Leucoanthocyanidin reductases 1 and 2 possess distinct, dual activities for regulating the composition and degree of polymerization of proanthocyanidins in grapevine.
Abstract
Proanthocyanidins (PAs) in grapevine (Vitis vinifera) are found mainly in berries, and their content and degree of polymerization are important for the mouth feel of red wine. However, the mechanism of PA polymerization in grapevine remains unclear. Previous studies in the model legume Medicago truncatula showed that 4β-(S-cysteinyl)-epicatechin (Cys-EC) is an epicatechin-type extension unit for nonenzymatic PA polymerization, and that leucoanthocyanidin reductase (LAR) converts Cys-EC into epicatechin starter unit to control PA extension. Grapevine possesses two LAR genes, but their functions are not clear. Here, we show that both Cys-EC and 4β-(S-cysteinyl)-catechin (Cys-C) are present in grapevine. Recombinant VvLAR1 and VvLAR2 convert Cys-C and Cys-EC into (+)-catechin and (−)-epicatechin, respectively, in vitro. The kinetic parameters of VvLARs are similar, with both enzymes being more efficient with Cys-C than with Cys-EC, the 2,3-cis conformation of which results in steric hindrance in the active site. Both VvLARs also produce (+)-catechin from leucocyanidin, and an inactive VvLAR2 allele reported previously is the result of a single amino acid mutation in the N terminus critical for all NADPH-dependent activities of the enzyme. VvLAR1 or VvLAR2 complement the M. truncatula lar:ldox double mutant that also lacks the leucoanthocyanidin dioxygenase (LDOX) required for epicatechin starter unit formation, resulting in increased soluble PA levels, decreased insoluble PA levels, and reduced levels of Cys-C and Cys-EC when compared to the double mutant, and the appearance of catechin, epicatechin, and PA dimers characteristic of the ldox single mutant in young pods. These data advance our knowledge of PA building blocks and LAR function and provide targets for grapevine breeding to alter PA composition.
Proanthocyanidins (PAs, also called condensed tannins) are oligomers or polymers of flavan-3-ol units. They are the second most abundant polyphenolic compounds in the plant kingdom after lignin and are present in the fruits, seeds, leaves, and bark of many plants (Dixon et al., 2005; Liu et al., 2016). PAs and their monomeric building blocks not only protect plants against stress (Scalbert, 1992; Dixon et al., 2005; Furlan et al., 2011) but also have beneficial health effects for humans and ruminant animals (Aerts et al., 1999; Bagchi et al., 2000; Middleton et al., 2000; Cos et al., 2004). In addition, grapevine (Vitis vinifera) berry skin and seed-derived PAs greatly affect the mouth-feel attributes of red wine—a more astringent or rougher sensation is associated with increased content, degree of polymerization, and galloylation of PAs (Peleg et al., 1999; Maury et al., 2001; Vidal et al., 2003). Understanding PA biosynthesis is important for the modification of PA traits via metabolic engineering or viticulture practice.
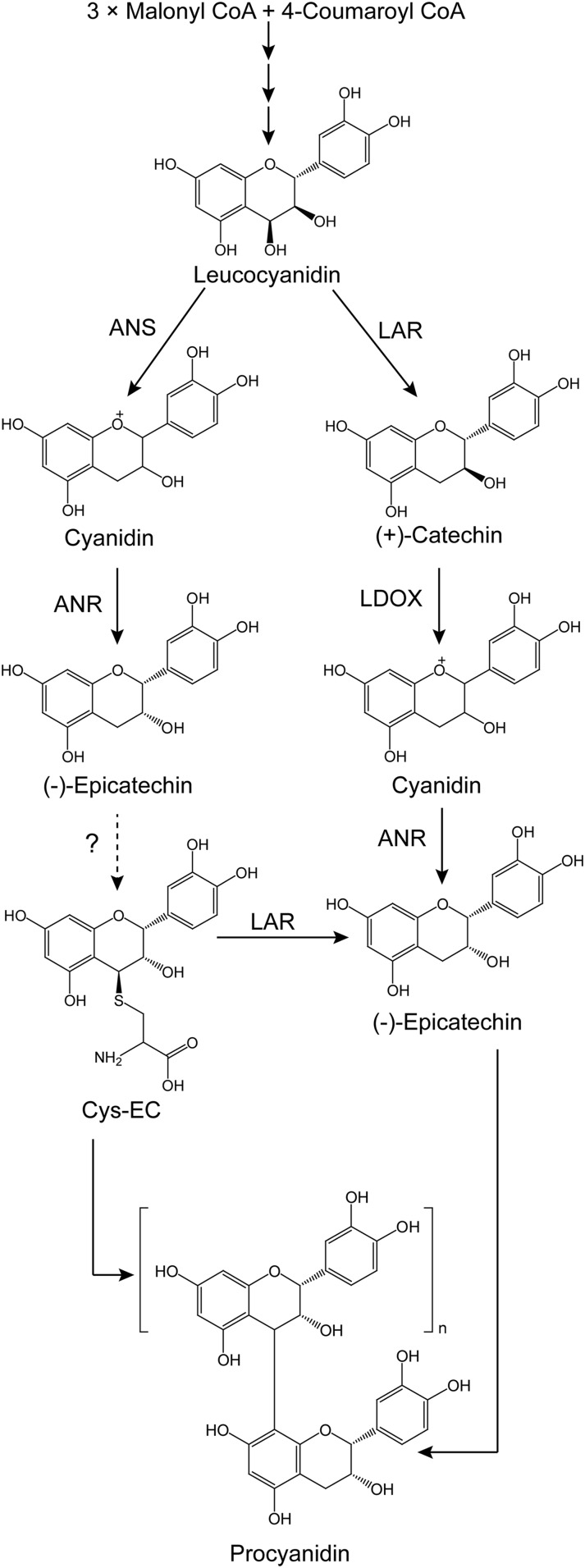
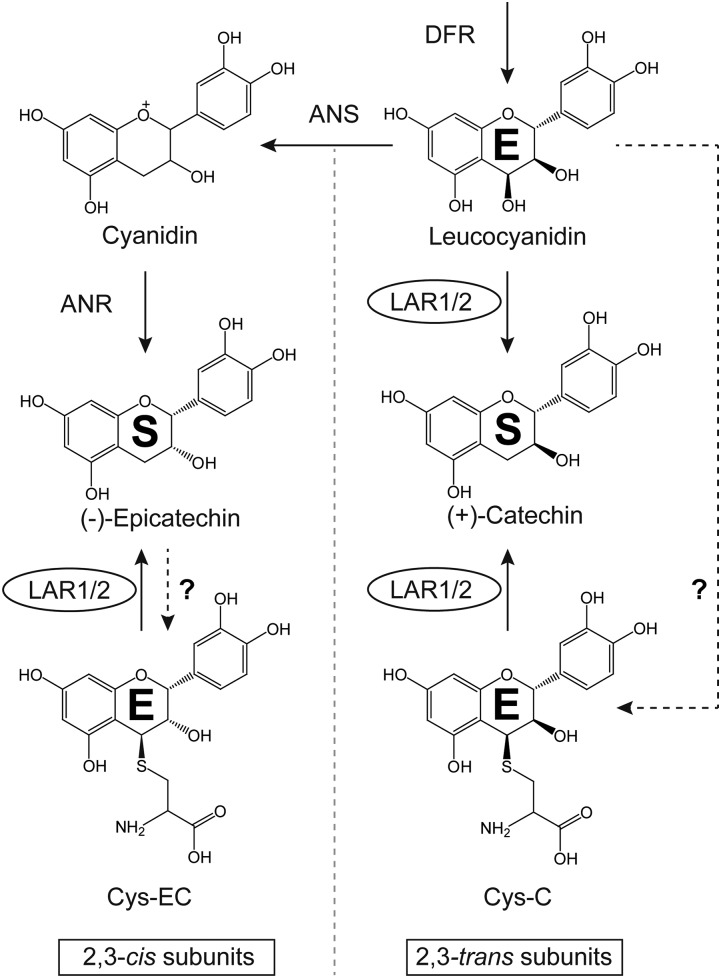
Flavan-3-ol monomers, primarily (−)-epicatechin or (+)-catechin, serve as the starter units for PA polymerization, which occurs when flavan-3-ol carbocations act as extension units by attacking the C8 position of the previous building block to lengthen the PA chains (Dixon et al., 2005; Liu et al., 2016). Both biochemical and genetic evidence indicate that anthocyanidin reductase (ANR), encoded by the BANYULS gene in Arabidopsis (Arabidopsis thaliana), converts cyanidin to (−)-epicatechin in the formation of PAs (Xie et al., 2003). (+)-Catechin, the trans isomer of (−)-epicatechin, is believed to be produced from leucocyanidin through the activity of leucoanthocyanidin reductase (LAR; Tanner and Kristiansen, 1993). LAR was first isolated from the tannin-rich legume Desmodium uncinatum, and the recombinant enzyme was shown to convert three differently hydroxylated leucoanthocyanidins to their corresponding 2,3-trans-flavan-3-ols (Tanner et al., 2003). Compared to D. uncinatum, Arabidopsis lacks a LAR ortholog, consistent with the presence of exclusively (−)-epicatechin-derived PAs in the seed coats of this species (Lepiniec et al., 2006). In vivo evidence of LAR function sometimes conflicts with the activity of producing (+)-catechin demonstrated in vitro. For example, overexpression of D. uncinatum LAR in tobacco (Nicotiana tabacum) and white clover (Trifolium repens) failed to generate (+)-catechin, although LAR activity was detectable in the leaf extracts of the transgenic plants (Tanner et al., 2003), and similar results have been reported with LARs from other plants (Pang et al., 2007; Ferraro et al., 2014; Wang et al., 2017). Expression of LARs from cacao (Theobroma cacao) or tea (Camellia sinensis) in tobacco resulted in greater accumulation of (−)-epicatechin than (+)-catechin (Liu et al., 2013; Pang et al., 2013). These paradoxical results have been explained recently by studies of LAR function in Medicago truncatula (Liu et al., 2016; Jun et al., 2018). This species possesses a highly expressed LAR, but (+)-catechin-derived PAs are only present in small amounts in young pods and seeds (Pang et al., 2007; Jun et al., 2018). M. truncatula LAR (MtLAR) is a bifunctional enzyme, catalyzing the reduction of leucocyanidin to produce (+)-catechin as well as cleavage of the potential PA extension unit precursor 4β-(S-cysteinyl)-epicatechin (Cys-EC) into Cys and (−)-epicatechin (starter unit; Fig. 1). The downstream enzyme leucoanthocyanidin dioxygenase (MtLDOX), a homolog of anthocyanidin synthase (ANS), converts (+)-catechin to cyanidin, which can be used by ANR to generate (−)-epicatechin as a PA starter unit (Jun et al., 2018; Fig. 1). Through these reactions, MtLAR can regulate the ratio of starter units to extension units, thereby controlling the degree of polymerization (DP) of PAs.
