Abstract
Mass spectrometry (MS)-based top-down proteomics characterizes complex proteomes at the intact proteoform level and provides an accurate picture of protein isoforms and protein post-translational modifications in the cell. The progress of top-down proteomics requires novel analytical tools with high peak capacity for proteoform separation and high sensitivity for proteoform detection. The requirements have made capillary zone electrophoresis (CZE)-MS an attractive approach for advancing large-scale top-down proteomics. CZE has achieved a peak capacity of 300 for separation of complex proteoform mixtures. CZE-MS has shown drastically better sensitivity than commonly used reversed-phase liquid chromatography (RPLC)-MS for proteoform detection. The advanced CZE-MS identified 6,000 proteoforms of nearly 1,000 proteoform families from a complex proteome sample, which represents one of the largest top-down proteomic datasets so far. In this review, we focus on the recent progress in CZE-MS-based top-down proteomics and provide our perspectives about its future directions.
Keywords: capillary zone electrophoresis, mass spectrometry, top-down proteomics, proteoform, post-translational modification, protein complex
1. Introduction
Top-down proteomics characterizes proteoforms in the cell. The word “proteoform” is used to represent all the protein species from the same gene due to gene-level variations, RNA-level alternative splicing, and protein-level post-translational modifications (PTMs) [1]. Another word “proteoform family” represents a group of proteoforms derived from the same gene [2]. Different proteoforms from the same gene can have drastically different functions [3–6]. We need to characterize proteomes in a proteoform-specific manner to understand protein function precisely.
Nanoflow reversed-phase liquid chromatography (nanoRPLC)-electrospray ionization (ESI)-mass spectrometry (MS) is typically employed for top-down proteomics. The advantages of RPLC include large sample loading capacity, wide separation windows, and good solvent compatibility with ESI-MS. The top-down proteomics community has made great effort on improving RPLC for proteoform separation [7–16]. The state-of-the-art RPLC-MS-based systems have achieved the identification of 3,000–5,000 proteoforms and about 1,000 proteoform families [7,8]. The proteome samples can be extremely complex, and it has been estimated that the human proteome contains over 1 million proteoforms [17]. Much better liquid-phase separation of proteoforms is vital for better proteome coverage with top-down proteomics.
Capillary zone electrophoresis (CZE)-MS has been suggested as an alternative approach for top-down proteomics. CZE employs a fused silica open tubular capillary for separation of analytes under an electric field based on their electrophoretic mobility that relates to analytes’ sizes and charges [18]. The typical inner diameter (i.d.) of the fused silica capillary used for CZE is in a range of 10–75 μm; the typical length of the capillary ranges from 20 to 100 cm. CZE-MS has several valuable features for top-down proteomics. First, CZE can reach a highly efficient separation of large biomolecules, e.g., proteoforms. As shown in equation (1),
| (1) |
the number of theoretical plates from CZE (N) only relates to the electrophoretic mobility of analytes (μ), the voltage applied across the capillary (V), and the analytes’ diffusion coefficient (D). Large biomolecules like proteoforms usually have low diffusion coefficients in solution, eventually leading to high separation efficiency in CZE. Our most recent data showed that CZE had up to one million theoretical plates for separation of proteoforms [19]. Second, CZE-MS has extremely high sensitivity for top-down characterization of proteins. In 1996, the McLafferty group achieved the detection of attomole amounts of intact proteins (less than 1 picogram in mass) using CZE-MS [20]. The Yates group has reported that CZE-MS approached comparable signal-to-noise ratios (S/N) to the widely used nanoRPLC-MS for characterization of intact proteins with 100-fold less sample consumption [21]. Third, CZE has the capability for high-resolution separation of protein complexes under native conditions [22]. This feature is unique to CZE and makes CZE-MS valuable for native top-down proteomics that aims to characterize endogenous protein complexes in the cell at a proteome-scale and in discovery mode [23,24]. Fourth, CZE separates analytes based on their sizes and charges; RPLC separates analytes based on their hydrophobicity. They are well-suited orthogonal techniques for proteoform separation. Coupling RPLC with CZE will drastically improve the peak capacity for proteoform separation.
Wide application of CZE-MS for large-scale top-down proteomics has been impeded by multiple factors. First, the stability and sensitivity of CE-MS interface have been major obstacles. Second, the separation window and sample loading capacity of CZE have been at least 10-folds narrower and 100-folds lower than that of RPLC-MS, respectively, which hampered the adoption of CZE-MS in large-scale top-down proteomics.
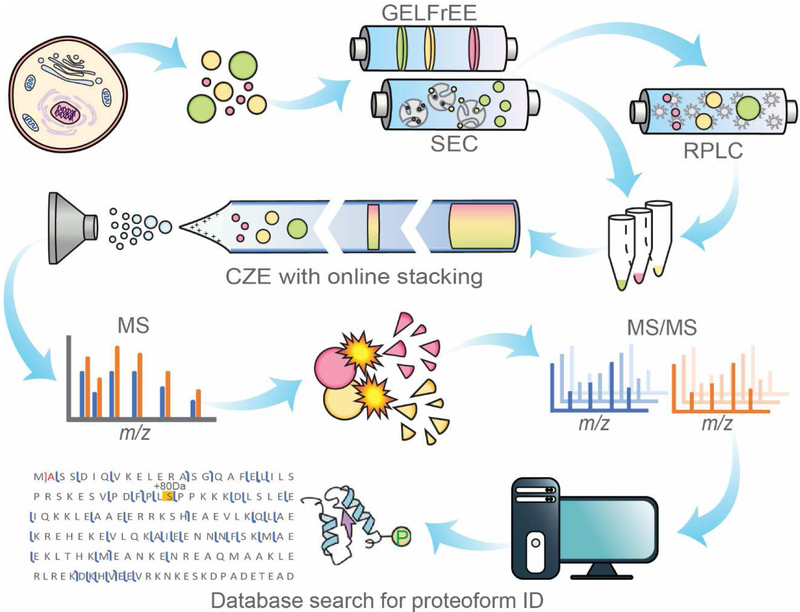
In last decade, the CE-MS interface as well as the sample loading capacity and separation window of CZE have been improved drastically. The advanced CZE-MS has become an attractive approach for large-scale top-down proteomics. Figure 1 shows the typical workflow of CZE-MS-based top-down proteomics. Complex proteoform mixtures extracted from cells are usually fractionated by their sizes first with gel eluted liquid fraction entrapment electrophoresis (GELFrEE) [7] or size exclusion chromatography (SEC). The S/N of a protein from MS decreases obviously with the increase of the protein mass [25]. It is essential to separate large proteoforms from small ones to advance the characterization of large proteoforms. The fractionated proteoforms can be further fractionated by their hydrophobicity using RPLC before CZE-MS analysis or can be analyzed by CZE-MS directly. Online sample stacking is typically utilized in CZE-MS to increase the sample loading capacity of CZE for more proteoform identifications (IDs). After MS analysis, the proteoforms can be isolated for fragmentation to achieve extensive backbone cleavages and generate sequence-informative fragments. Proteoforms are identified through a database search that matches the experimental masses of proteoforms and their fragments with the theoretical values derived from a proteome database. For instance, as shown in Fig. 1, one proteoform with one phosphorylation site (+80 Da) and N-terminal methionine removal was identified. Several bioinformatics tools are available for performing the database search, including but not limited to ProSight [26], TopPIC [27], Proteoform Suite [28], MASH Suite [29], and pTop [30].
Fig. 1.
Diagram of the typical workflow of top-down proteomics using CZE-MS/MS.
Several reviews about CZE-MS for bottom-up proteomics and characterization of intact proteins have been published [31–34]. In this review, we focus on the more recent progress of CZE-MS for top-down proteomics of complex proteomes. First, we summarized the technical improvement of CZE-MS for top-down proteomics regarding the CE-MS interface as well as sample loading capacity and separation window of CZE. Second, we discussed the online or offline coupling of other liquid-phase separation methods with CZE-MS for top-down proteomics. Third, we outlined the adoption of various gas-phase fragmentation methods in CZE-MS-based top-down proteomics. Fourth, we discussed CZE-MS for native top-down proteomics. In the end, we provided our conclusions and future perspectives in the field.
2. Advancing the CE-MS interface as well as sample loading capacity and separation window of CZE for top-down proteomics
2.1. CE-MS interface
ESI is the most popular ionization method for CZE-MS-based proteomics research. CE-MS requires an interface that can complete the electrical circuit for CE separation and provide voltage for ESI. Starting from the pioneering work of the Smith group in 1988 about the coaxial sheath-flow CE-MS interface [35], development of new CE-MS interfaces with higher sensitivity and stability has remained as a major trend in the CE-MS field. The Chen group and Mayboroda group performed comprehensive reviews of the CE-MS interface in 2008 and 2012 [36,37]. Here, we briefly discussed the most recent progress of CE-MS interface.
