian Toolbox (12), a data modeling software package based upon the precepts of Bayesian probability theory (13, 14) available for free download for noncommercial uses (http://bayesiananalysis.wustl.edu/). These analyses were performed in 2 different ways: (1) averaging the data and modeling the combined ROI data, indicated below as <ROI> and (2) modeling each voxel in the ROI independently and averaging the parameter estimates, indicated below as <Voxel>. Although not a focus of the test–retest studies herein, the spatial parameter-maps (parameter distributions) can provide additional valuable insight in the case of heterogeneous tissues (eg, large tumors with regions of central necrosis and hemorrhage versus metabolically active rims), whereas the <ROI> analysis assumes that the voxels all represent the same underlying quantity. The Bayes Toolbox “Analyze Image Pixel” package was used for voxel-wise parameter modeling, and the “Enter Ascii Model” package was used for ROI-averaged data modeling.
MR Signal Models.
The signal models (S) used were as follows:
For T1 determination via the variable FA (θ) method:
| (1) |
For T2 determination via the multispin-echo (ie, multi-TE) method:
| (2) |
For ADC determination via 3 b-vectors (bx, by, bz) and b0, the full tensor model was used to account for the diffusion weighting of the imaging gradients; however, the underlying free-diffusion model, equation (3), is valid for tissue water within the Gaussian phase approximation regime (15):
| (3) |
These signal models all share an underlying exponential character. The interested reader will find additional insight regarding Bayesian analysis of exponentially decaying signals in (16–20).
Although the DCE findings must be regarded as preliminary owing to the small number of paired test–retest determinations (n = 2 × 4), each signal time-course was quantified by 2 metrics: (1) the contrast agent extravasation rate constant, Ktrans, derived via the 3-parameter (Ktrans, Ve, Vp), 2-compartment exchange model, often referred to as the extended or modified Tofts model (21–26) and (2) the area under the time-course curve (AUC). For compartmental exchange modeling, the arterial input function was approximated using the reference region (RR) approach (27–30) in which muscle is chosen as the RR with fixed extended Tofts model parameters: RR Ktrans = 0.02 min−1 (31), RR Ve = 0.08 (31–33), and RR Vp = 0.02 (33). The underlying RR assumption is that muscle is well perfused, homogeneous, and responds identically to contrast agent on an exam-to-exam and mouse-to-mouse basis. Contrast agent relaxivity was assigned as 5.8 mM−1s−1, vide infra. Modeling (fitting the data) used the variable projection (VARPRO) formulation of the maximum likelihood parameter estimation approach (34).
The area under the DCE time-course curve (DCE-AUC) following contrast agent injection was calculated on a voxel-wise basis as follows. At each time point, the signal was expressed as the fractional enhancement in voxel intensity relative to the precontrast period, to account for signal variations between scans. To account for potential modest variations in the contrast agent injection, the signal was further normalized by the maximum fractional enhancement in the early time frames (n = 31-120) of the lateral dorsal (lumbosacral caudal) muscle in the 5 slices covering the central portion of the tumor. The AUC was expressed on a per-unit-time (s) basis by dividing the sum of the normalized signal over the postinjection frames by 810 (3 s/frame × 270 postinjection frames) and, in this sense, can be considered equal to the mean signal enhancement over the postinjection time window.
Statistical Analysis.
Standard statistical analysis—means, medians, standard deviations (SDs), coefficients-of-variation (CVs), Bland–Altman plots—of MR-derived parameters was performed using Matlab (MathWorks, Natick, MA) and Excel (Microsoft, Redmond, WA).
As noted by Hyslop and White in their report on estimating precision using duplicate measurements (35): “Precision is a concept for which there is no universally accepted metric. Reports of precision vary depending on the formula and inclusion criteria used to calculate them.” Herein, we wish to express the variability in duplicate (ie, test–retest) measurement pairs. Therefore, in addition to Bland–Altman plots (36, 37), we provide the within-subject SD (SDws) (38) and the within-subject CV (CVWS) (35), that is, the root-mean-square of the scaled relative differences. In assessing test–retest results for a given MRI-determined parameter, the absolute value of the difference between 2 paired repeat determinations is indicated by the symbol Δ. The within-subject standard deviation and CVWS are then given by equations (3) and (4):
| (4) |
where the sum is taken over the number of test–retest paired duplicate measurements (n), and
| (5) |
in which m is the mean of the 2 paired repeat determinations whose absolute difference is Δ, and the sum is taken over the n test–retest paired duplicate measurements.
The width of each voxel-wise parameter distribution is summarized as the “SD of the parameter distribution.” In this case, Δ for the distribution width test–retest (the difference between the standard deviations of the voxel-wise parameter distributions) is indicated by the symbol ΔDW.
In addition to metrics quantifying precision, assessing test–retest bias (significant difference from zero) is also of interest (Bland–Altman plots, vide infra). The 95% confidence interval for mean test–retest difference <d> was assigned as <d> ± (t × SEM) where t is the value of the t distribution with n − 1 degrees of freedom and SEM is the standard error of <d> (36, 37).
Results
Phantoms
Protocol Assessment via Single-Compartment Phantom Sample.
As expected, even with removal and repositioning of the phantom between measurements, the parameter variability across consecutive determinations of T1, T2, and ADC in the single-compartment homogeneous phantom was small (Table 2). CVs for <ROI> parameter values (n = 5) were 2.6, 3.7, and 3.9% for T1, T2, and ADC, respectively. Similar mean, SD, and CV values resulted from <ROI> and <Voxel> analysis. As will be seen, the phantom T1 and ADC CVs, whether <ROI> or <Voxel> based, are smaller (∼1.5 to 4.2×) than those obtained for PDX in vivo, while the phantom T2 CVs are somewhat larger (∼1.4×) than observed for PDX in vivo.
Table 2.
