Abstract
Background:
In TOPS trial, 4 or 7 days of Combivir (CBV; zidovudine/lamivudine) with maternal single dose nevirapine (sdNVP) significantly reduced the emergence of NVP resistance as determined by virus population genotype. To detect NVP resistance with greater sensitivity, we analyzed TOPS samples by allele-specific RT-PCR (ASP).
Methods:
In a random subset of women from each arm, plasma from before and 6 weeks after sdNVP were analyzed by ASP at codons 103, 181, 184, and 190.
Results:
Samples were analyzed from 27 women in the sdNVP arm and 24 each in the CBV 4 day (sdNVP/CBV4) and 7 day (sdNVP/CBV7) arms. ASP detected NVP-resistant variants in week 6 samples from 70% of women in the sdNVP arm, 29% in the sdNVP/CBV4 arm, and 33% in sdNVP/CBV7 arm (p<0.01 for sNVP/CBV4 or sdNVP/CBV7 vs. sdNVP; p=1.0 for sdNVP/CBV4 vs. sdNVP/CBV7). Lamivudine-resistance was detected by ASP in only 1 of 51 women who received CBV.
Conclusions:
Short-course CBV significantly reduced but did not eliminate the emergence of NVP resistance after sdNVP. NVP-resistant variants were detected in about one-third of women despite CBV, but the duration of persistence and clinical impact of these variants on response to antiretroviral therapy is uncertain.
Keywords: single-dose nevirapine, Combivir (zidovudine/lamivudine), HIV-1 drug resistance, allele-specific PCR
INTRODUCTION
A single dose of nevirapine (sdNVP), administered to HIV-infected women during labor and to their infants within 72 hours of birth, reduces peripartum mother-to-child transmission (MTCT) of HIV-1 [1–3]. Because of its long half-life and low genetic barrier to resistance, sdNVP produces measurable plasma levels of NVP for as long as 3 weeks that can select NVP-resistant variants. Such variants have been detected by virus population genotype in 19–75% of mothers and 33–87% of their infants who have received sdNVP [4–8]. Recent studies using more sensitive detection methods show that NVP-resistant variants can persist in plasma, as a minor fraction of the virus population not detected by virus population genotype, for up to 2 years after sdNVP [5, 7, 9]. These low frequency NVP-resistant variants can reduce the efficacy of subsequent NVP-containing antiretroviral therapy [10, 11]. A recent trial (A5208/OCTANE) comparing the efficacy of initial antiretroviral therapy (ART) of women exposed to sdNVP more than six months earlier showed that a lopinavir/ritonavir-containing regimen was significantly more effective than a NVP-containing regimen [12]. In A5208/OCTANE, detection of NVP resistance prior to antiretroviral therapy initiation, either by population genotype or by allele-specific PCR in genotype-negative samples, was associated with a significant increase in risk of virologic failure or death in women randomized to a NVP-containing regimen [13].
The emergence of NVP resistance can be reduced by a short course of antiretrovirals started simultaneously with sdNVP. For example, the Treatment Options Preservation Study (TOPS) trial compared the emergence of maternal and infant NVP resistance after exposure to one of three NVP-based therapies for the prevention of MTCT: sdNVP alone or sdNVP combined with short courses (4 or 7 days) of combivir (CBV: zidovudine and lamivudine) given to the mother and newborn infant [14]. Population genotype of plasma samples six weeks postpartum revealed significantly fewer women with NVP resistance in the sdNVP/CBV4 (11.7%) and sdNVP/CBV7 (7.3%) arms compared to those women who received sdNVP alone (59.2%; p<0.0001).
This landmark result changed the recommended standard of care to prevent HIV-1 MTCT from sdNVP alone to sdNVP with a 7 day course of CBV [15]. Nevertheless, suppression of NVP resistance by CBV was incomplete in the TOPS trial, as determined by virus population genotype, which reliably detects resistant variants that comprise more than 25% of the viral population. More sensitive methods, including allele-specific PCR [7, 16–18] and single-genome sequencing [19], detect clinically important resistant-variants that are missed by standard genotype. In the present study, therefore, we applied allele-specific PCR (ASP) assays with mutant detection limits of 0.1–0.3% [7, 16–18] to a random subset of women from each arm of the TOPS trial to better assess the extent of suppression of NVP resistance by CBV. We also applied single-genome sequencing (SGS) to a subset of samples tested by ASP to identify low-frequency NNRTI-resistant mutants at sites that ASP had not been designed to detect (e.g. codons 106, 108, 188) [19, 20].
METHODS
Patient Samples.
