Abstract
Maize genotypes can show different responsiveness to inoculation with Azospirillum brasilense and an intriguing issue is which genes of the plant are involved in the recognition and growth promotion by these Plant Growth-Promoting Bacteria (PGPB). We conducted Genome-Wide Association Studies (GWAS) using additive and heterozygous (dis)advantage models to find candidate genes for root and shoot traits under nitrogen (N) stress and N stress plus A. brasilense. A total of 52,215 Single Nucleotide Polymorphism (SNP) markers were used for GWAS analyses. For the six root traits with significant inoculation effect, the GWAS analyses revealed 25 significant SNPs for the N stress plus A. brasilense treatment, in which only two were overlapped with the 22 found for N stress only. Most were found by the heterozygous (dis)advantage model and were more related to exclusive gene ontology terms. Interestingly, the candidate genes around the significant SNPs found for the maize–A. brasilense association were involved in different functions previously described for PGPB in plants (e.g. signaling pathways of the plant's defense system and phytohormone biosynthesis). Our findings are a benchmark in the understanding of the genetic variation among maize hybrids for the association with A. brasilense and reveal the potential for further enhancement of maize through this association.
Introduction
Currently, major agro-systems are highly dependent on chemical fertilizers and pesticide inputs. One of the main strategies to develop sustainable agriculture in the face of natural resource scarcity and environmental impacts caused by the application of these products is the use of Plant Growth-Promoting Bacteria (PGPB) inoculants. These bacteria in association with plants may generate several benefits to the host, such as phytohormone biosynthesis, biological nitrogen fixation (BNF), and induction of resistance mechanisms. Consequently, there are positive effects on the enhancement of root traits, tolerance to abiotic stress, and defense against pathogens [1,2].
Azospirillum brasilense is a well-known PGPB marketed by several companies in South American countries (e.g. Brazil, Argentina, and Uruguay). It is used as a inoculant in some cereal crops such as maize and wheat [3]. Some studies have reported the influence of plant genotype on the degree of beneficial response to PGPB inoculation, including A. brasilense [4–6]. In this context, Genome-wide Association Studies (GWAS) is a powerful approach for the identification of genomic regions associated significantly with phenotypic trait variations and has been widely applied in the study of the genetic basis of plant–microbe interactions, including pathogens [7,8] arbuscular mycorrhizal fungi [9,10], and endogenous microbiomes [11]. As far as we know, only two GWAS studies were reported to PGPB. The first explored traits related to the BNF of Rhizobium tropici in a panel of 259 common beans [12]. The second evaluated shoot and root traits of 302 accessions of Arabidopsis thaliana inoculated with Pseudomonas simiae WCS417r [13]. However, GWAS studies related to genetic basis of cereals for the responsiveness to PGPB have not been reported so far, particularly for those with N-fixing ability.
Moreover, the growing of plants on unsterilized soil should be considered in studies concerning the relationship of plants with PGPB. The soil characteristics may influence this association, particularly due to the interaction of the inoculated strain with the soil microbiome. For instance, they might compete for resources and site, or show antagonist effects [14]. The understanding of the plants’ genetic basis related to PGPB and nitrogen (N) starvation is also crucial. It is known that changes in the diversity and the amount of the compounds released by the roots depend on the nutritional status, with consequences for the transcription of PGPB genes [15] and the composition of the plant-associated microbiome [16,17]. Furthermore, in tropical areas such as Africa and parts of South America, the soils are often N-limited and a significant proportion of maize production takes place under these conditions.
Another challenge is the heterosis (or hybrid vigor) of several maize traits [18–20]. Therefore, GWAS analyses should consider not only the additive marker effects but also the non-additive ones that might explain an important proportion of the variation in complex traits [21,22]. In this way, some authors speculate that the colonization of maize roots by beneficial microbes could be regulated by heterosis, due to hybrid plants supporting more numerous strains than their parental inbred lines [23,24]. In addition, studies of mechanisms underlying heterosis have shown changes, for example, in the expression patterns of hormone defense pathways and auxin biosynthesis [25], carbohydrate and nitrogen metabolism [26], besides increase of root and shoot biomass [27,28], which may also be related to plant responses to PGPBs [29–32]. However, this was not clearly elucidated. Thus, heterozygous (dis)advantage GWAS models [33,34] applied to the plant-related traits of the responsiveness to PGPB could provide additional information about the influence of heterosis concerning this association and help to identify candidate genes with heterotic performance under the inoculation conditions.
Knowledge about the genetic variation available and the genetic architecture of the traits involved in maize‒A. brasilense interaction is absent. However, this information can contribute to the understanding of its genetic base and how to apply it in plant breeding programs aimed at improving the germplasm for this association. Hence, we aimed with this study (i) to understand the genetic variation of maize in response to A. brasilense inoculation under low-N soil conditions, and (ii) to perform GWAS analyses using additive and heterozygous (dis)advantage models to identify Single Nucleotide Polymorphisms (SNPs) significantly associated with traits related to this association, the underlying candidate genes, and the importance of non-additive effects on the maize–A. brasilense association.
Materials and methods
Bacterial strain and inoculum
The bacterial strain A. brasilense Ab-V5 was selected from maize roots in Brazil and is registered by the Brazilian Ministry of Agriculture, Livestock and Food Supply (MAPA) for the inoculant production for maize, rice and wheat [3,35]. In addition, it is part of the Culture Collection of Diazotrophic and PGPB of Embrapa Soybean (Londrina, Paraná, Brazil). The bacterial inoculum of A. brasilense Ab-V5 was prepared in the Laboratory of Genetics of Microorganisms “Prof. João Lúcio de Azevedo” at ESALQ/USP, Piracicaba-SP, Brazil, and taken immediately to the experimental area. Bacterial inoculum was prepared by growing Ab-V5 in Dextrose Yeast Glucose Sucrose (DYGS) liquid medium [36] at 28°C with 150 rpm agitation. The inoculum concentration was adjusted to approximately 1 × 108 UFC mL–1 and transferred with a pipette into plastic bags containing the maize seeds of each genotype individually. Sowing was done about 30 min after inoculation.
Plant material and greenhouse experiments
The association panel was comprised of 118 single-cross hybrids from a diallel mating design between 19 tropical maize inbred lines with genetic diversity to nitrogen-use efficiency [37–39]. The plants were grown under semi-controlled conditions in a greenhouse located at the University of São Paulo, Brazil (22° 42' 39" S; 47° 38' 09" W, altitude 540 m), in two years: November–December 2016 and February–March 2017. A randomized complete block experimental design with three replications spatially arranged under two countertops was adopted in each season. Two main treatments were evaluated: N stress without bacterial inoculation and N stress plus A. brasilense inoculation. The decision to non-input N fertilizer was due to its reported negative effects on N fixation by diazotrophic bacteria [40,41]. In each plot, three seeds were sown at 3 cm depth in plastic pots of 3 L capacity containing unsterilized loam soil from an area not in agricultural use. Information about the soil chemical and physical characteristics is available in Vidotti et al. [42]. After germination, the seedlings were thinned singly. Only potassium chloride and single phosphate fertilizers were added to the soil according to the general crop demand. The average temperature was semi-controlled (20–33°C), and supplementation of luminosity was by fluorescent lamps to simulate a photoperiod of 12 h. The water supply was provided manually by pot, with the same amount applied to all of them and always maintaining a well-watered condition. During the conduction of the experiments, no insect or pathogen attack was detected, and pesticides were not used.
Approximately 35 days after the emergence, when most of the hybrids had reached the V7 stage (seven expanded leaves), plant height (PH, cm) was measured from the soil level to the insertion of the least expanded leaf. The shoot was harvested and dried in a forced-draft oven at 60°C for 72 h to determine the shoot dry mass (SDM, g). The soil particles in each root system were carefully removed with water and the individual storage was performed inside plastic pots in 25% ethanol solution for preservation. The root images acquired by an Epson LA2400 scanner (2400 dpi resolution) were analyzed by WinRHIZOTM (Reagent Instruments Inc., Quebec, Canada). This software provided the measurements of root average diameter (RAD, mm), root volume (RV, cm3), and the total length of a series of root diameter classes. The length of fragments with a diameter class less than or equal to 0.5 mm were considered as the lateral root length (roots from the axial roots—LRL, cm), while that with diameter classes greater than 0.5 mm were considered as axial root length (comprising crow, seminal and primary roots—ARL, cm) [43]. We determined the root dry mass (RDM, g) after drying the roots under the same conditions used for SDM measurement. This trait was used to calculate the specific root length (SRL, cm g–1) and specific root surface area (SRSA, cm2 g–1) dividing the total root length and the surface area by the RDM, respectively. Furthermore, the root/shoot ratio (RSR, g g–1) was obtained by dividing the RDM by the SDM. In total, 10 traits were evaluated and approximately 1416 root systems analyzed.
Phenotypic analyses
The analyses were conducted using Restricted Maximum Likelihood/Best Linear Unbiased Predictor (REML/BLUP) mixed models, by ASReml R package [44], considering the following model:
where y is the vector of phenotypic observations of the traits evaluated on maize hybrids; βE is the vector of fixed effects of year; βB is the vector of fixed effects of block within year; βC is the vector of fixed effects of countertop within block and year; βI is the vector of fixed effects of inoculation; βEI is the vector of fixed effects of inoculation × year interaction; uG is the vector of random effects of genotype, where ; uGE is the vector of random effects of genotype × year interaction, where ; uGI is the vector of random effects of genotype × inoculation interaction, where ; uGEI is the vector of random effects of genotype × year × inoculation interaction, where ; and ε is the vector of errors, where . XE, XB, XC, XI, XEI, ZG, ZGE, ZGI, and ZGEI are the respective incidence matrices related to each vector. The significance of fixed effects was tested using the Wald test implemented in the ASReml R package, while the significance of random effects was assessed by Likelihood Ratio Test (LTR) from the asremlPlus R package [45]. The variance components by treatment were estimated through reduced models disregarding the inoculation effect and its interaction with genotype. Broad-sense heritabilities were estimated as , where the is the genetic variance; is the genotype-by-year variance; is the error variance; j and r are the number of years and replications in each experiment, respectively.