Single-Compartment Phantom Test–Retest Variability
| Parameter | Spectroscopy True Value | Mean | SD | CV (%) | Bias (%) |
|---|---|---|---|---|---|
| T1<ROI>, sa | 0.921 ± 0.04 | 0.85 | 0.02 | 2.6 | −7.8 |
| T1<Voxel>, sb | 0.86 | 0.03 | 3.4 | −6.7 | |
| T2<ROI>, msa | 54 ± 3 | 52.1 | 1.9 | 3.7 | −2.6 |
| T2<Voxel>, msb | 52.2 | 1.9 | 3.5 | −2.8 | |
| ADC<ROI>, μm2/msa | 3.1 ± 0.1 | 3.0 | 0.1 | 3.9 | −1 |
| ADC<Voxel>, μm2/msb | 3.0 | 0.1 | 3.9 | −3.3 |
a<ROI>: Results from modeling averaged data from the entire phantom ROI over all five measurement repeats.
b<Voxel>: Results from modeling each phantom voxel independently and summarized over the five measurement repeats.
In addition to measurement precision, phantom samples offer an opportunity to assess accuracy (bias). MR spectroscopy–based measurements in the absence of imaging gradients provided “true value” determinations of T1, T2, and ADC for the single-compartment homogeneous phantom. The image-based determinations were within 8%, 3%, and 3% of the true value determinations for T1, T2, and ADC, respectively.
Six-Compartment Variable Relaxation Phantom.
The 6-compartment phantom contained homogeneous compartments, with T1 and T2 values bridging the expected range encountered for various tissues and over the DCE time course (Table 3).
Table 3.
T1 and T2 Determinations with the 6-Compartment Variable Relaxation Phantom
| MnCl2 Sample Concentration | IR MRSa | VFA (5 FA) <Voxel>b | VFA (18 FA) <Voxel>b | SE MRSc | ME 16 TEs × 16 ms <Voxel>d |
|---|---|---|---|---|---|
| mM | Mean T1, s, (SD) n = 5 | Mean T1, s, (SD) | Mean T1, s, (SD) | Mean T2, ms, (SD) n = 5 | Mean T2, ms, (SD) |
| 0.0125 | 3.63 (0.01) | 3.1 (0.5) | 3.2 (0.3) | 483 (2) | 250 (30) |
| 0.025 | 2.74 (0.01) | 2.6 (0.3) | 2.6 (0.2) | 281 (3) | 210 (20) |
| 0.050 | 2.14 (0.01) | 2.0 (0.2) | 2.0 (0.1) | 189 (6) | 140 (9) |
| 0.10 | 1.75 (0.01) | 1.2 (0.1) | 1.2 (0.1) | 113 (2) | 79 (4) |
| 0.20 | 0.744 (0.003) | 0.77 (0.07) | 0.75 (0.04) | 41.0 (0.4) | 45 (3) |
| 0.40 | 0.399 (0.005) | 0.42 (0.03) | 0.41 (0.02) | 15.0 (0.3) | 23 (2) |
aInversion-recovery (IR) MR spectroscopy (MRS) measurements of T1.
bVariable flip angle (VFA) T1 measurements across all phantom voxels with either 5 or 18 flip angles (FA).
cSpin-echo (SE) MRS measurements of T2.
dMulti-(Spin)-echo (ME) measurements of T2 across all phantom voxels.
Aqueous Mn2+ relaxivities (r1, r2) derived from these data (201.5 MHz; 37°C) via equation (5) were r1 = 5.8 ± 0.3 mM−1s−1 (R2 = 0.99) and r2 = 166 ± 14 mM−1s−1 (R2 = 0.98).
| (6) |
Using the DCE pulse-sequence parameters, the FA was varied to map signal (contrast) changes between compartments with differing T1 in the 6-compartment phantom. Conceptually, for an anticipated range (dynamic) of T1s, selection of the “optimal” FA will depend upon 2 competing/opposing considerations, with a desire to obtain: (1) the greatest T1-dependent signal amplitudes (ie, SNR) and (2) the greatest difference in T1-dependent signal amplitudes (ie, image contrast) for tissues with different T1s. Thus, the optimal/ideal FA would provide (1) maximal signal (tissue detectability) and (2) maximal tissue contrast (tissue differentiation) between tissues with different DCE time-course profiles. (We recognize that the dynamics of the time course itself provide a second contrast mechanism.) We qualitatively weighted these 2 factors equally for T1 values most representative of what would be anticipated for the various tissues in the FOV during the DCE time course. Although the region of optimality is rather broad, a nominal FA of 35° was selected for the in vivo PDX DCE protocol. This choice is at the upper end of the 25°–35° FA range recommended by the Quantitative Imaging Biomarkers Alliance (QIBA), but herein a contrast agent dose is leveraged that is substantially greater (∼10×) than QIBA suggests for clinical studies (39) (Figure 4).
Figure 4.
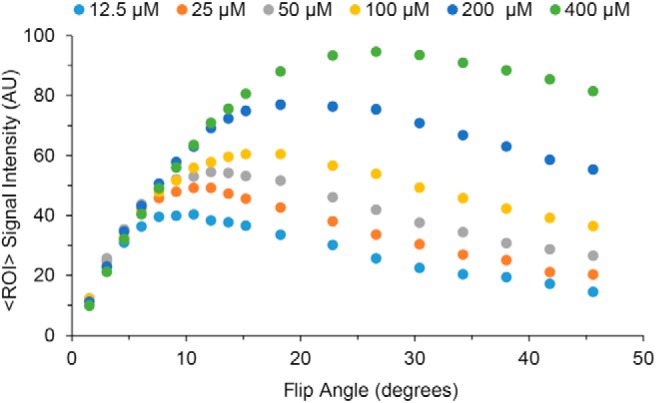
Exploring DCE with the 6-compartment relaxation phantom. Plots of the signal intensity of each tube as a function of flip angle (FA). The T1 values (Table 3) were chosen to bridge the expected range for tissues in vivo and during the DCE experiment.
