Abstract
Background
KIAA1199 has been reported to be associated with malignant progression and poor clinical outcomes in various human malignancies. However, its clinical role and molecular function remain unknown in papillary thyroid cancer (PTC).
Material/Methods
The Cancer Genome Atlas (TCGA) was used to investigate the expression profiles of KIAA1199 and miR-486-5p in PTC. Immunohistochemistry was used to validate the protein expression of KIAA1199 in PTC. The Weighted Gene Co-expression Network Analysis (WGCNA) and Gene Set Enrichment Analysis (GSEA) were used to explore the potential pathway underling KIAA1199 in PTC. In vitro and in vivo experiments were performed to investigate the biological role of KIAA1199 in PTC progression. Luciferase reporter assays and Western blot analysis were performed to determine whether KIAA1199 is a downstream target of miR-486-5p.
Results
We found that KIAA1199 was aberrantly elevated in PTC tissues compared with normal tissues, and upregulation of KIAA1199 was positively correlated with more advanced clinical variables. Additionally, bioinformatic analysis indicated that KIAA1199 was involved in cell migration and invasion. KIAA1199 silencing inhibited the invasive ability of PTC cells by affecting epithelial-mesenchymal transition (EMT) in vitro and in vivo. Furthermore, miR-486-5p was identified as an upstream microRNA that directly targets the 3′-UTR region of KIAA1199.
Conclusions
The miR-486-5p/KIAA1199/EMT axis might play a critical role in PTC invasion and metastasis and offers a potential therapeutic strategy for PTC.
MeSH Keywords: MicroRNAs, Neoplasm Metastasis, Thyroid Neoplasms
Background
Thyroid cancer is the most common malignancy in the endocrine system [1]. In recent decades, the incidence of thyroid cancer has been increasing steadily, largely due to advancements in diagnostic techniques for this disease [2]. Approximately 80% of diagnosed thyroid cancers worldwide are papillary thyroid cancer (PTC) [3]. Surgical resection along with radioactive iodine treatment is currently the standard procedure for the management of PTC [4]. However, due to certain aggressive PTC variants, lymph node metastasis or distant metastasis always occurs in PTC patients, who often exhibit poor responses to standard treatments and thus have poor clinical outcomes [5]. Thus, there is an urgent need for a better understanding of the molecular mechanisms behind PTC invasion and metastasis.
KIAA1199 was first identified by Satoko Abe et al. as a gene related to inner-ear function, as mutations in this gene can result in non-syndromic hearing dysfunction [6]. Recent studies have demonstrated that KIAA1199 also plays a critical role in tumor progression, especially invasion and metastasis. Upregulation of KIAA1199 expression was reported to be associated with cancer progression and to predict a poor prognosis in various cancers, including colorectal cancer [7,8], gastric cancer [9,10], breast cancer [11,12], and pancreatic cancer [13]. In gastric cancer, KIAA1199 can activate the Wnt/β-catenin signalling pathway and then upregulate matrix metalloproteinase (MMP) enzymatic activities, which contribute to epithelial-mesenchymal transition (EMT) [9]. However, the molecular roles of KIAA1199 remain unclear in papillary thyroid cancer.
The aberrant expression of miRNAs has been implicated in the development, progression, and prognosis of cancer [14]. miR-486-5p, widely documented to be a tumor-suppressive microRNA, inhibits proliferation and invasion in many types of cancers [15–17]. However, the clinical and biological role of miR-486-5p in PTC is still unclear.
In this study, we first determined the aberrant mRNA and protein expression of KIAA1199 in PTC. Then, we investigated the clinical significance and molecular role of KIAA1199 using in vitro and in vivo experiments. Finally, we identified specific sequences in the KIAA1199 3′-UTR that are directly targeted by miR-486-5p in the regulation of PTC invasion and metastasis.
Material and Methods
Data source and bioinformatic analysis
TCGA thyroid cancer level 3 mRNA-seq and miRNA-seq data were acquired from UCSC Xena (https://tcga.xenahubs.net), including 59 paired PTC and adjacent normal tissues along with clinicopathological data.
Normalization and differential expression analysis were performed with the Bioconductor package DESeq2 [18]. A heatmap was plotted using MeV [19]. Gene set enrichment analysis (GSEA) was performed to explore potential pathways with the “hallmark.all.v6.1.symbols.gmt” gene set [20]. The Weighted Gene Coexpression Network Analysis (WGCNA) R package [21] was used to evaluate gene significance (GS) and module membership (MM). The brown module, which was most significantly correlated with KIAA1199 expression, was selected for further study. Genes involved in the blue module were submitted to Metascape for Gene Ontology (GO) enrichment visualization [22].
