Inoculating Medicago truncatula roots with Aphanomyces euteiches spores leads to rapid upregulation of MtTPS10 and release of sesquiterpenes that may have a defense function against this oomycete.
Abstract
In nature, plants interact with numerous beneficial or pathogenic soil-borne microorganisms. Plants have developed various defense strategies to expel pathogenic microbes, some of which function soon after pathogen infection. We used Medicago truncatula and its oomycete pathogen Aphanomyces euteiches to elucidate early responses of the infected root. A. euteiches causes root rot disease in legumes and is a limiting factor in legume production. Transcript profiling of seedlings and adult plant roots inoculated with A. euteiches zoospores for 2 h revealed specific upregulation of a gene encoding a putative sesquiterpene synthase (M. truncatula TERPENE SYNTHASE 10 [MtTPS10]) in both developmental stages. MtTPS10 was specifically expressed in roots upon oomycete infection. Heterologous expression of MtTPS10 in yeast led to production of a blend of sesquiterpenes and sesquiterpene alcohols, with NMR identifying a major peak corresponding to himalachol. Moreover, plants carrying a tobacco (Nicotiana tabacum) retrotransposon Tnt1 insertion in MtTPS10 lacked the emission of sesquiterpenes upon A. euteiches infection, supporting the assumption that the identified gene encodes a multiproduct sesquiterpene synthase. Mttps10 plants and plants with reduced MtTPS10 transcript levels created by expression of an MtTPS10-artificial microRNA in roots were more susceptible to A. euteiches infection than were the corresponding wild-type plants and roots transformed with the empty vector, respectively. Sesquiterpenes produced by expression of MtTPS10 in yeast also inhibited mycelial growth and A. euteiches zoospore germination. These data suggest that sesquiterpene production in roots by MtTPS10 plays a previously unrecognized role in the defense response of M. truncatula against A. euteiches.
As sessile organisms, plants cannot evade attacks by different organisms, but they employ several strategies to defend themselves against detrimental organisms. To detect and counter pathogen attack, plants have evolved two interconnected layers of innate immunity (Jones and Dangl, 2006). The detection of highly conserved molecular signatures (e.g. pathogen-associated molecular patterns [PAMPs]) by membrane-associated receptors gives plants the ability to establish a defense response leading to PAMP-triggered immunity (PTI), resulting in a profound transcriptomic reprogramming (Monaghan and Zipfel, 2012). However, the signaling cascade leading to PTI can be disrupted by effectors produced by successful pathogens, thus promoting virulence (effector-triggered susceptibility; Fabro et al., 2011). Plants have evolved a second immune system that relies on effector recognition by specific proteins leading to effector-triggered immunity (Collier and Moffett, 2009).
In plants, PTI induces a variety of processes, among them the production of a huge diversity of secondary metabolites with a wide range of defense properties (Wink, 2018). In addition, exudation of secondary compounds by plant roots may affect the soil microbial community in their close vicinity and help plants to cope with various aggressors, including pathogens and herbivores. Furthermore, root exudates may also attract beneficial organisms to stimulate mutualistic symbioses, change the chemical and physical properties of the soil, or inhibit the growth of competing plant species and communicate with other species (Wink, 2003). More than 100,000 chemical compounds are produced by plants, and at least 1,700 of these are known to be volatile (Dudareva et al., 2013). Volatile organic compounds (VOCs) are defined as organic compounds with high vapor pressure at ambient temperatures (Loreto and Schnitzler, 2010). VOCs are involved in the plant’s response toward biotic factors (Sharifi et al., 2018), but also in its response to abiotic stress perception, such as physical damage, nutrient deficiency, salinity, drought, and ozone exposure (Shulaev et al., 2008; Dicke and Baldwin, 2010; Junker and Tholl, 2013).
VOCs emitted by plant shoots and flowers have been extensively described and characterized, whereas VOCs from the root systems have received comparatively less attention (Dudareva et al., 2013). Despite this, it may be expected that below-ground VOC-releasing responses are as common as above-ground responses (Erb et al., 2009). Most VOCs reported are largely lipophilic products and are mainly assigned to terpenoids, fatty acid derivatives, benzenoids, and phenylpropanoids, as well as various nitrogen- and sulfur-containing compounds (Dudareva et al., 2004). Among them, the terpenes constitute the largest class (Maimone and Baran, 2007), mostly represented by monoterpenes (C10) and sesquiterpenes (C15). All terpenes are derived from the C5 building blocks isopentenyl diphosphate and dimethylallyl diphosphate, which are produced by two alternative pathways: the mevalonate pathway, which is located in the cytosol, partially at the endoplasmic reticulum, and in peroxisomes and leads to sesquiterpenes, triterpenes, and polyterpenes; and the methylerythritol phosphate pathway located in plastids, which leads predominantly to monoterpenes and diterpenes (Lichtenthaler, 1999; Lange et al., 2000; Sapir-Mir et al., 2008). The structure of the terpene that is ultimately produced by the plant is determined by the activities of specific terpene synthases (TPSs), which form a large family (Dudareva et al., 2004). TPSs have been characterized from various plants. Analysis of the genome sequences of various seed plants showed that the number of full-length TPS-encoding genes ranges from ∼30 (e.g. in Arabidopsis [Arabidopsis thaliana]) to >60 in grapevine (Chen et al., 2011). In Medicago truncatula, an in silico analysis identified 23 putative full-length TPS-encoding genes, which all fall into the five established angiosperm TPS subfamilies, with a lineage-specific expansion of subfamily G (Parker et al., 2014).
M. truncatula a member of the Fabaceae, has been established as a model legume plant with a growing set of genetic and genomic tools (Rose, 2008). Medicago is a diploid species with a small, haploid genome consisting of eight chromosomes of ∼500 Mbp, is self-fertile, and has a short generation time. The current genome sequence of M. truncatula captures 94% of all M. truncatula genes (Young et al., 2011; Tang et al., 2014). To perform reverse-genetic approaches, a Medicago mutant population contacting a tobacco (Nicotiana tabacum) retrotransposon Tnt1 insertion was established, representing the largest collection of DNA-insertion mutants of all legumes (d’Erfurth et al., 2003). M. truncatula is used to obtain insights into legume-microbe interactions of agronomical relevance. As a legume, it interacts with rhizobia, leading ultimately to the formation of nitrogen-fixing structures known as nodules, but it can also establish a mutualistic interaction with arbuscular mycorrhizal fungi, which assist the plant in obtaining mineral nutrients from soil (Rose, 2008). Moreover, M. truncatula roots can be attacked by the oomycete Aphanomyces euteiches, which can infect other legume species (Gaulin et al., 2007) and is the most limiting factor for pea production in the United States and Europe (Hughes and Grau, 2014).
Primary disease symptoms of A. euteiches infected roots are water-soaked, softened brown lesions followed by root rotting, resulting in significant reduction of root biomass. Secondary symptoms like chlorosis, necrosis, and wilting of the foliage might follow, leading eventually to tremendous losses in yield (Hughes and Grau, 2014). The life cycle of A. euteiches includes asexual and sexual stages, and the infection starts by germination of oospores that have a thick protective wall and energy reserves allowing them to survive for many years in the soil. In the vicinity of a host root, oospores form a germ tube and a terminal zoosporangium, which may release >300 primary, biflagellate motile zoospores adhering to the host tissue. After root penetration, hyphae differentiate into haploid antheridia and oogonia, later producing diploid oospores (Hughes and Grau, 2014).
Upon infection with A. euteiches, M. truncatula roots respond with massive changes in transcript accumulation and protein composition, as analyzed recently for one to several days after infection (Colditz et al., 2004; Badis et al., 2015). In this article, we used a transcriptomic approach to investigate the very early response of M. truncatula roots to infection with A. euteiches. Using seedlings as well as adult plants grown in an aeroponic system, we analyzed the plant’s response 2 h after contact with the pathogen. Among the upregulated genes, we identified and characterized a gene encoding a sesquiterpene synthase (M. truncatula TERPENE SYNTHASE 10 [MtTPS10]), which was specifically expressed in roots after inoculation with the pathogen. Our experiments revealed that the enzyme catalyzes the production of a blend of sesquiterpenes in planta and of sesquiterpenes and sesquiterpene alcohols upon recombinant expression in yeast. We found that blocking TPS10 activity in planta by genetic approaches resulted in enhanced susceptibility of M. truncatula to A. euteiches, which might be a direct toxic effect of the sesquiterpenes on growth of the pathogen. Together, our data suggest that production of sesquiterpenes by MtTPS10 is a previously unrecognized role in the defense response of M. truncatula against the root pathogen A. euteiches.
RESULTS
Inoculation with A. euteiches for 2 h Alters the Transcriptome of Medicago Roots
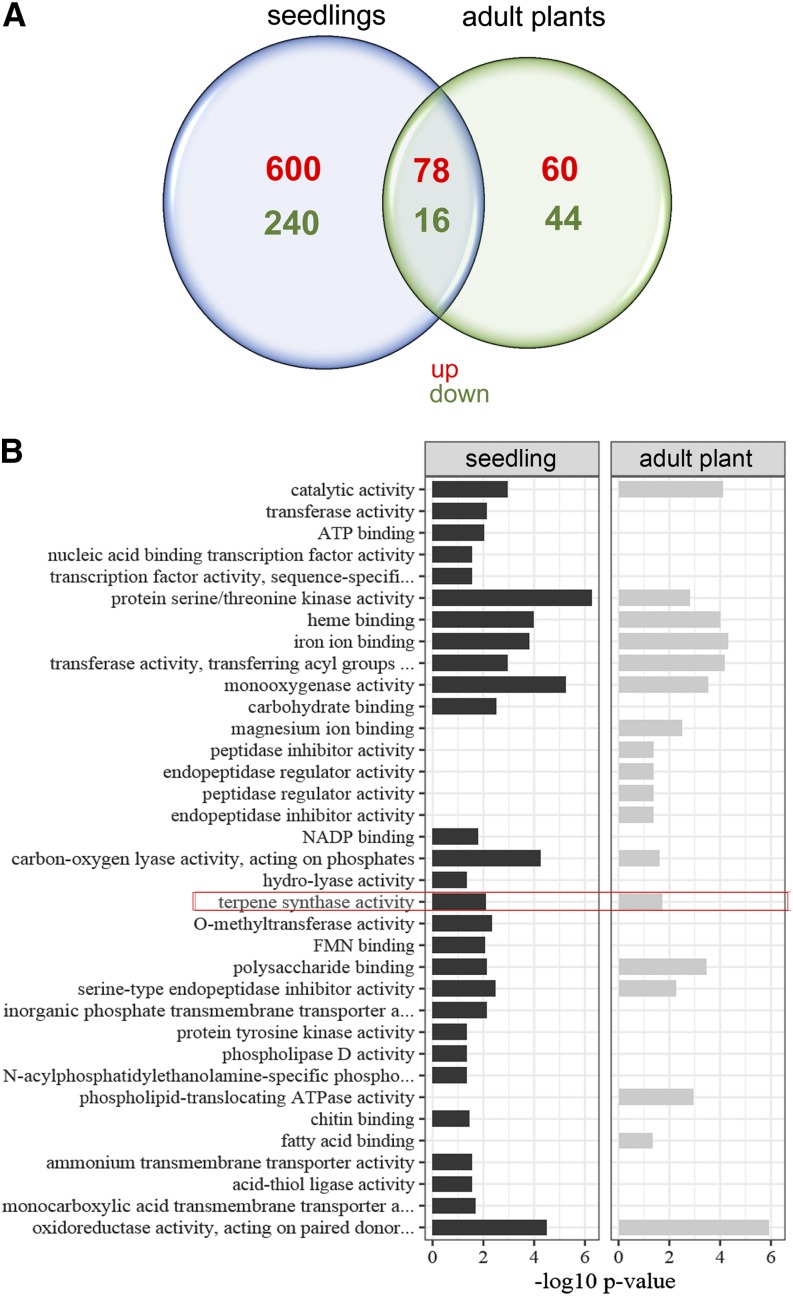
Previous work revealed that M. truncatula roots respond to inoculation with the oomycete pathogen A. euteiches with changes in transcript accumulation (Badis et al., 2015). To identify early induced genes, the best suitable early time point for a transcriptome analysis was specified by test microarrays using total RNA from seedlings at 0, 2, 6, and 10 h postinoculation (hpi) with A. euteiches zoospores. The transcriptomic data based on single biological replicates revealed that the 2 hpi dataset already contained a high number of genes that appeared to be differentially regulated. Therefore, further transcriptome analyses of M. truncatula roots infected with A. euteiches zoospores were done at 2 hpi. For this, seedlings grown in plates, as well as adult plants grown in an aeroponic system, were used independently to find differentially regulated genes in the developmental stage of the plant. On the one hand, 1-week-old seedlings were inoculated on a plate with 3,000 zoospores directly applied to the lower part of the root; on the other hand, roots of 6-week-old plants were inoculated by applying 105 zoospores to the complete root system. In both cases, mock-treated plants served as controls. Transcript profiling was conducted from four biological replicates using the Affymetrix MEDGene 1.FIRST Array Strip followed by robust multiarray analysis and Student’s t test. The comparison between mock treated and A. euteiches-inoculated roots in both developmental stages using a cut-off of at least a 2-fold difference in expression level revealed 1,038 differentially expressed genes (DEGs) in roots of both developmental stages (Fig. 1A; Supplemental Dataset S1). In both root types, about two-thirds of the DEGs were upregulated and one-third was downregulated. These numbers are similar to those obtained after infection for 24 h (Badis et al., 2015; Rey et al., 2016). The overlay between DEGs obtained in our approach and those obtained after infection for 24 h, however, is rather small, pointing to the fact that roots responded differently to the infection with A. euteiches at these two time points (Supplemental Fig. S1). The number of DEGs in seedling roots was almost four times higher than that in roots of adult plants, but there were 78 and 16 genes commonly upregulated and downregulated, respectively. The identified DEG could be assigned to 35 functional classes (Fig. 1B). The highest numbers of DEGs occurred in the classes “protein serine/threonine kinase activity,” “monooxygenase activity,” and “oxidoreductase activity.” Regarding production of secondary metabolites, the class “terpene synthase activity” showed significantly enhanced DEGs (Fig. 1B).