In Vivo TNBC PDX MRI
PDX Cohort MRI-Determined Parameter Summary.
Representative parameter maps for T1, T2, ADC, and DCE-AUC are shown in Figure 5 for a single transaxial slice through the center of the tumor for a single mouse. Characteristics of the entire cohort are summarized in Table 4. Citing parameter <ROI> values, which were similar to parameter <Voxel> values, tumor volume ranged from 31 to 318 μL with a mean ± SD of 174 ± 73 μL; T1 ranged from 1.4 to 2.3 seconds with a mean of 1.9 ± 0.2 seconds; T2 ranged from 56 to 68 milliseconds with a mean of 62 ± 3 milliseconds; ADC ranged from 0.59 to 0.83 μm2/ms with a mean of 0.71 ± 0.06 μm2/ms; DCE-Ktrans ranged from 0.01 to 0.06 min−1 with a mean of 0.025 ± 0.015 min−1; and DCE-AUC ranged from 0.18 to 1.4 au with a mean of 0.77 ± 0.30 au.
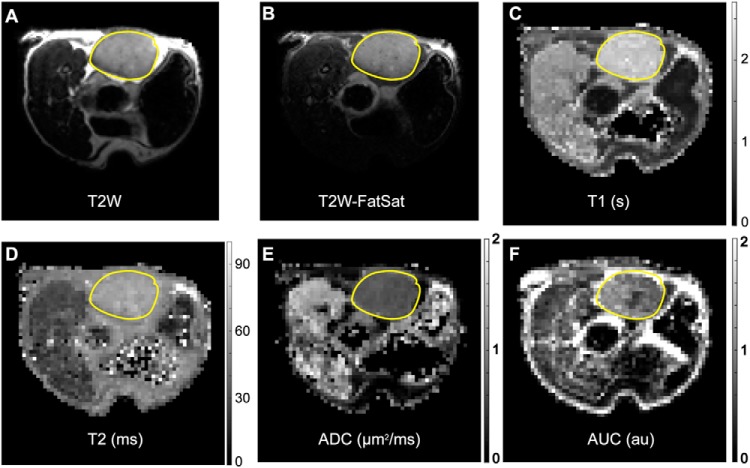
Figure 5.
Representative PDX T2-weighted (T2W) images and magnetic resonance (MR) parametric maps. T2W (A), T2W-FatSat (B), T1 (C), T2 (D), ADC (E), and DCE-AUC (F) parametric maps from a single central slice through a 206-μL tumor. T2W image intensity scaling is arbitrary. Parametric map intensities are given by vertical scale bars. The tumor is outlined with a yellow line. Isolated white or black voxels (specks) generally occur in anatomic regions of very low signal-to-noise ratio (SNR) under the protocol conditions and thus reflect a modeling (parameter estimation) failure. The hyperintense T2W fat signal is a known artifact of the fast spin-echo (FSE) protocol (45–47).
Table 4.
PDX Cohort MRI-Determined Parameter Summary
| Parameter | Mean | Median | SD | Range |
|---|---|---|---|---|
| Volume, μL | 174 | 177 | 73 | 31–318 |
| Volume, μL, for DCE subset | 235 | 249 | 62 | 145–299 |
| T1 <ROI>, sa | 1.86 | 1.88 | 0.22 | 1.4–2.3 |
| T1 <Voxel>, sb | 1.92 | 1.93 | 0.24 | 1.4–2.4 |
| T2 <ROI>, msa | 62 | 61 | 3.0 | 56–68 |
| T2<Voxel>, msb | 62 | 62 | 7.4 | 56–68 |
| ADC<ROI>, μm2/msa | 0.71 | 0.71 | 0.06 | 0.59–0.83 |
| ADC<Voxel>, μm2/msb | 0.73 | 0.73 | 0.08 | 0.59–0.88 |
| DCE-AUC<ROI>, aua | 0.77 | – | 0.30 | 0.18–1.4 |
| DCE-AUC<Voxel>, aub | 0.77 | 0.70 | 0.30 | 0.18–1.4 |
| DCE-Ktrans <ROI>, min−1 a | 0.025 | – | 0.015 | 0.01–0.06 |
| DCE-Ktrans <Voxel>, min−1 b | 0.035 | 0.027 | 0.031 | 0.00–0.10 |
aThe test and retest measurements were averaged and are summarized over all mice (n = 13 for T1, T2, ADC; or n = 4 for DCE-AUC, DCE-Ktrans).
b<ROI> measurements from modeling the averaged data over the entire tumor volume.
c<Voxel> measurements from modeling each tumor voxel independently, averaging across repeats, and summarized across all animals.
Dependence of MRI-Determined Parameters on PDX Volume.
Small-animal tumor models often use tumors in the 1- to 2-cm range. Approximating the tumor as a sphere of water, a 1-cm-diameter tumor would have a volume of 0.52 mL or ∼2.6% by weight of a 20-g mouse; a 2-cm-diameter tumor would have a volume of 4.2 mL or ∼20% by weight of a 20-g mouse. Such “large” tumors in small-animal models are often quite heterogeneous, with substantially necrotic and hemorrhagic centers and rims that remain metabolically active with continued growth. These characteristics are not generally representative of human breast tumors diagnosed in the clinic, where TNBC most commonly presents as a mass on mammography, most frequently round, oval, or lobular in shape, with indistinct margins, and no associated microcalcifications, irregular spiculated masses, or pleomorphic microcalcifications (40). The tumors used for the test–retest studies herein averaged 0.174 ± 0.073 mL in volume, representing <1% of the mouse's weight (∼20 g).