Cell culture and transfection
Two papillary thyroid cancer cell lines were purchased from ATCC and cultured in RPMI1640 media (Sigma-Aldrich). Transfection was performed with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. Nonsense RNAi was used as negative control. The efficiency of transfection was measured using Western blot analysis. A miR-486-5p mimic, negative control mimics, and siRNAs targeting KIAA1199 were purchased from Genechem (Shanghai, China). KIAA1199-targeted short hairpin RNAs (shRNAs) were designed according to siRNA and nsRNA sequences. Recombinant lentiviral particles were generated, and BCPAP cells were transfected with KIAA1199 or a negative control (sh-KIAA1199 or sh-scramble) for use in the animal study. To steadily overexpress miR-486-5p, recombinant lentiviruses containing the miR-486-5p precursor, as well as the negative control scramble sequences, were purchased from Genechem.
Luciferase reporter assay
A wild-type (WT) 3′-UTR of KIAA1199 cDNA was amplified by PCR and cloned into the XbaI and SacI sites of a pmirGLO dual-luciferase miRNA target expression vector. The mutant (Mut) sequence of the KIAA1199 3′-UTR was constructed according to the WT-KIAA1199 3′-UTR region with mutated nucleotides in specific sequences that can predictively bind to miR-486-5p. Vectors (WT-KIAA1199 3′-UTR or Mut-KIAA1199 3′-UTR with miR-486-5p mimic or miR-negative control) were transiently transfected into cells using Lipofectamine 2000 based on the user’s manuals. Luciferase activity was detected with the Dual-Luciferase Reporter Assay System after 48 h of transfection.
RNA extraction and qRT-PCR
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and qRT-PCR was performed according to the standard user’s manual. KIAA1199 primers used were:
forward 5′-CCAGGAATGTTGAATGTCT-3′,
reverse 5′-ATTGGCTCTTGGTGAATG-3′.
Western blot analysis
Western blot analysis was performed according to standard protocols. In brief, after separation in SDS-PAGE gels, protein was transferred on a PVDF membrane. After incubation with antibodies, membranes were washed in TBS-T. Then, membranes were incubated with secondary antibody. Blots were visualized using the ECL imaging system (Thermo Scientific). Antibodies against β-actin (#4970, CST, MA, USA, 1: 1000), KIAA1199 (NBP2–50336, Novus, USA, 1: 500), Vimentin (#5741, CST, CA, USA, 1: 1000), E-cadherin (sc-71008, Santa Cruz Biotechnology, CA, USA, 1: 1000), and Slug (#9585, CST, MA, USA, 1: 500) were used.
TMA and immunohistochemistry (IHC)
A tissue microarray (TMA) with 58 paired papillary thyroid cancer tissue dots and clinical annotations was used for protein expression validation. Immunohistochemistry (IHC) was performed to measure KIAA1199 protein expression in TMA samples. In brief, tissue sections on the TMA were deparaffinized and rehydrated through graded alcohol solutions. Endogenous peroxidase activity was blocked in 3% H2O2. Antigen retrieval was performed with 0.01 M citrate buffer (pH 6.0) and microwave heat treatment. IHC results for each dot were observed independently by 2 experienced pathologists. The staining intensity was classified into 4 grades: 0 points (no staining), 1 point (weak staining), 2 points (intermediate staining), or 3 points (strong staining). The product (percentage of positive regions multiplied by intensity points) was considered the staining score (range from 0 to 300).
Transwell and Matrigel assays
For the Transwell assay, 20 000 transfected PTC cells cultured in serum-free media were seeded in the upper chamber of an insert (8-μm pore size; Millipore, Billerica, MA, USA). Complete medium with 10% FBS was added to the lower chambers. After culturing for 24 h at 37°C in a 5% CO2 atmosphere, cells that had migrated through the membrane were fixed and stained, and then, for imaging and quantification by microscopy, cells were counted in at least 3 random fields to obtain an average count. For the Matrigel assay, 20 000 transfected cells were plated into the upper chamber on a Matrigel-coated membrane (BD Biosciences) in serum-free medium. The lower chambers also contained complete medium with 10% FBS. After a 48-h incubation, the cells were harvested under the same conditions as in the Transwell assay described above.
