In contrast to vertebrates, the function of TPX2 in plants is not essential; this is likely caused by the expansion of the TPX2 family, which has led to redundant and unique functions among its members.
Abstract
Aurora kinases are key regulators of mitosis. Multicellular eukaryotes generally possess two functionally diverged types of Aurora kinases. In plants, including Arabidopsis (Arabidopsis thaliana), these are termed α- and β-Auroras. As the functional specification of Aurora kinases is determined by their specific interaction partners, we initiated interactomics analyses using both Arabidopsis α-Aurora kinases (AUR1 and AUR2). Proteomics results revealed that TPX2-LIKE PROTEINS2 and 3 (TPXL2/3) prominently associated with α-Auroras, as did the conserved TPX2 to a lower degree. Like TPX2, TPXL2 and TPXL3 strongly activated the AUR1 kinase but exhibited cell-cycle–dependent localization differences on microtubule arrays. The separate functions of TPX2 and TPXL2/3 were also suggested by their different influences on AUR1 localization upon ectopic expressions. Furthermore, genetic analyses showed that TPXL3, but not TPX2 and TPXL2, acts nonredundantly to enable proper embryo development. In contrast to vertebrates, plants have an expanded TPX2 family and these family members have both redundant and unique functions. Moreover, as neither TPXL2 nor TPXL3 contains the C-terminal Kinesin-5 binding domain present in the canonical TPX2, the targeting and activity of this kinesin must be organized differently in plants.
The evolutionarily-conserved Aurora kinases are crucial players in various steps of the eukaryotic cell cycle. Vertebrate Auroras are functionally divided into AUR A and AUR B/C (Carmena and Earnshaw, 2003) based on their differential localization and regulation during the cell cycle. AUR A primarily acts on spindle microtubules (MTs) where it phosphorylates MT-associated proteins (MAPs) to enable spindle formation and proper dynamics (Barr and Gergely, 2007). AUR A is targeted to the centrosome and spindle MTs, where it is activated by the MAP TPX2 (for Targeting Protein of the Xenopus Kinesin-Like Protein2) in the frog Xenopus and other vertebrates (Wittmann et al., 2000; Kufer et al., 2002). AUR B, on the other hand, predominantly interacts with the chromosomal passenger complex (containing the inner centromere protein INCEN-P, Survivin, and Borealin). This complex associates with the centromeres in early stages of mitosis and translocates to MTs in the central spindle from anaphase through cytokinesis (for review, see Carmena and Earnshaw, 2003). The spatial compartmentalization and function of the Aurora kinases depends on their interaction partners (Carmena et al., 2009; Li et al., 2015). Remarkably, TPX2-dependent discrimination between AUR A and AUR B relies on a single amino acid close to the catalytic domain and a single amino acid change could functionally convert Aurora A into an Aurora B-like kinase (Bayliss et al., 2004; Fu et al., 2009).
TPX2 often mediates the interaction of proteins with spindle MTs, and it is widely accepted as an indispensable protein in mitosis (Gruss et al., 2002; Alfaro-Aco et al., 2017). Next to targeting, activating, and protecting AUR A from dephosphorylation and degradation, TPX2 makes a critical contribution to MT nucleation inside the mitotic spindle and to chromosome-induced MT assembly (Alfaro-Aco et al., 2017). A more recently reported function of TPX2 is its participation in the DNA damage response (Neumayer et al., 2014). In interphase, TPX2 interacts with IMPORTIN-α and IMPORTIN-β, which shuttle the AUR A-TPX2 complex to the nucleus. High RanGTP (Ras-related nuclear protein loaded with guanosine triphosphate) levels inside the nucleus mediate the dissociation of TPX2 from importins by binding of RanGTP to importin-β, thereby driving the accumulation of TPX2 inside the nucleus (Neumayer et al., 2014). In animal cells, a centrosomal pool of TPX2, which aids centrosome separation before nuclear envelope breakdown (NEBD), is generated by the phosphorylation of TPX2’s nuclear localization signal (NLS) by the Never In Mitosis A-Related Kinase9, which prevents its association with importins (Eibes et al., 2018). Upon NEBD, a high RanGTP concentration, and consequently, high levels of free TPX2, are maintained around the chromosomes due to the association of the RanGEF (Ras-related nuclear protein guanine nucleotide exchange factor) Regulator of chromosome condensation1 with chromatin. These RanGTP and TPX2 gradients create a positional cue that determines the site of TPX2-mediated MT nucleation (for review, see Neumayer et al., 2014).
In contrast to fungal and animal systems, very little has been learned regarding Aurora-dependent regulation of the cell division cycle as well as on its interaction partners and substrates in flowering plants (for review, see Weimer et al., 2016). Plant Aurora kinases can be classified into α-Aurora (AUR1 and AUR2) and β-Aurora (AUR3) in Arabidopsis (Arabidopsis thaliana), based on phylogenetic analysis, differences in subcellular localization and on their differential capacity to complement a weak double mutant in both Arabidopsis α-Auroras. The Arabidopsis AUR1 and AUR2 are functionally redundant and associate with the forming spindle and cell plate, whereas β-Aurora localizes to the centromeric region of mitotic chromosomes (Demidov et al., 2005; Kawabe et al., 2005; Van Damme et al., 2011). An aur1 aur2 double mutant shows defects in division plane orientation mainly during formative cell division in embryogenesis and early stages of lateral root development, suggesting α-Aurora’s critical function in establishing cellular asymmetry (Van Damme et al., 2011). Aurora kinases have also been implicated in mitotic and meiotic chromosome segregation in plants (Kurihara et al., 2006; Demidov et al., 2014) and in securing efficient cell cycle progression through phosphorylation of the MT-bundling protein MAP65-1 (Boruc et al., 2017).
Although a putative INCEN-P homolog termed “WYRD,” with deviating length and extremely poor sequence conservation to its animal counterpart, has been found (Kirioukhova et al., 2011), it is still unclear whether plants produce a chromosomal passenger complex. However, the Arabidopsis genome does contain a clear TPX2 homolog. The canonical TPX2 polypeptide includes an N-terminal hydrophobic Aurora-binding site, a central importin-binding domain, and a C-terminal TPX2 signature MT/kinesin-interacting region, all of which are conserved in the Arabidopsis TPX2 homolog (Vos et al., 2008; Zhang et al., 2017). Arabidopsis AUR1 colocalizes with TPX2 on the spindle MTs, was copurified with TPX2 from Arabidopsis cell cultures, and can phosphorylate TPX2 in vitro (Petrovská et al., 2012, 2013; Tomaštíková et al., 2015). Arabidopsis TPX2 can also bind to Xenopus importin-α in a RanGTP-dependent way (Vos et al., 2008). When antibodies raised against the human TPX2 were injected into the dividing stamen hair cells of the spiderwort Tradescandia virginiana, mitosis was blocked because of the inhibition of the formation of the prospindle, which is the bipolar-spindle–like MT array formed on the nuclear envelope (NE) at late prophase in plant cells (Vos et al., 2008). Furthermore, Arabidopsis TPX2 has MT nucleation capacity in vitro, and overexpression of TPX2 causes ectopic intranuclear MT nucleation in vivo that is independent of Aurora (Vos et al., 2008; Petrovská et al., 2013), suggesting that TPX2 is essential for bipolar spindle formation in plants, like in animals.
Study of multiple T-DNA insertional mutations revealed that homozygous tpx2 mutants did not exhibit obvious cell division or growth phenotypes. This finding implies that the function of canonical TPX2 may be shared with other related proteins. Next to the canonical TPX2, the Arabidopsis genome contains at least eight TPX-Like proteins (TPXLs), of which four bear predicted Aurora-binding motifs (Evrard et al., 2009; Tomaštíková et al., 2015), indicating that the TPX2 family expanded in plants. However, the function of these TPXLs, their connection with plant Aurora kinases, and the potential subfunctionalization of this protein family, remained up to now completely unknown. Here we present functional analyses of TPX2, TPXL2, and TPXL3 in Arabidopsis that fill some of these gaps in our knowledge.
RESULTS
TPXL2 and TPXL3 Are Interactors and Activators of AUR1 and AUR2
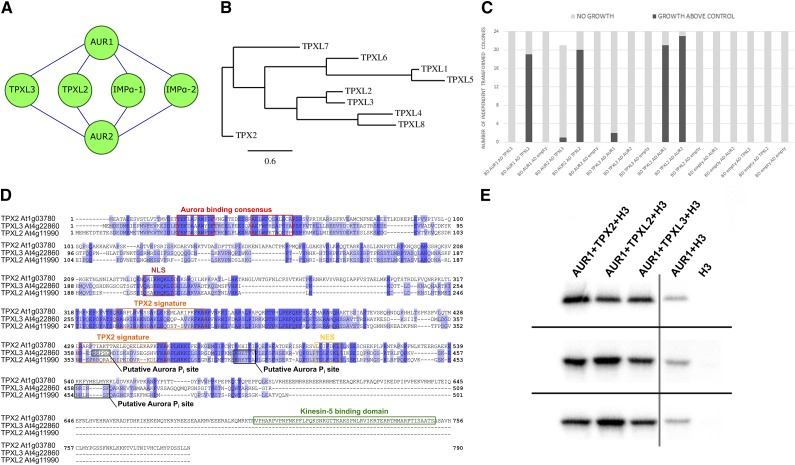
In the animal kingdom, Aurora kinases play critical roles in multiple cell division processes through interaction with partner proteins and phosphorylation of their substrates (Neumayer et al., 2014). To advance our knowledge on the function of AUR1 and AUR2 in Arabidopsis, we aimed to identify interactors by tandem affinity purification (TAP) coupled with mass spectrometry (MS) analyses, using AUR1 and AUR2 as the bait proteins. We performed two independent tandem affinity purification tag (TAP-tag) experiments using both N-terminal (NGSTEV), as well as C-terminal (CGSTEV) tagged AUR1 and AUR2, followed by MS through tandem matrix-assisted laser desorption/ionization–time of flight (Van Leene et al., 2011). From these experiments, we identified TPXL2, TPXL3, α-importin1 (IMPA1), and IMPA2 as interactors of both AUR1 and AUR2 (Fig. 1A; Supplemental Data Set S1A). Using the improved GSRHINO- and GSYELLOW-TAP tags fused C-terminally to AUR1 (CGSRHINO and CGSYELLOW), combined with the more sensitive Linear Trap Quadrupole (LTQ) Orbitrap Velos MS (Thermo Fisher Scientific; Van Leene et al., 2015), we not only confirmed these four proteins, but also identified seven additional interactors (Supplemental Data Set S1A). TPXL3 was consistently detected in all purifications with either AUR1 or AUR2 as the bait while TPXL2 was recovered in four out of six attempts. In contrast, TPX2 was only found in the Velos MS by using CGSRHINO- and CGSYELLOW-TAP–tagged AUR1. Whereas IMPA1 and IMPA2 were detected by both MALDI (Matrix Assisted Laser Desorption/Ionisation) and Velos with either AUR1 or AUR2, Velos MS revealed IMPA3, IMPA4, IMPA6, and three β-importins (karyopherin subunit-β, i.e. KPNB1–3). TPXL2 and TPXL3 sequences appear to be very similar to each other and cluster together (Fig. 1, B and D). We conclude that in contrast to TPX2, which we could only identify using the CGSrhino and CGSyellow tags coupled with the more sensitive LTQ detection, these TPXLs were also found in our experiments using the GSTEV tags combined with MALDI detection and therefore can be considered as bona-fide interactors of both AUR1 and AUR2. Except for TPX2, TPXL2, and TPXL3, our analysis did not identify other TPXLs using the quality criteria to filter interactors from TAP experiments (Van Leene et al., 2015).
