Abstract
Biological function arises from the interplay of proteins, transcripts, and metabolites. An ongoing revolution in miniaturization technologies has created tools to analyze any one of these species in single cells, thus resolving the heterogeneity of tissues previously invisible to bulk measurements. An emerging frontier is single cell multi-omics, which is the measurement of multiple classes of analytes from single cells. Here, we combine bead-based transcriptomics with microchip-based proteomics to measure intracellular proteins and transcripts from single cells and defined small numbers of cells. The transcripts and proteins are independently measured by sequencing and fluorescent immunoassays respectively, to preserve their optimal measurement modes, and linked by encoding the physical address locations of the cells into digital sequencing space using spatially patterned DNA barcodes. We resolve cell-type-specific protein and transcript signatures and present a path forward to scaling the platform to high-throughput.
Introduction
The advent of quantitative, high throughput single cell ‘omics’ technologies is transforming our understanding of biology by resolving the heterogeneity of tissues, from development1 to function2 to disease3. Single cell ‘whole’ transcriptomics methods, which combine next generation sequencing tools, microfluidic chips, and molecular barcoding4–7, have defined the paradigm with affordable benchtop implementations. Single cell proteomics methods provide excellent specificity including post-translational modification detection8–11, but remain reliant on antibodies12, which places practical limits on multiplexing. Further, different classes of proteins, from secreted to membrane-bound to cytoplasmic, can require different analysis platforms. An outstanding challenge is to combine single cell proteomic assays with transcriptomics to quantify both the direct protein effectors of cell function and the broader mRNA regulatory framework.
Proteins and transcripts are typically assayed for quantification using orthogonal signals (fluorescence and sequence reads). The transduction and amplification modes of these signals are generally incompatible for simultaneous assays, especially at the single cell level, but new solutions have emerged that coerce one signal into the other13–16. For example, groups have effectively “sequenced” surface proteins by tagging antibodies with DNA barcodes17–19 and analyzing both surface proteins and transcripts via sequencing. Splitting the contents of a single cell20–22 provides an alternative, albeit technically challenging method also capable of multi-omic analysis. New techniques in advanced molecular barcoding23, amplification, and expansion microscopy24 may soon allow proteins and mRNA to be simultaneously imaged in fixed single cells and tissues.
Here we report on a single cell method for simultaneous measurement of a panel of functional intracellular proteins plus whole transcriptome sequencing. The approach combines the single cell barcode chip (SCBC) platform for proteomics8, 9, 11 with bead-based droplet transcriptomics (Fig. 1)4, 6. In an SCBC, single cells are isolated within individual microchambers. After cell lysis, a panel of target proteins, whether secreted, cytoplasmic, or membrane-bound, is specifically captured using an antibody array and read out using a non-amplified fluorescent sandwich immunoassay. For transcriptomics, we add sequencing beads to the chambers and encode the physical microchamber location on to the beads with short DNA oligomers. After capturing transcripts from the lysed cell, the beads are pooled for sequencing and the DNA-encoded location of individual cell transcriptomes can be traced back to specific microchambers and the associated protein readouts.
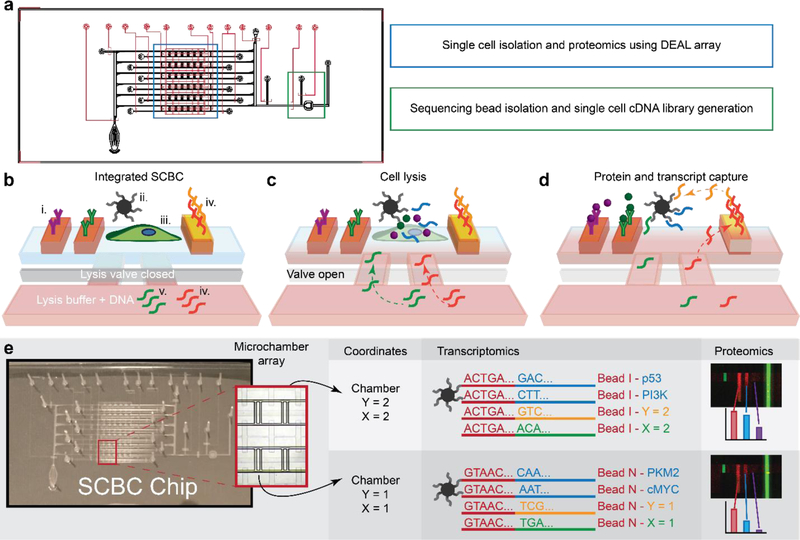
Figure 1.
Multi-omic SCBC schematic. a. The multi-omic SCBC combines the single cell proteomics SCBC with bead-based transcriptomics. The proteins are processed in the single cell microchambers, while sequencing beads are isolated for cDNA library generation before sequencing. b. Each microchamber in the SCBC contains (i.) an antibody barcode DEAL array, (ii.) sequencing beads, (iii.) cells, (iv.) X-coordinate ssDNA bound to the DEAL array, (v.) Y-coordinate ssDNA, and (vi.) DNA displacement oligos. c. Cell contents were released and location oligomers were introduced by opening a lysis valve. Note that the X-coordinate oligomer is released using a displacement strategy. d. After incubation, proteins were captured on the antibody DEAL array, and transcripts and location oligomers were captured on the sequencing beads. e. For these 6×6 chips, chambers were identified by their spatial coordinates on chip, and sequencing reads were sorted by their bead identifier sequence (red). These reads consisted of genes (blue), Y-coordinates (yellow), and X-coordinates (green), thus linking each bead identifier sequence to an X-Y location on chip. A spatial map of transcriptomes was generated and linked to proteomic measurements from each chamber.
Experimental
SCBC Mold and Device Fabrication
The SCBC is created in two steps. The first step involves construction and assembly of the molds and integrated microfluidic device used for executing the multi-omic SCBC experiments. The second step involves the flow patterning of the DNA barcodes that are used for assembling the DEAL arrays onto glass slides, and the flow patterning to encode the X-location of the cell microchambers.
Microfluidic Molds and Device Assembly.
Materials needed:
Sylgard 184 PDMS (Ellsworth Adhesives, Germantown WI), SU-8 2025, SU-8 Developer, SPR220–7.0, CD26 Developer (Microchem, Seattle WA), 0.5 mm hole punch, scotch tape.
The two-layer SCBC device for single cell isolation, proteomics, and transcriptomics (Fig. 1a) was fabricated as previously described9 with some modifications. Molds were photolithographically defined using SPR 220–7.0 for the flow layer (Fig. S1a) and SU-8 for the control layer (Fig. S1b, File S1). The channel height of the control layer was 40 microns and the channel height for the flow layer was 14 microns. Channel widths for the flow layer ranged from 100 to 400 microns wide.