Figure 1.
Comparative analysis of transcript accumulation in M. truncatula roots after treatment with A. euteiches zoospores for 2 h. A, Venn diagram showing the numbers of differentially expressed genes. Total RNA isolated from the roots of 1-week-old seedlings (blue) and 6-week-old plants (green) nontreated or treated with A. euteiches zoospores was subjected to transcript profiling using the Affymetrix M. truncatula GeneChip array. Numbers of significantly upregulated (red) and downregulated (green) genes in A. euteiches-treated roots in comparison to nontreated roots are shown (P ≤ 0.01, n = 3). B. Enriched GO molecular function terms (P < 0.05) associated with the core sets of differentially regulated genes in the treatment of seedlings and adult plants are shown with –log10 transformed P-values indicated on x axes (full data in Supplemental Dataset S1). The GO function “terpene synthase activity” is outlined in red. FMN, Flavin mono nucleotide.
Expression of Medtr5g073200 Is Induced upon Inoculation with Plant Oomycete Pathogens and Is Root Specific
Among the genes induced in both seedlings and adult plants, one gene (Medtr5g073200) attracted our attention, because it showed the highest induction, with a nearly 100-fold upregulation in seedlings upon infection with A. euteiches. Medtr5g073200 was annotated as a sesquiterpene synthase and was classified as MtTPS10, belonging to subfamily A of the terpene synthases in M. truncatula (Supplemental Fig. S2A), but it has not been characterized yet (Parker et al., 2014).
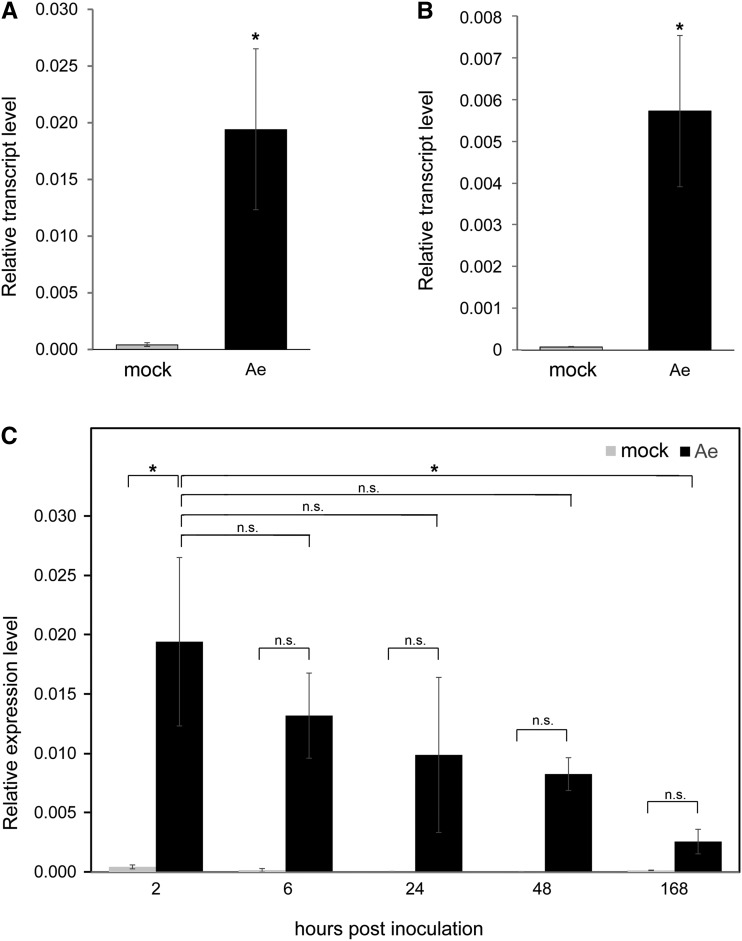
The induction of MtTPS10 in roots of adult plants was not as high as in seedlings, but it was still significant and more than 2-fold (Supplemental Dataset S1). The array data obtained for MtTPS10 were validated by reverse transcription quantitative PCR (RT-qPCR) using independent biological replicates for both seedlings and adult plants. In both developmental stages, significantly enhanced transcript levels could be observed 2 h after inoculation of roots with A. euteiches (Fig. 2, A and B). Again, transcript levels in adult plants were lower than in seedlings. The induction of MtTPS10 appeared to be specific for this TPS-encoding gene, since transcript levels of the most closely related genes, such as Medtr5g062230 (MtTPS5), Medtr6g008560, Medtr5g073260, Medtr5g094620, and Medtr6g039440, either did not increase upon infection or were below the detection limit (Supplemental Fig. S2B). MtTPS10 indeed belongs to genes that are induced early and transiently in A. euteiches-inoculated seedlings, because the transcript levels had already dropped at 6 hpi and further onward (Fig. 2C).
Figure 2.
Induction of MtTPS10 expression in M. truncatula roots by treatment with A. euteiches zoospores. Roots of 1-week-old seedlings (A and C) and 6-week-old plants (B) were treated with A. euteiches zoospores (Ae) or mock-treated with swamp-water (mock). MtTPS10 transcript levels were determined by RT-qPCR at 2 hpi (A and B) or at time points as indicated (C). Data are shown as the mean ± se. His-3-like was used as a reference gene and statistical analysis was done using Student’s t test (n ≥ 3 independent samples); *P < 0.05. n.s., not significant.
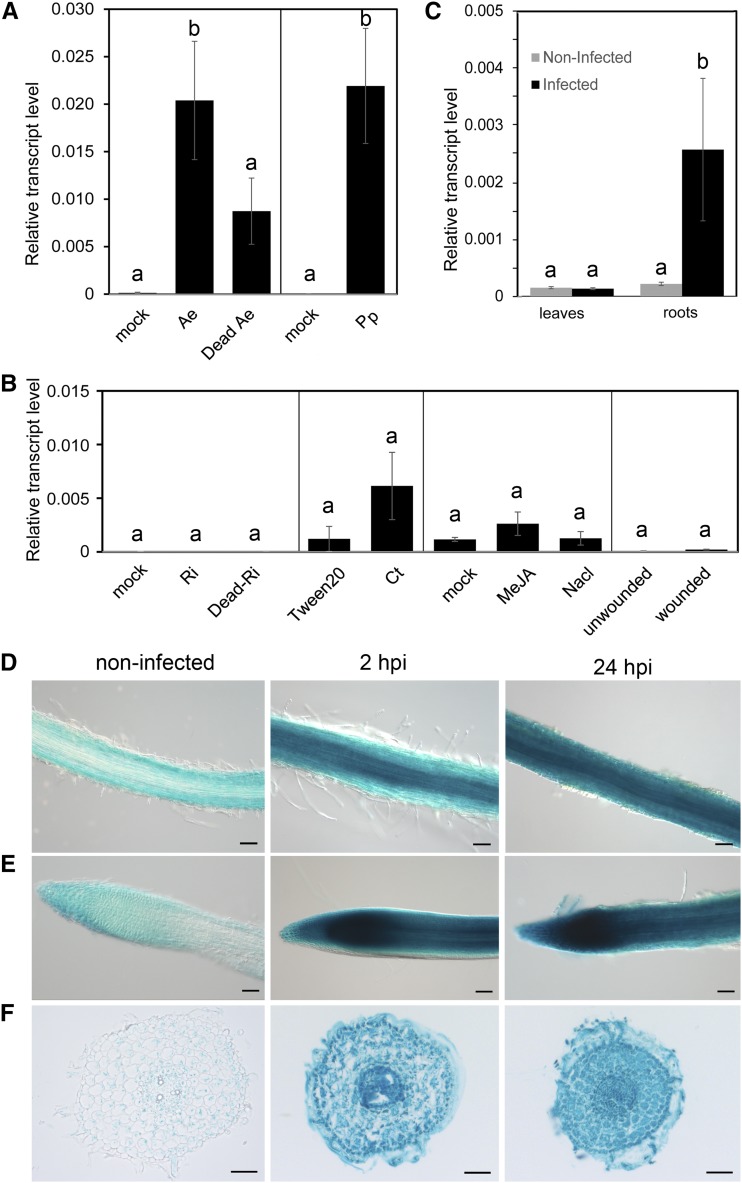
Next, it was tested whether MtTPS10 gene expression is specifically induced by inoculation with A. euteiches or whether inoculation with other soil-borne microorganisms or other treatments may also result in an enhanced transcript accumulation. In addition to inoculation with living zoospores of A. euteiches, seedlings were inoculated with dead zoospores of A. euteiches, zoospores of another oomycete pathogen, Phytophthora palmivora, spores of the fungal pathogen Colletrotrichum trifolii, or spores of the arbuscular mycorrhizal fungus Rhizophagus irregularis (Fig. 3, A and B). Analyzing the transcript accumulation by RT-qPCR showed that MtTPS10 is significantly induced by inoculation with living zoospores of A. euteiches, but not by application of dead zoospores (Fig. 3A). Infection with P. palmivora resulted in an increase of transcript levels similar to that from infection with A. euteiches, but neither inoculation with the true fungal root pathogen C. trifolii nor inoculation with the mutualistic symbiont R. irregularis led to a significant induction of MtTPS10 expression (Fig. 3, A and B). Also, no significant induction was detectable by application of salt stress, methyl jasmonate (MeJA), or wounding (Fig. 3B). All these treatments were effective, however, as confirmed by the enhanced transcript levels of respective response genes, such as CHALCONE SYNTHASE as a marker for C. trifolii infection (Jaulneau et al., 2010), CBF4 as a marker for salt stress (Li et al., 2011), and MtAOC1 as a marker for MeJA treatment and wounding (Isayenkov et al., 2005; Tretner et al., 2008; Supplemental Fig. S3). These data suggest that MtTPS10 is specifically induced upon infection by oomycetes.
