Abstract
Purpose
Next-generation sequencing (NGS) is rapidly replacing Sanger sequencing in genetic diagnostics. Sensitivity and specificity of NGS approaches are not well-defined, but can be estimated from applying NGS and Sanger sequencing in parallel. Utilizing this strategy, we aimed at optimizing exome sequencing (ES)–based diagnostics of a clinically diverse patient population.
Methods
Consecutive DNA samples from unrelated patients with suspected genetic disease were exome-sequenced; comparatively nonstringent criteria were applied in variant calling. One thousand forty-eight variants in genes compatible with the clinical diagnosis were followed up by Sanger sequencing. Based on a set of variant-specific features, predictors for true positives and true negatives were developed.
Results
Sanger sequencing confirmed 81.9% of ES-derived variants. Calls from the lower end of stringency accounted for the majority of the false positives, but also contained ~5% of the true positives. A predictor incorporating three variant-specific features classified 91.7% of variants with 100% specificity and 99.75% sensitivity. Confirmation status of the remaining variants (8.3%) was not predictable.
Conclusions
Criteria for variant calling in ES-based diagnostics impact on specificity and sensitivity. Confirmatory sequencing for a proportion of variants, therefore, remains a necessity. Our study exemplifies how these variants can be defined on an empirical basis.
Keywords: Genetic testing, Laboratory standards, Sensitivity, Specificity, exome sequencing
Introduction
Sanger sequencing has been the major technology for molecular diagnosis of inherited diseases. In genetically heterogeneous conditions, the associated “one gene after the other” strategy is costly and time-consuming. With the continuing increase in the number of candidate genes for many conditions,1 it also becomes more and more impractical. Next-generation sequencing (NGS) is therefore rapidly replacing Sanger sequencing not only in research settings, but also in routine genetic diagnostics. The corresponding approaches either target a predefined set of genes (panel sequencing), the exonic regions of all genes (exome-sequencing, ES), or the complete human genome (genome sequencing, GS).2
Initial applications of NGS suggested that an enrichment step as immanent to panel sequencing and ES introduces a significant amount of error which manifests as false positives and false negatives.3 For the implementation of NGS as a diagnostic test, determination (and optimization) of analytical specificity and sensitivity is therefore crucial, and orthogonal Sanger sequencing is the most straightforward corresponding approach.4
Early studies that addressed specificity of NGS-based diagnostics applied small gene panels to limited numbers of patients, and commonly observed a considerable fraction of false positives.5–8 Later studies on larger panels and more patients, however, frequently reported (close to) 100% specificity.9–13 Some authors therefore proposed that independent confirmation of NGS findings is unnecessarily redundant, and that panel sequencing can reliably be implemented as a stand-alone test.10,13,14 Similar conclusions have later been drawn for diagnostic ES.15,16 A very large recent study, however, (re-)raised some concerns toward this attitude: based on the analysis of 47 genes in ~20,000 patients with hereditary forms of cancer, Mu et al.17 suggested that up to 1.3% of gene panel–based candidate variants represent false positives. Based on the presumed negative effect on specificity, the authors therefore refrained from completely omitting Sanger-based confirmatory sequencing.17
The sensitivity aspect of NGS has apparently received much less attention. This may be due to the fact that, to properly determine sensitivity, NGS-derived as well as Sanger sequencing–derived data must be available for all (!) nucleotides analyzed. Naturally, this premise can only be met on a small scale. Of the few pertinent studies, most applied NGS-based resequencing of samples that had previously been Sanger-sequenced. Some of them found 100% sensitivity,11,14,18 but usually involved very small numbers of variants. Conceptually similar but larger studies did observe false negatives, and showed that overall sensitivity depends on certain thresholds applied during variant calling.6–9 In contrast to these initial efforts that focused on known variants, most subsequent reports on a combination of NGS and Sanger sequencing aimed at identifying (and verifying) novel family-specific variants. While this enabled estimating specificity (see also above), the sensitivity of NGS in these instances is unknown. This fact, however, is often not acknowledged (or not considered?) when claiming “high concordance” to Sanger sequencing for gene panels 13 and for clinical exomes.19 It was again Mu et al.17 who emphasized that the well-known interdependency of specificity and sensitivity needs to be accounted for also in NGS-based diagnostics: upon modeling 100% specificity for their set of ~8,000 panel-derived variants, they noticed a significant drop in sensitivity. They concluded that Sanger-based confirmatory sequencing for at least subsets of NGS-derived candidate variants is required to maintain high specificity and sensitivity in panel-based NGS. They also pointed out that such subsets can only be defined after having followed up very large numbers of variants, and that the corresponding criteria may be platform-specific.17
We set out to assess the performance of ES in a clinically heterogeneous diagnostic setting. We initially implemented very nonstringent criteria for variant calling to test whether the potential increase in diagnostic yield would outweigh the expected decrease in specificity. We subsequently utilized selected features of the >1,000 followed-up variants to generate an algorithm that reliably predicts true and false positives. We thereby minimized confirmatory Sanger sequencing load in an evidence-based and highly objective manner, while maintaining not only high specificity, but also high sensitivity.
