Dear Editor,
The measurement of malondialdehyde (MDA) content has long been used as a lipid peroxidation marker in studies related to oxidative stress and redox signaling, particularly in those studies focused on plant responses to abiotic and biotic stresses. A search for “malondialdehyde” and “plant*” in Scopus retrieves 9,000 publications from the last decade, with 1,221 of these published in 2018 (Scopus database, 2019). Unfortunately, however, there are major pitfalls in some of the current applications of this lipid peroxidation marker, including both (1) methodological aspects (a significant part of the scientific community still measures this compound using inadequate methodology), and (2) misinterpretation of results (mainly related to a misconception of oxidative stress, oxidative damage, and redox regulation).
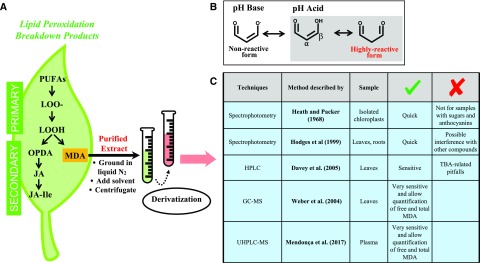
Life on our planet changed completely with the development of oxygenic photosynthesis by cyanobacteria ∼2.4 billion years ago (Harel et al., 2015). Higher oxygen tensions in the atmosphere brought about new opportunities for the diversification of complex life forms based on aerobic metabolism. Aerobic life implicitly afforded reactive oxygen species (ROS) a key role in the sophistication of acclimation mechanisms to a number of stresses, both of abiotic and biotic origin. Although ROS are part of aerobic life and play an essential role in stress acclimation and the regulation of plant development from germination to senescence, they are still today considered harmful molecules because of their high reactivity. Although ROS are indeed highly reactive and rapidly oxidize other target molecules leading to lipid peroxidation among many other biochemical reactions, it is important to keep in mind that both ROS production and lipid peroxidation are an intrinsic part of aerobic life and essential features of plant life. It is also important to bear in mind that lipid peroxidation not only is triggered by ROS, but can also result from increased lipoxygenase activity (e.g. as a result of a pathogen invasion or wounding attack). Therefore, both enzymatic and nonenzymatic lipid peroxidation processes may lead to the formation of MDA and other lipid peroxidation products in plants, such as jasmonates, which are an essential component of stress tolerance (Weber et al., 2004; Farmer and Mueller, 2013; Fig. 1A).
Figure 1.
Advantages and limitations of currently available methods for the measurement of MDA in plants. The most common extraction and analytical procedures for the analysis of MDA in plant samples are shown, indicating their advantages and limitations. (A) MDA is a secondary product of lipid peroxidation. (B) MDA exists in several different forms in aqueous solutions due to its pH-dependent tautometric chemical property. At higher pH than its pKa of 4.46, the enolic anion dominates, displaying a low chemical reactivity, whereas at lower pH (stress conditions), MDA appears in equilibrium between its protonated enol (⍺-β-unsaturated carbonyl) aldehyde form and the dialdehyde form: the high-reactive forms. (C) This chemical reactivity represents the basis for many MDA determination methods, which derivatize MDA prior to analysis and measure the complex formed by spectrophotometry, HPLC, or GC (in the latter two cases usually coupled to MS). LOOH, lipid hydroperoxide; OPDA, 12-oxo-phytodienoic acid; UHPLC, ultrahigh-performance liquid chromatography.