The sensitivity of sheath-flow interfaces could be improved by reducing the flow rate of sheath liquid. The Chen group developed a junction-at-the-tip type CE-MS interface in 2010 [38]. The design allows the sheath liquid solution driven by pressure flow through at a much lower flow rate compared to the coaxial sheath-flow CE-MS interface (nL/min vs. μ/min), leading to significantly higher sensitivity and a stable spray [38]. The junction-at-the-tip interface was noted as beneficial for its ease in providing stable spray in ESI. The interface-based CE-MS system has been applied for the analysis of proteins in several studies [39–41]. The Dovichi group reported the electro-kinetically pumped sheath flow interface in 2010 [42], and improved it further in 2013 and 2015 [43,44]. Figure 2 shows diagrams of the basic interface and its different generations. High potential applied in the sheath buffer reservoir produces electroosmotic flow (EOF) in the glass emitter, which pumps sheath liquid at nL/min flow rates through the emitter for ESI, leading to extremely high sensitivity [43]. Larger ESI emitter orifice and shorter distance between the capillary end and emitter orifice improve the robustness and sensitivity of the CE-MS interface. The improved electro-kinetically pumped sheath flow interface has been commercialized by CMP Scientific (http://www.cmpscientific.com). The interface-based CZE-MS has been used for bottom-up proteomics in many studies, and the topic has been well reviewed recently [31]. The Nemes group reported a tapered-tip metal emitter-based sheath-flow CE-MS interface with a low flow rate of sheath liquid (nL/min) [45]. They reported a 260-zmol limit of detection for angiotensin II using the interface-based CZE-MS system.
Fig. 2.
Diagrams of the basic design of the electrokinetically pumped sheath flow CE-MS interface (A) and its three different generations (B). Reproduced with permission [44]. Larger emitter orifice and shorter distance between the capillary end and emitter orifice improve the robustness and sensitivity of the CE-MS interface.
The potential drawback of sheath-flow CE-MS interfaces is significant sample dilution by the sheath liquid, which decreases the sensitivity of detection. To eliminate this effect, the Moini group developed a sheathless CE-MS interface using a porous capillary end as ESI emitter in 2007 [46]. The major benefit of the sheathless interfaces is the elimination of sample dilution by sheath liquid, thus leading to high sensitivity. The sheathless interface has been commercialized by Sciex and is used in the CESI 8000 and 8000 plus systems. The sheathless interface-based CZE-MS has been applied for top-down characterization of simple protein mixtures and complex proteomes [21,22,47–50].
2.2. Separation window of CZE
CZE typically employs a regular fused silica capillary for separation and the inner wall of the capillary is covered with silanol groups. When a high voltage is applied across the capillary filled with background electrolyte (BGE), an EOF will be produced inside the capillary due to the negatively charged inner wall and will push the analytes out of the capillary for detection quickly. Therefore, CE separation is typically fast with a separation window in a range of 1 to 30 min [43,45,51]. This feature makes CZE-MS attractive for high throughput analysis of relatively simple samples. However, the narrow separation window of CZE limits the number of MS/MS spectra that can be acquired during one run, leading to unsatisfied performance of CZE-MS for large-scale top-down proteomics. Boosting the separation window of CZE is crucial. The inner wall properties of fused silica capillaries could have a big impact on the separation window.
Several kinds of neutral and hydrophilic coatings, e.g., linear polyacrylamide (LPA) and hydroxypropyl cellulose (HPC), have been utilized to cover the capillary inner wall and eliminate the EOF in the CZE capillary, leading to wider separation windows. The neutral coatings could also help to suppress the protein adsorption on the capillary inner wall. The LPA coating is the most widely used neutral coating for CE-MS-based top-down proteomic studies. The preparation of the LPA coating has been well reviewed recently by the Dovichi group [31]. We recently published a detailed protocol about making the LPA-coated capillary for top-down proteomics [52] and the protocol is based on a procedure developed by the Dovichi group [53] with some modifications. We showed that the CZE-MS system with a LPA-coated capillary produced a 90-min separation window and a high peak capacity of nearly 300 for top-down proteomics of an Escherichia coli (E. coli) sample [54]. The separation window of the CZE-MS system is drastically wider than that of typical CZE-MS systems with uncoated capillaries [43,45,51].
Cationic coatings have also been used to coat the inner wall of the capillary for top-down proteomics [47,49,55,56]. In this case, the capillary inner wall has rich positive charges and these positive charges reduce the protein adsorption on the capillary inner wall because the proteins are also positively charged in an acidic BGE. After applying a negative potential across the capillary, the EOF towards the ESI tip will be generated. Proteins will migrate to the inlet of the capillary, but meanwhile will be pushed to the outlet of the capillary by the EOF [49,55,56]. Therefore, the migration rate of proteins can be abated, resulting in a wider separation window. However, the improvement of separation window using capillaries with cationic coatings is modest because of the strong EOF inside of the capillary.
Besides the covalently attached coatings described above, multiple-layer coatings that employ the successive multiple ionic-polymer layers (SMIL) have also been used to modify the capillary inner wall for protein separation with CZE [57,58]. However, more studies are needed to investigate the performance of the multiple-layer coatings for CZE-MS-based large-scale top-down proteomics.
2.3. Sample loading capacity of CZE
CZE employs an open-tubular capillary for separation without stationary phase meaning that the analytes cannot be trapped at the front end of the separation capillary like in RPLC, which results in a low loading capacity of CZE. The typical sample loading volume is less than 1% of the total capillary volume to obtain high separation efficiency. For a 1-meter-long capillary with a 50-μm i.d., the total capillary volume is about 2 μL, and the sample loading volume needs to be only 20 nL or lower. Sample loading volumes in CZE are orders of magnitude lower than that in RPLC, which makes the detection of low-abundance proteoforms in a complex sample challenging. The use of online sample preconcentration/stacking methods could help to improve the sample loading capacity.
Several preconcentration methods have been applied in CZE-MS-based top-down proteomics, including field-amplified sample stacking (FASS), transient isotachophoresis (tITP), and dynamic pH junction. In the following several paragraphs, we discussed the basic principles of the three online sample preconcentration methods and some top-down proteomic applications using these methods. We note that several other preconcentration methods have also been used in CZE for various applications [59].
FASS is a simple technique for sample stacking based on the idea that sample ions experience a dramatic decrease in velocity when migrating through a low-conductivity sample plug into a high-conductivity BGE zone and are stacked at the boundary between the sample and BGE zones. The addition of organic solvents, e.g., acetonitrile (ACN), in the sample buffer for lowering the conductivity of the sample zone is an efficient way to perform FASS [60,61].
Zhao et al. used FASS and an LPA-coated capillary for CZE-MS analysis of reduced monoclonal antibodies (mAbs) [61]. Reduced mAbs were dissolved in 35% (v/v) acetic acid with 50% (v/v) ACN and separated in a 5% (v/v) acetic acid BGE. The sample loading volume was 70 nL (approximately 7% of total capillary volume). The heavy chains and light chains of a two-antibody mixture were separated in 15 min. In addition, Zhao et al. also employed a 70% (v/v) acetic acid buffer as the sample buffer, which had much lower conductivity than the BGE that was 0.25% (v/v) formic acid, for top-down proteomics of a Mycobacterium marinum secretome [62]. About 120-nL protein sample corresponding to 12% of the total capillary volume was injected into an LPA-coated capillary for CZE-MS analysis in 60 min, leading to the identification of 22 proteoform families and 58 proteoforms from the secretome sample.
tITP requires the presence of a leading electrolyte (LE) and a terminating electrolyte (TE), whose electrophoretic mobility is higher and lower than the sample ions. At the beginning of a CZE separation, a plug of sample dissolved in a leading electrolyte (LE) and a plug of a terminating electrolyte (TE) are sequentially introduced into the capillary. After a voltage is applied, sample ions between LE and TE are arranged in the order of their mobility and are concentrated to achieve the same migrating velocity towards the outlet of the capillary. With tITP preconcentration, Li et al. were able to boost the sample loading volume of CZE to approximately 15% of the total capillary volume and identified 65 proteins in a top-down proteomic study of a Pseudomonas aeruginosa PA01 lysate using CZE-MS [55]. Han et al. employed RPLC fractionation and tITP-CZE-MS for top-down proteomics of Pyrococcus furiosus, resulting in the identification of 134 proteoform families and 291 proteoforms with the consumption of 270 ng of proteins [49]. tITP allowed an injection of a 13-cm-long sample plug into a 90-cm-long separation capillary with a cationic coating for CZE separation. Han et al. further applied the tITP-CZE-MS system for top-down characterization of subunits of a recombinant Dam1 complex [21]. The CZE-MS system enabled precise characterizations of the protein complex using only 2.5-ng protein material and showed 100-fold better sensitivity than nanoRPLC-MS regarding the mass of consumed protein material.
