Genes from the common garden plant montbretia enable a synthetic biology approach to the metabolic engineering of a plant anti-diabetic metabolite in Nicotiana benthamiana.
Abstract
The plant metabolite montbretin A (MbA) and its precursor mini-MbA are potential new drugs for treating type 2 diabetes. These complex acylated flavonol glycosides only occur in small amounts in the corms of the ornamental plant montbretia (Crocosmia × crocosmiiflora). Our goal is to metabolically engineer Nicotiana benthamiana using montbretia genes to achieve increased production of mini-MbA and MbA. Two montbretia UDP-dependent glycosyltransferases (UGTs), CcUGT1 and CcUGT2, catalyze the formation of the first two pathway-specific intermediates in MbA biosynthesis, myricetin 3-O-rhamnoside and myricetin 3-O-glucosyl rhamnoside. In previous work, expression of these UGTs in N. benthamiana resulted in small amounts of kaempferol glycosides but not myricetin glycosides, suggesting that myricetin was limiting. Here, we investigated montbretia genes and enzymes of flavonol biosynthesis to enhance myricetin formation in N. benthamiana. We characterized two flavanone hydroxylases, a flavonol synthase, a flavonoid 3′-hydroxylase (F3′H), and a flavonoid 3′5′-hydroxylase (F3′5′H). Montbretia flavonol synthase converted dihydromyricetin into myricetin. Unexpectedly, montbretia F3′5′H shared higher sequence relatedness with F3′Hs in the CYP75B subfamily of cytochromes P450 than with those with known F3′5′H activity. Transient expression of combinations of montbretia flavonol biosynthesis genes and a montbretia MYB transcription factor in N. benthamiana resulted in availability of myricetin for MbA biosynthesis. Transient coexpression of montbretia flavonol biosynthesis genes combined with CcUGT1 and CcUGT2 in N. benthamiana resulted in 2 mg g−1 fresh weight of the MbA pathway-specific compound myricetin 3-O-glucosyl rhamnoside. Additional expression of the montbretia acyltransferase CcAT1 led to detectable levels of mini-MbA in N. benthamiana.
Diabetes and obesity are diseases that are approaching globally epidemic proportions with tremendous human health and economic consequences. Type 2 diabetes, which is characterized by high blood glucose (Glc) levels, afflicts ∼6% of the population of the western world, making it the third most prevalent disease (Health Canada, 2019; The World Health Organization, 2019).
A major goal in treating diabetes and obesity is reducing high levels of blood Glc, which is a particular problem in patients with diets that are rich in starch and other sugars. Digestion of starch begins with the activity of salivary α-amylase. The main enzyme of starch degradation is the human pancreatic α-amylase (HPA), which produces linear and branched malto-oligosaccharides. These oligosaccharides are degraded to Glc and passed into the bloodstream by gut wall α-glucosidases. The α-glucosidase inhibitors Acarbose and Miglitol are two pharmaceuticals currently used for controlling blood Glc levels. These drugs cause the passage of undigested oligosaccharides to the lower gut, which commonly results in gastrointestinal upsets due to adverse osmotic effects and bacterial fermentation. Selective inhibitors of HPA, which act upstream in the starch degradation cascade by reducing the breakdown of starch into oligosaccharides, are emerging as alternatives for the treatment of type 2 diabetes. In a large-scale screen of 30,000 natural products for HPA inhibitors, the complex acylated flavonol glycoside montbretin A [MbA; myricetin 3-O-(6′-O-caffeoyl)-β-d-glycosyl 1,2-β-d-glucosyl 1,2-α-l-rhamnoside 4′-O-α-l-rhamnosyl 1,4-β-d-xyloside] was discovered as a potent (inhibitory constant [Ki] = 8 nm) and specific amylase inhibitor (Fig. 1; Tarling et al., 2008; Williams et al., 2015). The simpler MbA precursor mini-MbA is active with a Ki = 90 nm. MbA does not significantly affect human gut α-glucosidases or gut bacterial amylases and effectively controls blood Glc levels in Zucker Diabetic Fatty rats (Yuen et al., 2016). The promising preclinical studies prompt methods for producing large quantities of MbA for further drug development.
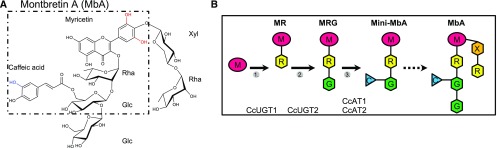
Figure 1.
Structure of MbA and schematic structures of metabolites in MbA biosynthesis. A, The building blocks of MbA are labeled: myricetin, rhamnose (Rha), glucose (Glc), xylose (Xyl), and caffeic acid. The dotted line marks the mini-MbA structure. Red hydroxyls mark groups specific to myricetin compared with kaempferol, and the blue hydroxyl marks the group specific to the caffeoyl compared with a coumaroyl moiety. B, Schematic of myricetin 3-O-α-l-rhamnoside (MR), myricetin 3-O-β-d-glucosyl 1,2-α-l-rhamnoside (MRG), myricetin 3-O-(6′-O-caffeoyl)-β-d-glucosyl 1,2-α-l-rhamnoside (mini-MbA), and MbA. Identified MbA biosynthesis steps and enzymes are labeled. C, Caffeic acid (blue); G, Glc (green); M, myricetin (pink); R, Rha (yellow); X, Xyl (orange).
MbA occurs in the corms (belowground bulb-like organs) of the ornamental plant montbretia (Crocosmia × crocosmiiflora, family Iridaceae), which is the only known source for this compound (Asada et al., 1988; Offen et al., 2006; Irmisch et al., 2018). MbA is present in the corms at low levels (Irmisch et al., 2018) that allow isolation of sufficient amounts for animal studies. However, much larger amounts will be required than can be harvested from the natural source for treatment of patients with type 2 diabetes as well as those diagnosed as prediabetic. The low amounts of MbA in montbretia corms, lack of agricultural production systems for this plant, and the destructive harvest of the belowground corms make it unrealistic to support MbA production via montbretia cultivation and harvest. Alternative avenues for MbA production are chemical synthesis and synthetic biology-enabled metabolic engineering. The complexity of the MbA molecule mitigates strongly against production by classical synthesis. Production by synthetic biology appears a viable approach, but it requires knowledge of the genes and enzymes of the MbA biosynthetic system.
The general flavonoid biosynthetic system (Fig. 2A) is widely conserved across diverse plant species (Schijlen et al., 2004; Martens et al., 2010). The first committed step of flavonoid biosynthesis is catalyzed by chalcone synthase (CHS). Chalcone isomerase (CHI) converts chalcone into the 4′OH-flavanone naringenin, which is the precursor for all other groups of flavonoids, including flavones, flavonols, flavandiols, anthocyanins, and proanthocyanidins (Martens et al., 2010). In the biosynthesis of flavonols, which includes the myricetin core of MbA, naringenin is hydroxylated by flavanone 3-hydroxylase (F3H) to form the 4′OH-dihydroflavonol dihydrokaempferol (DHK). F3H also converts the 3′4′OH-flavanone eriodictyol into dihydroquercetin (DHQ) and the 3′4′5′-OH flavanone pentahydroxyflavanone (PHF) into dihydromyricetin (DHM). Flavonol synthase (FLS) catalyzes the desaturation of dihydroflavonols to flavonols. F3H and FLS are members of the large enzyme family of 2-oxoglutarate-dependent dioxygenases (2OGDs). The flavonols kaempferol, quercetin, and myricetin differ by their hydroxylation pattern on the B-ring with 4′-, 3′4′-, or 3′4′5′-hydroxylation, respectively. Hydroxylation of the B-ring is produced by the cytochromes P450 (P450s) flavonoid 3′-hydroxylases (F3′Hs) and flavonoid 3′5’-hydroxylases (F3′5H).
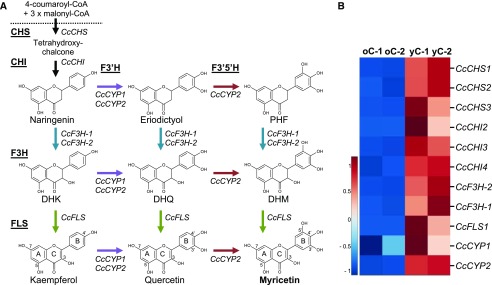
Figure 2.
General schematic of myricetin biosynthesis and transcript profiles of myricetin pathway genes in montbretia. A, Enzymes and metabolites of flavonol biosynthesis leading to the formation of the 3′4′5′-hydroxylated flavonol myricetin. B, Heat map showing relative transcript abundance of putative myricetin biosynthetic pathway genes in yC and oC of montbretia.
MbA is derived from the flavonol myricetin (Irmisch et al., 2018). The biosynthesis of MbA occurs in young developing corms and proceeds via the formation of MR, MRG, and mini-MbA (or MRG-Caff; Fig. 1; Irmisch et al., 2018). We recently discovered two UDP-dependent glycosyltransferases (UGTs), CcUGT1 (UGT77B2) and CcUGT2 (UGT709G2), and two BAHD-acyltransferases, CcAT1 and CcAT2, which catalyze the first three pathway-specific steps of MbA biosynthesis. CcUGT1 converts myricetin into MR, CcUGT2 converts MR into MRG, and CcAT1 and CcAT2 both convert MRG into MRG-Caff (mini-MbA; Irmisch et al., 2018). Transient coexpression of the two UGTs and CcAT1 in Nicotiana benthamiana resulted in the formation of small amounts of kaempferol 3-O-(6′-O-coumaroyl)-glucosyl rhamnoside, termed surrogate mini-MbA (Irmisch et al., 2018). In comparison with mini-MbA, the surrogate mini-MbA contained the 4′-hydroxylated flavonol kaempferol, instead of the 3′4′5′-hydroxylated flavonol myricetin, as the core flavonol structure. Instead of the caffeoyl moiety of mini-MbA, the surrogate mini-MbA had a coumaroyl moiety. These results demonstrated that N. benthamiana could be developed as a production system for MbA, but the availability of myricetin and caffeoyl-CoA may be limiting and require additional pathway engineering.