Materials and methods
Sample origin
Our study incorporates data on a total of 1,048 candidate genomic variants. They were derived from routine ES-based genetic workup of 773 individuals who had been diagnosed with presumably genetic diseases. Geographic origin of these patients was highly diverse (Europe, the Middle East, South and Central America, North America, other regions), and a significant fraction had a consanguineous background. Clinical conditions included (amongst others): (1) abnormalities of the nervous system, (2) abnormalities of the head and neck, (3) muscular hypotonia/weakness, (4) developmental and/or growth delay, (5) abnormalities of the eye, and (6) metabolic abnormalities. Patient samples were provided as EDTA blood or as dried blood spots on filter cards (CentoCard®). DNA was extracted as described previously.20
Exome sequencing
Exome capture was carried out with the Nextera Rapid Capture Exome Kit (Illumina, Inc., San Diego, CA). The kit covers 214,405 exons with a total size of about 37 Mb. Sequencing was done using either NextSeq500 or HiSeq4000 sequencers (Illumina) to produce 2 × 150-bp reads, and pooling up to nine exomes per lane. The bioinformatics pipeline was based on the 1000 Genomes Project data analysis pipeline,21 and on Genome Analysis Toolkits (GATK) best practice recommendations;22 it incorporated widely used open source software projects and was supplemented with custom-developed software (a list of relevant bioinformatics tools is provided in Supplementary Table 1). In short, raw sequencing data were first converted to standard fastq format using bcl2fastq (Illumina), and then aligned using Burrows–Wheeler Aligner (BWA) software.23 Alignments were converted to binary bam file format, sorted on the fly and de-duplicated. Variant calling utilized the GATK HaplotypeCaller (the approach for deriving the quality score is detailed in Supplementary File 1). Lower cutoffs were set to: frequency ≥7.5%, total number of reads ≥2, and phred-scaled quality score ≥20.
Sanger sequencing
The primary aim of our service is to provide informed genetic diagnosis. A “gene-hunting” aspect, i.e., the identification of novel disease genes, is therefore explicitly not part of our routine diagnostic pipeline. Confirmatory Sanger sequencing was therefore initiated only for ES-derived variants in established disease genes that were compatible with the primary clinical diagnosis. To this end, exons containing the candidate variant were amplified from genomic DNA, and resequenced bidirectionally.
Modeling the application of filtering criteria as commonly used in variant calling
The criteria for variant calling in panel-based NGS approaches vary widely. Thresholds for coverage (equivalent to number of reads for a given position) included 20×,18 30×,10 40×,9 and even 100×.13 Similarly wide ranges have been used for frequency, i.e., the fraction of reads for the candidate variant (15–30%).6,8 Cutoffs for the quality score have been reported as being at least 20 or 25,8,13 but frequently only the statement “high quality” is made.9 We eventually considered 20× coverage plus 20% frequency plus a quality score of 20 as representing a typical set of minimum values for variant calling in panel sequencing. For ES-based NGS, surprisingly, corresponding cutoffs are rarely reported. We therefore implemented those applied by Strom et al.,15 as their study, by Sanger following up selected ES-derived variants, is conceptually similar to ours. The thresholds thus considered are 5× coverage, 35% frequency, and a quality score of 139.
To model these criteria for our set of data, we identified the variants that would have met the filtering criteria and those that would not have met them. We then determined the fractions of Sanger-confirmed and Sanger–not confirmed variants in these groups.
