Abstract
Purpose
Circular RNAs (circRNAs) are emerging as promising biomarkers for various human malignancies. However, the application of circRNAs as non-invasive biomarkers in high-grade serous ovarian cancer (SOC) remains to be elucidated. Here, we aim to investigate the feasibility of using serum circSETDB1, a tumor-promoting circRNA generated from the SET domain bifurcated histone lysine methyltransferase 1 (SETDB1), known to be upregulated in SOC,as a biomarker for detecting SOC progression, predicting relapse, and evaluating the effectiveness of SOC treatment.
Methods
Serum circSETDB1 levels were measured using quantitative real-time RCR in 60 SOC patients (18 primary chemoresistance, 42 primary chemosensitive) and 60 healthy volunteers. Progression-free survival curve was calculated by Kaplan-Meier analysis. Diagnostic value was analyzed using receiver operating characteristic curve (ROC) method.
Results
Serum circSETDB1 expression is upregulated in SOC patients. Higher levels of circSETDB1 are positively associated with advanced clinical stage, lymph node metastasis of SOC patients. Notably, serum circSETDB1 levels are significantly increased in primary chemoresistance patients. Patients with higher levels of circSETDB1 have a shorter progression-free survival time. In addition, diagnostic value analyses revealed that serum circSETDB1 can distinguish patients with SOC from healthy volunteers as well as patients with primary chemoresistance from those with primary chemosensitivity.
Conclusion
Our data suggest that serum circSETDB1 may serve as a novel non-invasive biomarker for detecting SOC progression and predicting response to chemotherapy and relapse in high-grade serous ovarian cancer.
Keywords: circular RNA, circSETDB1, high grade serous ovarian cancer, chemotherapy, chemosensitivity, relapse
Introduction
High-grade serous ovarian cancer (SOC) is a serious gynecological malignancy with poor prognosis and high mortality.1–3 Optimal cytoreductive surgery combined with platinum-based combination chemotherapy is the first-line treatment. However, relapse and subsequent resistance to chemotherapy are responsible for high mortality rate of SOC.4 The identification of new biomarkers that can assist in monitoring chemotherapy response and identifying disease recurrence during routine treatment would help to improve outcomes of SOC patients.
Circular RNAs (circRNAs), a class of noncoding RNAs produced from back-splicing of precursor mRNA in eukaryotes, have been suggested to play critical roles in human diseases and show great potential as biomarkers and therapeutic targets.5–7 CircRNAs are characterized by 3′–5′ covalent RNA rings' closed-loop structure, resulting in RNA molecules that are more stable than linear RNAs.6,8 In particular, the stable circular structure of circRNAs lengthens their half-life, especially in cell-free samples (such as blood and urine), creating potential for the use of circRNAs as biomarkers in patient samples from noninvasive sources.6 Although increasing evidence indicates that aberrantly expressed circRNAs are involved in epithelial ovarian cancer development and progression, it remains undetermined whether circRNAs may be detected in serum samples and whether they are associated with clinical features and chemotherapy response as noninvasive biomarkers for SOC.9
Previous sequencing analyses have revealed an abnormal expression of hsa_circ_006352 in ovarian cancer.10 Hsa_circ_006352 is located at chr1q21.3: 150923840–150935195 with 1078 length in gene symbol SETDB1; thus, we nominate it as circSETDB1. As a proof of concept, circSETDB1 was selected for further investigation because its parental gene is involved in tumor growth and metastasis,11,12 epigenetic silencing,13 homologous recombination14,15 and signal transduction.16,17 Based on the currently available information, SETDB1 holds considerable value as a potential biomarker and therapeutic target for cancer.18 However, the specific role of circSETDB1 in high-grade serous ovarian cancer remains to be established.
In the present investigation, we aim to examine serum circSETDB1 expression level and its biological significance for detecting SOC progression, predicting relapse, and evaluating the effectiveness of SOC treatment.