For ASP analysis, we selected a random subset of women from each arm of the TOPS trial who had stored plasma samples available (>1.0 ml) from both pre-intervention and 6 weeks postpartum time points, with both samples having HIV-1 RNA >5,000 copies/ml. Selection was made regardless of the results of standard population genotype. A total of 75 women were randomly selected for ASP analysis. For SGS analysis, a random subset of 15 women, 5 from each treatment arm, were selected from the 75 women selected for ASP analysis. The TOPS trial was approved by the South African Medicines Control Council (Trial #1100.1413) and ethics committees of the Universities of the Witwatersrand, Pretoria, KwaZulu-Natal, and the Pharma-Ethics Independent Research Ethics Committee. The trial design was registered in the ClinicalTrials.gov register (#), and all participants provided written informed consent for HIV-1 resistance testing. Testing of samples by ASP and SGS was approved by the NIH Office of Human Subjects Protection.
Viral RNA extraction.
Viral RNA from plasma samples was extracted as previously described and resuspended in 40µl of 5mM Tris-HCl [7, 17, 18].
Standard Genotype.
Standard genotype analysis was performed on all specimens at an accredited laboratory in South Africa, employing the TruGen 334 HIV-1 genotyping kit and OpenGene DNA sequencing system (Bayer), and data were analyzed by using the Stanford database. The resistance testing laboratory was blinded to treatment allocation.
Allele-specific RT-PCR (ASP).
ASP testing was performed on pre-intervention and week 6 postpartum samples without knowledge of treatment assignment or results of population genotype. First-round, real-time RT-PCR amplification of a 637-bp fragment of pol including reverse transcriptase codons 22–234 was performed by using HIV-1 subtype C-specific primers with PCR product monitored by SYBR green fluorescence as published [7, 17–18]. All primers used were specifically designed to amplify subtype C virus found in South Africa (primer sequences in the Supplemental Table [18]). Viral RNA from each time point was tested in triplicate. No-template controls and HIV RNA standards (3 × 106 to 300 copies) were included in the same 96-well assay plate. The products of the first amplification were diluted to 107 total copies per reaction and second rounds of PCR were performed in parallel using allele-specific primers to amplify codon-specific mutant or wildtype sequences at codons 103, 181, 184, 190 and nonspecific primers to amplify all templates in the PCR reactions [primer sequences in Supplemental Table; 7, 17–18]. To confirm amplification specificity, PCR products were subjected to thermal denaturation analysis at 70–80°C, with readings taken every 0.1°C for 3 sec. Samples that did not yield the correct melting temperature (Tm) for the amplified product were excluded. Data from triplicate wells resulting in a coefficient of variation (CV) greater than or equal to 100% were repeated and reported as invalid if the CV remained > 100%. Results are reported as percent mutant or wildtype, determined by dividing the number of mutant or wild-type sequences amplified by the total number of sequences amplified. Values are normalized to the sum of the alleles detected. If the sum of the alleles detected was less than 25%, results are reported as “indeterminate” at that codon.
Single-genome sequencing (SGS).
SGS was performed on pre-intervention and week 6 postpartum samples as published [19]. PCR amplification products were sequenced by dideoxy-terminator sequencing (Applied Biosystems) in both directions using overlapping internal primers [19]. Sequences containing mixtures of more than one amplified genome were not included in analyses. The remaining single genome sequences from each plasma sample were aligned to an HIV-1 subtype C consensus using Sequencher software (Gene Codes Corporation). The goal was to sequence at least 45 genomes for each patient sample, giving 90% certainty of detecting a variant present at frequency of 5% or greater in the population. A mean of 51 genomes per patient sample was obtained. The aligned sequences were analyzed for mutations at drug resistance sites (IAS-USA 2007 definitions) using the Calibrated Population Resistance program available through the Stanford University HIV Drug Resistance Database [21]. Frequency estimates are calculated based on the number of sequences obtained in each plasma sample.
Statistical Methods.
Fishers exact test using 2 × 2 contingency tables were used to compute p-values for subgroup analysis of treatment arms. All p-values reported are two-sided.
RESULTS
Plasma samples (pre-intervention and 6 weeks postpartum) from 75 randomly selected women were analyzed by ASP; 27 in the sdNVP alone arm, and 24 each in the 4 day (sdNVP/CBV4) and 7 day (sdNVP/CBV7) CBV arms. SGS analysis was performed on plasma samples from 15 women, 5 in each arm.
Standard Genotype Analysis
Population genotype analysis had been performed for the primary endpoint of the TOPS trial on plasma samples obtained 6 weeks postpartum from the 75 women selected for ASP analysis. Population genotype detected NVP resistance in 18 (24%) of 75 women: 14 of 27 (52%) in the sdNVP alone arm, 4 of 24 (17%) in the sdNVP/CBV4, and 4 of 24 (17%) in the sdNVP/CBV7 arm.
