The combined data from in vivo regulation and in vitro binding analyses of transcription factor proteins and promoters identified a partial transcriptional regulatory network for the H2O2 response.
Abstract
Hydrogen peroxide (H2O2) is a common signal molecule initiating transcriptional responses to all the known biotic and abiotic stresses of land plants. However, the degree of involvement of H2O2 in these stress responses has not yet been well studied. Here we identify time-dependent transcriptome profiles stimulated by H2O2 application in Arabidopsis (Arabidopsis thaliana) seedlings. Promoter prediction based on transcriptome data suggests strong crosstalk among high light, heat, and wounding stress responses in terms of environmental stresses and between the abscisic acid (ABA) and salicylic acid (SA) responses in terms of phytohormone signaling. Quantitative analysis revealed that ABA accumulation is induced by H2O2 but SA is not, suggesting that the implied crosstalk with ABA is achieved through ABA accumulation while the crosstalk with SA is different. We identified potential direct regulatory pairs between regulator transcription factor (TF) proteins and their regulated TF genes based on the time-course transcriptome analysis for the H2O2 response, in vivo regulation of the regulated TF by the regulator TF identified by expression analysis of mutants and overexpressors, and in vitro binding of the regulator TF protein to the target TF promoter. These analyses enabled the establishment of part of the transcriptional regulatory network for the H2O2 response composed of 15 regulatory pairs of TFs, including five pairs previously reported. This regulatory network is suggested to be involved in a wide range of biotic and abiotic stress responses in Arabidopsis.
Hydrogen peroxide (H2O2) acts as a signal molecule for various stress responses, including the hypersensitive response (HR) and the systemic acquired response (SAR) stimulated by pathogen infection (Shirasu et al., 1997), ultraviolet (UV; Mackerness et al., 1999), drought (Miller et al., 2010), high light (HL; Karpinski et al., 1999), wounding (Orozco-Cardenas and Ryan, 1999), high temperature (Volkov et al., 2006), low temperature (van Buer et al., 2016), and anoxia (Banti et al., 2010). H2O2 mediates not only intracellular but also intercellular signaling to achieve systemic responses to biotic (Alvarez et al., 1998) and abiotic stress factors (Fryer et al., 2003; Baxter et al., 2014).
Exposure of plants to various stressors results in the activation of both H2O2 and phytohormone signaling, including salicylic acid (SA), jasmonic acid (JA), ethylene (ET), brassinosteroids, auxin, gibberellins, and abscisic acid (ABA; Baxter et al., 2014; Choudhury et al., 2017), and their possible crosstalk with H2O2 has been investigated. Of these, strong crosstalk between H2O2 and SA has been well established. H2O2 and SA make a positive feed-forward loop through mutual activation of their biosynthesis, and the regulatory loop is stimulated by both biotic and abiotic stressors (Herrera-Vásquez et al., 2015). This loop is a component of SAR during pathogen infection. Crosstalk with ABA has also been investigated. ABA and H2O2 make a feed-forward loop in guard cells to control stomatal closure. The loop has been established between ABA signaling and H2O2 accumulation: The ABA signal increases Respiratory Burst Oxidase Homolog expression leading to the production of H2O2, which in turn activates ABA signaling (Choudhury et al., 2017). Less is known about the putative crosstalk between other phytohormones.
The signals stimulated by H2O2 change the transcriptional profiles of plants. We previously reported that H2O2 treatment of seedlings activated 369 genes more than 3-fold (Yamamoto et al., 2004). In another report, 1,552 genes showed statistically significant changes in transcriptional response to HL stress by suppression of the peroxisomal peroxidase gene CATALASE 2, which turned out to be a major scavenger of H2O2 under stress (Vandenabeele et al., 2004). These reports, along with others, have revealed a large number of genes are regulated by H2O2, including those that are upregulated for reactive oxygen species (ROS) scavenging, small and large heat shock proteins (HSPs), defense from viruses and other pathogens, senescence-related proteins and other antistress proteins. This gene regulation is thought to be achieved by a so-called “transcriptional network” composed of a group of transcription factors (TFs) whose expression is regulated by each other. The network is supposed to be involved in recruiting multiple TFs to widen terminal responses and provide variation in induction kinetics and dose response. The actual “wiring diagram” within the regulatory network is largely unknown, so it is not understood how the H2O2 signal stimulates a large number of genes for antistress activities nor how early and late responses are generated.
Within the H2O2 regulatory network made of TFs, several examples of direct regulation have been reported: activation of ATERF71 (ARABIDOPSIS THALIANA ETHYLENE RESPONSE FACTOR 71), CRF5 and CRF6 (CYTOKININ RESPONSE FACTOR 5 and 6), and ATNFXL1 (ARABIDOPSIS THALIANA NF-X-LIKE 1) by ANAC017 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 017; Ng et al., 2013; Yamamoto et al., 2017), and also activation of RAP2.6 (RELATED TO APETALA2 6) by RRTF1 (REDOX RESPONSIVE TRANSCRIPTION FACTOR 1; Matsuo et al., 2015). These studies provide glimpses of a large regulatory network. Recently, large-scale analysis for the identification of a transcriptional regulatory network for the ABA response in etiolated Arabidopsis (Arabidopsis thaliana) seedlings has been conducted that was based on time-course transcriptome analyses of ABA response and comprehensive chromatin immunoprecipitation followed by sequencing (ChIP-seq) assays (Song et al., 2016). This pioneering work has opened up the way for mass identification of direct regulation in a network.
In this report, we have developed a strategy for medium-scale identification of direct regulation in a network using a combination of time-course transcriptome analyses, identification of in vivo regulation of a TF by a regulating TF using public and private transcriptome data of the mutants and/or overexpressors of the regulating factor, and identification of in vitro binding of the regulating factor protein to the promoter region of the regulated TF. Binding analysis between a TF and its putative target site in the promoter region was oriented by promoter prediction based on transcriptome data of mutants and/or overexpressors, and the success of this strategy is supported by our accurate and sensitive promoter prediction method (Yamamoto et al., 2011, 2017). This study provides insights into the transcriptional regulatory network for H2O2 signaling and also a strategy for medium-scale determination of protein-DNA interactions in a cost-effective manner.
RESULTS
Identification of H2O2-Regulated Genes by Microarray Analysis
To get an overview of the transcriptional response to H2O2, we performed time-course analysis of gene expression in shoots after spraying with H2O2. Quantitative analysis of H2O2 revealed a transient increase in accumulation at 1 min after spraying the shoots, but no drastic change was observed (Supplemental Fig. S1). Under these mild conditions, we harvested shoots at 1, 3, 6, 12, and 24 h after the H2O2 treatment and subjected them to microarray analysis using a custom-designed microarray with the Agilent long oligonucleotide probes (Hanada et al., 2013).
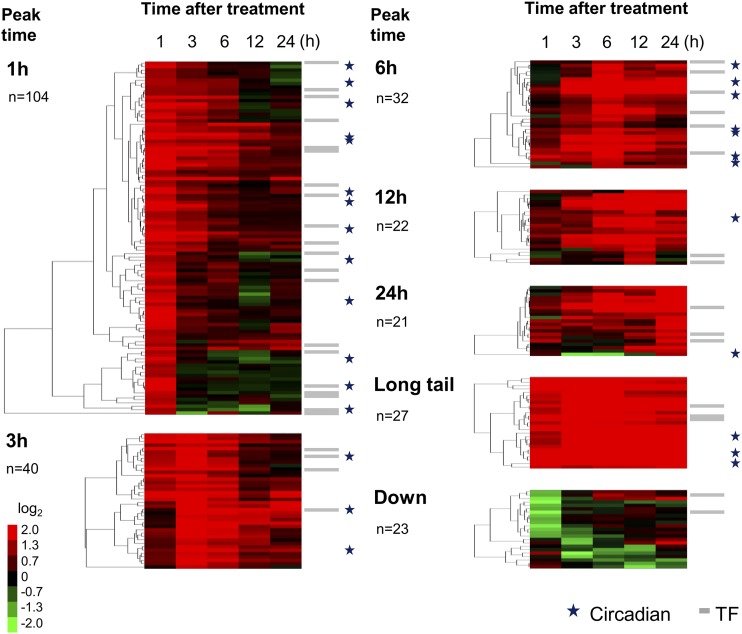
After evaluation of hybridization signals, we extracted genes with positive (fold change ≥ 2.5) and negative (≤0.4) responses that were statistically significant (Bayesian P value < 0.05; Supplemental Tables S1 and S2). TFs were further classified into seven groups according to the kinetic profiles (1, 3, 6, 12, 24 h peak, “Long tail,” and “Down,” Fig. 1; gene lists in Table 1 and Supplemental Table S1). As shown in the figure, more than half of the activated genes (144 out of 246) showed early responses, where the peak time point was 1 h or 3 h. Only 23 downregulated genes were identified, which was much fewer than the 246 upregulated genes.
Figure 1.
Groups of H2O2-responsive genes. H2O2-responsive genes (fold change ≥ 2.5 and ≤ 0.4 with statistical significance Bayesian P value < 0.05) were identified by triplicate microarray analysis and Cyber-T (Baldi and Long, 2001; Long et al., 2001; http://cybert.microarray.ics.uci.edu/) and divided into seven groups: upregulated gene groups (1, 3, 6, 12, 24 h, and Long tail), and a group for downregulated genes. Fold change is expressed after log2 transformation, as shown in the color bar, and subjected to hierarchical clustering (uncentered correlation, pairwise average linkage method). Gray bars and stars on the right of the color matrix indicate TF genes and circadian-regulated genes (identified by Covington and Harmer, 2007), respectively. The number of genes for each group is also shown.
Table 1. H2O2-responsive TFs.