Definition of potential classifiers
Numerous features were considered to be potentially informative for beforehand prediction of Sanger confirmation status for the candidate ES-derived variants. The majority of these features were analogous in nature, i.e., consisted of a continuum of quantitatively differing states. They included (1) the phred-based quality score (“quality”), (2) the total number of reads for the position in question (“read number”), (3) the fraction of reads for the variant allele (“frequency”), (4) the number of reads for the candidate variant allele (“variant reads”), (5) the number of reads for the reference allele (“reference reads”), and (6) the GC-content in the +/− 100 bp neighboring the position in question (“GC-content”). In addition, a couple of digital features, i.e., those with only two possible states, were considered. They included (7) suggested presence of the candidate variant in heterozygosity versus in hemi- or homozygosity (“zygosity”), (8) localization in exonic versus in intronic or UTR sequence (“localization”), (9) single nucleotide exchange versus. insertion or deletion (“type”), (10) predicted pathogenic versus other prediction (“prediction”), and (11) origin in a homopolymer region (>3 consecutive identical nucleotides) versus origin not in a homopolymer region (“homopolymer origin”). Supplementary Table 2 summarizes the selected features, describes in detail the possible states, and indicates the corresponding distributions.
Criteria for defining groups of variants that do and do not require Sanger sequencing
The above-listed 11 features were individually correlated with Sanger-based confirmation status. For the digital features, the overlap of each of the two values with confirmation status was recorded. The analogous features were used to generate receiver operating characteristics curves with respect to confirmation status. We defined two criteria for deeming diagnostic relevance to an observed correlation. First, the group defined by the correlation should maintain overall sensitivity at >99.75%. This threshold corresponds to a misclassification of a maximum of 2 (of 890) Sanger-confirmed variants from our data set. Second, not more than one misclassified variant should be present per group. Third, the number of variants per group should cover a minimum of 5% of all followed-up variants (n = 53 for our data set). The best of the above-defined correlations was chosen based on group size (large preferred over small). Following removal of the corresponding variants from the data set, the analysis was repeated on the remaining variants in an iterative manner until groups fulfilling the three criteria could no longer be generated.
Results
Stringent thresholds during variant calling reduce analytical sensitivity
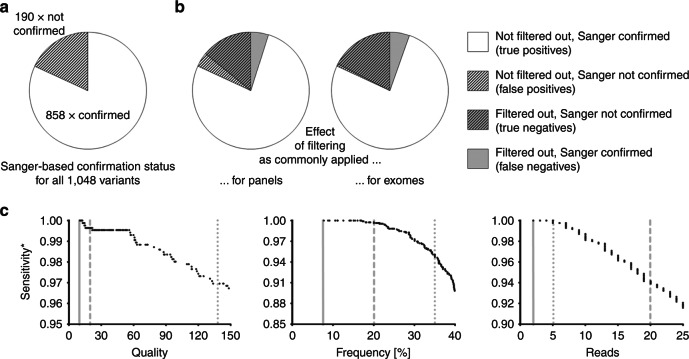
Our above-defined approach resulted in the following up of a total of 1,048 ES-derived variants (n = 735 identified on HiSeq400 versus n = 313 identified on NextSeq500) by Sanger sequencing. Of these, 858 and 190 were confirmed and not confirmed, respectively. This translates into a precision of 81.9% for our set of ES data (Fig. 1a).
Fig. 1. Performance of filtering criteria commonly used during variant calling in next-generation sequencing (NGS).
a Distribution of Sanger-confirmed vs. Sanger-rejected variants in our data set. b Modeling of the effects of standard panel- and exome sequencing (ES)-associated filtering cutoffs. c Contribution of individual parameters to the drop in sensitivity (stippled and dotted gray lines: common cutoffs for panel-based and ES-based approaches, respectively; solid gray lines: cutoffs initially used by the present study). The asterisk denotes that sensitivity as indicated here is not equivalent to the (unknown) overall sensitivity of the assay, but refers to our set of 1048 Sanger followed-up candidate variants
We were first interested in estimating the effect of having utilized very nonstringent criteria for variant calling. To this end, we derived two sets of modeled data by implementing more stringent thresholds as applied in previous panel- and ES-based approaches (see Materials and Methods for details).
Of all 1,048 variants, 849 survived filtering by the panel-associated criteria. Amongst them were 807 Sanger-confirmed variants, meaning that precision increased from 81.9 to 95.1% (=807/849). The remaining 51 Sanger-confirmed variants, however, had been filtered out. Sensitivity, therefore, decreased from 100% (as based on positive Sanger status) to 94.1% (=807/858). Using common ES-associated filtering criteria, 240 variants got removed, while 808 variants were retained. With 801 of the latter representing Sanger confirmed variants, precision rose to 99.3% (=801/808). Sensitivity, however, simultaneously dropped to 93.4% (=801/858) (Fig. 1b). Both sets of criteria, despite increasing precision, thus considerably reduce analytical sensitivity.