Chemically, MDA is a small and reactive organic molecule that occurs ubiquitously among eukaryotes, formed by three carbon molecules with two aldehyde groups at the carbon 1 and carbon 3 positions. MDA exists in different forms in aqueous solutions due to its pH-dependent tautomeric chemical property. At higher pH than its pKa of 4.46, the dominate form is the enolic anion, which displays low chemical reactivity. However, at lower pH (expected under oxidative stress conditions), MDA appears in equilibrium between its protonated enol (α-β-unsaturated carbonyl) aldehyde and the dialdehyde form (Fig. 1B). These tautomers produced in acidic pH are chemically reactive, and MDA and other molecules with α-β-unsaturated carbonyl groups are known, including reactive electrophile species and reactive carbonyl species. Reactive electrophile species are known for the electrophilic character of the β-carbon that reacts with an electron-donor (nucleophilic) atom, whereas reactive carbonyl species are known for the high reactivity of the ⍺-β-unsaturated carbonyl groups resulting from the peroxidation of triunsaturated fatty acids, mostly linoleic acid, which is the in vivo source of up to 75% of MDA in Arabidopsis (Arabidopsis thaliana) leaves (Weber et al., 2004; Farmer and Mueller, 2013). Formation of MDA can be induced nonenzymatically by ROS or enzymatically by the activity of lipoxygenase (Farmer and Mueller, 2013). In both cases, the quantification of primary lipid hydroperoxide products is difficult due to their instability and reactivity. For this reason, quantification of lipid peroxidation is usually estimated by measuring the concentration of secondary oxidation products derived from these initial hydroperoxides (Davey et al., 2005), which are mostly aldehydes, such as MDA (Fig. 1C). Interestingly, both free and bound MDA are found in plant samples and a significant increase in free MDA was observed in Arabidopsis leaves under oxidative stress conditions (Weber et al., 2004).
Several methods have been developed to assess MDA content using derivatization coupled with various separation and/or detection methods, which all take advantage of the electrophilic character of the MDA molecule. These methods include gas chromatography (GC), liquid chromatography (LC; with either UV or fluorescence detection) and mass spectrometry (MS; Fig. 1C). The thiobarbituric acid-reactive substances (TBARS) assay, first described five decades ago, is still the most commonly used method worldwide for both plant and animal samples. Heath and Packer (1968) described a very easy and quick method to estimate MDA generated by the polyunsaturated fatty acid (PUFA)-photoperoxidation process in isolated chloroplasts. This method is based on the electrophilic character of MDA, which binds readily at low pH and elevated temperature to the nucleophilic site of thiobarbituric acid (TBA). After extraction, plant extracts containing MDA are incubated with TBA at high temperatures, yielding an MDA(TBA)2 adduct of reddish color and green fluorescence with an absorbance maximum at 532 nm (Fig. 1). However, this rapid approach is particularly problematic when applied to plant samples that contain carbohydrates and anthocyanins (or other molecules rich in carbonyl groups), which are also susceptible to nucleophilic attack by TBA. These “artifacts” interfere with the absorbance measurements at 532 nm, resulting in an overestimation of MDA content (Taulavuori et al., 2001).
Various approaches have been proposed to solve the problems with the Heath and Packer (1968) method, mainly (1) subtraction of the nonspecific absorbance by spectrophotometry in the TBARS assay (Hodges et al., 1999) and (2) the resolution of the resulting derivatization complex using HPLC or GC coupled or not with MS detection (Weber et al., 2004; Davey et al., 2005; Mendonça et al., 2017). Due to its simplicity and low cost, the Hodges et al. (1999) spectrophotometric method has since been extensively used. It provides an improved estimate of endogenous MDA in plant samples containing sugars, anthocyanins, and other interfering compounds, since it corrects a limitation of the Heath and Packer (1968) method by including a control assay solution without TBA and subtracting the absorbance of this at 532 nm from that of assay samples with TBA. Indeed, in addition to Hodges et al. (1999), Taulavuori et