Figure 1.
Biosynthesis of proanthocyanidins in M. truncatula. CoA, coenzyme A. The dashed line with the question mark indicates a proposed reaction that has yet to be elucidated.
Two LAR orthologs from grapevine 'Shiraz' have been reported (Bogs et al., 2005), with different spatial and temporal patterns of transcript expression in developing berry skins, seeds, and leaves. Recombinant VvLAR1 has been shown to produce (+)-catechin, but there is still debate as to whether VvLAR2 has the ability to convert leucoanthocyanidins to 2,3-trans-flavan-3-ols in vitro (Bogs et al., 2005; Pfeiffer et al., 2006). Similarly, of the two Lotus corniculatus LARs, recombinant LcLAR1 but not LcLAR2 showed activity with leucocyanidin, questioning the potential function of the inactive enzyme (Paolocci et al., 2007). Additionally, in a quantitative trait locus study, most of the single nucleotide polymorphisms in the VvLAR1 exons are strongly associated with the mean degree of polymerization (mDP) of PAs (Huang et al., 2012). This suggests that at least VvLAR1 possibly has an additional function similar to that of MtLAR. Although VvLAR2 has a similar sequence and the same motifs as VvLAR1 (Bogs et al., 2005), it is uncertain whether VvLAR2 is redundant with VvLAR1 or whether it has a completely different function.
Here, we show that both VvLARs convert leucocyanidin to (+)-catechin and convert Cys-EC to (−)-epicatechin. However, they more efficiently convert 4β-(S-cysteinyl)-catechin (Cys-C), a potential (+)-catechin-type extension unit, to (+)-catechin. A key amino acid crucial for all the NADPH-dependent LAR activities was discovered through analysis of a null allele in 'Shiraz' grapevine. The two VvLARs complement the known MtLAR activities in the M. truncatula lar:ldox mutant, and their expression leads to decreased levels of Cys-C and Cys-EC in transgenic plants. Our findings suggest that VvLAR1 and VvLAR2 function similarly in controlling the degree of PA polymerization and support LAR as a target for molecular breeding to improve the PA composition of wine.
RESULTS
Cys-C Coexists with Cys-EC in Grapevine and May Function as a (+)-Catechin-Type PA Extension Unit
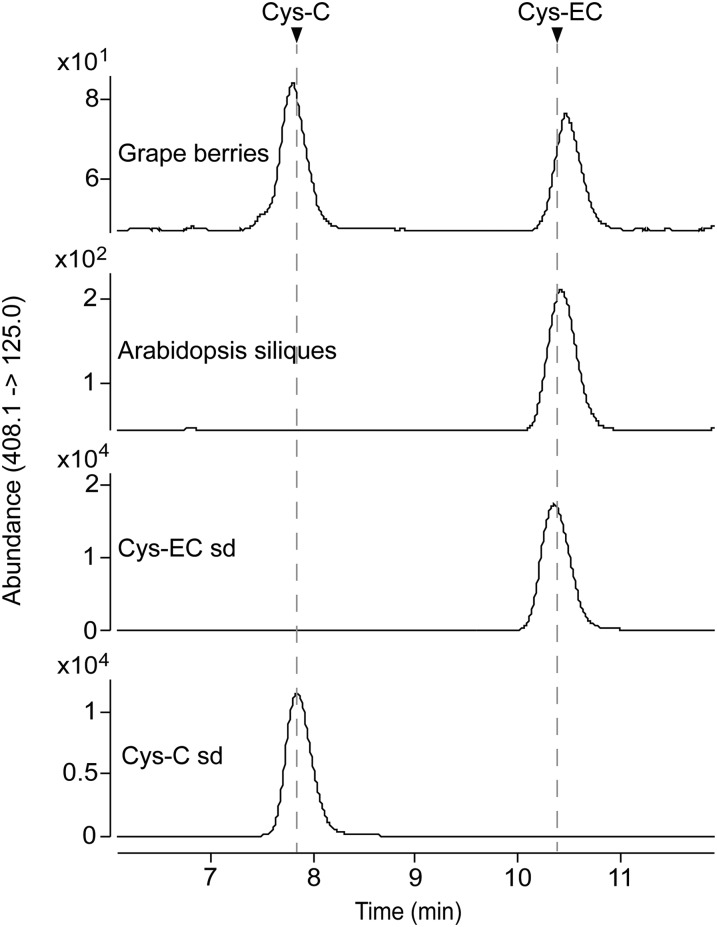
To study potential PA extension units in grapevine, we isolated soluble PAs from grapevine 'Cabernet Sauvignon' berries at veraison stage, as well as Arabidopsis siliques harvested 7 d after flowering. HPLC of the extracts with detection on a triple quadrupole mass spectrometer (HPLC-QqQ) revealed two compounds from grapevine with the same mass spectrum as Cys-EC (Liu et al., 2016), whereas only one was detected in Arabidopsis extracts, at the retention time of the later eluting compound from grapevine (Supplemental Fig. S1). Based on the polarity difference shown on reverse-phase HPLC, we speculated that the shared compound was Cys-EC and that the other compound in grapevine might be Cys-C. To confirm this, we synthesized Cys-EC and Cys-C standards and developed a multiple reaction monitoring (MRM) method using HPLC-QqQ to detect the two compounds. Reanalysis of the soluble PA fractions from grapevine and Arabidopsis revealed a peak at the same retention time as the Cys-EC standard in both extracts and a peak at the same retention time as Cys-C in the extract from grapevine only (Fig. 2). Cys-C can form a (+)-catechin carbocation, which could attack the PA starter unit at the C8 position under neutral conditions, because we have successfully synthesized procyanidin B4, C1, and C2 (with catechin as extension unit) using Cys-C with commercially available epicatechin and procyanidin dimers (Jun et al., 2018). The fact that grapevine PAs have both (−)-epicatechin and (+)-catechin-type extension units, whereas Arabidopsis only contains PAs made of (−)-epicatechin, suggests that Cys-C can be a (+)-catechin-type PA extension unit in grapevine.
Figure 2.
Cys-C coexists with Cys-EC in grapevine. Soluble PA extracts from grapevine ('Cabernet Sauvignon') skin and Arabidopsis siliques were analyzed using HPLC-QqQ by multiple reaction monitoring (MRM) in negative mode. The transition of mass to charge ratio (m/z) 408.1 to m/z 125.0 was used for the identification of Cys-C and Cys-EC. The arrows indicate the peaks for Cys-C and Cys-EC standards (sd).
