Abstract
Diagnostic genome-wide sequencing (exome or genome sequencing and data analysis for high-penetrance disease-causing variants) in acutely ill infants appears to be clinically useful, but the value of this diagnostic test should be rigorously demonstrated before it is accepted as a standard of care. This white paper was developed by the Paediatric Task Team of the Global Alliance for Genomics and Health’s Regulatory and Ethics Work Stream to address the question of how we can determine the clinical value of genome-wide sequencing in infants in an intensive care setting. After reviewing available clinical and ethics literature on this question, we conclude that evaluating diagnostic genome-wide sequencing as a comprehensive scan for major genetic disease (rather than as a large panel of single-gene tests) provides a practical approach to assessing its clinical value in acutely ill infants. Comparing the clinical value of diagnostic genome-wide sequencing to chromosomal microarray analysis, the current evidence-based standard of care, per case of serious genetic disease diagnosed provides a practical means of assessing clinical value. Scientifically rigorous studies of this kind are needed to determine if clinical genome-wide sequencing should be established as a standard of care supported by healthcare systems and insurers for diagnosis of genetic disease in seriously ill newborn infants.
Keywords: Exome sequencing, genome sequencing, Neonatal intensive care unit, Health policy
Introduction
Next-generation sequencing of an individual’s genome or exome with analysis of the data for high-penetrance disease-causing variants (genome-wide sequencing or GWS) is increasingly used for the clinical diagnosis of genetic disease.1,2 Diagnostic rates observed in reported patient series range from 25% to more than 60%, depending on how patients were selected for testing, the point in the diagnostic process at which the testing was performed, and how the sequencing and analysis were done. GWS has been shown to provide a higher diagnostic yield as a first-tier test than conventional clinical assessments and genetic testing in head-to-head comparisons in heterogeneous groups of pediatric patients with suspected genetic disorders.3–7 Some of the highest diagnostic rates reported to date have been in acutely ill infants tested in neonatal intensive care unit (NICU) or pediatric intensive care unit (PICU) settings.8–10
Making a specific genetic diagnosis in the newborn period is especially important because precise diagnosis can have critical implications for an infant’s health and wellbeing for her entire life.9–11 Most severe Mendelian diseases and pathogenic copy-number changes are manifest in infancy, and congenital anomalies and genetic disorders are the most frequent cause of death in infancy.12
However, there are thousands of individually rare genetic conditions and the presentation in babies may be nonspecific or differ from that observed in older patients. In addition, the general fragility of very small or premature infants may produce secondary multisystem involvement that obscures the primary pathology. These factors often make precise diagnosis of a rare genetic disease difficult or impossible in the newborn period without knowledge of the underlying pathogenic variant(s).
Diagnosis of a specific genetic disease in acutely ill infants may, therefore, be a particularly valuable clinical application of GWS. It has been suggested that rapid diagnostic GWS of acutely ill infants could be the breakthrough application that leverages widespread adoption of genomic medicine throughout the healthcare system.13
However, the complexity and cost of performing this test in critically ill newborns may be especially high because genome rather than exome sequencing, GWS on both parents as well as the affected child, and a very rapid turnaround time may be necessary.13 Moreover, the financial cost of the testing itself is insufficient to capture the full impact of clinical GWS. This is well illustrated by a recent study that compared children with suspected genetic disease who had initial chromosomal microarray analysis and subsequent diagnostic genome sequencing.14 The volumes and costs of medical interventions prompted by negative chromosomal microarray results and diagnostic GWS results were quantitatively similar but qualitatively different. Negative chromosomal microarray analysis sometimes led to additional laboratory investigations in pursuit of a diagnosis, while obtaining a diagnosis by genome sequencing often led to more visits to medical specialists or allied health professionals, reflecting changes in clinical management.