Myricetin is not a ubiquitous or abundant metabolite across the plant kingdom, due in part to the limited occurrence of F3′5′H, which is missing, for example, in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa; Tohge et al., 2013). Myricetin formation via DHM may be limited in cases where DHM is a poor substrate for FLS, such as in tomato (Solanum lycopersicum) or potato (Solanum tuberosum; Forkmann et al., 1986; Bovy et al., 2002; Tanaka et al., 2008). Research on flavonol biosynthesis has focused mostly on the more ubiquitous kaempferol and quercetin, while information about myricetin biosynthesis is missing. Here, we explore the genes and enzymes of myricetin biosynthesis in montbretia with the goal of engineering the availability of myricetin in N. benthamiana as a substrate for MbA production.
RESULTS
Transcripts for Flavonol Biosynthesis Genes Are Abundant in Montbretia Young Corms
We searched for montbretia transcripts of myricetin biosynthesis in the published corm transcriptome, which covers young corms (yC) and old corms (oC; Irmisch et al., 2018). A BLASTP analysis revealed three putative CHSs, CcCHS1, CcCHS2, and CcCHS3, and four putative CHIs, CcCHI1, CcCHI2, CcCHI3, and CcCHI4. We also identified two candidate F3Hs, CcF3H-1 and CcF3H-2, and a putative FLS, CcFLS. We found two candidate F3′H-like transcripts, CcCYP1 (CYP75B137) and CcCYP2 (CYP75B138), but no transcripts that were immediately obvious to encode a candidate F3′5′H. Since MbA biosynthesis occurs predominantly during the development of yC, while biosynthesis is lacking or minimal in oC (Irmisch et al., 2018), we compared transcript abundance of candidate myricetin biosynthesis genes between yC and oC using previously established differential expression analysis (Irmisch et al., 2018; Fig. 2B). CcCHI1 showed overall low expression and was excluded from further analysis. All other candidate genes showed higher transcript abundance in yC compared with oC (Fig. 2B).
CcF3H-1, CcF3H-2, and CcFLS Encode 2OGDs of Flavonol Biosynthesis
The full-length open reading frames (ORFs) of CcF3H-1, CcF3H-2, and CcFLS encode proteins of 372, 372, and 331 amino acids, respectively. CcF3H-1 and CcF3H-2 shared 93% identity on the amino acid level. CcF3H-1, CcF3H-2, and CcFLS belong to the class of 2OGDs and cluster with F3H or FLS from other plants (Supplemental Fig. S1). To test for F3H and FLS activity, CcF3H-1, CcF3H-2, and CcFLS were heterologously expressed from complementary DNAs (cDNA) in Escherichia coli. The three different purified proteins were tested with the two flavanone substrates naringenin and eriodictyol and the three dihydroflavonol substrates DHK, DHQ, and DHM. We detected products using liquid chromatography-mass spectrometry (LC-MS) and identified these based on mass and retention time compared with authentic standards. CcF3H-1 and CcF3H-2 showed the expected F3H activity, converting naringenin (mass-to-charge ratio [m/z] 271) into DHK (m/z 287) and eriodictyol (m/z 287) into DHQ (m/z 301; Fig. 3, A and B). CcFLS also showed some activity with naringenin and eriodictyol; however, the abundance of DHK and DHQ formed by FLS was less than 3% of product formed by CcF3H-1 or CcF3H-2 (Fig. 3, A and B). In addition to DHQ, CcFLS produced two unidentified peaks with m/z 303 when assayed with eriodictyol (Fig. 3B). In assays with the three dihydroflavonols, CcFLS, but not CcF3H-1 and CcF3H-2, showed flavonol synthase activity and converted DHK (m/z 287), DHQ (m/z 303), and DHM (m/z 319) into the respective flavonols kaempferol (m/z 285), quercetin (m/z 301), and myricetin (m/z 317; Fig. 3, C–E). Traces of kaempferol and quercetin formation in addition to the above-mentioned formation of DHK and DHQ were also observed when the flavanones naringenin and eriodictyol were used as substrates for CcFLS (Supplemental Fig. S2). No activity was observed with any of the empty vector controls.
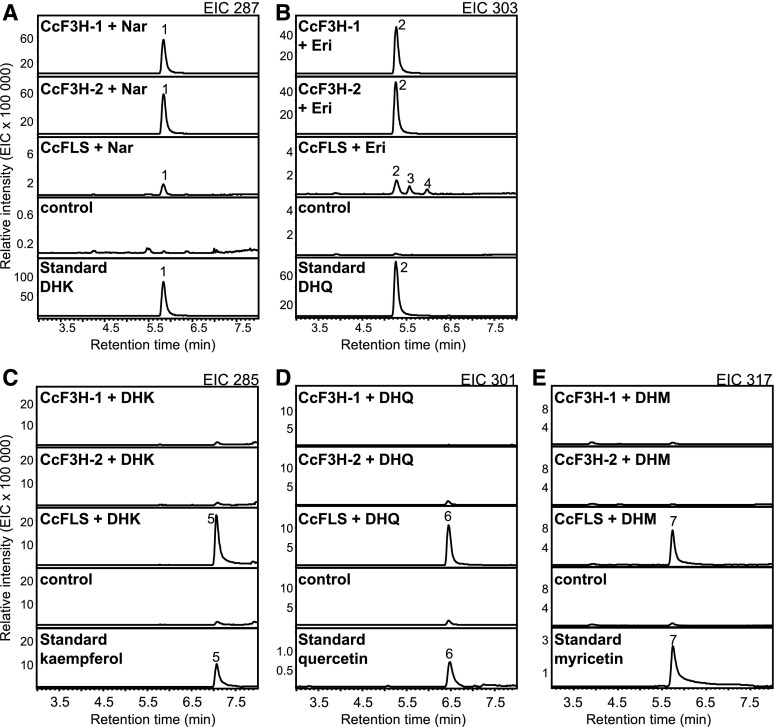
Figure 3.
Enzyme activity of CcF3H-1, CcF3H-2, and CcFLS. Enzymes were heterologously expressed in E. coli, and Ni-purified proteins were assayed for activity with the flavanones naringenin and eriodictyol (A and B), or the dihydroflavonols DHK, DHQ, and DHM (C–E). As controls, assays were performed with purified protein of E. coli transformed with the empty vector. Products were analyzed using LC-MS, and extracted ion chromatograms (EIC) are shown. Peak 1, DHK; peak 2, DHQ; peaks 3 and 4, unidentified; peak 5, kaempferol; peak 6, quercetin; peak 7, myricetin. Eri, Eriodictyol; Nar, naringenin.
CcCYP1 Encodes an F3′H and CcCYP2 Encodes an F3′5′H
The ORFs of CcCYP1 and CcCYP2 encode proteins of 508 and 527 amino acids, respectively, which share 60.5% amino acid identity. These two P450s fall into the CYP75B subfamily of the plant P450 family, which includes known F3′H of other species (Supplemental Fig. S3). To test CcCYP1 and CcCYP2 for functions in flavonoid 3′- or 5′-hydroxylations, we individually coexpressed the proteins with montbretia cytochrome P450 reductase (CcCPR1) in yeast (Saccharomyces cerevisiae) and used microsomes for enzyme assays. Both CcCYP1 and CcCYP2 catalyzed 3′-hydroxylations with naringenin (m/z 271), DHK (m/z 287), and kaempferol (m/z 285), leading to the formation of eriodictyol (m/z 287; peak 1), DHQ (m/z 303; peak 3), and quercetin (m/z 301; peak 5), respectively (Fig. 4).
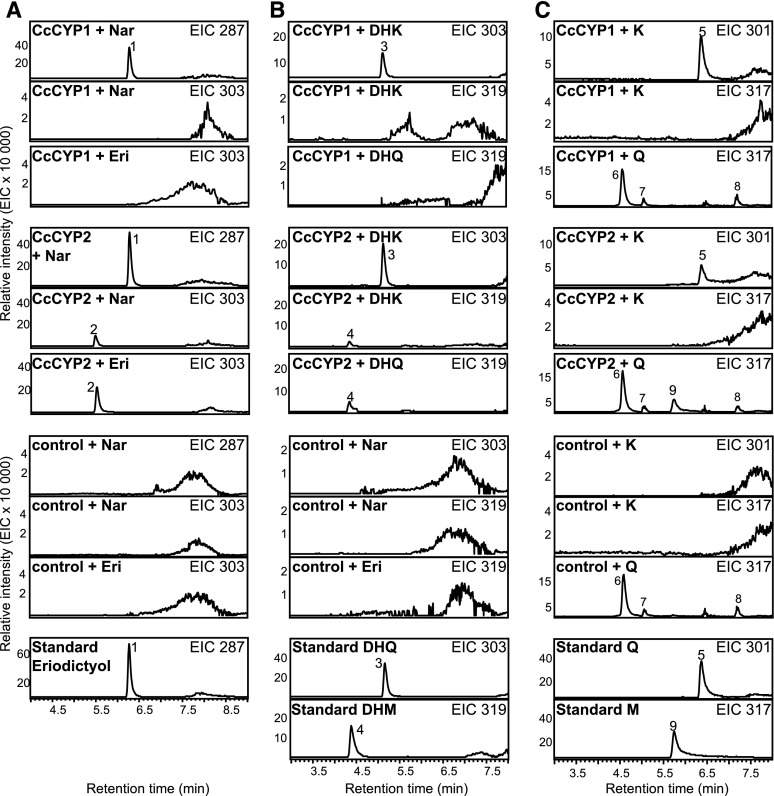
Figure 4.