Nevirapine (NVP) resistance in sdNVP alone arm
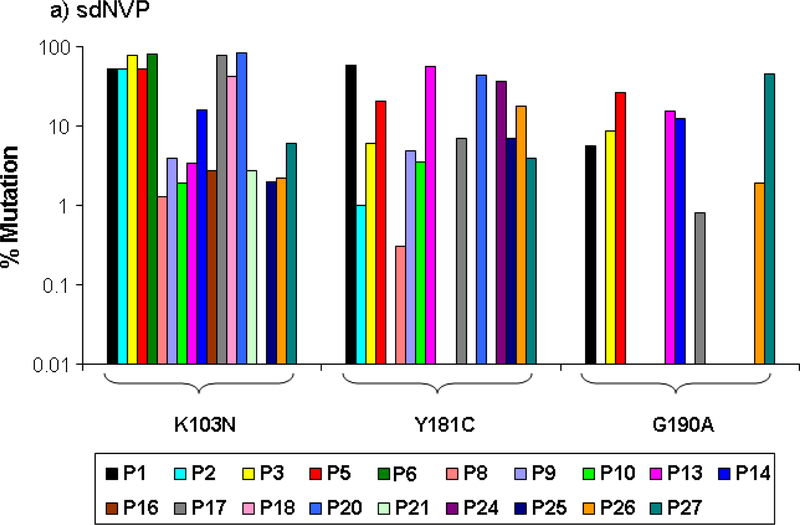
All pre-therapy samples were below the limit of detection for mutants at each codon tested (0.1% for codons 103 and 190 and 0.3% for codons 181 and 184). sdNVP alone resulted in the emergence of NVP resistance in the majority of patients 6 weeks postpartum with 19 of 27 (70%) and 3 of 5 (60%) having NVP resistance mutations as analyzed by ASP and SGS, respectively (Table 1, Figure 1a and 2a). The K103N mutation was detected most commonly (18 of 27 patients) with a median population frequency of 11% and a population frequency above 50% in 6 of 18 women (Table 1, Figure 1a). Although the G190A mutation was detected in fewer patients (8 of 27) and its population frequency was never above 50%, the median frequency of 11% was the same as for K103N. Y181C was detected in 13 patients and the median population frequency was 7%. Thirteen of 19 (68%) women with NVP resistance had multiple mutations detected; 7 of whom had all three NVP resistance mutations (103N, 181C and 190A).
Table 1.
Frequency of NVP mutations 103N, 181C, and 190A measure by allele-specific PCR in week-6 plasma samples.
| Treatment Arm | Number of Patients | % of Patients with NNRTI Resistance Mutations | 103N (AAC or AAT)a (0.1%)b | 181C (TGT) (0.3%) | 190A (GCA) (0.1%) | Any Mutation | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation Frequency | Mutation frequency | Mutation frequency | Mutation frequency | |||||||
| Median | Range | Median | Range | Median | Range | Median | Range | |||
| sdNVP | 27 | 70%d,e | 11% | 1.3–82% (18)c | 7% | 1–58% (13) | 11% | 0.8–45% (8) | 8.6% | 0.8–82% (19) |
| sdNVP + CBV 4 days | 24 | 29%d,f | 2% | 0.3–13% (5) | 1.6% | 0.8–49% (4) | 0.6% | 0.1–4% (4) | 1.5% | 0.1–49% (7) |
| sdNVP + CBV 7 days | 24 | 33%e,f | 0.7% | 0.4–10% (6) | 10.5% | 1.3–16% (3) | 0.6% | 0.3–1% (2) | 1% | 0.3–15.5% (8) |
| Combined | 75 | 45% | 3.8 | 0.3–82% (29) | 7% | 0.8–58% (20) | 3.8% | 0.1–45% (14) | 4.8% | 0.1–82.3% (34) |
The frequencies of 103N AAC and AAT mutations were combined.