Genotypic data
The Affymetrix® Axiom® Maize Genotyping Array [46] of 616,201 SNPs markers was used to genotype the parental inbred lines. Markers with call rate <95% and heterozygous loci on at least one individual were removed. Remaining missing data were imputed through the algorithms of Beagle 4.0 using the codeGeno function from the Synbreed R package [47]. The hybrid genotypes were obtained in silico from the genotypes of the corresponding parental inbred lines. After that, one more filter was applied to the matrix, eliminating SNPs with Minor Allele Frequency (MAF) ≤ 0.05. A final SNP set of 59,215 was obtained and used for the subsequent analyses.
GWAS analyses
Marker-trait association analyses were performed for the traits with significant inoculation effect. For these traits, the adjusted means for each hybrid were calculated by treatment (inoculated and non-inoculated), separately, considering the following model:
where y is the vector of phenotypic observations of the traits evaluated on maize hybrids; βE is the vector of fixed effects of year; βB is the vector of fixed effects of block within year; βC is the vector of fixed effects of countertop within block and year; βG is the vector of fixed effects of the genotype; βGE is the vector of fixed effects of genotype × year interaction; and ε is the vector of errors, where . XE, XB, XC, XG, and XGE are the respective incidence matrices for each vector. Density and box plots were used to compare the means between both treatments. In addition, the changes due to A. brasilense inoculation on the hybrid traits were calculated by Δ = M1−M2, where M1 is the adjusted mean under N stress plus A. brasilense and M2 is the adjusted mean under N stress.
Population structure was estimated by principal component analysis (PCA) using the genomic matrix through the SNPRelate R package [48]. The GWAS analyses were conducted by a Fixed and Random Model Circulating Probability Unification method thought the FarmCPU R package [49]. This statistical procedure considers the confounding effect between the testing marker and both kinship (K) and population structure (Q) as covariates to minimize the problem of false positive and false negative SNPs. The FarmCPU R package uses the FaST-LMM algorithm to calculate the K from selected pseudo-QTNs (Quantitative Trait Nucleotides) and not from the total SNP set, as the standard K. The threshold values were calculated by the p.threshold function of FarmCPU. This permutes the phenotypes to break any spurious relationship with the genotype. After obtaining a vector of the minimum p-values of each experiment, the 95% quantile value of the vector is recommended for p.threshold. Finally, quantile–quantile (Q–Q) plots were used to verify the fitness of the model, considering population structure and kinship as factors.
The additive and heterozygous (dis)advantage models were applied in GWAS analyses by using specific encodings for the SNP matrix. Concerning the additive SNP effect with two alleles (A1 and A2), the SNP matrix was coded by 0 (A1A1), 1(A1A2), and 2 (A2 A2), considering the A2 as the minor allele. In this context, the additive GWAS model assumes there is a linear change in the phenotype regarding the minor allele number of copies. On the other hand, in the heterozygous (dis)advantage GWAS model, the homozygous genotypes (A1A1 or A2A2) were assumed to have the same effect, while the heterozygous genotypes a different one, implying an increase or decrease of the effect on the trait. Therefore, the SNP matrix was coded by 0 (A1A1), 1 (A1A2), and 0 (A2A2) [33,34]. Box plots were then used to show the phenotype values by genotypes of the SNPs significantly associated with the traits.
The average linkage disequilibrium (LD) in the hybrid panel was investigated using the square allele frequency correlation coefficient r2 between all pairs of SNPs across the chromosomes using PLINK v.1.9 software [50]. The extension of LD decay was verified by plotting the r2 values against the physical distance of the SNPs. Moreover, the heterozygosity by hybrid and by SNP marker was estimated dividing the number of heterozygous loci by the total of SNP markers and maize genotypes, respectively.
Identification of candidate genes
The candidate genes associated with the significant SNPs were obtained from the B73 genome reference (version 4) in the MaizeGDB genome browser (https://www.maizegdb.org/). Complementary information was collected from the U.S. National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the Universal Protein Resource (http://www.uniprot.org/). Venn diagrams were constructed to summarize the number of candidate genes identified using the VennDiagram R package [51]. In addition, the sequences of the candidate genes were categorized functionally by Gene Ontology (GO) terms [52], disregarding those with hypothetical function. The terms were obtained using the Blast2GO software with the default parameters specified by the program [53] and were previously simplified using the GO Slim feature.
Results
The phenotypic effect of A. brasilense inoculation on the maize hybrids
Significant phenotypic differences among the 118 maize hybrids were observed for all traits evaluated, except PH and SDM (S1 Table). Furthermore, the genotypic performance for RDM, RV, RAD, SRL, SRSA, and RSR were affected significantly by the inoculation with A. brasilense; thus, only these traits were considered for subsequent analyses. In general, higher broad-sense heritabilities were found under inoculated treatment than non-inoculated (S1B Fig).
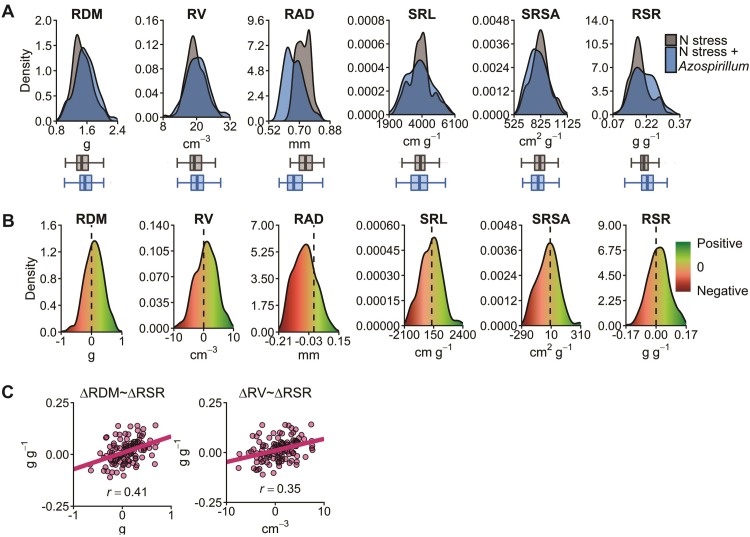
Regarding the density distribution of the adjusted means for all traits, larger phenotypic variances were found in the inoculated condition compared to the non-inoculated (Fig 1A). Overall, the inoculation increased the RDM, RV, RAD, and RSR while the opposite was observed for SRL and SRSA. Concerning the change due to inoculation (Δ) for all traits, a distribution close to normal was observed (Fig 1B). In this sense, most of the evaluated hybrids showed low responsiveness to A. brasilense. Moreover, a considerable portion of the genotypes showed negative responsiveness to A. brasilense; that is, a worse performance than the non-inoculated. The correlations between the ΔRDM and ΔRV with ΔRSR were 0.41 and 0.35, respectively (Fig 1C).
Fig 1. Density distributions and Pearson correlations of the phenotypic data.
(A) Density distribution and box plot of the maize hybrids adjusted means under N stress and N stress plus A. brasilense. (B) Density distribution of the Δ (difference between adjusted means of inoculated and non-inoculated treatments). (C) Pearson correlations between Δ values.
Population structure and LD decay
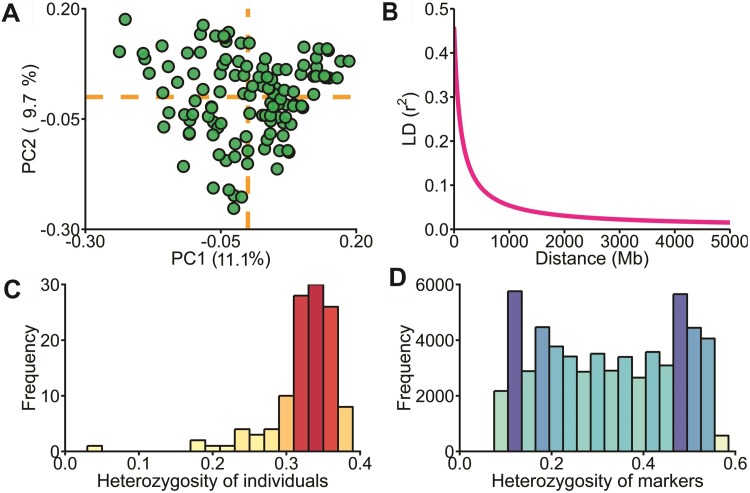
The genetic structure of the hybrid panel was accessed by PCA using 59,215 SNP markers (Fig 2A). The first two PCs captured a small percentage of the total variance (20.8%). In addition, the individuals had a wide distribution throughout the projection space, which indicates a weak structure among the genotypes. Moreover, a rapid decline in LD was observed (Fig 2B), with 121.7 kb extent when r2 reached 0.23 (half the maximum value). The average heterozygosity of hybrids was 0.32, ranging from 0.03 to 0.38 with most of the individuals presenting around 0.35 (Fig 2C). The low values found for some individuals indicate that some inbred lines used in the diallel crosses were of high genetic similarity. For the heterozygosity of markers, this value was also 0.32, varying from 0.10 to 0.61 (Fig 2D).