Figure 1.
Arabidopsis TPXL2 and TPXL3 are interactors and activators of AUR1 and AUR2. A, Cytoscape representation of the AUR1 and AUR2 interactions detected using the TAP-tag assay on the Arabidopsis cell suspension culture. Next to two members of the importin protein family, two TPXLs were copurified with both Aurora kinases. B, Phylogenetic tree of TPX2 and eight TPXLs showing the evolutionary relationship among these nine Arabidopsis proteins. TPXL2 and TPXL3 form a subfamily. C, Quantification of the Y2H assay (data shown in Supplemental Fig. S1). A total of 21–24 independent double-transformed yeast colonies from two independent yeast transformations were scored for their interaction based on their capacity for growth on selective medium. The obtained results confirm the interaction between both alpha Aurora kinases and TPXL2, whereas TPXL3 hardly interacts with the α-Aurora in this assay. D, Protein sequence alignment of TPX2, TPXL2, and TPXL3 showing that in contrast to TPX2, TPX2L and TPXL3 lack the Kinesin-5 binding domain. The Aurora binding consensus motif (red), NLS (light brown), nuclear export signal (NES, yellow), TPX2 signature (orange), Kinesin-5 binding domain (green), and the putative Aurora phosphorylation sites (Pi; black) are marked with colored boxes. E, In vitro kinase assay preformed in triplicates (top, middle, and bottom row) showing the phosphorylation level of recombinant Histone H3-6xHIS in the absence or presence of AUR1 and/or the N-terminal fragments of the TPX(L) proteins. The phosphorylation activity of recombinant Aurora1 is dramatically increased in the presence of the N-terminal TPX2, TPXL2, or TPXL3 fragments containing the predicted Aurora binding domain (amino acids 1–100). The gray line indicates the division between two parts of the kinase assay gel.
To take one step further from the TAP results, we tested interactions between AUR1 and AUR2 and both TPXLs using yeast two-hybrid (Y2H) analysis. The interaction between TPXL2 and both AUR1 and AUR2 could be confirmed in both directions using this system, yet, TPXL3 barely interacted with AUR1 and AUR2 in this assay (Fig. 1C; Supplemental Fig. S1)—likely reflecting differences between both TPXLs and/or the occurrence of counterselection against TPXL3 expression in this system.
TPXL2 and TPXL3 share a number of TPX2 motifs. They all bear the N-terminal Aurora-binding domain (to a certain extent the first of the two central TPX2 signature motifs; Vos et al., 2008), an NLS, and a nuclear export signal. Both TPXL2 and TPXL3, however, lack the C-terminal Kinesin-5-binding domain typical for TPX2 (Fig. 1D). Nevertheless, we did detect two and three putative Aurora phosphorylation sites within the last 150 amino acid fragments of TPXL2 and TPXL3, respectively (Fig. 1D).
The N-terminal part of canonical TPX2, containing the Aurora-binding domain, enhances the phosphorylation activity of the kinase (Tomaštíková et al., 2015). To test whether TPXL2 and TPXL3 are also activators of Aurora kinases, we performed in vitro kinase assays in triplicates in the absence and presence of recombinant proteins consisting of the first 100 amino acids of TPX2, TPXL2, and TPXL3. The activation of AUR1 kinase activity was demonstrated by the increased phosphorylation of recombinant Histone H3 in the presence of TPX(L) proteins when compared to AUR1 alone. Whereas recombinant AUR1 is capable of phosphorylating Histone H3 to some extent in vitro, this substrate is hyperphosphorylated in the presence of the N-terminal fragments of TPX2, TPXL2, or TPXL3 at similar capacities, indicating that all three TPX(L) proteins are potent activators of AUR1 (Fig. 1E). Our results indicate that at least in vitro, there is no major difference in activation of AUR1 by these three TPX(L) proteins (Fig. 1E).
TPX2, TPXL2, and TPXL3 Expression Is Correlated with Meristematic Activity
AUR1 and AUR2 expression is strongly correlated with cell division (Demidov et al., 2005; Van Damme et al., 2011) and both AUR1 and AUR2 show transcriptional coregulation with TPX2 and TPXL3 during the cell cycle (Supplemental Fig. S2A; Menges et al., 2003). To test whether the expansion of the plant TPX family could be associated with tissue-specific activation of aurora kinases, we analyzed their expression patterns in detail. We generated transcriptional reporter lines for TPX2, TPXL2, and TPXL3 with putative promoter sequences of 2630-, 2018-, and 1793-bp, respectively. The promoter activities of TPX2, TPXL2, and TPXL3 were detected in similar tissues, but overall those of TPX2 and TPXL3 seemed stronger than that of TPXL2 (Supplemental Fig. S2, B–D). All promoters showed high expression in shoot and root apical meristems, lateral root meristems, and in flowers. More specifically, they drove GUS expression in young pistils, ovules, and anthers (Supplemental Fig. S2, B–D). In developing siliques, staining was clearly detected in young seeds for all three promoters although again TPXL2 seemed to have a lower expression compared to the other ones. There was also expression detected in the vasculature of cotyledons and true leaves, again more conspicuous for TPX2 and TPXL3. Expression in the stomatal lineage cells was found only for promTPXL3 and promTPX2, while expression in the flower and silique abscission zone appeared to be more specific for promTPXL2.
The observed expression profiles, which correlated clearly with tissues enriched for actively dividing cells, are in agreement with the fact that these proteins are interactors and activators of the mitotic kinases of α-Aurora. The promTPX2, promTPXL2, and promTPXL3 activity showed clear overlap in dividing root tissues, as well as during embryo development, indicating that at least in these tissues, it would be unlikely for AUR1 and AUR2 to be selectively activated by the specific TPX family members studied here. The expansion of the TPX family, therefore, does not seem to be correlated with clear differences in expression domains.
TPX2, TPXL2, and TPXL3 Interact with AUR1 In Planta
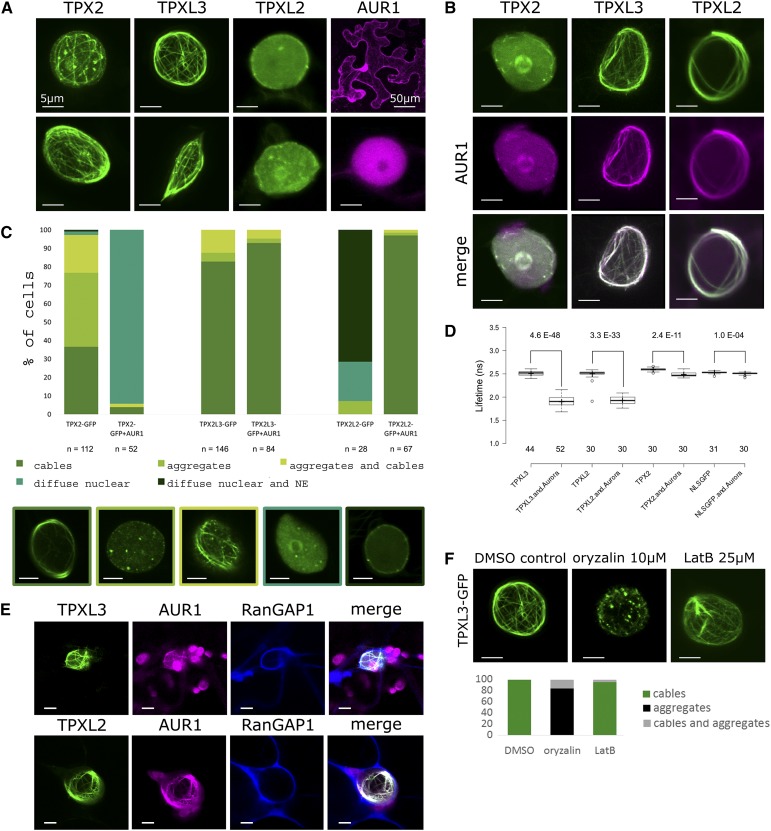
To corroborate the interactions among AUR1 and the three TPX2 family proteins in planta, AUR1 was transiently coexpressed with either TPX2, TPXL2, or TPXL3 in Nicotiana benthamiana leaves. The localization of the individual fluorescent fusions showed that the AUR1-RFP signal was diffuse in the cytoplasm and the nucleus in these epidermal pavement cells (Fig. 2A). TPX2-GFP and TPXL3-GFP overall formed nucleus-associated aggregates and filamentous structures, resembling a ball of yarn. In contrast, TPXL2 was more diffuse than filamentous, with the majority of the nuclei showing a pronounced localization close to the NE together with sporadic bright patches (Fig. 2, A and C). Coexpression of AUR1-RFP and GFP fusions of the three TPX family proteins led to distinct relocalization of AUR1. Instead of being cytoplasmic and nuclear, AUR1-RFP became restricted to the nucleus when it was coexpressed with either TPX2-, TPXL2-, or TPXL3-GFP (Supplemental Fig. S3). Surprisingly, we also observed changes in the nuclear localization of AUR1, TPX2, and TPXL2 when they were expressed simultaneously. When coexpressed with AUR1-RFP, TPX2-GFP mostly lost its filamentous localization, but shared diffused localization with AUR1. Coexpression of AUR1-RFP and TPXL2-GFP, on the other hand, altered the localization of both proteins to nucleus-associated filamentous cables (Fig. 2, B and C). TPXL3 retained its filamentous nuclear localization and recruited AUR1 to these cable-like structures. To determine on which side of the NE the observed cable-like structures formed, we used a NE marker, RanGAP1-BFP, which was coexpressed with AUR1-RFP and either TPXL2-GFP or TPXL3-GFP (Fig. 2E). The GFP and RFP signals colocalized with each other and were confined within the RanGAP1-delineated space, showing that these cable-like structures are intranuclear.
Figure 2.
TPX proteins interact with AUR1 in planta and initiate intranuclear MT nucleation/polymerization. A–C, Representative localizations (A and B) and quantification of the observed localization patterns (C, n = number of nuclei) for TPX2, TPXL3, and TPXL2 with and without AUR1 in N. benthamiana leaf epidermal cells. Single expression of TPX2 and TPXL3, but not TPXL2, causes the formation of intranuclear aggregates and cables resembling cytoskeletal filaments. AUR1 is diffuse nuclear and cytoplasmic (A and C). Coexpression of TPX2, TPXL2, and TPXL3 with AUR1 differently affects TPX/L localization. AUR1 has a negative impact on the bundling activity of TPX2, is recruited to the bundles formed by TPXL3 and activates the bundling activity of TPXL2 (B and C). Images below (C) represent the different classes of localizations observed. Left to right: cables (TPX2-GFP); aggregates (TPX2-GFP); aggregates and cables (TPX2-GFP); diffuse nuclear (TPX2-GFP and AUR1); diffuse nuclear and NE (TPX2L2-GFP). D, FLIM analysis of cotransformed N. benthamiana epidermal cells. Coexpression of TPX-GFP with AUR1-mRFP reduces the donor lifetime. The reduction in lifetime values is the most pronounced for TPXL2 and TPXL3 (from 2.5 to 1.9 ns); the lifetime decrease of TPX2 is less dramatic (from 2.59 to 2.48). The lifetime of NLS-GFP hardly changes when combined with AUR1 (from 2.53 to 2.51). Numbers represent Student’s t test P values (top) and the number of nuclei analyzed (bottom). E, Triple localization of TPXL3 or TPXL2 (green), AUR1 (magenta), and the NE marker RanGAP1 (blue) in N. benthamiana epidermal cells shows that the cable-like structures form inside the nuclei outlined by RanGAP1. F, The intranuclear cables marked by the TPX proteins are MTs. TPXL3-GFP localization to the cable-like intranuclear structures is sensitive to a 1-h treatment of 10-μm oryzalin (n = 25) and changes from intranuclear cables into a predominantly aggregated pattern in nuclei when compared to the DMSO control (n = 20). LatB does not affect the localization of TPXL3-GFP (1 H, 25 μM, n = 21). All scale bars = 5 μm, except for the top-right corner image in (A), which is = 50 μm.