Figure 3.
Expression of MtTPS10 is specifically induced by infection with oomycetes and is restricted to roots. (A) MtTPS10 transcript accumulation in 1-week-old Medicago A17 seedlings inoculated with A. euteiches zoospores (Ae), A. euteiches zoospores boiled for 10 min (dead Ae), and P. palmivora zoospores (Pp) for 2 h. Controls were treated with water (mock). B, MtTPS10 transcript accumulation was studied in 1-week-old M. truncatula A17 seedlings inoculated with spores of R. irregularis (Ri), spores of R. irregularis boiled for 10 min (dead Ri), and spores of C. trifolii (Ci), and in 1-week-old seedling roots treated with 100 mm NaCl or 50 µm MeJA or wounded by squeezing. All samples were analyzed 2 h after treatment. Control plants were either mock-treated, treated with Tween20, or left unwounded. C, Two-week-old M. truncatula A17 plants were infected with A. euteiches for 4 weeks and MtTPS10 transcript accumulation was analyzed in shoots and roots by RT-qPCR. Note that MtTPS10 transcripts were barely detected in leaves of either noninfected or infected plants. Data in A to C are shown as the mean ± se. His-3 like was used as the reference gene and statistical analysis was performed using ANOVA and Tukey’s honest significant difference (HSD) test (n = 3); lowercase letters a and b indicate significantly different values (P < 0.05). D to F, MtTPS10 is induced in all root cells after infection by A. euteiches. A 2-kb region of the MtTPS10 promoter fused to the GUS-encoding gene was transformed in M. truncatula roots via A. rhizogenes-mediated root transformation. Transformed roots were then treated with A. euteiches zoospores for 2 hpi and 24 hpi; controls were not treated. After treatment, roots were stained with X-Gluc and embedded in PEG for sectioning. Pictures were taken from roots (D), root tips (E), and cross sections of 10 µm thickness (F). Bars represent 100 µm (D and E) and 50 µm (F).
To address the organ specificity of MtTPS10 expression, Medicago plants were infected by A. euteiches for 4 weeks, and shoots and roots were analyzed independently. As shown in Figure 3C, MtTPS10 transcript accumulation was induced by A. euteiches infection in roots only. Transcript levels in both infected and uninfected shoots were at the detection limit and did not change. To gain insight into the cell and tissue specificity of MtTPS10 expression, a 2-kb fragment of the promoter upstream of the start codon was cloned to drive the GUS reporter gene. Medicago roots were transiently transformed with pTPS10::GUS to obtain chimeric plants (Mrosk et al., 2009). Transformed roots either noninoculated or inoculated with A. euteiches for 2 and 24 h were stained with 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc; Fig. 3, D–F). As visible by the blue coloration indicative of GUS activity, the promoter of MtTPS10 is highly active throughout the root, including at the root tip, whereas the basal activity without infection is very low, resulting in a faint staining of roots (Fig. 3, D and E). Moreover, as shown by cross sections of stained roots, the MtTPS10 promoter is active in all root cells after inoculation with A. euteiches (Fig. 3F). Summarizing these data, we conclude that the induction of the gene encoding the putative TPS10 seems to be restricted to roots, but is induced in all root cells. Moreover, MtTPS10 induction in roots is highly specific for infection by pathogenic oomycetes.
MtTPS10 Produces a Blend of Sesquiterpenes
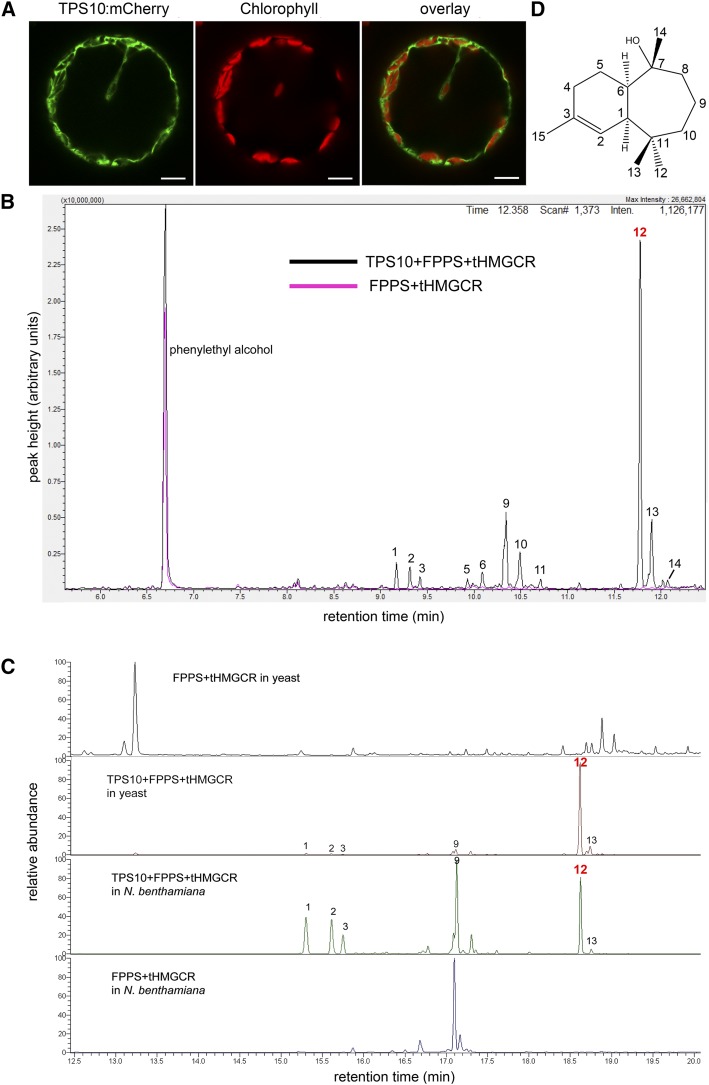
MtTPS10 shows high similarity to MtTPS5 (Medtr5g062230), which was characterized as a multiproduct sesquiterpene synthase (Arimura et al., 2008; Garms et al., 2010). This prompted us to investigate whether Medtr5g073200 indeed encodes an active sesquiterpene synthase. Sesquiterpene synthases typically use trans,trans-farnesyl diphosphate (FPP) as substrate and are located in the cytosol (Nagegowda, 2010). To determine the subcellular localization, MtTPS10 was fused to the gene encoding the fluorescent protein mCherry and transiently expressed in protoplasts of N. benthamiana under the control of a constitutive 35S Cauliflower mosaic virus promoter. The fusion protein was detectable in the cytosol surrounding the chloroplasts, which were visible by their red chlorophyll autofluorescence (Fig. 4A).
Figure 4.
MtTPS10 is a cytosolic protein and produces a blend of sesquiterpenes. A, N. benthamiana protoplasts were transformed with the MtTPS10 encoding sequence fused to mCherry. Fluorescence images were taken using confocal laser scanning microscopy. Note the localization of MtTPS10 (green) within the cytosol surrounding the chloroplasts visible by chlorophyll autofluorescence (red). Scale bars = 10 µm. B, Products of MtTPS10 identified after heterologous expression in S. cerevisiae are shown. MtTPS10 was expressed together with FPPS and HMGCR (tHMGCR) and the resulting products were analyzed using liquid-injection GC-MS (black line). Yeast cells expressing FPPS and HMGCR alone served as the control (pink line). C, Products of MtTPS10 identified after heterologous expression in S. cerevisiae and N. benthamiana leaves are shown. MtTPS10 was expressed together with FPPS and HMGCR. Yeast cultures and infiltrated leaves were analyzed using SPME-coupled GC-MS. Note that in B and C, several sesquiterpenes and sesquiterpene alcohols occurred upon expression of MtTPS10 and could be annotated by comparison of MS spectra based on library suggestions: (1) longipenene, (2) ylangene, (3) longicyclene, (5) farnesene, (6) α-himachalene (9) alloaromadendrene, (10) β-himachalene, (11) bisabolene, (12) himachalol, (13) α-bisabolol, and (14) shyobunol (D) The main product (peak 12) was purified, subjected to NMR analysis and identified as himachalol (Supplemental Fig. S2; Supplemental Table S1).
Next, MtTPS10 was expressed in yeast (Saccharomyces cerevisiae) to identify its products. To provide an adequate supply of FPP, a construct containing genes encoding the preceding enzymes FPP synthase (FPPS) and 3-hydroxy-3-methyl-glutaryl-coenzyme-A reductase (HMGCR) as well as MtTPS10, all under the control of Gal-inducible promoters, was transformed in the yeast S. cerevisiae. Analysis of yeast extracts by liquid-injection gas chromatography-mass spectrometry (GC-MS) revealed a complex pattern of at least 17 different sesquiterpenes and sesquiterpene alcohols, which were specifically found upon expression of MtTPS10 and were annotated by comparison of MS spectra in available library databases (Fig. 4B). In addition, MtTPS10 together with FPPS and HMGCR were expressed transiently in N. benthamiana leaves and the resulting products were analyzed using solid-phase microextraction (SPME) followed by GC-MS (Fig. 4C). In order to compare the TPS10 products obtained from yeast, the yeast cultures were also analyzed by the same method, and the compound spectra revealed high similarity and showed again production of sesquiterpenes and sesquiterpene alcohols with the same major compound (peak 12).
The major compound (Fig. 4B, peak 12), showing mass-to-charge ratio (m/z) values of 109, 119, and 204 and annotated as α-bisabolol (Supplemental Fig. S4A), was purified from yeast extracts and subjected to structural elucidation by NMR (Supplemental Fig. S4B; Supplemental Table S1). Based on one- and two-dimensional NMR as well as on comparison with literature data (Prakash et al., 1988; Agrawal et al., 1992), this compound was identified as himachalol (Fig. 4D).