Dynamic pH junction is also a widely utilized stacking technique in top-down CZE-MS studies. A simplified diagram of the dynamic pH junction method with a neutrally coated capillary is shown in Fig. 3A. The sample is usually dissolved in a basic buffer (i.e., ammonium bicarbonate, pH 8) and is injected into the separation capillary filled with an acidic BGE (i.e., 5%(v/v) acetic acid, pH 2.4). Both ends of the capillary are then immersed in the BGE vials and two pH boundaries form in the capillary. The analytes in the basic sample zone mostly have negative charges. After applying a positive potential at the injection end of the capillary for separation, the hydrogen protons start to titrate the basic sample zone gradually, and the pH boundary I starts to move towards the pH boundary II.
Fig. 3.
A simplified diagram of the dynamic pH junction method with a neutrally coated capillary (A) and a total ion current (TIC) electropherogram of a zebrafish brain sample after CZE-MS/MS analysis with a 1.5-meter-long LPA-coated capillary and the dynamic pH junction method [19] (B).
Meanwhile, the negatively charged analytes migrate towards the moving pH boundary I and are concentrated there eventually. Once the moving pH boundary I meets with the static pH boundary II, the analytes undergo a normal CZE separation. The dynamic pH junction method was invented by the Chen group in 2000 [63], and is a highly efficient method for online concentration of analytes, enabling the focusing of at least 95% of analytes injected into the capillary [64].
In 2014, the dynamic pH junction-based CZE-MS was investigated for bottom-up proteomics for the first time by the Dovichi group [65]. Our group systematically optimized the conditions of the dynamic pH junction-based CZE-MS for bottom-up proteomics in 2017 [66]. The optimized CZE-MS system approached a 140-min separation window and a microliter scale sample loading volume, opening the door of large-scale bottom-up proteomics using CZE-MS. More recently, we applied the dynamic pH junction-based CZE-MS system for deep and highly sensitive bottom-up proteomics of various complex proteome samples as well as for phosphoproteomics of human cancer cells [67–69].
We also systematically optimized the dynamic pH junction-based CZE-MS for large-scale top-down proteomics for the first time in 2017 [54]. The optimized CZE-MS system employed an LPA-coated capillary (50-μm i.d.), a 5% or 10% (v/v) acetic acid buffer (pH 2.4 or 2.2) as the BGE, and a 50-mM ammonium bicarbonate buffer (pH 8) as the sample buffer. We observed that the optimized dynamic pH junction method outperformed the FASS method for online concentration of proteins when the sample injection volume was higher than 5% of the total capillary volume. The optimized CZE-MS system reached a highly efficient separation of proteins even with a 1-μL sample injection into a 1-meter-long LPA coated capillary (~50% of the total capillary volume). The CZE-MS/MS system identified nearly 600 proteoforms in a single run using a Q-Exactive HF mass spectrometer (resolution as 120,000 at m/z 200). The number of proteoform IDs per CZE-MS/MS run was over three times higher than that reported in previous studies [49,55]. The single-shot CZE-MS/MS data was comparable to the top-down proteomics data reported by the Dovichi group using RPLC-CZE-MS/MS regarding the number of proteoform IDs [70]. The 600 proteoform IDs from a single CZE-MS/MS run is comparable to the data from a single RPLC-MS/MS run with a 21 T Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (resolution as 150,000–300,000 at m/z 400) [14]. The optimized dynamic pH junction-based CZE-MS/MS system has laid the foundation of large-scale top-down proteomics using CZE-MS/MS.
More recently, we further improved the dynamic pH junction based CZE-MS system using a 1.5-meter-long LPA coated capillary and achieved a 2-μL sample loading volume for top-down proteomics [19]. The CZE-MS/MS system produced a 180-min separation window for the analysis of a zebrafish brain sample and identified nearly 2,000 proteoforms in a single run, Fig. 3B. More importantly, these single-shot analyses only consumed hundreds of nanograms of proteins per run, which highlighted the potential of the CZE-MS/MS system for large-scale top-down proteomics of mass-limited samples, e.g., laser capture microdissected tissue samples and even single cells.
3. Coupling LC/electrophoresis prefractionation to CZE-MS for large-scale top-down proteomics
The advanced CZE-MS/MS itself has shown its capability for large-scale top-down proteomics. However, to improve the proteome coverage, multidimensional (MD) liquid-phase separation before MS and MS/MS analysis is typically required [71]. Various LC or electrophoresis methods have been coupled to CZE-MS offline or online to boost the peak capacity for proteoform separation, enabling large-scale top-down proteomic analysis of complex proteomes.
3.1. Offline LC/electrophoresis prefractionation-CZE-MS
The offline MD separations have simple system setup and can fully utilize the separation power of each dimension. RPLC, SEC and GELFrEE have been coupled to CZE-MS offline for top-down proteomics.
Coupling RPLC prefractionation to CZE-MS is one efficient approach. RPLC and CZE are orthogonal for proteoform separation. RPLC can desalt the proteoform samples and the RPLC eluates can be directly analyzed by CZE-MS after simple lyophilization and redissolution steps. Han et al. identified 291 proteoforms of 134 proteoform families from a Pyrococcus furiosus proteome using RPLC fractionation and CZE-MS/MS with a total sample consumption of 270 ng [49]. Zhao et al. coupled high-resolution RPLC fractionation to CZE-MS/MS for top-down proteomics of yeast cells and identified 580 proteoforms of 180 proteoform families [70]. The peak capacities of the two RPLC-CZE systems discussed above were estimated as about 500 and 800, respectively.
Size-based separation methods, e.g., SEC and GELFrEE, have also been coupled to CZE-MS/MS offline for top-down proteomics to improve the identification of large proteoforms. Li et al. identified 30 proteins with molecular weights higher than 30 kDa from a Pseudomonas aeruginosa PA01 cell lysate via coupling GELFrEE to CZE-MS/MS [55]. We coupled SEC fractionation to CZE-MS/MS for large-scale top-down proteomics of E. coli cells and identified 3,028 proteoforms of 387 proteoform families [72]. We identified 325 and 30 proteoforms larger than 20 kDa and 30 kDa, respectively. The SEC-CZE-MS/MS enabled the identification of proteoforms with N-terminal methionine removal, N-terminal truncations, signal peptide cleavage, and various PTMs including protein acetylation, methylation, S-thiolation, disulfide bonds, and lysine succinylation. For example, we detected two kinds of S-thiolation (S-glutathionylation and S-cysteinylation) for some E. coli proteins, and determined the relative abundance of the proteoforms from the same gene but with different S-thiolation PTMs, leading to a conclusion that E. coli cells cultured in the LB medium preferentially employed S-glutathionylation as a mechanism for thiol protection. The conclusion agreed well with an earlier top-down proteomic study [11].
Our group also coupled SEC-RPLC prefractionation to CZE-MS/MS for deep top-down proteomics of E. coli cells [73], Fig. 4. The SEC-RPLC-CZE platform achieved a high peak capacity of nearly 4,000 for proteoform separation. Nearly 6,000 proteoforms and 850 proteoform families were identified using the SEC-RPLC-CZE-MS/MS platform, and the data represents one of the largest top-down proteomic datasets reported so far. This study identified 52 proteoforms with molecular weights in a range of 30–52 kDa.
Fig. 4.
The SEC-RPLC-CZE-MS/MS system for large-scale top-down proteomics of E. coli cells [73]. Proteoforms extracted from E. coli cells were fractionated by SEC to 5 fractions. Each SEC fraction was further separated by RPLC into 20 fractions. All the fractions were analyzed by CZE-MS/MS and the data were processed by the TopPIC (Top-down mass spectrometry based Proteoform Identification and Characterization) software [27] for proteoform IDs. Reproduced with permission [73].
3.2. Online LC/CE-CZE-MS
Compared with the offline approach, the online approach can minimize the sample loss during sample transfer and increase throughput [74]. Various online LC/CE-CZE-MS systems were developed for proteomics more than 15 years ago [75–77].