Characterization of VvLARs with Cys-C and Cys-EC as Substrates
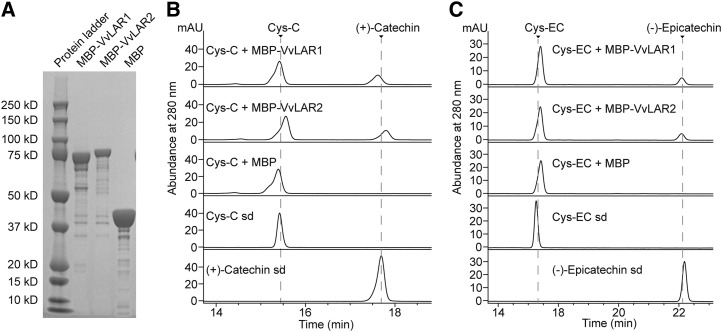
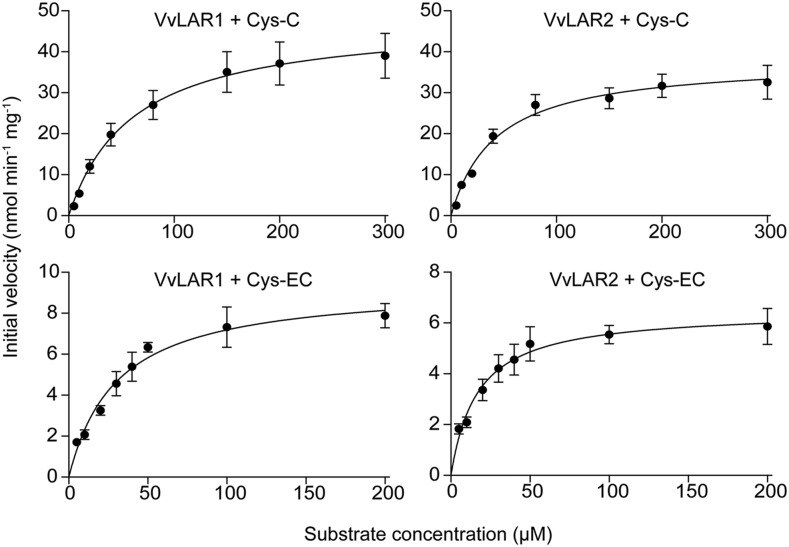
MtLAR can cleave Cys-EC into (−)-epicatechin and Cys (Liu et al., 2016). To determine whether Cys-EC or Cys-C are substrates for VvLARs, full-length VvLAR1 and VvLAR2 open reading frames (ORFs) from grapevine 'Cabernet Sauvignon' were separately subcloned into Escherichia coli expression vector pMal-c5x fused with a maltose-binding protein (MBP) tag at the N terminus. The MBP tag ORF alone was expressed in parallel as a negative control. Based on bioinformatic tool prediction, the molecular masses of MBP-VvLAR1, MBP-VvLAR2, and MBP are 80.53, 81.57, and 42.51 kD, respectively. MBP-VvLAR2 and MBP had molecular masses consistent with the prediction following separation on a Bis-Tris Plus Gel (Fig. 3A). However, MBP-VvLAR1 showed an apparent molecular mass of ∼75 kD (Fig. 3A), possibly due to instability at the C terminus (Maugé et al., 2010). The purified proteins were assayed using synthetic Cys-C or Cys-EC as substrate in the presence of NADPH, followed by HPLC-UV analysis. The empty vector control extract showed no product formation (Fig. 3, B and C), whereas both VvLAR1 and VvLAR2 proteins were able to convert Cys-C and Cys-EC to (+)-catechin and (−)-epicatechin, respectively (Fig. 3, B and C). Kinetic parameters (Table 1) were determined from plots of reaction velocity versus substrate concentration (Fig. 4). The Kcat (turnover number)/Km values for the two VvLARs with Cys-EC were similar and nearly 10 times that of MtLAR with Cys-EC (Liu et al., 2016). Although the Km values of both VvLAR1 and VvLAR2 for Cys-EC were lower than for Cys-C, the Kcat/Km values for both VvLARs with Cys-C were nearly the same and more than 20 times greater than with Cys-EC. The only obvious difference between the kinetics of the two enzymes was the lower Km values of VvLAR2 with both Cys-C and Cys-EC.
Figure 3.
Expression of recombinant VvLAR1 and VvLAR2 and assay with Cys-C and Cys-EC as substrates. A, Analysis of purified recombinant VvLAR1, VvLAR2, and MBP tag on a Bis-Tris Plus Gel. B and C, HPLC profiles of in vitro enzymatic reactions at 280 nm. The enzyme reactions were performed with 40 μm substrates, 250 μm NADPH, and 10 μg recombinant proteins for 1 h. Reactions with MBP tag alone were run as negative controls. The arrows indicate the peaks for Cys-C, Cys-EC, (+)-catechin, and (−)-epicatechin standards (sd).
Table 1. Kinetic parameters for VvLAR1 and VvLAR2 with Cys-C and Cys-EC as substrates.
Kinetic parameters were determined by fitting the initial velocity data to the Michaelis-Menten equation by nonlinear regression analysis using GraphPad Prism 7 software. Experimental data are presented in Figure 4.
| Enzyme | Substrate | Vmax (nmol min−1 mg−1) | Km (μm) | Kcat (min−1) | Kcat/Km (min−1 M−1) |
|---|---|---|---|---|---|
| VvLAR1 | Cys-EC | 9.37 ± 0.43 | 30.49 ± 3.91 | 0.74 | 2.43 × 104 |
| VvLAR2 | Cys-EC | 6.49 ± 0.28 | 16.80 ± 2.46 | 0.53 | 3.15 × 104 |
| VvLAR1 | Cys-C | 48.12 ± 2.66 | 61.21 ± 10.06 | 3.89 | 6.36 × 105 |
| VvLAR2 | Cys-C | 38.01 ± 1.53 | 42.40 ± 5.71 | 3.10 | 7.31 × 105 |
Figure 4.
Plots of initial velocity versus substrate concentration for recombinant VvLAR1 and VvLAR2 with Cys-C and Cys-EC as substrates. The enzyme reactions were performed in a total volume of 100 μL containing 250 μm NADPH, 4 μg VvLAR1 or VvLAR2, and indicated amounts of Cys-C or Cys-EC with 10 min incubation. The products were quantified using (+)-catechin as standard on HPLC at 280 nm. The experimental points are presented as the average of three technical replicates with sd as error bars.
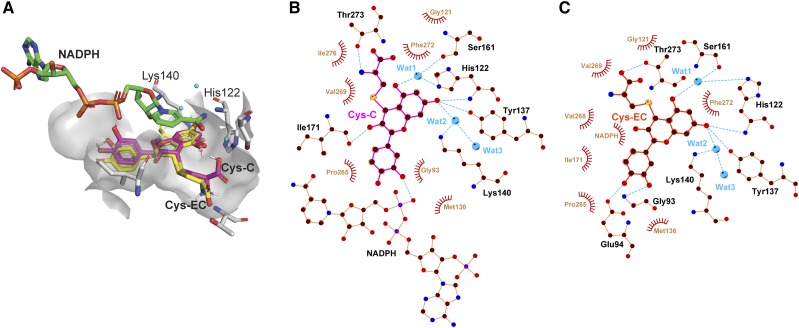
We then took advantage of the availability of the VvLAR1 crystal structure (PDB: 3I52) to perform molecular docking analysis in an attempt to explain the above differences in catalytic efficiencies. The two protein-substrate complexes were superimposed and are shown in Figure 5A. The benzopyran backbones of the two substrates have a very similar spatial position to that of catechin in the crystal structure (Maugé et al., 2010). The cysteinyl moieties were accommodated in the remaining cavity, which was spacious enough to allow the carboxyl group of the Cys to rotate nearly 180° (Fig. 5A). Schematic diagrams of protein-substrate interactions for Cys-C and Cys-EC are shown in Figure 5, B and C, respectively. The hydrogen-bonding networks between VvLAR1 and the phenolic OH5 and OH7 of the substrates suggested that the reductive cleavage of the carbon-sulfur bond at C4 might proceed by a similar “two-step catalytic mechanism” to that proposed for the reduction of leucocyanidin (Maugé et al., 2010), with His-122 and Lys-140 acting as acid-base catalysts though the water bridges and NADPH responsible for hydride transfer to C4 of Cys-C or Cys-EC. Based on the three-dimensional (3D) structure shown in Figure 5A, Cys-C was closer to NADPH than was Cys-EC, and the OH3 of Cys-EC might sterically restrict interaction between Cys-EC and NADPH to further limit the transfer of protons in the second step of the reaction, causing the observed lower Kcat/Km values in Table 1. The 3D structure of VvLAR2 was obtained by molecular modeling using the VvLAR1 structure as the template. Substrate docking analysis with VvLAR2 showed similar results to VvLAR1 regarding the interactions between enzyme and substrates (Supplemental Fig. S2).
Figure 5.
Molecular docking analysis of Cys-C and Cys-EC in the VvLAR1 active site. A, Superimposed 3D structures of Cys-C and Cys-EC docked with VvLAR1 (PDB ID: 3I52) active site. The ligands NADPH, Cys-C, and Cys-EC and catalytic residues His-122 and Lys-140 are shown as bond models, and the three water molecules are shown as blue spheres. B and C, ligand-protein interaction diagrams of VvLAR1 with Cys-C and Cys-EC as substrates. Water molecules (Wat) are shown as blue balls. Hydrogen bonds are shown as blue dotted lines, while the spoked arcs represent protein residues or NADPH making nonbonded contacts with Cys-C and Cys-EC.
