Genetic analysis revealed the role of pleiotropic gene action in shaping the root system architecture during domestication of common bean (Phaseolus vulgaris L.).
Abstract
Roots have been omitted from previous domestication analyses owing mostly to their subterranean nature. We hypothesized that domestication-associated changes in common bean (Phaseolus vulgaris) roots were due to direct selection for some aboveground traits that also affect roots, and to indirect selection of root traits that improved aboveground plant performance. To test this hypothesis, we compared the root traits of wild and domesticated accessions and performed a multistep quantitative trait locus (QTL) analysis of an intra-Andean recombinant inbred family derived from a landrace and a wild accession. Multivariate analysis of root traits distinguished wild from domesticated accessions and showed that seed weight affects many root traits of young seedlings. Sequential and methodical scanning of the genome confirmed the significant effect of seed weight on root traits and identified QTLs that control seed weight, root architecture, shoot and root traits, and shoot traits alone. The root domestication syndrome in the common bean was associated with genes that were directly selected to increase seed weight but had a significant effect on early root growth through a developmental pleiotropic effect. The syndrome was also associated with genes that control root system architecture and that were apparently the product of indirect selection.
Domestication produces a biological group that displays clear phenotypic and genetic differences from its wild ancestor (Garcia et al., 1997; Ross-Ibarra et al., 2007). However, plant root traits have not yet been considered as part of the “domestication syndrome” as described by Harlan (1992). A change in edaphic conditions imposed by early farmers on the ancestral wild populations may have imposed selection pressure resulting in allele frequency changes of root-specific genes. Also, functional equilibrium between root and shoot might have indirectly altered the root system due to direct selection of shoot traits.
Quantitative trait locus (QTL) analysis has been used effectively in the identification of genes associated with domestication (Burger et al., 2008; Olsen and Wendel, 2013), and it has also led to their isolation and molecular characterization (Doebley et al., 1997; Wang et al., 2005). QTL analysis using soil-free root phenotyping platforms has also identified genetic determinants of quantitative variation in root system architecture (RSA; Clark et al., 2013; Ron et al., 2013; Topp et al., 2013; Burton et al., 2014; Liang et al., 2014; Zurek et al., 2015; Ye et al., 2018). This work has led to the identification of genes that control root traits, some of which were previously identified through mutant analysis (Mouchel et al., 2004; Sergeeva et al., 2006). Although some studies have included progenies with wild accessions (Prince et al., 2015), as far as we know, none have directly addressed genes controlling root traits associated with domestication.
Phaseolus vulgaris, the common bean, was domesticated independently in Mesoamerica and the Andes ∼8,000–10,000 years ago (Kwak and Gepts, 2009; Rossi et al., 2009). The Mesoamerican and Andean gene pools are separated by partial reproductive isolation and exhibit significant inter- and intragene pool variation. Comparative analysis of wild and domesticated accessions previously identified several aboveground domestication-related traits, and genetic analysis of these traits has suggested the presence of a few major QTLs with large effects (Koinange et al., 1996).
The main objective of this study was to determine both the extent to which domestication had modified root traits of common bean and the genetic complexity of those traits through QTL analysis in an intra-Andean segregating progeny. We conducted a comprehensive phenotypic analysis using multivariate statistics and genetic characterization of 28 root and shoot traits to evaluate effects of domestication on RSA of common bean.
RESULTS
Comparative Root System Analysis of Wild and Domesticated Accessions of the Common Bean
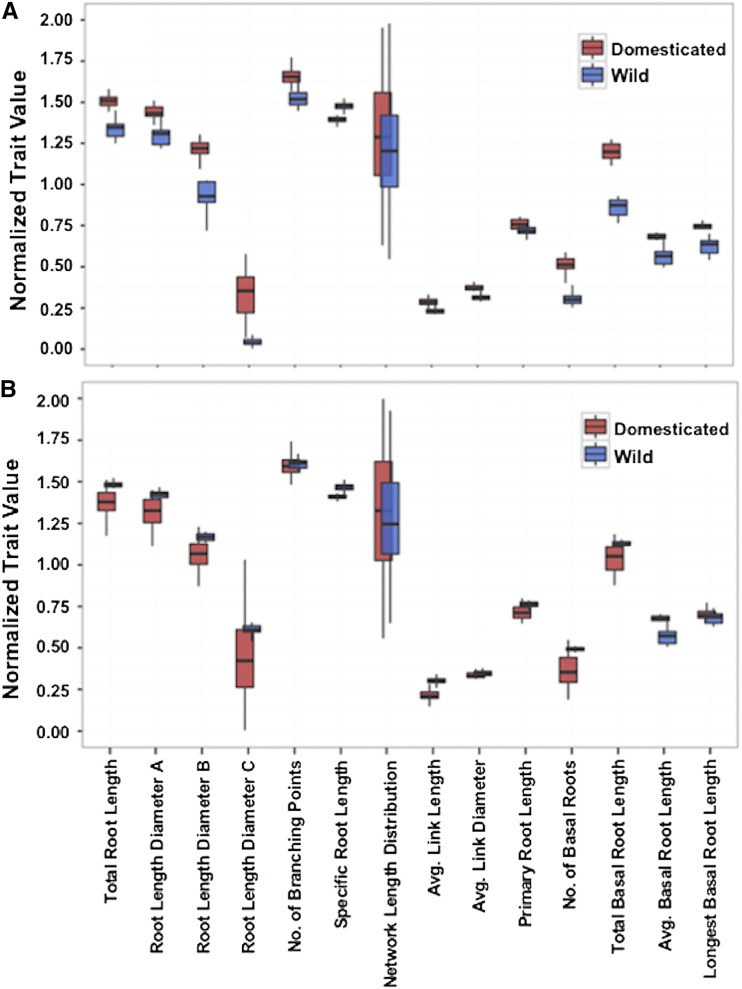
Statistical comparisons of wild and cultivated accessions revealed highly significant differences (P < 0.01) in the RSA of young seedlings (Fig. 1A; Supplemental Table S1). Cultivated accessions have significantly larger root systems than their wild counterparts and display marked differences in the distribution of root sizes (Supplemental Table S1). Domesticated beans have primary root lengths that are 1.27 times longer than those of wild accessions, but the increased number and length of basal roots significantly lowers the root apical dominance of domesticated beans. The primary root length of wild accessions is on average 47% of the total basal root length, whereas this relative proportion drops to 13% in domesticated accessions.
Figure 1.
Box plots of root trait of wild and domesticated accessions of the common bean. A and B, Trait values were normalized as log100 using three replicas of each of the 16 wild and 28 cultivated accessions, before (A) and after (B) correcting for the seed weight covariate.
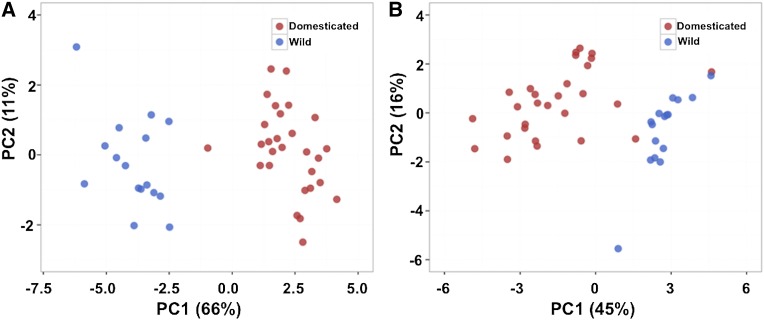
Principal component analysis (PCA) of root traits clearly shows that wild and domesticated genotypes form distinct clusters, suggesting selective forces applied during domestication significantly altered size and RSA (Fig. 2A; Supplemental Table S2). Principal component 1 (PC1) explained 66% of the variation, PC2 11%, and four additional PCs explained most of the remaining (20%) variation (Supplemental Table S2). All root traits contributed to variation explained by PC1, but PC2 had significant contributions from fewer traits (Supplemental Fig. S1). In contrast to other traits, basal root growth had a negligible contribution toward PC2. A negative correlation between specific root length and the other PC1 traits could be explained by the fact that as roots grow their spatial arrangement changes, resulting in a sparser distribution. This is also in agreement with the box plots pattern where the specific root length of the wild accessions has a pattern opposite to the other traits (Fig. 1). Negative correlations for other traits were observed in the remaining principal components, and these likely explain the hidden genetic variation contributed by the wild genotypes.