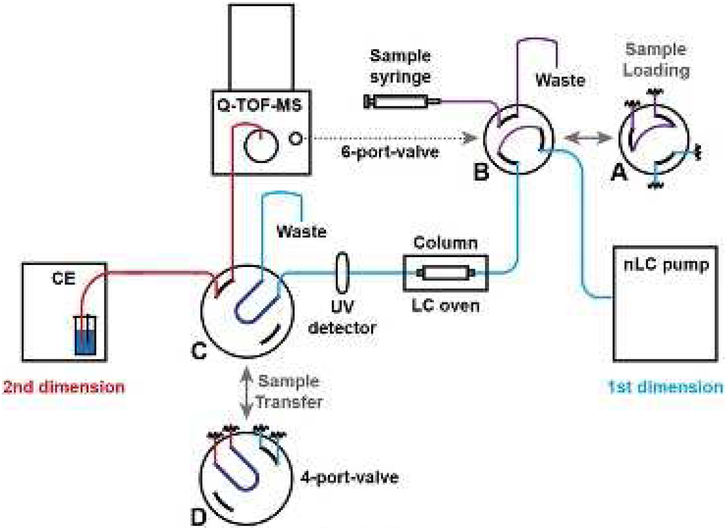
The Neusüß group recently online hyphenated three separation techniques to CZE-MS using a mechanical valve as the interface for top-down proteomics [78–80]. In a very recent study, the Neusüß group developed an online nanoRPLC-CZE-MS system using a 4-port valve for top-down characterization of intact proteins [80], Fig. 5. A protein mixture was first separated by the nanoRPLC and selected protein eluates were transferred into a 20-nL sample loop integrated in a 4-port valve. The protein eluate in the sample loop was further transferred to the CZE separation capillary through switching the 4-port valve and analyzed by CZE-MS. Four model proteins were separated and various glycoforms were identified with the online system. Their work demonstrated the capability of intact protein characterization via the online LC-CZE-MS system. However, more studies are certainly essential to evaluate the performance of the online system for top-down proteomics of complex proteomes.
Fig. 5.
Schematic diagram of an online nanoRPLC-CZE-MS system [80]. Reproduced with permission [80].
4. CZE-MS with various gas-phase fragmentation methods for top-down proteomics
Coupling LC fractionation to CZE-MS/MS has enabled the identification of 6,000 proteoforms from E. coli cells [73]. However, the comprehensive characterization of these proteoforms is still challenging because of the lack of extensive gas-phase fragmentation of proteoforms for accurate localization of PTMs. New fragmentation methods with better fragmentation of proteoforms are crucial.
Collision-based dissociation methods like collision-induced dissociation (CID) and higher-energy collisional dissociation (HCD) are universally used in the fragmentation of proteoforms [7,8,11,12,19,49]. The stepped HCD that combines fragment ions from several different HCD energies has also been evaluated for bottom-up proteomics and showed promising performance for enhancing sequence coverage of peptides [81]. However, CID and HCD have preferential cleavage of the most labile bonds, limiting the sequence coverage and labile PTM localization [82]. We recently employed CZE-MS/MS with HCD for large-scale top-down proteomics of E. coli cells and zebrafish brains, resulting in thousands of proteoform IDs [19]. However, for a large portion of the identified proteoforms, we cannot accurately localize the PTMs due to limited fragmentation coverage across proteoform backbones. Alternative gas-phase fragmentation methods are therefore vital for top-down proteomics moving forward.
Electron-based activation methods like electron transfer dissociation (ETD) [83] and activated ion ETD (AI-ETD) [84,85] are attractive alternatives to CID and HCD for proteoform fragmentation. AI-ETD is an improved ETD technique to minimize nondissociative electron transfer dissociation (ETnoD). Our group recently performed large-scale top-down proteomics of E. coli cells using SEC-CZE-MS/MS with AI-ETD on an Orbitrap Fusion Lumos mass spectrometer [72]. CZE-AI-ETD outperformed CZE-ETD and CZE-HCD regarding the number of proteoform IDs and expectation values (E-Values) of identified proteoforms. E-Values represent a non-linear transformation of numbers of fragment ions from proteoforms and are used to evaluate the confidence of proteoform IDs. The lower the E-value, the more confident the identification is. The data indicated that AI-ETD generated better proteoform fragmentation and higher quality of MS/MS compared to ETD and HCD. Our SEC-CZE-AI-ETD system identified 3,028 proteoforms and 387 proteoform families from E. coli cells. The data represents the largest top-down proteomic dataset with AI-ETD fragmentation.
Ultraviolet photodissociation (UVPD) has also been well recognized for enhancing proteoform fragmentation [86–88]. Because of the high energy of the photons (typically 193 nm), the activated proteins can have different pathways for fragmentation. Therefore, UVPD can produce a variety of fragment ions (a-, b-, c-, x-, y-, z-type ions), and obtain better protein fragmentation coverage compared to CID, HCD, and ETD. Near 100% backbone cleavage of a 29-kDa protein was achieved using UVPD with the 193 nm laser [87]. Very recently, we performed large-scale top-down proteomics of a zebrafish brain sample using SEC-CZE-MS/MS with UVPD (a 213-nm laser) on an Orbitrap Fusion Lumos mass spectrometer [89]. We identified 600 proteoforms and 369 proteoform families from the zebrafish brain sample. The work is the first example of CZE-UVPD for top-down proteomics. UVPD with 213-nm photons achieved good gas-phase fragmentation of proteoforms. For instance, 75% backbone cleavages were obtained for a 12-kDa protein.
Combination of different fragmentation methods has also been demonstrated as a valuable approach for extensive proteoform fragmentation. Zhao et al. showed that the combination of AI-ETD and HCD boosted the proteoform sequence coverage by more than 200% compared to HCD alone in a CZE-MS study [90]. Brunner et al. demonstrated that EThcD, which performs HCD fragmentation of all product ions after ETD reactions, outperformed ETD and HCD alone regarding the fragmentation coverage of a 17.5-kDa phosphoprotein, leading to a 75% sequence coverage of the protein [91].
5. CZE-MS for native top-down proteomics
Most of the top-down proteomics effort has been made to identify intact proteoforms using liquid-phase separations and ESI-MS operated under denaturing conditions, i.e., acidic aqueous buffers containing organic solvents. The denaturing top-down proteomic approach is powerful for the identification of many proteoforms from a complex proteome. However, it cannot provide direct information on endogenous protein complexes in the cell, which are formed through noncovalent interactions, including but not limited to protein-protein, protein-metal ion, and protein-small ligand interactions [23,24]. Since most of proteins in the cell form complexes to function, it is crucial to delineate protein complexes under near-physiological conditions at a global scale and in discovery mode, termed as native top-down proteomics.
First, native top-down proteomics requires native ESI for transferring protein complexes from liquid phase to gas phase for MS and MS/MS characterization. Native ESI-MS has been widely applied for characterization of various purified protein complexes via direct infusion [24,92–95]. Recently, Li et al. developed an integrated native ESI-MS and MS/MS workflow using FTICR with various gas-phase fragmentation methods to characterize protein subunit arrangements and identification of large protein complexes (up to 1.8 MDa in mass) in a single experiment [24].
Second, native top-down proteomics needs liquid-phase separation of protein complexes for characterization of complex proteomes. Skinner et al. employed off-line ion-exchange chromatography (IEX) or clear native GELFrEE prefractionation and direct infusion native ESI–MS/MS for native top-down proteomics of various mammalian cell and tissue samples [23]. They identified 125 protein complexes in discovery mode from 600 fractions.
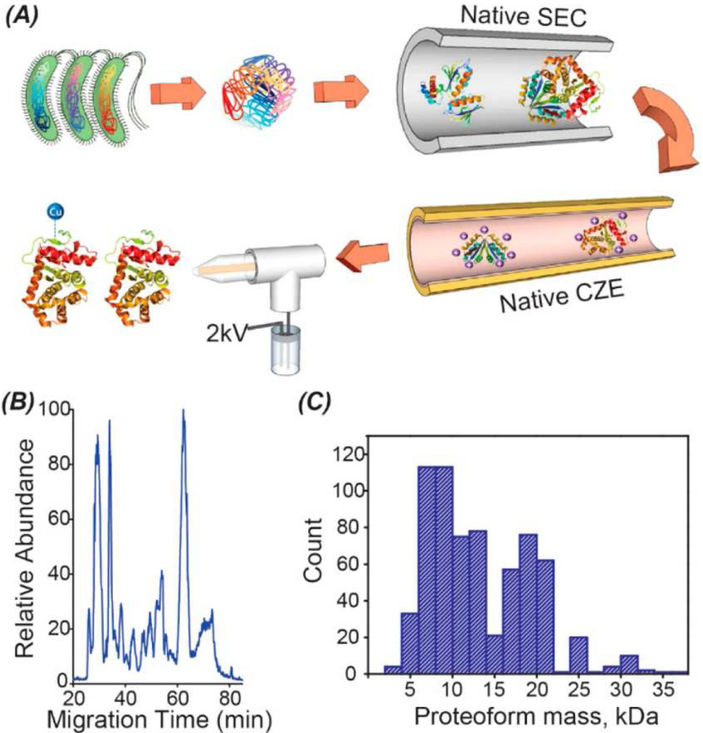
Third, online coupling of high-resolution separation to native ESI-MS is an efficient approach for native top-down proteomics. SEC, IEX and CZE separations have been online coupled to native-ESI-MS for analysis of simple protein complex samples [22,50,96,97]. CZE can achieve high separation efficiency for protein complexes at nL/min flow rates [22,50]. Coupling SEC and/or IEX fractionation to CZE-MS will be a powerful method for large-scale native top-down proteomics. Recently, we performed native top-down proteomics of E. coli cells using a native SEC-CZE-MS/MS approach and identified 672 proteoforms of 144 proteoform families and 23 protein complexes in discovery mode [98]. E. coli proteins extracted from cells under a native condition were first fractionated with native SEC, followed by native CZE-ESI-MS/MS analysis and database search for protein complex IDs, Fig. 6A. Native CZE-MS analysis of one SEC fraction produced a 60-min separation window, Fig. 6B. Proteoforms identified in the study had masses lower than 35 kDa, Fig. 6C. This study represents the first native top-down proteomic work of a complex proteome using online liquid-phase separation and native ESI-MS/MS.