All filtering parameters contribute to decreased sensitivity
The above calculations incorporated all filtering criteria simultaneously. We wondered whether the observed decreases in sensitivity could be explained by one parameter with a major influence, or whether parameters contribute more uniformly. We therefore analyzed the effects of quality, frequency, and read number separately. Regarding the panel-associated cutoffs, read number had the biggest effect on sensitivity. For the ES-associated cutoffs, however, the strongest contributions came from quality and frequency (Fig. 1c). The negative impact on sensitivity thus results from an additive effect rather than being traceable to only one of the filtering parameters.
Many variant-specific features are significantly associated with Sanger-based confirmation status
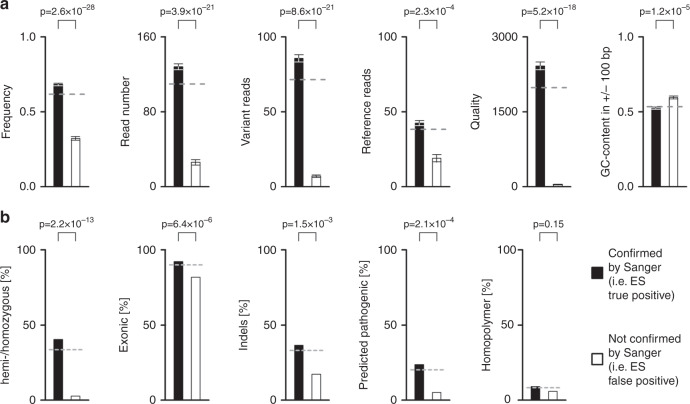
Having shown that standard filtering criteria are inappropriate for our data, we aimed at using our low-stringency data set to identify variant-specific features that are potentially more suitable for a priori prediction of Sanger confirmation status. For the analogous candidate features, we compared means of confirmed versus nonconfirmed variants. All differences were highly significant, with p values ranging from 2.3 × 10−4 (for “reference reads”) to 2.6 × 10−28 (for “frequency”) (Fig. 2a). For the digital features, we asked whether the two states are distributed differentially among confirmed versus nonconfirmed variants. The most striking difference was found for “zygosity”: homo- or hemizygous variants accounted for 40.4% of confirmed variants (347 of 858), but only 2.6% (5 of 190) of nonconfirmed variants (p = 2.2 × 10−13). Highly significant differences were also observed for “type of variant,” “localization within gene,” and “pathogenicity prediction.” Somewhat surprisingly, “homopolymer origin” was not significantly associated with confirmation status (Fig. 2b).
Fig. 2. Features of exome sequencing (ES)-derived candidate variants, and association with Sanger confirmation status.
Stippled gray lines indicate the corresponding values for all 1,048 variants. a Analogous features. Significance of differences between means (+/- SEM) was calculated using the two-sided Student’s t-test. b Digital features. Significance of differences was calculated using Fisher’s exact test
Several features can define diagnostically relevant subgroups of ES-derived variants, with “quality” being the most powerful predictor
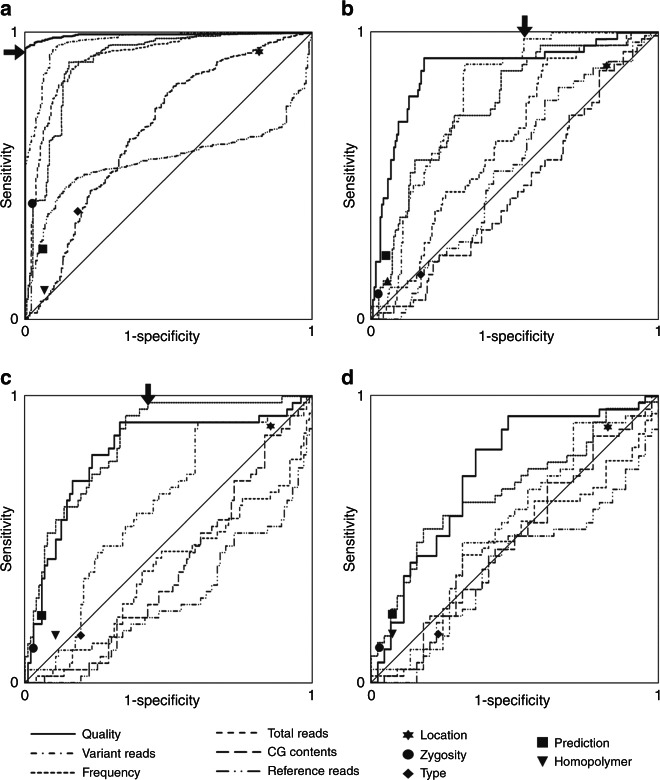
To analyze sensitivity and specificity for binary classifications according to the analogous features we generated receiver operating characteristic (ROC) curves. Consistent with the above statistical analyses (see Fig. 2a), areas under the curve were >0.5 for all features (Fig. 3a). More importantly, most features enabled the definition of thresholds which create subgroups that exclusively contain “Sanger confirmed” or “Sanger not confirmed” variants (Supplementary Table 3). By far the largest such subgroup was based on “quality”: a corresponding score of >215 unites 813 candidate ES-derived variants that all got confirmed by Sanger sequencing, and this figure corresponds to 94.8% of all such variants. Binary classification based on the digital features was unable to create groups that consist of “Sanger confirmed” or “Sanger not confirmed” exclusively (Supplementary Table 3; Fig. 3a, compare also Fig. 2b). Our analysis therefore suggested that 77.6% of all variants (813 of 1,048) do not require follow up by Sanger sequencing due to being a priori identifiable as true positives. In other words, Sanger sequencing load can be reduced to 22.4% by solely considering “quality.”