Mouse metastasis model
Animal studies were approved by Jiangsu Province Animal Ethics Committee and were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Ten male BALB/c nude mice (6 weeks old) were purchased from the Animal Center of the Chinese Academy of Science (Shanghai, China) and maintained in specific pathogen-free (SPF) conditions. Briefly, for the metastasis model, 3×106 BCPAP cells stably expressing shRNA-Scramble or shRNA-KIAA1199 were suspended in 100 μl PBS and injected intravenously into the tail vein of mice (5 mice in each group). Six weeks later, the mice were sacrificed and the lungs were harvested. Metastatic nodules on the lung surface were counted and hematoxylin and eosin (HE) staining was performed for histological examination.
Statistical analysis
Data are presented as the mean±S.D. Statistical analyses were conducted with SPSS Statistics tool, R software (version 3.5.1), and GraphPad Prism 8 software. We used the t test or one-way ANOVA to analyze differences. P<0.05 was considered statistically significant.
Results
Elevation of KIAA1199 is correlated with more advanced clinical variables
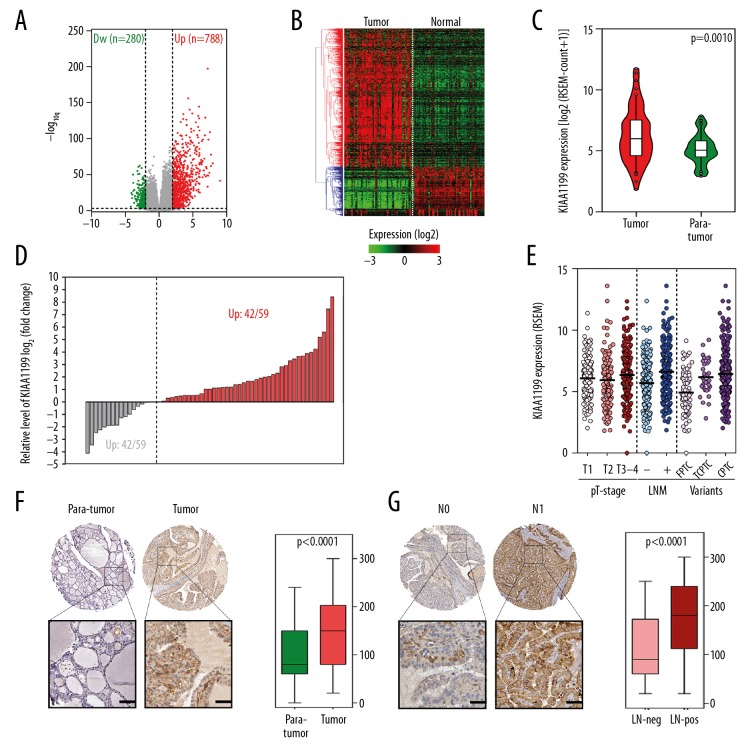
To identify potential oncogenes in PTC, we screened whole-genome RNA-seq data from the TCGA. Over 20 000 protein-coding genes were included in the DESeq2 analysis, and the volcano plot in Figure 1A shows significantly upregulated and downregulated genes that met the filtering criteria (|log2FC|>2, FDR<0.01). A heatmap based on these differentially expressed genes was plotted to show the detailed expression profile of 59 pairs of PTC and adjacent normal tissues (Figure 1B). KIAA1199 mRNA (read count) was significantly upregulated (p=0.0022) in cancer tissues (Figure 1C) and widely upregulated at a ratio of 42/59 compared with adjacent normal tissues (Figure 1D). Figure 1E shows that KIAA1199 was significantly upregulated in lymph node metastasis (LNM)-positive samples and in more malignant subtypes, but no significant difference was observed in tumor size. IHC analysis of the TMA indicated that KIAA1199 protein level was elevated in papillary thyroid cancer tissues compared to normal tissues (Figure 1F), especially in LNM-positive tissues (Figure 1G).
Figure 1.
Upregulation of KIAA1199 is correlated with more advanced clinical variables. (A) The volcano plot showed significantly upregulated and downregulated genes in PTC tissues. (B) The heatmap depicted the detailed expression profile of these genes in 59 pairs of PTC and adjacent normal tissues. (C) KIAA1199 mRNA was significantly upregulated in cancer tissues, and (D) was widely upregulated at a ratio of 42/59 compared with adjacent normal tissues. (E) KIAA1199 was significantly upregulated in lymph node metastasis (LNM)-positive samples and more malignant subtypes. (F) The KIAA1199 protein level was upregulated in PTC tissues, (G) especially in LNM-positive tissues.