The control layer was made with degassed PDMS (Sylgard 184) mixed at a ratio of 1 unit crosslinker to 8 units elastomer (1:8). The flow layer had two functional halves: the cell chambers and the microfluidic filter. For the cell chambers, softer PDMS was needed to allow valves to open and close easily. The microfluidic filter required stiffer PDMS to prevent the filter from collapsing or becoming blocked. To create the two different halves of the flow layer, softer 1:20 PDMS was poured over the half of the wafer mold with cell chambers and stiffer 1:8 PDMS was poured over the other half containing the filter (Fig. S1a). The wafer was placed on a spin coater and spun for 1 minute at 2000 RPM to create a PDMS flow layer with a height of 100 microns. Spin coating prevented the softer PDMS used for the cell chambers from mixing with the stiffer PDMS of the filter and vice versa to maintain separation between the two halves of the wafer mold. The height difference between the spin-coated softer and stiffer PDMS halves was negligible. The flow layer was cured for 15 minutes at 80 °C after spinning, and the control layer (~ 0.5 cm tall) was attached to the top of the flow layer.
The two-layer device was cured for 2 hours at 80 °C and lifted from the wafer, and access holes were punched into the device. In the flow layer (Fig. S1a, c), holes punched included the cell and bead inlet (1) and outlet, the reagent and wash inlet (2) and outlet (9), the individual row lysis buffer inlets (3–8) and outlets, and the transcriptomic inlet (10) and outlet. In the control layer (Fig. S1b, c), separate holes were required for valves regulating the inlets and outlets of the flow layer (1–4, 7–10, 12), as well as a cell chamber valve separating adjacent chambers (5), a lysis valve separating lysis buffer chambers from cell chambers (6), and a separation valve to isolate the cell chamber side of the chip from the microfluidic filter side (11). After the holes were punched they were flushed with water, the surfaces of the PDMS device were cleaned with scotch tape to enhance adhesion and remove particles, and the two-layer SCBC device was completed and ready for attachment to the glass slide.
Flow Patterning for DNA Barcode and X-Coordinate Oligo Addition.
Materials needed:
SuperChip polylysine-coated glass slides (Thermo Scientific, Federal Way WA), 0.1% (w/v) polylysine solution (Sigma, St. Louis MO), DNA oligomers (Table S1), BS3 crosslinker (Thermo Scientific 21580).
Every microchamber was surface-patterned with a DNA barcode which served two purposes. All barcode stripes except one were patterned with single-stranded (ss)DNA, and used for the spatially-selective assembly of DNA-labeled capture antibodies (the DEAL method)25. The remaining stripe was patterned with double-stranded (ds)DNA encoding the X-coordinate of the microchamber (Fig. S1d). Glass slides used for DNA-encoded antibody library (DEAL) arrays were prepared by flow patterning 20-micron wide DNA barcode lines as previously described9. A PDMS flow pattern with a winding snake-like pattern of parallel channels (File S1) was adhered to a SuperChip polylysine coated glass slide and cured for 2 hours at 80 °C. Polylysine solution was passed through the channels overnight, and amine-functionalized ssDNA oligomers (B-, C-, D-, I-, P-DNA, Table S1) were activated with BS3 crosslinker and flowed through the channel for 1 hour over ice. The device was removed from ice and incubated for 2 hours at room temperature to complete glass surface functionalization. The flow pattern was removed and the glass slide was washed in PBS-0.05% tween (PBST) and water.
A second flow patterning step was used to pattern X-coordinate ssDNA (X-Location Column 1–6, Table S1) on to the DNA barcode (P-DNA, Table S1). The second flow pattern consisted of individually addressable vertical channels (Fig. S1d, File S1). X-coordinate ssDNA was added to a solution of 0.5% BSA and 1mM MgCl2 in phosphate buffered saline (PBS) at a concentration of 10 nM and flowed through the second flow pattern, along with a Cy3 labeled DNA reporter to visually affirm that flow pattern molds were correctly aligned (I’-DNA, Table S1). The resulting base-paired X-coordinate dsDNA had 12 complementary base pairs with the DNA barcode (P-DNA), rendering it stable at room temperature (Fig. 1b, Tm = 38 °C). The channels were washed with PBST and the flow pattern was detached, creating a glass slide with a pattern of 3 ssDNA barcode lines and two dsDNA barcode lines, one containing the Cy3 fluorophore for visualization and one containing the X-coordinate dsDNA. The two-layer PDMS devices were aligned to the glass slide and cured at 80 °C for 2 hours to complete the multi-omic SCBC.
Multi-Omic SCBC Operation
The multi-omic SCBC is first primed and loaded on the day of measurements, and then split into three processing sections: cell lysis, proteomics, and transcriptomics. The multi-omic SCBC approach is illustrated in Fig. 1b–d. The completed SCBC contains an array of microchambers, each split into a cell chamber and lysis chamber and denoted by an X and Y DNA barcode (Fig. 1b–d, Fig. S1a–c). Cells and sequencing beads (10 μm diameter) are loaded into the cell chambers, and lysis buffer with two added ssDNA oligos, one variable and one constant, is loaded into the lysis chambers (Fig. 1b, Fig. S2a). The constant ssDNA is a displacement strand to displace one strand of the X-coordinate dsDNA from the barcode stripe, while the variable ssDNA is specific to each row of microchambers and encodes the Y-coordinates.
Finally, a computation pipeline analyzes proteomics and transcriptomics data separately before correlating the single-cell measurements. Full details of multi-omic SCBC operation are found in Table S3.
SCBC Chip Priming and Reagent Preparation.
Materials needed:
DNA oligomers (Table S1), antibodies (Table S2), Zeba 7K MWCO spin desalting columns (Thermo Scientific 89882), S-HyNic, S-4FB antibody-oligo crosslinkers (Trilink, San Diego CA), Amicon Ultra-4 centrifugal filter units (Millipore UFC801008, Burlington MA), diamond scribe.
To prepare the SCBC chip, the following reagents were prepared beforehand. Capture antibodies and DNA were purified and desalted with Zeba spin columns, conjugated to DNA (B’-, C’-, D’-DNA, Table S1) using the manufacturer’s protocols9 with S-HyNic and S-4FB, purified with FPLC (Superdex 200) and concentrated with Amicon Ultra-4 centrifugal filter units. The sequencing beads contained the same oligos as those reported previously4, but were custom manufactured using 10 micron diameter beads. Each oligo has a primer region, a bead identifier region unique to each bead, a random unique molecular identifier (UMI) for single molecule tracking and PCR bias correction, and a polyT tail for capturing polyadenylated transcripts (bead oligo, Table S1).