al. (2001) also compared the available spectrophotometric methods and clearly showed that MDA content is overestimated as a result of the absence of this correction in several plant tissues. However, because even such improved spectrophotometric measurements are not completely free of artifacts, alternative HPLC and GC techniques were introduced to specifically measure the MDA adducts formed, either with TBA or phenylhydrazines. The influence of possible interfering compounds in the spectrophotometric TBARS assay, such as high concentrations of stress-inducible compounds, was improved using reversed-phase HPLC coupled with UV detection, thus enhancing the specificity of MDA detection (Davey et al., 2005). However, specificity problems might still occur using this method, since other adducts formed could have the same retention time and be indistinguishable from MDA(TBA)2. One way to overcome this limitation is through the use of MS to identify peaks generated by GC after derivatization with pentafluorophenylhydrazine (Weber et al., 2004). Moreover, additional specific methods using other derivatizing chemicals to overcome the limitations of the TBA assay are currently available, among which the most sophisticated method (not yet applied to plant samples) comprises the sensitive and selective measurement of free and total plasmatic MDA using derivatization with 2,4-dinitrophenylhydrazine in ultra-HPLC (UHPLC) coupled to MS (Mendonça et al., 2017). This method solves the types of interference present in the aforementioned methods and can be used with any type of sample. Therefore, from a methodological point of view, two key aspects are essential considerations in MDA determination: (1) what type of sample will be analyzed and (2) what resources are available. The first method described by Heath and Packer (1968) is valid as a proxy of lipid peroxidation if isolated chloroplasts are used, in which neither sugars nor other interfering molecules are present. However, if the evaluation is of MDA in leaves or other plant tissues, the Heath and Packer (1968) method will overestimate MDA content, which will result not only from MDA-TBA adducts but also from the binding of TBA with other interfering compounds. This can be improved by the use of the Hodges et al. (1999) method to analyze MDA in leaves and other plant samples by spectrophotometry, although this method also has some limitations. Therefore, if the resources are available, it is better to use more specific and selective methods such as HPLC, UHPLC-MS, or GC-MS (Weber et al., 2004; Mendonça et al., 2017).
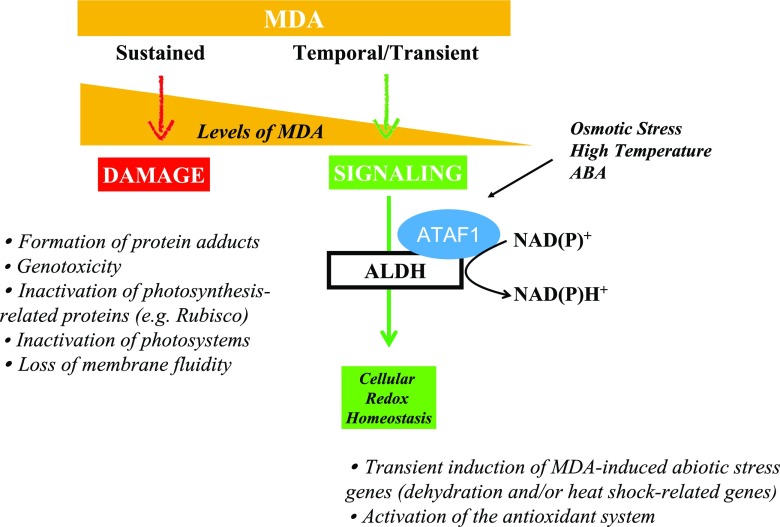
Last but not least is the question of how we interpret MDA results. Since the bulk of MDA derives from the lipid peroxidation of PUFAs in plant membranes in response to oxidative stress (via ROS and/or lipoxygenase), MDA content is widely used as an indicator of damage in plant membranes. This holds true if MDA levels remain high, irreparably modifying proteins and nucleic acids. However, if the elimination of MDA and redox signaling regulation work correctly, MDA increases may represent acclimation processes rather than damage, since MDA can exert a positive role by activating regulatory genes involved in plant defense and development (defense and reproduction) and granting cell protection under oxidative stress conditions. In this respect, it has been suggested that MDA may act as a protection mechanism rather than being an indicator of damage. For instance, in observations of salt-stressed rosemary plants, MDA increased transiently in leaves, an antioxidant system to dissipate ROS was efficiently activated, and no signs of oxidative damage were observed (Tounekti et al., 2011). Figure 2 illustrates how MDA can trigger damage or protection depending on its cellular concentration and dynamics. Genetic evidence suggests that membranes rich in PUFAs act as supramolecular antioxidants that capture ROS, thereby limiting damage to proteins. This process constantly generates lipid fragmentation products including MDA (Schmid-Siegert et al., 2016). Other benefits of MDA relating to the signaling and regulation of essential biological functions (e.g. meristem activity and flower opening) have recently been proposed (Schmid-Siegert et al., 2012; Muñoz et al., 2018). Aldehyde compounds, such as MDA, have indeed been described as both toxic molecules