Figure 2.
PCA using root trait means in wild and domesticated accessions of the common bean. A and B, The analysis was performed before (A) and after (B) the 3inclusion of seed weight as a covariate.
PCA results could be biased because seed weight was the main target of selection during domestication, and we have shown that seed reserves can significantly influence early heterotrophic growth (Singh et al., 2017). Thus, seed weight differences between wild and domesticated accessions could explain some of the root growth differences between the accessions as suggested by the significant phenotypic correlations between seed weight and many root traits, and root size traits in particular (Supplemental Tables S3 and S4). To address this point, we reanalyzed the trait differences through covariate analysis to remove the dependency of root trait variation on seed weight.
Domesticated accessions had lower root trait values than those of wild accessions after they were adjusted for seed weight (Fig. 1B). For example, total root length was reduced on average by 36% in the domesticated accessions. Overall, seed weight can significantly affect several root traits and should be considered as a covariate during further genetic analysis. Results from PCA using this covariate showed that the wild and domesticated clusters remained distinct from each other (Fig. 2B), although their positions were inverted along the PC1 axis. Nine PCs explained 99% of the variation, and unlike the previous PCA, PC1 explained only 45% of the variation. PC2 and PC3 explained ∼16% and 13% of the variation, respectively, while variation was evenly distributed among the remaining components (Supplemental Table S2). These results showed that domestication significantly altered various aspects of RSA because most root traits, without or with Seed Weight as covariate, show a relatively high degree of correlation as part of PC1, and that wild and cultivated accessions form distinct clusters along this axis. Moreover, genes controlling the targeted traits may be involved in adaptation to distinct edaphic conditions.
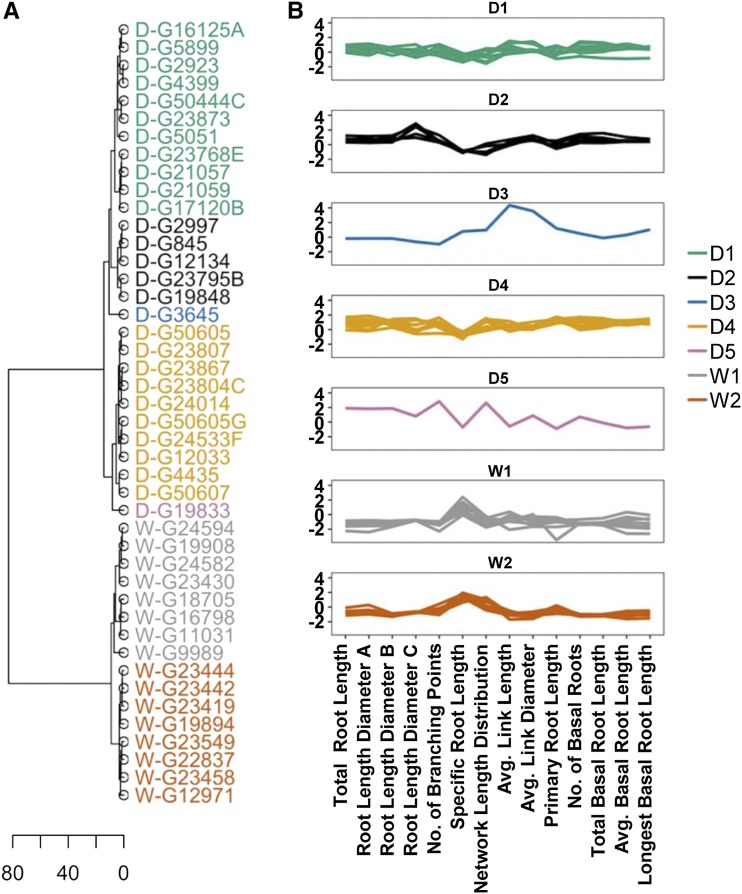
We further explored relationships between wild and domesticated accessions with hierarchical clustering which, in contrast to PCA, analyzes the extent of similarities among entries. This analysis detected a total of seven subgroups (threshold tree height = 8); five domesticated clusters (D1, D2, D3, D4, D5) and two wild clusters (W1, W2; Fig. 3A). Clusters D1 and D2 contained each Andean and Mesoamerican accessions, whereas cluster D4 had 10 Andean domesticated genotypes (Supplemental Table S5). Clusters D3 and D5 represented only one accession each from the Mesoamerican and Andean gene pools, respectively. Wild clusters W1 and W2 had Andean and Mesoamerican accessions. This analysis distinguished wild from domesticated accession and, to a limited extent, discriminated between gene pools. A parallel coordinate plot of normalized trait means showed unique root trait patterns that distinguish each cluster (Fig. 3B). An additional clustering analysis using seed weight as covariate revealed seven distinct groups (Supplemental Fig. S2a; Supplemental Table S5); six of them contained domesticated accessions and one contained only wild accessions. The clustering pattern was highly similar to the previous one, with the exception of cluster D4, which included one domesticated accession from two different previous clusters and a wild accession. In addition, clusters D3, D5, and D6 displayed gene pool specificity (Supplemental Table S5). A large amount of significant trait variation was apparent within and across clusters (Supplemental Fig. S2b).
Figure 3.
Hierarchical clustering of wild (W-) and domesticated (D-) accessions from the Mesoamerican and Andean gene pools based on 14 root traits. A, Dendrogram showing seven distinct clusters. Different genotype clusters were defined at a cut threshold of 8. The clustering height scale is shown at the bottom. B, Parallel coordinate plots showing mean normalized trait values of genotypes in each cluster.
Phenotypic Trait Segregation and Genetic Correlations among Root and Shoot Variables
Clustering of wild and cultivated beans by root traits suggested that genetic analysis of a segregating progeny may reveal the underlying genes. For this purpose, we conducted a QTL analysis of root traits using a recombinant inbred family (RIF) generated between a landrace and a wild accession from the Andean gene pool (Fig. 4; Supplemental Table S6). The frequency distributions of root and shoot traits (Supplemental Figs. S3 and S4) were continuous, indicating the quantitative nature of these traits. Transgressive behavior in all the traits, with the exception of seed and seed coat dry weights, indicated that each parent had genes that contributed to the traits in opposite directions. Broad-sense heritability estimates calculated for each of these traits (Supplemental Table S6) indicated the feasibility of identifying genes responsible for the phenotypic variation. In general, simple traits (weights and dimensions) had higher heritabilities than traits derived from a relationship of traits.
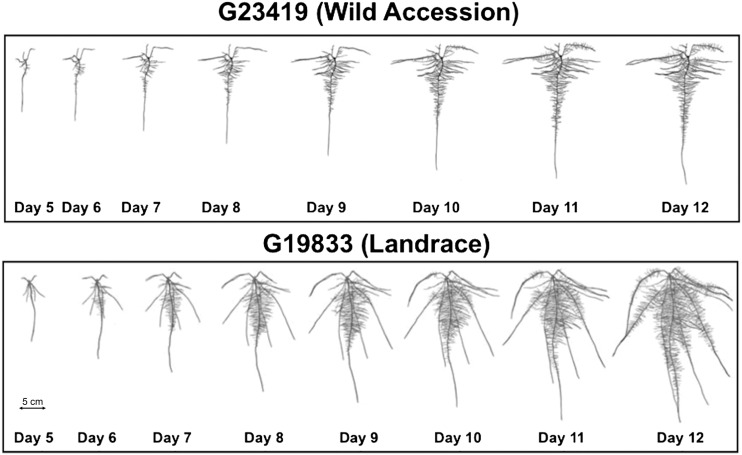
Figure 4.