The intranuclear interactions between AUR1 and the TPX proteins were assessed using fluorescence-lifetime imaging microscopy (FLIM) by comparing the lifetimes of GFP-tagged TPX2, TPXL2, and TPXL3 expressed individually, with those values when they were expressed together with AUR1-RFP (Fig. 2D). For TPXL2 and TPXL3, the lifetimes decreased from ∼2.5 ns (single infiltration) to 1.9 ns (double infiltration). This lifetime value drop indicates a direct interaction between AUR1 and TPXL2 or TPXL3. The lifetime reduction for TPX2 was much smaller than for the TPXLs (from 2.59 to 2.48 ns; Fig. 2D). As a control, we used NLS-GFP. As expected, the lifetime of NLS-GFP was similar in the absence or in the presence of AUR1 (from 2.53 to 2.51 ns). Although some energy transfer could be observed when both fluorophores were present in the nucleus, the statistical significance differed by several orders of magnitude compared to TPX2 and the TPXLs.
Next, we aimed to determine the nature of these cable-like structures using TPXL3-GFP as a representative marker in this in vivo experiment. N. benthamiana leaves infiltrated with the fluorescent fusion were subsequently injected with either oryzalin or Latrunculin B (LatB) to depolymerize MTs and actin microfilaments, respectively. Oryzalin reverted the TPXL3-GFP-labeled filaments to distinct nuclear foci. LatB, on the other hand, did not affect the structure of the TPXL3-GFP filaments (Fig. 2F). These results indicate that TPXL3, and very likely also TPX2 and TPXL2, associate with intranuclear MTs.
TPX2 Is Dispensable for AUR1 and AUR2 Localization and Spindle Formation
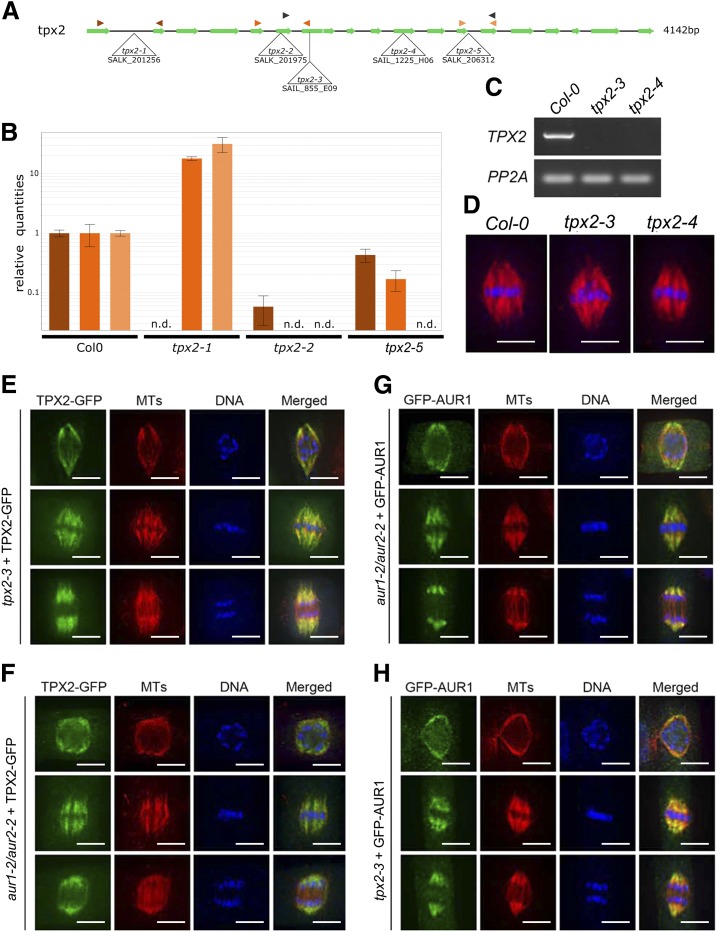
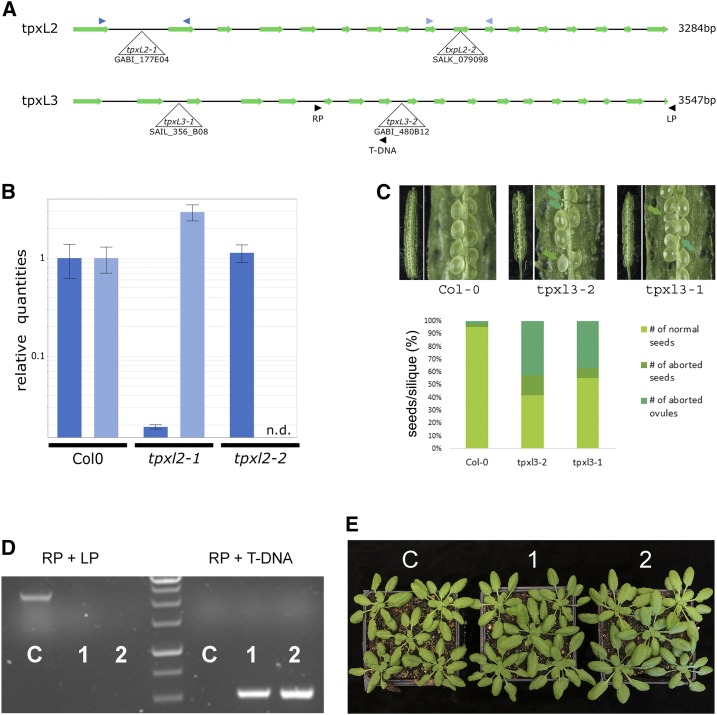
An earlier report stated that TPX2 was essential for spindle formation and there was no viable homozygous tpx2 mutant recovered when T-DNA insertional lines were screened (Vos et al., 2008). We aimed to determine the cause of such a lethality by examining community-generated T-DNA lines. To our surprise, homozygous mutants of five different T-DNA insertions in three exons and two introns of the TPX2 gene were recovered. None of these homozygous lines exhibited any aberrant morphological phenotype as they grew indistinguishably from the Col-0 control plants (Supplemental Fig. S4). Reverse transcription-quantitative PCR (RT-qPCR) analyses revealed that the expression of full-length TPX2 was absent in tpx2-1, tpx2-2, and tpx2-5 mutants (Fig. 3B). We also assayed TPX2 expression in the tpx2-3 and tpx2-4 mutants with insertions in the 7th and 11th exons, respectively (Fig. 3A). The RT-qPCR result showed that both were also likely knock-out mutants as the TPX2 mRNA was undetectable (Fig. 3C).
Figure 3.
Mutant analysis reveals TPX2 as nonessential. A, Schematic overview of the gene model for TPX2 with indication of the positions of the T-DNA insertion alleles analyzed. Primer pairs used for RT-qPCR and RT-PCR analysis are indicated by color-coded arrowhead pairs. B, RT-qPCR analysis of homozygous tpx2-1, tpx2-2, and tpx2-5 mutants using the three different primer pairs shown in (A). Error bars represent se. The graph shows the absence of full-length transcripts over the T-DNA positions (n.d., not detected). C, Using the primers marked by the black arrowheads in (A), RT-PCR of homozygous tpx2-3 and tpx2-4 mutants show the absence of transcript compared to wild type (Col-0). The constitutively expressed PP2A gene was set as the positive control. D, Similar spindle MT arrays are formed in metaphase cells in the control (Col-0) and homozygous tpx2-3 and tpx2-4 plants. The immunofluorescent images have MTs pseudo-colored in red and DNA in blue. E and F, TPX2-GFP localization upon expression in the null tpx2-3 mutant (E) and in homozygous aur1-2/aur2-2 double mutant cells (F). Representative cells are at late prophase (top row), metaphase (middle row), and anaphase (bottom row). TPX2-GFP is pseudo-colored in green, MTs in red, and DNA in blue. TPX2-GFP localizes to the prospindle MTs in prophase and K-fiber MTs at metaphase and anaphase. No obvious difference was detected between the control and aur1-2/aur2-2 mutant cells. G and H, GFP-AUR1 localization in complemented aur1-2/aur2-2 (G) and in tpx2-3 mutant cells (H). Representative cells are at late prophase (top row), metaphase (middle row), and anaphase (bottom row). GFP-AUR1 is pseudo-colored in green, MTs in red, and DNA in blue. GFP-AUR1 localizes to the prospindle MTs in prophase and K-fiber MTs at metaphase and anaphase. No obvious difference was detected between the control and tpx2 mutant cells. Images shown are representative examples chosen out of >10 individual images for each cell cycle phase. Scale bars = 5 μm.
Because TPX2 was implicated in spindle formation (Vos et al., 2008), we examined spindle morphology in the tpx2-3 and tpx2-4 mutants when compared to the control cells. Cells of all mutant lines formed mitotic spindles indistinguishable from those in the wild-type control cells. Metaphase spindles of the control, tpx2-3, and tpx2-4 cells had a fusiform appearance with chromosomes perfectly aligned at the metaphase plate (Fig. 3D). Therefore, both lines of evidence led to the conclusion that TPX2 is not essential for spindle assembly, or the growth and reproduction in Arabidopsis, a model representing dicotyledonous angiosperms.
Considering the fact that the tpx2 mutants did not show mitotic or developmental defects, we asked whether TPX2, by analogy to its primary function in animal cells, has a role in targeting plant Aurora kinases to the spindle. We therefore compared the localization of TPX2-GFP and GFP-AUR1, both expressed under the control of their native promotor, in their respective and complementary mutant backgrounds. Among the mitotic cells observed by immunolocalization, three representative stages were selected: late prophase (around the time of NEBD when the prospindle could be discerned), metaphase (chromosomes aligned in the middle of a well-established bipolar spindle), and anaphase (shortened kinetochore MT fibers). TPX2-GFP, expressed under the control of its native promoter, in the tpx2-3 mutant localized with a fusiform-shaped MT array on the NE (the prospindle), in late stages of prophase (Fig. 3E, top). It associated with spindle MTs, especially kinetochore (K)-fiber MTs, and became more pronounced toward spindle poles in both metaphase and anaphase ( Fig. 3E, middle and bottom). As the spindle localization of TPX2 is independent from Aurora A in animal cells (Kufer et al., 2002), we asked whether a similar phenomenon could be observed in plant cells by employing the aur1-2/aur2-2 double mutant. In the aur1-2/aur2-2 mutant background, the prospindle was often distorted and did not exhibit an obvious converging morphology (Fig. 3F, top row). However, TPX2-GFP still associated with MT bundles around the NE. At both metaphase and anaphase, TPX2-GFP localized with K-fiber MTs, similar to its localization in the control cells (Fig. 3F, middle and bottom rows, respectively). Therefore, we concluded that strongly reduced levels of α-Aurora do not prevent TPX2 association with spindle MTs in planta.