Enzyme activity of CcCYP1 and CcCYP2. The P450s CcCYP1 and CcCYP2 were individually coexpressed with CcCPR1 in S. cerevisiae. Isolated microsomes were assayed with the flavanones naringenin and eriodictyol (A), the dihydroflavonols DHK and DHQ (B), and the flavonols kaempferol and quercetin (C). As controls, assays were performed with microsomes prepared from S. cerevisiae transformed with the empty vector. Products were analyzed using LC-MS, and extracted ion chromatograms (EIC) are shown. Peak 1, Eriodictyol (Eri); peak 2, tentatively identified as PHF; peak 3, DHQ; peak 4, DHM; peak 5, quercetin (Q); peaks 6 to 8, unidentified; peak 9, myricetin (M). K, Kaempferol; Nar, naringenin.
In addition to the F3′H activity, CcCYP2 also showed 5′-hydroxylation activity, which identified this P450 as an F3′5′H. In assays with naringenin, CcCYP2 produced eriodictyol (m/z 287; peak 1) and a second product with m/z 303 (peak 2), tentatively identified as PHF (Fig. 4A; Supplemental Fig. S4). CcCYP2 converted DHK (m/z 287) into DHQ (m/z 303; peak 3) and DHM (m/z 319; peak 4). Kaempferol (m/z 285) was converted into quercetin (m/z 301; peak 5), but in this case no further 5′-hydroyxlation to myricetin (m/z 317) was observed (Fig. 4). We confirmed the 5′-hydroxylation activity of CcCYP2 in separate assays with the 3′4′-hydroxylated flavonoid substrates eriodictyol (m/z 287), DHQ (m/z 203), and quercetin (m/z 301), which resulted in PHF (m/z 303; peak 2), DHM (m/z 319; peak 4), and myricetin (m/z 317; peak 9), respectively, although myricetin formation was only observed with high substrate concentration.
No 5′-hydroxylation activity was observed in any of the assays with CcCYP1, and no product formation was observed when microsomes prepared from yeast expressing the CcCPR1 together with the empty vector were used (Fig. 4).
Transcript Expression Patterns of CcF3H-1, CcF3H-2, CcCYP1, CcCYP2, and CcFLS Support a Role in MbA Biosynthesis
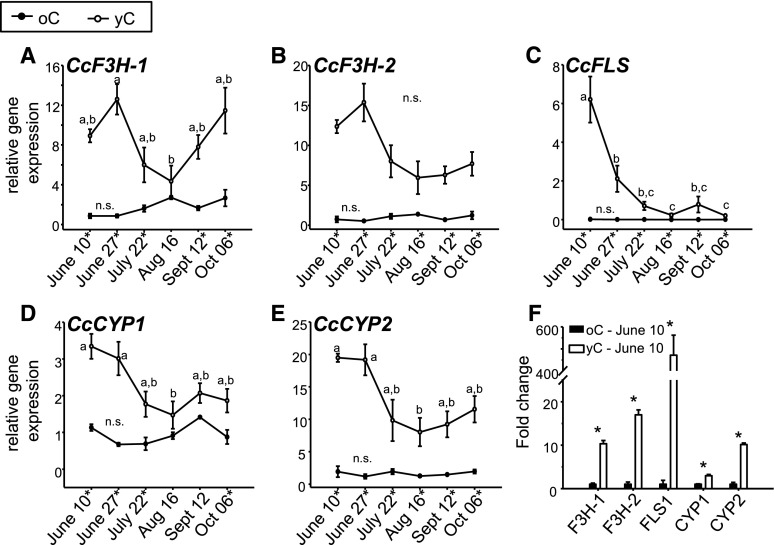
Following their functional characterization with in vitro assays, we measured transcript abundance of CcF3H-1, CcF3H-2, CcCYP1, CcCYP2, and CcFLS over a time course of corm development in yC and oC from early summer (June 10) to fall (October 6) using reverse transcription quantitative PCR (RT-qPCR). All genes showed significantly higher transcript abundance in yC compared with oC for the majority of sampling time points (Fig. 5). In general, transcript abundance was approximately three- to 17-fold higher in yC compared with oC. CcFLS showed up to 470-fold higher transcript levels in yC compared with oC (Fig. 5F). Transcript abundance in yC was generally higher in June and dropped toward August. This pattern was particularly pronounced with CcFLS (Fig. 5, C and F). Transcript abundance did not change significantly over time in oC. The patterns of differential transcript expression of CcF3H-1, CcF3H-2, CcCYP1, CcCYP2, and CcFLS in yC and oC over the time course of the growing season matched the previously reported patterns of MbA accumulation and CcUGT and CcAT transcript expression (Irmisch et al., 2018), supporting a role of these genes in myricetin and MbA biosynthesis.
Figure 5.
Transcript abundance of myricetin biosynthesis pathway genes in yC and oC. RNA was isolated from yC and oC harvested at six different time points of corm development. A to E, Transcript abundance was determined by RT-qPCR for CcF3H-1 (A), CcF3H-2 (B), CcFLS (C), CcCYP1 (D), and CcCYP2 (E). F, Fold change of myricetin biosynthesis pathway genes for yC relative to oC (set as 1) at the June 10 sampling time point. Means and se are shown (n = 3). Different letters above the data points indicate significant differences (Supplemental Table S4) between sampling time points, and n.s. indicates no significant difference. Asterisks indicate significant differences (Supplemental Table S4) between yC and oC for time points.
N. benthamiana Transiently Coexpressing Montbretia Myricetin Biosynthetic Pathway Genes and CcUGT1 Produce Myricetin 3-O-Rhamnoside
We previously showed that N. benthamiana leaves transiently expressing montbretia CcUGT1 produced small amounts of kaempferol 3-O-rhamnoside (KR) but not MR (Irmisch et al., 2018), suggesting that myricetin was limiting. Using transient coexpression of various combinations of montbretia myricetin biosynthetic pathway genes and CcUGT1, we tested if the flavonol pathway could be engineered to provide myricetin for heterologous MR production in N. benthamiana. MR is the first pathway-specific intermediate in MbA biosynthesis. We produced a set of Agrobacterium tumefaciens strains that were transformed with individual plasmids carrying the promoter-gene constructs 35Spro:CcCHS2, 35Spro:CcCHI2, 35Spro:CcF3H-2, 35Spro:CcCYP2, 35Spro:CcFLS, or 35Spro:CcUGT1. A. tumefaciens transformed with the enhanced GFP (eGFP) construct 35Spro:eGFP was used as a negative control. We used A. tumefaciens strains separately or in various different combinations to infiltrate N. benthamiana leaves (Table 1). We collected leaves that expressed eGFP, CcUGT1, or the four different gene combinations A to D 5 d after infiltration and analyzed methanol (MeOH)/water extracts by LC-MS/MS. The coexpression of CcUGT1 allowed us to perform targeted metabolite profiling for the flavonol rhamnosides KR (m/z 431), quercetin 3-O-rhamnoside (QR; m/z 447), and MR (m/z 463), which are not present in nontransformed N. benthamiana (Irmisch et al., 2018).
Table 1. Combinations of genes used for transient expression in N. benthamiana.
| Combinationa | Constructs |
|---|---|
| A | CcCHS-2 + CcCHI2 + CcUGT1 |
| B | CcFLS + CcF3H-2 + CcCYP2 + CcUGT1 |
| C | CcCHS2 + CcCHI2 + CcFLS + CcF3H-2 + CcCYP2 + CcUGT1 |
| D | CcCHS2 + CcCHI2 + CcFLS + CcF3H-2 + CcCYP2 |
| E | CcUGT1 + CcMYB1 |
| F | CcUGT1 + CcMYB2 |
| G | CcUGT1 + CcMYB3 |
| H | CcUGT1 + CcMYB4 |
| J | CcMYB4 + CcUGT1 + CcFLS |
| K | CcMYB4 + CcUGT1 + CcCYP2 |
| L | CcMYB4 + CcUGT1 + CcF3H-2 |
| M | CcMYB4 + CcUGT1 + CcFLS + CcCYP2 |
| N | CcMYB4 + CcUGT1 + CcFLS + CcF3H-2 |
| O | CcMYB4 + CcUGT1 + CcF3H-2 + CcCYP2 |
| P | CcMYB4 + CcUGT1 + CcFLS + CcF3H-2 + CcCYP2 |
| R | CcUGT1 + CcFLS + CcF3H-2 + CcCYP2 |
| S | CcMYB4 + CcFLS + CcCYP2 + CcUGT1 + CcUGT2 |
| T | CcMYB4 + CcFLS + CcCYP2 + CcUGT1 + CcUGT2 + CcAT1 |
Each combination is identified with a letter A to T. Each 35Spro:gene construct was separately transformed into A. tumefaciens. Different combinations were achieved by mixing different A. tumefaciens strains each carrying a different gene construct. The infiltration solution contained equal amounts of A. tumefaciens containing the listed gene constructs and A. tumefaciens pBIN:p19 for enhancement of gene expression.