WGCNA analysis indicates that KIAA1199 is involved in cell migration and invasion
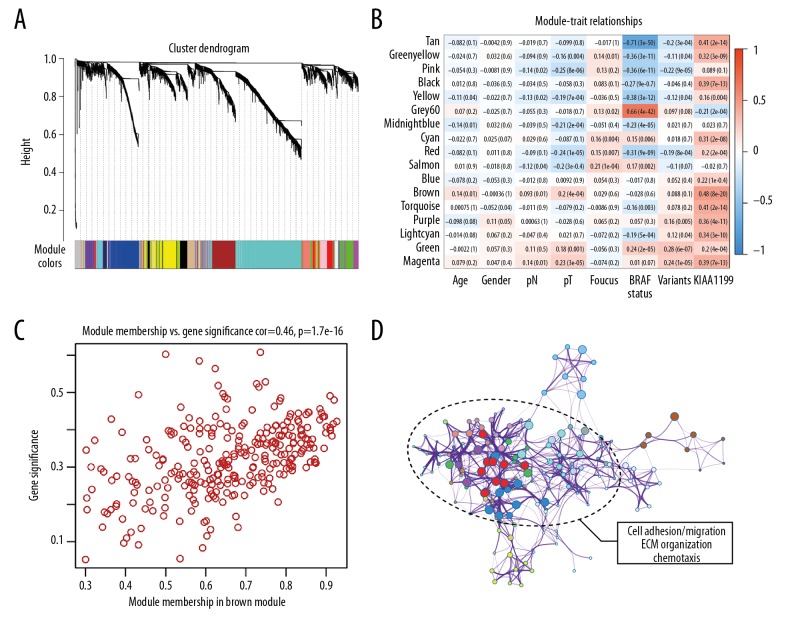
To construct a gene co-expression network, RNA-seq data from the whole genome of PTC samples were subjected to WGCNA. Genes were assigned to different modules by cluster dendrogram trees, and unassigned genes were categorized into the grey module (Figure 2A). A heatmap of the relationships between clinical traits and gene modules is shown in Figure 2B. We observed that the brown module was most significantly positively correlated with KIAA1199 expression. We then determined if gene significance and module membership exhibited a significant correlation (r=0.46, p=1.7e–16), and the result indicated that genes in the brown module were highly correlated with KIAA1199 (Figure 2C). Next, genes in the brown module were submitted to Metascape for GO enrichment visualization. As shown in Figure 2D, the major part of the network was labelled with “cell adhesion/cell migration/exocytosis/chemotaxis/ECM organization”, which are critical events in cancer invasion and metastasis.
Figure 2.
WGCNA analysis indicates that KIAA1199 is involved in cell migration and invasion. (A) Cluster dendrogram trees were constructed based on the whole-genome profiling data of TCGA. (B) Heatmap of the relationships between clinical traits and gene modules, and the brown module with highest correlation value (r=0.48, p=8e-20) was chosen for further study. (C) A significant correlation was found between module membership and gene significance of KIAA1199 in the brown module. (D) The major part of the KIAA1199-related network was labelled as “cell adhesion/cell migration/exocytosis/chemotaxis/ECM organization”, which are critical events in cancer invasion and metastasis.