Fig 2. Population structure and genomic data.
(A) Population structure of the 118 maize hybrids revealed by the first two principal components of 59,215 SNP markers. (B) LD decay across the whole genome. (C) and (D) heterozygosity of individuals and markers, respectively.
Marker-trait associations
The additive and heterozygous (dis)advantage GWAS models were used to dissect the genetic basis of the traits RDM, RV, RAD, SRL, and SRSA under N stress and N stress plus A. brasilense conditions, since for these traits the genotypes showed a differential performance due to the inoculation effect. Only the genetic relatedness (K matrix) was used as a covariate in all GWAS analyses.Thus, the population structure information was not included due to the increase in the deviation from the expected p-values showed by Q–Q plots (not presented). Furthermore, based on the LD decay for this hybrid panel, the gene annotation was performed within a 50 kb sliding window around each significant SNP.
Concerning the additive GWAS model, eight significant SNP-trait associations were revealed in the maize hybrids evaluated under N stress plus A. brasilense (Table 1 and S2 and S6A Figs). In general, at least one candidate gene was identified for each trait, which was located on the chromosomes 2, 4, 6, 7, and 9. In addition, using the same model but for N-stress treatment, one significant association was detected for each trait, totaling five candidate genes, which were located on chromosomes 2, 5, and 6 (Table 1 and S3 and S6B Figs). However, for chromosome 5, position 149998432, no candidate gene was found within the window considered. The results for RSR in both treatments were disregarded due to poor adjustment with the expected values shown by the Q–Q plot.
Table 1. List of candidate genes around of significant SNPs identified by additive GWAS model.
| Trait | Candidate gene | SNP | Chr. | Position | MAF | Effect | P-value | Gene annotation |
|---|---|---|---|---|---|---|---|---|
| N stress plus Azospirillum brasilense | ||||||||
| RDM | Zm00001d051881 | T/C | 4 | 173317340 | 0.14 | 0.20 | 5.50x10-10 | Protein BTR1 |
| RDM | Zm00001d035859 | T/C | 6 | 56793578 | 0.42 | -0.20 | 1.31x10-16 | Plastocyanin homolog 1 |
| RV | Zm00001d006108 | C/G | 2 | 198321726 | 0.43 | -3.61 | 1.61x10-21 | Hydroxyproline-rich glycoprotein family protein |
| RAD | Zm00001d013098 | A/G | 5 | 4668442 | 0.46 | 0.04 | 3.14x10-24 | Aldehyde oxidase 2 |
| RAD | Zm00001d046604 | T/C | 9 | 98488802 | 0.32 | 0.02 | 3.52x10-11 | (Z)-3-hexen-1-ol acetyltransferase |
| SRL | Zm00001d005892 | A/G | 2 | 191920029 | 0.48 | -386.6 | 1.40x10-09 | Ethylene-responsive transcription factor ERF109 |
| SRL | Zm00001d020747 | T/C | 7 | 131108804 | 0.34 | -357.7 | 1.98x10-08 | Aquaporin TIP4-1 |
| SRSA | Zm00001d052221 | A/G | 4 | 183616939 | 0.29 | 83.61 | 4.13x10-10 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| N stress | ||||||||
| RDM | Zm00001d013098 | A/G | 5 | 4668442 | 0.46 | 0.02 | 3.14x10-24 | Aldehyde oxidase2 |
| RV | Zm00001d038300 | A/T | 6 | 153844954 | 0.32 | -1.97 | 2.89x10-13 | Putative cytochrome P450 superfamily protein |
| RAD | Zm00001d005892 | A/G | 2 | 191920029 | 0.48 | 0.02 | 1.40x10-09 | Ethylene-responsive transcription factor ERF109 |
| SRL | Zm00001d002930 | A/G | 2 | 27052534 | 0.21 | 487.5 | 3.24x10-11 | Hypothetical protein |
| SRSA | - | A/C | 5 | 149998432 | 0.40 | 86.9 | 2.93x10-10 | There is no candidate gene in the region |
Description of candidate genes found for root dry mass (RDM), root volume (RV), root average diameter (RAD), specific root length (SRL), and specific root surface area (SRSA) evaluated in maize hybrids under N stress and N stress plus Azospirillum brasilense. Chr: Chromosome and MAF: Minor allele frequency.
Two candidate genes identified in the inoculated treatment were similar to those identified under N-stress treatment, but for different traits. In this sense, the candidate genes Zm00001d013098 and Zm00001d005892 were related to RAD and SRL under A. brasilense treatment, and to RDM and RAD under non-inoculated treatment, respectively.
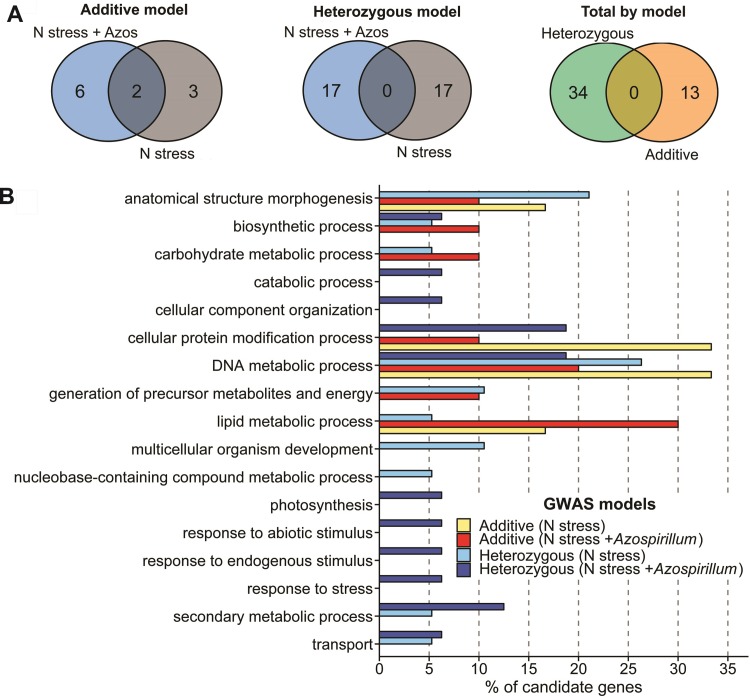
A higher number of significant associations were revealed using heterozygous (dis)advantage GWAS model. Seventeen significant SNPs were found associated with traits under N stress plus A. brasilense treatment located on chromosomes 1, 2, 3, 7, and 8 (Table 2 and S4 and S7 Figs). Several common candidate genes were found among the traits: Zm00001d029115 (RDM, RV, and RSR), Zm00001d037182 (RDM and RV), Zm00001d003312 (RV and RAD), and Zm00001d030590 (RAD and SRL). Under N stress, 17 significant associations were identified throughout the chromosomes 1, 2, 3, 4, 5, 6, and 9 (Table 2 and S5 and S8 Figs). For this model, any of the candidate genes were detected simultaneously for both inoculated and non-inoculated treatments (Fig 3A). No candidate genes were detected for chromosome 1 position 251090900 (RAD, inoculated treatment) and chromosome 3 position 165642810 (RDM, non-inoculated treatment).
Table 2. List of candidate genes around of significant SNPs identified by heterozygous (dis)advantage GWAS model.