Fig. 3. Iterative analyses aiming at the definition of classifiers that can predict true positive or false positive ES-derived candidate variants while maintaining high specificity and sensitivity.
Analogous features are depicted as receiver operating characteristic curves. Binary classifications according to digital features are indicated by filled symbols. a First round of analysis on all 1,048 variants. Arrow: “quality”-based binary classifier that correctly predicts status “confirmed” for 813 variants; 100% specificity is thus retained. b Second round of analysis on the 235 variants that remained after round 1. Arrow: “variant reads”–based binary classifier that correctly predicts status “not confirmed” for 87 of 88 variants. Overall sensitivity decreases to 99.9%. c Third round of analysis on the 147 variants that remained after round 2. Arrow: “frequency”-based binary classifier that correctly predicts status “not confirmed” for 59 of 60 variants. Overall sensitivity decreases to 99.8%. d A fourth round of analysis on the 87 variants remaining after round 3 does not define additional classifiers
Two iterative rounds of analysis define predictors for large groups of “Sanger not confirmed” variants
Having found that a “quality” score of >215 defines a large group of variants that do not require Sanger confirmation due to exclusively representing true positives, we next turned to the remaining 235 variants. Numerous features were, again, significantly associated with confirmation status (not shown). We therefore repeated the above analysis. The largest group that met our criteria for sensitivity and specificity was defined by “variant reads”: a value <3 separated 88 variants, of which 87 had not been confirmed by Sanger sequencing (Fig. 3b). By applying this threshold we increased the fraction of variants that do not require Sanger sequencing to 86.0% (901 of 1,048), and reduced Sanger sequencing load to 14.0% (147 of 1,048). The simultaneous drop in sensitivity (from 100 to 99.9%) resulted from the fact that one of the variants with only 2 reads for the candidate variant had actually been confirmed by Sanger sequencing. A third round of analysis using the 147 candidate variants that remained after round 2 showed that a frequency score ≤0.25 defines an additional 60 variants highly enriched for “Sanger not confirmed” variants (59 of 60) (Fig. 3c). By applying this finding, sensitivity minimally dropped further to 99.8%, but specificity remained at 100%. More importantly, the percentage of variants requiring Sanger sequencing got further reduced to 8.3% (87 of 1,048). A fourth round on the remaining 87 variants, despite still revealing significant associations of certain features with confirmation status (not shown), did not allow separation of further groups that fulfill our predefined criteria (Fig. 3d).