Time series root scans of the landrace (G19833) and the wild (G23419) accession.
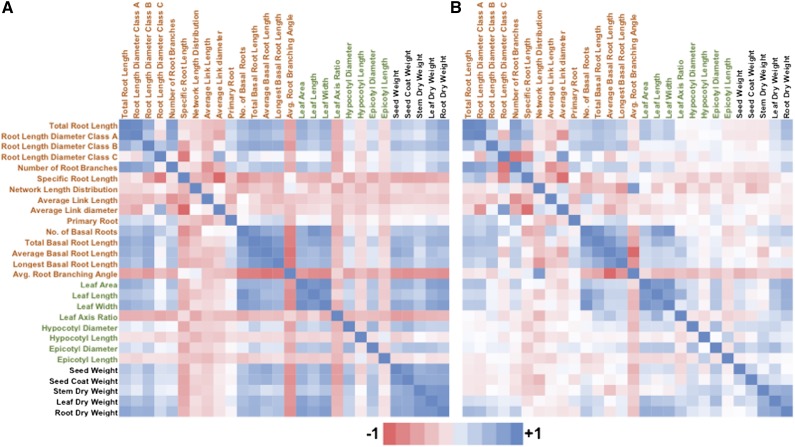
Genetic correlations obtained between trait pairs identified strong positive correlations among organ size traits, and negative genetic correlations between these and morphological traits (Fig. 5A). Correcting for the seed weight covariate significantly altered trait correlations (Fig. 5B). The strength of correlations between organ size and morphological traits was decreased by the seed weight covariate. However, correlations between shoot and root (SR) size traits remained strong in general. Correlations involving hypocotyl and epicotyl length were the least affected by the seed weight covariate. Overall, these results indicated that seed weight has variable effects on different traits.
Figure 5.
Genetic correlations among different root, shoot, seed, and dry weight traits of the RIF (n = 168; three replicas each), which were generated between the landrace (G19833) and the wild (G23419) accession. Correlations were calculated as described in the “Materials and Methods” section. A and B, Results obtained before (A) and after (B) the inclusion of seed weight as a covariate.
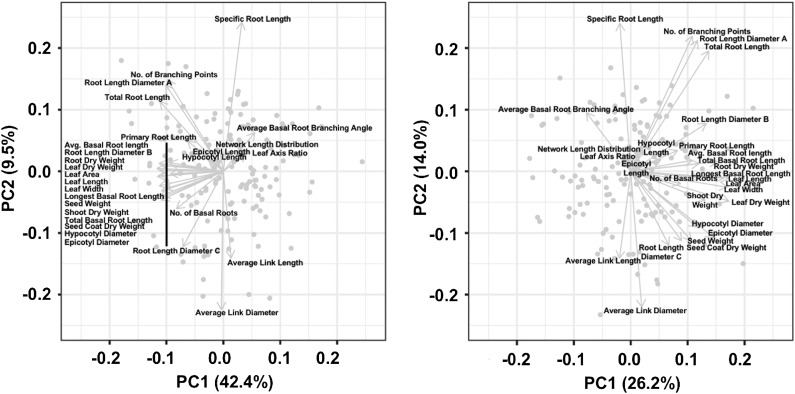
Multivariate analysis of root and shoot growth traits revealed 11 PCs contributing to 90% of the total variation in the RIF (Supplemental Table S7). PC1 explained 44% of the variation and had representation from almost all the traits except leaf axis ratio (Fig. 6, left; Supplemental Fig. S5). The remaining PCs explained the variation to a lesser extent and varied from 10% for PC2 to 2% for PC10. The relatively higher contribution of PC1 over the other PCs indicated a high level of correlation among different root and shoot variables and suggested common genetic determinants. When PCA was conducted after including seed weight as a covariate, PC1 explained only 26% of the total variation in the dataset (Fig. 6, right; Supplemental Table S7), suggesting again that most young seedling traits are affected by seed weight. In fact, some traits that appeared to cluster along with seed weight in the first biplot analysis (Fig. 6, left) showed a dispersed pattern when seed weight was used as covariate (Fig. 6, right).
Figure 6.
PCA using estimated means of root and shoot growth traits from the recombinant inbred population generated between the landrace (G19833) and the wild (G23419) accession. Biplots between first and second principal components before (left) and after (right) correcting for the seed weight covariate.
Construction and Characterization of the Linkage Map of the RIF Family
Genotyping-by-sequencing (GBS) produced an average of 225 million reads in each of two 96-genotype sequencing lanes, which yielded an average of 1.5 million reads per genotype after filtering. The alignment of the wild G23419 sequence reads against the G19833 reference genome (landrace parent) identified 1,984 single nucleotide polymorphisms (SNPs). Selection for high-quality SNPs present in 90% of the RIF yielded 905 informative markers. These were complemented by 75 locus-specific high-resolution melting PCR markers that were used to cover low-density marker sectors of the GBS map. We used 980 segregating markers for linkage analysis.
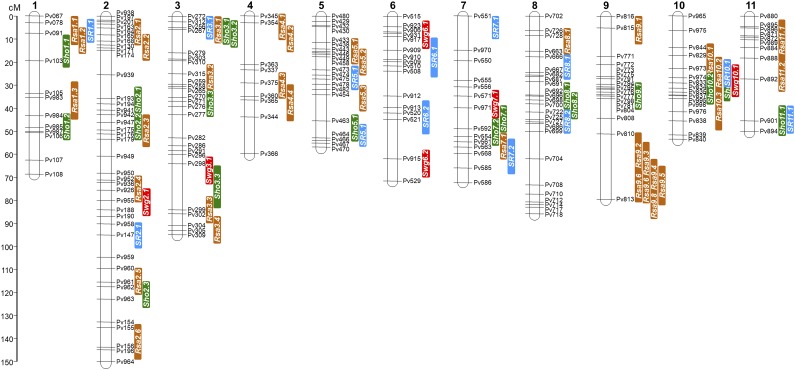
Linkage analysis identified 196 recombinationally unique markers distributed among the 11 linkage groups. The linkage map spans over a total distance of 827 cM, with an average of 75.2 cM per chromosome (Fig. 7; Supplemental Table S8). Chromosome lengths ranged from 49.6 cM (Chr04) to 149.8 cM (Chr02). The average intermarker distance ranged from 2.97 cM (Chr05) to 6.4 cM (Chr04), with an overall average of 4.2 cM. Chr02 contained 39 marker loci, the highest number of recombinationally unique loci, followed by Chr03 with 25 marker loci. Chr04 had the lowest number with only eight marker loci.
Figure 7.
Linkage map based on the RIF derived from a cross between the landrace G19833 and wild accession G23419. The map also marks genomic regions where quantitative analyses detected QTLs associated with RSA, SHO, RS, and SWG traits.
The GBS linkage map covered 94% of the P. vulgaris-sequenced genome (Supplemental Table S8), ranging in coverage from 98.8% for Chr08 to 81.8% for Chr06; low coverage of Chr06 might be due to its telocentric structure (Pedrosa et al., 2003; Bhakta et al., 2015).