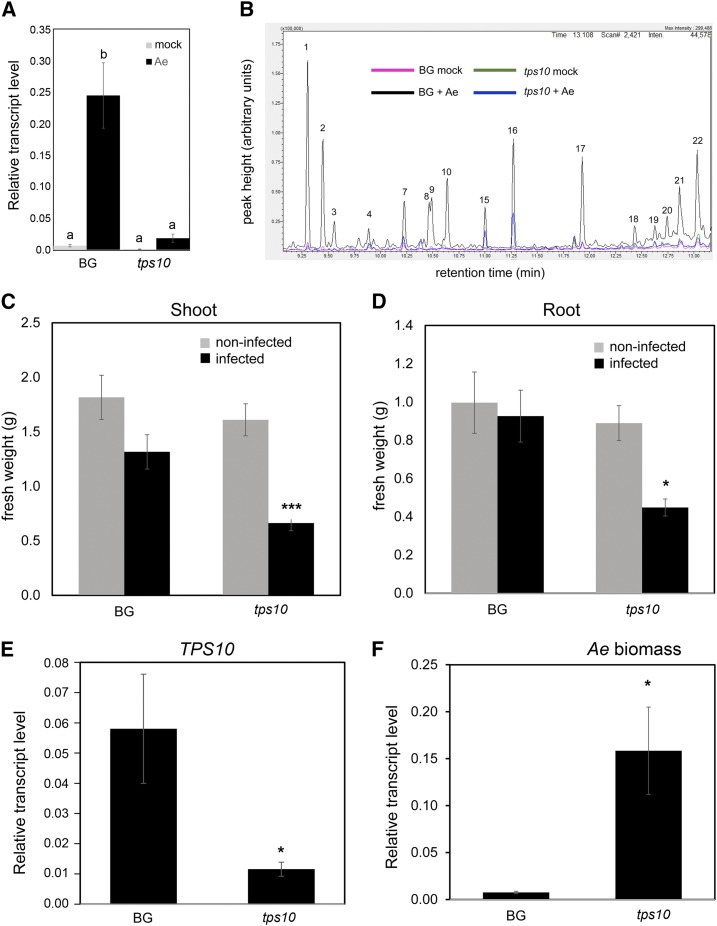
To elucidate the products of the MtTPS10 in M. truncatula, a mutant harboring a Tnt1 insertion within MtTPS10 (Supplemental Fig. S5A) was identified via a BLAST search from the M. truncatula mutant database provided by the Noble Research Institute (Tadege et al., 2008). The flanking sequence (223 bp) of the Tnt1 insertion from line NF10408_low_18 showed 100% identity to MtTPS10. The mutation was designated as tps10. Homozygous tps10 progeny were isolated from a self-pollinated heterozygous tps10/TPS10 individual. Homozygous TPS10 progeny of the same parent were also isolated and used in subsequent experiments as controls and named the background line (BG). The effect of the Tnt1 insertion on MtTPS10 expression was determined by RT-qPCR using RNA from noninfected and infected seedlings and primers binding upstream of the insertion site (Fig. 5A; Supplemental Fig. S5A). Similar to wild-type seedlings, BG seedlings responded to inoculation with A. euteiches with a strong MtTPS10 transcript accumulation at 2 hpi, whereas the tps10 mutant roots exhibited only basal levels of MtTPS10 transcripts, both with and without infection. Moreover, transcript levels of the genes closely related to MtTPS10, Medtr5g062230 and Medtr6g008560, were not affected by the Tnt1 insertion either in noninfected or in infected seedlings (Supplemental Fig. S6A). Therefore, the tps10 mutant can be regarded as a loss-of-function mutant. As expected, because of the restricted expression of MtTPS10 in infected roots, noninfected tps10 mutants did not show an obviously altered phenotype (Supplemental Fig. S5B).
Figure 5.
The Tnt1 insertion line NF10408 is defective in MtTPS10 expression, fails to produce sesquiterpene volatiles, and shows enhanced susceptibility to infection with A. euteiches. A, One-week-old seedlings of BG and tps10 lines were treated with A. euteiches zoospores for 2 h. Total RNA from roots was subjected to RT-qPCR. Transcript levels of MtTPS10 determined using primers binding upstream of the Tnt1 insertion site (see Supplemental Fig. S5A) were significantly reduced in roots of tps10 mutant plants compared to BG plants. Data are shown as the mean ± se. Actin was used as the reference gene and statistical analysis was done using ANOVA followed by Tukey’s honest significant difference (HSD) test (n = 3 independent samples); lowercase letters a and b indicate significantly different values (P < 0.05). B, Volatiles produced by roots of BG and tps10 mutant plants with and without infection by A. euteiches. Volatile compounds induced during the first 12 h after treatment with zoospores were adsorbed on PDMS strips and analyzed by thermal desorption-GC-MS. The labeled peaks correspond to (1) longipenene, (2) ylangene, (3) longicyclene, (4) longifolene, (7) isoshyobunone, (8) 7-epi-cis-sesquisabinene hydrate, (9) alloaromadendrene, (10) β-himachalene, (15) nerolidol, (16) fokienol, (17) ethyl iso-allocholate, (18) cholest-8-en-3-β-ol acetate, (19) cyclononasiloxane, (20) thunbergol, (21) nerolidyl propionate, and (22) heptasiloxane. The compounds representing peaks 1 to 3 and 9 and 10 were also identified in extracts from yeast expressing MtTPS10. Note that the major compound identified in yeast (himachalol [peak 12 in Fig. 4B]) is not detectable among root volatiles, possibly because of M. truncatularoot modification. C and D, Biomass of shoots (C) and roots (D) of BG and tps10 plants noninfected or infected by A. euteiches, as shown in C. tps10 mutant plants were strongly affected by infection with A. euteiches as visible by the diminished biomass of shoots and roots, whereas plants of the BG line did not show significant alterations in growth. Data are shown as the mean ± se (n = 18 independent samples). Statistical analysis was done using Tukey’s honest significant difference (HSD) test; ***P < 0.001; *P < 0.05. E and F, Two-week-old seedlings of BG and tps10 mutant lines were infected with A. euteiches zoospores and harvested 4 weeks later. Total RNA from roots was subjected to RT-qPCR. Transcript levels of MtTPS10 (E) were significantly reduced in roots of tps10 mutant plants compared to BG plants, whereas 5.8S rRNA of A. euteiches was strongly enhanced in tps10 roots in comparison to BG roots (F). Data are shown as the mean ± se. Actin was used as the reference gene and statistical analysis was done using ANOVA followed by Tukey’s honest significant difference (HSD) test (n = 3 independent samples); *P < 0.05.
Roots of 6-week-old tps10 and BG plants were infected with A. euteiches for 12 h, and VOCs were collected using Sorbstar sticks inserted in the root systems. Thermodesorption followed by GC-MS revealed the production of a complex sesquiterpene pattern emitted by BG roots that was almost missing in VOC collections of noninfected BG roots as well as in tps10 roots, both noninfected and infected (Fig. 5B). Among the sesquiterpenes missing in tps10 VOCs, five compounds annotated by MS spectrum comparison (Fig. 5B, peaks 1–3, 9, and 10) were identical to the compounds determined from yeast expressing MtTPS10. Interestingly, the main product of MtTPS10 expression in yeast and N. benthamiana leaves, himachalol, could not be identified among the root VOCs. This might hint to a conversion of the TPS10-produced sesquiterpene alcohols by other enzymes in roots of M. truncatula. The results from the expression of MtTPS10 in yeast and N. benthamiana and comparison of the profile of VOCs between BG and tps10 roots show that MtTPS10 is a multiproduct sesquiterpene synthase.
Mutants Affected in MtTPS10 Encoding Gene Show Enhanced Susceptibility to A. euteiches
Since the upregulation of MtTPS10 in wild-type roots 2 hpi with A. euteiches was accompanied by emission of volatile sesquiterpenes, we next tested whether the tps10 mutant would react differently to an infection with A. euteiches. Thus, 1-week-old tps10 and BG plants were inoculated with A. euteiches zoospores and analyzed 4 weeks later. Even after this long time of infection, differences in the MtTPS10 expression levels between BG and tps10 were detectable (Fig. 5E). Significant changes in transcript levels of the MtTPS10-related genes Medtr5g062230 and Medtr6g008560 were not detectable (Supplemental Fig. S6B). Infected tps10 and BG plants were compared in terms of shoot and root biomasses, which both were strongly reduced in tps10 plants (Fig. 5, C and D; Supplemental Fig. S5B) compared to BG plants. This prompted us to check whether the tps10 plants had an impaired resistance toward the pathogen. The colonization rates in both genotypes were determined by RT-qPCR quantification of the 5.8S ribosomal RNA (rRNA) of A. euteiches within the root tissue (Fig. 5F). Indeed, roots of tps10 mutant plants showed a significantly enhanced level of A. euteiches 5.8S rRNA, indicating a higher biomass of the oomycete within the roots. This suggests an impaired plant defense in tps10 plants in comparison to BG plants.
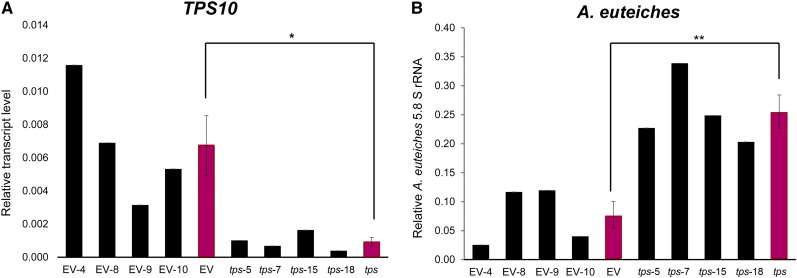
To confirm that the impaired defense in tps10 mutant plants was associated with loss of MtTPS10 function, we employed the artificial microRNA (amiR) technique to reduce MtTPS10 expression in Medicago roots. Expression of TPS10-amiR was driven by the constitutive ubiquitin promoter, and transgenic roots were identified by coexpression of Discosoma red fluorescent protein (Devers et al., 2013). Following inoculation with the pathogen for 4 weeks, controls created by expression of an empty vector showed variable, but detectable, transcript accumulation of MtTPS10 (Fig. 6A). By contrast, the MtTPS10 transcript accumulation was drastically reduced in plants expressing TPS10-amiR, but the expression of the closely MtTPS10-related genes Medtr5g062230 and Medtr6g008560 was not affected (Supplemental Fig. S6C). Most importantly, in comparison to the empty vector-transformed plants, the TPS10-amiR plants showed an enhanced infection by A. euteiches, as monitored by the amount of A. euteiches 5.8 S rRNA within roots (Fig. 6B). These data indicate that expression of TPS10-amiR mimics the tps10 mutant plants and that loss of TPS10 activity confers enhanced susceptibility of Medicago to A. euteiches.
Figure 6.
Expression of amiR targeting MtTPS10 (TPS10-amiR) reduced MtTPS10 expression and enhanced susceptibility to A. euteiches. M. truncatula cv R108 plants were transformed with an empty vector (EV) or a TPS10-amiR (tps) construct using A. rhizogenes-mediated root transformation and were subsequently infected with A. euteiches for 4 weeks. MtTPS10 transcript accumulation (A) and 5.8S rRNA of A. euteiches (B) were determined using RT-qPCR. Actin was used as the reference gene. Data from single transformed roots are shown. The red bars show the mean ± se for all four plants. Statistical analysis was performed using Tukey’s honest significant difference (HSD) test; *P < 0.05; **P < 0.01.