KIAA1199 promotes PTC invasion by influencing EMT
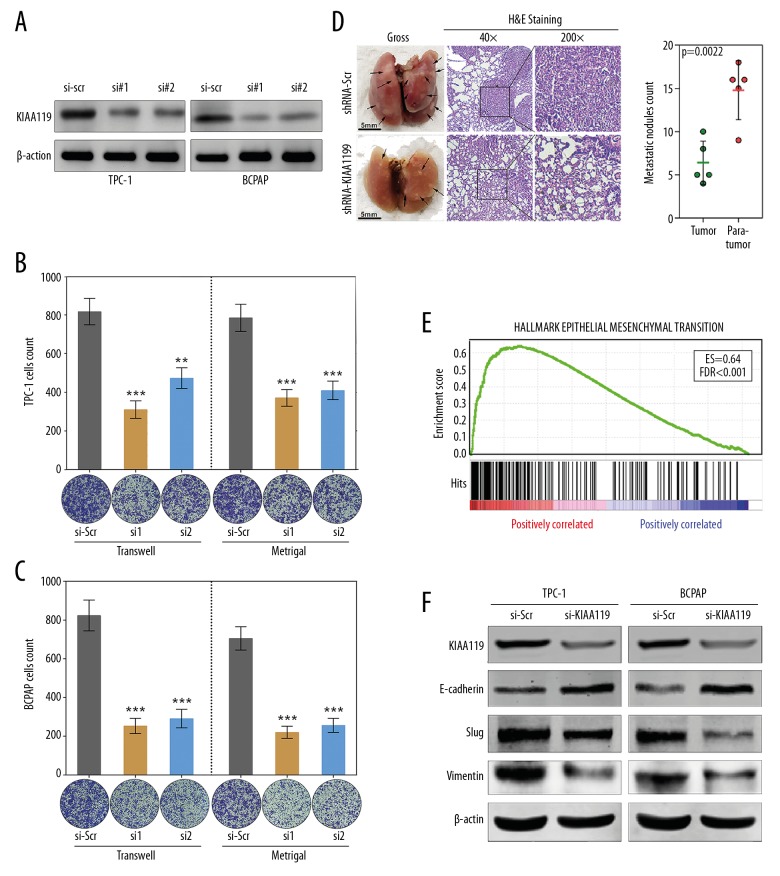
Two pairs of siRNAs were designed to inhibit KIAA1199 expression level in PTC cell lines (Figure 3A). Transwell and Matrigel assays indicated that silencing of KIAA1199 greatly impaired the migratory and invasive abilities of TPC-1 and BCPAP cells in response to siRNA-treatment compared with the group treated with siRNA-scramble (si-scr) (Figure 3B, 3C). To further validate the effect of KIAA1199 silencing on metastatic potential in vivo, we established a mouse metastasis model, as described in the Methods section above. We found that the incidence of lung metastasis was decreased in mice injected with BCPAP-shKIAA1199 cells compared with those injected with BCPAP-shScramble. Histological examination confirmed that silencing of KIAA1199 decreased the size and number of lung metastatic nodules (Figure 3D).
Figure 3.
KIAA1199 promotes PTC invasion by influencing EMT. (A) Two pairs of siRNAs were designed to knock down the KIAA1199 expression level in PTC cell lines. (B, C) Transwell and Matrigel assays revealed that silencing of KIAA1199 greatly reduces the migratory and invasive abilities of TPC-1 and BCPAP cells in response to siRNA-treatment compared with the cells treated with siRNA-scramble. (D) Representative images showing that the incidence of lung metastasis was decreased in mice injected with BCPAP-shKIAA1199 cells compared with those injected with BCPAP-shScramble (p=0.0022). (E) GSEA showed that EMT ranked first in the KIAA1199-related predictive outputs. (F) Knock down of KIAA1199 upregulated E-cadherin but downregulated Slug and Vimentin in TPC-1 and BCPAP cells.
PTC samples in the TCGA dataset with KIAA1199 expression <Q1 and >Q3 according to a quartile method were selected, and GSEA was performed based on RNA-seq data. Among all the outputs, EMT ranked first, with an enrichment score of 0.64 (Figure 3E). Thus, it is plausible to hypothesize that KIAA1199 promotes PTC progression via EMT. Then, the protein levels of EMT markers such as E-cadherin, Slug, and Vimentin were measured by Western blot. We observed that knockdown of KIAA1199 upregulated E-cadherin but downregulated Slug and Vimentin in TPC-1 and BCPAP cells (Figure 3F). These results indicated that KIAA1199 might exert its metastatic function by influencing EMT in PTC.