We note in passing that size comparisons are relative. Human female breast tumors range from barely detectable to 5 cm and larger. Typical human female breast volume is ∼550 mL, roughly a pound. Approximating the tumor as a perfect sphere of water, a 5-cm tumor would have a volume of 65 mL or 12% of the typical human breast volume or weight. The weight of the mouse fourth inguinal fat pad is ∼50 ± 10 mg (mean ± SD, n = 4). Thus, although only 30 μL (∼30 mg) of TNBC cells was implanted into the mouse mammary fat pad, the tumors used in our report had grown to a mean weight corresponding to 3.5× the mammary fat pad weight.
The dependence of MR parameters on PDX volume over the range of volumes examined (31–318 μL) was minimal and generally not statistically significant. There are only 2 statistically significant trends with increasing PDX volume: (1) T1<Voxel> showed a small decrease (−0.002 s/μL), as did (2) the voxel-wise T1 distribution width, as given by T1-CV<Voxel> (−0.03%/μL). Whether the weak dependence of these parameters on PDX volume reflects physiologic or microenvironment changes as tumors grow is unclear. The dependency may simply reflect the greater measurement precision/accuracy afforded by larger tumor volumes.
PDX MRI-Determined Parameter Test–Retest Variability
Test–retest variability in MRI-determined parameters is summarized in Table 5. Further assessment of DCE test–retest variability was not undertaken owing to the small number of paired DCE measurements (n = 4 × 2) and the substantial CVWS observed, the genesis of which remains to be ascertained.
Table 5.
PDX Cohort MRI-Determined Parameter Test–Retest Within-Subject Variability
| Parameter | Mean Δa | SDWS Δa | Mean ΔDWb | SDWS ΔDWb | CVWS Δa (%) | CVWS ΔDWb (%) |
|---|---|---|---|---|---|---|
| Volume, μL | 1.7 | 1.5 | – | – | 0.94 | – |
| T1<ROI>, s | 0.24 | 0.21 | – | – | 11 | – |
| T1<Voxel>, s | 0.18 | 0.16 | 0.06 | 0.05 | 8.6 | 22 |
| T2<ROI>, ms | 1.8 | 1.6 | – | – | 2.6 | – |
| T2<Voxel>, ms | 1.8 | 1.6 | 1.7 | 1.6 | 2.5 | 20 |
| ADC<ROI>, μm2/ms | 0.05 | 0.05 | – | – | 7.0 | – |
| ADC<Voxel>, μm2/ms | 0.05 | 0.04 | 0.03 | 0.03 | 5.8 | 16 |
| DCE-AUC<ROI>, au | 0.17 | 0.17 | – | – | 29 | – |
| DCE-AUC<Voxel>, au | 0.17 | 0.17 | 0.15 | 0.11 | 29 | 33 |
| DCE-Ktrans<ROI>, min−1 | 0.012 | 0.013 | – | – | 32 | – |
| DCE-Ktrans<Voxel>, min−1 | 0.017 | 0.016 | 0.012 | 0.01 | 44 | 28 |
aThe symbol Δ indicates the absolute value of the difference between two paired repeat determinations.
bThe width of each voxel-wise parameter distribution is summarized as the SD of the parameter distribution. In this case, Δ for the distribution width test-retest is indicated as ΔDW.
Figure 6 shows the full ROI test–retest T1-, T2-, and ADC-normalized voxel-wise distribution plots for the tumor displayed in Figure 5. The degree of test–retest voxel-wise distribution congruence shown is broadly representative of all 13 test–retest T1, T2, and ADC determinations. Bland–Altman plots (36, 37) are shown in Figure 7 for tumor volume and T1, T2, and ADC <ROI> means and in Figure 8 for voxel-wise distribution widths (voxel-wise SDs).
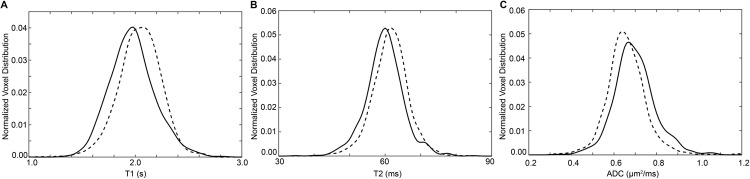
Figure 6.
Representative PDX test–retest full ROI-normalized voxel distribution plots. For the same 206-μL tumor shown in Figure 5, the test–retest-normalized voxel distributions for maps of T1 (A), T2 (B), and ADC (C) are overlaid to provide a visual representation of measurement reproducibility. Quantitative test–retest summary statistics for the entire cohort are listed in Table 5.
Figure 7.
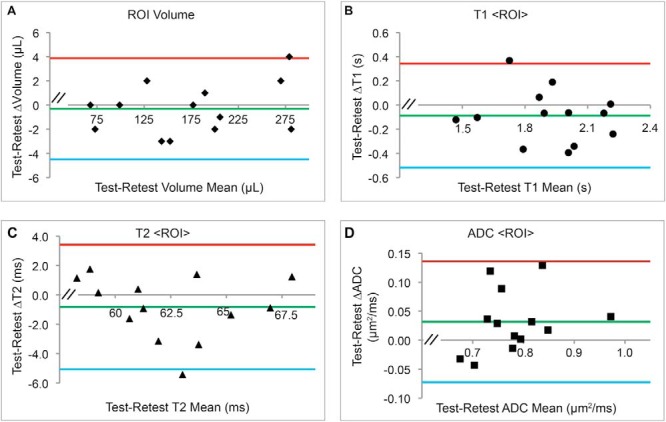
Bland–Altman plots for the test–retest cohort <ROI> Analysis. The <ROI>-determined mean of test and retest parameter values (x-axis) and difference in test and retest parameter values (y-axis) are shown for each tumor (volume (A), T1 (B), T2 (C), ADC (D)), together with overall mean of the test–retest differences (green lines) and 95% confidence limits (red and blue lines). Apparent biases were not statistically significant (P > .05).
Figure 8.