Analysis of Active and Inactive Alleles of VvLAR2 Reveals a Critical Residue for NADPH-Dependent LAR Activities
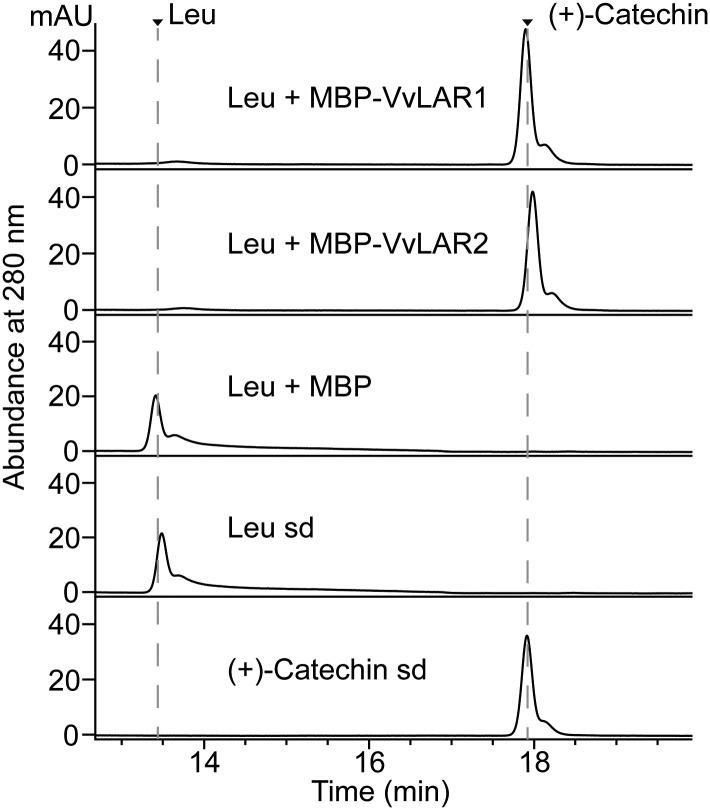
Previous studies confirmed that VvLAR1 converted leucocyanidin to (+)-catechin in vitro (Bogs et al., 2005; Pfeiffer et al., 2006) but were inconsistent as to the activity of VvLAR2 in vitro. In one study, coupled reactions with LAR from grapevine 'Regent' and apple dihydroflavanol reductase (DFR) could convert dihydroquercetin to (+)-catechin, whereas LAR from 'Shiraz' could not produce (+)-catechin from leucocyanidin (Bogs et al., 2005; Pfeiffer et al., 2006). Alignment of VvLAR2 sequences from grapevine cultivars 'Regent', 'Cabernet Sauvignon', and 'Shiraz' showed that there were three sites where 'Cabernet Sauvignon' and 'Shiraz' were identical but different from 'Regent', and none of these was in the proposed active site (Fig. 6). To address the activity of these different LAR proteins, MBP-VvLAR2 was incubated with leucocyanidin in the presence of NADPH. The MBP tag protein and MBP-VvLAR1 served as negative and positive controls, respectively. HPLC-UV analysis showed a peak with the same retention time as (+)-catechin in both the VvLAR1 and VvLAR2 reactions, with no product in the reaction with MBP tag alone (Fig. 7), confirming that both VvLAR1 and VvLAR2 from 'Cabernet Sauvignon' are able to convert leucocyanidin to (+)-catechin.
Figure 6.
Alignment of deduced amino acid sequences of LAR2 genes from three grapevine varieties—'Regent' (Re, DQ129686), 'Cabernet Sauvignon' (CS, MK726358) and 'Shiraz' (Sh, AJ865334). Sequence alignment was performed by Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and visualized by BoxShade (https://embnet.vital-it.ch/software/BOX_form.html). Identical amino acids are indicated by white letters on a black background and similar amino acids by white letters on a light gray background. The black asterisks indicate the sites where CS is the same as Sh but different from Re, whereas the black arrows indicate the sites where Sh is different from both Re and CS. Putative residues critical for activity are indicated by unfilled arrows.
Figure 7.
Assay of VvLARs from 'Cabernet Sauvignon' with leucocyanidin as substrate. The profiles of in vitro enzymatic reactions were analyzed using HPLC at 280 nm. The combinations of enzymes and substrate are shown along with the corresponding chromatograms. The reaction with MBP tag instead of VvLARs was run as negative control. The arrows indicate the peaks for leucocyanidin (Leu) and (+)-catechin standards (sd).
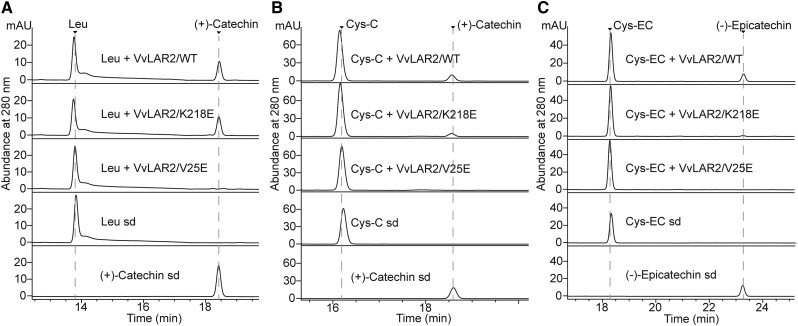
The Glu residues at positions 25 and 218 in 'Shiraz' VvLAR2 (VvLAR2-Sh) are substituted by Val and Lys, respectively, in the VvLAR2s of both 'Cabernet Sauvignon' (VvLAR2-CS) and 'Regent' (VvLAR2-Re; Fig. 6). To assess the reason for apparent lack of catalytic activity of VvLAR2-Sh, site-directed mutagenesis was performed on pMal-c5x harboring VvLAR2-CS to generate VvLAR2-CS/K218E and VvLAR2-CS/V25E, which were then expressed in E. coli. When incubated with leucocyanidin and NADPH, VvLAR2-CS and VvLAR2-CS/K218E produced (+)-catechin, whereas VvLAR2-CS/V25E showed no product formation (Fig. 8A), confirming that the Glu at position 25 accounts for the lack of activity of VvLAR2-Sh with leucocyanidin. Furthermore, both VvLAR2-CS and VvLAR2-CS/K218E converted Cys-C or Cys-EC to (+)-catechin or (−)-epicatechin in the presence of NADPH, although VvLAR2-CS/K218E exhibited lower activity (Fig. 8, B and C). VvLAR2-CS/V25E still yielded no products with either Cys-C or Cys-EC (Fig. 8, B and C), indicating that position 25 is crucial for all the activities of VvLAR2. Multiple sequence alignment of LARs demonstrated to show activity with leucocyanidin suggested that the amino acid at position 25 in active proteins does not have to be highly conserved but may need to be hydrophobic (Supplemental Fig. S3). The crystal structure of VvLAR1 shows that the N-terminal region forms a Rossmann fold to bind NADP(H) (Maugé et al., 2010). To investigate whether the V25E mutation affects NADPH binding, the amino acid sequences of VvLAR2-CS/V25E, together with VvLAR2-Sh, VvLAR2-CS/K218E, VvLAR2-CS, and VvLAR1, were submitted to the Cofactory V1.0 server (Geertz-Hansen et al., 2014) to identify the Rossmann folds in the proteins and predict their specificity for the cofactors. The output is shown in Table 2. VvLAR1 served as a positive control showing the predicted binding of NADP/NAD as cofactor. VvLAR2-CS/K218E and VvLAR2-CS were both able to bind NADP/NAD at the N terminus, whereas the software returned no cofactor prediction for VvLAR2-CS/V25E and VvLAR2-Sh. On the basis of the combined experimental and bioinformatic analysis results, we conclude that Glu 25 abolishes the cofactor binding of 'Shiraz' VvLAR2 to make it the nonfunctional enzyme found in nature.
Figure 8.
Assay of mutagenized proteins of VvLAR2 from 'Cabernet Sauvignon'. HPLC profiles of products from incubation of mutagenized VvLAR2 proteins with leucocyanidin (Leu), Cys-C, and Cys-EC are shown in A, B, and C, respectively. The combinations of enzymes and substrate are shown along with the corresponding chromatograms. The reactions with MBP-VvLAR2/wild type(WT) served as the positive controls. The arrows indicate the peaks for standards (sd).
Table 2. Identification of Rossmann folds and prediction of cofactor specificity for wild-type and mutant VvLAR2 proteins.
The amino acids used in the analysis are wild-type LAR2 from 'Cabernet Sauvignon' (VvLAR2-CS), mutagenized proteins of VvLAR2-CS (VvLAR2-CS/K218E and VvLAR2-CS/V25E), LAR2 from 'Shiraz' (VvLAR2-Sh), and LAR1 from 'Cabernet Sauvignon' (VvLAR1-CS), which served as a positive control. Each identified Rossmann fold sequence domain is associated with three neural network scores calculated by the Cofactor V.1.0 server (Geertz-Hansen et al., 2014). A score above 0.5 indicates that the domain is predicted to be specific for the particular cofactor. The prediction scores are followed by a summary of the predicted cofactor specificity. Multiple specificities are separated with a slash. The approximate Rossmann fold sequence boundaries are provided next to the summary.
| Sequence | Domain | FAD | NAD | NADP | Cofactor(s) | From | To |
|---|---|---|---|---|---|---|---|
| VvLAR2-CS | 1 | 0.149 | 0.362 | 0.757 | NADP | 19 | 64 |
| 2 | 0.000 | 0.616 | 0.121 | NAD | 62 | 113 | |
| VvLAR2-CS/K218E | 1 | 0.149 | 0.362 | 0.757 | NADP | 19 | 64 |
| 2 | 0.000 | 0.616 | 0.121 | NAD | 62 | 113 | |
| VvLAR2-CS/V25E | 1 | 0.027 | 0.239 | 0.498 | – | 21 | 113 |
| VvLAR2-Sh | 1 | 0.027 | 0.239 | 0.498 | – | 21 | 113 |
| VvLAR1-CS | 1 | 0.227 | 0.538 | 0.541 | NAD/NADP | 10 | 51 |
| 2 | 0.000 | 0.129 | 0.037 | – | 50 | 80 |
VvLAR1 and VvLAR2 Complement MtLAR Activities in M. truncatula lar:ldox Double Mutant
Developing seeds of M. truncatula use both (+)-catechin and (−)-epicatechin to generate epicatechin-based PA building blocks (Jun et al., 2018). Loss of function of MtLAR results in a decrease of soluble PAs and increase of insoluble PAs (Liu et al., 2016). MtLDOX functions downstream of MtLAR to convert (+)-catechin to cyanidin and MtANS functions in parallel with MtLDOX to generate cyanidin from leucocyanidin (Fig. 1). The lar:ldox double mutant shows the lar mutant phenotype (Jun et al., 2018), but the pathway removing (+)-catechin has also been removed, making this mutant background an ideal system to test VvLAR products in planta.