Sixty H2O2-responsive TFs (fold change ≥ 2.5 with statistical significance) identified by the microarray analysis and confirmed by RT-qPCR (see Supplemental Table S4 for details). The stress responses of these TFs are shown in the “Stress Response” column (see Supplemental Table S5–S7 for details). C, cold; D, Drought; H, heat; HL, high light; S, salt; UV, ultraviolet B; W, wounding; El, elicitor; Pa, pathogen; n.a., no data available in GeneChip Arabidopsis ATH1 microarray.
| AGI Code | Gene Name | Stress Response |
|---|---|---|
| 1 h Peak | ||
| AT1G02220 | ANAC003 | H, S, UV, El, Pa |
| AT1G20823 | — | C, D, UV, W |
| AT1G27730 | ZAT10 | C, D, H, S, UV, W, El, Pa |
| AT1G63840 | — | C, S, UV, W, El, Pa |
| AT1G77450 | ANAC032 | C, D, S, UV, W, Pa |
| AT1G80840 | WRKY40 | C, D, H, HL, S, UV, W, El, Pa |
| AT2G33710 | — | C, D, H, HL, S, UV, W, El, Pa |
| AT2G38470 | WRKY33 | C, D, H, HL, UV, W, El, Pa |
| AT3G12910 | — | C, D, H, HL, S, UV, W, El, Pa |
| AT3G16720 | ATL2 | C, D, UV, El, Pa |
| AT3G23240 | ERF1 | UV, Pa |
| AT3G44350 | ANAC061 | C, D, H, HL, S, UV, W, El, Pa |
| AT3G50260 | CEJ1 | C, D, UV, W, El, Pa |
| AT4G18880 | ATHSFA4a | C, UV, El |
| AT4G25470 | CBF2 | C, D, H, S, UV, W |
| AT5G04340 | ZAT6 | C, D, H, S, UV, W, El, Pa |
| AT5G08790 | ANAC081 | n.a. |
| AT5G39860a | PRE1 | C, H, S, UV, El, Pa |
| AT5G47230 | ERF5 | C, D, H, UV, El, Pa |
| AT5G51190 | ERF105 | C, D, H, HL, UV, El, Pa |
| AT5G62430a | CDF1 | C, H, UV |
| AT5G63790 | ANAC102 | C, D, UV, W, Pa |
| AT5G64810 | WRKY51 | n.a. |
| 3 h Peak | ||
| AT1G10170 | ATNFXL1 | UV, Pa |
| AT1G10585 | — | D, S, UV, W, El, Pa |
| AT1G19210 | — | C, W, Pa |
| AT1G22810 | — | HL, UV, W, Pa |
| AT1G26800a | — | C, H, Pa |
| AT1G32870 | ANAC013 | C, H, UV, Pa |
| AT1G71520 | — | n.a. |
| AT2G37430 | ZAT11 | C, D, HL, S, UV, W, El, Pa |
| AT2G47520 | ATERF71 | D, HL, S, UV, W, El, Pa |
| AT3G23230 | ERF98 | C, D, H, S, UV, El, Pa |
| AT3G24500a | ATMBF1c | C, H, HL, S, UV, W, El, Pa |
| AT4G34410 | RRTF1 | C, D, H, HL, S, W, El, Pa |
| AT5G05410 | DREB2A | C, D, H, HL, UV, Pa |
| AT5G62020a | ATHSFB2a | D, H, HL, UV, Pa |
| 6 h Peak | ||
| AT1G11100 | FRG5 | H, Pa |
| AT1G22985 | CRF7 | Pa |
| AT1G52890 | ANAC019 | C, D, S, UV, W, El, Pa |
| AT2G26150 | ATHSFA2 | C, D, H, HL, S, UV, W, El, Pa |
| AT2G40340b | ERF48 | n.a. |
| AT2G40350b | — | n.a. |
| AT2G42150 | — | C, Pa |
| AT3G28210 | SAP12 | C, D, H, S, UV, W, El, Pa |
| AT4G06746 | RAP2.9 | n.a. |
| AT4G15420 | — | C, H, UV, El, Pa |
| AT4G17490 | ERF6 | C, D, H, HL, S, UV, W, El, Pa |
| AT5G18270 | ANAC087 | UV, Pa |
| AT5G20910 | AIP2 | C, S, UV, W, El, Pa |
| AT5G64750 | ABR1 | n.a. |
| 12 h Peak | ||
| AT1G43160 | RAP2.6 | D, HL, S, UV, W, Pa |
| AT5G43620 | — | n.a. |
| AT5G64060 | ANAC103 | C, D, HL, UV, El, Pa |
| 24 h Peak | ||
| AT2G38250 | — | C, D, H, S, UV, W, El, Pa |
| AT2G38340 | DREB19 | C, D, H, HL, S, UV, W, El, Pa |
| AT3G23250 | MYB15 | C, D, S, UV, W, El, Pa |
| AT4G22070 | WRKY31 | S, UV, W, El, Pa |
| AT5G01380 | — | C, D, H, S, UV, W, Pa |
| Long tail | ||
| AT5G59820 | ZAT12 | C, D, H, S, UV, W, El, Pa |
Circadian-regulated gene identified by Covington and Harmer (2007).
AT2G40340 and AT2G40350 are detected by the same probe, 263823_s_at.
Because there are several reports of circadian oscillation of gene expression for H2O2-scavenging enzymes (Zhong et al., 1994; Zhong and McClung, 1996) and also accumulation of H2O2 (Lai et al., 2012), we assumed expression of most of the H2O2-regulated genes would show circadian oscillation. However, our results revealed that this is not the case, because only 28 out of the 246 H2O2-responsive genes match the circadian-regulated genes reported by Covington and Harmer (2007); see stars in Fig. 1). These results suggested that the degree of circadian regulation of H2O2-responsive genes is limited.
Gene ontology analysis on subcellular localization detected a high abundance of cell wall-related genes at 6 h and 24 h (Supplemental Fig. S2). They include some biotic stress-related genes, such as proteases and a protease inhibitor protein (Supplemental Table S3).
H2O2-Induced Accumulation of ABA and JA
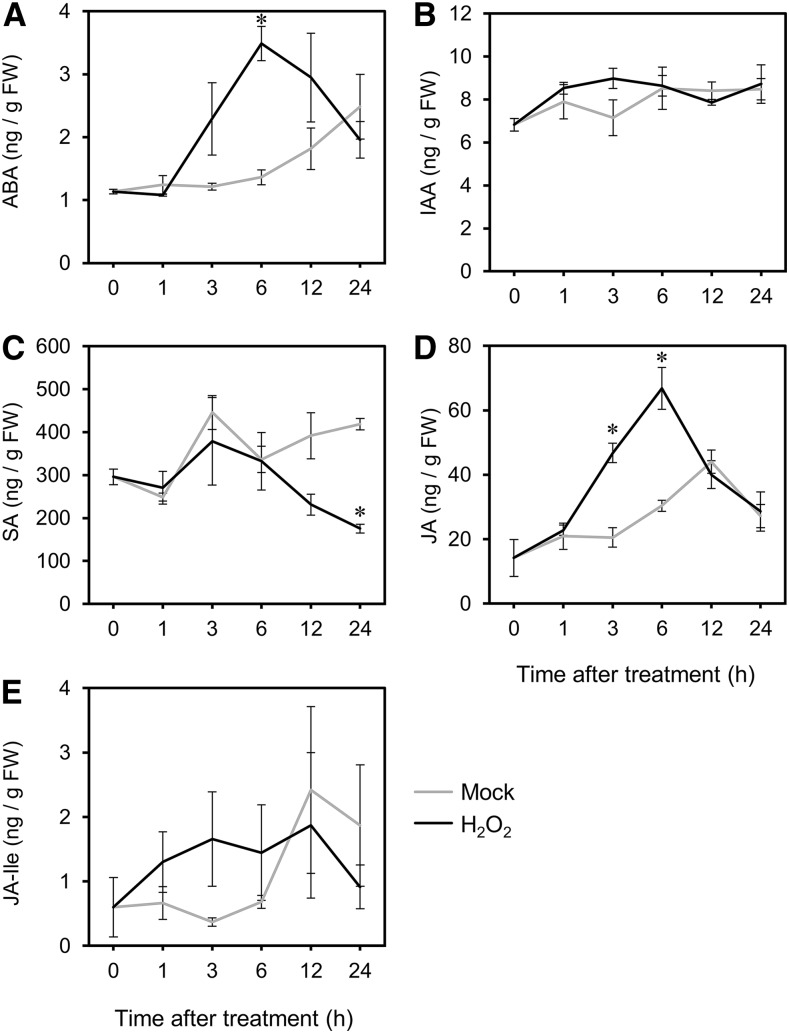
We investigated the effects of H2O2 on phytohormone signals. Firstly, we analyzed phytohormone accumulation after the H2O2 treatment. Accumulation of ABA, indole-3-acetic acid (IAA), SA, JA, and jasmonoyl-isoleucine (JA-Ile) in H2O2-treated shoots was subjected to quantitative analysis (Fig. 2). ABA (Fig. 2A) and JA (Fig. 2D) significantly increased at the peak time of 6 h. On the other hand, SA (Fig. 2C) did not show any significant increase, and a reduction was observed at 24 h. JA-Ile (Fig. 2E) and IAA (Fig. 2B) showed no significant changes after the treatment. These analyses indicate that H2O2 stimulates ABA and JA signaling at ∼6 h post-treatment.
Figure 2.
Change of endogenous hormone levels stimulated by H2O2 treatment. Quantification of phytohormones in shoots at each time point after the H2O2 and mock treatments (1, 3, 6, 12, and 24 h). A, ABA. B, IAA. C, SA. D, JA-Ile. E, Measurements were made on three independently processed samples. Line graphs and error bars represent average and se, respectively. An asterisk indicates statistical significance from the corresponding control at each time point based on the Student’s t test (P < 0.05). FW, fresh weight.