The N-terminal GFP-AUR1 fusion fully complemented the small dwarf and bushy growth phenotype of the homozygous aur1-2/aur2-2 double mutant in Arabidopsis (Supplemental Fig. S5), similar to what was reported for the C-terminal fusion (AUR1-GFP; Van Damme et al., 2011). In the aur1-2/aur2-2 double mutant background, the GFP-AUR1 fusion first associated with MT bundles in the prospindle (Fig. 3G, top row). Then it localized with K-fibers of both metaphase (Fig. 3G, middle row) and anaphase spindles (Fig. 3G, bottom row). The protein was not associated with MT bundles in the spindle midzone between two sets of sister chromatids (Fig. 3G, bottom row). The localization pattern is indistinguishable from that detected with the AUR1-GFP fusion in living cells (Demidov et al., 2005; Van Damme et al., 2011). We then examined GFP-AUR1 targeting in the tpx2-3 mutant cells. Unlike what has been reported in vertebrates (Kufer et al., 2002), GFP-AUR1 retained its localization pattern in the absence of TPX2 in prophase, metaphase, and anaphase cells (Fig. 3H, top, middle, and bottom rows, respectively). Therefore, we infer that AUR1 and AUR2 can associate with spindle MTs independently of the canonical TPX2 protein in Arabidopsis.
The Localization Pattern of TPXL2 and TPXL3 in Dividing Cells Differs from That of TPX2
Because of the dispensability of TPX2 for mitosis in Arabidopsis, we assessed the activities of TPXL2 and TPXL3 in mitotic cells because of their interaction with α-Aurora. First, we examined their subcellular localization in Nicotiana tabacum BY-2 cells as a reference system. The two fusion proteins (p35S::TPXL2-GFP and p35S::TPXL3-GFP) showed similar localization patterns in mitosis (Supplemental Fig. S6). Before cell division, both were detected close to the NE. Immediately before NEBD, the TPXL2 and TPXL3 signals appeared to accumulate outside of the NE, colocalizing with MTs marked by the MT-binding domain (MBD) at the prospindle (Supplemental Fig. S6). Next, TPXL2 and TPXL3 associated with the metaphase spindle and appeared more prominent on K-fiber MTs than on the bridging MTs, given the reduced overlap with the RFP-MBD signal at the center of the spindle. In anaphase, both TPXL2 and TPXL3 coincided with the shortening K-fiber MTs. In fact, they became increasingly concentrated toward the spindle poles in both metaphase and anaphase cells. But they clearly avoided the spindle midzone where increased MTs were developing into the bipolar phragmoplast MT array. A striking localization pattern was found during later stages of mitosis when the NE was reforming. Both TPXL2-GFP and TPXL3-GFP fusions heavily localized at the reforming daughter nuclei and eventually exhibited cage-like patterns. In contrast to the situation at prophase, these cage-like patterns did not overlap with the RFP-MBD MT marker, strongly suggesting that they were intranuclear. This cell cycle-dependent pattern spatially resembled the intranuclear cage-like structures observed in the N. benthamiana nuclei. During cytokinesis, both TPXL2-GFP and TPXL3-GFP were clearly absent from the phragmoplast MTs.
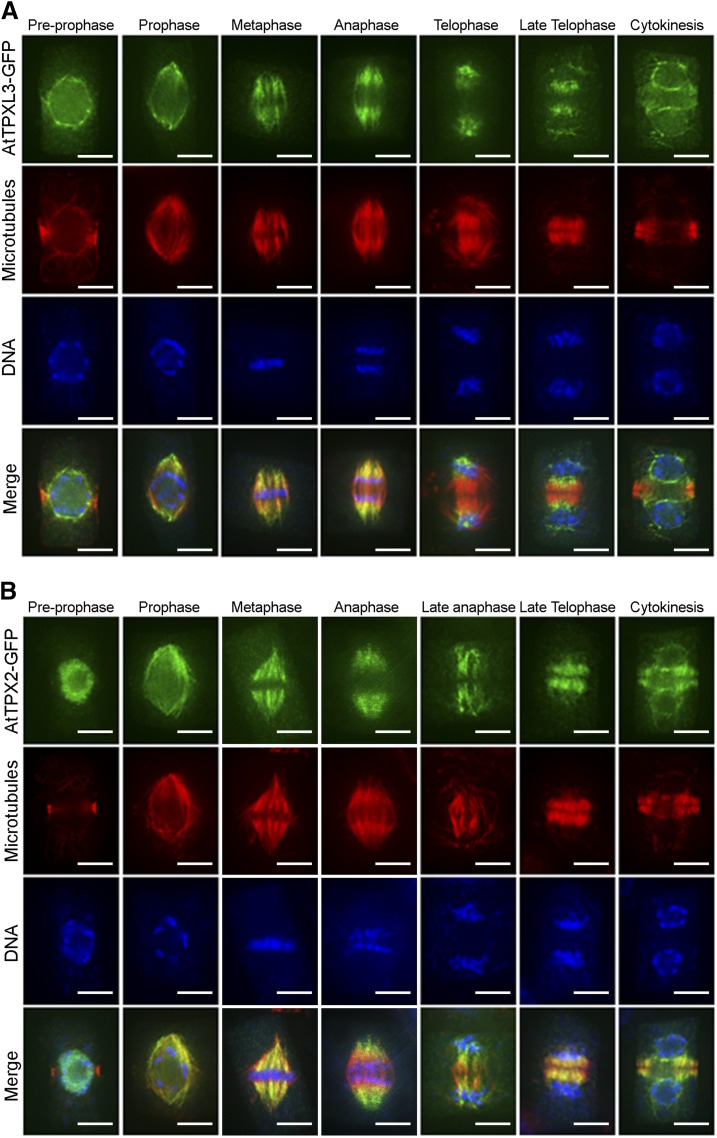
Because we understand the potential caveat of ectopic overexpression of GFP fusion proteins in a heterologous system, we aimed to determine the localization of TPXL2 and TPXL3 expressed at near-endogenous levels in planta. To do so, GFP fusion constructs were made using the corresponding genomic fragments including the putative promoters and coding sequences and transformed into wild-type Arabidopsis ecotype Col-0. Because the TPXL2-GFP signal was hardly detectable in our transformed plants, we focused our analysis on TPXL3-GFP. In fact, the localization pattern of TPXL3-GFP in dividing Arabidopsis root cells was similar to that in BY-2 cells. To gain high spatial resolution, the localization of TPXL3-GFP was determined in fixed cells with MTs and DNA as spatial and temporal references (Fig. 4A). After immunostaining with an anti-GFP antibody, TPXL3-GFP was detected prominently at the NE in cells with condensed preprophase bands (PPB), but was absent from the PPB, indicating that the protein was selectively associated with certain MTs. Toward late stages of prophase, when NEBD occurs and the PPB disappears, TPXL3-GFP strongly labeled the polar caps. Similar to BY-2, K-fiber MTs in both metaphase and anaphase spindles were labeled. After the chromosomes reached the spindle poles at telophase as marked by the nearly complete disappearance of K-fibers, TPXL3 became particularly prominent at the spindle poles where very few MTs were detected (Fig. 4A). At this stage, TPXL3 signal was also detected around the chromosome masses, although not as strongly as at the poles. It was, however, largely absent from the central spindle where MTs developed into a bipolar array and formed the phragmoplast. During cytokinesis, TPXL3 accumulated at the reforming NE of the daughter cells.
Figure 4.
TPX2 and TPXL3 localize differentially in dividing Arabidopsis root cells. A and B, TPXL3-GFP (A) and TPX2-GFP (B) localization in dividing Arabidopsis root cells obtained via immunofluorescence with antibodies against GFP (green), MTs (red), and DNA (blue) in cells from prophase to telophase. Before NEBD, TPXL3 largely accumulates at the NE, while TPX2 is mostly nuclear. Both proteins are not detectable at the PPB. After NEBD, both TPX2 and TPXL3 decorate the two “polar caps” of the prophase spindle as well as spindle MTs and the shortening K-fibers at anaphase. In late anaphase and telophase, when midzone MTs develop into the two mirrored sets of the early phragmoplast array, TPX2 association with the phragmoplast-forming MTs is much more pronounced than that of TPXL3, which remains heavily enriched at the former spindles poles and subsequently localizes to the reformed NE with a strong bias toward the part facing the reforming daughter nuclei. Scale bars = 5 μm.
We also compared the localization of TPXL3-GFP with that of TPX2-GFP in dividing root cells of Arabidopsis (Fig. 4B). Although the localization pattern during spindle formation up to anaphase was rather similar for TPXL3 and TPX2, we observed differences at the PPB stage. TPX2 remained primarily nuclear at this point, while TPXL3 accumulated at the NE. Furthermore, during telophase and cytokinesis, and in contrast to TPXL3, TPX2 did mark the phragmoplast MTs but its association to and its association with the NE of the daughter cells was not as prominent as that of TPXL3. In conclusion, our localization data in both BY-2 and Arabidopsis cells shows that regulatory mechanisms remain to be characterized that account for the overlaps as well as differences in the targeting of TPX2 as compared to TPXL2 and TPXL3. These differences mainly concern the capacities to associate with the NE before division and during cytokinesis.
TPXL3 Is Essential for Plant Development
To investigate the functions of TPXL2 and TPXL3 in plant development, two independent T-DNA insertional mutations were identified for each gene (Fig. 5A). For TPXL2, two homozygous mutants were recovered lacking full-length transcripts (Fig. 5B). Both mutants developed similarly, compared to the control plants. In addition, we could not identify any major developmental delay in plants where insertions in TPX2 and TPXL2 were combined [tpx2-3(−/−)/tpxl2-2(−/−); Supplemental Fig. S4]. In contrast to TPX2 and TPXL2, however, we failed to recover homozygous plants for either of the TPXL3 T-DNA insertional lines. Antibiotic selection revealed that offspring of tpxl3-1 Syngenta Arabidopsis Insertion Library (SAIL), confirmed by genotyping PCR to be heterozygous for the T-DNA) and offspring of tpxl3-2 (GABI-Kat, confirmed by genotyping PCR to be heterozygous for the T-DNA), segregated according to a 2:1 ratio of resistant versus sensitive plants (Table 1). Reciprocal back-cross experiments to Col-0, using both heterozygous tpxl3-1 and tpxl3-2 insertion lines, revealed transmission of the T-DNA via both the male and female, although a decrease in the female T-DNA transmission frequency was observed for tpxl3-1 (Table 1). A close inspection of the developing siliques of the self-pollinated tpxl3-1 and tpxl3-2 lines revealed that the mutant siliques were ∼20% shorter than Col-0 siliques (Col-0, 1.36 ± 0.061[sd] cm, n = 20; tpxl3-1, 1.15 ± 0.082[sd] cm, n = 30; tpxl3-2, 1.09 ± 0.08[sd] cm, n = 20). These siliques contained a high percentage of aborted ovules and some aberrant seeds (Fig. 5C), indicating defects during fertilization and/or during initial embryo development.