Materials and methods
Clinical specimens
A total of 60 serum samples were obtained from patients with primary SOC at the People’s Hospital of Zhengzhou University between January 2015 and October 2018. The diagnosis of SOC was confirmed by two pathologists separately and the tumor histologic classification was high-grade serous ovarian carcinoma. The median age of SOC patients was 58 years (range 49–66). Sixty age-matched healthy individuals were recruited through advertisements in our hospital as the control group. SOC patients and healthy participants did not receive surgery or chemotherapy or radiotherapy or biotherapy before blood collection. Clinical information of patients are summarized in Table 1. This study was conducted in accordance with the Declaration of Helsinki. This research was approved by the Life Science Ethics Review Committee of Zhengzhou University, and all patients provided written informed consent.
Table 1.
Clinical association between serum circ-SETDB1 expression levels and clinicopathologic characteristics of patients with high-grade serous ovarian cancer
| irc-SETDB1 expression | P-value | |||
|---|---|---|---|---|
| Total cases | High | Low | ||
| All cases | ||||
| Age | 60 | 32 | 28 | |
| ≤60 | 29 | 16 | 13 | 0.2192 |
| >60 | 31 | 16 | 15 | |
| FIGO stage | ||||
| I–II | 16 | 3 | 13 | 0.0010** |
| III–IV | 44 | 29 | 15 | |
| Lymph node metastasis | ||||
| Yes | 36 | 25 | 11 | 0.0001*** |
| No | 24 | 7 | 17 | |
| Residual tumor size | ||||
| ≤1 cm | 32 | 17 | 15 | 0.1256 |
| >1 cm | 28 | 15 | 13 | |
| Response to chemotherapy | ||||
| chemosensitive | 42 | 18 | 24 | 0.0002*** |
| chemoresistant | 18 | 14 | 4 | |
| CA-125 level | ||||
| >35 U/mL | 40 | 24 | 16 | 0.0554 |
| ≤35 U/mL | 20 | 8 | 12 | |
Notes: **P<0.01, ***P<0.001.
Abbreviations: FIGO, International Federation of Gynecology and Obstetrics; LGSC, low-grade serous carcinoma; HGSC, high-grade serous carcinoma.
Evaluation of response to chemotherapy
All patients received 6 cycles of platinum-taxane-combined chemotherapy after surgical resection. The evaluation criteria of patient’s response to chemotherapy was according to National Comprehensive Cancer Network (NCCN) Guidelines for epithelial ovarian cancer (version 2018). Primary chemosensitive group (n=42): complete remission and relapse ≥6 months after completing prior chemotherapy. Primary chemoresistant group (n=18): complete remission and relapse <6 months after completing prior chemotherapy or progression, stable, or persistent disease on primary chemotherapy. The patients were followed up for a median time of 24 months 13.5 (rang, 3–24 months) and the course documented. SOC patients were then divided into 2 groups: (i) evidence of disease group: evidence of disease including progression, stable, or persistent disease after primary surgery and platinum-taxane-combined chemotherapy (n=34) and (ii) no evidence of disease group: no evidence of disease after primary surgery and platinum-taxane-combined chemotherapy (n=26).
Serum collection and RNA extraction
Whole blood specimens were collected for serum circSETDB1 estimation. The serum was added with RNAlock Reagent (DP440, TIANGEN BIOTECH, Beijing, China) within 2 hrs of collection and stored at −80°C until further use. In addition to circSETDB1, the serum CA-125 levels were also evaluated. Total RNA was extracted and purified by using the RNAprep Pure Hi-Blood Kit (DP443, TIANGEN BIOTECH, Beijing, China) following the manufacturer’s instructions. 15 mL serum collected for RNA isolation, 100–150 ng yield per sample, was quantified by NanoDrop One C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Quantitative real-time PCR (qRT-PCR)
Total RNA was reverse transcribed by using FastKing gDNA Dispelling RT SuperMix (KR118, TIANGEN BIOTECH, Beijing, China) into a final volume of 20 μL (15 mins at 42°C, 3 mins at 95°C, and maintained at −20°C). Subsequent qRT-PCR experiments were performed using RealUniversal Color PreMix SYBR Green PCR Kit (FP201, TIANGEN BIOTECH, Beijing, China) in an ABI StepOnePlus Real-Time PCR system, with a 50 μL reaction consisting of 25 μL of 2× RealUniversal Premix, 0.3 uM of each of the forward and reverse primers, 5 μL 50XROX Reference Dye, and 5 ng of cDNA. The qPCR conditions were 95°C for 15 mins, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The primers used for circSETDB1 were forward: 5ʹ-TCCTGTGAAGCCTGAAGGAC-3ʹ and reverse: 5ʹ-TTAGTTGATGGCAGGCACAC-3ʹ. GAPDH was used as control to normalize the relative gene expression. The GAPDH primers for qPCR were forward: 5ʹ-AAAATCAAGTGGGGCGA,TGC-3ʹ and reverse: 5ʹ-GATGACCCTTTTGGCTCCCC-3ʹ. Relative gene expression was calculated using the comparative the 2−△△Ct method.