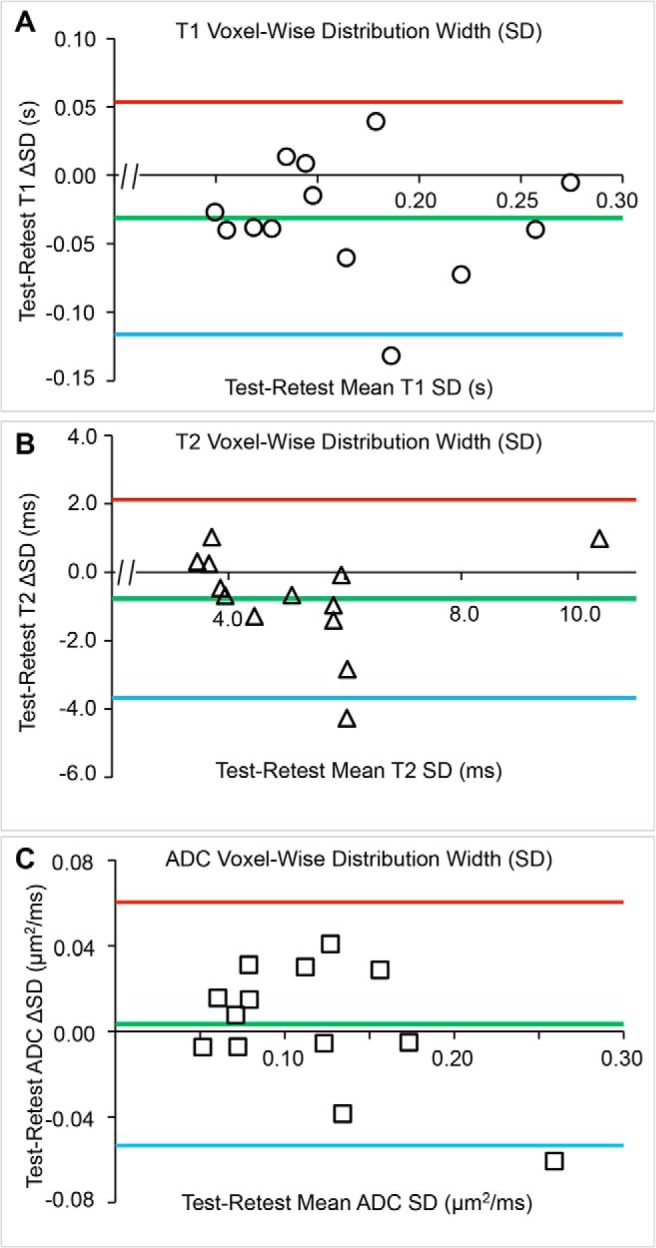
Bland–Altman plots for the test–retest cohort <Voxel> analysis. The voxel-wise-determined mean of test and retest parameter values (x-axis) and difference in test and retest parameter values (y-axis) are shown for each tumor (T1 (A), T2 (B), ADC (C)), together with the overall mean of the test–retest differences (green lines) and 95% confidence limits (red and blue lines). Apparent biases were not statistically significant (P > .05).
As anticipated, there is good concordance between tumor <ROI> and <Voxel> parameter mean test–retest results. Test–retest CVWS values are the smallest for tumor-volume determination, ∼1%, and then scale as T2 < ADC ∼ T1 ≪ DCE-AUC and DCE-Ktrans. Clearly, the DCE-AUC and DCE-Ktrans test–retest determinations showed the greatest variation of all the MRI parameter determinations, with CVWS in the 30%–40% range.
Discussion
Longitudinal monitoring of quantitative MRI parameter summary statistics (and, ultimately, voxel-wise distribution characteristics) as indices for predicting and assessing tumor therapeutic response requires knowledge of the test–retest precision of such metrics. In the clinic, initiating and modifying therapeutic intervention based on MRI metrics require knowledge of when an observed change in an MRI parameter(s) is likely to be significant. Supported through the National Cancer Institute Co-Clinical Imaging Resource Program initiative, this work is the first step toward such assessment with TNBC PDXs implanted in abdominal mammary fat pads. The measurement is challenged by the SNR achievable in a ∼60-minute multicontrast scanning protocol and by residual subject respiratory motion.
Herein, to suppress respiratory motion, several strategies have been used, including: (1) a half-cylindrical mouse cradle whose inner diameter is optimized for a snug fit with the mouse lying on its side, (2) a modified ridged plastic panel (10) to restrict respiratory motion to the chest area, away from the lower abdomen, (3) a judicious use of body taping to reduce residual motion (see Figure 1), and (4) for ADC mapping, where sensitivity to motion is extreme, respiratory gating of the data collection. The 2D slice-selective DCE protocol is particularly sensitive to motion, resulting in slice positional changes that disrupt the steady-state magnetization condition. This is graphically illustrated in Figure 2 for the cases of poor versus good motion control as achieved herein.
Obtaining sufficiently high SNR and spatial resolution is always challenging in MR imaging of mice, whose body volume is ∼3,000-fold less than that of the adult human. Although high magnetic fields and small-diameter RF receiver coils improve sensitivity, one is always faced with decisions regarding the trade-off of SNR versus spatial resolution versus data-acquisition scan time. Because our PDX-bearing mice experience unavoidable physiological stress, including therapy and repeated anesthesia and handling for MRI scanning, a 1-hour multicontrast scanning protocol was developed to provide sufficient throughput, SNR, and spatial resolution, while minimizing time under anesthesia. In addition, it was recognized that on a modern preclinical MRI scanner, image analysis often poses the more time-consuming component. Thus, to maximize throughput, standard multislice, Cartesian k-space-encoding pulse sequence protocols were used.