Figure 5.
Mutant analysis revealed TPXL2 as nonessential whereas TPXL3 is essential for embryo development. A, Schematic overview of the gene models for TPXL2 (top) and TPXL3 (bottom) with indication of the positions of the T-DNA insertion alleles analyzed. Primers used for RT-qPCR and genotyping analysis are indicated by color-coded arrowhead pairs. B, RT-qPCR analysis of homozygous tpxl2 mutants showing the absence of full-length transcripts over the T-DNA positions (n.d., not detected). Error bars = se. C, Representative silique pictures and quantification of seed development of selfed tpxl3-1 (+/−) and tpxl3-2 (+/−) plants grown together with control plants (Col-0). Both TPXL3 alleles show a high percentage of aborted ovules and some aborted seeds. The quantification shows the combined result of different siliques from different plants. For both Col-0 and tpxl3-2, 20 siliques were analyzed (from two different plants each), whereas for tpxl3-1, we analyzed 30 siliques (from three different plants). D and E, Introducing TPXL3-GFP into tpxl3-2(+/−) mutants allowed identification of homozygous tpxl3-2 mutants (two independent lines are shown; “1” and “2”), which develop similarly to the wild-type controls (“C”). Genotyping results revealed the absence of wild-type (LP + RP) fragment (∼2.5-kb) and the presence of the T-DNA specific fragment (RP + T-DNA; ∼0.5-kb) in two independent lines carrying the TPXL3-GFP transgene (“1” and “2”) in contrast to control plants (“C”).
Table 1. Segregation and back-cross segregation analysis of TPXL3 insertion lines.
Genotyping analysis for both TPXL3 insertion lines failed to recover homozygous mutants and segregation analysis showed that both tpxl3-1 and tpxl3-2 lines segregate in a 2:1 (Resistant/Sensitive [R/S]) ratio on antibiotic selection, hinting toward embryo lethality. Back-cross experiments to wild type revealed transmission of the mutant allele via both the male and female gametes, and a reduction in transmission of the tpxl3-1 mutant allele via the female gametes was observed.
| Segregation Analysis | ||||||
|---|---|---|---|---|---|---|
| Line | Insertion | Sensitive | Resistant | Total | Ratio R/S | χ2 |
| tpxl3-1 | SAIL_356B08 | 323 | 680 | 1,003 | 2.1 | |
| Expected (3:1) | 250.8 | 752.3 | 1,003 | 3 | 27.76 | |
| Expected (2:1) | 334.3 | 668.7 | 1,003 | 2 | 0.58 | |
| tpxl3-2 | GK_480B12 | 272 | 546 | 818 | 2.0 | |
| Expected (3:1) | 204.5 | 613.5 | 818 | 3 | 29.71 | |
| Expected (2:1) | 272.7 | 545.3 | 818 | 2 | 0.002 | |
| Back-Cross Segregation Analysis Wild-Type♀ × Mutant♂ | ||||||
| Line | Insertion | Mutant | Wild-Type | Total | Ratio Wild-Type/Mutant | χ2 |
| tpxl3-1 (+/−) | SAIL_356B08 | 73 | 85 | 158 | 1.16 | |
| Expected (1:1) | 79 | 79 | 158 | 1 | 0.91 | |
| tpxl3-2 (+/−) | GK_480B12 | 24 | 18 | 42 | 0.75 | |
| Expected (1:1) | 21 | 21 | 42 | 1 | 0.86 | |
| Back-Cross Segregation Analysis Wild-Type♂ × Mutant♀ | ||||||
| Line | Insertion | Mutant | Wild-Type | Total | Ratio Wild-Type/Mutant | χ2 |
| tpxl3-1 (+/−) | SAIL_356B08 | 44 | 101 | 145 | 2.30 | |
| Expected (1:1) | 72.5 | 72.5 | 145 | 1 | 22.41 | |
| tpxl3-2 (+/−) | GK_480B12 | 46 | 56 | 102 | 1.22 | |
| Expected (1:1) | 51 | 51 | 102 | 1 | 0.98 | |
To determine whether the lethality was linked to the identified insertion in the tpxl3-2 mutant and also to test the functionality of the TPXL3-GFP fusion protein described in Figure 4, we examined whether the tpxl3-2 mutation could be genetically suppressed. Using a primer pair specific for the TPXL3 locus, we were able to distinguish the native TPXL3 gene and the TPXL3-GFP transgene (Fig. 5, A and D). Indeed, we recovered two plants with homozygous tpxl3-2 background when the TPXL3-GFP fusion was expressed. The complemented plants developed similarly to controls (Fig. 5E). Taken together, in contrast to TPX2 and TPXL2, TPXL3 is an essential gene in Arabidopsis, and the TPXL3-GFP fusion used for determining TPXL3 localization (Fig. 4) is functional.
DISCUSSION
Our results showed that in Arabidopsis, representing dicotyledonous plants, the expansion of the TPX2 protein family has led to the selection of TPXL3 as an essential MAP while the canonical TPX2 has become dispensable. We also found that TPX2 and the two TPXLs examined here are in vivo partners of AUR1 and AUR2 and showed overlapping as well as differential localizations during mitosis. Thus our studies opened the door to further investigations of the relationship between TPX2 family proteins and AUR1 and AUR2 as well as how the functions of these MAPs are tied together with mitosis-dependent MT reorganization in plants.
The MAP TPX2, initially identified in Xenopus, is required for targeting a plus end-directed motor protein, Xenopus Kinesin-Like Protein2, to the minus ends of the spindle MTs. TPX2 is also necessary for MT nucleation around the chromosomes in response to a local RanGTP gradient (Wittmann et al., 1998, 2000; Gruss et al., 2001). Such TPX2-mediated MT nucleation from the chromosomes is essential for bipolar spindle formation in Hela cells (Gruss et al., 2002). Unlike vertebrates, however, the worm Caenorhabditis elegans and the fly Drosophila melanogaster lack a clear TPX2 ortholog (Ozlü et al., 2005; Goshima, 2011). Instead, the worm contains a protein harboring the Aurora binding domain of TPX2, but lacking the C-terminal kinesin binding domain. This invertebrate TPXL-1 protein shares with vertebrate TPX2 the capacity to activate Aurora A (Bayliss et al., 2003; Ozlü et al., 2005). More surprisingly, although the fly contains a protein with significant homology to TPX2, it lacks both the Aurora and the Kinesin-5 binding domains (Goshima, 2011). Arabidopsis does contain a clear TPX2 ortholog plus several related TPXLs. These TPXLs differ in the presence or absence of domains characterizing vertebrate TPX2, such as the Aurora binding domain, the TPX2 domain, or the Kinesin-5 binding domain (Goshima, 2011; Tomaštíková et al., 2015). In animal cells, the TPX2-Kinesin-5 (Eg5) interaction depends on the C-terminal Kinesin-5 binding domain of TPX2, and this domain is critical for the bipolar spindle organization and control of Eg5 activity (Ma et al., 2011). The finding that TPX2 is not essential in plants, therefore, points to a different regulation of Eg5 in this kingdom and a different mode of action of the TPXL in spindle assembly. Neither TPXL2 nor TPXL3 contain the C-terminal kinesin-binding domain and, thus, they are unlikely to act redundantly with TPX2 to control Eg5 activity.
In contrast to vertebrates, flies, and worms, plants contain both TPX2 and TPXLs and therefore possess an expanded family of TPX2 proteins. The closely related TPXL2 and TPXL3, especially TPXL3, were identified as the primary interactors of the AUR1 and AUR2 in vivo by our MS-based interactomic tests, and the interactions were recapitulated by independent assays. In contrast, TPX2 could only be identified using the more sensitive LTQ Orbitrap Velos. The interaction of AUR1-RFP and TPX2-GFP has previously been reported using coimmunoprecipitation from Arabidopsis culture cells upon co-overexpression of both proteins (Petrovská et al., 2012), indicating that the low detection frequency observed in our proteomics results was likely caused by the low abundance of TPX2 in cycling cells of the Arabidopsis culture or possibly by a weak interaction. These results are also consistent with the finding that TPXL3, but not TPX2 or TPXL2, is essential.
Next to TPXL2 and TPXL3, our TAP analyses using AUR1 as bait revealed several importin α- and β-proteins, known to facilitate nuclear import of various proteins as was recently shown for the auxin response protein BODENLOS (Herud et al., 2016). It is plausible that importins shuttle TPX2 and the TPXLs, and thereby also the Aurora kinases, to the nucleus. Aurora kinases are nuclear before cell division (Van Damme et al., 2011) and interaction between Arabidopsis TPX2 and importins was already shown before (Vos et al., 2008; Petrovská et al., 2013). Moreover, we observed a strong nuclear accumulation of Arabidopsis TPX2, TPXL2 and TPXL3 in N. benthamiana cells and the nuclear and cytoplasmic localization of AUR1 in N. benthamiana shifted to exclusively nuclear upon the overexpression of any one of the three TPX(L) proteins in this system.
Similar to TPX2 (Tomaštíková et al., 2015), the N-terminal part of TPXL2 and TPXL3 activates AUR1 kinase activity in vitro. Our findings, including the transcriptional coregulation of AUR1, AUR2, TPX2, and TPXL3, suggest that these two proteins not only act as targeting factors of AUR1 but also likely determine the phosphorylation of substrates at specialized locations. The differences in their localizations, especially between TPX2 and TPXL3 during later stages of the cell cycle and around the time of cell cycle exit, support the notion that different MT-associated proteins may be phosphorylated by AUR1 on different MT arrays. Currently, it is unclear how the localization differences are achieved. Because of the presence of the Kinesin-5-binding site in TPX2 but not TPXLs, TPX2 may specifically require Kinesin-5 for its localization. It would be interesting to test potential interaction(s) between TPX2 and the four isoforms of Kinesin-5 in Arabidopsis.
Our transient overexpression assays using TPX2, TPXL2, TPXL3, and AUR1 in N. benthamiana cells recapitulated the previously reported capacity of TPX2 to localize to or to generate intranuclear MTs in Arabidopsis cell cultures (Petrovská et al., 2013). The intranuclear cage could also be observed in stably transformed BY-2 cells expressing TPXL2-GFP and TPXL3-GFP, although only at NE reformation during cytokinesis. This might be attributed to the presence of specific modulators in cycling cells compared to differentiated leaf epidermal cells or to differences in expression levels between transient transformation and stable cell lines.
Previously, it was concluded that the formation of those intranuclear MT bundles by TPX2 did not require Aurora kinase activity and that they were not linked to a specific cell cycle phase (Petrovská et al., 2013). Here, differences in the generation of intranuclear MTs were found when different TPX(L) proteins were expressed. We also show that AUR1 affects the capacity of TPX2 and TPXL2 to bind to these MTs. Indeed, whereas TPX2 was capable of forming cage-like structures in the absence of AUR1, this capacity strongly declined in the presence of AUR1. This is in agreement with the observation that in cells with TPX2-localized MTs, the AUR1 signal was very low (Petrovská et al., 2013). For TPXL2, the opposite effect was observed as AUR1 coexpression had a pronounced positive effect on the recruitment of TPXL2 (and AUR1 itself) to these MTs. Taken together, these phenomena further support the hypothesis that there are nonoverlapping functions among the three proteins.