The variants that require Sanger sequencing lack specific characteristics
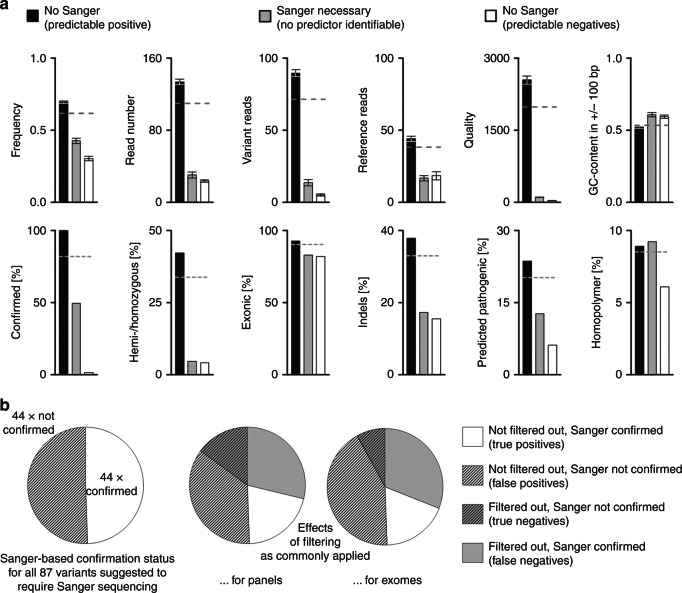
The above analyses defined three conceptually distinct groups of ES-derived candidate variants: (1) a group that does not require Sanger sequencing because every variant would get confirmed, (2) a group that does not require Sanger sequencing because the majority of the corresponding variants would not get confirmed, and (3) a group for which Sanger sequencing is required because attempts to predict confirmation status failed. These groups had been created based on iteratively applying thresholds for the three analogous features “quality,” “variant reads” and “frequency.” We were interested in how this impacted on overall composition of the three groups, especially as regards the features that had not been used to define these groups. A random distribution was observed for “homopolymer origin” and “GC-content within +/− 100 bp” (Fig. 4a). Interestingly, these were the features that had shown no association with confirmation status already in the first round of analysis (“homopolymer origin”; see Fig. 2b) or for which such an association had been comparatively weak and was not observed in the data derived from iterative steps 1–3 (“GC-content within +/− 100 bp”; Figs. 2a and 3). Strikingly, however, all other features showed a stepwise distribution of status (digital features) and mean values (analogous features), with the group that requires Sanger sequencing consistently ending up at the intermediate position (Fig. 4a). We also observed that this group is highly enriched for false positives and false negatives as resulting from the application of common filtering criteria in variant calling (Fig. 4b, compare Fig. 1a). The variants for which we found Sanger sequencing to be necessary do therefore indeed constitute a class that, in many respects, shows mixed contributions from the two other groups. This further underscores that their confirmation status is not predictable with any of the features at hand.
Fig. 4. Characteristics of the three groups created by our classifiers.
a Group-specific means (+/- SEM) for analogous features (upper panel), and intragroup distribution of states for digital features (lower panel). Note that for the majority of features, the variants that require Sanger sequencing occupy an intermediate position. b Performance of filtering criteria in the set of variants that require Sanger-based confirmation (compare Fig. 1a, b)
Discussion
The present paper reports on our experience with Sanger-based confirmatory sequencing of ES-derived variants in a clinically and ethnically heterogeneous diagnostic setting. Our strategy differed from that chosen in conceptually similar previous studies 13,15–17 in that very nonstringent criteria for variant calling were initially applied. Specifically, our lower cutoffs for “quality,” “read number,” and “frequency” were between 10 and 50% of those applied by others (Fig. 1c; see also Materials and Methods). We were aware that this approach was likely to decrease precision, but reasoned that it would enable us to address sensitivity issues at an hitherto unmatched scale.
Only 81.9% of the variants on our nonstringent candidate list got Sanger-confirmed (Fig. 1a). This finding corroborated the view that highly covered variants with a good quality score and frequencies around 50% or 100% do not require further confirmation.14,16 Having confirmed this precision-related aspect also in our data, we turned to the sensitivity issue. We started out by comparing our variant list to two lists generated with more stringent criteria. Rather unexpectedly, we found that >5% of the true positive variants were absent from these derived lists (Fig. 1b). Moreover, it appears that cutoffs for all three relevant features contribute to this phenomenon (Fig. 1c). In light of these observations, claims of a high concordance between NGS and Sanger sequencing in “high-cutoff studies” 10,13,19 need to be considered with caution. Along this line we note that our data support the assumption that laboratories reporting zero false positives may sacrifice sensitivity during variant calling.17 One explanation for this unsatisfactory situation may be a lack of awareness. Guideline papers have extensively discussed several aspects related to Sanger confirmation of NGS-derived variants 4,24–26, but at best have only shortly touched upon variant calling.27 This is, however, inherently related to the nature of such publications: they aim at providing general guidelines, while recognizing that specific details are dependent upon individual sequencing workflows. We believe that the present study will help to increase awareness for the fact that variant calling represents one of the more important pertinent details.