Statistical analysis
Data were presented as mean ± standard error of the mean (SEM). The differences between the groups were analyzed by Student’s t-test when two groups were compared. Receiver operating characteristic (ROC) curves and area under the curve (AUC) were constructed to determine the optimal values of circSETDB1, which provided high sensitivity and specificity. Analyses were performed with Graph-Pad Prism, version 6.0 (GraphPad Software, Inc., San Diego, CA, USA). Kaplan–Meier curves were constructed to determine patient progression-free survival rates using SPSS statistical package 17.0 (SPSS, Inc., Chicago, IL, USA). Patients who were lost to the follow-up or who died from causes unrelated to SOC were treated as censored events. The statistical differences in survival among subgroups were compared using the log-rank test. P-values of <0.05 were considered statistically significant.
Results
Serum circSETDB1 levels and clinical pathological features of SOC patients
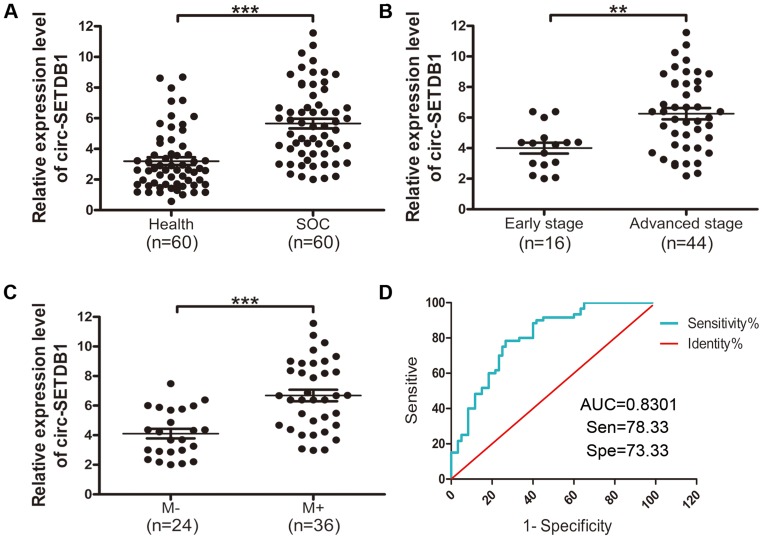
To investigate whether circSETDB1 could be served as a beneficial non-invasive biomarker for SOC, we analyzed circSETDB1 expression in 60 SOC patients' serum samples and 60 healthy volunteers’ serum samples. As shown in Figure 1, circSETDB1 expression was significantly up-regulated in SOC patients as compared to healthy volunteers (5.652±0.3131 vs 3.198±0.2551, P<0.0001, Figure 1A). We chose a cut-off value of 5.0 to divide the patients into those with low and high expression levels of circSETDB1. The associations between circSETDB1 expression levels and the clinical pathological features are illustrated in Table 1. Statistical analysis revealed that circSETDB1 expression levels in SOC serum specimens were positively associated with advanced clinical International Federation of Gynecology and Obstetrics (FIGO) stages (P=0.0010, Table 1, Figure 1B) and tumor lymph node metastasis (P<0.0001, Table 1, Figure 1C). No significant associations were found between circSETDB1 expression and age and residual tumor size (Table 1).