On the day of operation (Fig. S2a), the bottom of the glass device was first scored between the filter and the microchambers using a diamond scribe to facilitate later cleavage of the device without disturbing the valve connections. The valve attachments were then filled with water and the channels of the device were filled with 3% BSA in PBS. The valves were pressurized to allow water to fill the control layer channels and the device was incubated for 1 hour for blocking. After blocking, the microchambers were filled with a 1:20 dilution of the antibodies in a 1.5% BSA in PBS solution for 1 hour at 37 °C to allow complementary strands (e.g. Glass-B-DNA, Antibody-B’-DNA) to base pair to convert the ssDNA barcodes into DEAL antibody barcodes. Following antibody loading, one final blocking step of 1.5% BSA in PBST was done for 1 hour at room temperature, following by a wash of 100 µL PBST through the device.
Cell Loading and Lysis, and Bead Location Encoding and Collection.
Materials needed:
DNA oligomers (Table S1), sequencing beads (Chemgenes, Wilmington MA), cells, RNAse inhibitor (Lucigen 30281, Radnor PA), 6X saline sodium citrate (SSC, Ambion, Foster City CA).
Lysis buffer: 18 µL cell lysis buffer (Cell Signaling #9803, Danvers MA), 2 µL protease-phosphatase inhibitor (Cell Signaling #5872), 10 µL 20% Ficoll PM-400 (GE Health Care 17030010, Pittsburgh PA), 2µL RNAse inhibitor, 2 µL 2M DTT, 30 µL water, 6 µL 10 nM Cy3 labeled displacement DNA (P’-Displacement DNA, TableS 1).
A separate solution of lysis buffer was prepared for each row of the device. The lysis valve was closed to prevent any possible leakage of lysis buffer in to the cell chambers (Fig. S1b, valve 6). For each row, 1 µL of a 100 pM solution of the appropriate Y-coordinate ssDNA (Y-Location Row 1–6, Table S1) was added to 10 µL of lysis buffer and flowed through each row of lysis chambers before cell loading.
HEK and U87MG cells were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. After trypsinization, cells were washed twice with PBS and suspended in 0.01% BSA in PBS. A solution of 1 million cells and 4 million beads per mL was prepared, 1 µL of RNAse inhibitor was added to 20 µL of the cell/bead mixture, and the mixture was flowed through the device. When the appropriate cell loading was achieved, the cell chamber valve was closed to isolate the microchambers (Fig. S1b, valve 5) and the device was optically inspected by microscope to record the numbers of cells and beads in each chamber (Fig. 1e, Fig. 2a, Fig. S3a).
Figure 2.
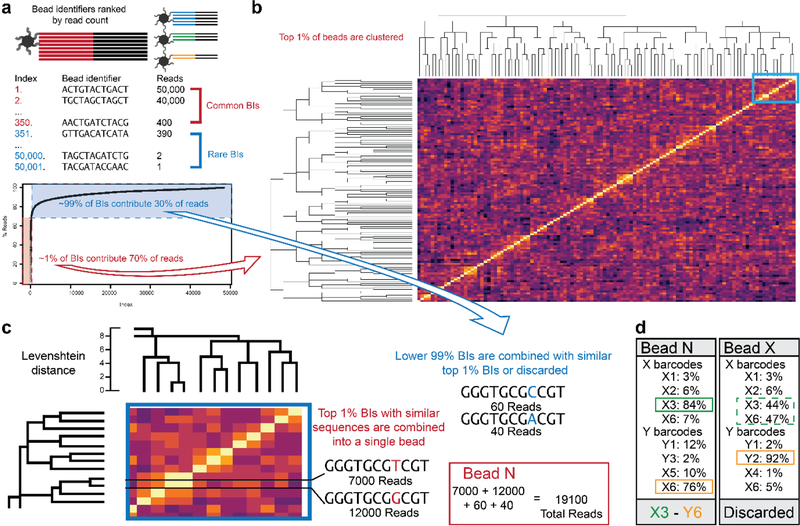
Digital bead identifier processing. a. Each bead identifier sequence (BI) is sorted by the frequency of its associated unique molecular identifiers (UMIs). b. A small fraction of BIs (~1–2%) contain approximately two-thirds of all UMIs, and these top BIs are clustered by their Levenshtein distance. c. Sequences with Levenshtein distance of 1 or less are grouped together as single beads and their UMIs pooled together. The remainder of the BIs are then grouped together with a top BI at Levenshtein distance 2 or less or discarded. d. Each of the grouped BIs and their pooled UMIs are analyzed for location barcode sequences. Beads that cannot be unambiguously assigned to a location are discarded to generate the complete digital location map of sequenced beads.
Cells were lysed by opening the lysis valve and allowing diffusion to mix the contents of the lysis and cell chambers, triggering 4 capture events (Fig. 1c). (1) Selected proteins are captured on the DEAL array. (2) Transcripts are captured on the sequencing beads. (3) One ssDNA oligo hybridizes to the sequencing beads, encoding the Y-coordinate of the cell chamber onto the beads. (4) A second ssDNA displaces the complementary strand of the dsDNA on the X-coordinate barcode stripe. The released strand is captured on the sequencing bead, thus encoding the X-coordinate of the bead (Fig. 1d). The displacement DNA in the lysis buffer was able to displace X-coordinate ssDNA from the chip during cell lysis due to the 21 base pair complementary region between P’-Displacement DNA and P-DNA, compared to just 12 between X-coordinate ssDNA and P-DNA. Cells were lysed on ice for 15 minutes with the valves open, followed by 15 minutes on ice with the valves closed and finally 2 hours at room temperature on a shaker to maximize analyte capture.
Following lysis and capture of proteins, transcripts, and X-Y coordinate ssDNA, the device was preconditioned by filling the upstream flow channels with SSC, the cell chamber isolation valve was opened, and sequencing beads were washed with SSC and collected at the filter (Fig. S2b). The filter contains 5 micron wide slits to allow solutions to pass through while retaining beads (Fig. S2c). After collection, the valve separating the two halves of the chip was closed and the chip was cleaved, first by carving a wedge of PDMS out of the device, then placing the SCBC on a foam piece and applying pressure to both sides to prevent delamination of the PDMS device from the glass beneath. With enough pressure, the chip was cleaved along the scored line with each side of the PDMS two-layer device remaining bound to the glass on both sides of the chip.