Sesquiterpenes Produced by MtTPS10 Inhibit Mycelial Growth and Zoospore Germination of A. euteiches
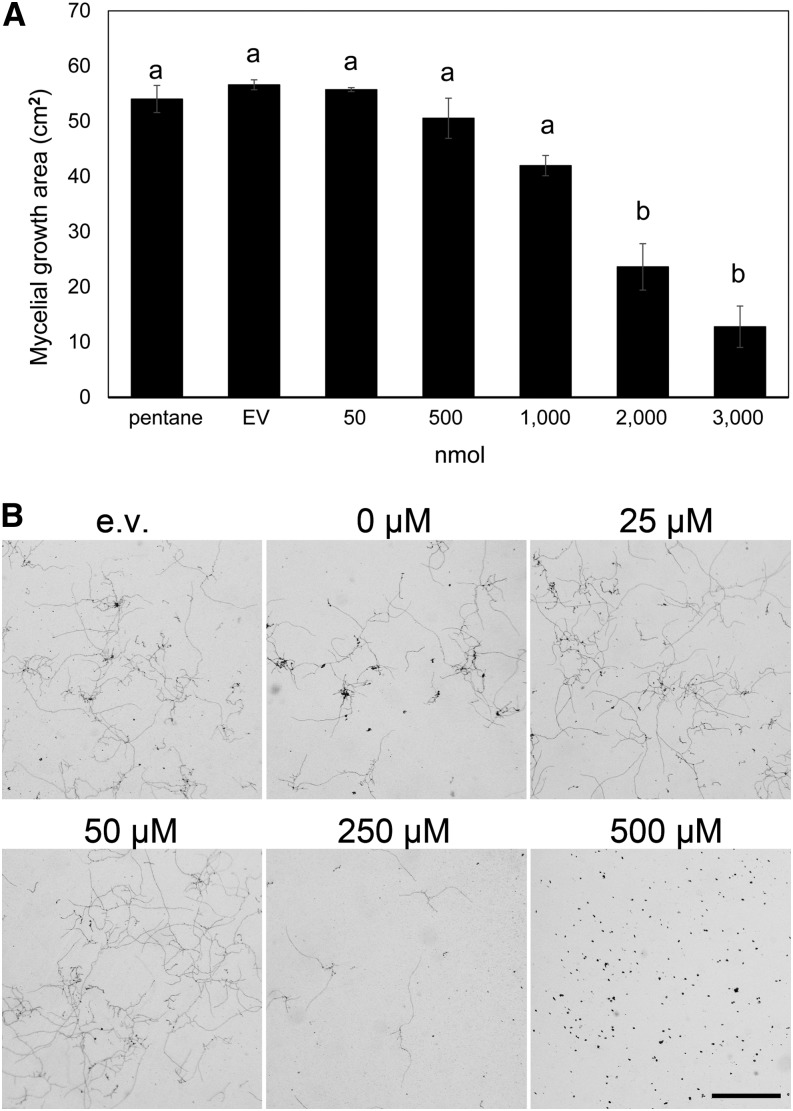
The enhanced susceptibility of plants with reduced MtTPS10 expression led to the question of whether the MtTPS10 products have direct effects on A. euteiches. To test this, we applied defined amounts of himachalol (plus minor compounds) produced in yeast to plates and monitored the mycelial growth of A. euteiches (Fig. 7A; Supplemental Fig. S7). After 1 week of growth, the mycelium covered almost the complete area of plates containing the solvent only or extracts from yeast expressing the empty vector. By contrast, application of 3,000 nmol himachalol inhibited mycelial growth up to 75%. In a second approach, the effect of MtTPS10 products on the germination of A. euteiches zoospores was analyzed. Zoospores were incubated with defined amounts of himachalol (plus minor products) produced in yeast for 1 h and transferred to cornmeal agar (CMA) plates for germination. Zoospores incubated with extracts from yeast expressing the empty vector germinated and produced a mycelium within 24 h, whereas zoospores incubated with a 250-µm himachalol solution produced fewer hyphae and zoospores incubated with a 500-µm himachalol solution did not germinate (Fig. 7B). Both the inhibitory effect on mycelial growth and the inhibition of zoospore germination indicate that the products of MtTPS10 might affect directly the pathogen A. euteiches, thereby contributing to protection of Medicago roots against A. euteiches.
Figure 7.
Products of MtTPS10 inhibit mycelial growth of A. euteiches. A, A. euteiches mycelial pieces were placed on plates and different amounts of the yeast extract containing MtTPS10 products (given in nmol himachalol per plate) were added at the edges of plates. Application of solvent only (pentane) or extracts from yeasts expressing the empty vector (EV) served as controls. After 1 week of mycelial growth, photographs were taken (Supplemental Fig. S4) and areas covered by mycelium were calculated using ImageJ. Data are shown as the mean ± se (n = 3 independent samples). Different letters indicate significant differences according to one-way ANOVA with Tukey’s honest significant difference (HSD) test, P < 0.05. B, Zoospores of A. euteiches were incubated for 1 h with different concentrations of yeast extract containing MtTPS10 products (himachalol plus some minor compounds) followed by plating on agar to allow germination. Pictures were taken 24 h later. Incubation with extracts from yeast expressing the empty-vector (EV) construct served as the control. Note the diminished hyphal growth at 250 µm himachalol and the complete inhibition of zoospore germination at 500 µm himachalol in the medium. Scale bar = 1 mm for all micrographs.
DISCUSSION
Previous studies have shown that roots of M. truncatula seedlings respond to inoculation with the pathogen A. euteiches for 24 h with a massive change in transcript accumulation (Badis et al., 2015). The main objective of this work was to elucidate the very early root responses to A. euteiches independently of the plant’s developmental stage. For this, a time point earlier than 24 hpi was chosen to uncover events before the penetration of the rhizodermis by the mycelium (Badis et al., 2015). The transcriptomes of seedlings and adult plants infected for 2 h were analyzed and searched for differentially regulated genes. The results revealed that the transcriptional reprogramming was substantially greater in seedling roots than in roots of adult plants, since there were about four times more genes differentially regulated in the seedling stage (Fig. 1A). On one hand, this could be attributed to different densities of zoospores that were applied to the roots. On seedlings, zoospores were applied directly onto the lower (young) part of the primary root, which is known from previous studies with symbiotic microorganisms to be the most responsive root tissue (Zipfel and Oldroyd, 2017). In contrast, the complete root system of adult plants was inoculated with the suspension of zoospores, which might lead to either a dilution effect in gene responsiveness or to less contact of the zoospores. On the other hand, seedlings might generally respond more strongly to the infection with A. euteiches, since in nature the infection frequently occurs during the early phases of seedling emergence (Hughes and Grau, 2014). Despite these differences, there were nearly 20% of DEGs found commonly regulated in roots of seedlings and adult plants. This suggests that a fast defense response might be activated upon recognition of the pathogen and that constitutively expressed defenses are not the only strategy used against A. euteiches. This was previously observed in transcriptome changes occurring at 24 hpi, where induction of genes involved in secondary metabolism was also most obvious, but concerned especially the flavonoid pathway, and was accompanied by enhanced levels of flavonoids (Badis et al., 2015). The comparison between both sets of DEGs obtained from seedling roots, those inoculated for 2 h (this study) and those inoculated for 24 h (Badis et al., 2015), revealed that a high number of A. euteiches regulated genes were not uncovered in the previous experiment.
Among the commonly upregulated genes in roots of seedlings and adult plants, the gene encoding a putative sesquiterpene synthase, MtTPS10, was found. MtTPS10 belongs to the subfamily A of TPSs in M. truncatula, which are primarily sesquiterpene producing (Parker et al., 2014). From the 23 TPSs that are encoded in the M. truncatula genome, four MtTPSs (sesquiterpene-producing MtTPS1, MtTPS3, and MtTPS5, and the monoterpene-producing MtTPS4) have been functionally characterized (Gomez et al., 2005; Arimura et al., 2008). Rapid expression of MtTPS1, MtTPS2, and MtTPS4 in herbivore-damaged leaves, as well as the specific expression of MtTPS11 in trichomes, points to their function in volatile plant defense and/or interactions of plants with herbivores (Gomez et al., 2005; Parker et al., 2014). However, no expression or functional data were available for most of the MtTPSs, including for MtTPS10 (Parker et al., 2014).
Transcript levels of MtTPS10 were very low in unstressed roots, but were induced in roots upon infection with A. euteiches. The expression of MtTPS10 appeared to be strong and very distinct for the early time point of the plant-pathogen interaction. MtTPS10 was significantly upregulated after 2 hpi, but at later time points the transcript levels were not significantly enhanced in comparison to controls (Fig. 2C). Transcript level analysis showed that different stress treatments, such as salt stress, wounding, and treatment with jasmonates, did not induce MtTPS10 transcripts. In addition, neither inoculation with the fungal pathogen C. trifolii nor that with the symbiotic fungus R. irregularis resulted in elevated transcript levels of MtTPS10. Even an in silico search using the Medicago truncatula Gene Expression Atlas (https://mtgea.noble.org/v3/) did not deliver other cues increasing MtTPS10 transcripts (data not shown). Therefore, it turned out that MtTPS10 expression was induced exclusively upon infection with oomycetes (Fig. 3, A and B). This might be attributed to the characteristics of cell wall components, which might act as PAMPs. However, both oomycetes, A. euteiches and P. palmivora, differ also in their cell wall composition. One specific characteristic of the Phytophthora genus is the presence of cellulose and the absence of chitin (Bartnicki-García, 1968), whereas it has been shown for A. euteiches that the dominating cell wall components are GlcNAc-based noncrystalline chitosaccharides and branched glucan-chitosaccharides (Badreddine et al., 2008). Among these components, fractions of the glucan-chitosaccharides were proposed to act as defense gene elicitors (Nars et al., 2013). However, this compound might not be an elicitor inducing MtTPS10, since (1) P. palmivora does not contain glucan-chitosaccharides, but application of P. palmivora zoospores lead to rapid induction of MtTPS10, and (2) dead (boiled) zoospores of A. euteiches containing cell walls and cell wall components were not able to significantly induce MtTPS10. It is tempting to speculate that the plant might recognize other molecules released specifically by oomycetes as PAMPs. Such molecules might either diffuse rapidly throughout the root tissue or might induce a secondary signal that is transduced into the inner part of the root, since induction of MtTPS10 occurred in all cells of the infected root (Fig. 3, D–F). However, the induction of MtTPS10 was root specific and was not detectable in leaves (Fig. 3C).
Expression of MtTPS10 in yeast and leaves of N. benthamiana resulted in production of several sesquiterpenes and sesquiterpene alcohols, with the main product identified by NMR as himachalol. Therefore, MtTPS10 can be regarded as a multiproduct terpene synthase like other identified monoterpene and sesquiterpene synthases, which generate substantial amounts of different products (Vattekkatte et al., 2018). For example, MtTPS5, a close relative of MtTPS10 (Supplemental Fig. S2A), catalyzes the formation of 27 different C15 hydrocarbon skeletons, with cubebol as the major product (Garms et al., 2010), and MtTPS1 yields (E)-β-caryophyllene and α-humulene (Arimura et al., 2008). This feature of TPSs—their ability to synthesize more than one product from a single substrate—may contribute to the high number of terpene carbon skeletons occurring in nature and may provide organisms with access to variable chemical defense in response to herbivores (e.g. in the case of MtTPS1 and MtTPS5) or pathogens (e.g. in the case of MtTPS10). Indeed, M. truncatula roots produced several sesquiterpenes upon infection with A. euteiches that were missing in a mutant defective in MtTPS10 expression (Fig. 5, A and B). Most of the TPS10 products produced in yeast could be identified also among the VOCs produced in roots of M. truncatula infected with A. euteiches. However, himachalol, the main product in yeast and N. benthamiana, could not be detected in roots. This led to the assumption that sesquiterpene alcohols produced by MtTPS10 might be further modified in Medicago. The modification of terpenes includes oxidations and transfer of glycosyl or acyl groups. These reactions can be carried out, for example, by cytochrome P450 monooxygenases, glycosyl-transferases, or acyl-transferases. A search for MtTPS10 coexpressed genes in A. euteiches-infected roots, however, has not delivered putative candidates so far, using either our dataset or public databases such as M. truncatula Expression Atlas (data not shown).