Figure 1.
Serum expression levels of circSETDB1 in SOC patients and its potential value for SOC diagnosis.
Notes: (A–C) qRT-PCR analysis of serum circSETDB1 expression in (A) SOC patients (n=60) and health volunteers (n=60), (B) SOC patients with early-stage (I, II) and advanced-stage (III, IV), (C) SOC patients with or without lymph node metastasis. Data are presented as the mean ± SEM. (D) ROC curve analysis of serum circSETDB1 for discriminating SOC patients from healthy controls. **P>0.01, ***P>0.001.
Abbreviations: SOC, high-grade serous ovarian cancer; ROC, receiver operating characteristic; AUC, curves and area under the curve; M+, lymph node metastasis, M−, non-lymph node metastasis.
To assess the diagnostic value of circSETDB1 as candidate biomarkers of SOC, we performed ROC curve analysis. As depicted graphically in Figure 1D, circSETDB1 had an AUC of 0.8031±0.03974 (95% CI=0.7252–0.8810) with sensitivity of 78.33% and specificity of 73.33% in separating the healthy volunteers from SOC patients (Figure 1D).
Serum circSETDB1 levels and primary response to chemotherapy
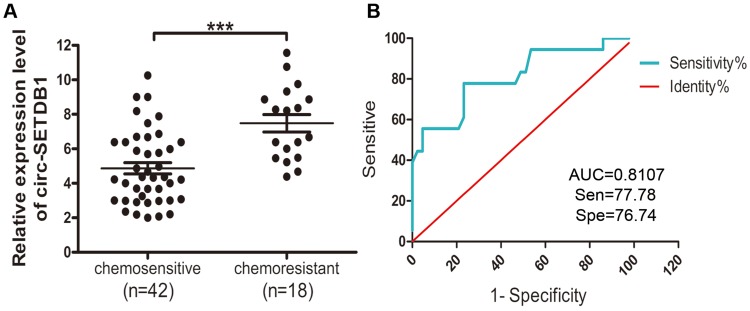
To further investigate the contribution of circSETDB1 to the chemotherapeutic response, we grouped 60 cases of SOC patients into primary chemosensitive group and primary chemoresistant group and conducted qRT-PCR on serum samples from patients. Statistical analysis revealed that serum circSETDB1 expression levels were elevated in primary chemoresistant patients compared with that in primary chemosensitive patients (P=0.0008, Figure 2A).
Figure 2.
Serum circSETDB1 expression level was associated with chemoresistance.
Notes: (A) qRT-PCR analysis of serum circSETDB1 expression in patients with chemoresistance (n=18) and patients with chemosensitive (n=42). Data are presented as the mean ± SEM. (B) ROC curve analysis of serum circSETDB1 for discriminating patients with primary chemoresistance from patients with chemosensitive. ***P>0.001.
Abbreviations: SOC, high-grade serous ovarian cancer; ROC, receiver operating characteristic; AUC, curves and area under the curve.
The prediction value of circSETDB1 as a specific and sensitive biomarker in identifying chemotherapy sensitivity of SOC was determined by ROC curve analysis. The data showed that circSETDB1 had an AUC of 0.8107±0.06424 (95% CI=0.6848–0.9367) with sensitivity of 77.78% and specificity of 76.74% in separating primary chemoresistant SOC patients from primary chemosensitive patients (Figure 2B).