The oryzalin-induced depolymerization of the TPXL3-positive intranuclear cables is in agreement with the published immunolocalization data (Petrovská et al., 2013), and shows that the cables are indeed MTs. It is tempting to speculate that the foci remaining in the presence of oryzalin reflect uncharacterized MT-organizing centers from which these cables polymerize. Alternatively, these punctae could also represent nuclear pores given the connection between TPXL and importins and the similarity of the pictures with previously published data (Wirthmueller et al., 2015). Although not addressed directly, it seems plausible, given the known function of TPX2 in MT-nucleation (Alfaro-Aco et al., 2017), that the intranuclear MT cables are generated as a consequence of ectopic TPX(L) expression and/or increased stabilization due to GFP-tagging. However, we cannot exclude that the formation of those cables is caused by TPX(L)-dependent altered tubulin import into the nucleus, which could drive MT polymerization in a concentration-dependent manner.
At least for TPX2, the position of the GFP appears to affect protein stability as GFP-TPX2 was reported to disappear already in late anaphase in Arabidopsis and BY-2 cells (Vos et al., 2008), while TPX2-GFP still labels the phragmoplast MTs (Fig. 3B). Whether the C-terminal tagging of TPXL2 and TPXL3 also stabilizes these proteins similar to TPX2 and whether this differs from the N-terminal tagging remains to be established as we failed to generate BY-2 cell cultures with visible expression of GFP-TPXL2 or GFP-TPXL3. The observed increase of MT-nucleation upon ectopic overexpression of TPX2-GFP, which has also been shown before in Arabidopsis suspension cultured cells (Petrovská et al., 2013), indicates that the tagging at least does not interfere with this feature of TPX2. Future testing for full functionality of the TPX2 fusions will necessarily require the identification of a mutant phenotype to revert, which will likely require higher order mutant combinations with other TPXL family members.
Nevertheless, the results with TPXL3-GFP in Arabidopsis that we present here show that the C-terminal tagging of TPXL3 does not affect its function as it allowed us to identify plants lacking the native TPXL3 gene. After cytokinesis, it is possible that intranuclear MTs are formed in Arabidopsis cells similarly to BY-2 cells and that this is a consequence of the extended stability of TPXL3, which is tolerated by the cells. Alternatively, intranuclear cage formation has a specific, yet unknown, function. This notion is supported by the discovery of intranuclear MT-like structures in the angiosperm Aesculus hippocastanum by electron microscopy (Barnett, 1991). Although speculative at this point, the formation of a MT cage inside the nucleus might aid to shape the daughter nuclei during NE reformation. On the other hand, it is possible that, similar to what happens during cold treatment, overexpression of TPX affects the nuclear pores’ capacity to export tubulin after NE reformation (Schwarzerová et al., 2006).
Our mutant analyses using independent T-DNA insertion lines showed that TPX2 and TPXL2 are redundant and dispensable for plant development, even in the double mutant combination, whereas TPXL3 is essential. For TPXL3, the clear 2:1 segregation ratio and back-cross experiments, which showed transfer of the T-DNA via the male and female for both insertion lines, indicate that the mutation does not block development of the gametes, but rather that homozygous mutants are lethal at the embryo stage. Given that promTPX2, promTPXL2, and promTPXL3 activity was clearly visible in developing embryos, the essential nature of TPXL3 is unlikely to be the result of differential expression between TPXL3 and TPX2. The analysis of the siliques of self-pollinated heterozygous tpxl3-1 and tpxl3-2 mutants did not reveal clear embryo abortion defects, but showed the presence of apparently unfertilized ovules and a minor fraction of aborted seeds instead, which superseded the expected ratio of 25% originating from homozygous lethality. How the presence of apparently unfertilized ovules and seed abortion can be reconciled with the observed segregation ratios pointing to embryo lethality as well as the capability of T-DNA transfer via the female remains enigmatic. One explanation might be that the lack of TPXL3 slows down mitotic progression of the female gametes, leading to asynchrony with the germinating pollen and, therefore, the lack of fertilization. While forced fertilization in the back-cross experiments might overcome this asynchrony, allowing for the observed T-DNA transfer via the female, this does not explain the observed segregation ratios for both T-DNA insertion lines, pointing to the equal fitness of the wild-type and mutant ovules. Future work is required to pinpoint the cause of the lethality of the tpxl3 homozygous mutants.
We hypothesize that AUR1 and AUR2 proteins require, next to TPX2, one or more of these TPX-like proteins as targeting and/or activation factors, simply because the plant Aurora kinases, similar to those in other model systems, will likely rely on other proteins for their defined localizations. The expansion of this protein family likely is associated with the specification of Aurora functions in different aspects of mitosis in a spatiotemporally regulated manner. Our findings also prompt us to hypothesize that TPX2 in plants may be largely devoted to MT nucleation in the spindle apparatus, as a redundant mechanism to those that are governed by the augmin and γ-tubulin complexes.
MATERIALS AND METHODS
Molecular Cloning
TAP Constructs, Y2H Constructs, and Fluorescent Fusions
Full-length open reading frames of TPX2, TPXL2, and TPXL3 were amplified via PCR using primers listed in Supplemental Table S1 and recombined into the pDONR221 entry vector (Invitrogen) by a BP reaction. The LR Gateway reaction resulted in fusions between the open reading frames and GFP, driven by the pr35S in the pK7FWG2 destination vector (Karimi et al., 2007).
Promoters of TPXL2, TPXL3, and TPX2 were cloned into the pDONR221 entry vector and recombined into the pBGWFS7,0 vector to generate the promoter::GFP-GUS fusions. The 35S::AUR1-mRFP construct was generated by the Gateway LR recombination using the AUR1 open reading frame (ORF) in pDONR207 (Van Damme et al., 2004a) with the pK7RWG2 vector (Karimi et al., 2007). For the Y2H assay, the genes of interest in pDONR221 (TPXL2 and TPXL3) or pDONR207 (AUR1 and AUR2; Van Damme et al., 2004b) were Gateway-recombined into pDEST22 (GAL4 AD fusion) and pDEST32 (GAL4 BD fusions) vectors. The 35S::RanGAP1-TagBFP2 construct was generated by Multisite LR reaction into pB7m34GW using pr35S in pDONRP4P1R, RanGAP1 in pDONR221 (Boruc et al., 2015), and TagBFP2 in pDONRP2P3R. To generate the TAP tag constructs for AUR1 and AUR2, TAG-ORF and ORF-TAG constructs (under control of the constitutive cauliflower tobacco mosaic virus 35S promoter) were generated by the Gateway recombination as previously described: N- and C-terminal GSTEV tag (Bürckstümmer et al., 2006; Van Leene et al., 2008) fusions with AUR1 and AUR2, and C-terminal GSRHINO tag (Van Leene et al., 2015) and GSYELLOW tag (Besbrugge et al., 2018) fusions with AUR1 were constructed.
Vectors for the Expression of GFP-AUR1 and TPX2-GFP in Corresponding Mutation Backgrounds
The putative promoter region plus the coding sequences were amplified using primers (Supplemental Table S1): IV32830F and IV32830R for AUR1, and I03780F and I03780R for TPX2 with Phusion DNA polymerase (Thermo Fisher Scientific). The fragments were cloned into the pENTR-d/TOPO vector (Thermo Fisher Scientific) according to the manufacturer’s instruction, resulting in pENTR-AUR1 and pENTR-TPX2, respectively. To produce the GFP-AUR1 fusion construct, the EGFP sequence was amplified using primers of GFP-AUR1F and GFP-AUR1R. The amplified fragment was cloned into a linearized pENTR-AUR1 plasmid via the Gibson Assembly method (New England BioLabs) to give rise to pENTR-GFP-AUR1. The final expression constructs were made by having pENTR-GFP-AUR1 and pENTR-TPX2 recombined with pGWB10 and pGWB4 (Nakagawa et al., 2007) using LR Clonase (Thermo Fisher Scientific). These constructs were introduced into the aur1-2/aur2-2 and tpx2-1 mutants by Agrobacterium-mediated DNA transformation via the standard floral dipping method.
Plant Growth and Transformation
Both the wild-type control and mutants are in the Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) background. All lines (except SAIL_855_E09 and SAIL_1225_H06) were grown under standard growth conditions at the Center for Plant Systems Biology in Ghent in continuous light on vertical plates containing half-strength Murashige and Skoog medium supplemented with 8 g/L of plant tissue culture agar and 1% (w/v) Suc. SAIL_855_E09 and SAIL_1225_H06, together with their control, were grown at 21°C with 16-h light and 8-h dark in growth chambers at the Controlled Environmental Facility on the campus of the University of CA in Davis.
Wild-type Col-0 and mutant plants were transformed using the floral dip protocol (Clough and Bent, 1998). The Arabidopsis mutant lines tpxl3-2 GABI_480B12 and tpxl2-1 GABI_177E04 were acquired from the GABI-Kat collection (Kleinboelting et al., 2012). Out of the 40 seeds received for the tpxl3-2 mutant, only eight seeds germinated. Out of these eight plants, three were identified as having the T-DNA (line 3, line 4, and line 7) and we continued working with line 7.
The Arabidopsis mutant lines tpxl2-2 SALK_079098, tpxl3-1 SAIL_356_B08, tpx2-1 SALK_201256, tpx2-2 SALK_201975, and tpx2-5 SALK_206312 were acquired from the Arabidopsis Biological Resource Center located at Ohio State University in Columbus, Ohio. Homozygous tpx2-3 and tpx2-4 mutant plants were isolated from the seed pools of SAIL_855_E09 (CS838303) and SAIL_1225_H06 (CS844904) lines, respectively, ordered from the Arabidopsis Biological Resource Center. For tpxl3-1, seven plants were genotyped and three were positive for the T-DNA band (line 1, line 4, and line 7). We continued working with those three lines for the segregation analysis. The aur1-2/aur2-2 double homozygous mutant was reported in Van Damme et al. (2011).
T-DNA transfer in the reciprocal back-cross experiments of heterozygous tpxl3-1 and tpxl3-2 mutants was quantified by antibiotic resistance associated with the SAIL and GABI-Kat T-DNA insertions of the offspring seedlings. Deviations from the expected theoretical segregation ratio for both selfed and crossed offspring seedlings were assessed via χ2 testing of observed versus theoretical values (3:1 and 2:1).
Wild-type Nicotiana benthamiana plants were grown under a normal light regime (14 h of light, 10 h of darkness) at 25°C and 70% relative humidity. N. benthamiana infiltration with Agrobacterium tumefaciens strain C58 was performed as described in Boruc et al. (2010). However, in addition, coexpression of the p19 protein from Tomato bushy stunt virus was used for suppression of transgene silencing (Voinnet et al., 2000). The quantification of the N. benthamiana localization data shown in Figure 2C is the combined result of 3–8 independent transformation events.
Stable Nicotiana tabacum BY-2 (N. tabacum ‘Bright Yellow-2’) cell culture transformation was carried out as described in Geelen and Inzé (2001). For the colocalization analysis, BY-2 cells were first transformed with the 35S::RFP-MBD construct (Van Damme et al., 2004b), selected on hygromycin, and then transformed again and screened for the presence of the kanamycin-resistant GFP-fusion constructs. Cells derived from several independently transformed calli were imaged for each construct.