Markers were not evenly distributed across the linkage map, showing some gaps. To investigate the nature of these gaps, we plotted the first derivative function of centimorgans over mega base pairs distances (Supplemental Fig. S6). These plots showed vast sectors of suppressed recombination corresponding to pericentromeric regions and also recombination hotspots distally located on the chromosomes, which were similar to the positions reported by Bhakta et al. (2015). The number of recombination hotspots as well as their recombination frequencies differed among chromosomes, with the highest centimorgans/mega base pairs peak detected on Chr10 followed by Chr02 and Chr07. Interestingly, Chr01 exhibited suppressed recombination over a region covering ∼38 Mbp where we found 51 markers with similar segregation patterns forming a single haplotype. This observation suggests that some structural differences may exist between the two parental genomes, which could be responsible for suppressed recombination in this wide genomic region.
Genetic Architecture of Domestication Syndrome in Early Seedlings of Common Bean
We conducted QTL analyses in multiple and progressive steps to identify chromosome regions that might have been targeted during domestication—directly or indirectly. We first used composite interval mapping (CIM) to scan the linkage map for QTLs associated with PCs. Out of 12 QTLs (Supplemental Table S9), four were associated with PC1 (ΣR2 = 0.4), five with PC2 (ΣR2 = 0.52), and three with PC3 (ΣR2 = 0.28). A second scan for the first three PCs, identified with Seed Weight covariate corrections, detected seven QTLs. Only three of these QTLs represented genomic regions that overlapped with those detected in the first scan, and the remaining four were new. These contrasting results highlight the strong effect that seed weight has on multiple traits of young seedlings.
Next, CIM analysis of 28 traits (Supplemental Table S10) identified 84 QTLs with log of odds (LOD) thresholds (P < 0.05) that ranged from 2.66 to 2.97 and LOD maxima that varied between 2.85 and 11.34 (Supplemental Table S11). The map distances covered by these QTLs varied between 3.7 and 34.1 cM; there was a relatively high correlation (R2 = 0.65) between map distance range and LOD maximum. No QTLs were detected for network length distribution. All the other traits were associated with 1–6 QTLs. The variation for each trait explained by the associated QTLs ranged from 0.08 for root dry weight to 0.47 for root length. QTLs were detected in all 11 chromosomes.
An alignment of all QTL-associated chromosome segments to the linkage map revealed 22 overlapping genomic sectors with 1–3 QTL sectors per chromosome (Supplemental Table S11). Several single QTL sectors had marginal LOD values and could be false positives, whereas others, like epicotyl length, had high LOD values (11.3). We detected segments on chromosomes 2, 5, and 7 with 9, 13, and 13 QTLs, respectively. Each of these large clusters had a QTL for Seed Weight, and the associated QTL displayed a large range of LOD values. Interestingly, these three sectors overlapped with sectors detected with PC-linked QTLs.
A new genome-wide scan using seed weight as covariate detected 80 QTLs, altering the previous QTL landscape. The variation explained by these QTLs ranged from 14% for the average basal root length to 45% for specific root length and seed coat dry weight (Supplemental Table S12). Removing the seed weight effect increased the number of QTL sectors. For instance, a single 19-cM multi-QTL sector was found on chromosome 1 before including the covariate, and a new QTL in that sector and two additional QTL sectors, outside the first one, were detected after including the covariate. A more pronounced effect was observed on chromosome 2 where three QTL sectors were detected without the seed weight covariate, and six sectors were detected after the inclusion of the covariate. Two of the six sectors have marginally significant QTLs, but the other four have LOD values that exceed the threshold by up to 7 units. A similar situation was observed for chromosome 5. Each one of these chromosomes carry one QTL for seed weight, which was exerting a significant pleiotropic influence on other traits before the seed covariate was applied. These findings indicated again that seed weight has a significant effect on the genetic analysis of several seedling traits.
We conducted a final genome scan with multiple interval mapping (MIM) using trait values after seed weight covariate correction. This analysis identified additional QTLs, repositioned most CIM-based QTLs, resolved some single CIM-QTL into two closely linked QTLs with opposing effects, and identified a few QTLs exerting additive-by-additive epistatic interactions (Cheverud and Routman, 1995). A total of 265 QTLs were detected for 26 traits with seed weight as covariate and with LOD values that ranged from 0.62 to 12.9 units. The lack of a reliable test of significance for MIM results led us to adopt a conservative ad hoc approach in which we selected a threshold that established the presence of a QTL if it was at least three times as likely as the threshold identified by CIM; that is, an LOD value of 0.5 above the CIM threshold of each trait.
The total number of QTLs with an additive effect was reduced to 142 (Supplemental Table S13). Ten of the 80 QTLs identified by CIM were not detected by MIM. MIM added 72 QTLs, four of which corresponded to two QTL pairs that have been resolved each from single QTLs detected by CIM; in each case the QTLs were closely linked and had opposing effects. The selected MIM QTL had LODs that ranged from 3.25 to 13.82, and 122 exceeded the CIM LOD threshold by 1.0 and 20 of them by 0.5 to 1. The combined QTL effect for each trait varied from 11.1% for the number of basal roots (1 QTL) to 84.7% for specific root length (7 QTLs). Other traits with a strong genetic component include leaf length, root length diameter b, seed weight, average link length, and stem dry weight. MIM also detected five epistatic interactions; of these, four were between QTL that had additive effects, and one interaction was between QTLs with and without an additive effect (Supplemental Table S13).
Correlations among traits and PC QTLs suggested the presence of pleiotropic QTL and/or QTL-linked clusters. To sort out these possibilities, all QTL-associated sectors, established by being within 1 LOD unit from that of the MIM-estimated peak position, were aligned to the linkage map to determine the extent to which they overlapped. Multiple trait (MT)-MIM was used to analyze QTLs within overlapping intervals to determine the likelihood of being pleiotropic or closely linked (Fig. 7; Supplemental Table S14). These analyses reduced the number of QTLs to 73; 35 of them were single trait QTLs, and 38 were found to be pleiotropic affecting between two and six traits. The QTLs were split into four groups: 35 RSA QTLs, 21 of which controlled single traits and the others controlled two (6), three (6), and four (2) traits; 18 shoot (SHO) QTLs found on nine chromosomes, 11 controlled single traits, and the others controlled two (5), three (1), and five (1) traits; 13 SR pleiotropic QTLs found on nine chromosomes; and six seed weight (SWG) QTLs.
DISCUSSION
Univariate, bivariate, and multivariate analyses of young seedling root traits clearly distinguished wild from domesticated accessions of the common bean. The high degree of correlation between most traits also suggested the presence of a root domestication syndrome. Differences in root-size traits between the groups raised the possibility that the 5-fold difference in seed weight between the groups could be responsible for the root trait differences. This is because bean cotyledons are the only source of nutrients during the early heterotrophic growth phase of seedlings. However, differences in root traits persisted even after correcting for the Seed Weight covariate. This observation indicated that other changes have taken place outside of the seed weight effect. The covariate correction also showed that most domesticated root traits have smaller values than those of the wild accessions. Overall, these comparisons indicated that domestication brought about genetic changes that directly affected root traits. Correlations between traits were reduced after correcting for seed weight, but were not eliminated, suggesting the presence of genetic correlations among some traits.
We conducted genetic analysis of young seedling traits that distinguishes wild from domesticated accessions with an intra-Andean RIF to avoid detection of genes responsible for intergene pool polymorphisms. This analysis was used to test our hypothesis that genes selected during domestication could be divided into three groups: one controlling shoot traits that were likely targets of direct selection; another controlling SR traits, also mostly targets of direct selection; and a third group that exclusively controls root traits, which should be considered targets of indirect selection.