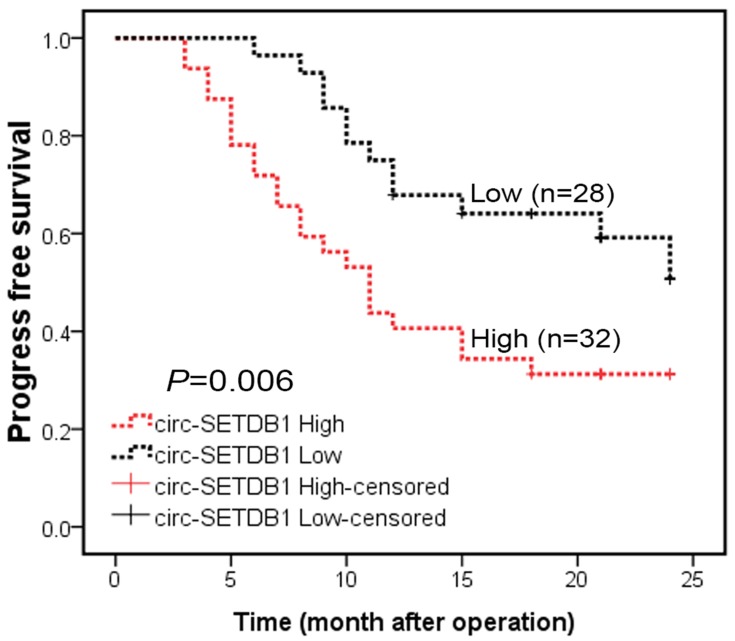
Serum circSETDB1 levels and progression-free survival
The correlations of progression-free survival and circSETDB1 levels were investigated using the Kaplan–Meier method. The data revealed that serum circSETDB1 levels were significantly correlated with progression-free survival of SOC patients. Patients with higher circSETDB1 levels had a mean progression-free survival of 13.2 months, whereas patients with lower circSETDB1 levers had a mean progression disease-free survival of 18.9 months (Log Rank =6.815, P=0.006; Figure 3).
Figure 3.
Serum circSETDB1 expression level was associated with progression-free survival.
Notes: Kaplan–Meier survival curves show that median progression-free survival is shorter in circSETDB1 high group than in circSETDB1 low group (13.2 vs 18.9 months, Log Rank =6.815, P=0.006).
Association of serum circSETDB1 levels with SOC tumor biomarker CA-125
The levels of CA-125 are established tumor marker of SOC patients. According to postoperative serum CA-125 levels, SOC patients were divided into two groups: CA-125-high and CA125-low. To determine the association between circSETDB1 and CA-125, serum circSETDB1 levels were analyzed in CA-125-high and CA125-low groups. We found an association of higher circSETDB1 levels with CA-125-high group, although the association was not statically significant (P=0.0554, Table 1).
Discussion
Circulating biomarkers are more acceptable than tissue biomarkers and have a greater value in clinical applications.19,20 Emerging evidence demonstrated the potential of circRNAs as serum biomarkers in various cancers.19,21,22 However, the application of circRNAs as non-invasive biomarkers in high-grade serous ovarian cancer (SOC) remains to be elucidated. In the present study, we evaluated the clinical significance of serum circSETDB1 level in SOC patients and demonstrated that circSETDB1 may serve as a promising biomarker for detecting SOC progression and predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer.
Recent studies have revealed that circRNAs were aberrantly expressed in ovarian serous cancer and aberrant expression of circRNAs contributed to the malignant cancer phenotypes.23,24 Ahmed et al performed paired-end RNA sequencing of primary sites, peritoneal and lymph node metastases from three patients with stage IIIC serous ovarian cancer and identified 67,580 circular isoforms in SOC.10 These candidate circRNAs show a more robust expression pattern across patients than mRNA forms indicating their suitability as biomarkers in highly heterogeneous cancer transcriptomes.10 Another two research groups investigate circRNA expression profiling in epithelial ovarian cancer (EOC) by high throughput sequencing; differential expression profiling analysis identified thousands of candidate circRNAs in clinical EOC samples compared with paired normal ovarian tissues indicate those differentially expressed circRNAs may participate in the pathogenesis of EOC.25,26 Although previous studies demonstrated that circRNAs are aberrantly expressed in epithelial ovarian cancer, it remains undetermined if circRNAs can be detected in serum samples and whether they are associated with clinical pathological features as noninvasive biomarkers for SOC. Here, we analyzed serum expression level of circSETDB1 in a cohort of health volunteers and SOC patients. Our results show that circSETDB1 was significantly upregulated in SOC serum samples compared with healthy samples. ROC analysis has shown that serum circSETDB1 had a high diagnostic accuracy in discriminating SOC patients from healthy controls. In addition, circSETDB1 was also significantly upregulated in subgroup of patients with lymph node metastasis as well as patients with advanced clinical stage. These results indicated that circSETDB1 may be a promising biomarker for SOC.