Multiproduct TPSs might confer upon the plant an advantage in the defense against pathogens by providing a blend of individual compounds, each with potential bioactivity, as has been shown for some individual sesquiterpenes that act as antimicrobial phytoalexins (Gomez et al., 2005). In addition, a role of the MtTPS10 products in defense is suggested by the strong and rapid induction of transcripts that occurred following infection by A. euteiches. Furthermore, M. truncatula mutants lacking MtTPS10 expression were more susceptible to infections with A. euteiches than the corresponding wild type (Fig. 5). In an independent approach, MtTPS10 expression was reduced by expressing an amiR construct targeting MtTPS10, which resulted in enhanced susceptibility of roots to A. euteiches (Fig. 6). This suggested a direct action of the emitted sesquiterpenes on the pathogen A. euteiches. It is well established that biosynthesis of phytoalexins at inhibitory concentrations is a defense response highly associated with resistance and relies on the correct location and time course of phytoalexin biosynthetic enzyme activities (Hammerschmidt, 1999; Sharifi et al., 2018). For instance, microbially induced terpenes from garlic exhibit negative effects on mycelial growth and on sclerotia production in Sclerotium cepivorum (Pontin et al., 2015), and the degradation product of a triterpene diol acts as an inducible defense compound from Arabidopsis, directly inhibiting the growth of the oomycete Pythium irregulare (Sohrabi et al., 2015). In addition, capsidiol, the major sesquiterpenoid phytoalexin produced in pepper (Capsicum annuum) and tobacco, is synthesized in tissues next to the site of invasion by fungi or oomycetes (Ahuja et al., 2012). The biological role of MtTPS10 products was further investigated by applying the products produced in yeast to mycelium and germinating zoospores. Indeed, the MtTPS10 products inhibited both mycelial growth and zoospore germination at concentrations in the micromolar range. Therefore, a biological role of the root-derived MtTPS10 products as antimicrobial phytoalexins is proposed. It should be noted, however, that the whole mixture of sesquiterpenes produced in yeast was used for these assays. Therefore, it is possible that only some individual compounds are responsible for this antimicrobial activity. Also, because himachalol cannot be detected in roots of Medicago, it is possible that either the in vitro effect visible on the mycelium and spore germination might not reflect the in planta situation at all or that derivatives of himachalol exhibit even stronger activity. Unfortunately, we have not been able so far to identify himachalol-derived products, which would be required to address this question.
In summary, our data showed a rapid induction of a gene encoding a sesquiterpene synthase (MtTPS10), which is directly involved in biosynthesis and emission of sesquiterpenes from infected roots. Application of these compounds to the pathogen inhibited its growth, whereas the absence of sesquiterpene production in roots of mutants resulted in enhanced plant susceptibility toward A. euteiches. A detailed understanding of the biological function of MtTPS10 will be of major interest because of its potential for control of the A. euteiches-induced root rot disease in agricultural systems.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Selection of TPS10 Mutant Plants
Medicago truncatula cv Jemalong A17 was used as reference for all the experiments, whereas cv R108 was used for root transformation. For germination, seeds were scarified using concentrated, anhydrous sulfuric acid for 5 min followed by intensive washing steps with distilled water and were then placed on 0.7% (w/v) water agar plates. After storage at 4°C for 3–4 d to break dormancy, seeds were incubated differently based on the final transfer either to pots or to plates. For use in pot experiments, seeds were incubated in the dark at room temperature for 1 day and in light for 1 day followed by transfer of single seedlings to pots (diameter of 13 cm) filled with expanded clay of 2- to 5-mm particle size (Original Lamstedt Ton; Fibo ExClay Deutschland). For use in plate experiments, aeroponics, and root transformation seeds were incubated at 12°C for 2 d. Seedlings were either transferred to plates (six to seven seedlings per plate) containing minimal medium (M medium; Bécard and Fortin, 1988) or were inserted in wetted rock wool placed over an aeroponic container filled with modified Strullu and Romand medium (Declerck et al., 2005) without vitamins and Suc. Plants were cultivated in a phytochamber or growth cabinet under long-day conditions at 40% humidity, with a light period of 16 h at 26°C and a dark period of 8 h at 20°C.
Nicotiana benthamiana plants were grown in growth chambers with a 16-h day/8-h night photoperiod and temperatures of 21°C and 18°C at day and night time, respectively.
For stress treatments, A17 seedlings were cultivated on M medium for 1 week and transferred to plates containing M medium supplied with 100 mm NaCl or 45 µm MeJA. As control, plants were transferred to M medium plates without additions. Two hours after transfer, the roots were harvested for subsequent analyses.
Plants of the Tnt1 insertion line NF10408_low_18 were screened for Tnt1 insertion in the TPS10 gene using PCR. For this, genomic DNA was isolated from 100 mg frozen plant material using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer instructions. To distinguish between the wild type and the mutant allele two PCR reactions were performed using GoTaq-DNA-Polymerase (Bio&Sell) and the primers (given in Supplemental Table S2) were used in 1+2 and 1+3 combinations.
Cultivation of Aphanomyces euteiches, Plate Assays, and Infection of Plants
Unless stated otherwise, A. euteiches strain ATCC201684 (kindly provided by Christoph Jacquet, University of Toulouse, France) was used for all infection studies. The strain was routinely subcultured on 1.7% (w/v) CMA (Sigma-Aldrich) in the dark at 24°C. Pieces of mycelium from a 10- to 15-d-old CMA plate were used for the production of zoospores in the dark at 24°C. Briefly, 15 to 20 mycelial explants were immersed in a 1:3 mixture of yeast tryptone medium (0.3% w/v yeast extract, 0.5% w/v tryptone) and swamp water in the dark at 24°C. After 3 d of mycelial growth, old medium was decanted, and the mycelial mats were washed several times with sterile tap water, resuspended in fresh tap water, and incubated overnight for zoospore release. The zoospores were counted in a counting chamber and appropriate volumes containing 105 zoospores and 3,000 zoospores were directly applied to the roots of adult plants and seedlings on plates, respectively. Infection was monitored by phenotypic characterization of plants and by determination of levels of A. euteiches 5.8 S rRNA using RT-qPCR.
Cultivation of Phytophthora palmivora, Colletrotrichum trifolii, and Rhizophagus irregularis and Inoculation of Plants
Fluorescently labeled P. palmivora LILI-td (accession P16830; Rey et al., 2015) expressing pTOR::tdTomato were cultivated on V8 vegetable juice agar plates as previously reported (Chaparro-Garcia et al., 2011). Seven-day-old plates of P. palmivora were flooded with 4°C sterile water and were incubated in the light at room temperature for 1 h to induce release of zoospores. For infection assays, suspensions of 5 × 104 spores per mL were used. Roots of 7-d-old Medicago seedlings grown on square plates containing 0.8% (w/v) agarose were inoculated with 50 μL of zoospore suspension per plant.
C. trifolii was cultivated on OMA plates (Werner et al., 2007) and grown at 23°C under UV light for sporulation. After 14 d of growth, C. trifolii spores were washed from the plates using 0.02% (v/v) Tween20 and collected in Eppendorf tubes. Of the collected spores, 105 were used for inoculating the roots of 7-d-old Medicago seedlings.
R. irregularis DAOM197198 was cultivated with Agrobacterium rhizogenes-transformed carrot roots in a bicompartment petri plate system as described (St-Arnaud et al., 1996). After 6–10 months of growth, spores were collected by dissolving agar pieces in 0.01 m citrate buffer (pH 6) at room temperature and with slow shaking in the dark overnight. Buffer was removed and spores were washed 2–3 times with water. Equal amounts of spores were used for plant inoculation.
RNA Isolation, Array Hybridization, and Evaluation
Total RNA was prepared from 100 mg root material using the RNeasy Plant MiniKit (Qiagen) according to the manufacturer’s instructions, followed by DNase digestion using RNase-free DNase (ThermoFisher Scientific). For probe synthesis, a GeneChip WT PLUS Reagent kit (Affymetrix) was used according to the manufacturer’s instructions, with 250 ng of total RNA. Hybridization of probes to the Affymetrix MEDGene 1.FIRST Array Strip and scanning of arrays were done using a GeneAtlas instrument (Affymetrix) according to the manufacturer’s instruction.
Microarray Data Analysis
Affymetrix exon microarrays were processed by the Bioconductor/R package XPS (Stratowa, 2018). XPS applies the robust multi-array average expression measure, which includes probe set summarization by median polish and quantile normalization. Expression values were obtained for transcript clusters and BG values were corrected using antigenomic probes. The dataset was filtered for unexpressed features by detection above BG calls. Features were retained if all signals of at least one replicate group were detected in either the control or treatment groups. Linear models were fitted for each feature using LIMMA (Ritchie et al., 2015) and p-values were adjusted by the false-discovery rate (FDR) procedure proposed by Benjamini and Hochberg (1995). Significant differentially expressed genes were identified using a significance level cutoff of FDR < 0.1. The normalized microarray data can be found under the accession number E-MTAB-7286 at Arrayexpress database (https://www.ebi.ac.uk/arrayexpress/). Affymetrix transcript clusters were mapped by using the best BLAST hit method to the M. truncatula MT 4.0 complementary DNA (cDNA) models. The corresponding MT 2.0 cDNA for each transcript cluster was parsed from the official Affymetrix annotation file and was used as the query for the BLASTN searches.
To compare the dataset obtained from seedlings with previously published data (Badis et al., 2015), the dataset with accession number GSE31198 was downloaded from the Gene Expression Omnibus and preprocessed with the Bioconductor package GEOquery using a significance level cutoff of FDR < 0.1 and Log2 fold change of ±1, similar to those used for the other datasets.
Gene Ontology (GO) terms were obtained using the Bioconductor/R package biomaRt, which interrogates the Plant Ensembl 39 database for the Medicago genome MedtrA17 4.0 build. Previously, mapping of transcript clusters to MedtrA17 transcript models was performed using the best-blast-hit method (National Center for Biotechnology Information BLAST+ version 2.3.0) using a minimal alignment length of 80% between hits and a sequence conservation of at least 98% at the nucleotide level. GO enrichment analysis was performed using the Bioconductor/R package topGO with Fisher statistics and the “elim” algorithm.
RT-qPCR
Total RNA was extracted from 100 mg roots and leaves as described above. Then, 0.5–1 µg RNA was reverse transcribed with the RevertAid first-strand cDNA synthesis kit (ThermoFisher Scientific). RT-qPCR was performed using a 5× QPCR Mix EvaGreen no ROX (Bio&Sell) in a real-time PCR instrument (BIO-RAD Laboratories). Specific primers listed in Supplemental Table S3 were designed using Primerfox software; their specificity was tested using BLAST search and validated by melt curve analysis. The following PCR conditions were used: initial denaturation at 95°C for 15 min, 39 cycles of denaturation at 95°C for 15 s, annealing, extension and plate read at 60°C for 30 s and 95°C for 10 s. The melt curve and plate read was performed from 65°C to 95°C, and data points were collected every 0.5°C with a 5 s hold between collections. The data were analyzed using CFX Manager software (Biorad) and normalized against housekeeping genes. Here, Medicago Histone3-like (MtHIS3L [Medtr4g097170]) and Medicago Actin (Medtr3g095530) were used for experiments with cv Jemalong A17 and cv R108, respectively. The resulting logarithmic values were converted using the equation 2−ΔCt. The data shown were summarized from at least three biological replicates and three technical PCR replicates.
Cloning of the TPS10-Encoding Gene and Creation of Fusion Constructs and amiR Constructs Targeting TPS10
The full-length coding sequence of TPS10 was amplified from M. truncatula cDNA using the primers (see Supplemental Table S4) and cloned into the level 0 vector pAGM4031 using the Golden Gate method (Werner et al., 2012). Depending on the final expression system (plant or yeast), suitable promoters and terminators were combined into the plant level 2 vector or the yeast level M vector.
For localization studies in N. benthamiana protoplasts, the full-length TPS10 encoding sequence without stop codon was C-terminally fused to mCherry and combined with the Cauliflower mosaic virus 35S promoter into the level 1 vector (pICH75055). For TPS10-promoter-GUS (pTPS10::GUS) fusions, 2 kb genomic region upstream from the start codon of TPS10 was amplified from M. truncatula genomic DNA using the primers listed in Supplemental Table S4 and cloned together with a GUS and a Discosoma red fluorescent protein module into a level 2 vector (pAGM4673). amiR constructs (TPS10-amiR) were created as described (Devers et al., 2013). For this, primers targeting TPS10 were designed using the WMD3 database (wmd3.weigelworld.org/; see Supplemental Table S4) and an overlapping PCR was performed using pB159b as the template and three primer combinations (TPS10_I_miR and TPS10_A_miR, TPS10_II_miR and TPS10_III_miR, and TPS10_IV_miR and TPS10_B_miR). The resulting PCR products were used as a template for the final PCR with the primers TPS10_A_miR and TPS10_B_miR. The final PCR product was subcloned into the pcr TOPO 2.1 cloning vector (ThermoFisher Scientific), digested, and ligated into the destination vector p9RFP_UBQ3 Expr.