Identification of Colocalized Putative Transcriptional Regulatory Elements for Crosstalk between H2O2 and Stress-Related Phytohormones of Biotic/Abiotic Stresses
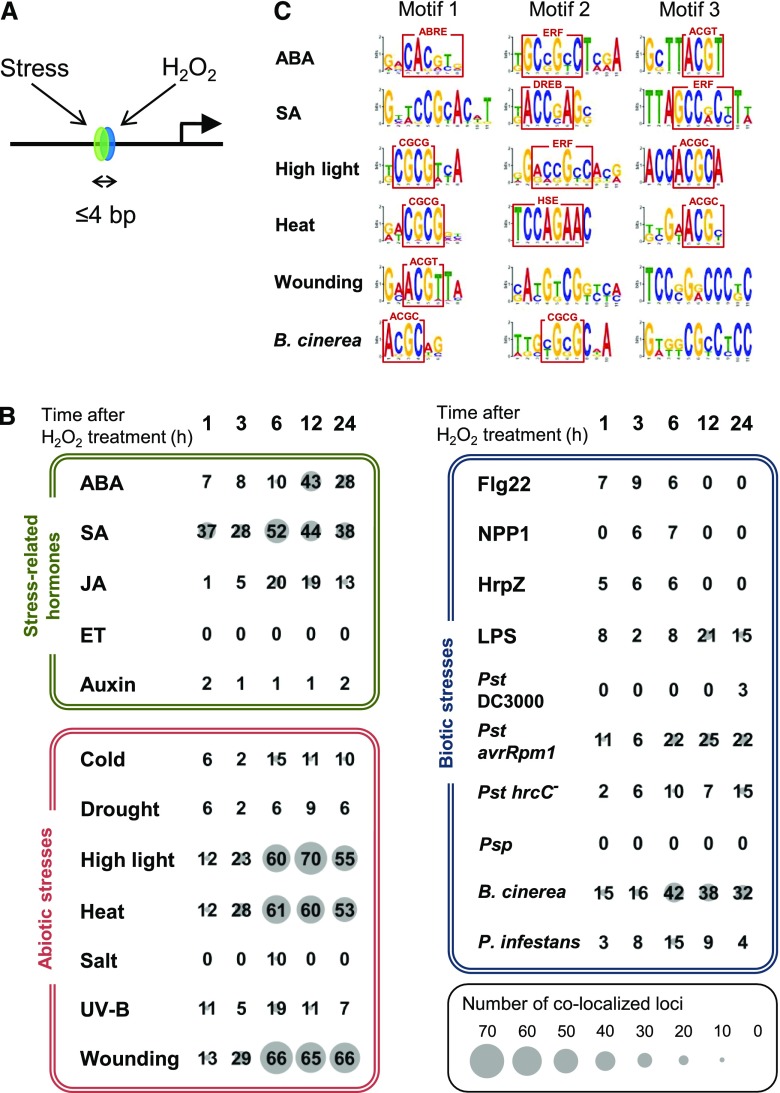
Next, we analyzed target promoter elements for H2O2 responses with the aid of crosstalk between stress-related phytohormones and stress responses. Conventionally, crosstalk between two distinct signals is detected by identification of coregulated genes of the two signals. Our analysis evaluates coregulated promoter elements by the two signals as illustrated in Figure 3A. This promoter element-based analysis excludes coregulated genes through two distinct promoter elements for each signal, and this forms a difference from conventional gene-based analysis. The advantage of this crosstalk analysis is that it allows more precise detection of crosstalk signals, excluding crosstalk responses of genes through two distinct regulatory elements and thus two distinct signals. Another advantage is the acquisition of the corresponding sequences of the promoter elements that receive a crosstalk signal. Using this method, we were able to evaluate the degree of crosstalk among phytohormone signals (Yamamoto et al., 2011). The detected colocalized elements are thought to receive a merged signal (model 1, Supplemental Fig. S3) or to form a junction for two distinct signals (model 2, Supplemental Fig. S3). In addition to the detection of crosstalk, there are two purposes of the “colocalization analysis.” One is the characterization of predicted H2O2-responsive elements using information about the crosstalk, and the other is an increase in accuracy through double prediction.
Figure 3.
Colocalization analysis of predicted cis-elements for H2O2 and other responses. A, Putative cis-elements identified by two independent predictions colocalizing within 4 bp (>3 bp overlap) were counted. B, The numbers in matrices indicate colocalized loci for two independent cis predictions. One prediction is for the H2O2 response, and the induction time after H2O2 treatment (h) used for the prediction is shown at the top (1–24). The second prediction includes three categories, stress-related hormone responses (green box), abiotic stress responses (red box), and biotic stress responses (blue box). The numbers at the cross point between the first horizontal prediction and the second vertical prediction represent loci of the corresponding crosstalk. The size of the gray circle behind a number gives a visual representation of the degree of colocalization. Time point of microarray data for promoter prediction was selected. ABA, 1 h; SA, 3 h; JA (treated with methyl jasmonate), 3 h; ET (treated with the metabolic precursor, 1-aminocyclopropane-1-carboxylic acid), 3 h; Auxin (treated with IAA), 3 h; Cold, 24 h; Drought, 1 h; High light, 3 h; Heat, 3 h; Salt, 3 h; UV-B, 3 h; Wounding, 3 h; flagellin22 (Flg22), 1 h; necrosis-inducing Phytophthora protein (NPP1), 1 h; harpinZ (HrpZ), 1 h; LPS (lipopolysaccharide), 4 h; Pst DC3000 (Pseudomonas syringae pv. tomato DC3000, 6 h; Pst avrRpm1 (P. syringae pv. tomato carrying an aviruent gene), 6 h; Pst hrcC− (P. syringae pv. tomato hrcC mutant), 24 h; Psp (P. syringae pv. phaseolicola), 6 h; B. cinerea, 18 h; P. infestans, 24 h. C, Sequences at the colocalization of double predictions were mixed for each second prediction, and subjected to motif extraction by Multiple Em for Motif Elicitation (http://meme-suite.org/tools/meme). Motifs in the red boxes are ABRE (Hattori et al., 2002), ERF (Hao et al., 1998), DREB (Sakuma et al., 2002), CGCG box (Yang and Poovaiah, 2002), HSE (Barros et al., 1992), core sequence recognized by bZIP family proteins (ACGT; Foster et al., 1994), and ACGC, which is a novel motif. See Supplemental Table S12 for accession numbers of microarray data using promoter prediction.
Microarray data of the H2O2 responses was subjected to promoter prediction according to a frequency comparison method (Yamamoto et al., 2011). In parallel, microarray data for phytohormone responses in Arabidopsis were retrieved from public databases and also subjected to promoter prediction. Abiotic and biotic stress responses were included in the analysis as well. After merging these two predictions, we identified colocalized loci in the promoter regions as illustrated in Figure 3A. As shown in Supplemental Figure S3, this analysis detects two types of signal crosstalk: merged signal of the crosstalk (model 1) and merging site of two independent signals (model 2). Although these two types cannot be distinguished, a large number reflects high crosstalk in the genome.
Figure 3B shows the number of colocalized loci between H2O2 responses of the time-course analysis and the stress-related hormones, and also biotic and abiotic stresses. H2O2 response after 1–24 h was individually used for prediction of promoter elements, so succession of crosstalk can be observed. For assistance viewing the degree of crosstalk, large numbers are accompanied with large gray circles (see scale in the figure). As shown in the green section of the figure, crosstalk between ET and auxin was negligible. In the case of SA, a high level of crosstalk was detected 1 h after the H2O2 treatment, and the high level continued until 24 h, which indicates that the crosstalk between H2O2 and SA signaling is durable. Crosstalk with ABA and JA was also detected. However, in contrast to that with SA, their crosstalk was transient. The peak time of the crosstalk between SA and JA appeared at 6 h, showing an earlier emergence than that with ABA (peak time = 12 h). We observed that accumulation of JA and ABA is induced by H2O2 and showed a peak time of 6 h (Fig. 2). Therefore, this crosstalk is suggested to be mediated by the accumulation of the corresponding phytohormones, while crosstalk with SA is achieved in a different manner.
Of the abiotic stresses, a high level of crosstalk was detected in HL, heat, and wounding, and crosstalk with cold, salt, and UV-B was much lower (red in Fig. 3B).
Biotic stresses were also subjected to crosstalk analysis as shown in the blue section in Figure 3B. Four elicitors were analyzed: three from bacteria—flagellin22, harpinZ, and lipopolysaccharide (LPS); and one from an oomycete—necrosis-inducing Phytophthora protein1. LPS showed the highest level of crosstalk. LPS has conserved molecular motifs from Gram-negative bacteria, called “pathogen-associated molecular patterns,” which trigger the plant’s innate immunity against nonhost pathogens. Because LPS induces SA and nitric oxide signals (Sun et al., 2012), the crosstalk with LPS may include part of the H2O2-SA signal.
We observed moderate crosstalk with the H2O2 response of the responses to Pseudomonas syringae pv. tomato (Pst) avrRpm1 and Pst hrcC−, both of which cause HR. Therefore, these are suggested to represent crosstalk with HR. Botrytis cinerea, a necrotic fungal pathogen causing SAR and no HR, induced the highest level of the crosstalk. This high crosstalk would reflect SAR. In contrast, we observed almost no crosstalk with the responses to P. syringae pv. phaseolicola (Psp) and Pst DC3000, suggesting no induction of immune response by the nonpathogenic Psp and suppression of the H2O2 response by pathogenic Pst DC3000. Phytophthora infestans, an oomycete pathogen, showed less crosstalk than B. cinerea and a comparable level with Pst avrRpm1 and Pst hrcC− (Fig. 3B).
Results shown in Figure 3B were obtained with one specific time point regarding the responses shown on the vertical axis, which include phytohormone and abiotic and biotic stress responses. Results including time-course responses were also prepared and are shown in Supplemental Figure S4. The crosstalk with the heat response was strong (Fig. 3B), and the analysis revealed that the greatest crosstalk starts between 6 h of the H2O2 response and 3 h of the heat response (Supplemental Fig. S4D). This later response time for H2O2 is also observed in the other strong crosstalk between H2O2 and HL (Supplemental Fig. S4C) and between H2O2 and wounding (Supplemental Fig. S4G). The physiological significance of the later response time for H2O2 is not clear.
Colocalization analysis identifies corresponding nucleotide sequences in the promoter region. We picked up sequences at the colocalized loci and extracted motifs by Multiple Em for Motif Elicitation (http://meme-suite.org/tools/meme). The top three motifs for the crosstalk with ABA, SA, HL, heat, wounding, and infection of B. cinerea were selected and are shown in Figure 3C. From these, several reported motifs have been identified: the core sequence of ABA-Responsive Element (ABRE), which is the major regulatory element for the ABA response (Hattori et al., 2002); motifs similar to binding sequences for ERF and Dehydration Responsive Element Binding (DREB) family proteins (Franco-Zorrilla et al., 2014); and ACGT, which is the core sequence for recognition of basic region-leucine zipper (bZIP) family proteins and has the potential to be a target site for PHYTOCHROME INTERACTING FACTOR and basic helix–loop–helix families (Yamamoto et al., 2011). The CGCG box is recognized by CAMTA/AtSR (CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR/ARABIDOPSIS THALIANA SIGNAL RESPONSIVE) family that is regulated by Ca2+ ions (Yang and Poovaiah, 2002). A motif related to the heat shock element (HSE; Barros et al., 1992) was also detected for crosstalk with the heat shock response. In addition to these motifs already reported, we also identified ones that have a core sequence ACGC (HL, Motif 3; heat, Motif 3; B. cinerea, Motif 1) and some others (SA, Motif 1; wounding, Motifs 2 and 3; B. cinerea, Motif 3). Our study suggests that all these motifs have the possibility to receive the crosstalk signals of H2O2 and another stress-related signal shown.