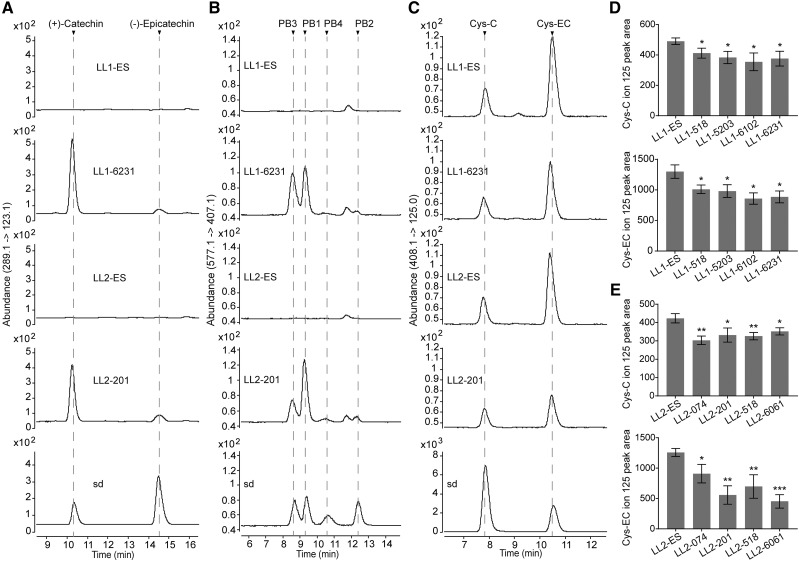
The VvLAR1 and VvLAR2 ORFs, driven by the cauliflower mosaic virus 35S promoter, were transformed into lar:ldox M. truncatula and four basta-resistant lines of each were selected for further analysis based on their high transgene expression (Fig. 9, A and B). Escape lines (the lines were able to grow on the selection medium but contained no binary vector) were used as controls. Transgenic and escape lines were planted side by side and pods were sampled at 4 d after pollination (DAP). Soluble PAs were extracted from the samples in 70% (v/v) acetone with 0.1% (v/v) acetic acid and were quantified by the dimethylaminocinnamaldehyde (DMACA) method. Compared with the controls, 35S:VvLAR1 and 35S:VvLAR2 lines showed an approximately 2-fold increase in soluble PA levels (Fig. 9, C and D). To quantify insoluble PAs, the residues left after extraction of soluble PAs were dried and heated in butanol-HCl. The insoluble PAs in both 35S:VvLAR1 and 35S:VvLAR2 lines showed at minimum a 20% decrease compared to the controls (Fig. 9, E and F), suggesting that both VvLARs complement the function of MtLAR in the control of PA size (with larger PAs being less soluble). To further investigate whether the increase in soluble PAs was due to an increase in low-molecular-weight PAs, the same soluble extracts used above were subjected to HPLC-QqQ analysis to measure levels of (+)-catechin/(−)-epicatechin, procyanidin B-type dimers, and Cys-C/Cys-EC. Neither catechin/epicatechin nor PA dimers were detected in pods of control lar:ldox plants (Fig. 10, A and B). In contrast, mutant plants transformed with VvLARs contained PA monomers and dimers with the same pattern as seen in the ldox single mutant (Jun et al., 2018; Fig. 10, A and B; Supplemental Figs. S4 and S5). The PA monomers consisted mainly of (+)-catechin and a small amount of (−)-epicatechin. Procyanidin B1 ((−)-epicatechin-(4β → 8)-(+)-catechin) and procyanidin B3 ((+)-catechin-(4α → 8)-(+)-catechin) were the major PA dimers in the transgenic pods, with lower levels of procyanidin B2 ((−)-epicatechin-(4β → 8)-(−)-epicatechin) and procyanidin B4 ((+)-catechin-(4α → 8)-(−)-epicatechin). Our results confirmed the presence of Cys-C along with Cys-EC in young pods of the M. truncatula lar:ldox mutant (Fig. 10C), with levels of both Cys conjugates decreasing on expression of VvLARs (Fig. 10, D and E). Together, our data indicate that both VvLARs can complement the function of MtLAR for conversion of the potential extension unit-generating Cys conjugates to catechin and epicatechin starter units, resulting in reduced PA extension.
Figure 9.
Analysis of transgene transcript levels and PA contents in 4 DAP pods from the M. truncatula lar:ldox mutant expressing VvLAR1 (LL1) or VvLAR2 (LL2). Nontransgenic escape lines (LL1-ES and LL2-ES) from transformations served as controls. Transcript levels of VvLAR1 and VvLAR2 from different transgenic lines are shown in A and B, respectively. Transcripts were determined by qPCR normalized relative to the expression of MtActin and MtTubulin. Soluble PA levels in the young pods from VvLAR1 transgenic lines (C) and VvLAR2 transgenic lines (D) were measured by using dimethylaminocinnamaldehyde (DMACA) reagent and expressed as epicatechin equivalents. Insoluble PA levels in the young pods from VvLAR1 transgenic lines (E) and VvLAR2 transgenic lines (F) were determined by the butanol-HCl method and expressed as procyanidin B1 equivalents. In the bar plots, data are shown as the mean ± sd (for n = 3 biologically independent samples); *P < 0.05, **P < 0.01, and ***P < 0.001 versus LL1-ES or LL2-ES, two-tailed Student’s t test. FW, fresh weight.
Figure 10.
HPLC-QqQ characterization of the soluble PA fraction in 4 DAP pods of the M. truncatula lar:ldox mutant expressing VvLAR1 (LL1) or VvLAR2 (LL2). Chromatograms for lar:ldox 35S:VvLAR1 line LL1-6231 and 35S:VvLAR2 line LL2-201 are presented here, and their corresponding control (escape) lines are denoted as LL1-ES and LL2-ES, respectively. Chromatograms for the other transgenic lines are presented in Supplemental Figures S4 and S5. A, MRM transition of m/z (289.1 → 123.1) showing that (+)-catechin and (−)-epicatechin accumulate in the young pods of 35S:VvLAR lines but are undetectable in controls. B, MRM transition of m/z (577.1 → 407.1) showing that procyanidin B1, B2, B3, and B4 (PB1 to PB4) accumulate in the young pods of 35S:VvLAR lines but are undetectable in controls. C, MRM transition of m/z (408.1 → 125.0) showing that Cys-C and Cys-EC in the young pods of 35S:VvLAR lines are less abundant than in their corresponding control plants. Cys-C and Cys-EC contents for young pods from each line were measured using the ion peak area at m/z 125.0. D, Cys-C and Cys-EC content in the young pods from control and 35S:VvLAR1 lines. E, Cys-C and Cys-EC content in the young pods from control and 35S:VvLAR2 lines. Data are shown as the mean ± sd (for n = 3 biologically independent samples); *P < 0.05, **P < 0.01, and ***P < 0.001 versus LL1-ES or LL2-ES, two-tailed Student’s t test.
DISCUSSION
Is Cys-C a (+)-Catechin-Type PA Extension Unit in Plants?
Cys-EC can act as a (−)-epicatechin-type extension unit for (4β → 8) B-type procyanidin polymerization under neutral pH (Liu et al., 2016) and has been detected in grapevine, Arabidopsis, and M. truncatula. Its trans isomer, Cys-C, is reported here to be also present in PA-accumulating tissues. Like Cys-EC, Cys-C can theoretically convert to a flavan-3-ol carbocation at around neutral pH and attack the C8 position to extend the growing PA chain. This model is supported by the in vitro synthesis of epicatechin-catechin dimer (procyanidin B4) and epicatechin-catechin-catechin trimer, by conjugation of Cys-C with (−)-epicatechin and procyanidin B4 under neutral pH (Jun et al., 2018). Cys-C was detected in grapevine and M. truncatula, but not in Arabidopsis. Correspondingly, grapevine and M. truncatula use both catechin (to a lesser extent in M. truncatula) and epicatechin as PA subunits (Huang et al., 2012; Jun et al., 2018), whereas Arabidopsis exclusively uses epicatechin as a PA building block (Lepiniec et al., 2006), supporting a role for Cys-C as a (+)-catechin-type extension unit in plants.
The source of Cys-C remains unclear. However, it is noteworthy that Cys-C is present in pods of the M. truncatula lar:ldox mutant, whereas (+)-catechin monomer is undetectable. This means that Cys-C biosynthesis does not rely on LAR and (+)-catechin. In the flavonoid pathway as currently understood, leucocyanidin is the only obvious (+)-catechin backbone donor for Cys-C other than (+)-catechin itself. For more than three decades, leucocyanidin has been considered as an intermediate to provide (+)-catechin-type extension units, because it can rapidly autopolymerize with (+)-catechin in vitro to produce mainly catechin-catechin dimer (procyanidin B3; Delcour et al., 1983). Unlike grapevine and M. truncatula, there is no LAR gene in the Arabidopsis genome (Lepiniec et al., 2006). Although leucocyanidin is very unstable and difficult to trap by analytical instruments, based on previous studies it can be inferred that leucocyanidin does not “leak out” of a coupled DFR/ANS in Arabidopsis (Liu et al., 2013; Ferraro et al., 2014; Wang et al., 2018). If leucocyanidin is available in Arabidopsis, the seeds should contain PAs with (+)-catechin as extension units, which is not observed. Moreover, overexpression of pea (Pisum sativum) LAR or three tea (Camellia sinensis) LARs in wild-type Arabidopsis does not result in synthesis of (+)-catechin, although LAR activity to generate (+)-catechin was detected in vitro (Ferraro et al., 2014; Wang et al., 2018), and expression of cacao LAR in the Arabidopsis ans mutant results in significant accumulation of (+)-catechin (Liu et al., 2013), indicating that ANS has a high affinity for leucocyanidin or associates with DFR very tightly. Based on the above, the absence of Cys-C in Arabidopsis might be due to the lack of availability of leucocyanidin in vivo due to channeling between DFR and ANS. Conversion of leucocyanidin to Cys-C, either chemically or enzymatically, could be a way to stabilize excess leucocyanidin in plants where this coupling was less tight and to provide a store of (+)-catechin-type extension units for further PA extension.