KIAA1199 is a direct target of miR-486-5p
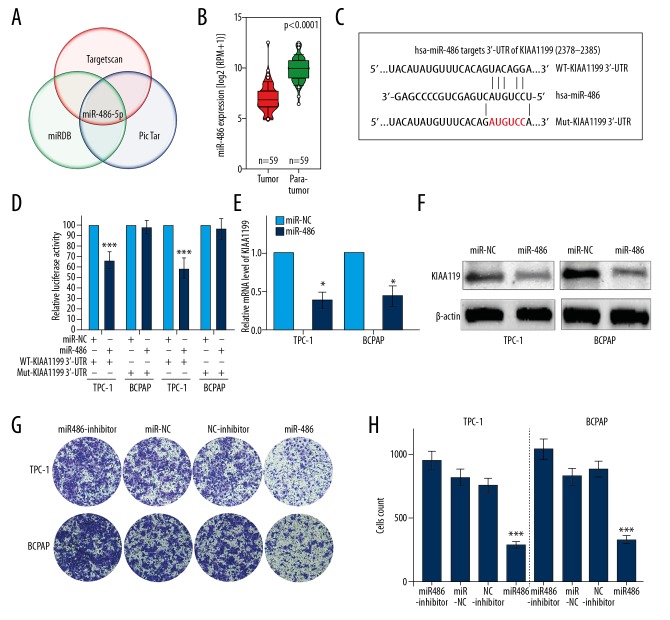
To identify potential microRNAs targeting KIAA1199, we analyzed the predictive results from the intersection of 3 predictive databases – TargetScan, miRDB, and PicTar. Among the overlapping candidates, miR-486-5p attracted our attention due to its high predictive score (Figure 4A). Then, we investigated the expression level of miR-486-5p in PTC, and observed in the TCGA miRNA-seq dataset that miR-486 expression was significantly downregulated in papillary thyroid cancer tissues compared to normal tissues (Figure 4B). As shown in Figure 4C, 2378-2385 of the KIAA1199 3′-UTR region was a predictive target sequence of miR-485-5p. Luciferase reporter assays were performed to validate whether miR-486-5p could bind to the predicted site described above. The data showed that miR-486-5p significantly inhibited luciferase activity in both TPC-1 and BCPAP cells with a reporter plasmid carrying the WT-KIAA1199 3′-UTR sequence. In contrast, we did not observe significant inhibition in cells with the reporter plasmid carrying a Mut-KIAA1199 3′-UTR (Figure 4D). Next, we measured the inhibitory effect of miR-486-5p on KIAA1199 expression in 2 PTC cell lines. As shown in Figure 4E and 4F, miR-486-5p overexpression significantly inhibited KIAA1199 mRNA and protein expression compared with the negative control group. Then, we investigated the invasion-promoting role of miR-486-5p in vitro. Matrigel assays indicated that miR-486-5p transfection markedly impaired the invasive ability of TPC-1 and BCPAP cells compared with the negative control group (Figure 4G, 4H).
Figure 4.
KIAA1199 is a direct target of miR-486-5p. (A) miR-486-5p was identified with a high predictive score in the intersection of 3 databases (TargetScan, miRDB, and PicTar). (B) TCGA data showed that miR-486 was significantly downregulated in cancer tissues compared to adjacent nontumoral tissues. (C) 2378-2385 of the KIAA1199 3′-UTR was a predicted sequence of miR-485-5p. (D) Luciferase reporter assays determined that miR-486-5p regulates KIAA1199 expression by directly binding to the predicted site in the 3′-UTR of KIAA1199. (E, F) qRT-PCR and Western blot analysis revealed the inhibitory effect of miR-486-5p on KIAA1199 expression in both cell lines. (G, H) A Matrigel assay showed that the invasive capacity of TPC-1 and BCPAP cells transfected with miR-486-5p was greatly impaired compared with that of the negative control group.
Discussion
Activation of invasion and metastasis is a critical hallmark of cancer, but the underlying mechanism remains unclear [23]. Although most PTC patients have a favorable prognosis, accumulating evidence has shown that regional invasion and lymph node or distant metastasis increase the risk of recurrence and a worse survival [24–26]. Therefore, a better understanding of tumor invasion and metastasis is essential to identify reliable biomarkers and potential therapeutic targets for PTC.
In our study, we revealed that KIAA1199 mRNA and protein expression is aberrantly and frequently elevated in PTC tissues compared with normal tissues. Furthermore, KIAA1199 expression is positively correlated with more advanced clinicopathological characteristics. A bioinformatic analysis indicated that KIAA1199 is involved in cell migration and invasion, and in vitro and in vivo experiments showed that KIAA1199 enhances PTC invasion and metastasis by influencing EMT.