The clinical value of diagnostic GWS in patients suspected of having a rare genetic disease must consider the downstream effects of making a genetic diagnosis, including the impact on care of the patient, emotional suffering for the patient and family, informed reproductive decision-making, societal costs and savings, and other important aspects of diagnostic GWS.15 Moreover, the value that the healthcare system places on achieving a diagnosis through GWS and the value families place on achieving such a diagnosis may be substantially different.16
Rapid diagnostic GWS in acutely ill infants also highlights a number of ethical issues related to the care of acutely ill newborn infants and to GWS more generally.17–19 The central ethical tenet of clinical pediatrics is that the best interests of the child are paramount, but determining the best interests of a severely ill infant may be challenging. For example, some of the benefits attributed to diagnostic GWS result from avoidance of high-intensity treatment and risky medical or surgical interventions in favor of palliative “comfort care” for infants who have uncontrollable suffering or whose prognosis is dismal.9–11,13,20 Is it in a baby’s best interests for his parents to find out that he has an untreatable genetic condition that has been fatal within the first few months of life in all previously reported cases? What if there are only three such cases reported in the entire world literature? What if the only available therapy is experimental and the child is likely to be severely impaired even if the therapy succeeds? Would the fact that the family has already lost two babies to the same disease be relevant? Such ethical concerns about GWS in newborn infants trouble the families of affected infants, healthcare providers, and insurers.19
Although the benefits of GWS in acutely ill infants do appear promising, the clinical value of this (or any other) diagnostic test should be rigorously demonstrated before it is accepted as a standard of care.21 Until the necessary evidence is available, it is impossible to say whether GWS is worth the associated costs and risks, and its adoption as a routine diagnostic service by publicly funded healthcare systems and private health insurers is likely to remain limited and inconsistent. This white paper was developed by the Paediatric Task Team of the Global Alliance for Genomics and Health’s Regulatory and Ethics Work Stream to address the question of how we can determine the clinical value of diagnostic GWS in acutely ill newborns.
Assessing the clinical value of GWS
The ACCE (Analytical validity, Clinical validity, Clinical utility, and associated Ethical, legal, and social implications) framework is the most widely used standard for evaluation of genetic tests for clinical use.22,23 Analytical validity is a measure of the accuracy with which a test determines a genotype of interest. It is a purely technical assessment that is independent of whether or not the genotype is likely to be pathogenic. In contrast, clinical validity is a measure of a test’s ability to diagnose a specific phenotype, such as a genetic disease, accurately. Clinical utility, a measure of the net benefit of the test in terms of clinical care, depends on factors like the availability of effective management if a particular disease is diagnosed as well as on the economic costs, accessibility, and operational impact of testing on the delivery of health services.
It has been argued that the clinical utility of a genetic test should also include consideration of ethical, legal, and social issues related to the diagnosis, prevention, or treatment of the disease that is being tested. Even this broad definition of clinical utility may not be fully inclusive of the overall costs and benefits of genetic testing: elements of “personal utility” may also need to be considered.15,24–27 As a practical matter, however, personal utility and social consequences are difficult to measure and have contributed little to the funding decisions healthcare systems and insurers have made regarding genetic testing to date.
The ACCE approach and others that organize or assess available evidence regarding the appropriateness of implementing particular genetic tests have recently been reviewed in detail by a committee of the US National Academy of Sciences.28 This expert committee pointed out that the ACCE framework was developed for single-gene tests and is difficult to apply to GWS. There are several reasons for this, including:
Every person’s genome differs by about 4 to 5 million nucleotides from the standard “reference genome”.29,30 As a consequence, the one or two variants that are actually causal for a Mendelian disease in an affected individual must be distinguished from the noise of millions of variants that are unrelated to the disease.
Clinical and bioinformatics analysis of exome-sequencing data provides a survey for disease-causing variants of all protein-coding genes simultaneously. Over 3400 different genes are currently known to be associated with Mendelian disease,31 and it would be extremely difficult to demonstrate analytical validity for any possible variant of every one of these genes. This problem is even greater for genome sequencing, which also detects structural variants that may involve multiple genes and noncoding regulatory regions.