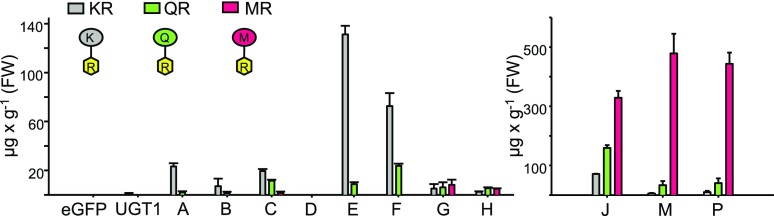
Small amounts of KR, but not QR or MR, were present when only CcUGT1 was expressed in N. benthamiana (Fig. 6; Supplemental Fig. S5A), confirming earlier work (Irmisch et al., 2018). Ninefold higher levels of KR and small amounts of QR were produced when CcUGT1 was coexpressed with CcFLS, CcF3H-2, and CcCYP2 (combination B; Fig. 6; Supplemental Fig. S5B). Coexpression of CcUGT1 with CcCHS2 and CcCHI2 (combination A) resulted in 30-fold more KR compared with CcUGT1 expression alone and twice the level of QR compared with combination B (Fig. 6). In combination C, where CcUGT1 was coexpressed with CcCHS2, CcCHI2, CcFLS, CcF3H-2, and CcCYP2, levels of KR were 26-fold higher compared with CcUGT1 expression alone and levels of QR were ninefold higher compared with combination A (Fig. 6; Supplemental Fig. S5, A and B). Importantly, expression of gene combination C also resulted in the formation of MR, albeit in small amounts of approximately 1 µg g−1 fresh weight (Fig. 6; Supplemental Fig. S5C). Controls expressing eGFP or myricetin biosynthetic pathway genes without CcUGT1 (combination D) did not yield detectable levels of flavonol rhamnoside production.
Figure 6.
Flavonol rhamnoside accumulation in N. benthamiana transiently expressing different combinations of montbretia myricetin biosynthetic pathways genes, CcMYB genes, and CcUGT1. N. benthamiana leaves were infiltrated with different combinations (Table 1) of A. tumefaciens transformed with plasmids carrying the promoter-gene constructs 35Spro:CcUGT1, 35Spro:CcCHI, 35Spro:CcCHS, 35Spro:CcF3H-2, 35Spro:CcFLS, 35Spro:CcCYP2, 35Spro:CcMYB1, 35Spro:CcMYB2, 35Spro:CcMYB3, and 35Spro:CcMYB4 or the eGFP gene. Leaves were collected at day 5 after infiltration. Metabolites were extracted with 50% MeOH, analyzed by LC-MS, and identified based on their fragmentation patterns and an authentic standard for MR. Quantification was conducted using an external MbA standard curve. Means and se of two or four biological replicates are shown. FW, Fresh weight.
Montbretia CcMYB3 and CcMYB4 Enhance DHM Biosynthesis in N. benthamiana
We used a BLASTP search with Arabidopsis MYB75 (Borevitz et al., 2000; Liu et al., 2015) and differential expression analysis in yC and oC to screen the montbretia transcriptome for MYB transcription factors (TFs) putatively involved in flavonoid biosynthesis. Four montbretia MYB-like TFs, CcMYB1, CcMYB2, CcMYB3, and CcMYB4, had, respectively, 60-, 46-, 169-, and 109-fold higher transcript abundance in yC compared with oC. Alignment with AtMYB75 showed that all four CcMYBs contained the conserved R2R3 domain (Supplemental Fig. S6). The most closely related Arabidopsis gene is AtMYB123, which is involved in proanthocyanidin biosynthesis (https://bioinformatics.psb.ugent.be/plaza/). To test if the montbretia MYB-TF could be used to enhance flavonol biosynthesis in N. benthamiana, we transformed A. tumefaciens with plasmids carrying the promoter-gene constructs 35Spro:CcMYB1, 35Spro:CcMYB2, 35Spro:CcMYB3, or 35Spro:CcMYB4, which were used for N. benthamiana leaf infiltration assays. Coexpression assays with CcUGT1 (combinations E–H; Table 1) allowed for flavonol rhamnoside screening without interference from endogenous N. benthamiana metabolites. The coexpression of CcUGT1 with CcMYB1 (combination E) or CcMYB2 (combination F) resulted in, respectively, 7-fold and 4-fold higher levels of KR and 0.8-fold and 2-fold higher levels of QR compared with expression of the gene combination C (Fig. 6). Expression of CcUGT1 with CcMYB1 (combination E) or CcMYP2 (combination F) did not result in detectable levels of MR. However, low levels of MR were observed when CcUGT1 was coexpressed with CcMYB3 (combination G) or CcMYB4 (combination H), while levels of KR were over 14-fold reduced compared with the results obtained with CcMYB1 (combination E) and CcMYB2 (combination F).
We also tested if N. benthamiana plants that transiently expressed CcMYB-TFs in combinations E to H had enhanced levels of flavonol precursors that accumulated as glucosides. Those precursors would not be accessible for CcUGT1 and would therefore have been missed in the screen for flavonol rhamnosides. We found pronounced m/z 481 peaks in extracts of plants expressing CcMYB3 or CcMYB4 and to a lesser extent in plants expressing CcMYB2 but not in plants expressing CcMYB1 (Supplemental Fig. S7A). The fragmentation pattern of m/z 481 peaks into m/z 319 (DHM minus Glc) led to the tentative identification of m/z 481 as DHM-glucosides possessing a Glc moiety at different positions of the DHM flavonoid core (Supplemental Fig. S7A). DHM is a direct precursor for the formation of myricetin (Fig. 2A), which makes the TFs CcMYB3 and CcMYB4 relevant for engineering of myricetin biosynthesis in combination with flavonol biosynthetic genes.
Extracts of plants expressing any of the four CcMYBs also revealed several m/z 449 peaks (Supplemental Fig. S7). The observed fragmentation patterns of m/z 449 suggested glucosides of DHK (m/z 287) and/or eriodictyol (m/z 287), which are respective precursors for kaempferol and quercetin. Plants expressing CcMYB2, CcMYB3, or CcMYB4 produced additional m/z 465 peaks tentatively identified as glucosides of DHQ (m/z 303) and/or PHF (m/z 303), precursors of quercetin and myricetin, respectively (Supplemental Fig. S7).
Transient Coexpression of CcMYB4 with CcFLS, CcCYP2, and CcUGT1 Boosts the Formation of Myricetin 3-O-Rhamnosides in N. benthamiana
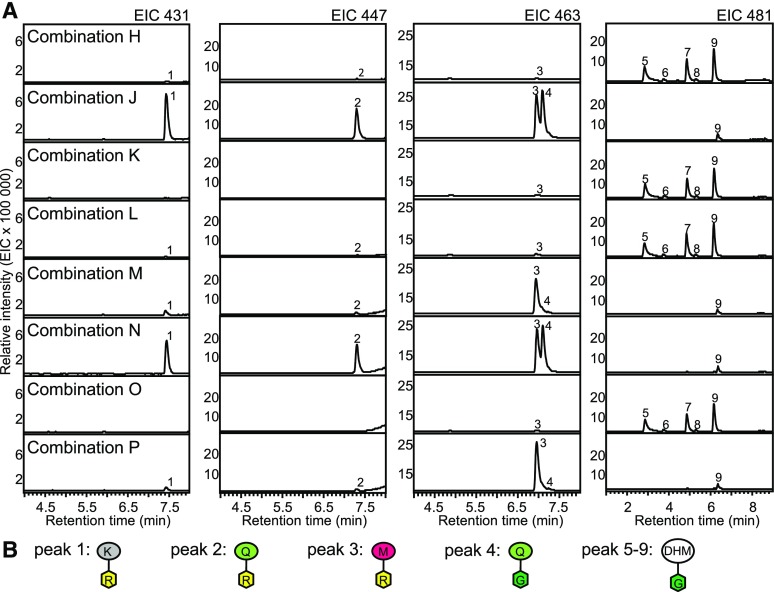
We tested expression of CcMYB4 in combination with different myricetin biosynthesis genes (Table 1) and CcUGT1 for enhanced MR formation in N. benthamiana. As described above, extracts of plants expressing combination H showed small amounts of m/z 463, m/z 431, and m/z 447, identified as MR and tentatively identified as KR and QR, respectively (Figs. 6 and 7). Small amounts of KR, QR, and MR were also detected for combination L, while combination K only showed MR (Fig. 7). Compared with the expression of CcMYB4 + CcUGT1 (combination H), flavonol rhamnoside levels increased over 40-fold by the additional expression of CcFLS (combination J; Figs. 6 and 7). In addition to KR, QR, and MR, quercetin 3-O-glucoside (QG) was detected with combination J. Finally, the further addition of CcCYP2 in combination M led to the dominant formation of the target metabolite MR and a major reduction in the formation of QR, KR, and QG (Figs. 6 and 7). MR levels in extracts of plants expressing combination M were about 100-fold higher compared with those expressing combination H. Additional coexpression of CcF3H-2 (combination P) did not seem to further affect flavonol rhamnoside levels compared with combination M (Figs. 6 and 7). Samples showing increased MR formation also showed a depletion in DHM-glucosides (m/z 481; Fig. 7).
Figure 7.
Formation of MR in N. benthamiana using montbretia genes. N. benthamiana leaves were infiltrated with different combinations of A. tumefaciens transformed with plasmids carrying the promoter-gene constructs 35Spro:CcUGT1, 35Spro:CcMYB4, 35Spro:CcF3H-2, 35Spro:CcFLS, and 35Spro:CcCYP2. Leaves were collected at day 5 after infiltration. Combinations used for infiltration can be found in Table 1. Metabolites were extracted with 50% MeOH, analyzed by LC-MS, and identified based on their fragmentation patterns and an authentic standard for MR and QG. The extracted ion chromatograms (EIC; A) and schematic structures of products (B) are shown. Peak 1, Tentatively identified as kaempferol 3-O-rhamnoside; peak 2, tentatively identified as quercetin 3-O-rhamnoside; peak 3, myricetin 3-O-rhamnoside; peak 4, quercetin 3-O-glucoside; peaks 5 to 9, tentatively identified as dihydromyricetin glucoside.
