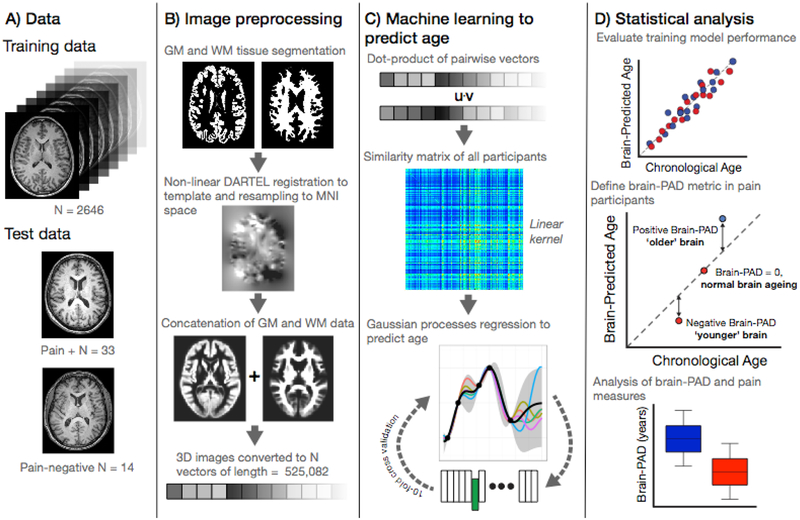
Figure 1.
Study methods.
A) Data used in the study comprising the ‘brain-age’ training sample of n=2,646 healthy individuals and the current experiment cohort (n=47) comprising those with chronic pain (n=33) and those without (n=14).
B) Image pre-processing, applied to all images, used SPM12 software to segment T1-weighted MRIs into gray and white matter probability maps. These were then spatially-normalized using DARTEL non-linear registration to a custom template in MNI152 space, with 1.5mm3 voxels, using 4mm spatial smoothing. These normalized 3D images were converted into 1D vectors and the gray and white matter vectors concatenated.
C) Machine learning age prediction involved generating a linear kernel by calculating the dot-product of all pairs of image vectors across all participants, resulting in a similarity matrix. The similarity matrix was used as input into a Gaussian Processes regression to predict chronological from the image vectors. The model trained on the full training set was then applied to the n=47 participants from the chronic pain study to generate a brain-predicted age value for each participant.
D) Statistical analysis was conducted to evaluate performance of the regression model performance using ten-fold cross-validation. Brain-predicted age difference (brain-PAD) was then calculated for the chronic pain study participants; whereby chronological age was subtracted from brain-predicted age. Brain-PAD was then used for subsequent statistical analysis of pain-related variables.