Chemoresistance and cancer relapse in epithelial ovarian cancer have become a burden in treating the disease effectively.4,27,28 The involvement of circRNAs in relation to chemoresistance in several types of cancers has also been reported. Abu et al developed a chemoresistance colorectal cancer cell line model and profiled the global circRNAs expression via microarray. There were 773 upregulated and 732 downregulated circRNAs between the chemoresistant and chemosensitive HCT-116 cells found. Among those circRNAs, hsa_circ_32883 was identified to be a promising biotarget for colorectal cancer chemoresistance.29 In gastric cancer (GC), the expression of circAKT3 was higher in cisplatin-resistant GC than in cisplatin-sensitive samples. Upregulation of circAKT3 in GC patients receiving cisplatin therapy was significantly associated with aggressive phenotype.30 Circular RNA-MTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis.31 In bladder cancer, the expression level of hsa_circ_0000285 was lower in cisplatin-resistant patients than in those cisplatin-sensitive, indicating that downregulation of hsa_circ_0000285 contributed to bladder cancer cisplatin resistance.32 Increasing evidence suggest that circRNAs' signatures may be useful prognostic and predictive factors for cancers.21,33 CircRNA_100876 was found to be significantly upregulated in esophageal squamous cell carcinoma tissues. Patients with high expression of circRNA_100876 had shorted overall survival time.34 Our investigation revealed that circSETDB1 expression level is higher in chemoresistant SOC patients than in chemosensitive patients. Furthermore, serum circSETDB1 level has a significant diagnostic value in discriminating patients with chemoresistance from patients with chemosensitivity. Moreover, patients with low circSETDB1 expression had longer progression disease-free survival time than those with high expression, thus defining circSETDB1 potential role in assessing clinical outcome of SOC patients.
CircRNAs are more stable than cognate linear transcripts and can be detected in urine samples and blood plasma.21 The unique circular structure makes circRNAs insensitive to ribonucleases and enables them to exist intact in various tissues and body fluids.35,36 These unique characteristics make circRNAs ideal candidates as noninvasive biomarkers for cancer diagnosis, prognosis, and treatment.35,37 Here, we show that circSETDB1, also known as hsa_circ_006352, can be used as a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and progression-free survival in SOC.
Our study has several limitations that must be considered in the interpretation of our results. First, the sample size applied in our single center study was limited. Further investigations are needed to expand and confirm this observation in a larger cohort of patients. Second, functional relevance is lacking. The regulatory mechanisms of circSETDB1 in ovarian cancer cell lines should be subsequently investigated. It is now widely acknowledged that no single biomarker will provide all the necessary information for diagnosis, prognosis, and thus development of therapeutic strategies for patients with ovarian cancer and other cancers.38–40 As a consequence, the combined circRNAs and/or clinical biomarkers may possess well diagnostic, prognostic efficacy for patients with SOC.