Engineered N. benthamiana Leaves Produce Intermediates in MbA Biosynthesis
N. benthamiana plants expressing the four montbretia cDNAs CcMYB4, CcFLS, CcCYP2, and CcUGT1 (combination M) had MR levels of 0.48 ± 0.07 mg g−1 fresh weight (Fig. 6). MR is the first pathway-specific intermediate in MbA biosynthesis. In addition to MR, the UV spectrum at 350 to 370 nm of extracts of combination M plants showed peaks that were not present in control plants expressing CcUGT1 alone or together with CcMYB4 (Supplemental Fig. S8A). To investigate the origin of those peaks, we performed a fragmentation pattern analysis of their main masses (Supplemental Fig. S8B). All peaks showed the loss of one or multiple 162 (presumably Glc) and/or 146 (presumably rhamnose) units and fragmented into m/z 316/317 (myricetin), which tentatively identified them as myricetin glycosides, and suggested off-target conversion of myricetin or MR by endogenous N. benthamiana glycosyltransferase activities.
We continued to build upon the combination M metabolic engineering of N. benthamiana with the additional expression of the second CcUGT gene of MbA biosynthesis in combination S (CcMYB4 + CcFLS + CcCYP2 + CcUGT1 + CcUGT2). Combination S resulted in the formation of 1.89 ± 0.23 mg g−1 fresh weight MRG (Supplemental Fig. S7A). The fourfold increase in product yield of MRG compared with MR production in combination M indicated that CcUGT2 enhances overall pathway flux into MbA biosynthesis. The UV spectrum at 350 to 370 nm of extracts of combination S leaves showed MRG as a major peak and additional peaks likely presenting myricetin glycosides, as indicated by their fragmentation patterns (Supplemental Fig. S7).
Next we added the third MbA pathway-specific gene, CcAT1, in combination T (CcMYB4 + CcFLS + CcCYP2 + CcUGT77B2 + CcUGT709G2 + CcAT1). We showed previously that CcAT1 uses caffeoyl-CoA to catalyze the formation of mini-MbA in montbretia, but when expressed in N. benthamiana, CcAT1 catalyzed the addition of a coumaroyl moiety most likely due to substrate availability (Irmisch et al., 2018). Expression of combination T in N. benthamiana led to the formation of 0.62 ± 0.17 mg g−1 fresh weight of a product with m/z 771, tentatively identified as myricetin 3-O-(6′-O-coumaroyl)-glucosyl 1,2-rhamnoside (MRG-Cou or mini-MbB; Supplemental Fig. S9). In addition to MRG-Cou, the UV spectrum at 350 to 370 nm showed a few other myricetin-based peaks (Supplemental Fig. S8). Compounds with m/z 949 and m/z 933 were tentatively identified as myricetin 3-O-(6′-O-coumaroyl)-glucosyl glucoside O-glucoside (MGG-Cou + Glc) and myricetin 3-O-(6′-O-coumaroyl)-glucosyl rhamnoside O-glucoside (MRG-Cou + Glc), respectively. Screening for m/z 787, which is diagnostic of mini-MbA (MRG-Caff), identified a product in plants expressing combination T that matched the retention time and fragmentation pattern of mini-MbA (Supplemental Fig. S9). Mini-MbA coeluted with another m/z 787 compound tentatively identified as myricetin 3-O-(6′-O-coumaroyl)-glucosyl glucoside (MGG-Cou). Mini-MbA (MRG-Caff) was about 15-fold less abundant compared with Mini-MbB (MRG-Cou).
DISCUSSION
Genes and Enzymes for Myricetin Biosynthesis in Montbretia
Flavonols are thought to be the most ancient group of flavonoids (Pollastri and Tattini, 2011). The biosynthesis of kaempferol and quercetin, which occurs in many plant species, including Arabidopsis (Lillo et al., 2008), is well characterized. Quercetin appears to be nearly ubiquitous in vascular plants, and genes for its biosynthesis are highly conserved (Pollastri and Tattini, 2011). In contrast, myricetin is less common and its biosynthesis is not well studied. For the purpose of engineering the myricetin-derived biosynthesis of MbA in N. benthamiana, we explored the montbretia corm transcriptome for a complete set of genes for myricetin biosynthesis (Fig. 2). The CcCHS and CcCHI genes represent the entry steps, and CcF3H-1, CcF3H-2, CcFLS, CcCYP1, and CcCYP2 cover the core metabolic grid of montbretia flavonoid biosynthesis. CcF3H-1, CcF3H-2, and CcFLS belong to the 2OGD gene family; CcCYP1 and CcCYP2 are members of the P450 gene family. Based on their differential spatial and temporal transcript profiles in developing yC relative to oC, the montbretia flavonol pathway genes CcF3H-1, CcF3H-2, CcFLS, CcCYP1, and CcCYP2 are likely involved in the formation of myricetin leading to MbA in yC. Functions of these five genes are supported by the results from assays with recombinant enzymes and transient expression in N. benthamiana (Fig. 2). Their expression in N. benthamiana led to the enhanced availability of myricetin for MbA biosynthesis.
The hydroxylation of the 3′-hydroxy group on the B-ring of flavonoids is typically catalyzed by P450s (F′3Hs) of the CYP75B subfamily, while members of the CYP75A subfamily (F3′5′Hs) catalyze 3′5′-hydroxylations (Seitz et al., 2006). In contrast to the 3′4′-hydroxylated flavonoids, 3′4′5′-hydroxylated flavonoids are not ubiquitous in plants, suggesting that genes encoding F3′5′H may have been lost in several major lineages in plant evolution (Seitz et al., 2006). For example, Arabidopsis has one gene for F3′H (CYP75B1) but no obvious genes for F3′5′H (Saito et al., 2013). CcCYP1 and CcCYP2 both fall into the CYP75B clade, and we would have a priori expected both of these P450s to be F3′Hs. Functional characterization confirmed CcCYP1 as F3′H. Unexpectedly, CcCYP2 had F3′5′H activity, which affords the 3′4′5′-hydroxylation pattern of myricetin. Most of the currently known F3′5′Hs appear to have evolved by gene duplication of an ancestral F3′H, prior to the separation of gymnosperms and angiosperms, which led to the two functionally distinct F3′5′H/CYP75A and F3′H/CYP75B clades (Supplemental Fig. S3; Seitz et al., 2006). However, in some members of Asteraceae (e.g. Callistephus chinensis and Pericallis cruenta; Supplemental Fig. S3), which lack CYP75A genes, F3′5′H appears to have evolved later within the CYP75B subfamily (Seitz et al., 2006). Apparently, such secondary evolution of F3′5′H functionality within CYP75B also occurred independently in montbretia, which belongs to Iridaceae, which is distant from Asteraceae. CcCYP2 was active with naringenin, eriodictyol, DHK, DHQ, and quercetin, similar to the broad substrate spectrum shown for F3′5′H from Gentiana triflora and petunia (Petunia hybrida; Tanaka et al., 1996; Brugliera et al., 1999), which is characteristic of flavonoid biosynthesis as a metabolic grid (Saito et al., 2013) or biosynthetic system, rather than a linear pathway.
In the flavonoid biosynthetic system, the dihydroflavonols are at the interface of flavonols and anthocyanidins. Flavonols are formed by FLS, while formation of anthocyanidins requires dihydroflavonol reductase (DFR). The substrate specificities of FLS and DFR determine the specific profiles of flavonols and anthocyanidins, respectively, of a given plant species. For example, in tomato and petunia, DFR is specific for DHM, leading to delphinidin-derived anthocyanins (Holton et al., 1993; Bovy et al., 2002). Previously characterized FLS uses DHK or DHQ as preferred substrates (Holton et al., 1993; Suzuki et al., 2000; Wellmann et al., 2002; Park et al., 2017). FLS in onion (Allium cepa) prefers DHQ over DHK, leading to quercetin glycosides as dominant flavonol glycosides (Park et al., 2017). In grapevine (Vitis vinifera), FLS specificity for DHQ explains quercetin as the major flavonol (Jeong et al., 2006). Transient expression of CcFLS in N. benthamiana led to the enhanced formation of kaempferol, quercetin, and myricetin, suggesting that CcFLS is active with DHK, DHQ, and DHM. We are aware of only one other study that tested FLS acting upon DHM, which reported that FLS from rose (Rosa hybrida) converted DHM into myricetin (Suzuki et al., 2000). However, rose FLS was 10-fold less efficient with DHM compared with DHQ, and since rose does not have F3′5′H activity, formation of DHM and myricetin may not occur in planta. To the best of our knowledge, this study in montbretia is the first to describe a system of myricetin biosynthesis. In montbretia, myricetin serves as the core flavonol building block for the complex MbA biosynthesis.
On the transcript level, the montbretia myricetin biosynthesis pathway genes CcF3H-1, CcF3H-2, CcFLS, CcCYP1, and CcCYP2 showed higher transcript abundance in developing yC compared with oC, matching the profile of MbA biosynthesis and accumulation (Irmisch et al., 2018). Of the myricetin and known MbA biosynthesis genes (Irmisch et al., 2018), CcFLS showed the most extreme differential transcript expression between yC and oC early in the growing season.