Some Late H2O2 Responses Require ABA Biosynthesis
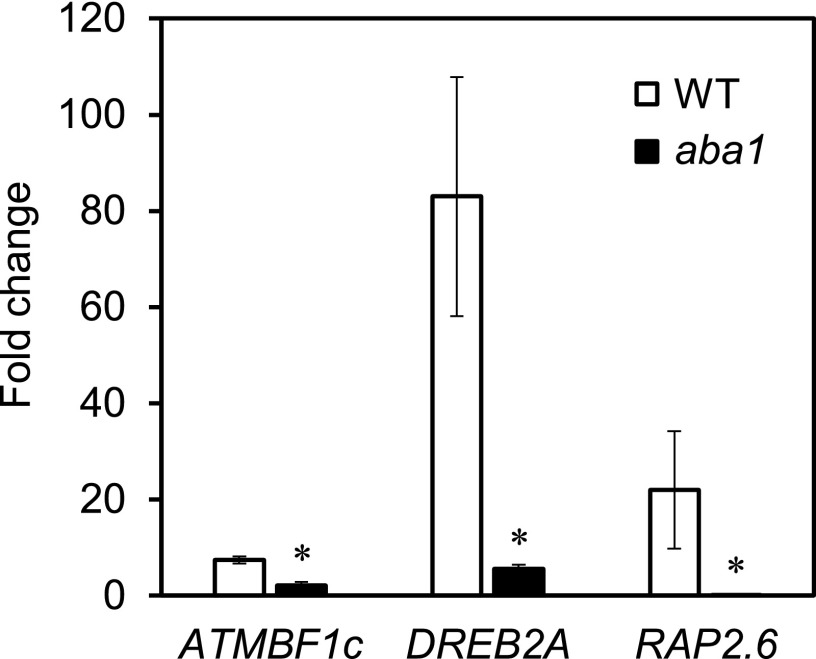
The crosstalk analysis (Fig. 3) suggests highest crosstalk with ABA occurs at 12 h after H2O2 treatment, and analysis of phytohormone revealed induction of ABA accumulation by H2O2 treatment with the peak time of 6 h (Fig. 2A), which precedes the crosstalk at 12 h. These results strongly suggest that H2O2 signal stimulates ABA accumulation, which causes ABA signaling and response in the late phase of H2O2 response. We then addressed this hypothesis using a mutant of ABA biosynthesis, aba1.
We examined expression of three genes showing crosstalk among H2O2 and ABA signals: ATMBF1c (ARABIDOPSIS THALIANA MULTIPROTEIN BRIDGING FACTOR 1c), DREB2A (DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2A), and RAP2.6. As shown in Figure 4, expression of all three genes was severely reduced in aba1 in response to H2O2 at 12 h after the treatment, demonstrating the requirement of ABA biosynthesis for the crosstalk. These results confirmed the hypothesis that the crosstalk with ABA signaling appearing at a late phase of the H2O2 response (12 h) is mediated via ABA accumulation, which precedes the crosstalk.
Figure 4.
Requirement of ABA biosynthesis for some H2O2 responses. Transcriptional responses of RAP2.6, DREB2A, and ATMBF1c to H2O2 treatments in wild type (WT) and ABA biosynthesis mutant (aba1) are shown. Seedlings were harvested 12 h after H2O2 treatment and subjected to RT-qPCR. Duplicate measurements were made for each of three independently prepared samples. Bar graphs and error bars represent average fold change and se, respectively. An asterisk indicates statistical significance from the corresponding wild-type control based on the Student’s t test (P < 0.05).
Degree of Involvement of HEAT SHOCK TRANSCRIPTION FACTOR A2 and NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 in H2O2 Responses
We furthered the colocalization analysis of promoter elements using gene expression data from knockout mutants of key regulators for heat shock and SA signaling.
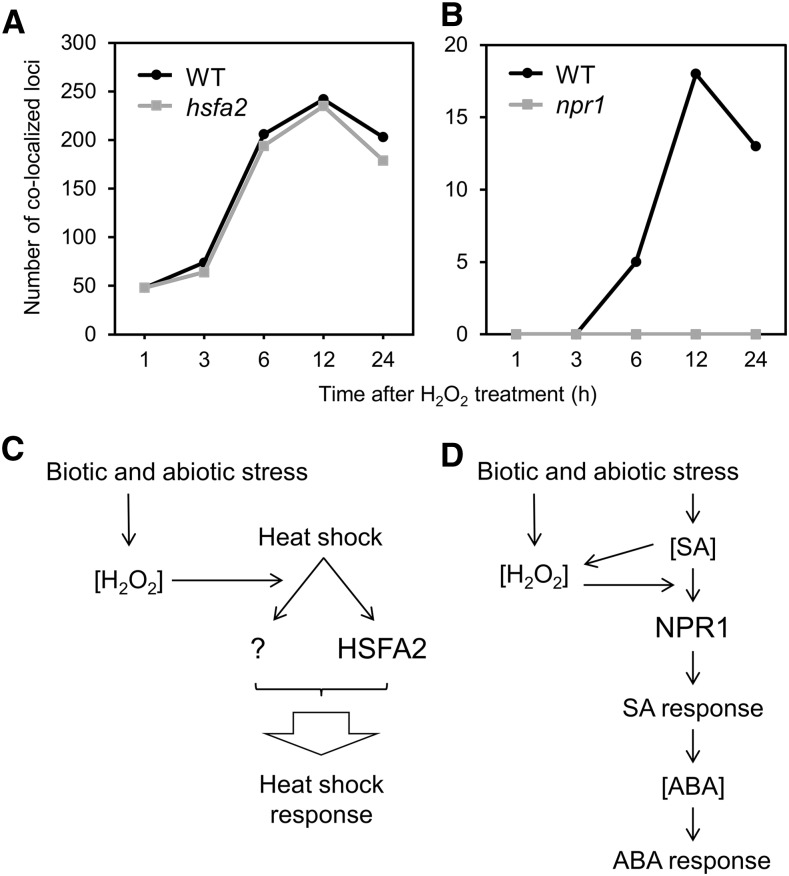
ATHSFA2 (ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTORA2) is one of the 22 Heat Shock Factor (HSF)s with no close homolog in the genome (AtHsf-04 in Fig. 4; Guo et al., 2008), and its expression is highly activated by heat stress (Schramm et al., 2008). Studies of knockout mutants show that it is indispensable for the suppression of ROS accumulation in mitochondria by heat shock (Zhang et al., 2009) and also for anoxia tolerance (Banti et al., 2010), and has only a partial requirement for heat shock induction of several other HSPs (Schramm et al., 2006). We analyzed colocalization of putative transcriptional regulatory elements between time-series responses to H2O2 and heat shock response; based on the publicly available microarray data, we obtained transcriptome data of hsfA2 and wild type under heat stress and then determined overrepresented, ATHSFA2 signal-associated octamers in the downregulated gene group in hsfA2 compared to wild type (Fig. 5A). As shown in Figure 5A, the number of colocalization loci in wild type increased until 12 h after H2O2 treatment, and a slight reduction was observed after 24 h. The number of colocalizations was almost the same in hsfA2 with a small reduction at 24 h. Little loss of the colocalized loci in the mutant suggests that ATHSFA2 is not involved in this crosstalk response as a major factor or is redundant with another HSF(s). A model of the crosstalk between H2O2 and heat shock responses is illustrated in Figure 5C. In this model, the H2O2 signal merges to a branch signal from heat shock stimulation to an unidentified factor designated as “?”, and ATHSFA2 receives another branch signal. Both branch signals promote heat shock responses. In this model, the heat shock signal does not include H2O2, but another model where H2O2 comes between the heat shock and the branch signals is also possible (not shown in the figure).
Figure 5.
Colocalization analysis of cis-regulatory elements in hsfa2 and npr1 mutants and models of crosstalk. A and B, Colocalization analysis of predicted cis-regulatory elements for H2O2 responses at the indicated time points and the heat shock response (A) or response to SA (B) were applied to wild type and hsfa2 (A) or npr1 (B) mutants. WT, wild type. C and D, The vertical axis shows the number of colocalized loci. Models for crosstalk between the H2O2- and HSFA2-associated heat stress responses (C) and NPR1-associated SA and ABA responses (D), based on colocalization analysis (D also includes the results of Figs. 2 and 3). See Supplemental Table S12 for accession numbers of microarray data of hsfa2 under heat shock stress and npr1 with SA treatment using promoter prediction.
NPR1 (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1) is the master switch for SA responses (Fan and Dong, 2002; Després et al., 2003; Johnson et al., 2003; Wu et al., 2012). We analyzed the effect of a NPR1 knockout mutation on the crosstalk between the H2O2 and SA signals. In contrast to the results of hsfA2, the lack of NPR1 resulted in complete loss of the colocalized promoter loci detected in wild type (Fig. 5B). These results suggest that the crosstalk between H2O2 and SA signaling completely depends on NPR1. A deduced model of the crosstalk is shown in Figure 5D. In the model, SA stimulates SA responses through NPR1, and the H2O2 signal merges upstream of NPR1, enabling the crosstalk signal to go through NPR1. Accumulation of SA occurs upstream of the merged point, based on the observation that there is no increase in SA accumulation after H2O2 stimulation (Fig. 2C). The crosstalk with ABA has a peak time of 12 h, which is later than the 6 h peak time of that with SA (Fig. 3B), so, in this model, the SA response stimulates the ABA response. This hierarchy between SA and ABA is shown by observation of the Arabidopsis response to infection by a pathogenic oomycete, Pythium irregulare, where infection-stimulated ABA accumulation requires SALICYLIC ACID INDUCTION DEFICIENT 2, a gene for SA biosynthesis (Adie et al., 2007). The ABA response is mediated through accumulation of ABA in the model, which is supported by the observation of ABA accumulation by H2O2 with a peak time of 6 h (Fig. 3B). The peak time of H2O2-SA in Figure 5B (12 h) is later than that in Figure 3B (6 h), probably because the samples utilized for microarray analysis were prepared under different conditions, including tissue age (7-d–old in Fig. 3B versus 2-week–old in Fig. 5B) and sampling time and concentration of applied SA (10 μm and 3 h in Fig. 3B versus 2 mm and 24 h in Fig. 5B).
Microarray Analysis and Reverse Transcription Quantitative PCR for identifying H2O2-responsive TFs
Microarray analysis and reverse transcription quantitative PCR (RT-qPCR) identified 60 TFs that reproducibly respond to H2O2, all of which are activated by the treatment (Supplemental Table S4). Because of this responsiveness, they are suggested to create a so-called “transcriptional network” that orchestrates expression of >250 terminal genes regulated by H2O2 (Supplemental Table S1). We decided to focus on this transcriptional network.
As H2O2 is involved in all the known stress responses of land plants, we wanted to address how many of the 60 TFs are actually involved in the stress responses and the stress-related phytohormone responses. With the aid of publicly available gene expression data, we investigated the stress responses of these TFs (Table 1; detailed information is shown in Supplemental Tables S5–S7. See “Materials and Methods” for sources of the microarray data). As shown in the table, most of the H2O2-responsive TFs also show responses to various stresses including cold (C), drought (D), high salinity (S), UV-B (UV), wounding (W), elicitors (El), and pathogen infection (Pa). These results indicate that the transcriptional network composed of H2O2-responsive TFs is actually involved in these biotic and environmental stress responses of Arabidopsis as a common network with small differences depending on the types of stress.