Fig. 6.
SEC-CZE-MS/MS for native top-down proteomics of E. coli cells [98]. The E. coli cells were lysed under a native condition and the extracted proteoforms were fractionated by native SEC, followed by native CZE-MS and MS/MS analysis for protein complex IDs (A). Native CZE-MS/MS analysis of one SEC fraction produced a 60-min separation window (B). The identified proteoforms had masses lower than 35 kDa (C). Reproduced with permission [98].
Native CZE-ESI-MS can also be useful for elucidating conformational changes of protein complexes. Shen et al integrated native CZE-ESI-MS/MS and in-capillary hydrogen/deuterium exchange (HDX) for real-time characterization of conformational states of protein complexes [41]. The flow-through micro-vial CE-MS interface was used in the work. A mixture of holo- and apo-myoglobin (Mb) was first separated with CZE and H/D exchanged in the capillary under a native condition. Then they migrated into the CE-MS interface filled with acidic modifier solution for quenching the HDX reaction and facilitating the subsequent ESI-MS and MS/MS analysis. They observed that apo-Mb had a higher deuteration level than holo-Mb, indicating that the absence of heme group led to more dynamic conformational fluctuations at helical regions of apo-Mb.
6. Conclusions and prospects
The drastic improvement in the CE-MS interface and CZE separation has made CZE-MS an important analytical tool for large-scale top-down characterization of proteoforms in the cell. It has been demonstrated that coupling MD-LC fractionation to CZE-MS is an efficient approach for deep top-down proteomics. The CZE separation and MD-LC fractionation can be further enhanced to achieve higher peak capacity for proteoform separation, better characterization of low-abundance proteoforms and large proteoforms, thus leading to deeper proteome coverage.
Extensive characterization of identified proteoforms from a complex proteome is still challenging. Combination of various gas-phase fragmentation techniques is necessary to boost the proteoform characterization. Coupling CZE separation to more advanced mass spectrometers equipped with multiple fragmentation methods will have the potential to advance top-down proteomics drastically. In addition, integration of bottom-up and top-down proteomic data is also beneficial to proteoform characterization [99].
Top-down proteomic analysis of mass-limited biological samples is still difficult. The current workflows typically require hundreds of micrograms of protein material extracted from cells for the identification of thousands of proteoforms. CZE-MS has great potential to advance top-down proteomics for mass-limited sample analysis. In a recent study, we demonstrated the identification of 800 and 2,000 proteoforms using CZE-MS with only submicrograms of proteins from an E. coli sample and a zebrafish brain sample, respectively [19].
Native top-down proteomics will be a new frontier in proteomic research. CZE-MS is a useful tool for native top-down proteomics because CZE can separate protein complexes with high efficiency [22,50], and because CZE-MS has achieved promising data for native top-down proteomics of a complex proteome [98]. The further advances in the field will require obvious improvement in CZE-MS. The peak capacity and sample loading capacity of CZE for separation of protein complexes need to be boosted. The mass spectrometers need to be advanced for highly sensitive detection and extensive fragmentation of large protein complexes.
Overall, CZE-MS will play an important role in advancing top-down proteomics for delineation of proteoforms and protein complexes in the cell. We expect that CZE-MS-based top-down proteomics will move from technical development to biological applications to understand the roles played by proteoforms and protein complexes in disease and development.
Highlights.
Technical improvement of CZE-MS for top-down proteomics is reviewed.
Coupling LC/CE fractionation to CZE-MS for top-down proteomics is presented.
CZE-MS with various fragmentation methods for top-down proteomics is reviewed.
Recent advances in CZE-MS for native top-down proteomics are discussed.
Future perspectives in CZE-MS-based top-down proteomics are provided.
Acknowledgments
We thank Prof. Heedeok Hong’s group (Department of Chemistry), Prof. Jose Cibelli and Mr. Billy Poulos (Department of Animal Science) at Michigan State University for providing biological materials for our studies. We thank Prof. Xiaowen Liu at Indiana University-Purdue University Indianapolis, Prof. Yansheng Liu at Yale University, and Prof. Joshua J. Coon at UW-Madison for their help on the project. We thank the support from the National Institute of General Medical Sciences, National Institutes of Health (NIH), through Grant R01GM125991.
Abbreviations
- PTM
post-translational modification
- ID
identification
- LPA
linear polyacrylamide
- HPC
hydroxypropyl cellulose
- EOF
electroosmotic flow
- E. coli
Escherichia coli
- FT-ICR
Fourier transform ion cyclotron resonance
- CID
collision-induced dissociation
- HCD
higher-energy collisional dissociation
- ETD
electron transfer dissociation
- UVPD
ultraviolet photodissociation
- mAb
monoclonal antibody
- HDX
hydrogen/deuterium exchange
- MD
Multi-dimensional
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
References
- [1].Smith LM, Kelleher NL, P. Consortium for Top Down, Proteoform: a single term describing protein complexity, Nat. Methods 10 (2013), 186–187. 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shortreed MR, Frey BL, Scalf M, Knoener RA, Cesnik AJ, Smith LM, Elucidating Proteoform Families from Proteoform Intact-Mass and Lysine-Count Measurements, J Proteome Res 15 (2016), 1213–1221. 10.1021/acs.jproteome.5b01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, Spirohn K, Begg BE, Duran-Frigola M, MacWilliams A, Pevzner SJ, Zhong Q, Trigg SA, Tam S, Ghamsari L, Sahni N, Yi S, Rodriguez MD, Balcha D, Tan G, Costanzo M, Andrews B, Boone C, Zhou XJ, Salehi-Ashtiani K, Charloteaux B, Chen AA, Calderwood MA, Aloy P, Roth FP, Hill DE, lakoucheva LM, Xia Y, Vidal M, Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing, Cell 164 (2016), 805–817. 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK, RNA splicing is a primary link between genetic variation and disease, Science 352 (2016), 600–604. 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Costa HA, Leitner MG, Sos ML, Mavrantoni A, Rychkova A, Johnson JR, Newton BW, Yee MC, De La Vega FM, Ford JM, Krogan NJ, Shokat KM, Oliver D, Halaszovich CR, CD. Bustamante, Discovery and functional characterization of a neomorphic PTEN mutation, Proc Natl Acad Sci U S A 112 (2015), 13976–13981. 10.1073/pnas.1422504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ntai I, Fornelli L, DeHart CJ, Hutton JE, Doubleday PF, LeDuc RD, van Nispen AJ, Fellers RT, Whiteley G, Boja ES, Rodriguez H, Kelleher NL, Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk, Proc Natl Acad Sci U S A 115 (2018), 4140–4145. 10.1073/pnas.1716122115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL, Mapping intact protein isoforms in discovery mode using top-down proteomics, Nature 480 (2011), 254–258. 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Catherman AD, Durbin KR, Ahlf DR, Early BP, Fellers RT, Tran JC, Thomas PM, Kelleher NL, Large-scale top-down proteomics of the human proteome: membrane proteins, mitochondria, and senescence, Mol. Cell. Proteomics 12 (2013), 3465–3473. 10.1074/mcp.M113.030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Durbin KR, Fornelli L, Fellers RT, Doubleday PF, Narita M, Kelleher NL, Quantitation and Identification of Thousands of Human Proteoforms below 30 kDa, J Proteome Res 15 (2016), 976–982. 10.1021/acs.jproteome.5b00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cai W, Tucholski T, Chen B, Alpert AJ, McIlwain S, Kohmoto T, Jin S, Ge Y, Top-Down Proteomics of Large Proteins up to 223 kDa Enabled by Serial Size Exclusion Chromatography Strategy, Anal. Chem 89 (2017), 5467–5475. 10.1021/acs.analchem.7b00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ansong C, Wu S, Meng D, Liu X, Brewer HM, Deatherage Kaiser BL, Nakayasu ES, Cort JR, Pevzner P, Smith RD, Heffron F, Adkins JN, Pasa-Tolic L, Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions, Proc Natl Acad Sci U S A 110 (2013), 10153–10158. 10.1073/pnas.1221210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shen Y, Tolic N, Piehowski PD, Shukla AK, Kim S, Zhao R, Qu Y, Robinson E, Smith RD, Pasa-Tolic L, High-resolution ultrahigh-pressure long column reversed-phase liquid chromatography for top-down proteomics, J. Chromatogr. A 1498 (2017), 99–110. 10.1016/j.chroma.2017.01.008. [DOI] [PubMed] [Google Scholar]