The range of FA and TE parameters used to collect the T1 and T2 data in the 2 phantoms was the same as used for the PDXs. The multiecho T2 measurement is time-efficient, and thus, the number of echoes (TE = ∼16 milliseconds) was set at 16 to quantify tissues with long T2 values (eg, bladder) in the FOV. The variable FA T1 measurement is not as time-efficient. The use of 5 FAs was found to provide moderately decreased, though sufficient, precision in T1 determination compared with the use of 18 FAs. As expected, the multiecho T2 measurement was the most accurate and precise when multiple e-foldings of the signal decay were captured by multiple echoes. At the extremes of long and short T2s examined in the 6-compartment phantom, the multiecho pulse sequence parameters that were used for in vivo PDX were far from optimal.
Tumor MRI parameter determinations showed different test–retest sensitivities to inevitable protocol nonidealities. T2W anatomical scanning is time-efficient and provides high spatial resolution and good tumor-to-background contrast. Thus, tumor volume determination via multislice T2W MRI was highly robust, with a ∼1% test–retest CVWS. T2-map determination using a multislice, multiecho protocol is also time-efficient and relatively insensitive to modest imperfections in pulse and transmit RF-fields. Thus, T2-map determinations were also robust, with a ∼3% test–retest CVWS (mean value over the tumor volume). The variable FA T1-map determination is sensitive to transmit RF-field imperfections, which results in a nonuniform FA over the FOV, and it suffers from limited dynamic range and SNR relative to more time-consuming (eg, inversion recovery) T1 protocols. In principle, the effect of transmit RF field imperfections can be mitigated by including a B1-field mapping protocol. This was not done herein because a transmit volume coil was used that produced a relatively homogeneous B1 field over the tumor volume. Nevertheless, despite these sensitivities to protocol imperfections, the T1-map determination was relatively robust, with a ∼9%–11% CVWS (mean value over the tumor volume).
The ADC map determination is sensitive to respiratory motion and the limited SNR at high b-values. Using lower b-values provides higher SNR at the expense of reduced dynamic range. Bito et al. (41) showed that the optimal b-value for a single ADC is that which results in ∼1 e-folding in the diffusion signal decay (ADC × b-value = ∼ 1). However, tissue ADC values vary depending upon tissue characteristics. Thus, providing good-quality ADC-map contrast between the PDX and surrounding abdominal tissues with different ADC values is also desired. Koc and Erbay (42) evaluated optimal b-values for differentiation of abdominal lesions and recommended values of 600 s/mm2 or higher. We anticipate animals in the second part of the coclinical trial, which will monitor PDX growth and therapeutic response, will ultimately be bearing substantially larger PDX with fluid-enriched necrotic regions (higher ADCs). Thus, we qualitatively chose a b-value of 850 s/mm2, somewhat higher than the recommended 600 s/mm2, to bridge between the ADCs for the more cellular-dense versus necrotic fluid-rich tissues. The resulting ADC maps showed a ∼6%–7% CVWS (mean value over the tumor volume).
The DCE protocol used herein was designed to provide strong DCE signal under high temporal resolution (3-second frames). In early exploratory studies, it was found that low doses of contrast agent provided low DCE SNR. The dose was adjusted to provide high DCE SNR, but not such a large dose as to induce DSC-like T2* effects with the short echo time (∼1.6 milliseconds) used. At the dose used, a strong muscle DCE signal was observed, useful for normalization regarding AUC and RR AIF estimation regarding Ktrans, and no apparent, frank effects on renal contrast agent clearance and vascular function were observed. Specifically, the “DCE signal” was near baseline at 1–2 h post administration.
While DCE findings should be considered preliminary owing to the small number of paired test–retest determinations, by far the least robust were the DCE-AUC and DCE-Ktrans determinations, yielding ∼30%–40% CVWS. The reason for this is unclear, but substantial DCE variability is a common finding (39, 43, 44). It seems unlikely that significant residual contrast agent remained in the tumor following the ∼16- to 20-h period between the paired test–retest determination, as the T1-maps did not show substantive test–retest bias. The DCE paired test–retest determination was carried out in the afternoon versus morning of consecutive days, and it is possible that animals were in different metabolic/physiologic states related to their normal diurnal cycle. No attempt was made to control for this. Contrast agent was administered via manual injection into the tail vein through catheters that were placed by a highly experienced small-animal procedure technologist. While a power injector might improve somewhat on within-animal bolus uniformity, the injected volumes were small and not highly variable. Further, the tumor DCE-AUC normalization to the maximum muscle DCE enhancement in the early time frames and the reference RR AIF estimation regarding Ktrans should have substantially mitigated effects of injection variability. Thus, while the genesis of the large CVWS observed with tumor DCE determinations remains unclear, it does motivate additional experiments with control-state (nontumor bearing) mice, and this will be the subject of future investigations.
Conclusion
Determination of PDX volume via manually drawn ROIs is highly robust, with a ∼1% test–retest CVWS. Mean T2 is also a robust determination, with a ∼3% test–retest CVWS. Mean T1 and ADC are less robust, with test–retest CVWS in the ∼6%–11% range. The least robust, in a test–retest sense, are the DCE-AUC and DCE-Ktrans, with test–retest CVWS in the ∼30%–40% range.
Ongoing work will assess methods to further quantify the shape/character of the voxel-wise parameter distributions (ie, methods of histogram analysis) and the robustness of these measures to test–retest evaluation.
Acknowledgments
U24 CA209837, Washington University Co-Clinical Imaging Research Resource, KI Shoghi, PI; Siteman Cancer Center Small-Animal Cancer Imaging Shared Resource (Cancer Center Support Grant P30 CA091842, TJ Eberlein, PI). We are grateful to Steve Baldwin for software enabling the compartment exchange modeling.
Disclosures: No disclosures to report.
Conflict of Interest: The authors have no conflict of interest to declare.