Proteomics Analysis.
Materials needed:
Antibodies (Table S2).
For the proteomics side of the chip, the device remained at room temperature. The entire half of the chip containing the cell chambers was protected from light for antibody development, incubated with a 1:20 dilution of detection antibodies in a solution of 1.5% BSA in PBS for 60 minutes, and washed with PBST for 30 minutes (Fig. S2b). The chip was imaged in the Genepix 4400 machine with laser power settings of 600 gain and 80% power for the 635 nm wavelength laser and 450 gain and 15% power for the 532 nm wavelength laser. The chip had two fluorescent alignment markers available, one added during X-coordinate ssDNA loading which spanned the entire chip, and another in the P’-Displacement DNA added during lysis. Each barcode strip was measured using three circular regions and mean values were used. Background values were calculated by the average of protein signals from chambers with zero cells for multi-dimensional analysis.
Transcriptomics Analysis.
Materials needed:
DNA oligomers (Table S1), Ampure XP beads (Beckman, Beverly MA), Nextera XT kit, MiSeq sequencing kit (Illumina, San Diego CA)
Reverse transcriptase solution: 2 µL Maxima H- reverse transcriptase, 10 µL reverse transcriptase buffer (Thermo Fisher, Waltham MA), 5 µL dNTP mix (Takara 4030, Mountain View CA), 10 µL 20% Ficoll PM-400, 1 µL RNAse inhibitor, 2 µL 0.1 mM template switch oligo solution (Table S1), 20 µL water.
TE-SDS solution: 10 mM Tris pH 8.0, 1 mM ethylene diaminotetraacetic acid (EDTA), 0.5 % sodium dodecyl sulfate (SDS).
TE-TW solution: 10 mM Tris pH 8.0, 1 mM EDTA, 0.01% Tween-20.
Exonuclease solution: 2.5 µL Exonuclease enzyme, 5 µL Exonuclease buffer (NEB M2093, Ipswich MA), 42.5 µL water.
PCR solution: 100 µL water, 100 µL KAPA HiFi Hotstart Readymix (Sigma Aldrich, St. Louis MO), 10 µL 10 µM SMART primer, 5 µL 10 µM location ssDNA oligomer primer (Table S1)
For the transcriptomics side of the chip, the separation valve was closed after bead collection and chip cleavage (Fig. S1b, valve 11). The microfluidic filter containing the beads was incubated in a reverse transcriptase solution by passing the solution slowly through the filter and incubating at room temperature for 30 minutes, and at 42 °C for 90 minutes (Fig. S2c). Following reverse transcription, the beads and cDNA libraries were washed sequentially with TE-SDS, TE-TW, and 10 mM Tris pH 8.0. Beads were then incubated with exonuclease solution for 45 minutes at 37 °C. Following washes with TE-SDS, TE-TW, and water, the chip was cleaved into approximately a 5 mm x 10 mm piece without allowing the PDMS to separate from the glass.
Following the final chip cleavage, the remainder of the chip containing the microfluidic filter and beads was added to a 2 mL microcentrifuge tube and submerged in PCR solution. Manual PCR was performed by cycling the tube through the following steps: 1. Melt 2 minutes @ 95 °C. 2. Chill 6 seconds in ice bath. 3. Anneal 2 minutes @ 64 °C. 4. Extend 5 minutes @ 72 °C. After 7 cycles, the tube was incubated at 72 °C for 10 minutes and 5 µL of the sample was used for PCR. The manual PCR library was then frozen at −80 °C to preserve for any further sequencing. Library preparation and sequencing proceeded similarly to Dropseq analysis4. Following 17 cycles of PCR in 50 µL of PCR solution, DNA libraries were purified using Ampure XP beads and incubated with 5 µL Tagmentation solution and 10 µL Tagmentation buffer for 5 minutes at 55 °C. The tagmented cDNA libraries were amplified using the Nextera PCR mix for 12 cycles using Nextera N70X indexing primers and P5-SMART primer (Table S1), and the amplified libraries were purified one final time. After Bioanalyzer and Qubit analysis, a 4 nM cDNA library was incubated with an equal volume of 0.2 M NaOH for five minutes to denature, and 10 µL of the denatured library was mixed with 990 µL HT1 buffer. 300 µL of the diluted library was mixed with 300 µL HT1 buffer and added to the Miseq sequencing cartridge, and 4 µL of custom sequencing primer (Custom Read 1 Primer, Table S1) was added to well 12 of the Miseq cartridge. The cDNA library was sequenced on an Illumina MiSeq machine.
Data Analysis and Computation Pipeline.
Materials needed:
R script (File S2), Dropseq tools (http://mccarrolllab.com/dropseq/), STAR alignment (https://github.com/alexdobin/STAR).
Following sequencing, transcripts were aligned and assigned to beads largely as suggested by the Dropseq tools available online. Instead of the typical Dropseq protocol used to correct errors in the bead identifier sequence, we developed an alternative method first to correct errors in the bead identifiers and then to determine the location of origin for each bead. We developed this method to perform the two bead clustering steps: the first by close mismatches in bead identifier sequences and UMIs, and the second by location, which required the formation of an external dictionary to group bead identifier sequences together. This method is detailed in the supplemental R code.
First, the most commonly appearing bead identifiers, either those that collectively account for 66% of the reads or those with more than 0.1% of the total reads sequenced each (Fig. 2a), were extracted and clustered based on their DNA sequences (Fig. 2b). We used the Levenshtein distance metric, which allows substitutions, insertions, and deletions. Alike beads that clustered at Levenshtein distance of 1 or less were assigned a common bead identifier. The remainder of the reads with different bead identifiers were only assigned to one of these common bead identifiers if they were similar in sequence (Fig. 2c).
The location-encoding ssDNA sequences (Table S1) for each of the common bead identifiers were then extracted, checked for sequence similarity, and counted. If the most frequent X or Y ssDNA sequence of a bead identifier comprised more than 60% of the total X or Y ssDNA sequences associated with that bead identifier and at least 10 total location ssDNA sequences were extracted from that bead, then that common bead identifier was considered to have passed filter (Fig. 2d). All beads that passed filter were then grouped into a location-specific bead identifier which was used to collect the reads originating from every bead that shared a common location. The Dropseq pipeline was then applied to this final location-compressed sequencing data.