Multiple Functions for VvLARs
Grapevine possesses two LARs—VvLAR1 and VvLAR2, which were believed to convert leucocyanidin to catechin, although one study failed to demonstrate any activity for VvLAR2 in vitro (Bogs et al., 2005). MtLAR, a homolog of VvLAR1 from M. truncatula, was recently shown to possess the additional ability of converting Cys-EC to (−)-epicatechin (Liu et al., 2016). Here, we show that VvLAR1 and VvLAR2 from 'Cabernet Sauvignon' can use both leucocyanidin and Cys-EC as substrates, in addition to generating (+)-catechin from Cys-C in vitro. Previous studies have provided genetic evidence to show that MtLAR can control PA extension, and hence mDP, by regulating the ratio of starter units to extension units (Liu et al., 2016). Overexpression of either VvLAR1 or VvLAR2 in the M. truncatula lar:ldox mutant led to increased soluble PA levels but decreased insoluble PA levels in young pods, suggesting that both VvLARs can also function in the control of PA extension in planta. As the ratio between soluble to insoluble PAs is positively correlated with the mDP of PAs (Pang et al., 2007; Liu et al., 2016; Jun et al., 2018), our results are consistent with the quantitative trait locus study showing that the VvLAR1 locus is associated with the mDP of PAs in grapevine (Huang et al., 2012).
Complementation of the M. truncatula lar:ldox mutant with VvLAR1 or VvLAR2 resulted in the same accumulation patterns of PA monomers and dimers as seen in the ldox single mutant (Jun et al., 2018), further confirming that VvLARs operate in the same position as MtLAR in the PA pathway. The lower levels of Cys-C and Cys-EC in the complemented mutant suggest the following scenarios. First, both Cys-C and Cys-EC can be directly used by VvLARs to produce catechin and epicatechin as PA starter units (Fig. 11). Second, the conversion of leucocyanidin to catechin by VvLARs may limit the upstream substrates for the synthesis of Cys-C and Cys-EC, which are likely derived respectively from leucocyanidin (as discussed above) and epicatechin (Liu et al., 2016; Fig. 11). Although kinetic parameters for leucocyanidin as substrate were not measured here due to its instability, we nevertheless conclude that VvLARs tend to mainly consume (+)-catechin-type extension units to produce (+)-catechin starter units, because Cys-EC is a poorer substrate for VvLARs than Cys-C due to steric hindrance in the enzyme active site. Such catalytic properties of VvLARs might help with maintaining 2,3-cis extension unit for PA polymerization—epi(gallo)catechins are the most common extension unit in grapevine (Bogs et al., 2005; Gagné et al., 2009; Huang et al., 2012).
Figure 11.
A revised model for the function of VvLARs. The figure demonstrates that both VvLAR1 and VvLAR2 channel three PA extension units (denoted as “E”) to starter units (denoted as “S”) directly and indirectly. Dashed lines with question marks indicate proposed pathways. On the one hand, VvLARs directly convert leucocyanidin and Cys-C to (+)-catechin and Cys-EC to (−)-epicatechin. On the other hand, by consuming leucocyanidin, VvLARs may not only prevent Cys-C synthesis, but also compete with the synthesis of cyanidin to regulate the flow to Cys-EC.
New Insights into PA Polymerization in Wine Grapevines
PAs in red wine greatly affect the mouth feel, and their astringency is positively correlated with their content, DP, and extent of galloylation (Peleg et al., 1999; Maury et al., 2001; Vidal et al., 2003). Although red grapevine berry is rich in PAs, only PA monomers and small oligomers (mDP < 8) are extractable during processing of different varieties (Mattivi et al., 2009). Thus, precise control of the soluble PA composition in grapevine is important for wine grapevine breeding, for which both VvLAR1 and VvLAR2 can now be useful markers for PA mDP.
Although genome editing offers a shortcut in obtaining crops with desired qualities, both vector-dependent and DNA-free CRISPR-associated RNA-guided endonuclease Cas9 systems for grapevine are still under development and most grapevine cultivars are recalcitrant for plant regeneration, which may be due to secretion of phenolics and other antioxidants (Osakabe et al., 2018). Thus, nowadays, breeding of new red-berried grapevine varieties with desired PA composition will still rely on traditional crossing methods. We found that 'Shiraz' VvLAR2 contains a key amino acid change of Val 25 to Glu 25, which causes inability to bind NADPH and thus abolishes all LAR activities using leucocyanidin, Cys-C, and Cys-EC. In both grapevine berry skins and seeds, VvLAR2 is highly expressed during the second half of the PA biosynthesis phase, when the VvLAR1 transcript level is very low (Bogs et al., 2005). It has been shown that mDP values of PA extracts from 'Shiraz' berry skins and wine are higher than those of 'Cabernet Sauvignon', although the mDP of PAs from the seeds of these two varieties is the same (Cosme et al., 2009; Mattivi et al., 2009; Hanlin et al., 2011). These findings suggest that the inactive VvLAR2 is likely associated with the formation of the higher DP PAs found in wine from 'Shiraz' grapevine berries. Thus, 'Shiraz' appears to be a suitable parent background for breeding to regulate LAR activities in grapevines, while preserving excellent quality at the same time. Transcriptional regulators of VvLAR1 and VvANR have been well studied, whereas which specific transcription factors regulate VvLAR2 remains unclear (Bogs et al., 2007; Terrier et al., 2009; Huang et al., 2014; Koyama et al., 2014). To better fine-tune PA biosynthesis in grapevines, regulation of VvLAR2 expression should now be addressed.
MATERIALS AND METHODS
Chemicals
(+)-Catechin and (−)-epicatechin were purchased from Sigma Aldrich. Procyanidin B1, B2, and B3 were purchased from Adooq BioScience. Procyanidin B4 was synthesized in vitro as described by Jun et al. (2018). 4β-(S-cysteinyl)-epicatechin and -catechin were chemically synthesized using procyanidin B2 and B3 with Cys under hot acidic conditions and purified as described by Liu et al. (2016). 3,4-cis-leucocyanidin was synthesized from in vitro (+)-dihydroquercetin (Sigma Aldrich) and purified using HPLC with the method described in Kristiansen (1984).
HPLC and HPLC-QqQ Analysis
HPLC analysis was carried out on an Agilent HP1100 system (Agilent) equipped with a HypersilGold 250 mm × 4.6 mm, 5 μm, C18 column (Thermo Fisher) and diode array detector, using the following gradient: solvent A (1% [w/v] phosphoric acid) and B (acetonitrile) at 1 mL min−1 flow rate, 0 to 5 min, 5% B; 5 to 10 min, 5% to 10% B; 10 to 25 min, 10% to 17% B; 25 to 35 min, 17% to 100% B; 35 to 45 min, 100% to 5% B. Data were collected at 280 nm. Quantification of (+)-catechin, (−)-epicatechin, Cys-C, and Cys-EC were based on the (+)-catechin standard curve.
HPLC-QqQ analysis was carried out on a 1290 Infinity II system (Agilent) equipped with a 6460 triple quadrupole mass spectrometer (Agilent). An XTerra 5 μm, 2.1 × 250 mm, C18 column (Waters) was used for separation. The elution program was as follows: solvent A (0.1% [v/v] formic acid in water) and solvent B (0.1% [v/v] formic acid in methanol); flow rate, 0.4 mL min−1; gradient, 0 to 1 min, 5% B; 1 to 2 min, 5% to 10% B; 2 to 17 min, 10% to 31% B; 17 to 18 min; 31% to 95% B; 19 to 20 min, 95% to 5% B; 20 to 23 min, 5% B. The spectrometer was set to MRM in negative mode. The detection parameters were optimized using flow injection analysis with Cys-EC, (+)-catechin and procyanidin B2 as standards. Ion source parameters were as follows: gas/sheath gas heater, 300°C/350°C; gas flow/sheath gas flow, 5/12 L/min; nebulizer, 30 psi; capillary, 12 V. MRM scan parameters were as follows: for Cys-C and Cys-EC, the transition of m/z (408.1→ 125.0) was monitored with Fragmentor (Frag) 94 V and collision energy (CE) 13 V; for (+)-catechin and (−)-epicatechin, the transition of m/z (289.1 → 123.1) was monitored with Frag 126 V and CE 33 V; for procyanidin B1, B2, B3, and B4, the transition of m/z (577.1 → 407.1) was monitored with Frag 136 V and CE 25 V; dwell 150 ms and cell acceleration 4 V were set for all the compounds. In the pilot study of screening the compounds with the same MS pattern as Cys-EC, analysis was carried out on an Agilent 1200 system (Agilent) equipped with a 6410 triple quadrupole mass spectrometer (Agilent) by negative MRM mode. HPLC elution program and mass spectrometer parameters can be found in Li et al. (2017), except Frag 125 V, CE 15 V; transitions of m/z 408.1 to m/z 125.0, m/z 161.0, and m/z 287.0 were used instead.