Acknowledgment
This work was supported by National Natural Science Foundation of China (grant no. 81572794 and U1204821) and Natural Science Foundation of the Henan Province of China (grant no. 182300410358).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Joly F, Ahmed-Lecheheb D, Kalbacher E, et al. Long-term fatigue and quality of life among epithelial ovarian cancer survivors: a GINECO case/control VIVROVAIRE I study. Ann Oncol. 2019;30:845–852. doi: 10.1093/annonc/mdz074 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3.Bowtell DD, Böhm S, Ahmed AA, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–679. doi: 10.1038/nrc4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan S, Coward JI, Bast RC, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 6.Li S, Han L. Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med. 2019;11:15. doi: 10.1186/s13073-019-0629-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36:152. doi: 10.1186/s13046-017-0624-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019. doi: 10.1002/1878-0261.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed I, Karedath T, Andrews SS, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7:36366–36381. doi: 10.18632/oncotarget.8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong CM, Wei L, Law CT, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. doi: 10.1002/hep.28304 [DOI] [PubMed] [Google Scholar]
- 12.Fei Q, Shang K, Zhang J, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun. 2015;6:8651. doi: 10.1038/ncomms9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tchasovnikarova IA, Timms RT, Matheson NJ, et al; GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alagoz M, Katsuki Y, Ogiwara H, et al. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015;43:7931–7944. doi: 10.1093/nar/gkv722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sridharan R, Gonzales-Cope M, Chronis C, et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q-Y, Ding L-W, Xiao J-F, et al. SETDB1 accelerates tumourigenesis by regulating the WNT signalling pathway. J Pathol. 2015;235:559–570. doi: 10.1002/path.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karanth AV, Maniswami RR, Prashanth S, et al. Emerging role of SETDB1 as a therapeutic target. Expert Opin Ther Targets. 2017;21:319–331. doi: 10.1080/14728222.2017.1279604 [DOI] [PubMed] [Google Scholar]
- 19.Vea A, Llorente-Cortes V, de Gonzalo-Calvo D. Circular RNAs in blood. Adv Exp Med Biol. 2018;1087:119–130. doi: 10.1007/978-981-13-1426-1_10 [DOI] [PubMed] [Google Scholar]
- 20.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, Zhang C, Lin J, Song X, Wang H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:1275–1283. doi: 10.2147/cmar.s166740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022 [DOI] [PubMed] [Google Scholar]
- 24.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027 [DOI] [PubMed] [Google Scholar]
- 25.Ning L, Long B, Zhang W, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53:2637–2646. doi: 10.3892/ijo.2018.4566 [DOI] [PubMed] [Google Scholar]
- 26.Teng F, Xu J, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;112:8–17. doi: 10.1016/j.biocel.2019.04.011 [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Ren F, Wu Q, et al. MicroRNA-497 inhibition of ovarian cancer cell migration and invasion through targeting of SMAD specific E3 ubiquitin protein ligase 1. Biochem Biophys Res Commun. 2014;449:432–437. doi: 10.1016/j.bbrc.2014.05.053 [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Du H, Liu H, Hu F, Liu G. SMAD specific E3 ubiquitin protein ligase 1 promotes ovarian cancer cell migration and invasion via the activation of the RhoA/ROCK signaling pathway. Oncol Rep. 2019;41:668–676. doi: 10.3892/or.2018.6836 [DOI] [PubMed] [Google Scholar]
- 29.Abu N, Hon KW, Jeyaraman S, et al. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11:875–884. doi: 10.2217/epi-2019-0042 [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18:71. doi: 10.1186/s12943-019-0969-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53:1752–1762. doi: 10.3892/ijo.2018.4485 [DOI] [PubMed] [Google Scholar]
- 32.Chi BJ, Zhao DM, Liu L, et al. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66:197–202. doi: 10.4149/neo_2018_180318N185 [DOI] [PubMed] [Google Scholar]
- 33.Tan H, Gan L, Fan X, Liu L, Liu S. Diagnostic value of circular RNAs as effective biomarkers for cancer: a systematic review and meta-analysis. Onco Targets Ther. 2019;12:2623–2633. doi: 10.2147/ott.s197537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao S, Chen G, Yan L, Li L, Huang X. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:7385–7394. doi: 10.2147/ott.s177524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolha L, Ravnik-Glavac M. Circular RNAs: biogenesis, function, and a role as possible cancer biomarkers. Int J Genomics. 2017;2017:1–19. doi: 10.1155/2017/6218353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 37.Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yousef GM, Fracchioli S, Scorilas A, et al. Steroid hormone regulation and prognostic value of the human kallikrein gene 14 in ovarian cancer. Am J Clin Pathol. 2003;119:346–355. doi: 10.1309/0ua57mnayv0mce9u [DOI] [PubMed] [Google Scholar]
- 39.Khan J, Wei JS, Ringnér M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]