Root and shoot traits targeted for genetic analysis displayed normal distributions and relatively high heritability values, indicating the feasibility of detecting underlying QTLs and important genetic gains to be achieved. With the exception of seed weight, all traits showed transgressive behavior revealing the presence of QTL alleles in each parent with opposing phenotypic effects. The linkage map constructed for QTL analysis had an average intermarker distance of 4.2 cM, which provided sufficient QTL power of detection, although the precision for localizing the QTL may have been decreased somewhat by that marker density (Stange et al., 2013).
QTL analysis of PCs and of root and shoot traits before and after correcting for seed weight revealed the strong effect that seed weight has on those young seedling traits. Thus, segregation of seed weight genes can lead to the detection of QTL artifacts when mapping growth and developmental traits of young seedlings. For example, CIM detected 14 QTLs on chromosome 5 before seed weight was used as covariate. However, only 11 of those QTLs were detected after the covariate correction, resulting in a net loss of three QTLs, but three new ones were gained with the covariate. Furthermore, 12 QTLs were detected in one sector of overlapping QTL ranges on chromosome 7, including one seed weight QTL, before covariate correction. This sector was resolved into five different QTL sectors after the covariate correction—the seed weight QTL range did not overlap with any of the other four QTL sectors. Introduction of the covariate also resulted in relevant QTL position changes. Similar changes were detected in other chromosomes, but changes were more dramatic for those with a seed weight QTL.
MIM analysis detected 70 additional QTLs using our ad hoc LOD threshold, bringing up the total to 142 QTLs. Bivariate and multivariate analysis, and the overlapping genetic ranges of various QTLs, suggested that some of these QTLs may have pleiotropic effects, whereas others belonged to clusters of closely linked QTLs. MT-MIM resolved these into 72 QTLs, half of which are single trait QTLs and the remaining appear to be pleiotropic. The MT-MIM QTL fell into one of four trait categories: SWG, SHO, SR, and RSA. The identification of these QTL provided evidence in support of our hypothesis. SWG and SHO are QTLs that control aboveground traits, which were subjected to direct selection during domestication, whereas RSA QTLs control traits that were selected indirectly during domestication as these traits are not visible and represent a product of “unconscious selection”—a term and concept first used by Darwin (1859) and considered by Heiser (1988)—which refers to selection of traits without “a predetermined purpose to improve the breed.” SR QTLs are clearly pleiotropic, as they appear to control both above- and below-ground traits. For instance, Sr2.1 on chromosome-2 controls Leaf Length and Basal Root Length, potentially through the control of cell division in both organs; although selection was exerted only on the leaf, it appears to have had an unintended effect on the length of the basal roots. The SR relationship has been framed under a functional equilibrium system with coordinating channels of chemical/hormonal communication (Wilson, 1988; Farrar and Jones, 2000; Bouteillé et al., 2012; Puig et al., 2012; Dignat et al., 2013).
Six SWG QTLs explained 70% of the variation of this trait and had LOD values that exceeded the threshold by 3–11 LOD units. These QTLs were clearly among the principal targets of direct selection during domestication and had significant pleiotropic effects that fall within both developmental and selectional pleiotropies, according to the classification proposed by Paaby and Rockman (2013). In the former, the phenotype selected for one stage of development (large seeds) has significant effects on traits expressed in subsequent developmental stages—germination and early seedling growth. Large seeded genotypes produce larger organs during heterotrophic growth, which increase the starting material for the subsequent exponential growth.
Selectional pleiotropy impacts on adaptation could be appreciated by examining certain root traits. Wild accessions have smaller root systems, less root branching, fewer and shorter basal roots than the domesticated counterparts, and comparatively greater root apical dominance than domesticated accessions. These root characteristics appear to condition adaptation to less favorable soil characteristics. For instance, the relatively long primary root allows the seedling to have access to water in lower soil horizons soon after germination, whereas the more horizontal basal roots provide the seedling with “top-soil foraging” capability for phosphorous acquisition (Lynch and Brown, 2001). These root traits do not appear to be of great adaptive value in the fertile alluvial soils where water is not a limiting factor, and where agriculture and domestication were initiated. Thus, larger root systems promoted by large seeds would be at a disadvantage in soils with limited water availability. The selection for larger plant organs, as reported for beans (Koinange et al., 1996; Araujo et al., 1997), could only be viable when optimum resources are available.
We acknowledge that the QTLs reported in this article, even those with high LOD values, were identified by statistical inference and for this reason they need to be validated experimentally. This task could be accomplished through Mendelization using recurrent selection and/or through comparative sequence analysis of the associated genomic regions to determine whether they possess signatures of selection due to domestication. The same word of caution applies to inferred pleiotropy, which may be the result of suppressed recombination. Methods designed to increase the resolution of QTLs (Xu et al., 2005; Heifetz and Soller, 2015) could be applied to address this issue. Finally, these results underscore the potential usefulness of wild germplasm as a potential source of adaptive root traits to stressful soil environments.
A number of studies have addressed the genetic control of the domestication syndrome (Koinange et al., 1996; Weeden, 2007; Wills and Burke, 2007; Kaga et al., 2008; Isemura et al., 2012), but none of them has directly focused on root growth and development. This study begins to fill the gap in knowledge about domestication-associated changes in various root traits. The work presented here documents a case of unconscious selection in which selection for increased seed weight had consequences in subsequent developmental stages in the common bean—an effect that could probably be extended to other domesticated species, in particular those that experienced a significant increase in seed weight. In contrast to other domestication studies that detected few genes with major effects on the shoot phenotype (Doebley et al., 1997; Wang et al., 2005; Olsen and Wendel, 2013), we have identified many more genes that appear to have smaller contributions to the domesticated root phenotype, perhaps suggesting that changes by indirect selection are more gradual and can accumulate more mutations over time.
MATERIALS AND METHODS
Plant Material
A set of wild and landrace accessions of the common bean from the Andean and Mesoamerican gene pools was phenotyped for various root growth traits (Supplemental Table S15). A RIF with 186 genotypes (F6:8) was generated from a cross between a landrace (G19833) from Northwestern Peru (6.27° S, 77.75° W) and a wild accession (G23419) from Central Peru (11.23° S, 75.53° W). The two genotypes exhibit significant phenotypic and gene expression differences at early seedling stage (Singh et al., 2017, 2018). The intra-Andean wild-landrace RIF excluded traits that may be found in modern cultivars and those controlling intergenepool variation. The RIF was generated minimizing any type of selection to maintain its expected diversity.
Plant Growth and Root Phenotyping Platform
Seeds were weighed and surface-sterilized with 50% commercial bleach for 5 min, rinsed in sterile deionized water, and imbibed overnight in the dark at 25°C. After removing the seed coat, seeds were germinated in paper rolls before the seedlings were transferred to root plates as described by Singh et al. (2017). A single tank held 16 plates (1 seedling/plate), and a maximum of four tanks were placed in a Conviron E15 growth chamber. Growth conditions were set at a photon flux density of 400 μmol m−2 s−1, a 12-h photoperiod with a coordinated 23°C/18°C thermoperiod, and 90% relative humidity. Thermochron data loggers were used to record temperature every 15 min, and photosynthetic active radiation was regularly monitored with a LiCor light meter. Details of the root phenotyping platform are described in Supplemental Figure S7.
Experimental Procedure and Trait Measurements
Root images were acquired nondestructively 12 d after transplantation using an Epson scanner. These images were stored as Tiff files and analyzed with three software packages: WinRhizo Pro-9a (Regent Canada), GiA Roots (Galkovskyi et al., 2012), and ImageJ (http://rsb.info.nih.gov/ij/). The bean root system comprises a primary root, several basal roots that arise from the upper part of the primary root, and the adventitious roots arising from the hypocotyl. The root-growing angle between primary and basal roots marks the spatial deployment of the roots and provides a distinct shape to every root system. Root framework data were manually acquired using ImageJ. A calibration image was used for scaling (pixels/cm) to estimate root lengths in centimeters
Data for various shoot traits were collected nondestructively using the same plant from which root measurements were taken. Hypocotyl and epicotyl lengths were measured using a graded ruler. Hypocotyl and epicotyl diameter were measured with a Vernier caliper (resolution of 0.1 mm). Leaf images were captured by photography using a custom-built platform with a built-in scale and were analyzed using ImageJ. Leaf image analysis provided area, length, width, and axis ratio.