Grapevine Materials and the Cloning of VvLAR1 and VvLAR2
The grapevine (Vitis vinifera) berries at veraison stage were sampled in 2016 on grapevine 'Cabernet Sauvignon' clone 169 in the experimental greenhouse of Shangzhuang Extension Center for Agricultural Technology in Haidian District, Beijing, China. The whole berries were finely grounded in liquid nitrogen for RNA extraction and soluble PA analysis. Total RNA was extracted using a Plant Total RNA Extraction Kit (Sigma-Aldrich). RNA content was measured with a NanoDrop 2000 (Thermo Fisher). One microgram of total RNA was used for reverse transcription with an M-MLV Reverse Transcriptase kit (Promega). Primers for cloning VvLAR1 and VvLAR2 were designed based on AJ865336.1 from GenBank and VIT_217s0000g04150 from Grape Genome Database (http://genomes.cribi.unipd.it/grape/) as shown in Supplemental Table S1. DNA fragments covering VvLAR1 or VvLAR2 ORFs were amplified using I-5 High-Fidelity DNA Polymerase (MCLAB) and subcloned into pMD19-T vector (Takara), resulting in pMD19-T-VvLAR1 and pMD19-T-VvLAR2, which were used for sequence confirmation.
In Vitro Expression of Recombinant VvLAR Proteins
The ORFs of the VvLAR1 and VvLAR2 genes were PCR amplified from pMD19-T-VvLAR1 and pMD19-T-VvLAR2, respectively, using Phusion High-Fidelity DNA Polymerase (New England Biolabs), and the primers listed in Supplemental Table S1. They were then subcloned into the NdeI and BamHI sites of the pMal-c5x expression vector (New England Biolabs), resulting in pMal-c5x-VvLAR1 and pMal-c5x-VvLAR2. VvLAR2 mutagenesis expression vectors pMal-c5x-VvLAR2/K218E and pMal-c5x-VvLAR2/V25E were constructed using pMal-c5x-VvLAR2 as a template with QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) exactly following the manufacturer’s protocol. The primers used in mutagenesis experiments are shown in Supplemental Table S1.
After confirmation by sequencing, the expression vectors were transformed into Escherichia coli strain Rosetta (DE3) pLysS competent cells (Novagen). Transformed bacteria were grown in LB liquid medium supplemented with 100 μg/mL carbenicillin and 0.2% (w/v) Glc to absorbance A600 of 0.7, and isopropyl β-d-1-thiogalactopyranoside was added at 0.3 mm to induce protein expression at 16°C. Bacteria were harvested after 16 h induction. MBP-tagged VvLAR proteins were purified with amylose resin (New England Biolabs) following the manufacturer’s protocol with slight modifications. In brief, bacteria were lysed by sonication on ice in column buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, 1 mm EDTA, v/v 1:5 50% [v/v] glycerol, and 10 mm β-mercaptoenthanol). The bacterial lysates were centrifuged at 12,000g for 15 min at 4°C. The amylose resin was incubated with the supernatant on a roller at 4°C for 2 h and then was washed with wash buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, and v/v 1:5 50% [v/v] glycerol). Finally, proteins were eluted by elution buffer (20 mm Tris-HCl, pH 7.4, 200 mm NaCl, v/v 1:5 50% [v/v] glycerol, and 10 mm maltose). Purified proteins were concentrated with Amicon Ultra-4 Centrifugal Filter-10K (Millipore), and aliquots were stored at −80°C. Protein concentration was first quantified using the Bradford method and corrected according to the proportion of band grayscale measured by Image J software (Schneider et al., 2012) on Bolt Bis-Tris Plus gels (Invitrogen) after electrophoresis.
Assay of LAR Activities
Enzyme reactions (100 μL) included 50 mm Tris-HCl buffer (pH 7.4), 250 μm NADPH, 40 μm Cys-C or Cys-EC, or 5 μL (∼50 μm) 2,3-cis-leucocyanidin and 10 μg recombinant enzymes. The reactions were carried out at 30°C for 1 h and terminated by addition of 50 μL methanol followed by vigorous vortexing. The resulting mixture was centrifuged at 12,000g at 4°C for 5 min and analyzed by HPLC as described above.
Kinetic constants for VvLARs with Cys-C or Cys-EC as substrates were determined as above in a 100-μL setup containing 4 μg of recombinant proteins and indicated amounts Cys-C or Cys-EC with 10 min incubation, and the product concentration was determined using (+)-catechin as a standard. Vmax and Km values were calculated by fitting to the Michaelis-Menten equation with GraphPad Prism 7 (GraphPad Software).
Molecular Docking
The crystal structure of VvLAR1 (PDB: 3I52) was acquired from the Protein Data Bank, while the 3D model of VvLAR2 was generated by SWISS-MODEL (Waterhouse et al., 2018) with VvLAR1 structure as a template. The water molecules in the crystal structures were deleted, except for the ones that had hydrogen bonds with the residues in the VvLAR1 active sites. The cofactor NADPH and water molecules for VvLAR2 were obtained by superposition of the VvLAR2 model onto the VvLAR1 structure, based on the conserved 3D conformation. Cys-C and Cys-EC structures were drawn in ChemAxon Marvin (ChemAxon). The charges of proteins and substrates (including NADPH) were assigned using Amberff14SB and Gasteiger force-field tools, respectively, in UCSF-Chimera (Pettersen et al., 2004). Autodock Vina (Trott and Olson, 2010) was used for the molecular docking of Cys-C and Cys-EC with VvLARs. The search box was the center coordinates (x, y, z) = (0.0, 2.3, 11.5) with the search volume 15.0 × 15.0 × 15.0 Å, which included the active sites. The 3D structures of docking were visualized with PyMol (Schrödinger), and the interactions between substrates and enzyme pockets were analyzed and visualized using LigPlot+ (Laskowski and Swindells, 2011).
Transformation of Medicago truncatula with VvLAR1 and VvLAR2
The construction of the M. truncatula R108 lar:ldox double mutant was described previously in Jun et al. (2018). The VvLAR1 or VvLAR2 ORFs were amplified with the primer pairs listed in Supplemental Table S1. The amplicons were cloned into entry vector pENTR/D-TOPO and then transferred into the Gateway plant binary vector pB7WG2D. The resulting basta-resistant vectors pB7WG2D-VvLAR1 or pB7WG2D-VvLAR2, with VvLAR1 or VvLAR2 driven by the 35S promoter, were transformed into Agrobacterium tumefaciens strain AGL1 by electroporation. Transgenic M. truncatula plants were generated by Agrobacterium-mediated transformation of leaf explants as described in the Medicago truncatula Handbook (https://www.noble.org/globalassets/docs/medicago-handbook/agrobacterium-tumefaciens.pdf). Only one spot of callus from each explant was transferred on SH9 medium for embryogenesis to ensure independent transformations. Total DNA from leaves of transgenic seedling candidates was isolated using the CTAB method (Clarke, 2009), and genotyping was performed by PCR using with primers listed in Supplemental Table S1. The plants were grown side by side in a growth chamber set at 16 h/8 h day/night cycle, 25°C.
RNA Isolation and Quantitative Reverse Transcription PCR Analysis of Transgenic Plants
Total RNA was extracted from frozen M. truncatula 4 DAP young pods using Trizol (Invitrogen) according to the manufacturer’s instructions. RNA was treated with TURBO DNA-free kit (Thermo Fisher) to remove DNA contamination and then quantified using a NanoDrop 2000 (Thermo Fisher). One microgram of total RNA was used for reverse transcription with SuperScript III Reverse Transcriptase (Thermo Fisher). The qPCR was set up with Applied Biosystems Power SYBR Green mix (Thermo Fisher) and performed using an Applied Biosystems QuantStudio 6 Flex Real-Time PCR system (Thermo Fisher). The Real-time PCR Data Markup Language format raw data from the qPCR equipment was imported into LinRegPCR software (Ruijter et al., 2009) to calculate PCR amplification efficiency and transcript level for each gene. MtActin and MtTubulin were used together as references (Pang et al., 2007; Liu et al., 2016). Primers for VvLAR1 and VvLAR2 were designed to distinguish between the two LAR transcripts using the method described elsewhere (Thornton and Basu, 2011) and are listed in the Supplemental Table S1.
Determination of PA Content
About 100 mg frozen samples were ground into powder in liquid nitrogen. The powder was extracted with 1 mL of proanthocyanidin extraction solvent (PES; 70% [v/v] acetone with 0.5% [v/v] acetic acid) by sonicating in ice water for 30 min. The resulting slurry was centrifuged at 12,000g for 5 min and supernatants were collected. The pellets were re-extracted for another two times and all the supernatants were pooled for further extraction of soluble PAs, and pellets were stored at −20°C for quantification of insoluble PAs. Equal volumes of chloroform were added to pooled supernatants, and the mixtures were vortexed vigorously and centrifuged at 5000g for 5 min, and the supernatants were further cleaned twice with chloroform and twice with hexane. The resulting aqueous phase (soluble PAs) was lyophilized and redissolved in 60 μL 50% (v/v) methanol. Soluble PAs were quantified by DMACA method (Pang et al., 2007), with slight modifications as follows: 2 μL soluble PA fractions were mixed with 100 μL of 0.2% (w/v) DMACA in methanol/HCl (1:1, v/v) on a 96-well plate, and the A640 was read after 4 min using a Synergy 2 Multi-Detection Microplate Reader (BioTek). (−)-Epicatechin was used as a standard and processed in parallel with experimental samples. The remaining samples were subjected to HPLC-QqQ analysis.