and gene activators (Tagnon and Simeon, 2017). Therefore, the role of these compounds as damagers or protectors depends upon production, scavenging, and signaling modulation and thus relies on the enzymatic activity of aldehyde dehydrogenases (ALDHs; Fig. 2). Under environmental stress or developmental signals, MDA is produced from the lipid peroxidation of PUFAs by ROS attack or the activation of lipoxygenases. When aldehyde levels increase, protein carbonylation occurs, and this may either result in defense signaling if MDA accumulates transiently, or trigger cell death if there is sustained accumulation of MDA and carbonylated proteins accumulate in the cells. ALDH expression is induced to control the level of aldehyde compounds (such as MDA) by oxidizing them to their corresponding carboxylic acids, reestablishing low cellular levels so that they may serve as signals rather than cause harm to the cell (Tagnon and Simeon, 2017). Indeed, ALDHs are also major contributors to cellular redox homeostasis (Missihoun et al., 2018), providing the reducing agent NADPH essential for both the antioxidant activity of the ascorbate-glutathione cycle and photosynthesis (Yalcinkaya et al., 2019). Interestingly, ALDH activity is induced by H2O2 and abscisic acid, and its expression is regulated by the transcription factor Arabidopsis thaliana activating factor 1; hence, ALDH expression is induced by H2O2 (Zhao et al., 2018). Transient increases in MDA can induce abiotic and biotic stress-related genes, as has been specifically shown in Arabidopsis (Weber et al., 2004). However, in heat-stressed plants, sustained increases in MDA accumulation can also modify and inactivate PSII core proteins and Rubisco (Yamauchi et al., 2008; Yamauchi and Sugimoto, 2010). A negative correlation between MDA content and electron transport was described, implying that the function of PSII might be damaged by MDA modification of PSII proteins in heat-stressed plants (Yamauchi et al., 2008;Yamauchhi and Sugimoto, 2010). MDA generation is clearly implicated in the symptoms of environmentally stressed plants, and therefore further studies of MDA accumulation are necessary to better understand the physiological functions of MDA for plant survival under stress.
Figure 2.
MDA in plants: damage or protection? The balance between MDA production, elimination, and signaling is an important feature of redox biology and may determine plant survival under stress. ABA, abscisic acid; ATAF1, Arabidopsis thaliana activating factor 1.
In conclusion, there is an urgent need in the scientific community to reduce common methodological pitfalls in the measurement of MDA content in plants and to improve the interpretation of results. It is essential that the scientific community as a whole operate at a level of precision far beyond that of the Heath and Packer (1968) method, evidence of whose inadequate use can be found in several recent examples in the literature, and carefully consider the optimal methodological approach for every scientific aim. Furthermore, it is very important that the MDA content in studies related to stress acclimation is correctly interpreted as a marker of lipid peroxidation, and that its limitations are considered alongside its advantages. In summary, to accurately estimate MDA content in plant samples and correctly interpret the results, we recommend that researchers do the following:
Run a pilot experiment on a reduced sample size to test for the most accurate method to estimate MDA in line with your research goals. For example, the basic method by Heath and Packer (1968) can be a proxy for estimating MDA in isolated chloroplasts, but not for measuring MDA in complex plant samples.
Choose the optimal methodology for your samples. Do not simply run the same protocol, as not all methods are appropriate for all sample types. Chromatography methods are especially recommended using adequate extraction processes, whereby the amount of material and solvent volume are adjusted in order to detect low MDA concentrations.
Whenever possible, quantify free and bound MDA if your experimental design needs it by using sensitive and selective methods beyond the TBARS assay.
Always keep in mind that whatever method is used, the determination of MDA will be an estimation of what the plant organ really contains and additional parameters must be used to confirm your results.