Dry weight measurements were taken from each seedling after the root scan. Seedlings were partitioned into leaves, roots, and stems. Seed coats were removed after seed imbibition and kept for dry weight measurement. Tissues were oven-dried at 60°C for 72 h Afterward, weights were obtained from the dry tissues equilibrated to room temperature. Root and shoot growth characters correspond to different aspects of organ size and morphology measurements (Supplemental Table S10).
Experimental Design and Statistical Analyses
Common bean (Phaseolus vulgaris) accessions and the RIF were grown and phenotyped using an incomplete block design. A subset of 168 recombinant inbred lines (RILs) was chosen at random, from the original 186 RIL set, to randomize across total 12 incomplete blocks in three growth chambers. Each incomplete block contained 16 genotypes (14 RILs and two parental lines). The experiments were replicated three times during May 2012, August 2012, and November 2012 using the same growth chambers and experimental conditions.
Quantitative trait measurements were first evaluated with univariate analyses using the following linear mixed-effect model:
where y corresponds to the response variable; μ is the overall mean; SW is the covariate of seed weight (when incorporated in calculations); Rep is a fixed effect of replicate or block; Rep.Iblock is a random effect of incomplete block within replicate; Chamber.Row is a random effect of row within the test chamber; RIL is the fixed effect of a recombinant inbred line; and ε is the residual term. Residuals were spatially modeled based on a separate autoregressive of order 1 error structure for the X and Y coordinates of each plot, which considers a spatial correlation in each of these directions. The above model was fitted using the software ASReml v3.0 (Gilmour et al., 2009), which estimates variance components based on a Residual Maximum Likelihood estimation method. Two types of models were fitted: with and without the SW covariate, and the significance of this covariate was evaluated using an approximate F-test (α = 0.05). Also, the significance of the spatial correlations on row and column was evaluated using the Bayesian Information Criteria and the likelihood ratio test (α = 0.05) to select the most parsimonious model. Best linear unbiased estimations of each of the RILs were used to predict RIL adjusted mean values. This approach provides the genetic means of individual traits corrected for all environmental variation, which were used to generate histograms from the RIL population, to provide the average phenotypic values of parentals and RILs, and to assess the trait frequency distributions. The trait distributions were tested for normality using the Shapiro-Wilk normality test (α = 0.05), and non-normal traits were normalized for quantitative genetic analysis. The same model presented above was also fitted with RIL as a random effect to calculate broad-sense heritability (H2 = genotype variance component/ [variance component + error variance component]) for each trait.
Later, a bivariate model was used to estimate genetic correlations among pairs of traits using the following linear model:
 |
where y corresponds to the vector of two stacked response variables; trait.SW is the fixed effect of covariate of seed weight within each trait; trait.Rep is a fixed effect of replicate nested within trait; trait.Rep.Iblock is a random effect of incomplete block within replicate for each trait; trait.Chamber.Row is a random effect of row within test chamber for each trait; trait.RIL is the fixed effect of a recombinant inbred line within trait; and ε is the residual term. The random terms of trait.Rep.iblock and trait.Chamber.Row were modeled using a diagonal variance-covariance matrix with a different variance component for each trait. The trait.RIL and residual terms were modeled using a heterogeneous correlation structure that contains a different variance for each trait and genetic and residual correlations between traits.
Finally, additional statistics were obtained to gain a better insight of the data, including rank correlations using the genetic estimates, PCAs, and hierarchical clustering, all of which were performed with the estimated means and using functions from the software package “R” 3.3.1 (http://www.r-project.org/).
DNA Extraction, GBS, and Linkage Map Construction
DNA extractions and library preparations for GBS (Elshire et al., 2011) were performed as described by Bhakta et al. (2015). Two GBS library pools, each containing 96 genotypes, were sequenced using the Illumina 2000 platform. Raw reads in “fastq” format were processed using the Tassel GBS pipeline (Glaubitz et al., 2014). RIL-specific sequencing reads and those of the parental lines were separated based on the unique barcode sequence. Sequence tags were aligned to the Phaseolus vulgaris reference sequence in the program Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) with the software Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) using the default settings (Langmead and Salzberg, 2012). SNPs were detected using the SNPcaller plugin, and duplicate SNPs were merged with the MergeDuplicateSNPs plugin in the software Tassel (https://www.maizegenetics.net/tassel). SNP variants were recorded in Variant Call Format and were further processed using the software “vcftools” (Danecek et al., 2011). Resulting SNPs were filtered for maximum amount of missing data (10%), minor allele frequency (10%), and minimum read depth of five.
PCR makers were developed to cover low-density marker regions in the GBS map. Reads from whole-genome sequencing of G23419 were aligned against low-density marker sectors in the reference genome to identify SNPs that could be detected through high-resolution melting of PCR products (Liew et al., 2004; Simko, 2016). Suitable SNP-flanking primers were designed to amplify short (<100 bps) sequences (Supplemental Table S16). PCR reactions were performed in 10-μL reaction volumes containing 1× PCR buffer, 200 μM of dNTPs, 1.5 mm of MgCl2, 0.1-μM forward and reverse primer each, 0.25 units of Taq DNA polymerase, 2 μM of Syto82, and 4 ng of DNA. PCR reaction conditions were: 95°C for 2 min, followed by 45 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. High resolution melting analysis of PCR products was carried out between 68°C and 90°C with temperature increase rate of 0.02°C/s in continuous acquisition mode.
A linkage map was constructed with the software Mapmaker 3.0 (Lander et al., 1987) as modified by Russell Malmberg (http://www.plantbio.uga.edu/007Erussell/) and described in Bhakta et al. (2015). Markers were sorted into the 11 linkage groups using an LOD of eight units and a maximum distance of 35 cM (Kosambi mapping function). Markers within each group were ordered by progressively lowering the linkage criteria to an LOD of 3 units.
QTL Analyses
Windows QTL Cartographer (Wang et al., 2007) was used for CIM (Zeng, 1993, 1994) and MIM (Kao et al., 1999; Zeng et al., 1999). CIM LOD thresholds used to identify QTLs at alpha = 0.05 were derived from 1,000 permutations for each trait (Churchill and Doerge, 1994). Forward and backward stepwise regressions (α = 0.05) were performed to select cofactors that accounted for genetic background variation. The analysis was performed using a 10-cM window size on either side of the interval being tested using a 1-cM walk speed. MIM analysis was applied to increase the sensitivity of QTL analysis, and the CIM results were used as the initial model. New QTLs were added to the model through recurrent searches using a minimum distance of 5 cM between QTLs until no new QTL was detected. QTLs with significant effects were retained in the model based on critical threshold values derived from likelihood ratio tests. The best QTL models, including optimized QTL map positions, were selected using Bayesian Information Criteria.
The significance of QTL-by-QTL interactions in the best genetic regression model were evaluated using likelihood ratio tests. Main and epistatic QTL effects falling below the critical threshold level were removed from the model. Main-effect QTLs that had significant epistatic interactions with other QTLs were retained if the significant threshold level was attained by the interactive effect. In the end, each QTL was optimized for their map position in the context of the position of other QTLs in the model; the position that conferred the maximum likelihood in the model was retained as the best QTL position.