| Trait | Candidate gene | SNP | Chr | Position | MAF | APHo | APHe | P-value | Gene annotation |
|---|---|---|---|---|---|---|---|---|---|
| N stress plus Azospirillum brasilense | |||||||||
| RDM | Zm00001d029115 | T/C | 1 | 58456902 | 0.09 | 1.34 | 1.51 | 1.92x10-10 | Protein strictosidine synthase-like |
| RDM | Zm00001d037182 | C/G | 6 | 114938556 | 0.10 | 1.40 | 1.26 | 1.07x10-11 | 12-oxo-phytodienoic acid reductase3 |
| RV | Zm00001d029115 | T/C | 1 | 58456902 | 0.09 | 17.75 | 20.31 | 4.37x10-13 | Protein strictosidine synthase-like |
| RV | Zm00001d032763 | A/G | 1 | 237658345 | 0.19 | 18.50 | 17.71 | 1.57x10-08 | Pre-mRNA-processing factor 19 homolog 2 |
| RV | Zm00001d003312 | T/G | 2 | 39796017 | 0.19 | 18.70 | 17.37 | 5.56x10-08 | 3-ketoacyl-CoA thiolase 2 peroxisomal |
| RV | Zm00001d037182 | C/G | 6 | 114938556 | 0.10 | 18.67 | 16.28 | 3.58x10-13 | 12-oxo-phytodienoic acid reductase3 |
| RAD | Zm00001d030590 | A/G | 1 | 146746338 | 0.17 | 0.66 | 0.69 | 3.02x10-13 | Hypothetical protein |
| RAD | - | T/G | 1 | 251090900 | 0.22 | 0.67 | 0.66 | 1.57x10-08 | There is no candidate gene in the region |
| RAD | Zm00001d003312 | T/G | 2 | 39796017 | 0.19 | 0.68 | 0.65 | 2.79x10-09 | 3-ketoacyl-CoA thiolase 2 peroxisomal |
| RAD | LOC103636767 | T/C | 8 | 14392135 | 0.18 | 0.66 | 9.68 | 5.99x10-11 | Formin-like protein 20 |
| SRL | Zm00001d030590 | A/G | 1 | 146746338 | 0.17 | 4364.2 | 4079.0 | 2.25x10-10 | Hypothetical protein |
| SRL | Zm00001d002736 | T/C | 2 | 20783203 | 0.18 | 4179.5 | 4433.6 | 2.52x10-09 | Carotenoid cleavage dioxygenase7 |
| SRL | Zm00001d008828 | T/C | 8 | 21875974 | 0.18 | 4146.3 | 4485.5 | 1.02x10-11 | Uncharacterized loci |
| SRSA | Zm00001d033957 | A/T | 1 | 27957495 | 0.20 | 656.8 | 680.9 | 1.86x10-08 | Helix-loop-helix DNA-binding domain containing protein |
| RSR | Zm00001d029115 | T/C | 1 | 58456902 | 0.09 | 0.20 | 0.26 | 9.11x10-12 | Protein strictosidine synthase-like |
| RSR | Zm00001d043812 | A/G | 3 | 210821486 | 0.20 | 0.21 | 0.20 | 6.28x10-1 | Isopentenyl transferase3B |
| RSR | Zm00001d020647 | T/C | 7 | 126412420 | 0.23 | 0.22 | 0.19 | 7.50x10-08 | Phospholipid:diacylglycerol acyltransferase 1 |
| N stress | |||||||||
| RDM | Zm00001d003331 | T/C | 2 | 40341681 | 0.22 | 1.41 | 1.32 | 9.16x10-11 | Putative WRKY transcription factor 34 |
| RDM | Zm00001d006036 | A/G | 2 | 195919131 | 0.27 | 1.40 | 1.34 | 3.55x10-08 | Heat shock 70 kDa protein 9 mitochondrial |
| RDM | - | T/C | 3 | 165642810 | 0.21 | 1.41 | 1.32 | 5.96x10-08 | There is no candidate gene in the region |
| RDM | Zm00001d044754 | A/T | 9 | 1340463 | 0.17 | 1.41 | 1.30 | 4.52x10-11 | Pyrophosphate—fructose 6-phosphate 1-phosphotransferase subunit beta 2 |
| RV | LOC109941493 | T/C | 1 | 162580315 | 0.12 | 17.59 | 20.17 | 7.31x10-16 | Plasma membrane ATPase 2-like |
| RV | Zm00001d036118 | T/C | 6 | 72999857 | 0.26 | 17.63 | 18.74 | 1.09x10-09 | Putative homeobox DNA-binding and leucine zipper domain family protein |
| RAD | Zm00001d006722 | T/C | 2 | 215259958 | 0.23 | 0.68 | 0.65 | 3.50x10-08 | Arabinosyltransferase RRA3 |
| RAD | Zm00001d037514 | T/C | 6 | 127764798 | 0.21 | 0.68 | 0.65 | 1.49x10-09 | Putative uncharacterized mitochondrial protein |
| SRL | LOC100279630 | T/C | 1 | 4994455 | 0.15 | 4117.5 | 4646.4 | 8.56x10-17 | MADS-box transcription factor family protein |
| SRL | Zm00001d029134 | T/C | 1 | 59906568 | 0.25 | 4401.4 | 4138.5 | 5.25x10-09 | CW-type Zinc Finger |
| SRL | Zm00001d029247 | T/C | 1 | 63585160 | 0.12 | 4151.8 | 4632.4 | 2.23x10-14 | ARM repeat superfamily protein |
| SRL | Zm00001d029385 | T/C | 1 | 68562928 | 0.25 | 4429.5 | 4115.7 | 1.74x10-07 | AAA-type ATPase family protein |
| SRL | Zm00001d030287 | T/C | 1 | 119697532 | 0.25 | 4194.0 | 4343.3 | 2.67x10-08 | Protein CLT2 chloroplastic |
| SRL | Zm00001d013070 | T/C | 5 | 4219053 | 0.25 | 4300.5 | 4239.4 | 7.64x10-09 | Transcription factor MYB98 |
| SRL | Zm00001d037596 | A/T | 6 | 130932567 | 0.20 | 4445.7 | 4013.6 | 1.78E-07 | RING/U-box superfamily protein |
| SRSA | LOC100279630 | T/C | 1 | 4994455 | 0.15 | 636.8 | 739.5 | 1.99x10-13 | MADS-box transcription factor family protein |
| RSR | Zm00001d051886 | C/G | 4 | 173630271 | 0.15 | 0.18 | 0.22 | 1.04x10-13 | Putative MATE efflux family protein |
Description of candidate genes found for root dry mass (RDM), root volume (RV), root average diameter (RAD), specific root length (SRL), and specific root surface area (SRSA) evaluated in maize hybrids under N stress and N stress plus Azospirillum brasilense. Chr: Chromosome, MAF: Minor allele frequency, APHo: Average phenotype of individuals with homozygous genotype, and APHe: Average phenotype of individuals with heterozygous genotype.
Fig 3. Candidate genes found by different GWAS models for N stress and N stress plus Azospirillum brasilense.
(A) Venn diagrams showing the unique and shared significant SNPs. (B) Enriched GO terms.
In total, 47 significant SNP-trait associations were found, where 25 were related to traits under N stress plus A. brasilense and 22 to N stress. Regarding the models, 13 significant associations were identified by using the additive GWAS model and 34 the heterozygous (dis)advantage model. There was no candidate gene shared between them (Fig 3A). Finally, the nature of the SNP effect on the traits, positive or negative, was independent of the treatment or GWAS model (S6, S7, and S8 Figs).
The categorization of candidate genes sequences according to biological process using the Blast2GO software showed that just one biosynthetic category was present in all treatments (Fig 3B). Moreover, in general, the candidate genes found by additive GWAS model tended to be mainly enriched for terms such as “DNA metabolic process” and “lipid metabolic process”. In turn, those found by the heterozygous (dis)advantage model showed more exclusive biological functions; for example, “catabolic process”, “cellular component organization”, “response to stress”, and “secondary metabolic process”. Comparing the inoculated and non-inoculated treatments, a different pattern of categorization was seen between them, especially for the candidate genes found by the heterozygous (dis)advantage model.
Discussion
Genotypic variation of maize to A. brasilense under nitrogen stress
One of our aims was to evaluate the genetic variability of the responsiveness of maize hybrids to the inoculation with the PGPB A. brasilense and the genetic control of related traits to this effect. The few studies that have reported the differential responsiveness among maize genotypes to A. brasilense inoculation were based on a smaller number of hybrids or inbred lines [5,6,54]. Moreover, as far as we know, our report has evaluated the largest number of maize genotypes for their association with PGPB.
In general, Azospirlillum spp. promotes several benefits and changes in maize, including phytohormone production of auxins, cytokinins, and gibberellins [55,56], plant growth and yield [35,55], contents of secondary metabolites [57], photosynthetic potential [1], anatomical pattern (e. g. metaxylem vessel elements) and architecture of roots [31,58], N2 fixation [6], fertilizer-N recovery [59], tolerance of abiotic stresses (e. g. N limitation and drought conditions) [55,60]. In this work, the inoculation of A. brasilense under N stress promoted significant change in maize performance for six root-related traits: RDM, RV, RAD, SRL, SRSA, and RSR. Some studies have also shown the positive effect of the inoculation of Azospirillum spp. on RDM, RV, and the promotion of thinner root growth [55,61,62], but our study reports for the first time the response in SRL and SRSA.
Our results did not show pronounced differences among the distributions of adjusted means of the hybrids under N stress and N stress plus A. brasilense. However, we observed a significant variation in the delta (the difference between inoculated and non-inoculated treatments), including some of the maize hybrids showing negative effects on the traits due to the inoculation. This result shows that adding only one PGPB to the microbiome is enough to expand the range of maize plant responses under low-N stress. This may be because microbes alter the plant functioning and confer different characteristics to the host plant, reinforcing the emerging idea of the holobiont being a unit of selection that possess a larger variability to be explored in plant breeding [63–65].
Studies reporting a decrease in the phenotypic traits of host plants due to the inoculation with PGPBs, such as A. brasilense, are not common in the literature [66]. One possibility is that the genotypes with a negative response to inoculation can have more unfavorable alleles related to the association with A. brasilense. For example, triggering plant defense responses incurs an energetic cost [67], which may lead to a reduction in resources to root system development, causing a worse growth than only the N-stress condition would already entail. In addition, similarly to the plant–endophyte interactions, the “balanced antagonism theory” applies to the plant–PGPB relationships [68,69]; then, phenotypic plasticity in the host plants may vary from mutualism to antagonism depending on the plant genotype, the environmental conditions, and the bacterial strain.
Another explanation for the negative responsiveness is because the effect of A. brasilense on the plant can vary according to the concentration of the inoculant [66,70]. In general, plant hormones are stimulatory only at certain concentrations, which should not exceed the stimulatory threshold specific to each plant genotype [71]. The higher concentration of A. brasilense in the root environment may increase the release of plant hormones that consequently inhibit root growth [66]. Thus, considering the number of genotypes evaluated, the concentration of the inoculant used in our experiment could be unfavorable for some of them, even at the recommended dose.
On the other hand, the reduction in root traits due to inoculation would not necessarily be a negative factor for the plant. Under abiotic stress conditions, such as low N supply and drought, high root/shoot ratios are common [72,73]. In this sense, we found moderate positive correlations between the ΔRDM and ΔRV root traits and ΔRSR, which indicates that under A. brasilense inoculation some plant genotypes could reduce the investment in root growth in order to allocate it to shoot development. However, further studies are needed to better understand the influence of inoculation with this PGPB on the distribution of dry matter between roots and shoots.