Engineering Myricetin Biosynthesis and Effects of CcMYBs in N. benthamiana
We had previously shown that myricetin is limiting for the use of N. benthamiana as a synthetic biology host for MbA production (Irmisch et al., 2018). Expression of the first two MbA pathway-specific genes, CcUGT1 and CcUGT2, in N. benthamiana resulted in the formation of small amounts of kaempferol glucosyl rhamnoside but not MRG (Irmisch et al., 2018). Here, we showed that coexpression of three montbretia genes, CcMYB4, CcFLS, and CcCYP2, facilitated the availability of myricetin in N. benthamiana as a substrate for CcUGT1.
MYB-TFs are known to effect flavonoid biosynthesis in several plant species (Liu et al., 2015). Expression of different montbretia MYB-TFs in N. benthamiana resulted in the formation of different flavonols or flavonol precursors. CcMYB1 mainly enhanced 4′-hydroxy flavonoid biosynthesis, and CcMYB2 enhanced 3′- and 4′-hydroxy flavonoid biosynthesis. In contrast, CcMYB3 and CcMYB4 led to higher levels of 3′4′5′-hydroxy flavonoid. The accumulation of the 3′4′5′-hydroxylated dihydroflavonol DHM in N. benthamiana expressing CcMYB3 or CcMYB4 made these two MYB-TFs particularly interesting for improving myricetin formation. Endogenous N. benthamiana FLS appeared to be inefficient at converting DHM to myricetin, as is also known for other plant species (Forkmann et al., 1986; Bovy et al., 2002; Tanaka et al., 2008). Additional coexpression of CcFLS enabled the efficient conversion of DHM into myricetin, and coexpression of CcCYP2 also helped increase flux into the 3′4′5′-hydroxylated myricetin.
The four montbretia MYB-TFs enhanced 3′OH-flavonoid biosynthesis, but only CcMYB3 and CcMYB4 appeared to enhance the F3′5′H step in N. benthamiana. In G. triflora, GtMYB3 interacts with GtbHLH1 to activate expression of F3′5′H (Nakatsuka et al., 2008). In general, many R2R3-MYB-TFs rely on interaction with a bHLH-TF to regulate flavonoid biosynthesis (Lloyd et al., 1992; Borevitz et al., 2000; Liu et al., 2015). For example, coexpression of the maize (Zea mays) TF C1 and the bHLH LC in tomato increased kaempferol levels, while plants expressing either C1 or LC did not (Bovy et al., 2002). Arabidopsis MYB11, MYB12, and MYB111 and maize P1 do not require a bHLH protein (Grotewold et al., 1994; Stracke et al., 2007). CcMYBs have conserved amino acids for bHLH interaction (Grotewold et al., 2000); thus, it is possible that they interact with an endogenous N. benthamiana factor.
Engineering of MR, MRG, and Mini-MbA Biosynthesis in N. benthamiana
Transient expression in N. benthamiana has been developed as a useful system for testing gene functions and reconstructing the biosynthesis of complex metabolic pathways. For example, Lau and Sattely (2015) achieved the biosynthesis of an etoposide aglycone, the precursor for the chemotherapeutic etoposide, by transient expression of 10 genes from mayapple (Podophyllum hexandrum) in N. benthamiana. In the case of MbA, harvesting leaves of engineered N. benthamiana will have practical and economic advantages over harvesting of montbretia corms, which are the belowground storage organs and required for vegetative reproduction. N. benthamiana leaves are easily accessible; they produce a larger volume of faster growing biomass per plant than montbretia corms, and MbA biosynthesis will not be limited to a short developmental period, as is the case with the annual corm growth cycle in montbretia.
Coexpression of CcMYB4, CcFLS, and CcCYP2 with CcUGT1 or with both CcUGT1 and CcUGT2 led to the formation of the MbA pathway-specific intermediates MR and MRG, respectively. Levels of MRG produced in N. benthamiana are comparable to the level of MbA found in montbretia corms. Additional expression of CcAT1 resulted in the formation of small amounts of mini-MbA in N. benthamiana. Mini-MbA is the first intermediate in the MbA pathway that can be used as a potent (Ki = 90 nm) HPA inhibitor. Although three additional UGT steps are required to complete MbA biosynthesis, which remain to be discovered, the results presented here are promising for our goal to develop N. benthamiana as a production system for the antidiabetic compound MbA. However, in addition to MR and MRG, we also observed other myricetin-derived glycosides, likely formed by endogenous N. benthamiana UGTs. Such off-target metabolites may affect MbA yields and purification. We noticed that coexpression of CcMYB4, CcFLS, and CcCYP2 with both CcUGT1 and CcUGT2 increased target product yield fourfold, compared with coexpression of CcMYB4, CcFLS, and CcCYP2 with only CcUGT1. This result suggests that additional MbA pathway genes may drive biosynthesis further towards target as opposed to off-target compounds. For reference, in the production of the anticancer glucoraphanin in N. benthamiana using 13 genes from Arabidopsis, the expression of two additional Arabidopsis genes resulted in decreased levels of off-target products and an increase in product yield (Mikkelsen et al., 2010; Crocoll et al., 2016).
Beyond engineering of myricetin biosynthesis as the core for MbA in N. benthamiana, as described here, additional work is required to enhance the availability of caffeoyl-CoA as the substrate for the CcAT reaction to increase yield toward mini-MbA instead of mini-MbB. CcAT uses caffeoyl-CoA to produce mini-MbA, while it produces mini-MbB with coumaroyl-CoA. Internal π-stacking between myricetin and the caffeoyl moiety of MbA is essential for HPA inhibition (Williams et al., 2015). In a crystal structure of the MbA-HPA complex, HPA catalytic side chains interact directly with the caffeoyl hydroxyl groups. The caffeoyl meta-hydroxyl group of MbA accounts for 1,000-fold more effective HPA inhibition compared with MbB and MbC, analogs of MbA in which the meta-hydroxyl is replaced by a hydrogen or a methoxyl, respectively (Williams et al., 2015).
CONCLUSION
By exploring montbretia myricetin biosynthesis genes and MYB-TFs, we generated an essential combination of genes to engineer myricetin biosynthesis in N. benthamiana plants. Relevant amounts of the MbA precursor MRG were produced in N. benthamiana when montbretia myricetin biosynthesis genes were transiently coexpressed with CcUGT1 and CcUGT2. Additional coexpression of CcAT1 led to the formation of mini-MbA, a potential antidiabetic compound and precursor of MbA. This work confirms the potential of N. benthamiana as a heterologous production host for synthetic biology of the antidiabetic compound MbA.
MATERIALS AND METHODS
Plant Material, Sampling, RNA Extraction, and cDNA Synthesis
Montbretia (Crocosmia × crocosmiiflora var ‘Emily McKenzie’) plants were obtained and maintained as previously described (Irmisch et al., 2018). oC and newly developing corms (yC) were harvested at six different time points in 2016 as previously described (Irmisch et al., 2018). RNA isolation and cDNA biosynthesis were performed as previously described (Kolosova et al., 2004; Irmisch et al., 2018).
Identification and Isolation of Target Genes
The previously published montbretia corm transcriptome assembly (Irmisch et al., 2018) was used to identify candidate genes for enzymes of myricetin biosynthesis, including CHS, CHI, F3H, FLS, F3′H, and F3′5′H. A BLASTP search against the montbretia translated protein database was conducted (for accession numbers of query proteins, see Supplemental Table S1). Results were filtered using a reciprocal BLASTP search against the nonredundant protein database of the National Center for Biotechnology Information (NCBI). Selected montbretia sequences were used for a reciprocal BLASTP analysis to obtain a full set of target proteins. Phylogenetic analysis was used to support sequence annotations. Closely related sequences were clustered at a cutoff of 98% amino acid identity, using CD-HIT (version 4.6.1) to collapse possible allelic variants (Fu et al., 2012). In addition, short sequences (200 or fewer amino acids) were removed, and sequences were manually reviewed to remove likely chimeric or misassembled genes. For MYB-TFs, Arabidopsis (Arabidopsis thaliana) MYB75 was used in a BLASTP search against the montbretia translated protein database. Candidates were selected with 60% or greater amino acid identity to AtMYB75, at least eightfold higher transcript abundance in yC compared with oC, and a minimal length of 220 amino acids. CcMYB-TFs, CcF3H, CcFLS, CcCYP1, and CcCYP2 were amplified from cDNA attained from young corm material (June 10, 2016) and cloned into the pJET1.2/blunt vector (Thermo Fisher Scientific) for sequencing (Supplemental Table S2). CcCPR1 with 70% identity to AtCPR2 (X66017) was amplified.
Sequence Analysis and Phylogenetic Tree Reconstruction
An amino acid alignment of CcCYP1 and CcCYP2 and other published CYP75As and CYP75Bs (Supplemental Table S3) was constructed using the MUSCLE algorithm (gap open, −2.9; gap extend, 0; hydrophobicity multiplier, 1.5; clustering method, upgmb) implemented in MEGA6 (Tamura et al., 2011). Based on this alignment, a phylogenetic tree was estimated using the neighbor-joining algorithm (Poisson model) in MEGA6. A bootstrap resampling analysis with 1,000 replicates was performed to evaluate the tree topology. Following the same procedure, a phylogenetic tree of CcFLS, CcF3H-1, CcF3H-2, and other published FLS and F3H enzymes (Supplemental Table S3) was constructed. An alignment of CcMYB1 to CcMYB4 with published MYB-TFs was constructed using CLC and the ClustalW algorithm.
Generation of Transcript Abundance Heat Map
Transcript expression data in counts per million for manually curated genes were extracted from the transcriptome assembly (Supplemental Data Set S1; Irmisch et al., 2018). The heat map was generated with the R package heatmaply.