Using electroporation technique, cells of the A. rhizogenes strain ARqua1 were transformed with either pTPS10::GUS or TPS10-amiR constructs, and cells of Agrobacterium tumefaciens strain GV3101 were transformed with either 35S::TPS10, 35S::tHMGCR, or 35S::FPPS.
Heterologous Expression of TPS10 in Saccharomyces cerevisiae and Leaves of N. benthamiana and Product Identification by GC-MS
For expression in S. cerevisiae the TPS10 encoding sequence was combined in one vector together with the genes from N. benthamiana encoding the preceding biosynthetic enzymes FPPS and HMGCR according to Scheler et al. (2016). All genes were under the control of Gal inducible promoters. S. cerevisiae orotidine-5′-phosphate decarboxylase was used as a selection marker for positively transformed cells. Omitting TPS10 (empty vector) and expression of FPPS and HMGCR alone served as a negative control. The expression vector was transformed in S. cerevisiae strain INVSc1 (genotype MATa his3D1 leu2 trp1-289 ura3-52; ThermoFisher Scientific) and positive clones were selected by plating on uracil-free synthetic medium (1 g·L−1 synthetic drop-out medium without uracil, 6.7 g·L−1 yeast nitrogen base without amino acids, and 20 g·L−1 microagar).
The products were extracted according to Scheler et al. (2016) and determined using liquid-injection GC-MS QP2010 SE (Shimadzu) coupled to an AOC 5000 Sample Injection System and QP2010 Ultra mass spectrometer with electron ionization at 70 eV. Chromatographic separation was performed on a RXI-5il MS capillary column (30 m × 0.25 mm; Shimadzu) using a splitless Injection System and an injection volume of 1 µL. Data analysis was done by device-specific GC-MS Postrun Analysis (Shimadzu).
A. tumefaciens strains containing the respective constructs were cultured according to Schreiber et al. (2019) and diluted to an OD600 of 0.4. The constructs were mixed in a 1:1 ratio before injection into mature leaves of 4-week-old N. benthamiana plants using a needleless syringe. After 3 d, infiltrated leaves were harvested and directly subjected to SPME (SPME Fiber Assembly 65 µm polydimethylsiloxane/divinylbenzene, Supelco 57902-U) coupled with GC-MS (Trace GC Ultra gas chromatograph, ThermoFisher Scientific).The chromatographic separation was performed on a ZB-5ms capillary column (30 m × 0.32 mm; Phenomenex) and the GC oven temperature ramp was as follows: 50°C for 1 min, 50°C to 150°C with 7°C/min, and 150°C to 300°C with 25°C/min. Mass spectrometry was performed at an ionization of 70 eV and m/z was measured in a full scan mode from 50 to 450. The data were analyzed using device-specific Xcalibur software (ThermoFisher Scientific). For comparison with sesquiterpenes produced by yeast expressing MtTPS10 , yeast cultures were also directly subjected to SPME-coupled GC-MS.
Peak Separation and Identification by HPLC and NMR
To yield high amounts of the major product of TPS10 for structural elucidation, the protocol described by Scheler et al. (2016) was used with the following modifications. Cells grown for 24 h at 30°C in a 5 mL yeast peptone dextrose (YPD) preculture medium (2% [w/v] Tryptone and 1% [w/v] yeast extract) containing 2% (w/v) Glc were pelleted, resuspended in 250 mL fresh YPD medium (with 2% [w/v] Glc) in a 1 L flask and grown for 24 h. Transgene expression was induced by resuspending the cells in 250 mL YPD containing 2% (w/v) Gal. Additionally, 20 silica tubes of 1- to 2-cm length were added for product adsorption. After 24 h of cultivation, cultures and silica tubes were extracted separately with 100 and 50 mL pentane, respectively. Finally, both the extracts were mixed and evaporated to full dryness using a rotatory evaporator. The dried extract was redissolved in methanol for separation by liquid chromatography-MS, and a compound representing the major peak was characterized by NMR analysis. For use in bioassay experiments, the concentration of himachalol was determined using a standard curve plot with transfarnesol as the standard and the concentration ranged from 10 to 100 µm. For each concentration, three replicates were measured by GC-MS.
The dried yeast extract was dissolved in 5 mL methanol and was subjected to HPLC-atmospheric-pressure chemical ionization-MS analysis. The peaks were separated using a YMC-Pack ODS-A column (YMC; 150 × 10 mm inner diameter, 120 Å pore size, and 5-µm particle size) and an Agilent 1260 Infinity Quaternary Pump (Agilent), including an autosampler and a fraction collector. For elution, a gradient of 30% (v/v) methanol to 100% over 15 min was used. The flow rate was maintained at 800 µL/min and the column temperature was maintained at 25°C. An Agilent MSD Single 6120 Single Quadrupole mass spectrometer was used to detect the metabolites and was operated in positive mode. The operation parameters for liquid chromatography-atmospheric-pressure chemical ionization were as follows: ion spray voltage was 3,500 V; nebulizing gas was used at 35 pounds-force per square inch gauge; the drying gas temperature was 250°C; and scanning was performed over a mass range of 150–300 m/z.
NMR spectra of the purified compound were recorded on a Varian/Agilent VNMRS 600 NMR spectrometer operating at a proton NMR frequency of 599.829 MHz, using a 5 mm inverse detection cryoprobe. The sample was dissolved in 0.75 mL CD3OD (99.96% deuterium) with a final concentration of ∼20 mmol·L−1. 1H NMR spectra were recorded with a digital resolution of 0.37 Hz/point, a pulse width of 2.2 μs (30°), a relaxation delay of 0.27 s, and an acquisition time of 2.73 s, and the number of transients was 40. Two-dimensional NMR (gDQCOSY, zTOCSY, ROESYAD, gHSQCAD, gHMBCAD) and selective excitation (zTOCSY1D, ROESY1D) spectra were recorded using standard CHEMPACK 8.1 pulse sequences and parameter sets implemented in Varian VNMRJ 4.2A spectrometer software. Mixing times of 0.25 s (ROESYAD), 0.3 s (ROESYAD1D), and 80 ms (zTOCSY, zTOCSY1D) were used. The heteronuclear single-quantum coherence experiment was optimized for 1JCH = 146 Hz with DEPT-like editing and 13C-decoupling during acquisition time. The heteronuclear multiple bond correlation experiment was optimized for a long-range coupling of 8 Hz; a two-step 1JCH filter was used (130–165 Hz). 1H and 13C chemical shifts are referenced to internal tetramethylsilane (0 µL L-1).
Root VOC Collection
Plants were grown in pots for 6 weeks (as described above) and then were separated from the substrate and placed in sterile 500 mL Erlenmeyer flasks containing filter paper. Depending on the root architecture, two to four Sorbstar sticks (Restek GmbH) were inserted in the roots, infected with 105 zoospores per plant, and inoculated for 12 h for VOC release. After 12 h, Sorbstar sticks were collected and stored at –20°C in GC vials until analysis. The root VOCs were measured using a Thermo-Desorption System (TD-20) coupled to a GCMS QP2010 SE (Shimadzu), a 48-sample Autosampler, and a QP2010 Ultra mass spectrometer with Electron Ionization at 70 eV. Chromatographic separation was performed on a RXI-5il MS capillary column (30 m × 0.25 mm; Shimadzu) using split injection mode. The GC Column oven temperature ramp was as follows: 50°C for 1 min, 50°C to 300°C at a rate of 15°C min−1, 300°C to 320°C at a rate of 20°C min−1, and 320°C for 2 min. MS was performed in a full scan mode from 35 to 500 m/z. Data analysis was done by device-specific GCMS Postrun Analysis.
Expression of TPS10-mCherry Fusion in N. benthamiana Protoplasts
The third and fourth leaves of 4-week-old, well developed N. benthamiana plants were used for protoplast isolation according to Sheen (2002). The resulting protoplasts were suspended to 100,000 protoplasts/mL. Polyethylene glycol (PEG)-mediated transformation (Sheen, 2002) was performed using 5–10 µg plasmid DNA for 10,000 protoplasts. Transformed protoplasts were incubated overnight at room temperature followed by monitoring of GFP and chlorophyll fluorescence using a Confocal Laser Scanning Microscope (LSM780, Zeiss).
Root Transformation and Histochemical GUS Staining
A. rhizogenes ARqua1 cells carrying pTPS10::GUS and TPS10-amiR constructs were used to transform roots of M. truncatula cv A17 and cv R108, respectively, as described in Boisson-Dernier et al. (2001). TPS10-promoter activity was visualized using X-Gluc as the substrate. Transformed roots were incubated under vacuum infiltration with the GUS staining solution (100 mm Na2H2PO4, pH 7; 10 mm Na2EDTA, pH 7; 0.5 mm K3Fe(CN)6; 0.5 mm K4Fe(CN)6; 2 mm X-Gluc; and 0.1% [v/v] Triton X-100) for a few minutes to allow penetration of staining solution into the roots and then incubated for 2 h at 37°C. For histological analyses, GUS-stained roots were embedded in PEG 1500 according to Tretner et al. (2008) and cross sectioned. Stained roots and sections of 10 µm thickness were analyzed using a Stemi2000 (Zeiss) and an AxioImager (Zeiss), respectively.
A. euteiches Mycelial Growth and Zoospore Germination Assays
Mycelial growth was monitored by assays performed on CMA plates. For this, a mycelial agar block was put into the middle of a plate and incubated for 1 week. Different concentrations of extracts from yeast expressing MtTPS10 were applied at one edge. Extract from yeast expressing the empty vector and pentane served as controls. For each concentration, at least three biological repeats were made and the photographs were taken when the mycelia in control plates were fully grown. For zoospore germination assays, extract from MtTPS10-expressing yeast was diluted with swamp water to obtain different concentrations. Then, 100 µL of zoospores (∼7,000 zoospores) were added to 100 µL of diluted extracts. After incubation for 1 h in the dark, 100 µL of the suspension was added to CMA plates and incubated for 24 h in the dark. Pictures were taken with a Nikon Macroscope AZ100 multizoom (Nikon Instruments).
Statistical Analyses
All experiments were done with at least three independent biological replicates. Depending on the experiment, either a one-factorial or a two-factorial ANOVA was performed using R software (R Core Team; https://www.r-project.org/). Detailed descriptions of statistical tests performed and their p-values are provided in the figure legends.
Accession Numbers
Normalized microarray data from this article can be found in the Arrayexpress database (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-7286.
Supplemental Data
The following supplemental information is available:
Supplemental Figure S1. Comparative analysis of transcript accumulation in roots of M. truncatula seedlings after treatment with A. euteiches zoospores.
Supplemental Figure S2. Phylogenetic tree of MtTPS gene families and transcript levels of genes closely related to MtTPS10.
Supplemental Figure S3. Transcript accumulation of stress-responsive genes in M. truncatula roots.
Supplemental Figure S4. MS and NMR spectra of himachalol.
Supplemental Figure S5. Tnt1 insertion into the gene encoding MtTPS10 and phenotype of BG and tps10 mutant plants without and with infection by A. euteiches.
Supplemental Figure S6. Transcript levels of MtTPS10-related genes (Medtr5g062230 [MtTPS5] and Medtr6g008560) in roots of tps10 mutant plants and roots expressing an amiR targeting MtTPS10.
Supplemental Figure S7. Products of MtTPS10 inhibit mycelial growth of A. euteiches.
Supplemental Table S1. NMR data of himachalol.
Supplemental Table S2. Primers used for identification of Tnt1 insertion into MtTPS10.
Supplemental Table S3. Primers used for RT-qPCR.