The continuous phenotypic variation and the moderate estimates of heritability for the traits related to the maize responsiveness to A. brasilense suggest the influence of several genes of small effect and a strong environmental influence. In summary, these results reinforce the complex PGPB × plant × environment interactions. Furthermore, they show the possibility of improving plants to be more efficient by the association with PGPB.
Candidate genes related to the maize responsiveness to A. brasilense
To the best of our knowledge, this is the first report employing GWAS to assess the genetic architecture of the association of maize with A. brasilense. Several candidate genes related to the maize responsiveness to A. brasilense were detected. Considering the panel size used in our study, possibly the power of our GWAS analyses has been low and only theose SNPs with more greater effect have been identified [74]. Korte & Farlow [75] suggest that a way to mitigate the small sample effect is to account for large phenotypic variability. Thus, as we used hybrids rather than inbred lines, a series of different allelic combinations can occur, increasing the genetic variants with heterozygous loci and thereby allowing the finding of better results in GWAS analysis [76]. This is reflected in the number of significant SNPs identified by the heterozygous (dis)advantage model, which was about three times higher than the additive model. Consequently, given the high number of candidate genes found, we focused our discussion mainly on those with functions more related to the treatments of this study.
It is known that the colonization of host plants by beneficial microbes depends on their ability to manipulate defense-related pathways [4]. In this study, the candidate gene Zm00001d051881 (additive model) was found, which codes the protein Binding to ToMV RNA 1 (BTR1). This is involved in the defense against Tomato Mosaic Virus (TOMV) RNA, with possible indirect effects on the host’s innate immunity [77]. In addition, Zm00001d052221 (additive model) codes the tetratricopeptide repeat (TPR)-like protein superfamily, which is determinant for the transduction of signals mediated by plant hormones and able to activate the plant’s defense response. For example, TPR is related to the quantitative resistance of soybean to Fusarium graminearum [78]. Another candidate gene is the ethylene-responsive transcription factor ERF109 (Zm00001d005892, additive model), which besides being involved in ethylene-activated abiotic stress responses [79], induces the expression of defense-related genes promoting a positive modulation of the response against pathogen infections [80]. Gene Zm00001d029115, identified for two traits using the heterozygous (dis)advantage model, codes the protein strictosidine synthase-like, known to play a key role in the alkaloid biosynthesis pathway. These chemical compounds function as protection against pathogenic microorganisms and herbivorous animals. In addition, the improvement in alkaloid content in the roots has been observed with A. brasilense inoculation in medicinal plants [81], but there are no reports about its induction in cereal crops.
The modulation of plant hormones and related signaling pathways by A. brasilense are aspects also frequently reported [2,61]. For example, we found Zm00001d013098 (additive model) corresponding to Aldehyde oxidase 2, which is a key enzyme in the final step of abscisic acid (ABA) biosynthesis. In addition, it performs the final catalytic conversion of indole-3-acetaldehyde (IAAld) in indole-3-acetic acid (AIA) in different tryptophan-dependent auxin biosynthesis pathways [82]. Moreover, we found the candidate gene for 12-oxo-phytodienoic acid reductases (Zm00001d037182, heterozygous model), which are key enzymes in the control of jasmonate (JA) biosynthesis in plants such as maize [83] and wheat [83]. Among other functions, this phytohormone orchestrates defense and growth responses [84].
Some studies show modulation of the induction and emission of plant volatiles by plant-associated microorganisms, including PGPBs and Rhizobia [85,86]. In turn, these chemicals have an important role, especially on the induction of resistance in plants against insects and pathogens [87,88]. We found the Zm00001d046604 candidate gene (additive model) corresponding to (Z)-3-hexen-1-ol acetyltransferase. This enzyme is involved in the green leaf volatile biosynthetic process that is derived from the lipoxygenase pathway [89]. In agreement with this finding, A. brasilense negatively affects the attraction of the pest insect Diabrotica speciose to maize by inducing higher emissions of the volatile (E)-β-caryophyllene. Therefore, the validation of this candidate gene in further studies could help to understand better the role of plant defense against pests induced by A. brasilense.
Regarding the candidate genes related to abiotic stress mitigation, we found Zm00001d020747 (additive model), encoding Aquaporin TIP4-1. Under N deficiency, this plant transporter is up-regulated in Arabidopsis [90] and it is induced by rhizobial and arbuscular mycorrhizal fungi symbiosis [91]. In both cases, its function is related to the N delivery between plant compartments.
We found a candidate gene directly involved in plant root growth encoding hydroxyproline-rich glycoprotein family protein (Zm00001d006108, additive model), a protein family from plant cell walls classified as arabinogalactan-proteins (AGPs), extensins (EXTs), and proline-rich proteins (PRPs). It plays a key role in several processes of plant development such as root elongation and root biomass, especially under stress conditions [92]. Additionally, AGPs are exuded in the rhizosphere and help in communicating with soil microbes, participate in the signaling cascade modulating the plant’s immune system, and are required for root colonization by symbiotic bacteria [93]. Another, LOC103636767 (heterozygous model), corresponds to formin-like protein 20, which is involved in cytoskeleton movement and secondary cell wall formation [94].
The major part of N in the leaf is allocated to the chloroplast proteins, and deficiency in this nutrient leads to a reduction in photosynthetic efficiency [95]. The Zm00001d035859 candidate gene (additive model) found in our study is related to the Plastocyanin homolog 1, a protein involved in the transfer of electrons in the photosystem I. In accordance with this result, the inoculation of the PGPB Burkholderia sp. in Arabidopsis thaliana leads to modification in the expression of this protein [96]. Moreover, it is involved in the response of maize to N deficiency [97].
Although the candidate genes found for the N-stress treatment were not the main focus of this study, many of them have been previously described due to their direct or indirect relation to plant responses to abiotic stress conditions. The LOC109941493 gene (heterozygous model) encodes the plasma membrane ATPase 2-like, this being an ion pump in the plant cell membrane important for root growth and architecture under different nitrogen regimes [98]. In addition, Zm00001d006722 (heterozygous model) is related to arabinosylation of extensin proteins that contribute to root-cell hair growth, these being specialized in the absorption of nutrients [99]. The Zm00001d013098 and Zm00001d038300 genes (additive model) corresponding to Aldehyde oxidase2 and Ethylene-responsive transcription factor ERF109, respectively, were the only candidate genes shared between both treatments. Their functions described above, related to ABA and AIA biosynthesis and the ethylene-activated signaling pathway, are also frequently reported in N availability and hormone interactions [100,101]. Moreover, this suggests that the regulation and signalization of these hormones in the plant can be involved in the cross talk between A. brasilense and N stress in maize. Therefore, besides these results indicating that the stress applied in our experiment was effective, they also could be helpful in further studies to understand better the genetic control of root traits under N stress in the early stages of plant development for improving tolerance in maize.
Some of the candidate genes found by the heterozygous (dis)advantage GWAS model were identified for more than one trait; this was not observed using the additive model. For these, the effect on phenotypes was always in the same direction; for example, the candidate gene Zm00001d029115 (protein strictosidine synthase-like) had a negative effect on both RDM and RV. Possibly, this occurred because these pleiotropic candidate genes were only found between RDM, RV, and RAD traits which showed a positive correlation among them.
Additive and heterozygous (dis)advantage GWAS models
GWAS analyses using non-additive models are common in human and animal studies [102–104]. However, few studies have been reported using plant species [21,22]. In our study, most of the significant SNPs were identified by heterozygous (dis)advantage GWAS analyses and none were detected by the additive model, which demonstrates how important it is to study the non-additive effects on the genetic variability of maize responsiveness to both A. brasilense and N stress. This was also evident through the results of GO terms, where an increase in exclusive biological functions was verified. This indicates that the PGPB provide the plant with a broader spectrum of internal activities, which may be an advantage for growth in stressful environments, such as N deficiency, with possible consequences in plant evolutionary potential.
Furthermore, our results showed that heterozygous genotypes can have advantages or disadvantages on the root traits (both treatments) depending on the allelic combinations that are formed by the parental crossing. Thus, the strategy of use of SNP‒trait associations found by heterozygous loci in breeding programs depends on the effect of the heterozygous genotype. This is a challenge to plant breeders because during hybrid development the allele combination should be predicted by parental selection in order to benefit its association with PGPB. In this sense, further studies underlying these candidate genes are required to understand better the biological mechanisms of heterotic performance, in comparison to homozygous, in the presence of these PGPB. For those providing an advantage, the alleles should be improved separately in different heterotic groups for their subsequent combination in the mating process. On the other hand, when the heterozygous genotypes are a disadvantage, one or other allele should be improved simultaneously in both heterotic groups in order to obtain homozygous genotypes in hybrids.
Conclusions
Our study modeling additive and heterozygous (dis)advantage effects in GWAS analyses revealed 25 candidate genes for the responsiveness of maize to A. brasilense, with key roles particularly in plant defense, hormonal biosynthesis, signaling pathways, and root growth providing insights into their complex genetic architecture. In this context, non-additive effects contribute substantially to the maize phenotypic variation in response to the inoculation and are related to a wider spectrum of biological functions. Together, these findings allow to be started the marker-assisted selection and genome editing in breeding programs for the development of maize hybrids that can take advantage of this association more efficiently. Finally, our results also represent a benchmark in the identification of homologous genes in important related species, such as rice and wheat, besides advancing the understanding of the genetic basis of plant–PGPB interactions.
Supporting information
Wald Test for fixed effects and Likelihood Ratio Test for random effects from the joint diallel analysis of 118 maize hybrids evaluated under N stress and N stress plus Azospirillum brasilense treatments.