Experimental Identification of the Transcriptional Network among H2O2-Responsive TFs
We wanted to identify a potential network composed of the 60 H2O2-responsive TFs that is the basis for the actual H2O2-responsive networks, which appear under various physiological conditions, such as different developmental stages, cell types and also background environmental conditions, in addition to the different types of stressors. In this report, we set two criteria with which to identify potential, direct regulatory pairs from the TFs involved in the network. One criterion is evidence of in vivo regulation by a regulator TF in mutants and/or overexpressors of the regulator TF. The second is evidence of in vitro direct binding by the regulator TF to promoter DNA of the regulated TF. In vitro binding is not influenced by the physiological conditions of the plant, and thus gives simpler results than in vivo binding analysis, such as ChIP assays. We consider that this can be an advantage of in vitro analysis. Results of in vitro binding analysis reveal the potential for binding activity that occurs with modulated forms under various physiological conditions.
Identification of In Vivo Regulation and In Vitro Binding Assays among H2O2-Responsive TFs
Because identification of the complete network is difficult, we decided to focus on 38 early responsive TFs, which have peak times of 1 h, 3 h, and long tail, as potential regulators of the TF network. As for the potential regulated side of TFs, all the 60 genes were considered. Microarray data of mutants and/or overexpressors of the 38 TFs from public databases were surveyed and corresponding data sets were obtained for CBF2 ( C-REPEAT BINDING FACTOR 2), ERF5 and ERF6, ZAT10 (ZINC FINGER OF ARABIDOPSIS THALIANA 10), WRKY40 (WRKY DNA-BINDING PROTEIN 40), RRTF1, ATMBF1c, and DREB2A (see “Materials and Methods” for source of the microarray data). We subjected microarray data of the double mutant of ERF5 and ERF6 to the analysis because these genes are functionally redundant (Moffat et al., 2012). Changes in gene expression of the 60 TFs in the mutants and/or overexpressors are summarized in Supplemental Table S8. Of the 60 TFs, these data detect in vivo regulation by those responding to early responsive TFs. Supplemental expression analysis of knockout mutants of ATHSFA4a and CEJ1 (COOPERATIVELY REGULATED BY ETHYLENE AND JASMONATE 1) by RT-qPCR was also included to examine expression of the 60 TFs. The summarized data are shown in Table 2. Only positive regulation was detected for CBF2, RRTF1, DREB2A, ATHSFA4a, and CEJ1 while both positive and negative regulation was detected for ZAT10, WRKY40, and ATMBF1c. The figure shows that these early responsive TFs are actually involved in the regulation of the early and late responsive TFs. It is not clear, however, if each regulatory pair of TFs presented in the figure shows direct regulation or not.
Table 2. In vivo regulation among H2O2-responsive TFs.
The effects of knockout mutants and overexpressors of the TFs are summarized in terms of gene expression of the H2O2-Regulated TFs (details are shown in Supplemental Table S8). P, positive regulation; N, negative regulation; P/N, positive and negative regulation depending on microarray data; —, no significant change in mutants/overexpressors; n.a., no available information.
| H2O2–Regulated TFs | CBF2 | ERF5 and ERF6 | ZAT10 | WRKY40 | RRTF1 | ATMBF1c | DREB2A | ATHSFA4aa | CEJ1a | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak Time | AGI Code | TF | |||||||||
| 1 h Peak | AT1G02220 | ANAC003 | — | — | — | P | — | — | — | P | — |
| AT1G20823 | — | — | — | — | — | — | P | — | — | — | |
| AT1G27730 | ZAT10 | — | — | n.a. | N | P | — | P | — | — | |
| AT1G63840 | — | — | — | — | N | — | — | — | P | — | |
| AT1G77450 | ANAC032 | — | — | — | — | P | — | — | n.a. | n.a. | |
| AT1G80840 | WRKY40 | — | — | P | n.a. | P | P | — | — | — | |
| AT2G33710 | — | — | — | — | — | — | — | — | P | — | |
| AT2G38470 | WRKY33 | — | — | P | — | — | P | — | P | — | |
| AT3G12910 | — | — | — | — | — | — | — | — | n.a. | n.a. | |
| AT3G16720 | ATL2 | — | — | — | — | — | — | — | n.a. | n.a. | |
| AT3G23240 | ERF1 | — | — | — | — | — | — | — | P | — | |
| AT3G44350 | ANAC061 | — | — | — | — | — | P | — | — | — | |
| AT3G50260 | CEJ1 | P | — | — | — | P | P | — | — | n.a. | |
| AT4G18880 | ATHSFA4a | — | — | — | N | — | — | — | n.a. | — | |
| AT4G25470 | CBF2 | n.a. | — | N | N | — | P | — | — | P | |
| AT5G04340 | ZAT6 | P | — | — | N | — | — | — | P | — | |
| AT5G08790 | ANAC081 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | — | — | P | |
| AT5G39860 | PRE1 | — | — | — | N | — | — | — | n.a. | n.a. | |
| AT5G47230 | ERF5 | — | n.a. | — | N | — | P | — | — | — | |
| AT5G51190 | ERF105 | — | — | — | P | — | P | — | n.a. | n.a. | |
| AT5G62430 | CDF1 | — | — | N | — | — | — | — | n.a. | n.a. | |
| AT5G63790 | ANAC102 | — | — | — | — | — | — | — | P | — | |
| AT5G64810 | WRKY51 | — | — | — | — | — | — | — | P | — | |
| 3 h Peak | AT1G10170 | ATNFXL1 | — | — | — | N | — | — | P | — | — |
| AT1G10585 | — | — | — | N | — | P | N | — | — | P | |
| AT1G19210 | — | — | — | — | P | — | P | — | n.a. | n.a. | |
| AT1G22810 | — | — | — | — | — | — | P | — | — | P | |
| AT1G26800 | — | — | — | P | — | — | — | — | n.a. | n.a. | |
| AT1G32870 | ANAC013 | — | — | — | — | — | — | — | — | — | |
| AT1G71520 | — | — | — | — | — | — | — | — | n.a. | n.a. | |
| AT2G37430 | ZAT11 | — | — | — | — | — | — | — | — | — | |
| AT2G47520 | ATERF71 | — | — | — | — | — | — | — | — | — | |
| AT3G23230 | ERF98 | — | — | P | — | — | P | — | — | — | |
| AT3G24500 | ATMBF1c | — | — | — | N | — | n.a. | P | — | P | |
| AT4G34410 | RRTF1 | — | — | — | — | n.a. | P | — | — | — | |
| AT5G05410 | DREB2A | — | — | — | — | — | — | n.a. | — | P | |
| AT5G62020 | ATHSFB2a | — | — | — | — | — | P | P | — | — | |
| 6 h Peak | AT1G11100 | FRG5 | — | — | — | — | — | — | — | n.a. | n.a. |
| AT1G22985 | CRF7 | — | — | — | — | — | — | — | — | — | |
| AT1G52890 | ANAC019 | — | — | P | — | P | — | — | n.a. | n.a. | |
| AT2G26150 | ATHSFA2 | — | — | — | — | — | — | — | P | P | |
| AT2G40340 | ERF48 | — | — | — | — | — | — | — | — | — | |
| AT2G40350 | — | — | — | — | — | — | — | — | — | — | |
| AT2G42150 | — | — | — | — | — | — | — | — | — | P | |
| AT3G28210 | SAP12 | — | — | P | N | — | — | — | P | — | |
| AT4G06746 | RAP2.9 | — | — | — | — | — | — | — | — | — | |
| AT4G15420 | — | — | — | — | P | — | — | — | — | — | |
| AT4G17490 | ERF6 | — | n.a. | — | P/N | P | P | — | — | — | |
| AT5G18270 | ANAC087 | — | — | — | P/N | — | — | — | — | — | |
| AT5G20910 | AIP2 | — | — | — | — | — | — | P | — | — | |
| AT5G64750 | ABR1 | — | — | — | — | — | — | — | — | P | |
| 12 h Peak | AT1G43160 | RAP2.6 | P | — | — | P | P | — | — | — | — |
| AT5G43620 | — | — | — | — | — | — | — | — | — | — | |
| AT5G64060 | ANAC103 | — | — | — | N | — | — | — | — | — | |
| 24 h Peak | AT2G38250 | — | — | — | — | — | — | — | — | — | — |
| AT2G38340 | DREB19 | — | — | — | N | — | — | — | — | — | |
| AT3G23250 | MYB15 | — | — | — | P/N | — | P | — | — | — | |
| AT4G22070 | WRKY31 | — | — | — | — | — | P | — | n.a. | n.a. | |
| AT5G01380 | — | — | — | P | — | — | — | — | — | P | |
| Long Tail | AT5G59820 | ZAT12 | — | — | P | — | — | P | — | P | — |
Expression data of the knockout mutants of ATHSFA4a and CEJ1 with H2O2 treatment were obtained by our RT-qPCR analysis.
To identify direct regulation among TFs, we examined in vitro binding of a regulator TF to the promoter of the other TF in the regulatory pair detected in vivo. The binding assay was done using AlphaScreen (PerkinElmer Japan), where biotinylated oligo DNA and a FLAG-tagged DNA binding protein are subjected to homogenous assays in 384-well microtiter plates. In the assay, the binding signal is obtained as luminescence (see “Materials and Methods”). To obtain some scalability of experimental size, we developed an experimental strategy for preparation of biotinylated DNA probes (as illustrated in Supplemental Fig. S5) and FLAG-tagged TF proteins. This strategy dose not necessitate the purchase of specific biotinylated oligo DNA, which is the most expensive element of the experiment.
The microarray data of the mutants and the overexpressors were utilized for predicting target sites in the promoter region. Our microarray data of the H2O2 responses were also used for double detection. In total, 161 probe sequences from the promoter region of the regulated TFs were prepared for AlphaScreen.
Nine FLAG-tagged TF proteins were synthesized in vitro using a wheat germ system (Matsuo et al., 2015) from corresponding complementary DNA (cDNA; Mitsuda et al., 2010). The 161 biotinylated and double-stranded probes were prepared using a homemade DNA polymerase (see “Materials and Methods”). In total, 258 combinations were subjected to AlphaScreen in triplicated assays. Positive combinations were further subjected to mutation analysis to confirm sequence-specific binding to the predicted site. During the analysis, we noticed that sequence-independent binding of some TFs to probe DNA depends on a possible hairpin structure of the probe as shown in Supplemental Figure S6. In this example, mutations of two distinct sites both resulted in loss of binding activity, and thus we could not identify a binding sequence in a positive probe. We noticed that both mutations possibly disrupt a potential hairpin structure of the probe. This type of apparent structure-dependent binding is excluded from the positive results of AlphaScreen. Other combinations where the binding site in the probe was not identified by mutation analysis were also excluded from the positive results. After removing these possible false positives, we identified 11 binding pairs of TFs and probes, in addition to two binding pairs of TFs to their own promoter fragments (Fig. 6; Supplemental Table S9).