Footnotes
- PDXs
- Patient-derived xenografts
- ADC
- apparent diffusion coefficient
- DCE
- dynamic contrast-enhanced
- CVWS
- within-subject test–retest coefficient-of-variation
- TNBC
- triple-negative breast cancer
- MRI
- magnetic resonance imaging
- T1W
- T1-weighted
- T2W
- T2-weighted
- RF
- radiofrequency
- TR
- repetition time
- SNR
- signal-to-noise ratio
- TE
- echo time
- FOV
- field of view
- FA
- flip angle
- AUC
- area under the time-course curve
- SDs
- standard deviations
References
- 1. Sulaiman A, Wang L. Bridging the divide: preclinical research discrepancies between triple-negative breast cancer cell lines and patient tumors. Oncotarget. 2017;8:113269–113281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes SL, Sorace AG, Loveless ME, Whisenant JG, Yankeelov TE. Correlation of tumor characteristics derived from DCE-MRI and DW-MRI with histology in murine models of breast cancer. NMR Biomed. 2015;28:1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barnes SL, Sorace AG, Whisenant JG, McIntyre JO, Kang H, Yankeelov TE. DCE- and DW-MRI as early imaging biomarkers of treatment response in a preclinical model of triple negative breast cancer. NMR Biomed. 2017;30:e3799. [DOI] [PubMed] [Google Scholar]
- 4. Beeman SC, Shui YB, Perez-Torres CJ, Engelbach JA, Ackerman JJ, Garbow JR. O2 -sensitive MRI distinguishes brain tumor versus radiation necrosis in murine models. Magn Reson Med. 2016;75:2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duan C, Yang R, Yuan L, Engelbach JA, Tsien CI, Rich KM, Dahiya SM, Johanns TM, Ackerman JJH, Garbow JR. Late effects of radiation prime the brain microenvironment for accelerated tumor growth. Int J Radiat Oncol Biol Phys. 2019;103:190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, Ding L, Griffith OL, Miller C, Larson D, Fulton RS, Harrison M, Mooney T, McMichael JF, Luo J, Tao Y, Goncalves R, Schlosberg C, Hiken JF, Saied L, Sanchez C, Giuntoli T, Bumb C, Cooper C, Kitchens RT, Lin A, Phommaly C, Davies SR, Zhang J, Kavuri MS, McEachern D, Dong YY, Ma C, Pluard T, Naughton M, Bose R, Suresh R, McDowell R, Michel L, Aft R, Gillanders W, DeSchryver K, Wilson RK, Wang S, Mills GB, Gonzalez-Angulo A, Edwards JR, Maher C, Perou CM, Mardis ER, Ellis MJ. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuperwasser C, Chavarria T, Wu M, Magrane G, Gray JW, Carey L, Richardson A, Weinberg RA. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101:4966–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garbow JR, McIntosh C, Conradi MS. Actively decoupled transmit-receive coil-pair for mouse brain MRI. Concepts Magn Reson Part B Magn Reson Eng. 2008;33B:252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thangavel K, Saritaş EÜ. Aqueous paramagnetic solutions for MRI phantoms at 3 T: a detailed study on relaxivities. Turk J Elec Eng & Comp Sci. 2017;25:2108–2121. [Google Scholar]
- 10. Kim H, Samuel S, Totenhagen JW, Warren M, Sellers JC, Buchsbaum DJ. Dynamic contrast enhanced magnetic resonance imaging of an orthotopic pancreatic cancer mouse model. J Vis Exp. 2015;98:52641–52646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. [DOI] [PubMed] [Google Scholar]
- 12. Quirk JD, Bretthorst GL, Garbow JR, Ackerman JJH. Magnetic resonance data modeling: the Bayesian analysis toolbox. Concepts Magn Reson Part A.2019;47A:e21467. [Google Scholar]
- 13. Bayes T. An essay towards solving a problem in the doctrine of chances. 1763. MD Comput. 1991;8:157–171. [PubMed] [Google Scholar]
- 14. McGrayne SB, Merlington L. The Theory That Would Not Die: How Bayes' Rule Cracked the Enigma Code, Hunted Down Russian Submarines, and Emerged Triumphant from Two Centuries of Controversy. Yale University Press; 2012. [Google Scholar]
- 15. Yablonskiy DA, Sukstanskii AL. Theoretical models of the diffusion weighted MR signal. NMR Biomed. 2010;23:661–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bretthorst GL. How accurately can parameters from exponential models be estimated? A Bayesian view. Concepts Magn Reson Part A. 2005;27A:73–83. [Google Scholar]
- 17. Bretthorst GL, Hutton WC, Garbow JR, Ackerman JJH. Exponential parameter estimation (in NMR) using Bayesian probability theory. Concepts Magn Reson Part A. 2005;27A:55–63. [Google Scholar]
- 18. Neil JJ, Bretthorst GL. On the use of Bayesian probability theory for analysis of exponential decay data: an example taken from intravoxel incoherent motion experiments. Magn Reson Med. 1993;29:642–647. [DOI] [PubMed] [Google Scholar]
- 19. d'Avignon DA, Bretthorst GL, Holtzer ME, Holtzer A. Site-specific thermodynamics and kinetics of a coiled-coil transition by spin inversion transfer NMR. Biophys. J. 1998;74:3190–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. d'Avignon DA, Bretthorst GL, Holtzer ME, Holtzer A. Thermodynamics and kinetics of a folded-folded' transition at valine-9 of a GCN4-like leucine zipper. Biophys. J. 1999;76:2752–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Demsar F, Roberts TPL, Schwickert HC, Shames DM, Mann JS, Tomazic S, Brasch RC. MRI mapping of microvascular permeability and tissue blood volume. Pflugers Arch. 1996;431:R263–R264. [DOI] [PubMed] [Google Scholar]
- 22. Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7:91–101. [DOI] [PubMed] [Google Scholar]