Mutant Genotyping and Complementation
The tpx2-1, tpx2-2, tpx2-5, tpxl2-1, tpxl2-2, tpxl3-1, and tpxl3-2 mutations were identified using the genotyping primers listed in Supplemental Table S1 by combining either the left primer (LP) or right primer (RP) with the T-DNA specific primer of the SALK, SAIL, or Gabi-Kat collection. The tpx2-3 mutation was detected by PCR using 838303RP and GLB3 and tpx2-4 by using 844902RP and GLB3. Gene-specific primer pairs spanning the T-DNA insertions for tpx2-3 were 838303LP and 838303RP and a combination of 844902LP and 844902RP for tpx2-4. The complementation of homozygous tpxl3-2 mutants was performed by transformation of a heterozygous tpxl3-2 mutant with the genomic fragment of TPXL3 fused to GFP via floral dip. To do so, the fragment was amplified by using the primer pairs IV22860F and IV22860R and cloned into the pENTR-d/TOPO vector followed by an LR reaction with the pGWB4 destination vector. Transformants were selected on hygromycin, grown in soil, and 16 of them carrying the tpxl3-2 mutation were kept and allowed to self-pollinate. In the progeny of these 16 plants, we screened for complemented mutant plants by genotyping until two were recovered that expressed TPXL3-GFP in the tpxl3-2 homozygous background.
Histochemical Analysis
Seedlings or flowering stems were stained in multiwell plates (Falcon 3043; Becton Dickinson). GUS assays were performed as described in Beeckman and Engler (1994). Briefly, samples were fixed in ice-cold 80% (v/v) acetone, washed three times in P buffer, and stained for 3–5 h at 37°C. Next, three washes in P buffer were followed by immersion in 100% ethanol. Samples mounted in lactic acid were observed and photographed with a stereomicroscope (BX51 microscope; Olympus) or with a differential interference contrast microscope (Leica).
TAP Tag Assay
Transformation of Arabidopsis cell suspension cultures was performed by Agrobacterium cocultivation and transgenic culture regeneration. TAP of protein complexes, further processing, MS analysis, data analysis, and background filtering was done as described before: GSTEV purifications were analyzed on a model no. 4800 MALDI TOF/TOF Proteomics Analyzer (AB SCIEX; Van Leene et al., 2011), see Supplemental Data Sets S1A and S1B for details), GSRHINO and GSYELLOW purifications were analyzed on a LTQ Orbitrap Velos (Thermo Scientific) as described before (Van Leene et al., 2015; Besbrugge et al., 2018; see Supplemental Data Sets S1A and S1C for details).
Y2H Assay
For the Y2H assay, plasmids encoding the baits (pDEST32) and preys (pDEST22) were transformed into the yeast strain PJ69-4A (MATa; trp1-901, leu2-3,112, ura3-52, his3-200, gal4D, gal80D, LYS2TGAL1-HIS3, GAL2-ADE2, and met2TGAL7-lacZ) by the lithium acetate method (Gietz et al., 1992). Cotransformed yeast cells were selected on synthetic dextrose (SD) plates containing 20-g/L select agar (Invitrogen) without Leu (pDEST32) and without Trp (pDEST22). Individual colonies were grown in liquid SD medium lacking Leu and Trp for 3 d at 30°C with shaking at 200 rpm. Liquid cultures were diluted five times in SD lacking Leu, Trp, and His, and 10 μL was spotted on SD plates. Interactions between proteins were scored visually by examining growth on the SD −Leu −Trp and −His plates after incubation at 30°C for 3 d. The results shown in Figure 1 and Supplemental Figure S1 represent the scored growth of 21–24 individual double transformed yeast colonies from two independent yeast transformations (repeat 1 and repeat 2).
Plasmid Construction for Expression of Recombinant Proteins in Escherichia coli
Coding sequences of the N-terminal putative Aurora binding domain (amino acids 1–100) of AtTPXL2 and AtTPXL3 were cloned as described in Petrovská et al. (2012) using primers listed in Supplemental Table S1 (TPXL2forand TPXL2rev, and TPXL3for and TPXL3rev). For expression of recombinant 6His-tagged protein, AtTPXL2- and AtTPXL3 fragments were subcloned into the Gateway expression vector pET55DEST (Novagen) and AtAurora1 was cloned as described in Demidov et al. (2009).
Production of Recombinant Proteins
GST-Aurora1 was expressed in E. coli C-43 (Lucigen) strain and purified under native conditions as described in Demidov et al. (2009). The Aurora binding domains of TPX2, TPXL2, and TPXL3 as well as Arabidopsis histone H3 proteins were expressed in E. coli BL-21 and purified under denaturing conditions according to Tomaštíková et al. (2015).
In Vitro Kinase Assay
Purified recombinant proteins were desalted in kinase buffer using 7kDa Molecular Weight Cut-off Zeba Spin Columns (Thermo Fisher Scientific) and processed as described in Tomaštíková et al. (2015). Briefly, 0.25 µg of AtAurora1 alone (as a control) or with the same amount of TPX(L) were incubated at 30°C, for 30 min with kinase buffer and 0.1 mm of ATP for activation of the kinases. Subsequently, [32P]ATP and substrate (recombinant variant of Arabidopsis histone H3 ∼10 µg) were added and incubated for an additional 60 min at 30°C, as described in Tomaštíková et al. (2015). Kinase reactions were mixed with 2x Laemmli buffer, separated on 12% SDS-PAGE, stained with Coommasie Brilliant Blue, dried and subjected to autoradiography. Signals were detected using an FLA 5100 phosphor imager (Fujifilm) during overnight exposion. The experiment was performed in triplicates (biological repeats).
Immunofluorescence Microscopy
Root meristematic cells were processed for immunofluorescence microscopy as described in Lee and Liu (2000). A rabbit anti-GFP antibody and the DM1A mouse monoclonal anti-α–tubulin antibody (both from Sigma-Aldrich) were used for the detection of GFP fusion proteins and tubulin/MTs in fixed root cells.
Sequence Alignment and Phylogenetic Analyses
The following reference sequences of proteins were used to generate the protein sequence alignment and the phylogenetic tree in Figure 1: AT1G03780 (TPX2), AT3G01015 (TPXL1), AT4G11990 (TPXL2), AT4G22860 (TPXL3), AT5G07170 (TPXL4), AT5G15510 (TPXL5), AT5G37478 (TPXL6), AT5G44270 (TPXL7), AT5G62240 (TPXL8). Protein sequence alignment was performed using the program “Clustal Omega” (https://www.ebi.ac.uk/Tools/msa/clustalo/; EMBL-European Bioinformatics Institute) and processed with the program “Jalview” (http://www.jalview.org/; Waterhouse et al., 2009). The phylogenetic tree was built using the “phylogeny” tool (www.phylogeny.fr). The tool uses the “MUSCLE” multiple sequence alignment (EMBL-EBI) and “PhyML” (http://www.atgc-montpellier.fr/phyml/) for the construction of phylogenetic trees. The “TreeDyn” viewer (http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi?task_type=treedyn) was used to visualize the tree (Dereeper et al., 2008, 2010). The tree was rooted on TPX2.
RT-qPCR Expression Analysis
Seven-day–old seedlings were frozen in liquid nitrogen and ground. RNA was extracted using standard Trizol-chloroform extraction and genomic DNA was removed by DNase treatment. Reverse transcription was performed using the ImProm-II reverse transcription system (Promega) according to the manufacturer’s instructions. qPCR was performed on the LC480 (Roche) and results were analyzed using Qbase+ (Biogazelle; https://www.qbaseplus.com/). Primer sets spanning the T-DNA positions for tpx2-1, tpx2-2, tpx2-5, tpxl2-1, and tpxl2-2 as well as normalization primers (CDKA and Actin4) are listed in Supplemental Table S1. The values presented are the average of three technical repeats and the experiment was performed twice with similar results starting from independent seedlings and independent RNA extraction.
RNA Extraction and RT-PCR (SAIL_855_E09 and SAIL_1225_H06)
Total RNA was isolated from 3-d–old seedlings using the PureLink RNA Mini Kit (Invitrogen) according to the manufacturer’s instructions. Reverse transcription was performed with the iScript cDNA Synthesis Kit (Bio-Rad). The primers used for RT-PCR were TPX2F and TPX2R. The PP2A transcript was used as a positive control as described in Czechowski et al. (2005). PCR products that resulted from 35 amplification cycles were analyzed by agarose gel electrophoresis.
Confocal Laser Scanning Microscopy
Fluorescence was analyzed with an inverted FluoView confocal microscope (FV1000; Olympus), equipped with a 60± water-corrected objective (n.a. 1.2); or an model no. LSM710 laser scanning confocal module mounted on an inverted Axiovert system (Carl Zeiss). Fluorescence was imaged in a multichannel setting with 488- and 543-nm excitation light for GFP and RFP excitation, respectively. Emission fluorescence on the Olympus system was captured in the frame-scanning mode via a 500- to 550-nm and 560- to 660-nm band-pass emission windows for GFP and mRFP fluorescence, respectively. Imaging on the LSM710 used 40× C-Plan (water) or 63× Plan-Apo (oil) objectives, and the GFP signal was excited by a 488-nm argon laser, and images were acquired using the ZEN software package attached to the confocal system. Confocal images were processed with ImageJ (www.imagej.nih.gov/ij).
Førster Resonance Energy Transfer-FLIM
The donor FLIM was determined by time-correlated single-photon counting in N. benthamiana epidermal cells transiently transfected with the proteins of interest fused to either eGFP (donor) or mRFP (acceptor).
Images were collected using an LSM Upgrade Kit (PicoQuant) attached to a FluoView confocal microscope (FV1000; Olympus) equipped with a Super Apochromat 60× UPLSAPO water immersion objective (n.a. 1.2) and a time-correlated single-photon counting module (Timeharp 200; PicoQuant). A pulsed picosecond diode laser (PDL 800-B; PicoQuant) with an output wavelength of 440 nm at a repetition rate of 40 MHz was used for donor fluorescence excitation. Laser power was adjusted to avoid average photon counting rates exceeding 10,000 photons/s to prevent pulse pile-up. Samples were scanned continuously for 1 min to obtain appropriate photon numbers for reliable statistics for the fluorescence decays. A dichroic mirror DM 458/515 and a band pass filter BD520/32-25 were used to detect the emitted photons using a Single Photon Avalanche Photodiode (SPAD; PicoQuant). Fluorescence lifetimes were calculated using the software package SymPhoTime (v5.3.2.2; PicoQuant). Selected areas of the images corresponding to single nuclei (n ≥ 30 cells from several independent transformations) were fitted by either a single-monoexponential fitting for a donor-alone sample or a biexponential fitting for a combined donor-acceptor sample including the measured instrument response function. The measured instrument response function was determined using an erythrosine B solution as described for the samples but with a lower count rate (1,000 photons/s). The lifetimes τ for a series of measurements were presented in a boxplot in Figure 2D showing the median (center lines), mean, sd, and the outliers (dots). Significance between donor-alone and donor-with acceptor samples was checked using Student’s t test, assuming a two-tailed distribution and unequal variance.
Primers Used
All primers used in this study can be found in Supplemental Table S1.
Quantification
Box plot graphs were generated using the web tool “BoxplotR” (Spitzer et al., 2014).
Accession Numbers
The Arabidopsis Information Resource (TAIR) locus identifiers for the genes mentioned in this study are At1g03780 for TPX2, At4g11990 for TPXL2, At4g22860 for TPXL3, At4g32830 for AUR1, and At2g25880 for AUR2.