Heterologous Expression of FLS and F3H Enzymes in Escherichia coli
The complete ORFs of CcFLS, CcF3H-1 and CcF3H-2 were cloned as BsaI fragments into the pASK-IBA37 vector (IBA). The E. coli TOP10 strain (Invitrogen) was used for heterologous protein expression. Cultures were grown at 21°C, induced at OD600 = 0.5 with 200 µg L−1 anhydrotetracycline (Sigma-Aldrich), and subsequently placed at 18°C and grown for another 20 h. Cells were collected by centrifugation and disrupted by five freeze and thaw cycles in chilled extraction buffer (50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm dithiothreitol [DTT], 2% [v/v] glycerol, 150 mm NaCl2, 20 mm imidazole, 1× Pierce protease inhibitor [EDTA free; Thermo Fisher Scientific], 25 units of Benzonase Nuclease [Merck], and 0.2 mg mL−1 lysozyme). Cell fragments were removed by centrifugation at 14,000g, and the lysate was directly loaded onto an Ni-NTA agarose column (Qiagen). Protein was eluted with elution buffer (10 mm Tris-HCl, pH 7.5, 500 mm imidazole, 1 mm DTT, and 10% [v/v] glycerol) and desalted into assay buffer (10 mm Tris-HCl, pH 7.5, 1 mm DTT, and 10% [v/v] glycerol) through an Illustra NAP-5 column (GE Healthcare). Enzyme concentrations were determined using UV absorption at 280 nm.
Heterologous Expression of P450 Enzymes in the Yeast Saccharomyces cerevisiae
The complete ORFs of the putative montbretia P450 enzymes CcCYP1 and CcCYP2 were cloned into the pESC-Leu-2d vector and the ORF of montbretia CPR1 was cloned into the pESC-His vector following the User Cloning method (Ro et al., 2008). For expression, the resulting P450 constructs were each cotransferred with CcCPR1 into the yeast strain BY4741 (GE Life Sciences). CcCPR1 (NADPH-cytochrome P450 oxidoreductase) serves as an electron donor for the P450s. For gene expression, a single yeast colony was used to inoculate a starting culture in 30 mL of selective dropout medium, which was grown overnight at 28°C and 180 rpm. One OD of this culture (approximately 2 × 107 cells mL−1) was used to inoculate 100 mL of YPDE medium (1% [w/v] yeast extract, 2% [w/v] bacto-peptone, 5% [v/v] ethanol, and 2% [w/v] dextrose), which was grown for 32 to 35 h, induced by transferring the cells into YPG medium (1% [w/v] yeast extract, 2% [w/v] bacto-peptone, and 2% [w/v] Gal), and cultures were grown for another 15 to 18 h at 28°C. Cells were harvested and yeast microsomes were isolated according to the procedures described by Pompon et al. (1996) and Urban et al. (1994) with minor modifications. Briefly, the cultures were centrifuged (7,500g, 10 min, and 4°C), the supernatant was decanted, and the pellet was resuspended in 30 mL of TEK buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 100 mm KCl) and then centrifuged again. The cell pellet was resuspended in 2 mL of TES buffer (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 600 mm sorbitol, 10 g L−1 bovine serum fraction V protein, and 1.5 mm β-mercaptoethanol) and transferred to a 50-mL conical tube. Glass beads (acid-washed glass beads, 425–600 µm; Sigma-Aldrich) were added to fill the complete volume of the cell suspension. Yeast cell walls were disrupted by five cycles of 1 min of shaking by hand and subsequent chilling on ice for 1 min. The crude extract was recovered by washing the glass beads four times with 5 mL of TES. The combined washing fractions were centrifuged (7,500g, 10 min, and 4°C), and the supernatant was transferred and centrifuged again (100,000g, 60 min, and 4°C). The resulting microsomal protein fraction was homogenized in 2 mL of TEG buffer (50 mm Tris-HCl, 1 mm EDTA, and 30% [w/v] glycerol) using a glass homogenizer (Potter-Elvehjem, Fisher Scientific). Aliquots were stored at −20°C and used for protein assays.
Enzyme Assays
To test cloned montbretia P450 enzymes CcCYP1 and CcCYP2 for F3′H and F3′5′H activity, yeast microsomes harboring recombinant P450 protein and CcCPR1 were incubated in separate assays with the potential substrates naringenin, eriodictyol, DHK, DHQ, or kaempferol for 20 min at 25°C and 300 rpm. Assays were conducted in glass vials containing 200 µL of the reaction mixture (75 mm sodium phosphate buffer, pH 7.4, 10 µm substrate, 1 mm NADPH, and 2 µL of the prepared microsomes). Higher substrate concentrations and longer incubation times as well as higher volumes of microsomes did not affect substrate preference. To test activity with quercetin as the substrate, reactions were incubated for 60 min and 500 µm quercetin. All assays were stopped by placing on ice after the addition of an equal volume of MeOH. Reaction mixtures containing microsomes prepared from BY4741 transformed with the empty vector (pESC-Leu2d) and CcCPR1 served as negative controls. To test CcFLS, CcF3H-1, and CcF3H-2 for 2OGD activity, enzyme assays in 200 μL of assay buffer containing 5 μg of protein, 100 μm substrate, 0.5 mm ascorbate, 0.25 mm FeCl3, and 1 mm oxoglutarate were performed in a Teflon-sealed, screw-capped 1-mL gas chromatography glass vial. Naringenin, eriodictyol, DHK, DHQ, or DHM served as substrates. Assays were incubated for 60 min at 25°C with slight shaking and stopped by placing assay vials on ice after the addition of an equal volume of MeOH. Reaction products of all assays were analyzed using LC-MS/MS as described below.
RT-qPCR
cDNA syntheses were conducted with 650 ng of total RNA using the dsDNase and Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For RT-qPCR, the cDNA was diluted 1:5 with water. For the amplification of target gene fragments with a length of about 150 bp, primer pairs were designed having a melting temperature of 60°C or greater, GC content of 45% to 60%, and primer length of 20 to 25 nucleotides (Supplemental Table S2). Primer specificity was confirmed by agarose gel electrophoresis, melting curve analysis, standard curve analysis, and sequence verification of cloned PCR amplicons. Samples were run in duplicate using SsoFast EvaGreen Supermix (Bio-Rad). The following PCR conditions were applied for all reactions: initial incubation at 95°C for 30 s followed by 40 cycles of amplification (95°C for 5 s and 60°C for 10 s). Melting curve data were recorded at the end of cycling from 55°C to 95°C. All assays were performed with the same PCR machine (Bio-Rad CFX96; Bio-Rad) on optical 96-well plates. RT-qPCR analyses were performed with three biological replicates for each of the six different time points for yC and oC samples (June 10, June 27, July 22, August 16, September 12, and October 6). The genes for serin-incooperator (MEP) and the zinc-finger protein (ZF) were used as reference genes as described previously (Irmisch et al., 2018; Supplemental Table S2).
Transient Expression of Montbretia Genes in Nicotiana benthamiana
For expression in N. benthamiana, the coding regions of CcCHS2, CcCHI2, CcF3H-2, CcFLS, CcCYP2, CcMYB1, CcMYB2, CcMYB3, CcMYB4, CcUGT1 (MG938542), CcUGT2 (MG938543), and CcAT1 (MH365462) were cloned individually into a pCAMBIA2300U vector. After sequence verification, pCAMBIA vectors carrying the individual montbretia cDNAs, eGFP, or the pBIN:p19 plasmid were separately transferred into Agrobacterium tumefaciens strain C58pMP90. One milliliter of overnight cultures (220 rpm and 28°C) was used to inoculate 10 mL of Luria-Bertani medium containing 50 µg mL−1 kanamycin, 25 µg mL−1 rifampicin, and 25 µg mL−1 gentamicin for overnight growth. The following day the cultures were centrifuged (4,000g and 5 min) and cells were resuspended in infiltration buffer (10 mm MES, pH 5.6, 10 mm MgCl2, and 100 µm acetosyringone) to a final OD600 of 0.5. After shaking for 3 h at room temperature, different combinations of equal volumes of individual transformed A. tumefaciens (35Spro:gene) as shown in Table 1 were prepared for leaf infiltration. All combinations included A. tumefaciens pBIN:p19 for enhancement of gene expression. Leaves of 4-week-old N. benthamiana plants were infiltrated with A. tumefaciens solution using a 1-mL needle-free syringe to gently push the solution into the abaxial surface. Infiltrated leaves were labeled with tape and harvested 5 d after infiltration. Four biological replicates were used and analyzed for the final combinations M, P, S, and T, and two biological replicates were used for screening of all other combinations. Leaf material was directly frozen in liquid nitrogen and stored at −80°C until further analysis. Plant material was ground in liquid nitrogen into a fine powder, and 100 mg was extracted with 1 mL of 50% (v/v) MeOH for 2 h at room temperature. The extract was analyzed using LC-MS/MS.