The variants that can be detected by diagnostic GWS are associated with more than 5000 different phenotypes,31 and additional Mendelian conditions and disease-associated genes are being discovered almost every day. Demonstrating clinical validity for the genetic variants associated with each of these diseases would be very challenging because most of the conditions are rare and our knowledge of their penetrance, phenotypic spectra, and natural history is incomplete.21
Determining the clinical utility of GWS for each of these thousands of different genetic disorders on a gene-by-gene or disease-by-disease basis would be even more difficult. Clinical utility is dependent, among other things, on the availability and efficacy of medical interventions, which are nonspecific and of unproven benefit for most rare genetic conditions.21,32
All of these factors apply to assessment of the clinical value of diagnostic GWS in critically ill infants. This assessment is also colored by ethical issues related to the testing of such patients9,11,20,33 and further complicated by the technical and analytical complexity (and consequent cost) of using diagnostic GWS in a NICU or PICU setting. These considerations might constitute an insurmountable barrier to assessing the clinical value and appropriateness of diagnostic GWS if it were an enormous panel of single-gene tests. It is not: it is a broad scan for high-penetrance genetic variants that can cause disease. Analysis of diagnostic GWS as a single test, rather than as thousands of different tests done at once, simplifies consideration of all aspects of clinical value.
Performing genome-wide surveys for pathogenic genetic alterations in patients suspected of having serious genetic disorders has been widely used clinically for more than 50 years—initially as cytogenetic analysis of solid-stained chromosomes, then as banded chromosomal analysis, and more recently as chromosomal microarray analysis. Treating diagnostic GWS as a genome-wide scan for high-penetrance disease-causing variants rather than as a large panel of single-gene tests provides a practical approach to assessing its clinical value in critically ill infants.
Some practical solutions
Empirical studies of variants identified by clinical GWS using an orthogonal method, most often Sanger sequencing, have demonstrated very high rates of confirmation, implying excellent analytical validity, or at least specificity34–36 of the test. More generally, analytical validity can be established for GWS by using reference DNA specimens, such as those available through the Genome In A Bottle Consortium.37 Each of these specimens includes millions of presumably benign polymorphic single-nucleotide or structural variants that have been characterized in detail by several analytical technologies. Although comparison of GWS results with these reference standards does not establish sensitivity or specificity for every individual disease gene, such an analysis does provide a robust assessment of analytical validity across the entire genome.
The clinical validity of GWS can be evaluated directly for a few rare diseases against standard clinical tests for these disorders, either in samples with known pathogenic variants or prospectively for samples submitted for clinical testing. For this assessment to be relevant to the question at hand, the diseases chosen would have to be ones that present in infants hospitalized in a NICU or PICU. Extrapolating the findings to genetic disease in general would require an assumption that the small subset actually evaluated is representative of all genetic diseases that present in infancy. This assumption is unlikely to be correct, given our current limited knowledge of the penetrance and phenotypic variability of most genetic conditions, and thus the clinical validity for diagnostic GWS obtained in this way would have to be considered a maximal estimate.
An holistic approach to assessing the clinical value of GWS in acutely ill newborn infants
Petrikin and his associates33 have speculated that rapid diagnostic WGS in critically ill infants may substantially reduce mortality. If this were true, a single important outcome (mortality) could be used to determine the clinical value of diagnostic GWS in infants in an intensive care setting (Fig. 1). It is important to note, however, that there are two different ways that making a precise diagnosis in a critically ill infant might influence the child’s survival. Definitive diagnosis of an untreatable condition that carries a very poor prognosis might enable intensive medical support to be withdrawn and comfort care instituted, in accordance with the family’s wishes. This would accelerate early mortality among newborns who would otherwise die later, after futile efforts to alter the inevitable course of their disease (red portion of the GWS curve in Fig. 1). The second possible effect of diagnosing a specific genetic defect in an acutely ill newborn infant is that it would permit effective precision treatments to be provided and access to promising experimental treatments for some of these infants much earlier in the course of their disease. These interventions may be lifesaving, reducing overall mortality among patients who receive diagnostic WGS and dramatically improving the quality of life in surviving children (purple portion of the curve with GWS in Fig. 1). Examples from our own experience of lifesaving therapeutic interventions that were instituted because a specific genetic diagnosis was made include bone marrow transplantation, administration of riboflavin (vitamin B2) in very large doses,38 and repurposing an available drug for treatment of overexpression of a cell signaling pathway.39
Fig. 1. Hypothetical 180-day mortality of severely ill infants with suspected genetic disease without diagnostic genome-wide sequencing (GWS) (green curve) or with the benefit of diagnostic GWS (red and purple curves).