- [13].Fornelli L, Durbin KR, Fellers RT, Early BP, Greer JB, LeDuc RD, Compton PD, Kelleher NL, Advancing Top-down Analysis of the Human Proteome Using a Benchtop Quadrupole-Orbitrap Mass Spectrometer, J Proteome Res 16 (2017), 609–618. 10.1021/acs.jproteome.6b00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Anderson LC, DeHart CJ, Kaiser NK, Fellers RT, Smith DF, Greer JB, LeDuc RD, Blakney GT, Thomas PM, Kelleher NL, Hendrickson CL, Identification and Characterization of Human Proteoforms by Top-Down LC-21 Tesla FT-ICR Mass Spectrometry, J Proteome Res 16 (2017), 1087–1096. 10.1021/acs.jproteome.6b00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schaffer LV, Rensvold JW, Shortreed MR, Cesnik AJ, Jochem A, Scalf M, Frey BL, Pagliarini DJ, Smith LM, Identification and Quantification of Murine Mitochondrial Proteoforms Using an Integrated Top-Down and Intact-Mass Strategy, J Proteome Res 17 (2018), 3526–3536. 10.1021/acs.jproteome.8b00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Riley NM, Sikora JW, Seckler HS, Greer JB, Fellers RT, LeDuc RD, Westphall MS, Thomas PM, Kelleher NL, Coon JJ, The Value of Activated Ion Electron Transfer Dissociation for High-Throughput Top-Down Characterization of Intact Proteins, Anal. Chem 90 (2018), 8553–8560. 10.1021/acs.analchem.8b01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, Costello CE, Cravatt BF, Fenselau C, Garcia BA, Ge Y, Gunawardena J, Hendrickson RC, Hergenrother PJ, Huber CG, Ivanov AR, Jensen ON, Jewett MC, Kelleher NL, Kiessling LL, Krogan NJ, Larsen MR, Loo JA, Ogorzalek Loo RR, Lundberg E, MacCoss MJ, Mallick P, Mootha VK, Mrksich M, Muir TW, Patrie SM, Pesavento JJ, Pitteri SJ, Rodriguez H, Saghatelian A, Sandoval W, Schluter H, Sechi S, Slavoff SA, Smith LM, Snyder MP, Thomas PM, Uhlen M, Van Eyk JE, Vidal M, Walt DR, White FM, Williams ER, Wohlschlager T, Wysocki VH, Yates NA, Young NL, Zhang B, How many human proteoforms are there?, Nat. Chem. Biol 14 (2018), 206–214. 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jorgenson J, Lukacs K, Capillary zone electrophoresis, Science 222 (1983), 266–272. 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- [19].Lubeckyj RA, Basharat AR, Shen X, Liu X, Sun L, Large-Scale Qualitative and Quantitative Top-Down Proteomics Using Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry with Nanograms of Proteome Samples, J. Am. Soc. Mass Spectrom. 30 (2019), 1435–1445. https://link.springer.com/article/10.1007/s13361-019-02167-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Valaskovic GA, Kelleher NL, McLafferty FW, Attomole protein characterization by capillary electrophoresis mass spectrometry, Science 273 (1996), 1199–1202. 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- [21].Han X, Wang Y, Aslanian A, Fonslow B, Graczyk B, Davis TN, Yates JR 3rd, In-line separation by capillary electrophoresis prior to analysis by top-down mass spectrometry enables sensitive characterization of protein complexes, J Proteome Res 13 (2014), 6078–6086. 10.1021/pr500971h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nguyen A, Moini M, Analysis of major protein-protein and protein-metal complexes of erythrocytes directly from cell lysate utilizing capillary electrophoresis mass spectrometry, Anal. Chem 80 (2008), 7169–7173. 10.1021/ac801158q. [DOI] [PubMed] [Google Scholar]
- [23].Skinner OS, Haverland NA, Fornelli L, Melani RD, Do Vale LHF, Seckler HS, Doubleday PF, Schachner LF, Srzentic K, Kelleher NL, Compton PD, Top-down characterization of endogenous protein complexes with native proteomics, Nat. Chem. Biol 14 (2018), 36–41. 10.1038/nchembio.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li H, Nguyen HH, Ogorzalek Loo RR, Campuzano IDG, Loo JA, An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes, Nat Chem 10 (2018), 139–148. 10.1038/nchem.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Compton PD, Zamdborg L, Thomas PM, Kelleher NL, On the scalability and requirements of whole protein mass spectrometry, Anal. Chem 83 (2011), 6868–6874. 10.1021/ac2010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, Early BP, Bluhm EJ, Babai S, Kelleher NL, ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry, Nucleic Acids Res. 35 (2007), W701–706. 10.1093/nar/gkm371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kou Q, Xun L, Liu X, TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization, Bioinformatics 32 (2016), 3495–3497. 10.1093/bioinformatics/btw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cesnik AJ, Shortreed MR, Schaffer LV, Knoener RA, Frey BL, Scalf M, Solntsev SK, Dai Y, Gasch AP, Smith LM, Proteoform Suite: Software for Constructing, Quantifying, and Visualizing Proteoform Families, J Proteome Res 17 (2018), 568–578. 10.1021/acs.jproteome.7b00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cai W, Guner H, Gregorich ZR, Chen AJ, Ayaz-Guner S, Peng Y, Valeja SG, Liu X, Ge Y, MASH Suite Pro: A Comprehensive Software Tool for Top-Down Proteomics, Mol. Cell. Proteomics 15 (2016), 703–714. 10.1074/mcp.O115.054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sun RX, Luo L, Wu L, Wang RM, Zeng WF, Chi H, Liu C, He SM, pTop 1.0: A High-Accuracy and High-Efficiency Search Engine for Intact Protein Identification, Anal. Chem 88 (2016), 3082–3090. 10.1021/acs.analchem.5b03963. [DOI] [PubMed] [Google Scholar]
- [31].Zhang ZB, Qu YY, Dovichi NJ, Capillary zone electrophoresis-mass spectrometry for bottom-up proteomics, Trac-Trends in Analytical Chemistry 108 (2018), 23–37. 10.1016/j.trac.2018.08.008. [DOI] [Google Scholar]
- [32].Sun L, Zhu G, Yan X, Dovichi NJ, High sensitivity capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry for the rapid analysis of complex proteomes, Curr. Opin. Chem. Biol 17 (2013), 795–800. 10.1016/i.cbpa.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].DeLaney K, Sauer CS, Vu NQ, Li L, Recent Advances and New Perspectives in Capillary Electrophoresis-Mass Spectrometry for Single Cell “Omics”, Molecules 24 (2018). 10.3390/molecules24010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haselberg R, de Jong GJ, Somsen GW, CE-MS for the analysis of intact proteins 2010–2012, Electrophoresis 34 (2013), 99–112. 10.1002/elps.201200439. [DOI] [PubMed] [Google Scholar]
- [35].Smith RD, Barinaga CJ, Udseth HR, Improved electrospray ionization interface for capillary zone electrophoresis-mass spectrometry, Anal. Chem 60 (1988), 1948–1952. 10.1021/ac00169a022. [DOI] [Google Scholar]
- [36].Maxwell EJ, Chen DD, Twenty years of interface development for capillary electrophoresis-electrospray ionization-mass spectrometry, Anal. Chim. Acta 627 (2008), 25–33. 10.1016/j.aca.2008.06.034. [DOI] [PubMed] [Google Scholar]
- [37].Ramautar R, Heemskerk AA, Hensbergen PJ, Deelder AM, Busnel JM, Mayboroda OA, CE-MS for proteomics: Advances in interface development and application, J Proteomics 75 (2012), 3814–3828. 10.1016/j.jprot.2012.04.050. [DOI] [PubMed] [Google Scholar]
- [38].Maxwell EJ, Zhong X, Zhang H, van Zeijl N, Chen DD, Decoupling CE and ESI for a more robust interface with MS, Electrophoresis 31 (2010), 1130–1137. 10.1002/elps.200900517. [DOI] [PubMed] [Google Scholar]
- [39].Zhong X, Maxwell EJ, Ratnayake C, Mack S, Chen DD, Flow-through microvial facilitating interface of capillary isoelectric focusing and electrospray ionization mass spectrometry, Anal. Chem 83 (2011), 8748–8755. 10.1021/ac202130f. [DOI] [PubMed] [Google Scholar]