Figure 6.
Direct binding of TF to promoter fragment. FLAG-tagged TF proteins and biotinylated and unbiotinylated DNA probes were used in AlphaScreen assays to detect direct binding in vitro. Detected signals were normalized with those of the control samples with the corresponding unbiotinylated probes. A gene name in the probe ID indicates the source promoter of the probes. Relative AlphaScreen signals were calculated as the ratio of biotinylated-probe signal to unbiotinylated-probe signal. The values represent the averages and ses of triplicate experiments. A vertical dotted line shows a threshold value (relative AlphaScreen signal = 10.0). The positive control shows assays of the RRTF1 protein and the target sequence probe (Matsuo et al., 2015). The negative control shows assays with no template control of protein synthesis.
Based on the binding analysis with mutation probes (Supplemental Fig. S7; Supplemental Table S10), we identified the target sites of the TFs as DREB2A, RRTF1, WRKY40, ZAT10, ERF5, CBF2, and CEJ1, as shown in Supplemental Table S10. The table also shows reported target sequences of some of the TFs, and our results are consistent with these reports.
Table 3 summarizes the results of in vivo regulation and in vitro direct binding. Of the 84 pairs of detected via in vivo regulation as shown in Table 2, we subjected 55 pairs to in vitro binding analysis (Table 3). Among them, 11 pairs showed positive results in the binding analysis, suggesting that these 11 are examples of direct regulation. Additionally, one case of binding of a TF to its own promoter was detected for WRKY40. As its in vivo autoregulation has not been detected due to technical limitations of the microarray analysis, this case of autoregulation remains speculative. In addition, regulation of ERF6 by ERF5 also remains speculative as ERF5 and ERF6 are a redundant pair, although we did detect binding of the ERF6 promoter by ERF5.
Table 3. Identification of direct regulation among H2O2-activated TFs.
A summary of the in vivo transcriptional regulation identified (“Regulation”) and in vitro binding analysis (“Binding”) is shown. If both results are positive, direct regulation is judged as positive, and indicated on the figure with a check mark (“Direct Regulation”). As expression data for self-regulation is not available, direct regulation could not be judged as positive and is indicated with a question mark. P, positive regulation; N, negative regulation; P/N, positive and negative regulation in multiple microarray data; n.a., no available information.
| Upstream TF | Downstream Promoter | Regulation | Binding | Direct Regulation |
|---|---|---|---|---|
| CBF2 | CEJ1 | P | Yes | ✓ |
| ZAT6 | P | Yes | ✓ | |
| RAP2.6 | P | Yes | ✓ | |
| CBF2 | n.a. | No | ||
| ERF5 | ERF6 | n.a. | Yes | ? |
| ZAT10 | ERF98 | P | No | |
| ZAT12 | P | Yes | ✓ | |
| WRKY40 | WRKY40 | n.a. | Yes | ? |
| ERF6 | P/N | No | ||
| ATMBF1c | N | No | ||
| SAP12 | N | No | ||
| ANAC087 | P/N | No | ||
| RAP2.6 | P | No | ||
| ANAC103 | N | No | ||
| DREB19 | N | No | ||
| ATMBF1c | ANAC061 | P | No | |
| WRKY33 | P | No | ||
| WRKY40 | P | No | ||
| ERF98 | P | No | ||
| ERF6 | P | No | ||
| RRTF1 | P | No | ||
| ATMBF1c | n.a. | No | ||
| ZAT12 | P | No | ||
| RRTF1 | ZAT10 | P | No | |
| WRKY40 | P | No | ||
| ERF6 | P | Yes | ✓ | |
| RRTF1 | P | No | ||
| AT1G10585 | P | No | ||
| RAP2.6 | P | Yes | ✓ | |
| DREB2A | ZAT10 | P | Yes | ✓ |
| ATMBF1c | P | Yes | ✓ | |
| DREB2A | n.a. | No | ||
| ATHSFA4a | ERF1 | P | No | |
| AT2G33710 | P | No | ||
| ZAT6 | P | No | ||
| AT1G63840 | P | No | ||
| ANAC003 | P | No | ||
| ANAC102 | P | No | ||
| WRKY33 | P | No | ||
| WRKY51 | P | No | ||
| SAP12 | P | No | ||
| ATMBF1c | P | No | ||
| ATHSFA2 | P | No | ||
| ZAT12 | P | No | ||
| CEJ1 | CBF2 | P | No | |
| ZAT10 | P | No | ||
| ANAC081 | P | No | ||
| AT1G10585 | P | Yes | ✓ | |
| ATMBF1c | P | Yes | ✓ | |
| AT1G22810 | P | Yes | ✓ | |
| AT2G42150 | P | No | ||
| ABR1 | P | No | ||
| ATHSFA2 | P | No | ||
| AT5G01380 | P | No | ||
| DREB2A | P | No |
DISCUSSION
Transcriptional Response to H2O2 Is In Part Achieved by SA and ABA Signaling
In this study, we conducted time-course analysis of the transcriptional response to H2O2 treatment in Arabidopsis shoots. The treatment did not cause severe oxidative damage such as chlorosis, but caused a mild and transient increase in endogenous H2O2 (Supplemental Fig. S1). The increase ended 5 min after the treatments but triggered a long-lasting signal(s) detected even 24 h after the treatments. More than 250 genes showed activation or repression, demonstrating a great influence on plant gene expression. The induction kinetics of the activated genes revealed that their peak times varied from 1 to 24 h, showing that the H2O2 response is a mixture of rapid and slow responses.
The transcriptional response to H2O2 is in part achieved by some phytohormone signals. Our results show that H2O2 treatment increases the accumulation of ABA and JA and stimulates crosstalk with SA signaling without the accumulation of SA. Galvez-Valdivieso et al. (2009) suggested that the ROS signal stimulated by HL treatments activates the ABA response through ABA biosynthesis, based on the observation of ABA biosynthesis mutants. This is consistent with our observation of H2O2-stimulated ABA accumulation that preceded the crosstalk with ABA signaling (summarized in Fig. 5D), and of suppressed gene expression of H2O2-responsive TFs in aba1 mutants (Fig. 4). Mutual activation between H2O2 and SA signals is well known in the pathogen response (Herrera-Vásquez et al., 2015). Therefore, this mutual interplay making a positive feedback loop may cause the observed durable, nontransient crosstalk between H2O2 and SA (Fig. 3B). Although the positive feedback loop is reported to include mutual activation of the production of H2O2 and SA (Herrera-Vásquez et al., 2015), our results did not detect induction of SA accumulation by H2O2 until at least 24 h after the treatment. Therefore, activation of the SA signal by H2O2 in our case did not induce activation of SA accumulation, but there was stimulation of the SA signal after SA accumulation and before NPR1 activity (Fig. 5D). Our results, taken together with previous reports, suggest that the positive feedback loop of H2O2 and SA is established by the different levels of the SA signal induced by H2O2, including biosynthesis and signaling of SA.
Promoter Analysis Revealed Crosstalk between H2O2 and Stress Signals with Putative Transcriptional Regulatory Elements
The H2O2 signal is involved in various stress responses of land plants (Baxter et al., 2014). Our promoter analysis estimated the degree of crosstalk in several stress responses. In terms of environmental stress responses, a high degree of crosstalk was observed for HL, heat, and wounding stress, and there was less with cold, drought, salt, and UV-B (Fig. 3B). Cold, HL, and UV-B cause ROS production, which causes damage, while drought, heat, salt, and wounding stimulate enzymatic ROS biosynthesis, generating the stress signal. However, this does not explain the high degree of crosstalk with the stress response to HL, heat, and wounding.
Of the three strains of Pst, almost no crosstalk was observed for the pathogenic Pst DC3000, while moderate levels were detected for HR-inducing Pst avrRpm1, which is nonpathogenic (Debener et al., 1991), and also for the nonpathogenic Pst hrcC−, whose ability to inject the effector is disabled (Boch et al., 2002). This comparative analysis suggests that pathogenic Pst DC3000 suppresses almost all the defensive H2O2 responses, including HR and SAR in Arabidopsis, and the crosstalk observed in Pst avrRpm1 and Pst hrcC− corresponds to HR. No crosstalk was observed in a nonhost bacterial pathogen, Psp, suggesting that it does not elicit either SAR or HR. The latter suggestion is consistent with the observation of HR deficiency in Arabidopsis plants after Psp infection (Yu et al., 1993).
B. cinerea, a necrotrophic fungal pathogen, showed the highest level of crosstalk among the analyzed responses to bacterial and fungal pathogens and elicitors. B. cinerea uses penetration hyphae to enter the plant, which then stimulate the HR of the host plant (Govrin et al., 2006). The HR cannot block the invasion of B. cinerea, and the pathogen propagates by taking nutrition from the dead plant cells produced by the HR (Govrin and Levine, 2000), resulting in infection through necrotrophic progression. Because of these features, the high crosstalk observed in B. cinerea is suggested to include not only the HR but also the SAR and wounding responses, which show high crosstalk with the H2O2 responses (red section in Fig. 3B). P. infestans, an oomycete pathogen, showed less crosstalk than B. cinerea and a comparable level with Pst avrRpm1 and Pst hrcC−. P. infestans also causes an HR in infected Arabidopsis tissue due to induction of nonhost resistance (Huitema et al., 2003).
Involvement of H2O2 in response to many stressors raises the question of how specificity of the responses is guaranteed using a common signaling molecule (Mittler et al., 2011). Our promoter analysis detected a variety of putative transcriptional regulatory elements that are suggested to receive “specific” H2O2 signals, including H2O2/ABA, H2O2/SA, H2O2/HL, H2O2/heat, H2O2/wounding, and H2O2/B. cinerea infection (Fig. 3C). However, these results are not sufficient to base a practical model on, so more studies are necessary to understand the molecular mechanisms for specific H2O2 responses under different stresses. The predicted target sequence motifs for these specific responses, as shown in Figure 3C, will be key for future studies in this area.
In Figure 3C, there are several motifs containing CGCG for response to HL, heat, and B. cinerea infection. Our previous studies on the HL-inducible ELIP2 promoter revealed that a CGCG-related element designated as Element C (TACGCGCG) in the promoter is not necessary for the environmental stress response (HL, UV-B, and cold) but is involved in constitutive expression in the shoot apical meristem (Hayami et al., 2015). These results suggest the CGCG box is not an element receiving stress signals, but it modifies the stress response so as to express in weak tissues that need to be constitutively protected. Our promoter prediction also detects such elements as long as they are enriched in a selected promoter group (Yamamoto et al., 2011), so this hypothetical role is not in conflict with our cis prediction. However, the CGCG box is also reported as a target element of CAMTA/AtSR family proteins that are regulated by Ca2+ (Yang and Poovaiah, 2002), and regulation of CAMTA by Ca2+ suggests that the element also receives a Ca2+ signal, which is regulated by environmental cues. The latter point of view suggests that the CGCG box receives stress signals, which are repressed by pathogen infection (Galon et al., 2008), or activation by cold stress (Doherty et al., 2009), drought stress (Pandey et al., 2013), aluminum stress (Tokizawa et al., 2015), and several other types of stresses (Benn et al., 2014). These points of view can be consistent with each other if the CGCG box is composed of multiple motifs.