The above-discussed first part of our study suggested that candidate lists should be extended by variants that, on average, are more likely to be false positives. This clearly argues against complete omission of confirmatory sequencing as frequently proposed 10,13–16, but does not necessarily imply that all variants require confirmation. In the second part, we therefore wanted to define criteria that are able to predict confirmation status. In contrast to a more or less intuitive listing of rather conservative round values for one or two standard features,26 or an a priori exclusion of certain classes of variants,15 we aimed at utilizing all information available, and to thereby derive an evidence-based conclusion. By considering a variety of NGS-derived, nucleotide-specific, variant-associated features, we constructed a decision tree that, in our data set, is associated with 100% specificity and 99.8% sensitivity (Fig. 5). Especially the latter value is much higher than the ~94% that would be achieved by standard criteria (compare Fig. 1b). That this improvement comes at a price, i.e. confirmatory sequencing for 8.3% of all variants, appears more than justified. We have meanwhile updated our ES-associated diagnostic pipeline accordingly. We are, however, also Sanger sequencing 10% of the variants regarded to not actually require this step. This quality assurance measure will reveal whether the chosen cutoffs remain valid over time. It will also allow for revalidations after substantial changes to the workflow such as implementation of a new sequencing platform.
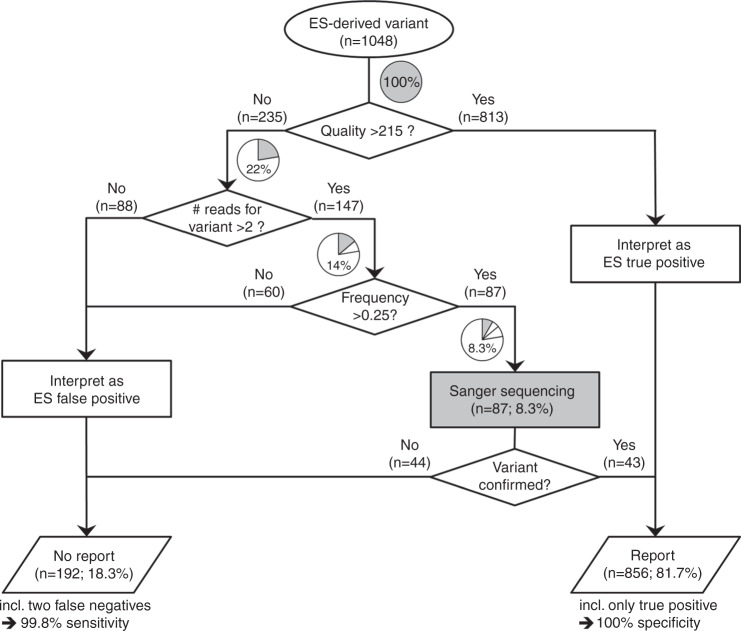
Fig. 5. Workflow for following up on identification of a candidate variant by exome sequencing (ES).
Note that this workflow is based on the data set presented in the present study, and is also specific to it (corresponding numbers of variants are indicated in parentheses). Gray sectors and corresponding percentages in pie charts indicate the remaining Sanger sequencing load
The present study was based on the standardized collection of data over a long period of time. Retrospective analysis then enabled conclusions that are of immediate relevance for both cost (i.e., Sanger sequencing load) and quality (specificity, sensitivity) of our diagnostic service. We thereby followed suggestions as to a reevaluation of NGS-associated policies once a meaningful amount of data and experience are available.4,26,27 Due to platform and operator characteristics likely playing a role,17,24 our very cutoffs should, however, not be simply adopted by other labs. Instead we recommend that, in analogy to the strategy applied here, an initial test period be combined with a subsequent evaluation step.
Electronic supplementary material
Acknowledgements
We acknowledge Christian Beetz (Jena University Hospital, Jena, Germany) for support in the preparation of the manuscript.
Disclosure
The authors declare no conflicts of interest.
Electronic supplementary material
The online version of this article (10.1038/s41436-018-0016-6) contains supplementary material, which is available to authorized users.