- [40].Wang L, Bo T, Zhang Z, Wang G, Tong W, Da Yong Chen D, High Resolution Capillary Isoelectric Focusing Mass Spectrometry Analysis of Peptides, Proteins, And Monoclonal Antibodies with a Flow-through Microvial Interface, Anal. Chem 90 (2018), 9495–9503. 10.1021/acs.analchem.8b02175. [DOI] [PubMed] [Google Scholar]
- [41].Shen Y, Zhao X, Wang G, Chen DDY, Differential Hydrogen/Deuterium Exchange during Proteoform Separation Enables Characterization of Conformational Differences between Coexisting Protein States, Anal. Chem (2019). 10.1021/acs.analchem.9b00558. [DOI] [PubMed] [Google Scholar]
- [42].Wojcik R, Dada OO, Sadilek M, Dovichi NJ, Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis, Rapid Commun. Mass Spectrom. 24 (2010), 2554–2560. 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- [43].Sun L, Zhu G, Zhao Y, Yan X, Mou S, Dovichi NJ, Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests, Angew. Chem. Int. Ed. Engl 52 (2013), 13661–13664. 10.1002/anie.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sun L, Zhu G, Zhang Z, Mou S, Dovichi NJ, Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests, J Proteome Res 14 (2015), 2312–2321. 10.1021/acs.jproteome.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Choi SB, Zamarbide M, Manzini MC, Nemes P, Tapered-Tip Capillary Electrophoresis Nano-Electrospray Ionization Mass Spectrometry for Ultrasensitive Proteomics: the Mouse Cortex, J. Am. Soc. Mass Spectrom. 28 (2017), 597–607. 10.1007/s13361-016-1532-8. [DOI] [PubMed] [Google Scholar]
- [46].Moini M, Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip, Anal. Chem 79 (2007), 4241–4246. 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- [47].Haselberg R, Ratnayake CK, de Jong GJ, Somsen GW, Performance of a sheathless porous tip sprayer for capillary electrophoresis-electrospray ionization-mass spectrometry of intact proteins, J. Chromatogr. A 1217 (2010), 7605–7611. 10.1016/j.chroma.2010.10.006. [DOI] [PubMed] [Google Scholar]
- [48].Haselberg R, de Jong GJ, Somsen GW, Low-flow sheathless capillary electrophoresis-mass spectrometry for sensitive glycoform profiling of intact pharmaceutical proteins, Anal. Chem 85 (2013), 2289–2296. 10.1021/ac303158f. [DOI] [PubMed] [Google Scholar]
- [49].Han X, Wang Y, Aslanian A, Bern M, Lavallee-Adam M, Yates JR 3rd, Sheathless capillary electrophoresis-tandem mass spectrometry for top-down characterization of Pyrococcus furiosus proteins on a proteome scale, Anal. Chem 86 (2014), 11006–11012. 10.1021/ac503439n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Belov AM, Viner R, Santos MR, Horn DM, Bern M, Karger BL, Ivanov AR, Analysis of Proteins, Protein Complexes, and Organellar Proteomes Using Sheathless Capillary Zone Electrophoresis - Native Mass Spectrometry, J. Am. Soc. Mass Spectrom. 28 (2017), 2614–2634. 10.1007/s13361-017-1781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Moini M, Martinez B, Ultrafast capillary electrophoresis/mass spectrometry with adjustable porous tip for a rapid analysis of protein digest in about a minute, Rapid Commun. Mass Spectrom. 28 (2014), 305–310. 10.1002/rcm.6786. [DOI] [PubMed] [Google Scholar]
- [52].McCool EN, Lubeckyj R, Shen X, Kou Q, Liu X, Sun L, Large-scale Top-down Proteomics Using Capillary Zone Electrophoresis Tandem Mass Spectrometry, J Vis Exp (2018), e58644 10.3791/58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhu G, Sun L, Dovichi NJ, Thermally-initiated free radical polymerization for reproducible production of stable linear polyacrylamide coated capillaries, and their application to proteomic analysis using capillary zone electrophoresis-mass spectrometry, Talanta 146 (2016), 839–843. 10.1016/j.talanta.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lubeckyj RA, McCool EN, Shen X, Kou Q, Liu X, Sun L, Single-Shot Top-Down Proteomics with Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry for Identification of Nearly 600 Escherichia coli Proteoforms, Anal. Chem 89 (2017), 12059–12067. 10.1021/acs.analchem.7b02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li Y, Compton PD, Tran JC, Ntai I, Kelleher NL, Optimizing capillary electrophoresis for top-down proteomics of 30–80 kDa proteins, Proteomics 14 (2014), 1158–1164. 10.1002/pmic.201300381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bush DR, Zang L, Belov AM, Ivanov AR, Karger BL, High Resolution CZE-MS Quantitative Characterization of Intact Biopharmaceutical Proteins: Proteoforms of Interferon-beta1, Anal. Chem 88 (2016), 1138–1146. 10.1021/acs.analchem.5b03218. [DOI] [PubMed] [Google Scholar]
- [57].Bekri S, Leclercq L, Cottet H, Polyelectrolyte multilayer coatings for the separation of proteins by capillary electrophoresis: Influence of polyelectrolyte nature and multilayer crosslinking, J. Chromatogr. A 1399 (2015) 80–87. https://www.sciencedirect.com/science/article/pii/S0021967315005956?via%3Dihub [DOI] [PubMed] [Google Scholar]
- [58].Dawod M, Arvin NE, Kennedy RT, Recent advances in protein analysis by capillary and microchip electrophoresis, Analyst 142 (2017) 1847–1866. https://pubs.rsc.org/en/content/articlelanding/2017/AN/C7AN00198C#!divAbstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Simpson SL Jr., Quirino JP, Terabe S, On-line sample preconcentration in capillary electrophoresis. Fundamentals and applications, J. Chromatogr. A 1184 (2008), 504–541. 10.1016/j.chroma.2007.11.001. [DOI] [PubMed] [Google Scholar]
- [60].Sun L, Knierman MD, Zhu G, Dovichi NJ, Fast top-down intact protein characterization with capillary zone electrophoresis-electrospray ionization tandem mass spectrometry, Anal. Chem 85 (2013), 5989–5995. 10.1021/ac4008122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhao Y, Sun L, Knierman MD, Dovichi NJ, Fast separation and analysis of reduced monoclonal antibodies with capillary zone electrophoresis coupled to mass spectrometry, Talanta 148 (2016), 529–533. 10.1016/j.talanta.2015.11.020. [DOI] [PubMed] [Google Scholar]
- [62].Zhao Y, Sun L, Champion MM, Knierman MD, Dovichi NJ, Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry for top-down characterization of the Mycobacterium marinum secretome, Anal. Chem 86 (2014), 4873–4878. 10.1021/ac500092q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Britz-McKibbin P, Chen DDY, Selective Focusing of Catecholamines and Weakly Acidic Compounds by Capillary Electrophoresis Using a Dynamic pH Junction, Anal. Chem 72 (2000), 1242–1252. 10.1021/ac990898e. [DOI] [PubMed] [Google Scholar]
- [64].Wang L, MacDonald D, Huang X, Chen DD, Capture efficiency of dynamic pH junction focusing in capillary electrophoresis, Electrophoresis 37 (2016), 1143–1150. 10.1002/elps.201600008. [DOI] [PubMed] [Google Scholar]
- [65].Zhu G, Sun L, Yan X, Dovichi NJ, Bottom-up proteomics of Escherichia coli using dynamic pH junction preconcentration and capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry, Anal. Chem 86 (2014), 6331–6336. 10.1021/ac5004486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chen D, Shen X, Sun L, Capillary zone electrophoresis-mass spectrometry with microliter-scale loading capacity, 140 min separation window and high peak capacity for bottom-up proteomics, Analyst 142 (2017), 2118–2127. 10.1039/c7an00509a. [DOI] [PubMed] [Google Scholar]
- [67].Chen D, Shen X, Sun L, Strong cation exchange-reversed phase liquid chromatography-capillary zone electrophoresis-tandem mass spectrometry platform with high peak capacity for deep bottom-up proteomics, Anal. Chim. Acta 1012 (2018), 1–9. 10.1016/j.aca.2018.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen D, Ludwig KR, Krokhin OV, Spicer V, Yang Z, Shen X, Hummon AB, Sun L, Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Large-Scale Phosphoproteomics with the Production of over 11,000 Phosphopeptides from the Colon Carcinoma HCT116 Cell Line, Anal. Chem 91 (2019), 2201–2208. 10.1021/acs.analchem.8b04770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Yang Z, Shen X, Chen D, Sun L, Microscale Reversed-Phase Liquid Chromatography/Capillary Zone Electrophoresis-Tandem Mass Spectrometry for Deep and Highly Sensitive Bottom-Up Proteomics: Identification of 7500 Proteins with Five Micrograms of an MCF7 Proteome Digest, Anal. Chem 90 (2018), 10479–10486. 10.1021/acs.analchem.8b02466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhao Y, Sun L, Zhu G, Dovichi NJ, Coupling Capillary Zone Electrophoresis to a Q Exactive HF Mass Spectrometer for Top-down Proteomics: 580 Proteoform Identifications from Yeast, J Proteome Res 15 (2016), 3679–3685. 10.1021/acs.jproteome.6b00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Yuan H, Jiang B, Zhao B, Zhang L, Zhang Y, Recent Advances in Multidimensional Separation for Proteome Analysis, Anal. Chem 91 (2019), 264–276. 10.1021/acs.analchem.8b04894. [DOI] [PubMed] [Google Scholar]