Interpret your results with caution, taking into account that spectrophotometric methods have some limitations, whereas if you need a very precise determination it is better to use more specific and sensitive techniques (in particular chromatography coupled with MS).
References
- Davey MW, Stals E, Panis B, Keulemans J, Swennen RL (2005) High-throughput determination of malondialdehyde in plant tissues. Anal Biochem 347: 201–207 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol 64: 429–450 [DOI] [PubMed] [Google Scholar]
- Harel A, Karkar S, Cheng S, Falkowski PG, Bhattacharya D (2015) Deciphering primordial cyanobacterial genome functions from protein network analysis. Curr Biol 25: 628–634 [DOI] [PubMed] [Google Scholar]
- Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198 [DOI] [PubMed] [Google Scholar]
- Hodges DM, DeLong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611 [DOI] [PubMed] [Google Scholar]
- Mendonça R, Gning O, Di Cesaré C, Lachat L, Bennett NC, Helfenstein F, Glauser G (2017) Sensitive and selective quantification of free and total malondialdehyde in plasma using UHPLC-HRMS. J Lipid Res 58: 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missihoun TD, Kotchoni SO, Bartels D (2018) Aldehyde dehydrogenases function in the homeostasis of pyridine nucleotides in Arabidopsis thaliana. Sci Rep 8: 2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P, Briones M, Munné-Bosch S (2018) Photoinhibition and photoprotection during flower opening in lilies. Plant Sci 272: 220–229 [DOI] [PubMed] [Google Scholar]
- Schmid-Siegert E, Loscos J, Farmer EE (2012) Inducible malondialdehyde pools in zones of cell proliferation and developing tissues in Arabidopsis. J Biol Chem 287: 8954–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Siegert E, Stepushenko O, Glauser G, Farmer EE (2016) Membranes as structural antioxidants: Recycling of malondialdehyde to its source in oxidation -sensitive chloroplast fatty acids. J Biol Chem 291: 13005–13013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopus database (2019) www.scopus.com (February 28, 2019)
- Tagnon MD, Simeon KO (2017) Aldehyde dehydrogenases may modulate signaling by lipid peroxidation-derived bioactive aldehydes. Plant Signal Behav 12: e1387707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulavuori E, Hellström E-K, Taulavuori K, Laine K (2001) Comparison of two methods used to analyse lipid peroxidation from Vaccinium myrtillus (L.) during snow removal, reacclimation and cold acclimation. J Exp Bot 52: 2375–2380 [DOI] [PubMed] [Google Scholar]
- Tounekti T, Vadel AM, Oñate M, Khemira H, Munné-Bosch S (2011) Salt-induced oxidative stress in rosemary plants: Damage or protection? Environ Exp Bot 71: 298–305 [Google Scholar]
- Weber H, Chételat A, Reymond P, Farmer EE (2004) Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J 37: 877–888 [DOI] [PubMed] [Google Scholar]
- Yalcinkaya T, Uzilday B, Ozgur R, Turkan I (2019) The roles of reactive carbonyl species in induction of antioxidant defence and ROS signalling in extreme halophytic model Eutrema parvulum and glycophytic model Arabidopsis thaliana. Environ Exp Bot 160: 81–91 [Google Scholar]
- Yamauchi Y, Sugimoto Y (2010) Effect of protein modification by malondialdehyde on the interaction between the oxygen-evolving complex 33 kDa protein and photosystem II core proteins. Planta 231: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Furutera A, Seki K, Toyoda Y, Tanaka K, Sugimoto Y (2008) Malondialdehyde generated from peroxidized linolenic acid causes protein modification in heat-stressed plants. Plant Physiol Biochem 46: 786–793 [DOI] [PubMed] [Google Scholar]
- Zhao J, Missihoun TD, Bartels D (2018) The ATAF1 transcription factor is a key regulator of aldehyde dehydrogenase 7B4 (ALDH7B4) gene expression in Arabidopsis thaliana. Planta 248: 1017–1027 [DOI] [PubMed] [Google Scholar]