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Observed Y2H interactions among TPXL2, TPXL3, and AUR1 and AUR2.
Supplemental Figure S2. Expression analysis of promTPX2, promTPLXL2, and promTPXL3.
Supplemental Figure S3. Coexpression of Aurora 1 (magenta) and either TPX2, TPXL2, or TPXL3 (green) restricts the colocalization of Aurora 1 to the nuclear structures in N. benthamiana leaves.
Supplemental Figure S4. Homozygous tpx2 and tpxl2 plants lack any obvious macroscopic mutant phenotype.
Supplemental Figure S5. GFP-AUR1 complements the bushy growth phenotype of the aur1-2/aur2-2 double mutant.
Supplemental Figure S6. Subcellular localization of TPX2, TPXL2, and TPXL3 in BY-2 cells.
Supplemental Table S1. List of primers used.
Supplemental Data Set S1. List of identified α-Aurora interactors using TAP.
Acknowledgments
The authors thank Pavla Binarova for sharing material.
Footnotes
This work was supported by the Research Foundation of Flanders (grant no. G029013N to D.V.D. and T.B.), the U.S. National Science Foundation (grant no. MCB–1616462 to B.L.), the Federal Ministry of Education and Research, Germany (“HaploTools” project to D.D.), the U.S. Department of Agriculture, the National Institute of Food and Agriculture (Agricultural Experiment Station Hatch Project no. CA-D-PLB-2260-H to B.L.). the European Regional Development Fund project "Plants as a tool for sustainable global development" (no. CZ.02.1.01/0.0/0.0/16_019/0000827 to E.D.T.), the Agency for Innovation by Science and Technology of Belgium (fellowship no. 121110 to M.V.D.), and the European Research Council Starting Grant PROCELLDEATH (project no. 639234 to M.K.N.)..
Articles can be viewed without a subscription.
References
- Alfaro-Aco R, Thawani A, Petry S (2017) Structural analysis of the role of TPX2 in branching microtubule nucleation. J Cell Biol 216: 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett J. (1991) Microtubules in interphase nuclei of Aesculus hippocastanum L. Ann Bot (Lond) 68: 159–165 [Google Scholar]
- Barr AR, Gergely F (2007) Aurora-A: The maker and breaker of spindle poles. J Cell Sci 120: 2987–2996 [DOI] [PubMed] [Google Scholar]
- Bayliss R, Sardon T, Vernos I, Conti E (2003) Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell 12: 851–862 [DOI] [PubMed] [Google Scholar]
- Bayliss R, Sardon T, Ebert J, Lindner D, Vernos I, Conti E (2004) Determinants for Aurora-A activation and Aurora-B discrimination by TPX2. Cell Cycle 3: 404–407 [PubMed] [Google Scholar]
- Beeckman T, Engler G (1994) An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Report 12: 37–42 [Google Scholar]
- Besbrugge N, Van Leene J, Eeckhout D, Cannoot B, Kulkarni SR, De Winne N, Persiau G, Van De Slijke E, Bontinck M, Aesaert S, et al. (2018) GSyellow, a multifaceted tag for functional protein analysis in monocot and dicot plants. Plant Physiol 177: 447–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E (2010) Functional modules in the Arabidopsis core cell cycle binary protein-protein interaction network. Plant Cell 22: 1264–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Griffis AH, Rodrigo-Peiris T, Zhou X, Tilford B, Van Damme D, Meier I (2015) GAP activity, but not subcellular targeting, is required for Arabidopsis RanGAP cellular and developmental functions. Plant Cell 27: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Weimer AK, Stoppin-Mellet V, Mylle E, Kosetsu K, Cedeño C, Jaquinod M, Njo M, De Milde L, Tompa P, et al. (2017) Phosphorylation of MAP65-1 by Arabidopsis Aurora kinases is required for efficient cell cycle progression. Plant Physiol 173: 582–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Carmena M, Earnshaw WC (2003) The cellular geography of Aurora kinases. Nat Rev Mol Cell Biol 4: 842–854 [DOI] [PubMed] [Google Scholar]
- Carmena M, Ruchaud S, Earnshaw WC (2009) Making the Auroras glow: Regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 21: 796–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A (2005) Identification and dynamics of two classes of Aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D, Hesse S, Tewes A, Rutten T, Fuchs J, Ashtiyani RK, Lein S, Fischer A, Reuter G, Houben A (2009) Aurora1 phosphorylation activity on histone H3 and its cross-talk with other post-translational histone modifications in Arabidopsis. Plant J 59: 221–230 [DOI] [PubMed] [Google Scholar]
- Demidov D, Lermontova I, Weiss O, Fuchs J, Rutten T, Kumke K, Sharbel TF, Van Damme D, De Storme N, Geelen D, et al. (2014) Altered expression of Aurora kinases in Arabidopsis results in aneu- and polyploidization. Plant J 80: 449–461 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibes S, Gallisa-Sune N, Rosas-Salvans M, Martinez-Delgado P, Vernos I, and Roig J (2018) Nek9 phosphorylation defines a new role for TPX2 in Eg5-dependent centrosome separation before nuclear envelope breakdown. Curr Biol 28: 121-129 [DOI] [PubMed] [Google Scholar]
- Evrard JL, Pieuchot L, Vos JW, Vernos I, Schmit AC (2009) Plant TPX2 and related proteins. Plant Signal Behav 4: 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Bian M, Liu J, Jiang Q, Zhang C (2009) A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci USA 106: 6939–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geelen DN, Inzé DG (2001) A bright future for the bright yellow-2 cell culture. Plant Physiol 127: 1375–1379 [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G. (2011) Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS One 6: e28120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104: 83–93 [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Wittmann M, Yokoyama H, Pepperkok R, Kufer T, Silljé H, Karsenti E, Mattaj IW, Vernos I (2002) Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat Cell Biol 4: 871–879 [DOI] [PubMed] [Google Scholar]
- Herud O, Weijers D, Lau S, Jürgens G (2016) Auxin responsiveness of the MONOPTEROS-BODENLOS module in primary root initiation critically depends on the nuclear import kinetics of the Aux/IAA inhibitor BODENLOS. Plant J 85: 269–277 [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe A, Matsunaga S, Nakagawa K, Kurihara D, Yoneda A, Hasezawa S, Uchiyama S, Fukui K (2005) Characterization of plant Aurora kinases during mitosis. Plant Mol Biol 58: 1–13 [DOI] [PubMed] [Google Scholar]
- Kirioukhova O, Johnston AJ, Kleen D, Kägi C, Baskar R, Moore JM, Bäumlein H, Gross-Hardt R, Grossniklaus U (2011) Female gametophytic cell specification and seed development require the function of the putative Arabidopsis INCENP ortholog WYRD. Development 138: 3409–3420 [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer TA, Silljé HH, Körner R, Gruss OJ, Meraldi P, Nigg EA (2002) Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol 158: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D, Matsunaga S, Kawabe A, Fujimoto S, Noda M, Uchiyama S, Fukui K (2006) Aurora kinase is required for chromosome segregation in tobacco BY-2 cells. Plant J 48: 572–580 [DOI] [PubMed] [Google Scholar]
- Lee YR, Liu B (2000) Identification of a phragmoplast-associated kinesin-related protein in higher plants. Curr Biol 10: 797–800 [DOI] [PubMed] [Google Scholar]
- Li S, Deng Z, Fu J, Xu C, Xin G, Wu Z, Luo J, Wang G, Zhang S, Zhang B, et al. (2015) Spatial compartmentalization specializes the function of Aurora A and Aurora B. J Biol Chem 290: 17546–17558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P (2011) TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol 195: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA (2003) Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol Biol 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Neumayer G, Belzil C, Gruss OJ, Nguyen MD (2014) TPX2: Of spindle assembly, DNA damage response, and cancer. Cell Mol Life Sci 71: 3027–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlü N, Srayko M, Kinoshita K, Habermann B, O’Toole ET, Müller-Reichert T, Schmalz N, Desai A, Hyman AA (2005) An essential function of the C. elegans ortholog of TPX2 is to localize activated Aurora A kinase to mitotic spindles. Dev Cell 9: 237–248 [DOI] [PubMed] [Google Scholar]
- Petrovská B, Cenklová V, Pochylová Z, Kourová H, Doskočilová A, Plíhal O, Binarová L, Binarová P (2012) Plant Aurora kinases play a role in maintenance of primary meristems and control of endoreduplication. New Phytol 193: 590–604 [DOI] [PubMed] [Google Scholar]
- Petrovská B, Jerábková H, Kohoutová L, Cenklová V, Pochylová Ž, Gelová Z, Kocárová G, Váchová L, Kurejová M, Tomastíková E, et al. (2013) Overexpressed TPX2 causes ectopic formation of microtubular arrays in the nuclei of acentrosomal plant cells. J Exp Bot 64: 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzerová K, Petrásek J, Panigrahi KC, Zelenková S, Opatrný Z, Nick P (2006) Intranuclear accumulation of plant tubulin in response to low temperature. Protoplasma 227: 185–196 [DOI] [PubMed] [Google Scholar]
- Spitzer M, Wildenhain J, Rappsilber J, Tyers M (2014) BoxPlotR: A web tool for generation of box plots. Nat Methods 11: 121–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaštíková E, Demidov D, Jeřábková H, Binarová P, Houben A, Doležel J, Petrovská B (2015) TPX2 protein of Arabidopsis activates Aurora Kinase 1, but not Aurora Kinase 3 in vitro. Plant Mol Biol Report 33: 1988–1995 [Google Scholar]
- Van Damme D, Bouget FY, Van Poucke K, Inzé D, Geelen D (2004a) Molecular dissection of plant cytokinesis and phragmoplast structure: a survey of GFP-tagged proteins. Plant J 40: 386–398 [DOI] [PubMed] [Google Scholar]
- Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inzé D, Geelen D (2004b) In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol 136: 3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E (2011) Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23: 4013–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Witters E, Inzé D, De Jaeger G (2008) Boosting tandem affinity purification of plant protein complexes. Trends Plant Sci 13: 517–520 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Persiau G, Van De Slijke E, Geerinck J, Van Isterdael G, Witters E, De Jaeger G (2011) Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol Biol 754: 195–218 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Eeckhout D, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Vercruysse L, Dedecker M, Verkest A, Vandepoele K, et al. (2015) An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc 10: 169–187 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103: 157–167 [DOI] [PubMed] [Google Scholar]
- Vos JW, Pieuchot L, Evrard JL, Janski N, Bergdoll M, de Ronde D, Perez LH, Sardon T, Vernos I, Schmit AC (2008) The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell 20: 2783–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Demidov D, Lermontova I, Beeckman T, Van Damme D (2016) Aurora kinases throughout plant development. Trends Plant Sci 21: 69–79 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L, Roth C, Fabro G, Caillaud MC, Rallapalli G, Asai S, Sklenar J, Jones AM, Wiermer M, Jones JD, et al. (2015) Probing formation of cargo/importin-α transport complexes in plant cells using a pathogen effector. Plant J 81: 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I (1998) Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J Cell Biol 143: 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I (2000) TPX2, a novel Xenopus MAP involved in spindle pole organization. J Cell Biol 149: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Roostalu J, Surrey T, Nogales E (2017) Structural insight into TPX2-stimulated microtubule assembly. eLife 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]