Limit of detection of mutant frequency
Number of patients with measurable mutation by ASP
P=0.005 for comparison of treatment arms sdNVP and sdNVP + CBV 4 days
P=0.012 for comparison of treatment arms sdNVP and sdNVP + CBV 7 days
P=1.0 for comparison of treatment arms sdNVP + CBV 4 and sdNVP + CBV 7 days
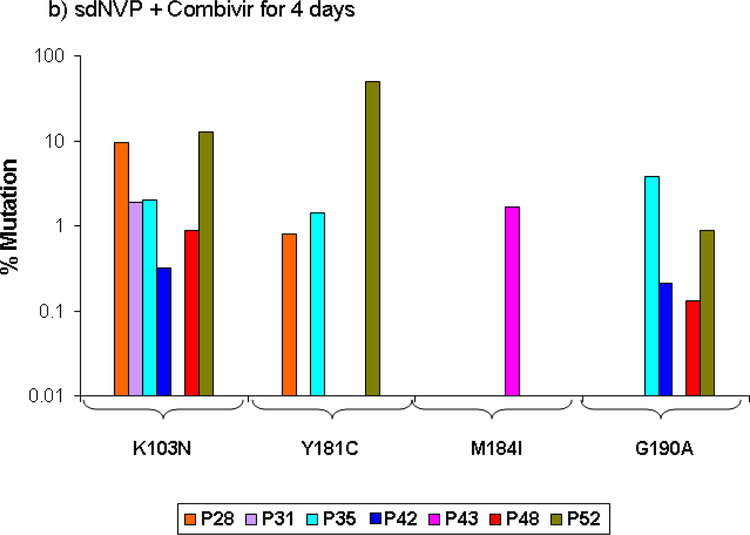
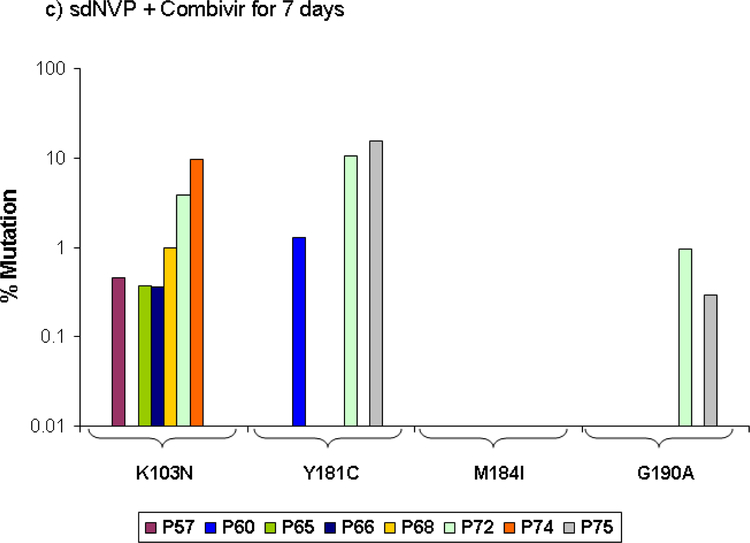
Figure 1:
Population frequency of NVP resistance mutations at the specified codons detected by ASP in samples obtained 6 weeks after sdNVP determined by ASP at codons 103, 181, and 190 in women receiving sdNVP alone (panel A), sdNVP plus CBV for 4 days (panel B) or for 7 days (panel C). Vertical bars show data from individual women.
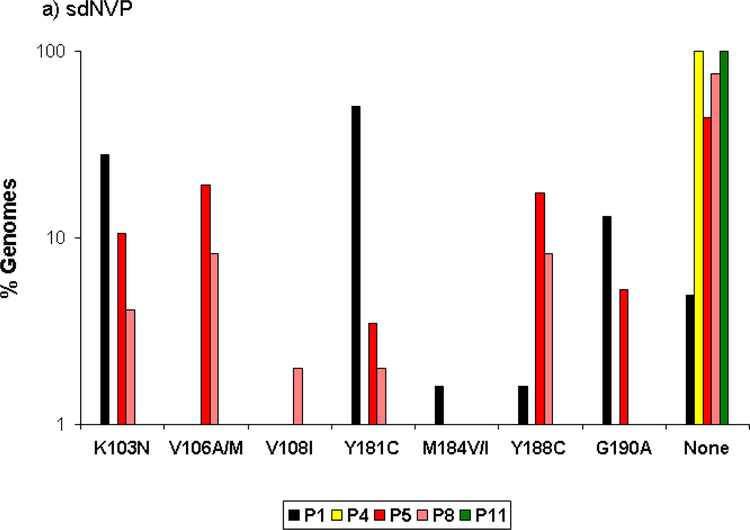
Figure 2:
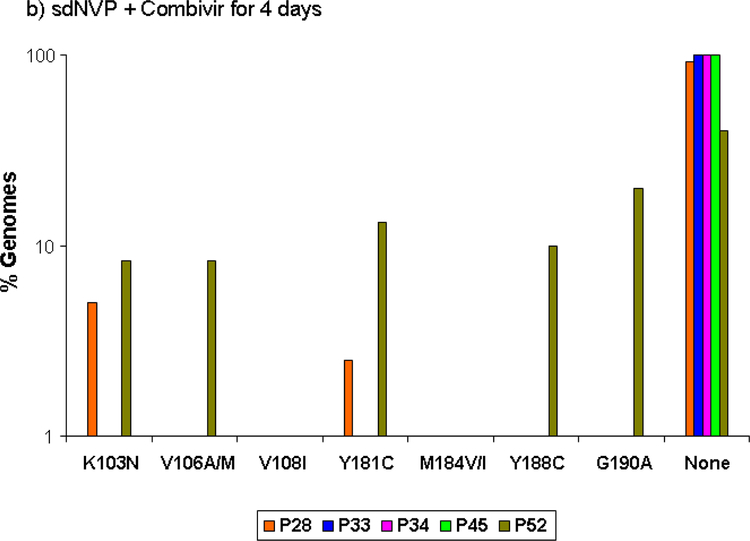
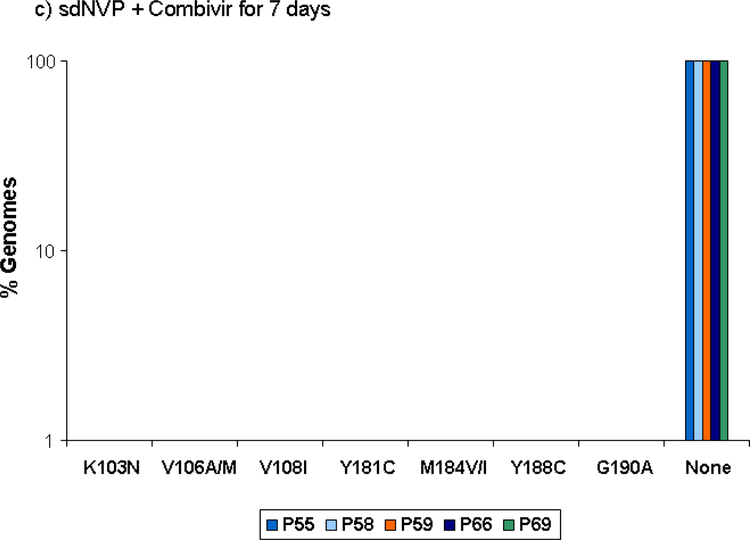
Proportion of genomes showing the specified NVP and lamivudine resistance mutations detected by SGS in samples obtained 6 weeks after sdNVP in women receiving sdNVP alone (panel A), sdNVP plus CBV for 4 days (panel B) or for 7 days (panel C). Vertical bars show data from individual women. None indicates genomes with no resistance mutations.
SGS revealed that viral variants with mutations at codons 103, 106, 181, 188, and 190 comprised a substantial fraction of the circulating virus population in 3 of 5 women 6 weeks after sdNVP, although generally not exceeding 30% (Figure 2a). The total percentage of genomes with any drug resistance mutation was 45%. Mutations at codons 106 (A/M) and 188 (C), which were not analyzed by ASP, were detected in approximately 10% of genomes from 2 of 5 patients who received sdNVP alone. Mutation at codon 108 (I) was detected at low frequency (2%) in 1 of 5 patients. None of the mutations listed above (103, 106, 108, 181, 184, 188, 190) were linked on the same genome in any of the patients. The two patients (#4 and 11; Figure 2a) without NVP-resistant genomes detected by SGS also did not have mutations detected by ASP.
NVP and NRTI resistance in the sdNVP/CBV4 arm