Insoluble PA content was analyzed by the butanol/HCl method (Pang et al., 2007), with slight modifications. The pellets after PES extraction were lyophilized and 1 mL of butanol/HCl (95:5, v/v) was added. The mixtures were sonicated in ice water for 30 min for resuspension followed by heating at 95°C for 1 h. The mixtures were then allowed to cool down to room temperature and were centrifuged at 12,000g for 5 min. One hundred microliters supernatant was added on a 96-well plate, and the A550 was measured using a Synergy 2 Multi-Detection Microplate Reader (BioTek). Procyanidin B1 was used as standard and processed in parallel with experimental samples.
Statistical Analysis
Statistical analysis was carried out by GraphPad Prism 7 (GraphPad Software) with two-tailed Student’s t test. All the measurements were visualized as the average of three biological replicates with sd as error bars on the bar plots.
Accession Numbers
The sequences of the two LARs from grapevine 'Cabernet Sauvignon' can be found in GenBank under the accession numbers VvLAR1, MK726357; VvLAR2, MK726358.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Screening of compounds with the same MS pattern as Cys-EC in grape and Arabidopsis.
Supplemental Figure S2. Molecular docking analysis of Cys-C and Cys-EC in the modeled VvLAR2 active site.
Supplemental Figure S3. Alignment of deduced amino acid sequences of functionally characterized LARs.
Supplemental Figure S4. HPLC-QqQ chromatograms for PA monomers, dimers, Cys-C, and Cys-EC in 35S:VvLAR1 M. truncatula lar:ldox plants not shown in the body of the paper.
Supplemental Figure S5. HPLC-QqQ chromatograms for PA monomers, dimers, Cys-C, and Cys-EC in 35S:VvLAR2 M. truncatula lar:ldox plants not shown in the body of the paper.
Supplemental Table S1. Sequences of primers used in this work.
Acknowledgments
The authors thank Xirong Xiao for assistance with transgenic plant construction, Dr. Christophe Cocuron for advice on optimizing mass spectrometry parameters, and Dr. Antonella Longo for suggestions about molecular docking.
Footnotes
This work was supported by the University of North Texas and the China Scholarship Council (grant 201706350125 to K.Y.).
Articles can be viewed without a subscription.
References
- Aerts RJ, Barry TN, McNabb WC (1999) Polyphenols and agriculture: Beneficial effects of proanthocyanidins in forages. Agric Ecosyst Environ 75: 1–12 [Google Scholar]
- Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray SD, Kuszynski CA, Joshi SS, Pruess HG (2000) Free radicals and grape seed proanthocyanidin extract: Importance in human health and disease prevention. Toxicology 148: 187–197 [DOI] [PubMed] [Google Scholar]
- Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139: 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD. (2009) Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb Protoc 2009: t5177. [DOI] [PubMed] [Google Scholar]
- Cos P, De Bruyne T, Hermans N, Apers S, Berghe DV, Vlietinck AJ (2004) Proanthocyanidins in health care: Current and new trends. Curr Med Chem 11: 1345–1359 [DOI] [PubMed] [Google Scholar]
- Cosme F, Ricardo-Da-Silva JM, Laureano O (2009) Tannin profiles of Vitis vinifera L. cv. red grapes growing in Lisbon and from their monovarietal wines. Food Chem 112: 197–204 [Google Scholar]
- Delcour JA, Ferreira D, Roux DG (1983) Synthesis of condensed tannins. Part 9. The condensation sequence of leucocyanidin with (+)-catechin and with the resultant procyanidins. J Chem Soc, Perkin Trans 1: 1711–1717 [Google Scholar]
- Dixon RA, Xie D-Y, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165: 9–28 [DOI] [PubMed] [Google Scholar]
- Ferraro K, Jin AL, Nguyen T-D, Reinecke DM, Ozga JA, Ro D-K (2014) Characterization of proanthocyanidin metabolism in pea (Pisum sativum) seeds. BMC Plant Biol 14: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan C, Motta L, Santos D (2011) Tannins: What do they represent in plant life? In Petridis GK, eds, Tannins: Types, Foods Containing, and Nutrition. Nova Science Publishers, Hauppauge, NY, pp 251–263 [Google Scholar]
- Gagné S, Lacampagne S, Claisse O, Gény L (2009) Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol Biochem 47: 282–290 [DOI] [PubMed] [Google Scholar]
- Geertz-Hansen HM, Blom N, Feist AM, Brunak S, Petersen TN (2014) Cofactory: Sequence-based prediction of cofactor specificity of Rossmann folds. Proteins 82: 1819–1828 [DOI] [PubMed] [Google Scholar]
- Hanlin RL, Kelm MA, Wilkinson KL, Downey MO (2011) Detailed characterization of proanthocyanidins in skin, seeds, and wine of Shiraz and Cabernet Sauvignon wine grapes (Vitis vinifera). J Agric Food Chem 59: 13265–13276 [DOI] [PubMed] [Google Scholar]
- Huang Y-F, Doligez A, Fournier-Level A, Le Cunff L, Bertrand Y, Canaguier A, Morel C, Miralles V, Veran F, Souquet J-M, et al. (2012) Dissecting genetic architecture of grape proanthocyanidin composition through quantitative trait locus mapping. BMC Plant Biol 12: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YF, Vialet S, Guiraud JL, Torregrosa L, Bertrand Y, Cheynier V, This P, Terrier N (2014) A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol 201: 795–809 [DOI] [PubMed] [Google Scholar]
- Jun JH, Xiao X, Rao X, Dixon RA (2018) Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat Plants 4: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Koyama K, Numata M, Nakajima I, Goto-Yamamoto N, Matsumura H, Tanaka N (2014) Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J Exp Bot 65: 4433–4449 [DOI] [PubMed] [Google Scholar]
- Kristiansen KN. (1984) Biosynthesis of proanthocyanidins in barley: Genetic control of the conversion of dihydroquercetin to catechin and procyanidins. Carlsberg Res Commun 49: 503–524 [Google Scholar]
- Laskowski RA, Swindells MB (2011) LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51: 2778–2786 [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li S-Y, He F, Zhu B-Q, Wang J, Duan C-Q (2017) Comparison of phenolic and chromatic characteristics of dry red wines made from native Chinese grape species and Vitis vinifera. Int J Food Prop 20: 2134–2146 [Google Scholar]
- Liu C, Wang X, Shulaev V, Dixon RA (2016) A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat Plants 2: 16182. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shi Z, Maximova S, Payne MJ, Guiltinan MJ (2013) Proanthocyanidin synthesis in Theobroma cacao: Genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol 13: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattivi F, Vrhovsek U, Masuero D, Trainotti D (2009) Differences in the amount and structure of extractable skin and seed tannins amongst red grape varieties. Aust J Grape Wine Res 15: 27–35 [Google Scholar]
- Maugé C, Granier T, d’Estaintot BL, Gargouri M, Manigand C, Schmitter JM, Chaudière J, Gallois B (2010) Crystal structure and catalytic mechanism of leucoanthocyanidin reductase from Vitis vinifera. J Mol Biol 397: 1079–1091 [DOI] [PubMed] [Google Scholar]
- Maury C, Sarni-Manchado P, Lefebvre S, Cheynier V, Moutounet M (2001) Influence of fining with different molecular weight gelatins on proanthocyanidin composition and perception of wines. Am J Enol Vitic 52: 140–145 [Google Scholar]
- Middleton E Jr., Kandaswami C, Theoharides TC (2000) The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol Rev 52: 673–751 [PubMed] [Google Scholar]
- Osakabe Y, Liang Z, Ren C, Nishitani C, Osakabe K, Wada M, Komori S, Malnoy M, Velasco R, Poli M, et al. (2018) CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat Protoc 13: 2844–2863 [DOI] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Abeysinghe ISB, He J, He X, Huhman D, Mewan KM, Sumner LW, Yun J, Dixon RA (2013) Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol 161: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolocci F, Robbins MP, Madeo L, Arcioni S, Martens S, Damiani F (2007) Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis. Structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiol 143: 504–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg H, Gacon K, Schlich P, Noble AC (1999) Bitterness and astringency of flavan-3-ol monomers, dimers and trimers. J Sci Food Agric 79: 1123–1128 [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pfeiffer J, Kühnel C, Brandt J, Duy D, Punyasiri PA, Forkmann G, Fischer TC (2006) Biosynthesis of flavan 3-ols by leucoanthocyanidin 4-reductases and anthocyanidin reductases in leaves of grape (Vitis vinifera L.), apple (Malus x domestica Borkh.) and other crops. Plant Physiol Biochem 44: 323–334 [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalbert A. (1992) Tannins in woods and their contribution to microbial decay prevention. In Plant Polyphenols, Basic Life Sciences, Vol 59 Springer, Boston, pp 935–952 [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner GJ, Kristiansen KN (1993) Synthesis of 3,4-cis-[3H]leucocyanidin and enzymatic reduction to catechin. Anal Biochem 209: 274–277 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR (2003) Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278: 31647–31656 [DOI] [PubMed] [Google Scholar]
- Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149: 1028–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B, Basu C (2011) Real-time PCR (qPCR) primer design using free online software. Biochem Mol Biol Educ 39: 145–154 [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ (2010) AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S, Francis L, Guyot S, Marnet N, Kwiatkowski M, Gawel R, Cheynier V, Waters EJ (2003) The mouth-feel properties of grape and apple proanthocyanidins in a wine-like medium. J Sci Food Agric 83: 564–573 [Google Scholar]
- Wang P, Zhang L, Jiang X, Dai X, Xu L, Li T, Xing D, Li Y, Li M, Gao L, et al. (2018) Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta 247: 139–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018) SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1): W296–W303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D-Y, Sharma SB, Paiva NL, Ferreira D, Dixon RA (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]