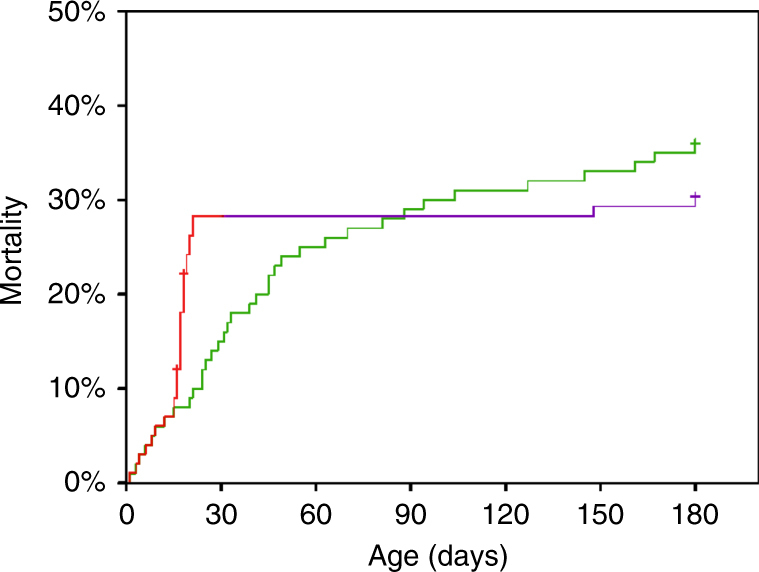
Note that diagnostic GWS is postulated to have two opposing effects on mortality: In infants diagnosed as having a condition that is lethal early in infancy, diagnostic GWS permits substitution of comfort care for intensive support, which results in earlier death (red portion of the curve). Some other infants with potentially lethal conditions are diagnosed as having a treatable condition, and the institution of precision therapy provides a reduction in mortality (purple portion of the curve). Figure redrawn after Petrikin et al., 2015 (ref. 33)
An overall reduction in infant mortality would only occur if the prolongation of life in healthier infants who receive precision therapy outweighs the accelerated mortality that occurs in infants who are diagnosed with lethal conditions and changed from intensive care to comfort care early in life (Fig. 1). These two different kinds of interventions influence infant mortality in opposite directions, and might exert opposing effects on costs, as well. Earlier transition to comfort care and consequent earlier death in infants with a grim prognosis would be expected to produce substantial healthcare cost savings. Provision of a lifesaving treatment to an acutely ill newborn infant might either reduce or increase healthcare costs, depending on the effectiveness and expense of the treatment and on the residual symptoms and limitations that persist after treatment is begun.
Death can only be used as the primary outcome for a health technology assessment when the background mortality is very high over a relatively short period of time, as is the case for newborn infants hospitalized in NICUs or PICUs. A statistically significant improvement in, say, 6-month survival in the GWS arm of a randomized diagnostic trial designed to test this possibility against conventional diagnostic evaluation might provide compelling evidence of clinical value, given the financial, systemic, and personal costs of caring for acutely ill infants with serious genetic diseases. Such studies would necessarily include patients who have a range of clinical problems and who are found to have a variety of specific genetic conditions, each of which has its own prognosis, characteristic clinical course, potential therapeutic interventions, and effects on quality of life for the affected individual and family. Using mortality as an outcome circumvents this clinical heterogeneity, which confounds the disease-specific outcome metrics used to assess clinical validity and clinical utility of single-gene tests.40–43
The use of mortality as a measure of the clinical value of GWS also has important limitations. To begin with, even if mortality could be used to compare diagnostic GWS with conventional genetic testing, mortality is unlikely to be useful for other important clinical comparisons—e.g., whether genome sequencing is better than exome sequencing or a turnaround time of 48 h is better than 2 weeks, despite the higher costs. Moreover, there are few circumstances outside of an intensive care unit in which mortality would be frequent enough to be used as an outcome measure in a study of the clinical value of genetic or genomic testing.
Are there alternative outcomes that could be used for this purpose? Improvements in treatment or earlier decisions to pursue comfort care after a precise diagnosis might be reflected in an easily measurable metric like NICU or PICU length of stay. An obvious advantage of using length of stay as an outcome measure is that it is amenable to conventional economic analysis because NICU or PICU care is so costly.38 A major disadvantage is that length of stay in an intensive care unit considers only a small portion of the life, benefits achieved, and costs incurred by surviving patients and their families.