Single cell and single chamber transcript data was analyzed using the Seurat package26. Additional R code was written to process proteomics data, including background subtraction and normalization to single cell averaged protein measurements for multi-cell chambers. Pairwise gene-protein correlations were performed for every gene-protein pair for each data set, but few correlations were significant given the size of the data set associated with each chip. Transcript data sets for pairwise correlations were processed by choosing only those genes that appeared at least 5 times in at least 1 cell, appeared more than a total of 15 times across all cells on a chip, and had a variance of more than 2. For the aggregated chip data set, following log-normalization, scaling, and variable gene analysis using the default Seurat parameters, the 5 chip data sets and 2 resampling data sets were aggregated and variable genes were chosen by only those variable genes from individual data sets that appeared in each sample, except for one of the HEK data sets which was smaller than the others.
To obtain variable genes for bulk transcriptomic chip analysis, 5 transcriptomic data sets available from the Gene Expression Omnibus (GEO) were selected, GSE79133 (HEK 1), GSM2794663 (U87 1 replicate 1), GSM2794664 (U87 1 replicate 2), GSM2486332 (HEK 2), GSE89164 (HEK 3), and GSM2333485 (U87 2). These were treated as single cells in Seurat, variable genes were extracted, and a separate Seurat analysis was performed using only these genes as measured in the multi-omic SCBC dataset. Further details are available in the R code. Other average bulk transcriptome measurements were obtained from the Human Protein Atlas (www.proteinatlas.org)27.
Bulk Proteomics
To perform bulk proteomics, a DEAL array chip with PDMS wells was incubated with DNA-antibody conjugates for 1 hour at 42 °C. After washing the chip, one million cells of each type were lysed in the multi-omic SCBC lysis buffer or standard SCBC lysis buffer9, 11 for 10 minutes on ice. The lysate was sonicated for 45 seconds, then centrifuged at 14000 rcf for 10 minutes, added to the bulk proteomics chip, and incubated for 1 hour. After 1 hour of incubation with detection antibodies at room temperature and 30 minutes washing in PBST, the PDMS wells were removed and the device was imaged.
Results and Discussion
Multi-Omic Chip Design and Operation
The multi-omic SCBC operating parameters were closely aligned to standard proteomic SCBC chips to maintain near-optimal proteomic measurements. This design choice was dictated by the difference between non-amplified protein signals and amplified transcript signals. However, certain steps were modified for multi-dimensional analysis. First, sequencing beads were 10, and not 30 microns in diameter. Standard Dropseq methods use larger beads4, but smaller beads allow smaller channels to be used, reducing bead settling and clogging issues and facilitating valve sealing. The use of location tagging also alleviates the loss of primers when using smaller beads by allowing multiple beads to be used in each chamber.
The cell lysis and capture step also required a compromise to balance the instability of transcripts and the kinetics of antibody binding of proteins, with protein capture typically requiring more time. The multi-omic SCBC used protein optimized lysis buffer with additional RNAse inhibitor, DTT, and low incubation temperatures to limit transcript degradation. Switching to the modified lysis buffer resulted in similar protein capture compared to the previously reported SCBC lysis buffer9. The modified buffer was important for generating high-quality transcriptome data compared to standard Dropseq buffer, considering the extended lysis time (Fig. S3). There are a variety of other lysis buffers available28 which may achieve similar results. Previous iterations of the protein-only SCBC also used a sonication step that was omitted to prevent shearing of transcripts and the displacement of beads into microfluidic dead space.
The next critical step was bead collection and chip splitting. The gold standards of measurement for different analytes are rarely complementary, thus requiring some degree of separation or splitting, in this case of proteins from transcripts. Previous multi-omic methods required this separation to occur on a cell-by-cell basis, which necessitated the use of either complex microfluidic mixers and valves22 or large dilutions of cellular contents to pipet-friendly volumes20, placing a ceiling on throughput. By capturing a cell’s proteins on an antibody barcode at an X-Y location, and its transcripts on a bead labeled with X-Y location barcodes, we were able to split the contents of a chip collectively at a single step while retaining the correlated measurements of each cell. Another compromise is made at this step, using 6X SSC instead of the typical PBST9 to collect beads and wash unbound analytes from cell chambers.
Separating proteins from transcripts is a crucial step which allows reverse transcription of bound transcripts to proceed at recommended temperatures without destabilizing protein-antibody interactions. Here we chose to intervene at the macro level to split the chip directly, as opposed to collecting the beads through a fluidic outlet and performing PCR off-chip19. This is necessary for analyzing smaller numbers of cells where each cell and bead is essential to the analysis, and allows us to perform micro-scale enzymatic reactions and avoid the loss of beads during transfer off of the chip.
A significant amount of error correction and filtering was applied in digital space. The most common errors observed were single base pair deletions in bead identifiers and high variability in the 12th base pair of the bead identifier. Due to our unorthodox on-chip cDNA generation and manual PCR procedures, we anticipate that insertions and deletions occur more frequently, making Levenshtein distance a more appropriate metric than Hamming distance, which counts the distance between sequences by substitutions only. In our analysis of bead identifiers, we used 11 base pairs, which provides more than enough information diversity for the number of beads analyzed. In fact, the likelihood of duplicate or similar bead identifiers was very low given the small number of beads per chip (several hundred). We apply computationally intensive clustering to only 1–2% of bead identifiers and apply coarser clustering to the remainder of reads to collect all reads efficiently and quickly. After computational analysis, approximately half of the beads are discarded due to ambiguous location assignments.
Single Cell Isolation and Multi-Cell Chambers
We designed experiments to test whether the multi-omic SCBC could successfully capture both proteins and transcripts from single cells. The cell loading and isolation step effectively analyzes a bolus of injected cells, with the bolus size effectively limited by traditional tissue culture and cell purification methods. In principle, far fewer cells are needed as the microfluidic volume of the device is much less than 1 µL. However, we did not seek to optimize the device for highly efficient cell capture and loading. At this proof-of-concept phase, we generated a data set from 5 chips with 39 single cells, 102 total cell chambers and two cell types of different origins, HEK and U87, to provide contrast in transcriptomes. Each SCBC contained a DEAL barcode to assay for the levels of 3 proteins, pyruvate kinase isozyme M2 (PKM2), the c-Myc transcription factor, and pyruvate dehydrogenase kinase (PDHK) 1. These proteins are primarily localized to the cytosol, nucleus, and mitochondria respectively, allowing SCBC analysis to span the range of subcellular compartmentalization. While most microchambers contained 0 or 1 cell, a significant number contained 2 or more cells (Fig. 3a, Fig. S4a). The large ratio of beads to cells (4:1) loaded in the chip ensured that an excess of sequencing beads was available in each chamber.
Figure 3.