In the TOPS study, the addition of CBV for 4 days after sdNVP significantly reduced the proportion of patients with NVP resistance detected by standard genotype by about 5-fold from 59% with sdNVP alone to 12% with sdNVP/CBV4 [14]. ASP analysis confirmed that CBV for 4 days significantly reduced the proportion of women with NVP resistance detected, from 70% in the sdNVP arm to 29%, or about 2.5 fold (p=0.005). In addition, the median population frequency of NVP-resistant mutants decreased from 11% to 2% (Table 1). Five of the 7 women with NVP resistance had multiple NVP-resistance mutations detected by ASP (Figure 1b). Only 2 of 5 patients analyzed by SGS had NVP resistance mutations (Figure 2b), and the remaining 3 patients were negative by both SGS and ASP. The total percentage of genomes having NVP resistance mutations detected by SGS was 13.5% compared with 45.2% in the sdNVP alone arm. Lamivudine-resistant variants were detected by ASP in only 1 of 24 patients with a frequency of 2% M184I. This patient did not have NNRTI resistance. The M184V mutation was not detected by ASP in any patient. SGS did not detect M184I/V or other NRTI resistance mutations including thymidine analog mutations (TAMs) in postpartum samples.
NVP and NRTI resistance in the sdNVP/CBV7 arm
CBV for 7 days also significantly reduced the proportion of patients from 70% with sdNVP alone to 33% (Table 1) with sdNVP/CBV7 (p=0.012), but the extent of reduction in resistance was less than that observed by population genotype (59.2% for sdNVP alone vs. 7.3% for sdNVP/CBV7 [14]). There was no significant difference in the proportion of women with NVP-resistant variants detected by ASP between the 4 and 7 day CBV arms (Table 1; p=1.0). However, only 2 of 8 patients with NVP resistance detected by ASP in the sdNVP/CBV7 arm had multiple NVP-resistance mutations detected compared with 5 of 7 in the sdNVP/CB4 arm (Figure 1c), although this difference was not significant (p=0.13). NVP resistance mutations were not detected by SGS in any of the 5 women analyzed in the sdNVP/CBV7 arm, but one had low frequency NVP-resistant variants detected by ASP at codon 103 (0.4%). Lamivudine-resistance mutations were not detected by ASP and neither lamivudine- nor other NRTI-resistance mutations were detected by SGS (Figure 2c).