Other outcomes that might be considered as part of the assessment are quality-adjusted life years (QALYs), a measure of survival time adjusted by the quality of that life,44,45 or disability-adjusted life years (DALYs), a measure of duration of survival adjusted for severity of disability.46,47 Unfortunately, both QALYs and DALYs are difficult to estimate for infants and are unknown for the vast majority of rare genetic diseases.48 Moreover, estimating overall QALYs or DALYs for a group of critically ill infants when almost every baby has an entirely different diagnosis (and the group selected for study differs from every other group of similar size) is challenging. The use of QALYs or DALYs in infants also raises important ethical issues regarding how to value the duration and quality of life in young children in comparison with adults and in individuals with qualitatively different kinds of disabilities and diseases (e.g., a serious cardiac malformation versus severe intellectual disability).49
Comparisons have been made in other clinical contexts between diagnostic GWS and current standard-of-care genetic testing using cost per case diagnosed as the outcome.7,8,38,43,50,51 For example, the current standard of care in the evaluation of children with intellectual disability is to perform chromosomal microarray analysis52 in combination with additional genetic tests suggested by the clinical features.53,54 Comparing GWS to conventional genetic testing on a per-case diagnosed basis has the advantage of permitting straightforward comparisons of diagnostic yields and testing costs. In addition, it can be done as two simultaneous undertakings in a single group of patients, providing a perfect control that mitigates the effect of clinical and genetic heterogeneity.
It is important to note, however, that determining the diagnostic yield for GWS is not the same thing as establishing its clinical value or even its clinical validity. This is because most of the thousands of different rare genetic diseases that have been described cannot be diagnosed by clinical means alone, i.e., there is no “gold standard” method of establishing the presence (or absence) of the disease without a genetic test. For most rare genetic diseases, demonstration of the presence of a pathogenic variant that has (or of pathogenic variants that have) previously been shown to produce a phenotype consistent with the clinical features found in a patient is necessary to establish a diagnosis. In other words, the diagnosis depends on genotype–phenotype correlation, which requires the use of clinical expertise by specialist physicians.55 Such clinical judgment is necessarily subjective, so it is critically important, in a comparison of the clinical value of GWS with conventional genetic testing, that all diagnoses are made in a consistent fashion and without knowledge of the test used to identify the disease-associated genotype. The use of a multidisciplinary consensus panel may be useful in this process.38,56,57
There are other issues that might also confound a head-to-head comparison of diagnostic rates achieved by GWS and conventional genetic testing in critically ill infants. These include the fact that different diagnoses may be made in the two arms of the study, with GWS being more sensitive for most variants that cause Mendelian disease and conventional testing being more sensitive for some others (e.g., trinucleotide repeat expansions). In addition, GWS analysis pipelines identify more than one Mendelian diagnosis in about 5% of cases, sometimes producing a “blended” phenotype, sometimes with each variant producing its own separate characteristic phenotype, and sometimes with a secondary finding that is completely unrelated to the indication(s) for testing.58,59
Nevertheless, comparing the benefits and costs of diagnostic GWS with the current standard of care per case of serious genetic disease identified holds promise as a practical means of assessing clinical value in a NICU or PICU setting, and in other contexts as well. For a comparative study to provide the robust evidence that is needed, both costs and benefits will have to be to be collected in a rigorous prospective manner, and the results will need to be analyzed comprehensively.
Conclusions
On the basis of the foregoing considerations, we conclude that:
Diagnostic genome-wide sequencing shows great promise for improving the management of acutely ill infants in a NICU or PICU setting.
Before diagnostic GWS can be considered to be a standard of care, the test must first be shown to have analytical validity and clear clinical value.
Evaluating diagnostic GWS as a comprehensive scan for major genetic disease provides a practical approach to assessing its clinical value in acutely ill infants.
The analytical validity of diagnostic GWS can be assessed using standard reference specimens and data.
Total mortality over a defined period of time might serve as a compelling summary measure to assess the clinical value of diagnostic GWS in comparison with conventional genetic testing in acutely ill infants. This will only be possible if lifesaving treatments enabled by precision diagnosis are frequent enough. Mortality is unlikely to be useful as an outcome measure in comparing incremental changes in diagnostic GWS, such as different sequencing technologies or improvements in bioinformatics software.