Supplemental Table S4. Primers used for DNA constructs involving MtTPS10 (Medtr5g073200).
Supplemental Dataset S1. Differentially regulated genes in roots of seedlings and adult plants 2 hpi of infection with A. euteiches.
Acknowledgments
We thank Hagen Stellmach (Leibniz Institute of Plant Biochemistry) for help in protoplast transformation and Sylvestre Marillonnet (Leibniz Institute of Plant Biochemistry) for help in Golden Gate cloning and for providing vectors. Christoph Jacquet (University of Toulouse) and Holger Deising (University of Halle) are acknowledged for providing the A. euteiches strain ATCC201684 and for help in culturing C. trifolii, respectively. Franziska Krajinski-Barth (University of Leipzig) is acknowledged for providing vectors containing the amiR backbone. We thank Anja Ehrlich (Leibniz Institute of Plant Biochemistry) and Andrej Frolov (Leibniz Institute of Plant Biochemistry) for their support in purification of himachalol from yeast extracts.
Footnotes
This work was supported by a grant from the Leibniz Association (SAW-PAKT “Chemical Communication in the Rhizosphere”) and by the BRAVE project funded by the ERASMUS MUNDUS Action 2 program of the European Union.
Articles can be viewed without a subscription.
References
- Agrawal P, Frahm A, Schneider M (1992) Structure establishment by two-dimensional NMR spectrocopy of the product obtained in the reaction of MCPBA and himachalol and revised 1H NMR assignments for himachalol. Magn Reson Chem 30: 1079–1083 [Google Scholar]
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Arimura G, Garms S, Maffei M, Bossi S, Schulze B, Leitner M, Mithöfer A, Boland W (2008) Herbivore-induced terpenoid emission in Medicago truncatula: Concerted action of jasmonate, ethylene and calcium signaling. Planta 227: 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis Y, Bonhomme M, Lafitte C, Huguet S, Balzergue S, Dumas B, Jacquet C (2015) Transcriptome analysis highlights preformed defences and signalling pathways controlled by the prAe1 quantitative trait locus (QTL), conferring partial resistance to Aphanomyces euteiches in Medicago truncatula. Mol Plant Pathol 16: 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badreddine I, Lafitte C, Heux L, Skandalis N, Spanou Z, Martinez Y, Esquerré-Tugayé M-T, Bulone V, Dumas B, Bottin A (2008) Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches. Eukaryot Cell 7: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki-García S. (1968) Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu Rev Microbiol 22: 87–108 [DOI] [PubMed] [Google Scholar]
- Bécard G, Fortin JA (1988) Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol 108: 211–218 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14: 695–700 [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Wilkinson RC, Gimenez-Ibanez S, Findlay K, Coffey MD, Zipfel C, Rathjen JP, Kamoun S, Schornack S (2011) The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen phytophthora infestans in Nicotiana benthamiana. PLoS One 6: e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Colditz F, Nyamsuren O, Niehaus K, Eubel H, Braun H-P, Krajinski F (2004) Proteomic approach: Identification of Medicago truncatula proteins induced in roots after infection with the pathogenic oomycete Aphanomyces euteiches. Plant Mol Biol 55: 109–120 [DOI] [PubMed] [Google Scholar]
- Collier SM, Moffett P (2009) NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci 14: 521–529 [DOI] [PubMed] [Google Scholar]
- d’Erfurth I, Cosson V, Eschstruth A, Lucas H, Kondorosi A, Ratet P (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34: 95–106 [DOI] [PubMed] [Google Scholar]
- Declerck S, Strullu D, Fortin A (2005) In vitro culture of mycorrhizas, Vol Vol 4 Springer-Verlag Berlin Heidelberg, Berlin [Google Scholar]
- Devers EA, Teply J, Reinert A, Gaude N, Krajinski F (2013) An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol 13: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicke M, Baldwin IT (2010) The evolutionary context for herbivore-induced plant volatiles: Beyond the ‘cry for help’. Trends Plant Sci 15: 167–175 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E, Gershenzon J (2004) Biochemistry of plant volatiles. Plant Physiol 135: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Klempien A, Muhlemann JK, Kaplan I (2013) Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol 198: 16–32 [DOI] [PubMed] [Google Scholar]
- Erb M, Lenk C, Degenhardt J, Turlings TCJ (2009) The underestimated role of roots in defense against leaf attackers. Trends Plant Sci 14: 653–659 [DOI] [PubMed] [Google Scholar]
- Fabro G, Steinbrenner J, Coates M, Ishaque N, Baxter L, Studholme DJ, Körner E, Allen RL, Piquerez SJ, Rougon-Cardoso A, et al. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog 7: e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garms S, Köllner TG, Boland W (2010) A multiproduct terpene synthase from Medicago truncatula generates cadalane sesquiterpenes via two different mechanisms. J Org Chem 75: 5590–5600 [DOI] [PubMed] [Google Scholar]
- Gaulin E, Jacquet C, Bottin A, Dumas B (2007) Root rot disease of legumes caused by Aphanomyces euteiches. Mol Plant Pathol 8: 539–548 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Cox MM, Bede JC, Inoue K, Alborn HT, Tumlinson JH, Korth KL (2005) Lepidopteran herbivory and oral factors induce transcripts encoding novel terpene synthases in Medicago truncatula. Arch Insect Biochem Physiol 58: 114–127 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R. (1999) Phytoalexins: What have we learned after 60 years? Annu Rev Phytopathol 37: 285–306 [DOI] [PubMed] [Google Scholar]
- Hughes TJ, Grau CR (2014) Aphanomyces root rot or common root rot of legumes. https://www.apsnet.org/edcenter/disandpath/oomycete/pdlessons/Pages/Aphanomyces.aspx (March 1, 2019)
- Isayenkov S, Mrosk C, Stenzel I, Strack D, Hause B (2005) Suppression of allene oxide cyclase in hairy roots of Medicago truncatula reduces jasmonate levels and the degree of mycorrhization with Glomus intraradices. Plant Physiol 139: 1401–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulneau V, Cazaux M, Wong Sak Hoi J, Fournier S, Esquerré-Tugayé MT, Jacquet C, Dumas B (2010) Host and nonhost resistance in Medicago-Colletotrichum interactions. Mol Plant Microbe Interact 23: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Junker RR, Tholl D (2013) Volatile organic compound mediated interactions at the plant-microbe interface. J Chem Ecol 39: 810–825 [DOI] [PubMed] [Google Scholar]
- Lange BM, Rujan T, Martin W, Croteau R (2000) Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA 97: 13172–13177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang Y, Hu X, Shen X, Ma L, Su Z, Wang T, Dong J (2011) Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol 11: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 50: 47–65 [DOI] [PubMed] [Google Scholar]
- Loreto F, Schnitzler J-P (2010) Abiotic stresses and induced BVOCs. Trends Plant Sci 15: 154–166 [DOI] [PubMed] [Google Scholar]
- Maimone TJ, Baran PS (2007) Modern synthetic efforts toward biologically active terpenes. Nat Chem Biol 3: 396–407 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Mrosk C, Forner S, Hause G, Küster H, Kopka J, Hause B (2009) Composite Medicago truncatula plants harbouring Agrobacterium rhizogenes-transformed roots reveal normal mycorrhization by Glomus intraradices. J Exp Bot 60: 3797–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda DA. (2010) Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett 584: 2965–2973 [DOI] [PubMed] [Google Scholar]
- Nars A, Lafitte C, Chabaud M, Drouillard S, Mélida H, Danoun S, Le Costaouëc T, Rey T, Benedetti J, Bulone V, et al. (2013) Aphanomyces euteiches cell wall fractions containing novel glucan-chitosaccharides induce defense genes and nuclear calcium oscillations in the plant host Medicago truncatula. PLoS One 8: e75039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MT, Zhong Y, Dai X, Wang S, Zhao P (2014) Comparative genomic and transcriptomic analysis of terpene synthases in Arabidopsis and Medicago. IET Syst Biol 8: 146–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontin M, Bottini R, Burba JL, Piccoli P (2015) Allium sativum produces terpenes with fungistatic properties in response to infection with Sclerotium cepivorum. Phytochemistry 115: 152–160 [DOI] [PubMed] [Google Scholar]
- Prakash O, Roy R, Kulshreshtha D (1988) Two-dimensional NMR analysis of himachalol and its correlation with other sesquiterpenoids of the himachalane series. Magn Reson Chem 26: 47–50 [Google Scholar]
- Rey T, Chatterjee A, Buttay M, Toulotte J, Schornack S (2015) Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytol 206: 497–500 [DOI] [PubMed] [Google Scholar]
- Rey T, Laporte P, Bonhomme M, Jardinaud M-F, Huguet S, Balzergue S, Dumas B, Niebel A, Jacquet C (2016) MtNF-YA1, a central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Front Plant Sci 7: 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R. (2008) Medicago truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: Past, present and future. Funct Plant Biol 35: 253–264 [DOI] [PubMed] [Google Scholar]
- Sapir-Mir M, Mett A, Belausov E, Tal-Meshulam S, Frydman A, Gidoni D, Eyal Y (2008) Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol 148: 1219–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler U, Brandt W, Porzel A, Rothe K, Manzano D, Božić D, Papaefthimiou D, Balcke GU, Henning A, Lohse S, et al. (2016) Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat Commun 7: 12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber T, Prange A, Hoppe T, Tissier A (2019) Split-TALE: A TALE-based two-component system for synthetic biology applications in planta. Plant Physiol 179: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi R, Lee S-M, Ryu C-M (2018) Microbe-induced plant volatiles. New Phytol 220: 684–691 [DOI] [PubMed] [Google Scholar]
- Sheen J. (2002) A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/ (March 1, 2019)
- Shulaev V, Cortes D, Miller G, Mittler R (2008) Metabolomics for plant stress response. Physiol Plant 132: 199–208 [DOI] [PubMed] [Google Scholar]
- Sohrabi R, Huh J-H, Badieyan S, Rakotondraibe LH, Kliebenstein DJ, Sobrado P, Tholl D (2015) In planta variation of volatile biosynthesis: An alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell 27: 874–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Arnaud M, Hamel C, Vimard B, Caron M, Fortin JA (1996) Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res 100: 328–332 [Google Scholar]
- Stratowa C. (2018) xps: Processing and analysis of Affymetrix oligonucleotide arrays including exon arrays, whole genome arrays and plate arrays. R package version 1.40.0. https://rdrr.io/bioc/xps/ (March 1, 2019)
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tang H, Krishnakumar V, Bidwell S, Rosen B, Chan A, Zhou S, Gentzbittel L, Childs KL, Yandell M, Gundlach H, et al. (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretner C, Huth U, Hause B (2008) Mechanostimulation of Medicago truncatula leads to enhanced levels of jasmonic acid. J Exp Bot 59: 2847–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattekkatte A, Garms S, Brandt W, Boland W (2018) Enhanced structural diversity in terpenoid biosynthesis: Enzymes, substrates and cofactors. Org Biomol Chem 16: 348–362 [DOI] [PubMed] [Google Scholar]
- Werner S, Sugui JA, Steinberg G, Deising HB (2007) A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Microbe Interact 20: 1555–1567 [DOI] [PubMed] [Google Scholar]
- Werner S, Engler C, Weber E, Gruetzner R, Marillonnet S (2012) Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs 3: 38–43 [DOI] [PubMed] [Google Scholar]
- Wink M. (2003) Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64: 3–19 [DOI] [PubMed] [Google Scholar]
- Wink M. (2018) Plant secondary metabolites modulate insect behavior-steps toward addiction? Front Physiol 9: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Oldroyd GED (2017) Plant signalling in symbiosis and immunity. Nature 543: 328–336 [DOI] [PubMed] [Google Scholar]