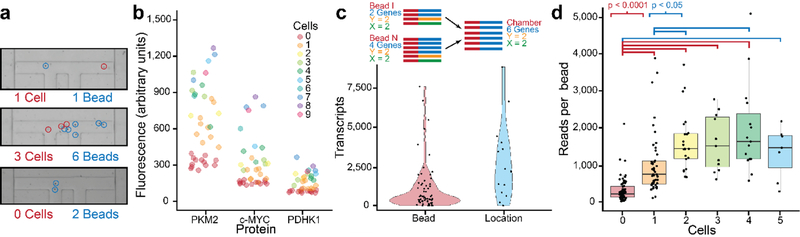
Proteomic and transcriptomic quality metrics. a. Microchambers were stochastically loaded with cells and beads. The density was such that some chambers had no cells, but nearly every chamber had beads. b. Raw protein measurements from a chip show that adding cells increases the protein levels proportionally. Zero-cell chambers represented a clear background. c. Transcriptome measurements were clustered by bead and then by X-Y coordinates. This allowed reads from multiple beads in a chamber to be condensed to give a single chamber transcriptome. d. A plot of reads per bead versus the number of cells in the chamber shows that numbers of reads increases with numbers of cells, up to a few cells, after which the trend flattens.
We first ascertained that the recorded levels of proteins from a given microchamber correlated with the number of cells in that microchamber (Fig. 3b, Fig. S4b). Raw protein values are plotted, showing the background fluorescence and variance of zero-cell chambers. For zero-cell chambers, the background fluorescence values exhibit no dependence on the number of neighboring cells, confirming that there is effective isolation between chambers for proteomic measurements (Fig. S4c). With large numbers of beads or cells, a stringing effect is observed as cellular material attaches to beads, making them difficult to collect and causing clogging, so we focus on microchambers containing 5 or fewer cells here to show the dose-dependence of analyte capture.
Transcript reads were first grouped by the bead identifier sequence specific to each bead. By then matching the bead identifier sequence of each bead with its sequenced X-Y coordinate oligos, we were able to link transcript reads from different beads to a shared microchamber (Fig. 3c). In Fig. 3d we present the numbers of reads per bead for microchambers with varying numbers of cells. Similar to the protein assays, we find a dose-dependence correlation up to about 3 cells per microchamber. There are also a small number of reads found on chambers with no cells, possibly captured during the bead pooling step. This demonstrates that both protein and transcriptome assays can be independently measured and traced back to the cell microchamber where those analytes were released.
Transcriptome Analysis
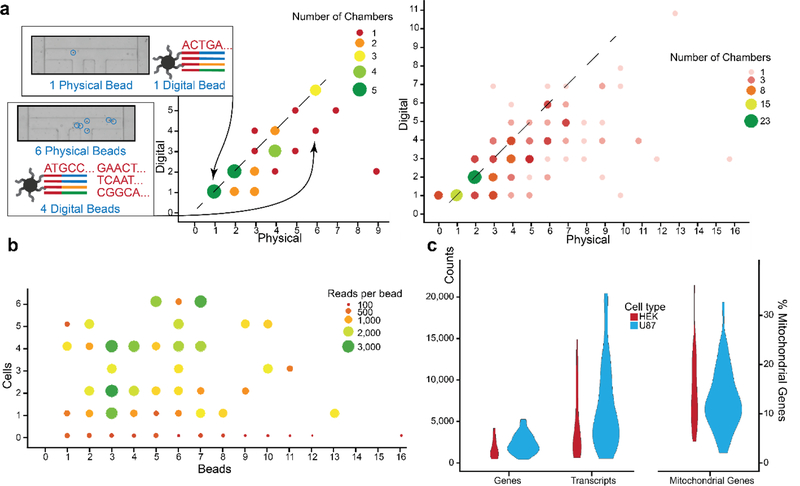
We then tested the quality of transcriptomic data captured from the SCBC as it related to other bead based methods4. We determined that for each chamber, the number of “digital” beads identified by sequencing bead identifiers was similar to the number of “physical” beads observed by microscope analysis (Fig. 4a). For each chamber with observed physical beads, digital beads were also identified, meaning no cell transcriptomes were lost. We observed that beads were lost more frequently when many beads were present, likely due to poor capture rates and clogging. Overall, 68% of physical beads were paired with digital sequencing data, with most unmatched digital beads being from chambers with >5 beads. Only 3.5% of beads were falsely tagged (e.g. 4 digital beads in a chamber with 3 physical beads), with 1% of digital beads being identified in chambers with no physical beads. These discrepancies can occur by poor bead collection, unequal distribution of transcripts across many beads within a chamber, transfer of barcodes between beads during bead collection, and PCR and synthesis errors.
Figure 4.
Bead extraction by sequencing. a. Beads observed by microscope analysis are compared to the digital location map of sequenced beads for one chip (left) and all chips (right). For many chambers, the number of physical beads matches the number of digital beads assigned to that chamber’s location (diagonal line). In some multi-bead chambers, the number of digital sequenced beads is fewer than physical beads, especially as the bead count increases. Rarely are there more digital beads than physical beads, and every chamber with physical beads has at least one digital bead. b. The number of sequencing reads per bead is relatively insensitive to the number of beads in the chamber and the number of cells in the chamber above one. This suggests that there was an excess of transcripts, and that each bead is bound to near-capacity. Some sequencing background was found on all beads. c. U87 cells were sequenced at more depth than HEK cells for genes (U87: mean 2332, CV 52%; HEK: mean 1710, CV 69%;) and UMIs (U87: mean 6729, CV 76%; HEK: mean 4326, CV 96%) detected per cell. Mitochondrial genes were detected at high proportions on SCBC chips (U87: mean 12.8%, CV 48%; HEK: mean 14.5%, CV 52%). Overall, sequencing metrics for the multi-omic SCBC are similar to other high throughput single cell sequencing methods, with higher mitochondrial gene counts.
After cDNA generation, the SCBC protocol diverges from typical Dropseq methods by the manual PCR step. We resampled manual PCR solutions to show that manual PCR-derived transcriptomes were reproducible (Fig. S4d). A general concern among all bead-based transcriptomics methods is the relatively low yield of transcripts per cell. We observed that the amount of reads per bead is insensitive to both the number of beads in a chamber and the number of cells above 1 (Fig. 4b). This suggests that each bead is saturated with transcripts below its theoretical capacity and most transcripts go uncaptured. Hierarchical clustering of the pooled RNA-seq data showed that transcriptome measurements were not clustered by their position on the chip, suggesting that there was little if any crossover between chambers (Fig. S4e).