LC-MS Analysis
LC was performed on an Agilent 1100 HPLC device (Agilent Technologies) with an Agilent ZORBAX SB-C18 column (50 × 4.6 mm, 1.8-μm particle size; Merck) using aqueous formic acid (0.2% [v/v]; mobile phase A) and acetonitrile plus formic acid (0.2% [v/v]; mobile phase B). The elution profile for in vitro enzyme activity assays was as follows: 0 to 0.5 min, 95% A; 0.5 to 5 min, 5% to 40% B in A; 5 to 7 min, 90% B in A; and 7.1 to 10 min, 95% A. The elution profile for N. benthamiana leaf extracts was as follows: 0 to 0.5 min, 95% A; 0.5 to 5 min, 5% to 20% B in A; 5 to 7 min, 90% B in A; and 7.1 to 10 min, 95% A. The flow rate was 0.8 mL min−1 at a column temperature of 50°C. LC was coupled to an Agilent MSD Trap XCT-Plus mass spectrometer equipped with an electrospray operated in negative ionization mode (capillary voltage, 4,000 eV; temperature, 350°C; nebulizing gas, 60 p.s.i.; dry gas, 12 L min−1) and an Agilent 1100 Diode Array Detector (detection, 200–700 nm; J&M Analytik). MS/MS was used to monitor daughter ion formation. The LC/MSD Trap Software 5.2 (Bruker Daltonik) was used for data acquisition and processing. Metabolites were quantified using an MbA standard curve. Compounds were identified using their retention times, molecular masses, and specific fragmentation patterns and by use of authentic standards if they were available. Authentic standards for eriodictyol, naringenin, DHK, DHM, myricetin, and MR were from Sigma-Aldrich. Other authentic standards used were DHQ (Sequoia Research Products), quercetin and kaempferol (TRC), myricetin 3-O-glucoside (Extrasynthese), and MbA and mini-MbA (Williams et al., 2015).
Statistical Analysis
To test for significant differences in gene expression patterns in yC and oC at different time points (June to October 2016), two-way ANOVA was performed followed by Tukey’s test using SigmaPlot 11.0 for Windows (Systat Software; Supplemental Table S4). Data for CcF3H-1, CcF3H-2, CcFLS, CcCYP1, and CcCYP2 gene expression were transformed (log, log, ^0.25, ^0.25, and ^0.3, respectively) in order to meet statistical requirements. Student’s t test was used to test for significant fold change differences between oC and yC harvested on June 10 (Supplemental Table S4).
Accession Numbers
Nucleotide sequences were deposited in GenBank with the accession numbers MK562522 (CcF3H-1), MK562523 (CcF3H-2), MK562524 (CcFLS1), MK562521 (CcCYP2; CYP75B138), MK562520 (CcCYP1; CYP75B137), MK562529 (CcCPR), MK562525 (CcMYB1), MK562526 (CcMYB2), MK562527 (CcMYB3), and MK562528 (CcMYB4). Accession numbers for all other genes/proteins used in this work are indicated in Supplemental Tables S1 and S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogeny of montbretia CcF3H-1, CcF3H-2, and CcFLS.
Supplemental Figure S2. Bifunctional enzyme activity of CcFLS.
Supplemental Figure S3. Phylogeny of montbretia CcCYP1 and CcCYP2.
Supplemental Figure S4. Mass spectrum of m/z 303 produced by CcCYP2.
Supplemental Figure S5. Flavonol rhamnoside detection in N. benthamiana transiently expressing montbretia flavonol biosynthetic pathway genes and CcUGT1.
Supplemental Figure S6.. Sequence alignment of montbretia CcMYB-TF with other characterized MYB-TFs.
Supplemental Figure S7.. Dihydroflavonol glycosides in N. benthamiana transiently expressing montbretia CcMYB-TF and CcUGT1.
Supplemental Figure S8. MR, MRG, and MRG-Cou production in N. benthamiana transiently expressing montbretia MbA biosynthesis genes.
Supplemental Figure S9. Mini-MbA production in N. benthamiana.
Supplemental Table S1. NCBI identification numbers of protein sequences used for BLASTP analysis.
Supplemental Table S2.. Oligonucleotides used in this study.
Supplemental Table S3. NCBI identification numbers of protein sequences utilized for phylogenetic reconstruction.
Supplemental Table S4. Statistical analysis of myricetin biosynthesis pathway gene expression.
Supplemental Data Set S1. Transcriptome sequences of myricetin biosynthesis pathway genes used for heat map generation.
Acknowledgments
We thank Stephen G. Withers for insightful discussions and collaboration, Gary Brayer for providing montbretia corms, Carol Ritland for project management, and David Nelson for cytochrome P450 nomenclature support.
Footnotes
The research was supported by funds to J.B. from the GlycoNet Network of Centres of Excellence, Canada, and the Natural Sciences and Engineering Research Council of Canada (Discovery Grant). S.I. was supported by the Alexander von Humboldt Foundation through a Feodor Lynen Research Fellowship.
Articles can be viewed without a subscription.
References
- Asada Y, Hirayama Y, Furuya T (1988) Acylated flavonols from Crocosmia crocosmiiflora. Phytochemistry 27: 1497–1501 [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al. (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugliera F, Barri-Rewell G, Holton TA, Mason JG (1999) Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J 19: 441–451 [DOI] [PubMed] [Google Scholar]
- Crocoll C, Mirza N, Reichelt M, Gershenzon J, Halkier BA (2016) Optimization of engineered production of the glucoraphanin precursor dihomomethionine in Nicotiana benthamiana. Front Bioeng Biotechnol 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkmann G, Vlaming P, Spribille R, Wiering H, Schram AW (1986) Genetic and biochemical studies on the conversion of dihydroflavonols to flavonols in flowers of Petunia hybrida. Z Naturforsch 41: 179–186 [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97: 13579–13584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Canada(2019) http://www.hc-sc.gc.ca
- Holton TA, Brugliera F, Tanaka Y (1993) Cloning and expression of flavonol synthase from Petunia hybrida. Plant J 4: 1003–1010 [DOI] [PubMed] [Google Scholar]
- Irmisch S, Jo S, Roach CR, Jancsik S, Man Saint Yuen M, Madilao LL, O’Neil-Johnson M, Williams R, Withers SG, Bohlmann J (2018) Discovery of UDP-glycosyltransferases and BAHD-acyltransferases involved in the biosynthesis of the antidiabetic plant metabolite montbretin A. Plant Cell 30: 1864–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong ST, Goto-Yamamoto N, Hashizume K, Esaka M (2006) Expression of the flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci 170: 61–69 [Google Scholar]
- Kolosova N, Miller B, Ralph S, Ellis BE, Douglas C, Ritland K, Bohlmann J (2004) Isolation of high-quality RNA from gymnosperm and angiosperm trees. Biotechniques 36: 821–824 [DOI] [PubMed] [Google Scholar]
- Lau W, Sattely ES (2015) Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349: 1224–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillo C, Lea US, Ruoff P (2008) Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ 31: 587–601 [DOI] [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8: 689–708 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Walbot V, Davis RW (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258: 1773–1775 [DOI] [PubMed] [Google Scholar]
- Martens S, Preuss A, Matern U (2010) Multifunctional flavonoid dioxygenases: Flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 71: 1040–1049 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Olsen CE, Halkier BA (2010) Production of the cancer-preventive glucoraphanin in tobacco. Mol Plant 3: 751–759 [DOI] [PubMed] [Google Scholar]
- Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, Shimada N, Yamamura S, Nishihara M (2008) Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol 49: 1818–1829 [DOI] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kim DH, Lee JY, Ha SH, Lim SH (2017) Comparative analysis of two flavonol synthases from different-colored onions provides insight into flavonoid biosynthesis. J Agric Food Chem 65: 5287–5298 [DOI] [PubMed] [Google Scholar]
- Pollastri S, Tattini M (2011) Flavonols: Old compounds for old roles. Ann Bot 108: 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Ro DK, Ouellet M, Paradise EM, Burd H, Eng D, Paddon CJ, Newman JD, Keasling JD (2008) Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Schijlen EGW, Ric de Vos CH, van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65: 2631–2648 [DOI] [PubMed] [Google Scholar]
- Seitz C, Eder C, Deiml B, Kellner S, Martens S, Forkmann G (2006) Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′,5′-hydroxylase in the Asteraceae family. Plant Mol Biol 61: 365–381 [DOI] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Tsuda S, Fukui Y, Fukuchi-Mizutani M, Yonekura-Sakakibara K, Tanaka Y, Kusumi T (2000) Molecular characterization of rose flavonoid biosynthesis genes and their application in Petunia. Biotechnology and Biotechnological Equipment 14: 56–62 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Yonekura K, Fukuchi-Mizutani M, Fukui Y, Fujiwara H, Ashikari T, Kusumi T (1996) Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora. Plant Cell Physiol 37: 711–716 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Tarling CA, Woods K, Zhang R, Brastianos HC, Brayer GD, Andersen RJ, Withers SG (2008) The search for novel human pancreatic alpha-amylase inhibitors: High-throughput screening of terrestrial and marine natural product extracts. ChemBioChem 9: 433–438 [DOI] [PubMed] [Google Scholar]
- The World Health Organization(2019) http://www.who.int
- Tohge T, Watanabe M, Hoefgen R, Fernie AR (2013) The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol Biol 48: 123–152 [DOI] [PubMed] [Google Scholar]
- Urban P, Werck-Reichhart D, Teutsch HG, Durst F, Regnier S, Kazmaier M, Pompon D (1994) Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast: Kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur J Biochem 222: 843–850 [DOI] [PubMed] [Google Scholar]
- Wellmann F, Lukacin R, Moriguchi T, Britsch L, Schiltz E, Matern U (2002) Functional expression and mutational analysis of flavonol synthase from Citrus unshiu. Eur J Biochem 269: 4134–4142 [DOI] [PubMed] [Google Scholar]
- Williams LK, Zhang X, Caner S, Tysoe C, Nguyen NT, Wicki J, Williams DE, Coleman J, McNeill JH, Yuen V, et al. (2015) The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif. Nat Chem Biol 11: 691–696 [DOI] [PubMed] [Google Scholar]
- Yuen VG, Coleman J, Withers SG, Andersen RJ, Brayer GD, Mustafa S, McNeill JH (2016) Glucose lowering effect of montbretin A in Zucker Diabetic Fatty rats. Mol Cell Biochem 411: 373–381 [DOI] [PubMed] [Google Scholar]