Identified Transcriptional Regulatory Network of H2O2 Response Based on In Vivo and In Vitro Data
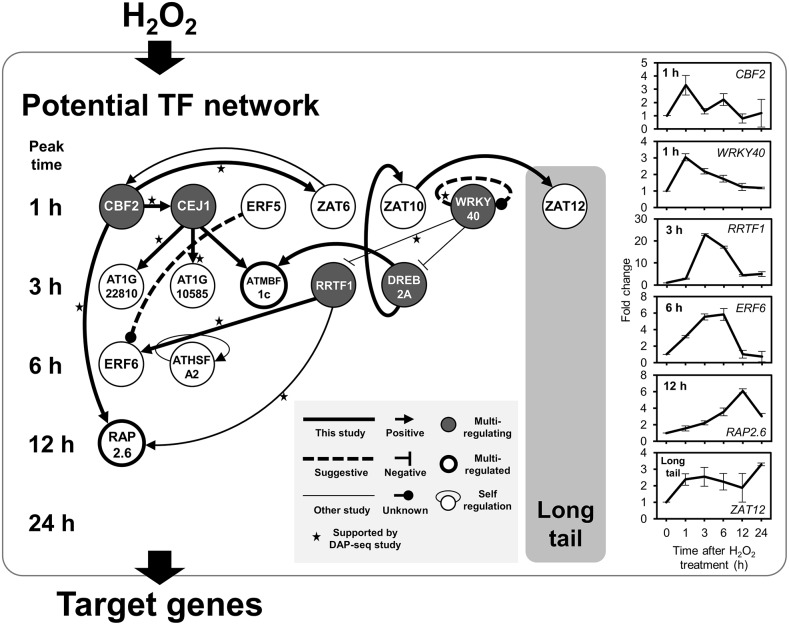
Based on the experimentally identified direct regulation pairs (Table 3), a potential regulatory network of H2O2-responsive TFs is illustrated in Figure 7. A thick solid line indicates direct regulation detected in this study, and a thick dotted line shows speculative autoregulation not supported by in vivo evidence. The figure also includes direct regulation reported in previous studies indicated by thin lines [ZAT6 to CBF2 (Shi et al., 2014); WRKY40 to DREB2A (Shang et al., 2010); WRKY40 to RRTF1 (Pandey et al., 2010); RRTF1 to RAP2.6 (Matsuo et al., 2015); autoregulation of ATHSFA2 (Liu et al., 2013)]. Thirty-four H2O2-responsive TFs that are not involved in the regulatory pairs detected are omitted from the illustration. We also reported results of in vitro DNA-protein binding, called DAP-seq (O’Malley et al., 2016), and binding pairs of consistent results are shown with stars in the figure. Although we should be careful about thresholds of positive binding for each report, eight pairs out of 15 showing direct regulation pairs are consistent with the DAP-seq analysis using partial proteins containing DNA binding domains (stars in Fig. 7), providing reasonably good support of the illustrated network.
Figure 7.
Transcriptional regulatory network activated by H2O2. A directional line means that an upstream TF protein directly regulates gene expression of its downstream TF. Positive and negative regulation was judged based on gene expression data of each TF knockout and/or overexpressor (Supplemental Table S8). A dotted line shows potential autoregulation which is only supported by the data from in vitro binding analysis. Five examples of direct regulation of a TF protein to a target promoter have been reported in previous studies: ZAT6 to CBF2 (Shi et al., 2014); WRKY40 to DREB2A (Shang et al., 2010) and to RRTF1 (Pandey et al., 2010); RRTF1 to RAP2.6 (Matsuo et al., 2015), which was confirmed in this study; and ATHSFA2 autoregulation (Liu et al., 2013). Stars indicate the regulations are supported by in vitro bindings detected in the cistrome study using DAP-seq (O’Malley et al., 2016). Line graph shows expression pattern of representative TF gene in the network. Average fold change and se were obtained from triplicate microarray data.
Figure 7 shows some TFs receive more than one regulation, and they are highlighted with thick-rimmed circles in the figure (ATMBF1c and RAP2.6). They represent hubs and thus have important roles in the network. Five TFs identified by dark gray coloring (CBF2, CEJ1, WRKY40, RRTF1, and DREB2A) regulate more than one TF and also represent hubs.
Induction profiles of some TFs are shown on the right in Figure 7. Comparison of the profiles of RRTF1 and RAP2.6 shows that RAP2.6 has a later induction. RRTF1 is a positive regulator of RAP2.6, so the later induction of RAP2.6 suggests a time lag after the activation peak of RRTF1 before that of RAP2.6. In addition to the time-lagged regulations, simultaneous regulations are also found, as in the case of regulation of ERF6 by RRTF1, both of which have the peak time of 3 h. RRTF1 is negatively regulated by WRKY40, which is transiently induced with a peak time of 1 h. Therefore, the induction profile of RRTF1 suggests it is regulated by WRKY40 so as to be suppressed in the early phase of induction.
What is not shown in the network map is the initial trigger for transcriptional regulation after H2O2 treatment and the involvement of SA and ABA. It also does not include the target genes of the TFs shown, which are directly involved in physiological responses to H2O2, including the genes for ROS scavengers, HSPs, and other factors directly involved in the stress response of land plants.
Until recently, a transcriptional regulatory network based on coexpression data had been proposed. Using multiple transcriptome data sets, a coexpressed gene group is assumed to be regulated by TFs in the group. This method has been applied to predict a transcriptional network for the response to oxidative stress (Vermeirssen et al., 2014). Using this approach, which has been used for a decade, to construct a network is convenient because it requires only transcriptome data, a massive amount of which is available in public databases. However, it does not use experimental evidence of direct regulation between a regulator and a regulated gene. Therefore, network structures deduced by this methodology are limited in their application, such as in silico mining of potentially pivotal, and thus physiologically important, genes for environmental adaptation.
Recently, determination of a regulatory network based on experimental evidence has been developed for the ABA response in etiolated seedlings of Arabidopsis (Song et al., 2016). This was essentially achieved based on two lines of evidence: time-course transcriptome analysis and determination of target sites of the 21 TFs by in vivo ChIP-seq analysis. The latter data were utilized for the identification of a regulator TF and its target genes.
Our approach introduced in this report is restricted to TF–TF regulation and is based on a combination of data for in vivo regulation of a downstream TF expression by an upstream TF and in vitro binding of the upstream TF to the downstream TF promoter, so there are some differences from the approach used by Song et al. (2016). Our medium-scale in vitro assays are compact and less expensive, so they are applicable to studies on nonmodel organisms as well, which is an advantage of our approach.
According to our promoter prediction, we detected 258 putative combinations of 9 TF proteins and their target sites in TF promoters. Experimental analysis with mutated DNA probes verified 20 sequence-specific binding pairs (Fig. 6; Supplemental Fig. S7; Supplemental Tables S9 and S10). Regarding rate of proven direct regulations over predictions, 16 protein–probe pairs were verified out of 132 putative combinations by the double prediction, giving a success rate of ∼12%. In the case of single predictions for ATHSFA4a and CEJ1, success rate was as low as 3.2% (4/126). Together with our new strategy for AlphaScreen using inexpensive preparation of biotinylated double-strand DNA probes, it is now possible to determine target sites of TFs on a medium scale, if guided by the double prediction. This scalability of our method enables genomic studies for nonmodel organisms.
Figure 7 summarizes 15 TF–TF pairs showing direct regulation among H2O2-responsive TFs, including five reported and 10 newly identified pairs. One feature of the revealed regulatory network is the presence of multiple regulators for one TF. We suggest that this feature contributes to shaping the kinetics and dose response of gene expression for each gene in the network.
We wondered how many of the pairs are actually coexpressed, which is a feature used to detect a regulatory network. Therefore, we consulted ATTED-II, an Arabidopsis coexpression database (Obayashi et al., 2007), and found that only three pairs (ZAT10–ZAT12, RRTF1–ERF6, and DREB2A–ATMBF1c) out of 14, excluding one self-regulation, showed positive or negative coexpression (r > 0.5 or < −0.5) in either or both orientations. One possible reason for this infrequent coexpression among regulation and regulated TFs may be that the regulated TF is often controlled by multiple regulator TFs (Fig. 7). The expression profile of the regulated TF in focus would be different from either regulator. These results demonstrate limited detection of a network based on coexpression profiles.
Figure 7 does not cover the very start of the transcriptional network, because the corresponding factor is received at the protein level, and thus gene regulation is not involved in the initial process. One such starter is ANAC017. A recent report demonstrated that the ANAC017 protein localizes to the endoplasmic reticulum, and upon accumulation of H2O2 in a plant cell, it translocates to the nucleus and acts as a starter of transcriptional activation (Ng et al., 2013). Target TFs of this starter are expected to be regulated at the level of gene expression, and of 60 TFs, ATERF71 (AT2G47520) is reported as a direct target of ANAC017. This is based on the observation of reduced expression in anac017 and binding of ANAC017 protein to the promoter region of ATERF71 (Ng et al., 2013; Yamamoto et al., 2017). Unfortunately, we failed to identify any downstream targets of ANAC017, and thus could not add ANAC017 and ATERF71 to Table 3, leaving starters of the network shown in Figure 7 unidentified.
One feature of ROS signaling in land plants is its participation in various stress signaling pathways. This led to the idea of ROS being convergence points of biotic and abiotic stress signaling to achieve a common response (Fujita et al., 2006). Using this assumption, ROS are proposed to have a role in the synthetic response to a combination of abiotic stresses (Choudhury et al., 2017), where stimulation by two weak stressors results in a strong stress response. Our comparative transcriptome analysis as shown in Table 1 is consistent with this idea, and suggests common machinery after H2O2 accumulation for transcriptional responses to various biotic and abiotic stressors. To understand the mechanism of the synthetic response to a combination of stresses, further studies are necessary, including analysis of accumulation profiles of H2O2 and of dose responses of the network itself and its terminal responses.