Transcriptomes generated by the multi-omic SCBC averaged approximately 2000 unique genes and 6000 unique molecules for performance which, although not state-of-the-art for single cell RNA-seq5, is similar to reported Dropseq and related applications2, 29, 30, with relatively high mitochondrial gene levels, likely due to the physical stresses of cell handling during the chip operation (Fig. 4c). Increased mitochondrial gene levels may also be a product of the long lysis times, which allows cytosolic transcripts to degrade and creates a delayed release effect for mitochondrial transcripts. The lysis times are a consequence of the manual operation of the microchip, and could be reduced via automating certain steps.
Cell Type Specific Signatures
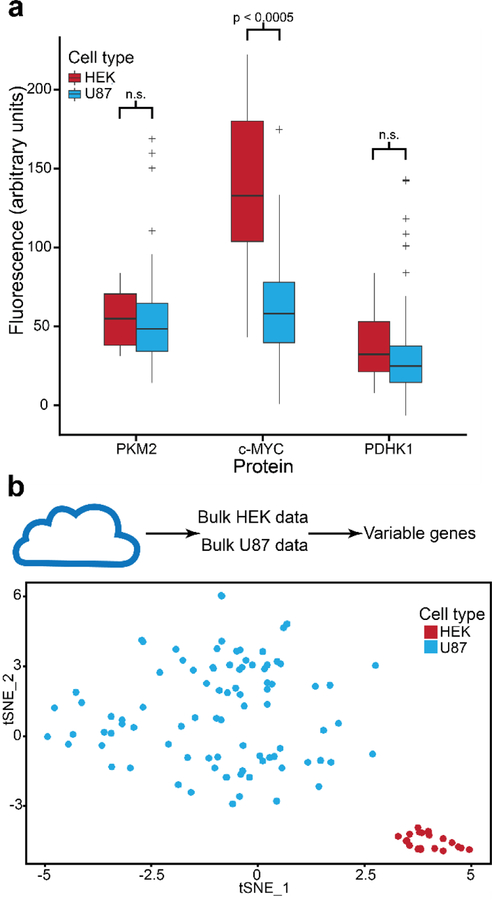
We then tested the platform to determine whether it could resolve the unique biology associated with different cell types. Each cell type measured had a unique protein signature, with higher expression of c-MYC in HEK cells (Fig. 5a). The same relative protein signature is reproduced in bulk proteomic measurements (Fig. S5a). Some variation is seen between same-cell-type chips due to the small number of cells per chip (Fig. S5b).
Figure 5.
Cell type identification with the multi-omic SCBC. a. The aggregated data gathered from chips analyzing HEK and U87 cells showed that each cell type has a specific protein signature (student’s t-test), with increased c-MYC protein signals from HEK cells. b. Using variable genes from public data sets of HEK and U87 cells, SCBC transcriptomes clustered by cell type (t-SNE).
To determine cell type-specific transcriptome signatures, we used public bulk transcriptomic data sets (3 HEK, 2 U87) to select 247 variable genes. We then carried out dimensional reduction of SCBC transcriptomes using these variable genes and we found that the HEK and U87 cells were easily distinguished (Fig. 5b). The coefficient of variation between chips within one cell type is substantially lower than the coefficient of variation between all chips, demonstrating that cell-type specific transcriptome variation is dominant over any batch effects between chips (Fig. S5c–e). Focusing on the subset of chambers containing only single cells, we observe that the cells continue to cluster by cell type with minimal batch effects despite the limited size of the data set (Fig. S5f, g). Using the full transcriptomic gene set, cell-type-specific transcriptome signatures were still present with 148 highly variable genes (Fig. S6a). Additional clustering effects such as mitochondrial gene levels were more readily observed (Fig. S6b).
The measurements of the 3 proteins and their corresponding transcripts were uncorrelated between single cells, but reflected bulk level trends. Public databases show that PKM2 transcripts are expressed at the highest level (HEK: 1104.7 transcripts per million (TPM), U87: 3637.6 TPM), followed by c-MYC (HEK: 175.7 TPM, U87: 59.3 TPM) and PDHK1 (HEK: 10.6 TPM, U87: 70.9 TPM)27, and these relative c-MYC and PDHK1 cell type-specific abundances were reflected in bulk protein data (Fig. S5a). In accordance with these values, we detected PKM2 transcripts at approximately 6 counts per cell (HEK: 298.4 TPM, U87: 1209.5 TPM) and c-MYC at ~0.3 counts per cell (HEK: 0 TPM, U87: 30.9 TPM). Note that for rare transcripts such as PDHK1, which we did not detect in single cell transcriptomes, we were still able to obtain PDHK1 protein measurements. Increasing both the cell count and the numbers of proteins assayed would likely be necessary to reveal significant protein-transcript correlations, and accompanying biological insight (Fig. S6c).
Conclusions
Using the multi-omic SCBC, we demonstrated that single cell proteomic and transcriptomic data sets can be captured in their native measurement medium, using fluorescent sandwich immunoassays for proteins8, 9 and bead based sequencing for transcripts4, 5. These assays could be traced back to a particular location that contained a single microscopically imaged cell, or a defined small colony of cells. Compared to methods that convert mRNA to protein signals or vice versa, our method has certain advantages. First, there is no clear limitation on the types of proteins that are assayed, and post-translational modification detection11 and certain metabolite assays9 should also be possible. Second, the mRNA measurements are not limited by the multiplex ceiling of proteins13, 14, 16. Further, proteins measured as DNA barcodes do not overwhelm the lower abundance transcript measurements17, 18. On the multi-omic SCBC, both measurements were obtained in a “one-pot” measurement step without requiring any manipulation of sub-cellular volumes.
However, there are a number of engineering challenges that need to be addressed to make the platform high-throughput and straightforward to operate. For example, two perpendicularly oriented flow patterning steps would permit X- and Y-location barcodes to be surface patterned, along with the antibody array for protein capture. Within a Cytoseq7 or Seq-well31 format of microwell arrays, such a design should permit a much higher number of cells to be analyzed at the proteomic and transcriptomic level, although new cell lysis chemistries would likely be required. We note that the original SCBC proteomics platform32 required high operator skill and yielded relatively small numbers of analyzed cells with modest protein multiplexing capacity. This compares to the commercial hands-free SCBC platform in which the multiplexing capacity and the numbers of cells and distinct biospecimens analyzed are both dramatically increased. In the near term, the throughput of the modest sized SCBC used here should be increased through scaling the chip design, and further simplification of the workflow, perhaps by removing valve requirements, would improve the dissemination of SCBC methods. With a larger single cell data set, a myriad of bioinformatics tools becomes effective for further analysis including hierarchy inference33, multi-modal analysis26, and deeper phenotypic classification34–36.