- [72].McCool EN, Lodge JM, Basharat AR, Liu X, Coon JJ, Sun L, Capillary Zone Electrophoresis-Tandem Mass Spectrometry with Activated Ion Electron Transfer Dissociation for Large-scale Top-down Proteomics, J. Am. Soc. Mass Spectrom. (2019). 10.1007/s13361-019-02206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].McCool EN, Lubeckyj RA, Shen X, Chen D, Kou Q, Liu X, Sun L, Deep Top-Down Proteomics Using Capillary Zone Electrophoresis-Tandem Mass Spectrometry: Identification of 5700 Proteoforms from the Escherichia coli Proteome, Anal. Chem 90 (2018), 5529–5533. 10.1021/acs.analchem.8b00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Magdeldin S, Moresco JJ, Yamamoto T, Yates JR 3rd, Off-Line Multidimensional Liquid Chromatography and Auto Sampling Result in Sample Loss in LC/LC-MS/MS, J Proteome Res 13 (2014), 3826–3836. 10.1021/pr500530e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lewis KC, Opiteck GJ, Jorgenson JW, Sheeley DM, Comprehensive on-line RPLC-CZE-MS of peptides, J. Am. Soc. Mass Spectrom. 8 (1997), 495–500. 10.1016/S1044-0305(97)00009-3. [DOI] [Google Scholar]
- [76].Zhang J, Hu H, Gao M, Yang P, Zhang X, Comprehensive two-dimensional chromatography and capillary electrophoresis coupled with tandem time-of-flight mass spectrometry for high-speed proteome analysis, Electrophoresis 25 (2004), 2374–2383. 10.1002/elps.200405956. [DOI] [PubMed] [Google Scholar]
- [77].Michels DA, Hu S, Schoenherr RM, Eggertson MJ, Dovichi NJ, Fully automated two-dimensional capillary electrophoresis for high sensitivity protein analysis, Mol. Cell. Proteomics 1 (2002), 69–74. 10.1074/mcp.t100009-mcp200. [DOI] [PubMed] [Google Scholar]
- [78].Hühner J, Neusüß C, CIEF-CZE-MS applying a mechanical valve, Anal. Bioanal. Chem 408 (2016), 4055–4061. 10.1007/s00216-016-9498-8. [DOI] [PubMed] [Google Scholar]
- [79].Jooß K, Hühner J, Kiessig S, Moritz B, Neusüß C, Two-dimensional capillary zone electrophoresis-mass spectrometry for the characterization of intact monoclonal antibody charge variants, including deamidation products, Anal. Bioanal. Chem 409 (2017), 6057–6067. 10.1007/s00216-017-0542-0. [DOI] [PubMed] [Google Scholar]
- [80].Jooß K, Scholz N, Meixner J, Neusüß C, Heart-cut nano-LC-CZE-MS for the characterization of proteins on the intact level, Electrophoresis 40 (2019), 1061–1065. 10.1002/elps.201800411. [DOI] [PubMed] [Google Scholar]
- [81].Diedrich JK, Pinto AF, Yates JR 3rd, Energy dependence of HCD on peptide fragmentation: stepped collisional energy finds the sweet spot, J. Am. Soc. Mass Spectrom. 24 (2013), 1690–1699. https://link.sprinqer.com/article/10.1007%2Fs13361-013-0709-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH, Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns, Anal. Chem 77 (2005), 5800–5813. 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Riley NM, Coon JJ, The Role of Electron Transfer Dissociation in Modern Proteomics, Anal. Chem 90 (2018), 40–64. 10.1021/acs.analchem.7b04810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Riley NM, Westphall MS, Coon JJ, Activated Ion Electron Transfer Dissociation for Improved Fragmentation of Intact Proteins, Anal. Chem 87 (2015), 7109–7116. 10.1021/acs.analchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Rush MJP, Riley NM, Westphall MS, Coon JJ, Top-Down Characterization of Proteins with Intact Disulfide Bonds Using Activated-Ion Electron Transfer Dissociation, Anal. Chem 90 (2018), 8946–8953. 10.1021/acs.analchem.8b01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dang X, Young NL, Ultraviolet photodissociation enhances top-down mass spectrometry as demonstrated on green fluorescent protein variants, Proteomics 14(2014), 1128–1129. 10.1002/pmic.201400114. [DOI] [PubMed] [Google Scholar]
- [87].Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS, Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation, J. Am. Chem. Soc 135 (2013), 12646–12651. 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Cleland TP, DeHart CJ, Fellers RT, VanNispen AJ, Greer JB, LeDuc RD, Parker WR, Thomas PM, Kelleher NL, Brodbelt JS, High-Throughput Analysis of Intact Human Proteins Using UVPD and HCD on an Orbitrap Mass Spectrometer, J. Proteome Res. 16 (2017), 2072–2079. 10.1021/acs.jproteome.7b00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].McCool EN, Chen D, Li W, Liu Y, Sun L, Capillary zone electrophoresis-tandem mass spectrometry using ultraviolet photodissociation (213 nm) for large-scale top-down proteomics, Anal. Methods 11 (2019), 2855–2861. https://pubs.rsc.org/en/content/articlepdf/2019/ay/c9av00585d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhao Y, Riley NM, Sun L, Hebert AS, Yan X, Westphall MS, Rush MJ, Zhu G, Champion MM, Mba Medie F, Champion PA, Coon JJ, Dovichi NJ, Coupling capillary zone electrophoresis with electron transfer dissociation and activated ion electron transfer dissociation for top-down proteomics, Anal. Chem 87 (2015), 5422–5429. 10.1021/acs.analchem.5b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brunner AM, Lössl P, Liu F, Huguet R, Mullen C, Yamashita M, Zabrouskov V, Makarov A, Altelaar AF, Heck AJ, Benchmarking multiple fragmentation methods on an orbitrap fusion for top-down phospho-proteoform characterization. Anal. Chem 87 (2015), 4152–4158. https://pubs.acs.org/doi/10.1021/acs.analchem.5b00162 [DOI] [PubMed] [Google Scholar]
- [92].Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJ, Studying 18 MDa virus assemblies with native mass spectrometry, Angew. Chem. Int. Ed 52 (2013), 4020–4023. 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Quintyn RS, Zhou M, Yan J, Wysocki VH, Surface-Induced Dissociation Mass Spectra as a Tool for Distinguishing Different Structural Forms of Gas-Phase Multimeric Protein Complexes, Anal. Chem 87 (2015), 11879–11886. 10.1021/acs.analchem.5b03441. [DOI] [PubMed] [Google Scholar]
- [94].Susa AC, Xia Z, Williams ER, Native Mass Spectrometry from Common Buffers with Salts That Mimic the Extracellular Environment, Angew. Chem. Int. Ed 56 (2017), 7912–7915. 10.1002/anie.201702330. [DOI] [PubMed] [Google Scholar]
- [95].Marty MT, Hoi KK, Gault J, Robinson CV, Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs, Angew. Chem. Int. Ed 55 (2016), 550–554. 10.1002/anie.201508289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Muneeruddin K, Nazzaro M, Kaltashov IA, Characterization of intact protein conjugates and biopharmaceuticals using ion-exchange chromatography with online detection by native electrospray ionization mass spectrometry and top-down tandem mass spectrometry, Anal. Chem 87 (2015), 10138–10145. 10.1021/acs.analchem.5b02982. [DOI] [PubMed] [Google Scholar]
- [97].Muneeruddin K, Thomas JJ, Salinas PA, Kaltashov IA, Characterization of small protein aggregates and oligomers using size exclusion chromatography with online detection by native electrospray ionization mass spectrometry, Anal. Chem 86 (2014), 10692–10699. 10.1021/ac502590h. [DOI] [PubMed] [Google Scholar]
- [98].Shen X, Kou Q, Guo R, Yang Z, Chen D, Liu X, Hong H, Sun L, Native Proteomics in Discovery Mode Using Size-Exclusion Chromatography-Capillary Zone Electrophoresis-Tandem Mass Spectrometry, Anal. Chem 90 (2018), 10095–10099. 10.1021/acs.analchem.8b02725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Dai Y, Shortreed MR, Scalf M, Frey BL, Cesnik AJ, Solntsev S, Schaffer LV, Smith LM, Elucidating Escherichia coli Proteoform Families Using Intact-Mass Proteomics and a Global PTM Discovery Database, J Proteome Res 16 (2017), 4156–4165. 10.1021/acs.jproteome.7b00516. [DOI] [PMC free article] [PubMed] [Google Scholar]