Summary outcomes, such as hospital length of stay, QALYs, or DALYs, which have been used for cost-effectiveness studies in other contexts, are incomplete or difficult to obtain as metrics for comprehensive assessment of diagnostic GWS in infants in an intensive care setting.
Comparing diagnostic GWS with the current standard of care per case of serious genetic disease diagnosed can provide a practical means of assessing clinical value. Such comparisons should be designed and carried out in a scientifically rigorous manner through collaboration of clinical investigators, health services researchers, and health economists. These studies are needed to determine if clinical GWS should be supported by healthcare systems and private insurers as the standard of care for diagnosis of genetic disease in seriously ill newborn infants.
Acknowledgements
We are grateful to Denise Avard, Nick Dragojlovic, Robin Hayeems, Muin Khoury, Stephen Kingsmore, Karine Sénécal, and Danya Vears for their critical reviews of this manuscript and suggestions for its improvement. This work was supported by CanSHARE, which is jointly funded by Genome Canada, the Canadian Institutes of Health Research, Genome Quebec, and Genome British Columbia.
Disclosure
The authors declare no conflicts of interest.
References
- 1.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370:2418–25. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 4.Stavropoulos DJ, Merico D, Jobling R, et al. Whole-genome sequencing expands diagnostic utility and improves clinical management in paediatric medicine. npj Genom Med. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anazi S, Maddirevula S, Faqeih E, et al. Clinical genomics expands the morbid genome of intellectual disability and offers a high diagnostic yield. Mol Psychiatry. 2017;22:615–24. doi: 10.1038/mp.2016.113. [DOI] [PubMed] [Google Scholar]
- 6.Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2017;20:435–43. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissers LELM, van Nimwegen KJM, Schieving JH, et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet Med. 2017;19:1055–63. doi: 10.1038/gim.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168–265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willig LK, Petrikin JE, Smith LD, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 2015;3:377–87. doi: 10.1016/S2213-2600(15)00139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017;171:e173438. doi: 10.1001/jamapediatrics.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Diemen CC, Kerstjens-Frederikse WS, Bergman KA, et al. Rapid targeted genomics in critically ill newborns. Pediatrics. 2017;140:e20162854. doi: 10.1542/peds.2016-2854. [DOI] [PubMed] [Google Scholar]
- 12.Murphy SL, Xu J, Kochanek KD, Curtin SC, Arias E. Deaths: final data for 2015. Natl Vital Stat Rep. 2017;66:1–73. [PubMed] [Google Scholar]
- 13.Kingsmore SF, Petrikin J, Willig LK, Guest E. Emergency medical genomes: a breakthrough application of precision medicine. Genome Med. 2015;7:82. doi: 10.1186/s13073-015-0201-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayeems RZ, Bhawra J, Tsiplova K, et al. Care and cost consequences of pediatric whole genome sequencing compared to chromosome microarray. Eur J Hum Genet. 2017;25:1303–12. doi: 10.1038/s41431-017-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ACMG Board of Directors. Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2015;17:505–7. doi: 10.1038/gim.2015.41. [DOI] [PubMed] [Google Scholar]
- 16.Stivers T, Timmermans S. The actionability of exome sequencing testing results. Sociol Health Illn. 2017;39:1542–56. doi: 10.1111/1467-9566.12614. [DOI] [PubMed] [Google Scholar]
- 17.Deem MJ. Whole-genome sequencing and disability in the NICU: exploring practical and ethical challenges. Pediatrics. 2016;137(suppl 1):S47–55. doi: 10.1542/peds.2015-3731I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel LA, Pereira S, McGuire AL. Potential psychosocial risks of sequencing newborns. Pediatrics. 2016;137(suppl):S24–S29. doi: 10.1542/peds.2015-3731F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson DJ, Barnett C, Savulescu J, Newson AJ. Genomic intensive care: should we perform genome testing in critically ill newborns? Arch Dis Child Fetal Neonatal Ed. 2016;101:F94–8. doi: 10.1136/archdischild-2015-308568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith LD, Willig LK, Kingsmore SF. Whole-exome sequencing and whole-genome sequencing in critically ill neonates suspected to have single-gene disorders. Cold Spring Harb Perspect Med. 2016;6:a023168. doi: 10.1101/cshperspect.a023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoury MJ. No shortcuts on the long road to evidence-based genomic medicine. JAMA. 2017;318:27–28. doi: 10.1001/jama.2017.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Secretary’s Advisory Committee on Genetic Testing. Enhancing the oversight of genetic tests: recommendations of the SACGT Secretary’s Advisory Committee on Genetic Testing; 2000. Bethesda, Maryland: National Institutes of Health.