Supplementary Material
Acknowledgements
This study was assisted by the facilities of the Center for High Throughput Single Cell Monitoring and Engineering, the Millard and Muriel Jacobs Genetics and Genomics Laboratory, and the Institute for Systems Biology Sequencing Core. Paul Rivaud, Devdoot Majumdar, Sylvia Plevritis, and Lior Pachter contributed helpful discussions. Q.L., K.L.T., and S.J. were participants of the Caltech Summer Undergraduate Research Fellowship program. The research was funded by U54 NSBCC grant 1U54 CA199090–01, the Jean Perkins Foundation, and the Ben and Catherine Ivy Foundation. A.M.X. is supported by a Ruth L. Kirschstein F32 Postdoctoral Fellowship F32CA213966.
Footnotes
Accession Numbers
Transcriptome data was deposited in the Sequence Read Archive under Bioproject PRJNA472815.
Competing Interests
JRH is a founder and board member of Isoplexis, a company that is seeking to commercialize certain aspects of the SCBC platform.
References
- 1.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, Ounadjela JR, Shuga J, Wang X, Lim DA, West JA, Leyrat AA, Kent WJ and Kriegstein AR, Science, 2017, 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A and Sanes JR, Cell, 2016, 166, 1308–1323 e1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A and Garraway LA, Science, 2016, 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N and Martersteck EM, Cell, 2015, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ and Bielas JH, Nat Commun, 2017, 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA and Kirschner MW, Cell, 2015, 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan HC, Fu GK and Fodor SP, Science, 2015, 347, 1258367. [DOI] [PubMed] [Google Scholar]
- 8.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A and Heath JR, Nat Med, 2011, 17, 738–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue M, Wei W, Su Y, Kim J, Shin YS, Mai WX, Nathanson DA and Heath JR, Journal of the American Chemical Society, 2015, 137, 4066–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD and Nolan GP, Science, 2011, 332, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, Ikegami S, Gu Y, Herrmann K, Johnson D, Ding X, Hwang K, Kim J, Zhou J, Su Y, Li X, Bonetti B, Chopra R, James CD, Cavenee WK, Cloughesy TF, Mischel PS, Heath JR and Gini B, Cancer Cell, 2016, 29, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendall SC and Nolan GP, Nat Biotechnol, 2012, 30, 639–647. [DOI] [PubMed] [Google Scholar]
- 13.Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP and Gherardini PF, Nat Methods, 2016, 13, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genshaft AS, Li S, Gallant CJ, Darmanis S, Prakadan SM, Ziegler CG, Lundberg M, Fredriksson S, Hong J, Regev A, Livak KJ, Landegren U and Shalek AK, Genome Biol, 2016, 17, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Buggenum JA, Gerlach JP, Eising S, Schoonen L, van Eijl RA, Tanis SE, Hogeweg M, Hubner NC, van Hest JC, Bonger KM and Mulder KW, Sci Rep, 2016, 6, 22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrigucci R, Bushkin Y, Radford F, Lakehal K, Vir P, Pine R, Martin D, Sugarman J, Zhao Y, Yap GS, Lardizabal AA, Tyagi S and Gennaro ML, Nat Protoc, 2017, 12, 1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert P, Nat Methods, 2017, 14, 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S and Klappenbach JA, Nat Biotechnol, 2017, 35, 936–939. [DOI] [PubMed] [Google Scholar]
- 19.Shahi P, Kim SC, Haliburton JR, Gartner ZJ and Abate AR, Sci Rep, 2017, 7, 44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albayrak C, Jordi CA, Zechner C, Lin J, Bichsel CA, Khammash M and Tay S, Mol Cell, 2016, 61, 914–924. [DOI] [PubMed] [Google Scholar]
- 21.Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, Fredriksson S, Assarsson E, Lundberg M, Nelander S, Westermark B and Landegren U, Cell Rep, 2016, 14, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han L, Zi X, Garmire LX, Wu Y, Weissman SM, Pan X and Fan R, Scientific Reports, 2014, 4, 6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eng C-HL, Shah S, Thomassie J and Cai L, Nature Methods, 2017, 14, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin R, Feng Q, Li P, Zhou P, Wang R, Liu Z, Wang Z, Qi X, Tang N, Shao F and Luo M, Nature Methods, 2018, 15, 275. [DOI] [PubMed] [Google Scholar]
- 25.Bailey RC, Kwong GA, Radu CG, Witte ON and Heath JR, J Am Chem Soc, 2007, 129, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler A, Hoffman P, Smibert P, Papalexi E and Satija R, Nature Biotechnology, 2018, 36, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J and Pontén F, Science, 2015, 347. [Google Scholar]
- 28.Ståhlberg A, Thomsen C, Ruff D and Åman P, Clinical Chemistry, 2012, 58, 1682–1691. [DOI] [PubMed] [Google Scholar]
- 29.Stephenson W, Donlin LT, Butler A, Rozo C, Bracken B, Rashidfarrokhi A, Goodman SM, Ivashkiv LB, Bykerk VP, Orange DE, Darnell RB, Swerdlow HP and Satija R, Nature Communications, 2018, 9, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickel DE, Ypsilanti AR, Pla R, Zhu Y, Barozzi I, Mannion BJ, Khin YS, Fukuda-Yuzawa Y, Plajzer-Frick I, Pickle CS, Lee EA, Harrington AN, Pham QT, Garvin TH, Kato M, Osterwalder M, Akiyama JA, Afzal V, Rubenstein JLR, Pennacchio LA and Visel A, Cell, 2018, 172, 491–499.e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gierahn TM, Wadsworth Ii MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC and Shalek AK, Nature Methods, 2017, 14, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin YS, Ahmad H, Shi Q, Kim H, Pascal TA, Fan R, Goddard WA 3rd and Heath JR, Chemphyschem, 2010, 11, 3063–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anchang B, Hart TDP, Bendall SC, Qiu P, Bjornson Z, Linderman M, Nolan GP and Plevritis SK, Nat. Protocols, 2016, 11, 1264–1279. [DOI] [PubMed] [Google Scholar]
- 34.Haghverdi L, Lun ATL, Morgan MD and Marioni JC, Nature Biotechnology, 2018, 36, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiselev VY, Yiu A and Hemberg M, Nature Methods, 2018, 15, 359. [DOI] [PubMed] [Google Scholar]
- 36.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA and Trapnell C, Nat Methods, 2017, 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.