Comparison of Population Genotype and ASP for NVP Resistance
As noted earlier, among the 75 women studied, 18 (24%) had NVP resistance detected by population genotype. Of these 18 women, 15 (83%) had NVP resistance detected by ASP and 3 had mutations detected at codons not examined by ASP (106, 108, 188). Among the women who were negative for NVP resistance by standard genotype, the proportions with NVP resistance detected by ASP were 31% (5 of 16), 15% (3 of 20), and 24% (5 of 21) in the sdNVP, sdNVP/CB4, and sdNVP/CB7 arms, respectively. Overall, the proportions of women with NVP resistance detected by either population genotype or ASP were 74% (20 of 27), 37.5% (9 of 24), and 33% (8 of 24), in the sdNVP, sdNVP/CB4, and sdNVP/CB7 arms, respectively.
DISCUSSION
Prevention of MTCT of HIV-1 continues to be a major challenge in resource-poor settings where antiretroviral therapy (ART) for pregnant women is not routinely available [1]. In some settings, sdNVP alone is still used to prevent HIV-1 transmission to infants [3], even though NVP-resistant variants are selected in the majority of women and these resistant variants can decrease the effectiveness of subsequent ART with NVP-containing regimens [10, 11]. Thus, reducing the emergence of NVP resistance after sdNVP is an important goal. In the TOPS trial, initiation of short course CBV at the same time as sdNVP significantly reduced the proportion of women with new NVP resistance mutations detected by population genotype analysis 6 weeks post-therapy [14]. Specifically, 59.2%, 11.7%, and 7.3% of women in the sdNVP, sdNVP/CBV4, and sdNVP/CBV7 arms, respectively, had NVP resistance detected. In the present study, we used more sensitive methods - ASP and SGS - to identify NVP resistance that could have been missed by population genotype analysis. Testing for NVP resistance by more sensitive methods is important because the presence of low frequency NNRTI-resistant variants, below the detection limit of population genotype, is strongly associated with failure of NNRTI-containing regimens [11, 13, 22–26].
Our results corroborate the main finding of the TOPS trial [14] and provide additional insights into the efficacy of short course CBV in preventing NVP resistance. ASP analysis revealed a significant, albeit smaller, reduction in the frequency of NVP resistance with 4 or 7 days of CBV. To illustrate, by population genotype analysis, only 11.7% and 7.3% of women in the 4 and 7 day CBV arms had NVP resistance detected. By contrast, 29% and 33% of women in the 4 and 7 day CBV arms had NVP resistance detected by ASP, respectively, and 37.5% and 33%, respectively, with the two methods combined. More frequent detection of mutants by ASP is not surprising given it’s higher sensitivity that standard genotype, but the emergence of NVP-resistant mutants in about one-third women in both the CBV 4 and 7 day arms was not anticipated.
Although the SGS analyses were restricted to 15 patients, the results are also consistent with the findings of the TOPS trial, with both 4 and 7 days of CBV showing suppression of NVP resistance compared with sdNVP alone, and 7 days of CBV showing greater suppression of NVP resistance than 4 days of CBV (Figure 2). In addition, SGS and ASP results were consistent with each other (Figures 1 and 2). For the codons analyzed by ASP, all mutations detected by SGS were also detected by ASP at roughly similar frequencies. There was only one instance (patient #66) in which a mutation was detected by ASP, but not by SGS. In this instance, the mutant frequency was <1% and well below the sensitivity of 45 single genome sequences, which is 90% sensitivity for mutants at 5% frequency. Conversely, there were 4 women who had drug-resistant mutants detected by SGS at sites that were not included in the ASP assay (106, 108, and 188), although each of these women had NVP resistance mutations detected by ASP at codons 103, 181, or 190. Only 9 of the 22 mutations detected by SGS were detected by standard sequencing (data not shown), which is consistent with prior reports showing improved sensitivity of SGS compared with population genotype [16, 19]. Additionally, patients #8 and #28 had multiple mutations detected by SGS and ASP but population genotype failed to detect any resistance mutations.
ASP analysis revealed no significant difference between the 4 and 7 day CBV arms in the proportion of women with NVP-resistant variants (p=1.0). However, 5 of 7 patients receiving sdNVP/CBV4 had multiple NVP mutations detected by ASP compared to only 2 of 8 patients receiving sdNVP/CBV7; however, this difference was not significant (p=0.13).