KIAA1199 was first identified as a novel large, long cDNA in the Human Unidentified Gene Encoded (HUGE) protein database [27]. Mutations in KIAA1199 were reported to be associated with loss of function of non-syndromic hearing [6]. Subsequently, some researchers reported that KIAA1199 is a novel hyaluronan-binding protein (HYBID) involved in hyaluronan depolymerization [28]. Recently, it was reported that tumor hypoxia-induced aberrant overexpression of KIAA1199 enhances tumor invasive capacity; thus, this protein is also known as cell migration-inducing protein (CEMIP) [29]. Some recently published studies have revealed the clinical and biological roles of KIAA1199 in cancer progression. KIAA1199 expression is aberrantly upregulated in gastric cancer [9], colon cancer [7,8], breast cancer [11,12], and pancreatic cancer [13]. In cervical cancer, KIAA1199 upregulation can be induced by human papillomavirus infection, and its expression is also aberrantly upregulated in pre-cancerous lesions [30]. Furthermore, KIAA1199 was reported to act as a predictor of poor clinical outcomes in lung cancer [31], pancreatic cancer [13], and colon cancer [7].
The malignant progression of PTC is considered to be a comprehensive event that includes a gene expression network and alterations in the tumor microenvironment, in which microRNAs play critical roles [32]. From the intersection of predictive results from 3 databases, we identified miR-486-5p as a promising upstream microRNA, which specifically targets KIAA1199 with the highest predictive score. Previously, Ma et al. reported that miR-486-5p inhibits cell growth of PTC by targeting fibrillin-1 [33]. We validated the miR-486-5p expression profile in PTC and revealed its downregulation in PTC tissues compared with adjacent normal tissues. Subsequently, by directly targeting the 3′-UTR region of KIAA1199, we demonstrated its invasion-suppressive role in PTC. In previous studies, KIAA1199 was reported to interact with many microRNAs such as miR-29c [34], miR-216a [35] and miR-600 [36], and we present here the first evidence that KIAA1199 enhances PTC invasion and metastasis by promoting EMT and that KIAA1199 is regulated by the tumor-suppressor miR-486-5p, which provides more clues to the regulation network of KIAA1199.
Conclusions
In summary, our study demonstrates the biological function of the miR-486-5p/KIAA1199/EMT axis, which provides strong evidence for the invasive ability of KIAA1199 in PTC. KIAA1199 might serve as a potential therapeutic target in PTC, and further studies are required to investigate the plausibility and potential pathways involved in KIAA1199-mediated cell invasion and metastasis.
Data availability
Normalized expression of KIAA1199 and miR-486-5p in PTC tissues and corresponding adjacent normal tissues in the TCGA database can be downloaded from the following website: https://xenabrowser.net/. The original data from the PTC TMA can be downloaded from the following website: http://www.superchip.com.cn/.
Footnotes
Source of support: Departmental sources
Conflicts of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388(10061):2783–95. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 3.Sherman SI. Thyroid carcinoma. Lancet. 2003;361(9356):501–11. doi: 10.1016/s0140-6736(03)12488-9. [DOI] [PubMed] [Google Scholar]
- 4.American Thyroid Association (ATA) Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid Cancer. Cooper DS, Doherty GM, Haugen BR, et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19(11):1167–214. doi: 10.1089/thy.2009.0110. [DOI] [PubMed] [Google Scholar]
- 5.Pellegriti G, Scollo C, Lumera G, et al. Clinical behavior and outcome of papillary thyroid cancers smaller than 1.5 cm in diameter: study of 299 cases. J Clin Endocrinol Metab. 2004;89(8):3713–20. doi: 10.1210/jc.2003-031982. [DOI] [PubMed] [Google Scholar]
- 6.Abe S, Usami S, Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters’ cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J Hum Genet. 2003;48(11):564–70. doi: 10.1007/s10038-003-0079-2. [DOI] [PubMed] [Google Scholar]
- 7.Tiwari A, Schneider M, Fiorino A, et al. Early insights into the function of KIAA1199, a markedly overexpressed protein in human colorectal tumors. PLoS One. 2013;8(7):e69473. doi: 10.1371/journal.pone.0069473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenkamp-Demtroder K, Maghnouj A, Mansilla F, et al. Repression of KIAA1199 attenuates Wnt-signalling and decreases the proliferation of colon cancer cells. Br J Cancer. 2011;105(4):552–61. doi: 10.1038/bjc.2011.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia S, Qu T, Wang X, et al. KIAA1199 promotes migration and invasion by Wnt/beta-catenin pathway and MMPs mediated EMT progression and serves as a poor prognosis marker in gastric cancer. PLoS One. 2017;12(4):e0175058. doi: 10.1371/journal.pone.0175058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Yu T, Li W, et al. The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene. 2019;38(17):3134–50. doi: 10.1038/s41388-018-0642-0. [DOI] [PubMed] [Google Scholar]