References
- 1.Boycott KM, Vanstone MR, Bulman DE, et al. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–91. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 2.Pinto AM, Ariani F, Bianciardi L, et al. Exploiting the potential of next-generation sequencing in genomic medicine. Expert Rev Mol Diagn. 2016;16:1037–47. doi: 10.1080/14737159.2016.1224181. [DOI] [PubMed] [Google Scholar]
- 3.Tewhey R, Nakano M, Wang X, et al. Enrichment of sequencing targets from the human genome by solution hybridization. Genome Biol. 2009;10:R116. doi: 10.1186/gb-2009-10-10-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rehm HL, Bale SJ, Bayrak-Toydemir P, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci USA. 2010;107:21104–9. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amstutz U, Andrey-Zurcher G, Suciu D, et al. Sequence capture and next-generation resequencing of multiple tagged nucleic acid samples for mutation screening of urea cycle disorders. Clin Chem. 2011;57:102–11. doi: 10.1373/clinchem.2010.150706. [DOI] [PubMed] [Google Scholar]
- 7.Berg JS, Evans JP, Leigh MW, et al. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: implications for application to clinical testing. Genet Med. 2011;13:218–29. doi: 10.1097/GIM.0b013e318203cff2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra64. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivakumaran TA, Husami A, Kissell D, et al. Performance evaluation of the next-generation sequencing approach for molecular diagnosis of hereditary hearing loss. Otolaryngol Head Neck Surg. 2013;148:1007–16. doi: 10.1177/0194599813482294. [DOI] [PubMed] [Google Scholar]
- 10.Sikkema-Raddatz B, Johansson LF, de Boer EN, et al. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–42. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]
- 11.Chin EL, da Silva C, Hegde M. Assessment of clinical analytical sensitivity and specificity of next-generation sequencing for detection of simple and complex mutations. BMC Genet. 2013;14:6. doi: 10.1186/1471-2156-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Judkins T, Leclair B, Bowles K, et al. Development and analytical validation of a 25-gene next generation sequencing panel that includes the BRCA1 and BRCA2 genes to assess hereditary cancer risk. BMC Cancer. 2015;15:215. doi: 10.1186/s12885-015-1224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baudhuin LM, Lagerstedt SA, Klee EW, et al. Confirming variants in next-generation sequencing panel testing by Sanger sequencing. J Mol Diagn. 2015;17:456–61. doi: 10.1016/j.jmoldx.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 14.McCourt CM, McArt DG, Mills K, et al. Validation of next generation sequencing technologies in comparison to current diagnostic gold standards for BRAF, EGFR and KRAS mutational analysis. PLoS ONE. 2013;8:e69604. doi: 10.1371/journal.pone.0069604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strom SP, Lee H, Das K, et al. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med. 2014;16:510–5. doi: 10.1038/gim.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck TF, Mullikin JC, Program NCS, et al. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem. 2016;62:647–54. doi: 10.1373/clinchem.2015.249623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mu W, Lu HM, Chen J, et al. Sanger confirmation is required to achieve optimal sensitivity and specificity in next-generation sequencing panel testing. J Mol Diagn. 2016;18:923–32. doi: 10.1016/j.jmoldx.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Jones MA, Bhide S, Chin E, et al. Targeted polymerase chain reaction-based enrichment and next generation sequencing for diagnostic testing of congenital disorders of glycosylation. Genet Med. 2011;13:921–32. doi: 10.1097/GIM.0b013e318226fbf2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton A, Tetreault M, Dyment DA, et al. Concordance between whole-exome sequencing and clinical Sanger sequencing: implications for patient care. Mol Genet Genom Med. 2016;4:504–12. doi: 10.1002/mgg3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trujillano D, Bertoli-Avella AM, Kumar Kandaswamy K, et al. Clinical exome sequencing: results from 2819 samples reflecting 1000 families. Eur J Hum Genet. 2017;25:176–82. doi: 10.1038/ejhg.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Lu J, Yu J, et al. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 2013;23:833–42. doi: 10.1101/gr.146084.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gargis AS, Kalman L, Berry MW, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–6. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthijs G, Souche E, Alders M, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016;24:1515. doi: 10.1038/ejhg.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hegde M, Santani A, Mao R, et al. Development and validation of clinical whole-exome and whole-genome sequencing for detection of germline variants in inherited disease. Arch Pathol Lab Med. 2017;141:798–805. doi: 10.5858/arpa.2016-0622-RA. [DOI] [PubMed] [Google Scholar]
- 27.Aziz N, Zhao Q, Bry L, et al. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med. 2015;139:481–93. doi: 10.5858/arpa.2014-0250-CP. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.