Several QTLs were detected with short spans along the chromosomes. To discern between QTLs with pleiotropic effects and those tightly linked, we used MT-MIM algorithm in Windows QTL Cartographer. First, we manually identified overlapping sectors of QTLs associated with different root and shoot traits and pooled them to perform MT-MIM analysis. Pairwise MT-MIM tests were performed for all QTLs within a specific QTL cluster to estimate whether colocalized QTLs had significantly different map positions (linkage) or not (pleiotropy). The significance of close linkage or pleiotropy was determined using likelihood ratio test statistics. A minimum LOD threshold of 3 units was used to consider significantly better fit of close linkage model. Pleiotropy was assumed when the LOD score fell below the threshold; optimal map positions were obtained for a single pleiotropic QTL.
Accession Numbers
The molecular marker data and accompanying marker-flanking sequences are provided in Supplemental Table S16. Marker and QTL data have been submitted to the Legume Information System (Dash et al., 2016).
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. PCA of root traits from wild and domesticated accessions.
Supplemental Figure S2. Hierarchical clustering of wild (W-) and domesticated (D-) accessions.
Supplemental Figure S3. Frequency distribution histograms of root traits from the RIF.
Supplemental Figure S4. Frequency distribution histograms of shoot traits from the RIF.
Supplemental Figure S5. PCA of root and shoot traits from the RIF.
Supplemental Figure S6. A scatterplot showing the distribution of recombination hotspots.
Supplemental Figure S7. Details of the 2D root phenotyping platform.
Supplemental Table S1. Statistical comparisons of root traits between wild and domesticated accessions of common bean.
Supplemental Table S2. Summary of PCA of various root traits in a set of wild and domesticated common bean genotypes.
Supplemental Table S3. Phenotypic correlations between root traits, including seed weight, measured in a set of wild and domesticated accessions of the common bean—no seed weight covariate adjustment.
Supplemental Table S4. Phenotypic correlations between root traits, including seed weight, measured in a set of wild and domesticated accessions of the common bean—with seed weight covariate adjustment.
Supplemental Table S5. Hierarchical clustering analysis of different wild and domesticated accessions using root trait values before and after applying seed weight covariate adjustment.
Supplemental Table S6. Mean trait values of various root and shoot traits of G19833 and G23419 and their heritability estimates based on data from their RIF.
Supplemental Table S7. Summary of PCA using root and shoot growth traits of the RIF generated between the landrace G19833 and the wild accession G23419.
Supplemental Table S8. Descriptors of the linkage map constructed with the RIF generated between the landrace G19833 and the wild accession G23419.
Supplemental Table S9. Results obtained with CIM of principal components before and after adjusting the data with the seed weight covariate.
Supplemental Table S10. List of root, leaf, stem, and seed traits. Root traits were measured in the collection of wild and landrace accessions and in the RIF.
Supplemental Table S11. CIM analysis of various root and shoot traits; data were analyzed without using seed weight as a covariate.
Supplemental Table S12. CIM analysis of various root and shoot traits; data were analyzed using seed weight as a covariate.
Supplemental Table S13. MIM analysis of root and shoot traits using data adjusted with the seed weight covariate.
Supplemental Table S14. MIM-based assessment of pleiotropy over linkage among closely linked QTLs.
Supplemental Table S15. List of wild and domesticated genotypes used for preliminary analysis of various root growth and architectural traits.
Supplemental Table S16. List of SNP markers.
Supplemental Table S1. Statistical comparisons of root traits between wild and domesticated accessions of common bean. The se (in parentheses) of the mean (bold) is listed below the means.
Supplemental Table S2. Summary of PCA of various root traits in a set of wild and domesticated common bean genotypes. PCA analysis was conducted separately with and without seed weight as a covariate.
Supplemental Table S3. Phenotypic correlations between root traits, including seed weight, measured in a set of wild and domesticated accessions of the common bean—no seed weight covariate adjustment. The bottom-left sector of the matrix contains the R2 values, and the top right sector of the matrix contains the corresponding P values.
Supplemental Table S4. Phenotypic correlations between root traits, including seed weight, measured in a set of wild and domesticated accessions of the common bean—with seed weight covariate adjustment. The bottom-left sector of the matrix contains the R2 values, and the top right sector of the matrix contains the corresponding P values.
Supplemental Table S5. Hierarchical clustering analysis of different wild and domesticated accessions using root trait values before and after applying seed weight covariate adjustment.
Supplemental Table S6. Mean trait values of various root and shoot traits of G19833 and G23419 and their heritability estimates based on data from their RIF.
Supplemental Table S7. Summary of PCA using root and shoot growth traits of the RIF generated between the landrace G19833 and the wild accession G23419. PCA analyses were conducted separately using datasets analyzed before and after adjusting for the seed weight covariate.
Supplemental Table S8. Descriptors of the linkage map constructed with the RIF generated between the landrace G19833 and the wild accession G23419. The physical information for SNP markers were obtained from the P. vulgaris (v1.0), and coordinates of the pericentromeric regions from the Legume Information System (Dash et al., 2016).
Supplemental Table S9. Results obtained with CIM of principal components before and after adjusting the data with the seed weight covariate. Threshold LODs (0.05) were estimated after 1,000 random permutations. QTLs with overlapping chromosome segments are marked with the same superscript letter.
Supplemental Table S10. List of root, leaf, stem, and seed traits. Root traits were measured in the collection of wild and landrace accessions and in the RIF. Leaf and stem traits were only measured in the mapping population.
Supplemental Table S11. CIM analysis of various root and shoot traits; data were analyzed without using seed weight as a covariate. The threshold values for significance (α = 0.05) were obtained from CIM data after 1,000 permutations for each trait.
Supplemental Table S12. CIM analysis of various root and shoot traits; data were analyzed using seed weight as a covariate. The negative or positive additivity values indicate whether the G23419 (wild) or G19833 (landrace) alleles added value to the trait, respectively. The threshold values for significance (0.05) were obtained from CIM data after 1,000 permutations for each trait.
Supplemental Table S13. MIM analysis of root and shoot traits using data adjusted with the seed weight covariate. Results from CIM analyses were used as starting models. The negative or positive additivity values indicate whether the G23419 (wild) or G19833 (landrace) alleles added value to the trait, respectively. The threshold values for identifying QTLs exceeded by 0.5 those determined for CIM analysis. The map interval for MIM QTLs corresponded to segments that were within 1 LOD of the QTL peak. Effects are labeled as “A” for additive, and “AA” for epistatic interactions of the additive-by-additive type. Interacting QTLs are marked with the same superscript.
Supplemental Table S14. MIM-based assessment of pleiotropy over linkage among closely linked QTLs. QTLs were assigned to one of four categories: RSA, SR, SHO, and SWG.
Supplemental Table S15. List of wild and domesticated genotypes used for preliminary analysis of various root growth and architectural traits.
Supplemental Table S16. List of SNP markers. The position of each marker in the linkage map, the coordinates of the genomic sequence, and 100-base sequence including the SNP are listed.
Acknowledgments
We are thankful to Dr. Marcio Resende for his critical reading of the article and his valuable suggestions.
Footnotes
This work was supported in part by the National Science Foundation (grant no. IOS-0920145).
Articles can be viewed without a subscription.