- 11.Jami MS, Hou J, Liu M, et al. Functional proteomic analysis reveals the involvement of KIAA1199 in breast cancer growth, motility and invasiveness. BMC Cancer. 2014;14:194. doi: 10.1186/1471-2407-14-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evensen NA, Kuscu C, Nguyen HL, et al. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst. 2013;105(18):1402–16. doi: 10.1093/jnci/djt224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koga A, Sato N, Kohi S, et al. KIAA1199/CEMIP/HYBID overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Pancreatology. 2017;17(1):115–22. doi: 10.1016/j.pan.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Zheng X, Li W, et al. Serum miR-486-5p as a diagnostic marker in cervical cancer: with investigation of potential mechanisms. BMC Cancer. 2018;18(1):61. doi: 10.1186/s12885-017-3753-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi Y, Lu X, Chen J, et al. Downregulated miR-486-5p acts as a tumor suppressor in esophageal squamous cell carcinoma. Exp Ther Med. 2016;12(5):3411–16. doi: 10.3892/etm.2016.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He Y, Liu J, Wang Y, et al. Role of miR-486-5p in regulating renal cell carcinoma cell proliferation and apoptosis via TGF-beta-activated kinase 1. J Cell Biochem. 2019;120(3):2954–63. doi: 10.1002/jcb.26900. [DOI] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu VT, Gottardo R, Raftery AE, et al. MeV+R: Using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol. 2008;9(7):R118. doi: 10.1186/gb-2008-9-7-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tripathi S, Pohl MO, Zhou Y, et al. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–35. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Schneider DF, Chen H, Sippel RS. Impact of lymph node ratio on survival in papillary thyroid cancer. Ann Surg Oncol. 2013;20(6):1906–11. doi: 10.1245/s10434-012-2802-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu MH, Shen WT, Gosnell J, Duh QY. Prognostic significance of extranodal extension of regional lymph node metastasis in papillary thyroid cancer. Head Neck. 2015;37(9):1336–43. doi: 10.1002/hed.23747. [DOI] [PubMed] [Google Scholar]
- 26.Lang BH, Wong KP, Cheung CY, et al. Evaluating the prognostic factors associated with cancer-specific survival of differentiated thyroid carcinoma presenting with distant metastasis. Ann Surg Oncol. 2013;20(4):1329–35. doi: 10.1245/s10434-012-2711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kikuno R, Nagase T, Nakayama M, et al. HUGE: A database for human KIAA proteins, a 2004 update integrating HUGEppi and ROUGE. Nucleic Acids Res. 2004;32(Database issue):D502–4. doi: 10.1093/nar/gkh035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida H, Nagaoka A, Kusaka-Kikushima A, et al. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci USA. 2013;110(14):5612–17. doi: 10.1073/pnas.1215432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evensen NA, Li Y, Kuscu C, et al. Hypoxia promotes colon cancer dissemination through up-regulation of cell migration-inducing protein (CEMIP) Oncotarget. 2015;6(24):20723–39. doi: 10.18632/oncotarget.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shostak K, Zhang X, Hubert P, et al. NF-kappaB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun. 2014;5:5232. doi: 10.1038/ncomms6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng F, Lei J, Zhang X, et al. Overexpression of KIAA1199: An independent prognostic marker in nonsmall cell lung cancer. J Cancer Res Ther. 2017;13(4):664–68. doi: 10.4103/jcrt.JCRT_61_17. [DOI] [PubMed] [Google Scholar]
- 32.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X, Wei J, Zhang L, et al. miR-486-5p inhibits cell growth of papillary thyroid carcinoma by targeting fibrillin-1. Biomed Pharmacother. 2016;80:220–26. doi: 10.1016/j.biopha.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Yu T, Li W, et al. The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene. 2019;38(17):3134–50. doi: 10.1038/s41388-018-0642-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Zhao L, Shen Q, et al. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int J Cancer. 2017;140(10):2298–309. doi: 10.1002/ijc.30656. [DOI] [PubMed] [Google Scholar]
- 36.Sun J, Hu J, Wang G, et al. LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):106. doi: 10.1186/s13046-018-0771-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Normalized expression of KIAA1199 and miR-486-5p in PTC tissues and corresponding adjacent normal tissues in the TCGA database can be downloaded from the following website: https://xenabrowser.net/. The original data from the PTC TMA can be downloaded from the following website: http://www.superchip.com.cn/.