Our findings are also generally consistent with the important report by Farr et al. (27) showing suppression of NVP resistance by CBV for 7 days in an observational study conducted in Malawi. In that study, NVP resistance at 6 weeks postpartum, as detected by population genotype combined with ASP, was reduced from 74% with sdNVP alone to 10% with sdNVP + CBV for 7 days. Our study extends this finding to a randomized clinical trial and provides a comparison of 4 vs. 7 days of CBV. Of note, the frequency of NVP resistance detected in both the 4 and 7 day CBV arms (37.5% and 33%, respectively) in our study was considerably higher than the 10% frequency reported by Farr et al. after 7 days of CBV (10%). Possible explanations for the discordant outcomes are differences in the patient population studied, the study design (observational vs. randomized), medication adherence and the methods used to detect low frequency variants. Regarding the latter possibility, the mutant detection limit for ASP used in our study was 0.1–0.3% vs. 0.5–1.0% reported by Farr et al. (27).
We found that only 1 of 51 women receiving sdNVP/CBV4 had a lamivudine-resistance mutation (M184I). The absence of this mutation in the baseline sample from the same patient implies that it was selected by CBV. The absence of M184V or other NRTI mutations in women from either the 4 or 7 day CBV arms may be due to the shorter half-life of the active forms of lamivudine and zidovudine; or alternatively, that NRTI-resistant variants decline more rapidly than NVP-resistant mutants after clearance of drug. By contrast, NVP has a very long half-life and the K103N mutation appears to have little effect on HIV-1 replicative fitness, resulting in persistence of variants after drug is cleared.
Our findings raise important additional questions. For example, the long-term effects of the NVP-resistant variants we detected on subsequent treatment outcome are incompletely defined. Two initial studies suggested that the effects of NVP resistance were limited to those women who initiated ART within 6 months after receiving sdNVP [10, 28]. By contrast, a more recent study by Coovadia et al. suggested that women who developed K103N mutations after sdNVP had a significantly higher chance of virologic failure after initiating ART 18–36 months after sdNVP [11]. Similarly, a study using ASP in 26 women who received sdNVP, found minor NNRTI variants in 86% of the women who failed subsequent NVP-containing therapy compared to 32% of women who continued to respond [29]. Finally, A5208/OCTANE Trial 1 was designed to compare protease inhibitor-based therapy using lopinavir/ritonavir (LPV/r) to NVP-based therapy in women with prior sdNVP. The trial showed LPV/r + tenofovir and emtricitabine (TDF/FTC) to be superior to NVP + TDF/FTC (p=0.001) for initial ART of women with prior sdNVP more than 6 months earlier [12]. Population genotype analysis of stored pretherapy plasma samples revealed NVP resistance mutations in 13% of women in the NVP arm and 73% of these women reached a primary endpoint of virologic failure or death, but this association explained less than half of the primary endpoints in the NVP arm. Among the 201 women without NVP resistance by standard genotype, 70 (35%) had NVP-resistant variants detected by ASP. Among these 70 women, a primary study endpoint occurred in 12 of 38 (32%) women in the NVP arm versus 3 of 32 (9%) in the LPV/r arm (HR 3.84 [95% CI 1.06–13.9]; p=0.040). NVP-resistant variants at frequencies >1% were significantly associated with the primary study endpoint in the NVP arm (p=0.003), but not in the LPV/r arm [13]. These findings suggest that the persistence of NVP-resistant variants at frequencies as low as 1% of the virus population can negatively influence subsequent treatment response.
This level of NVP-resistant mutant (>1%) was found in 21% of women in the current study 6 weeks after receiving CBV for 4 or 7 days. A critical issue to be resolved is how long such low frequency mutants persist after sdNVP with 4 or 7 days of CBV and whether they persist at levels that would influence subsequent treatment response.
In conclusion, by applying highly sensitive methods to detect resistance, we have shown that short-course CBV therapy significantly reduces but does not eliminate the selection of NVP-resistant variants. The persistence and ultimate clinical impact of low frequency NVP-resistant variants after sdNVP with short-course CBV or other antiretroviral regimens remains to be determined. Additional strategies are needed to maximize prevention of NVP resistance and optimize subsequent response to antiretroviral therapy in women who have received sdNVP.
Supplementary Material
ACKNOWLEDGEMENTS
Funding: Boehringer Ingelheim, the manufacturer of nevirapine, funded the 1100.1413 trial. Allele-specific PCR and single-genome sequencing were supported by the National Cancer intramural program and contracts from SAIC to J.M.C (#HHSN261200900346P) and J.W.M (#25XS119). J.M.C was a Research Professor of the American Cancer Society with support from the FM Kirby Foundation. S.P. is funded in part by the Swedish Research Council.
Footnotes
Presented in part at the XIVth International HIV Drug Resistance Workshop: Basic Principles and Clinical Implications, June 2005, Quebec, Canada.