- 23.Haddow J, Palomaki G. ACCE: a model process for evaluating data on emerging genetic tests. In: Khoury MJ, Little J, Burke W, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. Oxford: Oxford University Press; 2003. pp. 217–33. [Google Scholar]
- 24.Grosse SD, Khoury MJ. What is the clinical utility of genetic testing? Genet Med. 2006;8:448–50. doi: 10.1097/01.gim.0000227935.26763.c6. [DOI] [PubMed] [Google Scholar]
- 25.Bombard Y. Translating personalized genomic medicine into clinical practice: evidence, values, and health policy. Genome. 2015;58:491–7. doi: 10.1139/gen-2015-0145. [DOI] [PubMed] [Google Scholar]
- 26.Bunnik EM, Janssens ACJW, Schermer MHN. Personal utility in genomic testing: is there such a thing? J Med Ethics. 2015;41:322–6. doi: 10.1136/medethics-2013-101887. [DOI] [PubMed] [Google Scholar]
- 27.Kohler JN, Turbitt E, Lewis KL, et al. Defining personal utility in genomics: a Delphi study. Clin Genet. 2017;92:290–7. doi: 10.1111/cge.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Academies of Sciences, Education, and Medicine. An evidence framework for genetic testing. 2017. Washington, DC: The National Academies Press. 10.17226/24632. Accessed 1 June 2017. [PubMed]
- 29.Auton A, Abecasis GR, Altshuler DM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Telenti A, Pierce LCT, Biggs WH, et al. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA. 2016;113:11901–6. doi: 10.1073/pnas.1613365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.OMIM. OMIM gene map statistics. https://www.omim.org/statistics/geneMap. Accessed 18 January 2018.
- 32.Phillips KA, Deverka PA, Sox HC, et al. Making genomic medicine evidence-based and patient-centered: a structured review and landscape analysis of comparative effectiveness research. Genet Med. 2017;19:1081. doi: 10.1038/gim.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrikin JE, Willig LK, Smith LD, Kingsmore SF. Rapid whole genome sequencing and precision neonatology. Semin Perinatol. 2015;39:623–31. doi: 10.1053/j.semperi.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linderman MD, Brandt T, Edelmann L, et al. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med Genom. 2014;7:20. doi: 10.1186/1755-8794-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strom SP, Lee H, Das K, et al. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med. 2014;16:510. doi: 10.1038/gim.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beck TF, Mullikin JC, Biesecker LG. Systematic evaluation of Sanger validation of next-generation sequencing variants. Clin Chem. 2016;62:647 LP–654. doi: 10.1373/clinchem.2015.249623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zook JM, Catoe D, McDaniel J, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. doi: 10.1038/sdata.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Bel KL, Ragotte RJ, Saferali A, et al. JAK1 gain-of-function causes an autosomal dominant immune dysregulatory and hypereosinophilic syndrome. J Allergy Clin Immunol. 2017;139:2016.e5. doi: 10.1016/j.jaci.2016.12.957. [DOI] [PubMed] [Google Scholar]
- 39.Chang SM, Matchar DB, Smetana GW, Umscheid CA, editors. Methods guide for medical test reviews. Rockville, MD: AHRQ (US Agency for Healthcare Research and Quality); 2012. [PubMed] [Google Scholar]
- 40.National Institute for Health and Care Excellence. Guide to the methods of technology appraisal 2013. 2013. Available at https://www.nice.org.uk/process/pmg9/resources/guide-to-the-methods-of-technology-appraisal-2013-pdf-2007975843781. Accessed 27 May 2016. [PubMed]
- 41.Sanders GD, Neumann PJ, Basu A, et al. Recommendations for conduct, methodological practices, and reporting of cost-effectiveness analyses: second panel on cost-effectiveness in health and medicine. JAMA. 2016;316:1093–103. doi: 10.1001/jama.2016.12195. [DOI] [PubMed] [Google Scholar]