(DOCX)
(A) Pearson correlation between maize hybrid adjusted means under N stress and N stress plus A. brasilense. (B) Estimates of variance components (: genotypic variance; : genotype-by-year variance; : error variance) and broad-sense heritabilities.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) N stress plus Azospirillum brasiliense and (B) N stress.
(TIF)
(TIF)
(TIF)
Acknowledgments
We thank the team at the Allogamous Plant Breeding and Microbiology Laboratories for technical and scientific support. We also thank Mariangela Hungria from Embrapa Soybean for the supply of Azospirillum brasilense Ab-V5 strain.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by São Paulo Research Foundation (FAPESP) process: 2015/01188-9, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) - Finance Code 001, and National Council Coordination for the Scientific and Technological Development (CNPq).
References
- 1.Fukami J, Ollero FJ, Megías M, Hungria M. Phytohormones and induction of plant-stress tolerance and defense genes by seed and foliar inoculation with Azospirillum brasilense cells and metabolites promote maize growth. AMB Express. 2017; 7: 153 10.1186/s13568-017-0453-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukami J, Ollero FJ, de la Osa C, Valderrama-Fernández R, Nogueira MA, Megías M, et al. Antioxidant activity and induction of mechanisms of resistance to stresses related to the inoculation with Azospirillum brasilense. Arch Microbiol. 2018; 200: 1191–1203. 10.1007/s00203-018-1535-x [DOI] [PubMed] [Google Scholar]
- 3.Cassán F, Diaz-Zorita M. Azospirillum sp. in current agriculture: From the laboratory to the field. Soil Biol Biochem. 2016; 103: 117–130. 10.1016/j.soilbio.2016.08.020 [DOI] [Google Scholar]
- 4.Carvalho TLG, Ballesteros HGF, Thiebaut F, Ferreira PCG, Hemerly AS. Nice to meet you: genetic, epigenetic and metabolic controls of plant perception of beneficial associative and endophytic diazotrophic bacteria in non-leguminous plants. Plant Mol Biol. 2016; 90: 561–574. 10.1007/s11103-016-0435-1 [DOI] [PubMed] [Google Scholar]
- 5.Cunha FN, Silva NF da, Ribeiro Rodrigues C, Alves Morais W, Ferreira Gomes FH, Filho Lopes, Luiz C, et al. Performance of different genotypes of maize subjected to inoculation with Azospirillum brasilense. African J Agric Res. 2016; 11: 3853–3862. 10.5897/AJAR2016.11496 [DOI] [Google Scholar]
- 6.Brusamarello-Santos LC, Gilard F, Brulé L, Quilleré I, Gourion B, Ratet P, et al. Metabolic profiling of two maize (Zea mays L.) inbred lines inoculated with the nitrogen fixing plant-interacting bacteria Herbaspirillum seropedicae and Azospirillum brasilense. Aroca R, editor. PLoS One. 2017; 12: e0174576 10.1371/journal.pone.0174576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosas JE, Martínez S, Blanco P, Pérez de Vida F, Bonnecarrère V, Mosquera G, et al. Resistance to Multiple Temperate and Tropical Stem and Sheath Diseases of Rice. Plant Genome. 2017; 11: 1–13. 10.3835/plantgenome2017.03.0029 [DOI] [PubMed] [Google Scholar]
- 8.Genissel A, Confais J, Lebrun M-H, Gout L. Association Genetics in Plant Pathogens: Minding the Gap between the Natural Variation and the Molecular Function. Front Plant Sci. 2017; 8: 8–11. 10.3389/fpls.2017.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehnert H, Serfling A, Enders M, Friedt W, Ordon F. Genetics of mycorrhizal symbiosis in winter wheat (Triticum aestivum). New Phytol. 2017; 215: 779–791. 10.1111/nph.14595 [DOI] [PubMed] [Google Scholar]
- 10.De Vita P, Avio L, Sbrana C, Laidò G, Marone D, Mastrangelo AM, et al. Genetic markers associated to arbuscular mycorrhizal colonization in durum wheat. Sci Rep. 2018; 8: 10612 10.1038/s41598-018-29020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wallace JG, Kremling KA, Kovar LL, Buckler ES. Maize Leaf Microbiome. Phyt. J. 2018; 2:208–224. 10.1094/PBIOMES-02-18-0008-R [DOI] [Google Scholar]
- 12.Kamfwa K, Cichy KA, Kelly JD. Genome-wide association analysis of symbiotic nitrogen fixation in common bean. Theor Appl Genet. 2015; 128: 1999–2017. 10.1007/s00122-015-2562-5 [DOI] [PubMed] [Google Scholar]
- 13.Wintermans PCA, Bakker PAHM, Pieterse CMJ. Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol Biol. 2016; 90: 623–634. 10.1007/s11103-016-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pieterse CMJ, de Jonge R, Berendsen RL. The Soil-Borne Supremacy. Trends Plant Sci. 2016;21: 171–173. 10.1016/j.tplants.2016.01.018 [DOI] [PubMed] [Google Scholar]
- 15.Carvalhais LC, Dennis PG, Fan B, Fedoseyenko D, Kierul K, Becker A, et al. Linking Plant Nutritional Status to Plant-Microbe Interactions. PLoS One. 2013; 8: e68555 10.1371/journal.pone.0068555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castrillo G, Teixeira PJPL, Paredes SH, Law TF, De Lorenzo L, Feltcher ME, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017; 543: 513–518. 10.1038/nature21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes EA, Lana UGP, Quensen JF, Oliveira CA, Guo J, Guimar LJM, et al. Root-associated microbiome of maize genotypes with contrasting phosphorus use efficiency. Phyt. J. 2018; 2: 129–137. 10.1094/PBIOMES-03-18-0012-R [DOI] [Google Scholar]
- 18.Yang J, Mezmouk S, Baumgarten A, Buckler ES, Guill KE, McMullen MD, et al. Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLOS Genet. 2017; 13: e1007019 10.1371/journal.pgen.1007019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Coffey L, Garfin J, Miller ND, White MR, Spalding EP, et al. Genotype-by-environment interactions affecting heterosis in maize. PLoS One. 2018; 13: e0191321 10.1371/journal.pone.0191321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Yang Q, Gao L, Zhang M, Ni Z, Zhang Y. Identification of Heterosis-Associated stable QTLs for ear-weight-related traits in an elite maize hybrid by Zhengdan 958 Design III. Front Plant Sci. 2017; 8: 1–10. 10.3389/fpls.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnafous F, Fievet G, Blanchet N, Boniface MC, Carrère S, Gouzy J, et al. Comparison of GWAS models to identify non-additive genetic control of flowering time in sunflower hybrids. Theor Appl Genet. 2018; 131: 319–332. 10.1007/s00122-017-3003-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monir MM, Zhu J. Dominance and Epistasis Interactions Revealed as Important Variants for Leaf Traits of Maize NAM Population. Front Plant Sci. 2018; 9: 1–10. 10.3389/fpls.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picard C, Bosco M. Maize heterosis affects the structure and dynamics of indigenous rhizospheric auxins-producing Pseudomonas populations. FEMS Microbiol Ecol. 2005; 53: 349–357. 10.1016/j.femsec.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 24.Picard C, Bosco M. Heterozygosis drives maize hybrids to select elite 2,4-diacethylphloroglucinol-producing Pseudomonas strains among resident soil populations. FEMS Microbiol Ecol. 2006; 58: 193–204. 10.1111/j.1574-6941.2006.00151.x [DOI] [PubMed] [Google Scholar]
- 25.Groszmann M, Gonzalez-Bayon R, Lyons RL, Greaves IK, Kazan K, Peacock WJ, et al. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc Natl Acad Sci. 2015; 112: E6397–E6406. 10.1073/pnas.1519926112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Wang H, Li K, Wu Y, Liu Z, Huang C. Genome-wide proteomic profiling reveals the role of dominance protein expression in heterosis in immature maize ears. Sci Rep. 2017; 7: 16130 10.1038/s41598-017-15985-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko DK, Rohozinski D, Song Q, Taylor SH, Juenger TE, Harmon FG, et al. Temporal Shift of Circadian-Mediated Gene Expression and Carbon Fixation Contributes to Biomass Heterosis in Maize Hybrids. PLOS Genet. 2016; 12: e1006197 10.1371/journal.pgen.1006197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paschold A, Marcon C, Hoecker N, Hochholdinger F. Molecular dissection of heterosis manifestation during early maize root development. Theor Appl Genet. 2010; 120: 383–388. 10.1007/s00122-009-1082-6 [DOI] [PubMed] [Google Scholar]
- 29.Hardoim PR, de Carvalho TLG, Ballesteros HGF, Bellieny-Rabelo D, Rojas CA, Venancio TM, et al. Genome-wide transcriptome profiling provides insights into the responses of maize (Zea mays L.) to diazotrophic bacteria. Plant Soil. Plant and Soil; 2019; 10.1007/s11104-019-03939-9 [DOI] [Google Scholar]
- 30.Planchamp C, Glauser G, Mauch-Mani B. Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front Plant Sci. 2015; 5: 1–10. 10.3389/fpls.2014.00719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calzavara AK, Paiva PHG, Gabriel LC, de Oliveira ALM, Milani K, Oliveira HC, et al. Associative bacteria influence maize (Zea mays L.) growth, physiology and root anatomy under different nitrogen levels. Plant Biol. 2018; 20: 870–878. 10.1111/plb.12841 [DOI] [PubMed] [Google Scholar]