DISCLOSURE STATEMENT
Competing Interests: M. Hopley, D. B. Hall, and P. Robinson are employed by Boehringer Ingelheim, the sponsor of the 1100.1413 study, and D. Mayers was employed by Boehringer Ingelheim at the time of the study. J. A. McIntyre and G. E. Gray have received research funding, travel grants and speaker’s honoraria from Boehringer Ingelheim and Glaxo SmithKline. J.W. Mellors is a consultant for Merck, Gilead Sciences, and RFS Pharma and owns share options in RFS Pharma. S. Palmer, V.F. Boltz, J.Y. Chow, J.M. Coffin, F. Maldarelli, and N. A. Martinson declare no competing interests.
REFERENCES
- 1.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 1999; 354:795–802. [DOI] [PubMed] [Google Scholar]
- 2.Jackson JB, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet 2003; 362:859–68. [DOI] [PubMed] [Google Scholar]
- 3.Arrive E, Dabis F. Prophylactic antiretroviral regimens for prevention of mother-to-child transmission of HIV in resource-limited settings. Curr Opin HIV AIDS 2008; 3:161–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eshleman SH, Mracna M, Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). AIDS 2001; 15:1951–7. [DOI] [PubMed] [Google Scholar]
- 5.Flys T, Nissley DV, Claasen CW, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis 2005; 192:24–9. [DOI] [PubMed] [Google Scholar]
- 6.Johnson JA, Li JF, Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis 2005; 192:16–23. [DOI] [PubMed] [Google Scholar]
- 7.Palmer S, Boltz V, Martinson N, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A 2006; 103:7094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinson NA, Morris L, Gray G, et al. Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine. J Acquir Immune Defic Syndr 2007; 44:148–53. [DOI] [PubMed] [Google Scholar]
- 9.Flys TS, Donnell D, Mwatha A, et al. Persistence of K103N-containing HIV-1 variants after single-dose nevirapine for prevention of HIV-1 mother-to-child transmission. J Infect Dis 2007; 195:711–5. [DOI] [PubMed] [Google Scholar]
- 10.Jourdain G, Ngo-Giang-Huong N, Le Coeur S, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med 2004; 351:229–40. [DOI] [PubMed] [Google Scholar]
- 11.Coovadia A, Hunt G, Abrams EJ, et al. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin Infect Dis 2009; 48:462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockman S, Hughes M, McIntyre J, et al. Lopinavir/ritonavir- versus nevirapine-based antiretroviral therapy in women with and without prior single-dose nevirapine use. N Engl J Med 2010; 363: 1499–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci USA 2011, in press [DOI] [PMC free article] [PubMed]
- 14.McIntyre JA, Hopley M, Moodley D, et al. Efficacy of short-course AZT plus 3TC to reduce nevirapine resistance in the prevention of mother-to-child HIV transmission: a randomized clinical trial. PLoS Med 2009; 6:e1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. Antiretroviral drugs for treating pregnant women and preventing HIV infections in infants. Recommendations for a public health approach Geneva: World Health Organization; 2010. [PubMed] [Google Scholar]
- 16.Halvas EK, Aldrovandi GM, Balfe P, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol 2006; 44:2612–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer S, Boltz V, Maldarelli F, et al. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS 2006; 20:701–10. [DOI] [PubMed] [Google Scholar]
- 18.Boltz VF, Maldarelli F, Martinson N, et al. Optimization of allele-specific PCR using patient-specific HIV consensus sequences for primer design. J Virol Methods 2010; 164:122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 2005; 43:406–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearney M, Palmer S, Maldarelli F, et al. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS 2008; 22:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gifford RJ, Liu TF, Rhee SY, Kiuchi M, Hue S, Pillay D, Shafer RW. The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 2009; 25:1197–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS Med 2008; 5(7):e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simen BB, Simons JF, Hullsiek KH, et al. Low abundance drug-resistant viral variants in chronically infected HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199:693–701. [DOI] [PubMed] [Google Scholar]
- 24.Paredes R, Lalama CM, Ribaudo HJ, et al. Preexisting minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 2010; 201:662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halvas EK, Wiegand A, Boltz VF, et al. Low frequency nonnucleoside reverse transcriptase inhibitor–resistant variants contribute to failure of efavirenz-containing regimens in treatment-experienced patients. J Infect Dis 2010; 201:672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heneine W When do minority drug-resistant HIV-1 variants have a major clinical impact? J Infect Dis 2010; 201:649. [DOI] [PubMed] [Google Scholar]
- 27.Farr S, Nelson JAE, Ng’ombe TJ, et al. Addition of 7 days of zidovudine plus lamivudine to peripartum single-dose nevirapine effectively reduces nevirapine resistance postpartum in HIV-infected mothers in Malawi. J Acquir Immune Defic Syndr 2010; 54:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med 2007; 356:135–47. [DOI] [PubMed] [Google Scholar]
- 29.Rowley CF, Boutwell CL, Lee EJ, et al. Ultrasensitive detection of minor drug-resistant variants for HIV after nevirapine exposure using allele-specific PCR: clinical significance. AIDS Res Hum Retroviruses 2010; 26:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.