- 42.Schwarze Katharina, Buchanan James, Taylor Jenny C, Wordsworth Sarah. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genetics in Medicine. 2018;20(10):1122–1130. doi: 10.1038/gim.2017.247. [DOI] [PubMed] [Google Scholar]
- 43.Stark Z, Lunke S, Brett GR, et al. Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med. E-pub ahead of print 15 March 2018. [DOI] [PubMed]
- 44.Weinstein MC, Torrance G, McGuire A. QALYs: the basics. Value Health. 2009;12(suppl 1):S5–9. doi: 10.1111/j.1524-4733.2009.00515.x. [DOI] [PubMed] [Google Scholar]
- 45.Wouters OJ, Naci H, Samani NJ. QALYs in cost-effectiveness analysis: an overview for cardiologists. Heart. 2015;101:1868–73. doi: 10.1136/heartjnl-2015-308255. [DOI] [PubMed] [Google Scholar]
- 46.Gold MR, Stevenson D, Fryback DG. HALYs and QALYs and DALYs, oh my: similarities and differences in summary measures of population health. Annu Rev Public Health. 2002;23:115–34. doi: 10.1146/annurev.publhealth.23.100901.140513. [DOI] [PubMed] [Google Scholar]
- 47.Sassi F. Calculating QALYs, comparing QALY and DALY calculations. Health Policy Plan. 2006;21:402–8. doi: 10.1093/heapol/czl018. [DOI] [PubMed] [Google Scholar]
- 48.Stark Zornitza, Schofield Deborah, Martyn Melissa, Rynehart Luke, Shrestha Rupendra, Alam Khurshid, Lunke Sebastian, Tan Tiong Y., Gaff Clara L., White Susan M. Does genomic sequencing early in the diagnostic trajectory make a difference? A follow-up study of clinical outcomes and cost-effectiveness. Genetics in Medicine. 2018;21(1):173–180. doi: 10.1038/s41436-018-0006-8. [DOI] [PubMed] [Google Scholar]
- 49.Pettitt D, Raza S, Naughton B, et al. The limitations of QALY: a literature review. J Stem Cell Res Ther. 2016;6:1000334. [Google Scholar]
- 50.Monroe GR, Frederix GW, Savelberg SMC, et al. Effectiveness of whole-exome sequencing and costs of the traditional diagnostic trajectory in children with intellectual disability. Genet Med. 2016;18:949–56. doi: 10.1038/gim.2015.200. [DOI] [PubMed] [Google Scholar]
- 51.Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19:867–74. doi: 10.1038/gim.2016.221. [DOI] [PubMed] [Google Scholar]
- 52.Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moeschler JB, Shevell M. Comprehensive evaluation of the child with intellectual disability or global developmental delays. Pediatrics. 2014;134:e903 LP–e918. doi: 10.1542/peds.2014-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mithyantha R, Kneen R, McCann E, Gladstone M. Current evidence-based recommendations on investigating children with global developmental delay. Arch Dis Child. 2017;102:1071 LP–1076. doi: 10.1136/archdischild-2016-311271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson BJ, Miller FA, Rousseau F, Adam S, Friedman JM. Controversy and debate on clinical genomics sequencing. J Clin Epidemiol. 2017;92(suppl C):4–17. doi: 10.1016/j.jclinepi.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 56.Lazaridis KN, Schahl KA, Cousin MA, et al. Outcome of whole exome sequencing for diagnostic odyssey cases of an individualized medicine clinic: the Mayo Clinic experience. Mayo Clin Proc. 2016;91:297–307. doi: 10.1016/j.mayocp.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 57.Tan T, Dillon O, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. 2017;171:855–62. doi: 10.1001/jamapediatrics.2017.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balci TB, Hartley T, Xi Y, et al. Debunking Occam’s razor: diagnosing multiple genetic diseases in families by whole-exome sequencing. Clin Genet. 2017;92:281–9. doi: 10.1111/cge.12987. [DOI] [PubMed] [Google Scholar]
- 59.Posey JE, Harel T, Liu P, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]