- 32.Di Salvo LP, Ferrando L, Fernández-Scavino A, García de Salamone IE. Microorganisms reveal what plants do not: wheat growth and rhizosphere microbial communities after Azospirillum brasilense inoculation and nitrogen fertilization under field conditions. Plant Soil. Plant and Soil; 2018; 1–13. 10.1007/s11104-017-3548-7 [DOI] [Google Scholar]
- 33.Tsepilov YA, Shin S-Y, Soranzo N, Spector TD, Prehn C, Adamski J, et al. Nonadditive Effects of Genes in Human Metabolomics. Genetics. 2015; 200: 707–718. 10.1534/genetics.115.175760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goyette P, Boucher G, Mallon D, Ellinghaus E, Jostins L, Huang H, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015; 47: 172–9. 10.1038/ng.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hungria M, Campo RJ, Souza EM, Pedrosa FO. Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil. 2010; 331: 413–425. 10.1007/s11104-009-0262-0 [DOI] [Google Scholar]
- 36.Rodrigues Neto J, Malavolta VA Jr, Victor O. Meio simples para o isolamento e cultivo de Xanthomonas campestris pv. citri Tipo B. Summa Phytopathol. 1986; 12: 32. [Google Scholar]
- 37.Mendonça LDF, Granato ÍSC, Alves FC, Morais PPP, Vidotti MS, Fritsche-Neto R. Accuracy and simultaneous selection gains for N-stress tolerance and N-use efficiency in maize tropical lines. Sci Agric. 2017; 74: 481–488. 10.1590/1678-992x-2016-0313 [DOI] [Google Scholar]
- 38.Lanes ECM, Viana JMS, Paes GP, Paula MFB, Maia C, Caixeta ET, et al. Population structure and genetic diversity of maize inbreds derived from tropical hybrids. Genet Mol Res. 2014; 13: 7365–7376. 10.4238/2014.September.12.2 [DOI] [PubMed] [Google Scholar]
- 39.Morosini JS, Mendonça LDF, Lyra DH, Galli G, Vidotti MS, Fritsche-Neto R. Association mapping for traits related to nitrogen use efficiency in tropical maize lines under field conditions. Plant and Soil; 2017; 421: 453–463. 10.1007/s11104-017-3479-3 [DOI] [Google Scholar]
- 40.Kox MAR, Lüke C, Fritz C, van den Elzen E, van Alen T, Op den Camp HJM, et al. Effects of nitrogen fertilization on diazotrophic activity of microorganisms associated with Sphagnum magellanicum. Plant and Soil; 2016; 406: 83–100. 10.1007/s11104-016-2851-z [DOI] [Google Scholar]
- 41.Zeffa DM, Fantin LH, Santos OJAP dos, Oliveira ALM de, Canteri MG, Scapim CA, et al. The influence of topdressing nitrogen on Azospirillum spp. inoculation in maize crops through meta-analysis. Bragantia. 2018; 77: 493–500. 10.1590/1678-4499.2017273 [DOI] [Google Scholar]
- 42.Vidotti MS, Matias FI, Alves FC, Pérez-Rodríguez P, Beltran GA, Burgueño J, et al. Maize responsiveness to Azospirillum brasilense: Insights into genetic control, heterosis and genomic prediction. PLoS One. 2019; 14: e0217571 10.1371/journal.pone.0217571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trachsel S, Kaeppler SM, Brown KM, Lynch JP. Maize root growth angles become steeper under low N conditions. F Crop Res. 2013; 140: 18–31. 10.1016/j.fcr.2012.09.010 [DOI] [Google Scholar]
- 44.Butler DG, Cullis BR, Gilmour AR, Gogel BJ. Analysis of Mixed Models for S–language Environments: ASReml-R Reference Manual. 2009; Available: https://vsn.klever.co.uk/downloads/asreml/release3/asreml-R.pdf
- 45.Brien C. asremlPlus: Augments the Use of “ASReml-R” in Fitting Mixed Models. R Packag Version 20–12. Available: https://cran.r-project.org/web/packages/asremlPlus/asremlPlus.pdf
- 46.Unterseer S, Bauer E, Haberer G, Seidel M, Knaak C, Ouzunova M, et al. A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genomics. 2014; 15: 823 10.1186/1471-2164-15-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet. 2016; 98: 116–126. 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012; 28: 3326–3328. 10.1093/bioinformatics/bts606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Huang M, Fan B, Buckler ES, Zhang Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016; 12: 100 10.1371/journal.pgen.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007; 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011; 12: 35 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K D, t SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM SG. Gene Ontology: tool for the unification of biology. Nat Genet. 2000; 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21: 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 54.Rozier C, Erban A, Hamzaoui J, Prigent-Combaret C, Comte G, Kopka J, et al. Xylem Sap Metabolite Profile Changes During Phytostimulation of Maize by the Plant Growth-Promoting Rhizobacterium, Azospirillum lipoferum CRT1. J Postgenomics Drug Biomark Dev. 2016; 6: 1–10. 10.4172/2153-0769.1000182 [DOI] [Google Scholar]
- 55.Zeffa DM, Perini LJ, Silva MB, de Sousa NV, Scapim CA, Oliveira ALM de, et al. Azospirillum brasilense promotes increases in growth and nitrogen use efficiency of maize genotypes. PLoS One. 2019; 14: e0215332 10.1371/journal.pone.0215332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen AC, Travaglia CN, Bottini R, Piccoli PN. Participation of abscisic acid and gibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botany. 2009; 87: 455–462. 10.1139/B09-023 [DOI] [Google Scholar]
- 57.Walker V, Bertrand C, Bellvert F, Moënne-Loccoz Y, Bally R, Comte G. Host plant secondary metabolite profiling shows a complex, strain-dependent response of maize to plant growth-promoting rhizobacteria of the genus Azospirillum. New Phytol. 2011; 189: 494–506. 10.1111/j.1469-8137.2010.03484.x [DOI] [PubMed] [Google Scholar]
- 58.Rozier C, Hamzaoui J, Lemoine D, Czarnes S, Legendre L. Field-based assessment of the mechanism of maize yield enhancement by Azospirillum lipoferum CRT1. Sci Rep. 2017; 7: 1–12. 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martins MR, Jantalia CP, Reis VM, Döwich I, Polidoro JC, Alves BJR, et al. Impact of plant growth-promoting bacteria on grain yield, protein content, and urea-15 N recovery by maize in a Cerrado Oxisol. Plant and Soil; 2018; 422: 239–250. 10.1007/s11104-017-3193-1 [DOI] [Google Scholar]
- 60.Curá JA, Franz DR, Filosofía JE, Balestrasse KB, Burgueño LE. Inoculation with Azospirillum sp. and Herbaspirillum sp. bacteria increases the tolerance of maize to drought stress. Microorganisms. 2017; 5: 41 10.3390/microorganisms5030041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spaepen S, Bossuyt S, Engelen K, Marchal K, Vanderleyden J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014; 201: 850–861. 10.1111/nph.12590 [DOI] [PubMed] [Google Scholar]
- 62.Chamam A, Wisniewski-Dyé F, Comte G, Bertrand C, Prigent-Combaret C. Differential responses of Oryza sativa secondary metabolism to biotic interactions with cooperative, commensal and phytopathogenic bacteria. Planta. 2015; 242: 1439–1452. 10.1007/s00425-015-2382-5 [DOI] [PubMed] [Google Scholar]
- 63.Gopal M, Gupta A. Microbiome selection could spur next-generation plant breeding strategies. Front Microbiol. 2016; 7: 1–10. 10.3389/fmicb.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hohmann P, Messmer MM. Breeding for mycorrhizal symbiosis: focus on disease resistance. Euphytica. 2017;213: 1–11. 10.1007/s10681-017-1900-x [DOI] [Google Scholar]
- 65.Nogales A, Nobre T, Valadas V, Ragonezi C, Döring M, Polidoros A, et al. Can functional hologenomics aid tackling current challenges in plant breeding? Brief Funct Genomics. 2016;1 5: 288–297. 10.1093/bfgp/elv030 [DOI] [PubMed] [Google Scholar]
- 66.Fukami J, Nogueira MA, Araujo RS, Hungria M. Accessing inoculation methods of maize and wheat with Azospirillum brasilense. AMB Express. 2016; 6: 3 10.1186/s13568-015-0171-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosier A, Bishnoi U, Lakshmanan V, Sherrier DJ, Bais HP. A perspective on inter-kingdom signaling in plant–beneficial microbe interactions. Plant Mol Biol. 2016; 90: 537–548. 10.1007/s11103-016-0433-3 [DOI] [PubMed] [Google Scholar]
- 68.Fesel PH, Zuccaro A. Dissecting endophytic lifestyle along the parasitism/mutualism continuum in Arabidopsis. Curr Opin Microbiol. 2016; 32: 103–112. 10.1016/j.mib.2016.05.008 [DOI] [PubMed] [Google Scholar]
- 69.Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005; 109: 661–686. 10.1017/S095375620500273X [DOI] [PubMed] [Google Scholar]
- 70.German MA, Burdman S, Okon Y, Kigel J. Effects of Azospirillum brasilense on root morphology of common bean (Phaseolus vulgaris L.) under different water regimes. Biol Fertil Soils. 2000; 32: 259–264. 10.1007/s003740000245 [DOI] [Google Scholar]