- 23. Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HBW, Lee T-Y, Mayr NA, Parker GJM, Port RE, Taylor J, Weisskoff RM. Estimating kinetic parameters from dynamic contrast-enhanced t1-weighted MRI of a diffusable tracer: Standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. [DOI] [PubMed] [Google Scholar]
- 24. Roberts HC, Roberts TPL, Brasch RC, Dillon WP. Quantitative measurement of microvascular permeability in human brain tumors achieved using dynamic contrast-enhanced MR imaging: correlation with histologic grade. AJNR Am J Neuroradiol. 2000;21:891–899. [PMC free article] [PubMed] [Google Scholar]
- 25. Sourbron SP, Buckley DL. On the scope and interpretation of the Tofts models for DCE-MRI. Magn Reson Med. 2011;66:735–745. [DOI] [PubMed] [Google Scholar]
- 26. Luypaert R, Sourbron S, de Mey J. Validity of perfusion parameters obtained using the modified Tofts model: A simulation study. Magn Reson Med. 2011;65:1491–1497. [DOI] [PubMed] [Google Scholar]
- 27. Kovar DA, Lewis M, Karczmar GS. A new method for imaging perfusion and contrast extraction fraction: Input functions derived from reference tissues. J Magn Reson Imaging. 1998;8:1126–1134. [DOI] [PubMed] [Google Scholar]
- 28. Yang C, Karczmar GS, Medved M, Stadler WM. Estimating the arterial input function using two reference tissues in dynamic contrast-enhanced MRI studies: Fundamental concepts and simulations. Magn Reson Med. 2004;52:1110–1117. [DOI] [PubMed] [Google Scholar]
- 29. Riabkov DY, Bella EVRD. Estimation of kinetic parameters without input functions: analysis of three methods for multichannel blind identification. IEEE Trans Biomed Eng. 2002;49:1318–1327. [DOI] [PubMed] [Google Scholar]
- 30. Yankeelov TE, Luci JJ, Lepage M, Li R, Debusk L, Lin PC, Price RR, Gore JC. Quantitative pharmacokinetic analysis of DCE-MRI data without an arterial input function: a reference region model. Magn Reson Imaging. 2005;23:519–529. [DOI] [PubMed] [Google Scholar]
- 31. Yankeelov TE, Cron GO, Addison CL, Wallace JC, Wilkins RC, Pappas BA, Santyr GE, Gore JC. Comparison of a reference region model with direct measurement of an AIF in the analysis of DCE-MRI data. Magn Reson Med. 2007;57:353–361. [DOI] [PubMed] [Google Scholar]
- 32. Donahue KM, Weisskoff RM, Parmelee DJ, Callahan RJ, Wilkinson RA, Mandeville JB, Rosen BR. Dynamic Gd-DTPA enhanced MRI measurement of tissue cell volume fraction. Magn Reson Med. 1995;34:423–432. [DOI] [PubMed] [Google Scholar]
- 33. Faranesh AZ, Kraitchman DL, McVeigh ER. Measurement of kinetic parameters in skeletal muscle by magnetic resonance imaging with an intravascular agent. Magn Reson Med. 2006;55:1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hernando D, Haldar JP, Sutton BP, Ma J, Kellman P, Liang Z-P. Joint estimation of water/fat images and field inhomogeneity map. Magn Reson Med. 2008;59:571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hyslop NP, White WH. Estimating precision using duplicate measurements. J Air Waste Manage. 2009;59:1032–1039. [DOI] [PubMed] [Google Scholar]
- 36. Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb). 2015;25:141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. Statistician. 1983;32:307–317. [Google Scholar]
- 38. Synek V. Evaluation of the standard deviation from duplicate results. Accredit Qual Assur. 2008;13:335–337. [Google Scholar]
- 39. DCE MRI Technical Committee. DCE MRI Quantification Profile, Quantitative Imaging Biomarkers Alliance. Version 1.0. Reviewed Draft. QIBA, July 1, 2012. Available from: https://qibawiki.rsna.org/images/1/12/DCE-MRI_Quantification_Profile_v1.0.pdf.
- 40. Yang WT, Dryden M, Broglio K, Gilcrease M, Dawood S, Dempsey PJ, Valero V, Hortobagyi G, Atchley D, Arun B. Mammographic features of triple receptor-negative primary breast cancers in young premenopausal women. Breast Cancer Res. Treat. 2008;111:405–410. [DOI] [PubMed] [Google Scholar]
- 41. Bito Y, Hirata S, Yamamoto E. Optimum gradient factors for apparent diffusion coefficient measurement. Proceedings of the 3rd Scientific Meeting of Soc Magn Reson (now ISMRM), 1995; p. 913. [Google Scholar]
- 42. Koc Z, Erbay G. Optimal b value in diffusion-weighted imaging for differentiation of abdominal lesions. J Magn Reson Imaging. 2014;40:559–566. [DOI] [PubMed] [Google Scholar]
- 43. Ng CS, Waterton JC, Kundra V, Brammer D, Ravoori M, Han L, Wei W, Klumpp S, Johnson VE, Jackson EF. Reproducibility and comparison of DCE-MRI and DCE-CT perfusion parameters in a rat tumor model. Technol Cancer Res Treat. 2012;11:279–288. [DOI] [PubMed] [Google Scholar]
- 44. He D, Zamora M, Oto A, Karczmar GS, Fan X. Comparison of region-of-interest-averaged and pixel-averaged analysis of DCE-MRI data based on simulations and pre-clinical experiments. Phys Med Biol. 2017;62:N445–N59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henkelman RM, Hardy PA, Bishop JE, Poon CS, Plewes DB. Why fat is bright in RARE and fast spin-echo imaging. J Magn Reson Imaging. 1992;2:533–540. [DOI] [PubMed] [Google Scholar]
- 46. Mulkern RV, Packard AB, Gambarota G. Field dependence of the bright fat effect in fast spin echo imaging: theory and experiment. Proc Intl Soc Mag Reson Med. 2003;11:1107. [Google Scholar]
- 47. Stables LA, Kennan RP, Anderson AW, Gore JC. Density matrix simulations of the effects of J coupling in spin echo and fast spin echo imaging. J Magn Reson. 1999;140:305–14. [DOI] [PubMed] [Google Scholar]