References
- Araujo AP, Teixeira G, De Almeida DL (1997) Phosphorus efficiency of wild and cultivated genotypes of common bean (Phaseolus vulgaris L.) under biological nitrogen fixation. Soil Biol Biochem 6: 951–957 [Google Scholar]
- Bhakta MS, Jones VA, Vallejos CE (2015) Punctuated distribution of recombination hotspots and demarcation of pericentromeric regions in Phaseolus vulgaris L. PLoS One 10: e0116822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouteillé M, Rolland G, Balsera C, Loudet O, Muller B (2012) Disentangling the intertwined genetic bases of root and shoot growth in Arabidopsis. PLoS One 7: e32319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JC, Chapman MA, Burke JM (2008) Molecular insights into the evolution of crop plants. Am J Bot 95: 113–122 [DOI] [PubMed] [Google Scholar]
- Burton AL, Johnson JM, Foerster JM, Hirsch CN, Buell CR, Hanlon MT, Kaeppler SM, Brown KM, Lynch JP (2014) QTL mapping and phenotypic variation for root architectural traits in maize (Zea mays L.). Theor Appl Genet 127: 2293–2311 [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ (1995) Epistasis and its contribution to genetic variance components. Genetics 139: 1455–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RT, Famoso AN, Zhao K, Shaff JE, Craft EJ, Bustamante CD, McCouch SR, Aneshansley DJ, Kochian LV (2013) High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ 36: 454–466 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. ; 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27: 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (1859) On the Origin of Species by Means of Natural Selection. Murray, London [Google Scholar]
- Dash S, Campbell JD, Cannon EKS, Cleary AM, Huang W, Kalberer SR, Karingula V, Rice AG, Singh J, Umale PE, et al. (2016) Legume information system (LegumeInfo.org): A key component of a set of federated data resources for the legume family. Nucleic Acids Res 44(D1): D1181–D1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignat G, Welcker C, Sawkins M, Ribaut JM, Tardieu F (2013) The growths of leaves, shoots, roots and reproductive organs partly share their genetic control in maize plants. Plant Cell Environ 36: 1105–1119 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JF, Jones DL (2000) The control of carbon acquisition by roots. New Phytol 147: 43–53 [Google Scholar]
- Galkovskyi T, Mileyko Y, Bucksch A, Moore B, Symonova O, Price CA, Topp CN, Iyer-Pascuzzi AS, Zurek PR, Fang S, et al. (2012) GiA Roots: Software for the high throughput analysis of plant root system architecture. BMC Plant Biol 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia EH, Pena-Valdivia CB, Rogelio-Aguirre JR, Muruaga JSM (1997) Morphological and agronomic traits of a wild population and improved cultivar of common bean (Phaseolus vulgaris L.). Ann Bot 79: 207–213 [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml User Guide Release 3.0. VSN International Ltd, Hemel Hempstead, United Kingdom, https://www.vsni.co.uk/software/asreml-r/ [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR. (1992) Crops and Man. American Society of Agronomy, Madison, WI [Google Scholar]
- Heifetz EM, Soller M (2015) Targeted Recombinant Progeny: A design for ultra-high resolution mapping of quantitative trait loci in crosses between inbred or pure lines. BMC Genet 16: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser CB. (1988) Aspects of unconscious selection and the evolution of domesticated plants. Euphytica 37: 77–81 [Google Scholar]
- Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T, Jo U, Vaughan DA, Tomooka N (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS One 7: e41304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A, Isemura T, Tomooka N, Vaughan DA (2008) The genetics of domestication of the azuki bean (Vigna angularis). Genetics 178: 1013–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinange EMK, Singh SP, Gepts P (1996) Genetic control of domestication syndrome in common bean. Crop Sci 36: 1037–1045 [Google Scholar]
- Kwak M, Gepts P (2009) Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor Appl Genet 118: 979–992 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Yu Y, Yang H, Xu L, Dong W, Du H, Cui W, Zhang H (2014) Inheritance and QTL mapping of related root traits in soybean at the seedling stage. Theor Appl Genet 127: 2127–2137 [DOI] [PubMed] [Google Scholar]
- Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C (2004) Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 50: 1156–1164 [DOI] [PubMed] [Google Scholar]
- Lynch JP, Brown KM (2001) Topsoil foraging—an architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237 [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Wendel JF (2013) A bountiful harvest: Genomic insights into crop domestication phenotypes. Annu Rev Plant Biol 64: 47–70 [DOI] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV (2013) The many faces of pleiotropy. Trends Genet 29: 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa A, Vallejos CE, Bachmair A, Schweizer D (2003) Integration of common bean (Phaseolus vulgaris L.) linkage and chromosomal maps. Theor Appl Genet 106: 205–212 [DOI] [PubMed] [Google Scholar]
- Prince SJ, Song L, Qiu D, Maldonado Dos Santos JV, Chai C, Joshi T, Patil G, Valliyodan B, Vuong TD, Murphy M, et al. (2015) Genetic variants in root architecture-related genes in a Glycine soja accession, a potential resource to improve cultivated soybean. BMC Genomics 16: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig J, Pauluzzi G, Guiderdoni E, Gantet P (2012) Regulation of shoot and root development through mutual signaling. Mol Plant 5: 974–983 [DOI] [PubMed] [Google Scholar]
- Ron M, Dorrity MW, de Lucas M, Toal T, Hernandez RI, Little SA, Maloof JN, Kliebenstein DJ, Brady SM (2013) Identification of novel loci regulating interspecific variation in root morphology and cellular development in tomato. Plant Physiol 162: 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Bitocchi E, Bellucci E, Nanni L, Rau D, Attene G, Papa R (2009) Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evol Appl 2: 504–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J, Morrell PL, Gaut BS (2007) Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc Natl Acad Sci USA 104(Suppl 1): 8641–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJB, Bentsink L, Vonk J, van der Plas LHW, Koornneef M, Vreugdenhil D (2006) Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA 103: 2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I. (2016) High-resolution DNA melting analysis in plant research. Trends Plant Sci 21: 528–537 [DOI] [PubMed] [Google Scholar]
- Singh J, Clavijo Michelangeli JA, Gezan SA, Lee H, Vallejos CE (2017) Maternal effects on seed and seedling phenotypes in reciprocal F1 hybrids of the common bean (Phaseolus vulgaris L.). Front Plant Sci 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Zhao J, Vallejos CE (2018) Differential transcriptome patterns associated with early seedling development in a wild and a domesticated common bean (Phaseolus vulgaris L.) accession. Plant Sci 274: 153–162 [DOI] [PubMed] [Google Scholar]
- Stange M, Utz HF, Schrag TA, Melchinger AE, Würschum T (2013) High-density genotyping: An overkill for QTL mapping? Lessons learned from a case study in maize and simulations. Theor Appl Genet 126: 2563–2574 [DOI] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nussbaum-Wagler T, Li B, Zhao Q, Vigouroux Y, Faller M, Bomblies K, Lukens L, Doebley JF (2005) The origin of the naked grains of maize. Nature 436: 714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC, http://statgen.ncsu.edu/qtlcart/WQTLCart.htm [Google Scholar]
- Weeden NF. (2007) Genetic changes accompanying the domestication of Pisum sativum: Is there a common genetic basis to the ‘domestication syndrome’ for legumes? Ann Bot 100: 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills DM, Burke JM (2007) Quantitative trait locus analysis of the early domestication of sunflower. Genetics 176: 2589–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JB. (1988) A review of evidence on the control of shoot:root ratio in relation to models. Ann Bot 61: 433–449 [Google Scholar]
- Xu Z, Zou F, Vision TJ (2005) Improving quantitative trait loci mapping resolution in experimental crosses by the use of genotypically selected samples. Genetics 170: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Roorkiwal M, Valliyodan B, Zhou L, Chen P, Varshney RK, Nguyen HT (2018) Genetic diversity of root system architecture in response to drought stress in grain legumes. J Exp Bot 69: 3267–3277 [DOI] [PubMed] [Google Scholar]
- Zeng Z-B. (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90: 10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z-B. (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z-B, Kao CH, Basten CJ (1999) Estimating the genetic architecture of quantitative traits. Genet Res 74: 279–289 [DOI] [PubMed] [Google Scholar]
- Zurek PR, Topp CN, Benfey PN (2015) Quantitative trait locus mapping reveals regions of the maize genome controlling root system architecture. Plant Physiol 167: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]