- 71.Duca DR, Rose DR, Glick BR. Indole acetic acid overproduction transformants of the rhizobacterium Pseudomonas sp. UW4 Antonie Van Leeuwenhoek. Springer International Publishing; 2018; 111: 1645–1660. 10.1007/s10482-018-1051-7 [DOI] [PubMed] [Google Scholar]
- 72.Wang C, Liu W, Li Q, Ma D, Lu H, Feng W, et al. Effects of different irrigation and nitrogen regimes on root growth and its correlation with above-ground plant parts in high-yielding wheat under field conditions. F Crop Res. 2014; 165: 138–149. 10.1016/j.fcr.2014.04.011 [DOI] [Google Scholar]
- 73.Xu W, Cui K, Xu A, Nie L, Huang J, Peng S. Drought stress condition increases root to shoot ratio via alteration of carbohydrate partitioning and enzymatic activity in rice seedlings. Acta Physiol Plant. 2015; 37 10.1007/s11738-014-1760-0 [DOI] [Google Scholar]
- 74.Yan J, Warburton M, Crouch J. Association Mapping for Enhancing Maize (L.) Genetic Improvement. Crop Sci. 2011; 51: 433 10.2135/cropsci2010.04.0233 [DOI] [Google Scholar]
- 75.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013; 9: 29 10.1186/1746-4811-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H, Xu C, Liu X, Guo Z, Xu X, Wang S, et al. Development of a multiple-hybrid population for genome-wide association studies: theoretical consideration and genetic mapping of flowering traits in maize. Sci Rep. 2017; 7: 40239 10.1038/srep40239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huh SU, Paek K-H. Plant RNA binding proteins for control of RNA virus infection. Front Physiol. 2013; 4: 1–5. 10.3389/fphys.2013.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng P, Gedling CR, Patil G, Vuong TD, Shannon JG, Dorrance AE, et al. Genetic mapping and haplotype analysis of a locus for quantitative resistance to Fusarium graminearum in soybean accession PI 567516C. Theor Appl Genet. 2017; 130: 999–1010. 10.1007/s00122-017-2866-8 [DOI] [PubMed] [Google Scholar]
- 79.Klay I, Gouia S, Liu M, Mila I, Khoudi H, Bernadac A, et al. Ethylene Response Factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018; 274: 137–145. 10.1016/j.plantsci.2018.05.023 [DOI] [PubMed] [Google Scholar]
- 80.Sun X, Yu G, Li J, Liu J, Wang X, Zhu G, et al. AcERF2, an ethylene-responsive factor of Atriplex canescens, positively modulates osmotic and disease resistance in Arabidopsis thaliana. Plant Sci. 2018; 274: 32–43. 10.1016/j.plantsci.2018.05.004 [DOI] [PubMed] [Google Scholar]
- 81.Rai A, Kumar S, Bauddh K, Singh N, Singh RP. Improvement in growth and alkaloid content of Rauwolfia serpentina on application of organic matrix entrapped biofertilizers (Azotobacter chroococcum, Azospirillum brasilense and Pseudomonas putida). J Plant Nutr. 2017; 40: 2237–2247. 10.1080/01904167.2016.1222419 [DOI] [Google Scholar]
- 82.Tivendale ND, Ross JJ, Cohen JD. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014; 19: 44–51. 10.1016/j.tplants.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Yuan G, Yuan S, Duan W, Wang P, Bai J, et al. TaOPR2 encodes a 12-oxo-phytodienoic acid reductase involved in the biosynthesis of jasmonic acid in wheat (Triticum aestivum L.). Biochem Biophys Res Commun. 2016; 470: 233–238. 10.1016/j.bbrc.2016.01.043 [DOI] [PubMed] [Google Scholar]
- 84.Koo AJ. Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochem Rev. 2018; 17: 51–80. 10.1007/s11101-017-9510-8 [DOI] [Google Scholar]
- 85.Dicke M. Plant phenotypic plasticity in the phytobiome: a volatile issue. Curr Opin Plant Biol. 2016; 32: 17–23. 10.1016/j.pbi.2016.05.004 [DOI] [PubMed] [Google Scholar]
- 86.Sharifi R, Lee S-M, Ryu C-M. Microbe-induced plant volatiles. New Phytol. 2017; 220: 684–691. 10.1111/nph.14955 [DOI] [PubMed] [Google Scholar]
- 87.Disi JO, Zebelo S, Kloepper JW, Fadamiro H. Seed inoculation with beneficial rhizobacteria affects European corn borer (Lepidoptera: Pyralidae) oviposition on maize plants. Entomol Sci. 2018; 21: 48–58. 10.1111/ens.12280 [DOI] [Google Scholar]
- 88.Ding Y, Huffaker A, Köllner TG, Weckwerth P, Robert CAM, Spencer JL, et al. Selinene Volatiles Are Essential Precursors for Maize Defense Promoting Fungal Pathogen Resistance. Plant Physiol. 2017; 175: 1455–1468. 10.1104/pp.17.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.D’Auria JC, Pichersky E, Schaub A, Hansel A, Gershenzon J. Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J. 2007; 49: 194–207. 10.1111/j.1365-313X.2006.02946.x [DOI] [PubMed] [Google Scholar]
- 90.Liu L-H, Ludewig U, Gassert B, Frommer WBF, Wirén N Von. Urea Transport by Nitrogen-Regulated Tonoplast Intrinsic Proteins in Arabidopsis. Plant Physiol. 2003; 133: 1220–1228. 10.1104/pp.103.027409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding L, Lu Z, Gao L, Guo S, Shen Q. Is Nitrogen a Key Determinant of Water Transport and Photosynthesis in Higher Plants Upon Drought Stress? Front Plant Sci. 2018; 9: 1–12. 10.3389/fpls.2018.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kavi Kishor PB. Role of proline in cell wall synthesis and plant development and its implications in plant ontogeny. Front Plant Sci. 2015; 6: 1–17. 10.3389/fpls.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguema-Ona E, Vicré-Gibouin M, Cannesan M-A, Driouich A. Arabinogalactan proteins in root–microbe interactions. Trends Plant Sci. 2013; 18: 440–449. 10.1016/j.tplants.2013.03.006 [DOI] [PubMed] [Google Scholar]
- 94.Oda Y, Hasezawa S. Cytoskeletal organization during xylem cell differentiation. J Plant Res. 2006; 119: 167–177. 10.1007/s10265-006-0260-8 [DOI] [PubMed] [Google Scholar]
- 95.Ding L, Wang KJ, Jiang GM, Biswas DK, Xu H, Li LF, et al. Effects of Nitrogen Deficiency on Photosynthetic Traits of Maize Hybrids Released in Different Years. Ann Bot. 2005; 96: 925–930. 10.1093/aob/mci244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Timm CM, Pelletier DA, Jawdy SS, Gunter LE, Henning JA, Engle N, et al. Two Poplar-Associated Bacterial Isolates Induce Additive Favorable Responses in a Constructed Plant-Microbiome System. Plant Sci. 2016; 7: 10 10.3389/fpls.2016.00497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luo B, Tang H, Liu H, Shunzong S, Zhang S, Wu L, et al. Mining for low-nitrogen tolerance genes by integrating meta-analysis and large-scale gene expression data from maize. Euphytica. 2015; 206: 117–131. 10.1007/s10681-015-1481-5 [DOI] [Google Scholar]
- 98.Młodzińska E, Kłobus G, Christensen MD, Fuglsang AT. The plasma membrane H + -ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol Plant. 2015; 154: 270–282. 10.1111/ppl.12305 [DOI] [PubMed] [Google Scholar]
- 99.Velasquez SM, Ricardi MM, Dorosz JG, Fernandez P V., Nadra AD, Pol-Fachin L, et al. O-Glycosylated Cell Wall Proteins Are Essential in Root Hair Growth. Science. 2011;332: 1401–1403. 10.1126/science.1206657 [DOI] [PubMed] [Google Scholar]
- 100.Khan MIR, Trivellini A, Fatma M, Masood A, Francini A, Iqbal N, et al. Role of ethylene in responses of plants to nitrogen availability. Front Plant Sci. 2015;6: 1–15. 10.3389/fpls.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ristova D, Carré C, Pervent M, Medici A, Kim GJ, Scalia D, et al. Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Sci Signal. 2016; 9 10.1126/scisignal.aaf2768 [DOI] [PubMed] [Google Scholar]
- 102.Abri MA, Posbergh C, Palermo K, Sutter NB, Eberth J, Hoffman GE, et al. Genome-Wide Scans Reveal a Quantitative Trait Locus for Withers Height in Horses Near the ANKRD1 Gene. J Equine Vet Sci. 2018; 60: 67-73.e1. 10.1016/j.jevs.2017.05.008 [DOI] [Google Scholar]
- 103.Lee Y-S, Shin D, Song K-D. Dominance effects of ion transport and ion transport regulator genes on the final weight and backfat thickness of Landrace pigs by dominance deviation analysis. Genes Genomics. 2018; 40: 1331–1338. 10.1007/s13258-018-0728-7 [DOI] [PubMed] [Google Scholar]
- 104.Tsairidou S, Allen AR, Pong-Wong R, McBride SH, Wright DM, Matika O, et al. An analysis of effects of heterozygosity in dairy cattle for bovine tuberculosis resistance. Anim Genet. 2018; 49: 103–109. 10.1111/age.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wald Test for fixed effects and Likelihood Ratio Test for random effects from the joint diallel analysis of 118 maize hybrids evaluated under N stress and N stress plus Azospirillum brasilense treatments.
(DOCX)
(A) Pearson correlation between maize hybrid adjusted means under N stress and N stress plus A. brasilense. (B) Estimates of variance components (: genotypic variance; : genotype-by-year variance; : error variance) and broad-sense heritabilities.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) root dry mass, (B) root volume, (C) root average diameter, (D) specific root length, (E) specific root surface area, and (F) root/shoot ratio.
(TIF)
(A) N stress plus Azospirillum brasiliense and (B) N stress.
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.