MATERIALS AND METHODS
Plant Material
For microarray experiments in Arabidopsis (Arabidopsis thaliana; wild type, Col-0) and RT-qPCR in T-DNA knockout lines and the wild-type control, seeds were grown in a hydroponic culture system (Hieno et al., 2013) for 10 d. Briefly, 120 Arabidopsis seeds were soaked in 0.5 mL of sterile distilled water in 1.5-mL microfuge tubes, kept at 4°C for 2 d, and then placed on a nylon mesh (PE-50, 2,500 holes per square inch; 50-mesh; Filter-net) held in a plastic photo slide mount (Fuji Photo). The mounts were floated on 1/5 × MGRL nutrients (at pH 5.6; Fujiwara et al., 1992) and grown at 22°C under continuous light (30 ∼ 40 μE · m−2 · s−1). T-DNA knockout lines of H2O2-responsive TFs, CEJ1 (GK-310C06-015791) and ATHSFA4a (GK-18H12-013603) were obtained from The Arabidopsis Biological Resource Center (http://arabidopsis.org), and their genotypes were confirmed by PCR with a pair of gene-specific and T-DNA–specific primers (Supplemental Table S11).
Treatment of shoots with H2O2 was performed by spraying with 100 mm H2O2 or sterile distilled water as a control, and the shoots were harvested at each time point. It should be noted that treatments with spraying cause much milder effects than with submerging tissue into H2O2 solution of the same concentration. Harvested samples were immediately frozen in liquid nitrogen and crushed into a powder with zirconium dioxide beads using a vertical shaker (ShakeMaster Neo v1.0; Bio Medical Science). Ground samples were stored at −80°C until extraction.
For RT-qPCR experiments in the aba1 mutant (CS25407/N25407, a progeny line of SALK_059469) and the wild-type control, 50 seeds were grown on round deep plastic plates (20 × 90 mm, FG-2090; NIPPON Genetics) containing 25 mL germination medium (consisting of: half-strength Murashige and Skoog [MS] salts [Wako Pure Chemical]; 1% [w/v] Suc, 0.8% [w/v] agar, 1 × Gamborg’s B5 Medium Vitamin Mixture [Research Products International]; and 2.56 mm 2-(n-morpholino) ethanesulfonic acid [MES]-KOH buffer, at pH 5.6, for 10 d at 22°C under continuous light [30 ∼ 40 μE · m−2 · s−1]). H2O2 treatment was performed as described in "Plant Material" for microarray experiments. Then, 12 h after treatment, shoots were frozen in liquid nitrogen and then harvested and crushed in the same manner as described in "Plant Material" for microarray experiments. Ground samples were stored at −80°C until extraction.
Quantification of H2O2 and Plant Hormones
Quantification of H2O2 in shoots was performed using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher Scientific), according to the manufacturer’s protocol. Tissues of ∼100 mg fresh weight were crushed into a powder as described in "Plant Material" for microarray experiments. The frozen powder was dissolved in 900 μL of 1× reaction buffer (50 mm sodium phosphate, at pH 7.4), and 10 μL of the dissolved sample was diluted with 490 μL of 1× reaction buffer, of which 50 μL was used for the assay. Reaction mixtures (50 μL of the sample and 50 μL of Amplex Red reagent/HRP working solution) were incubated in 96-well plates (OptiPlate-96 F; PerkinElmer Japan) with excitation of 531 nm and emission of 579 nm. H2O2 concentrations were calculated using standard curves, according to the manufacturer’s protocol. Samples were triplicated for calculation of averages and ses.
Plant hormones (ABA, IAA, SA, JA, JA-Ile) in shoots were determined according to Mikami et al. (2016). In brief, H2O2-treated samples (∼350 mg fresh weight) were collected and subjected to extraction in 80% (v/v) acetonitrile and 1% (v/v) acetic acid containing stable isotope-labeled compounds for internal standards. Hormones were analyzed with a model no. 6410 Triple Quad LC/MS System (Agilent Technologies) equipped with a ZORBAX Eclipse XDB-C18 column and XDB-C8 Guard column (Agilent Technologies), and peak areas were determined using MassHunter Workstation software (vB.04.00; Agilent Technologies). Samples were triplicated for calculation of averages and ses.
Microarray Analysis
Total RNA was extracted according to Zhao et al. (2009). Microarray analyses were performed using an Agilent custom array (4 × 44k) containing probes for 26,254 annotated genes and an additional 7,901 small open reading frames (sORFs; Hanada et al., 2013). Three biological replicates for each treatment were analyzed to confirm by a Quick Amp Labeling Kit, two-color, or Low Input Quick Amp Labeling Kit, two-color (Agilent Technologies), following the manufacturer’s protocols. Labeled probes were purified with a RNeasy Mini Kit (Qiagen), then hybridized with a Gene Expression Hybridization Kit (Agilent Technologies), following the manufacturer’s protocol, and scanned using an Agilent DNA Microarray Scanner with Scan Control v6.3.1 (G2565BA; Agilent Technologies). Three independent raw data were processed with the software GeneSpring 12.5 (Agilent Technologies). Filtering microarray data for quality control was carried out using the default settings of GeneSpring to remove “Compromised” and thus nonuniform spots, saturated spots, and population outliers. Additional data filtering was performed by Cyber-T (Baldi and Long, 2001; Long et al., 2001; http://cybert.microarray.ics.uci.edu/) to calculate the Bayesian P value for the identification of statistical significance (P value < 0.05).
Hierarchical clustering of log2 transformed microarray data, after classification into 1, 3, 6, 12, and 24 h and “long tail” groups according to their peak time of induction, was achieved using the software Cluster 3.0 (de Hoon et al., 2004; http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm; uncentered correlation, pairwise average linkage method) and visualized using the software Java TreeView 3.0 (Keil et al., 2016; https://bitbucket.org/TreeView3Dev/treeview3/).
Microarray data from the public databases used are listed in Supplemental Table S12.
Gene Expression Analysis by RT-qPCR
Total RNA was extracted using Sepasol-RNA I Super G (Nacalai Tesque) following the manufacturer’s protocol, subjected to LiCl precipitation and subsequent isopropanol precipitation, and dissolved in RNase-free water. The concentrations of the extracted RNA samples were measured with a spectrophotometer (BioSpectrometer Basic; Eppendorf), and 500 ng of total RNA was used to synthesize first-strand cDNA by ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO), according to the manufacturer’s protocol. The reverse transcription products (10 μL) were diluted to one-half with water and used as templates for RT-qPCR performed using SYBR Premix Ex Taq II (Tli RNase H Plus; TaKaRa). RT-qPCR reaction mixtures were prepared in a total volume of 10 μL containing 5 μL of 2 × SYBR Premix, 0.4 μL of 10 μM each of forward and reverse primers (0.4 μm final concentration), 0.8 μL of the cDNA template, and 3.4 μL of H2O. Gene-specific primers used for analysis are listed in Supplemental Table S11. The reactions were performed using Thermal Cycler Dice Real Time System II (TP900l; TaKaRa) under the following conditions: 95°C for 30 s, 40 cycles of two-step thermal cycling composed of 95°C for 5 s and 60°C for 60 s, and one cycle of 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. The relative standard curve method was used for the quantification of mRNA expression. cDNA standard curves were prepared using the threshold cycles with a serial dilution series (one-half, one-fourth, one-eigth, one-sixteenth, and one-thirty-second). Results are reported as the average ± se of at least three samples from three or four independent experiments.
Prediction of cis-Elements and Colocalization Analysis
Public microarray data listed in Supplemental Table S12 were used for prediction of cis-elements (Yamamoto et al., 2011) based on 25k promoter sequences that were 1,000 bp-long for protein-coding genes (Tokizawa et al., 2017). Putative regulatory sequences were extracted by evaluation of relative appearance ratio (RAR) for each octamer sequence, which is a ratio of appearance of an octamer in a promoter set of a responsive gene group to that in total promoters of the genome. Continuous octamers (Yamamoto et al., 2011) and also bipartite octamers with a spacer, which have a potential of cis-elements in Arabidopsis (Yamamoto et al., 2017), were applied for the extraction. Octamers with high RAR values (RAR ≥ 3) were extracted as putative cis-elements.
Colocalization analysis of cis-prediction was performed as described before (Yamamoto et al., 2011). Two independent dye-swap microarray data of H2O2 response described before were used for promoter prediction. Two predictions, H2O2 response and biotic/abiotic stress response, were applied to the responsive promoters to detect overlapping loci whose distance was 4 bp or less in the promoter region.
Motifs referred to in Figure 3C are ABRE (Hattori et al., 2002), ERF (Hao et al., 1998), DREB (Sakuma et al., 2002), CGCG box (Yang and Poovaiah, 2002), HSE (Barros et al., 1992), and core sequence recognized by bZIP family proteins (ACGT; Foster et al., 1994).
Protein–DNA Binding Assays by AlphaScreen
Three independent microarray data obtained from time-course experiments after the H2O2 treatment were used to identify H2O2-responsive TFs with ≥ 2.5-fold change. Identified TFs were filtered by Cyber-T (Baldi and Long, 2001; Long et al., 2001; http://cybert.microarray.ics.uci.edu/) utilized to calculate the Bayesian P value for the identification of statistical significance (P value < 0.05). If a P value of an identified TF was 0.05 or over, RT-qPCR analysis was used for confirmation. A > 2.5 fold change in TF expression was regarded as H2O2-responsive. Target promoters of a TF are listed in Supplemental Table S9.
Downstream regulation of H2O2-related TFs were identified based on public microarray data for knockout mutants or overexpressors of the seven TFs, CBF2, ERF5, ZAT10, WRKY40, RRTF1, ATMBF1c, and DREB2A, from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/; Supplemental Table S12). Positive and negative regulation was identified by fold change ≥ 2.0 or ≤ 0.5 and statistical significance with Bayesian P value < 0.05 using the software program Cyber-T (Baldi and Long, 2001; Long et al., 2001); http://cybert.micriarray.ics.uci.edu/). Target sites for each TF were predicted according to our previous report (Yamamoto et al., 2011), using microarray data of H2O2 and mutants and/or overexpressors of the TF. Probes were designed for colocalized sites (≤ 4 bp) of H2O2 and TF-targeting sequences (Fig. 3A; Supplemental Fig. S5) to enable more accurate prediction rather than each single target site. There was no microarray data available for two TFs, ATHSFA4a and CEJ1. For these, positive or negative regulation was identified by RT-qPCR analysis, and probes were designed based on only H2O2 microarray data.
Next, 5′ biotinylated double-strand DNA probes were synthesized using T7 polymerase (exo−; Sequenase 2.0; Affymetrix/Thermo Fisher Scientific). The templates used were 60 base single-strand DNA fragments containing 44 bases of promoter sequences and 16 bases of the primer binding site corresponding to the 5′-biotinylated universal primer sequence from the CIP7 (AT4G27430) intron (Supplemental Fig. S5; Supplemental Table S11).
Probe-specific single-stranded DNA containing an annealing site for the biotinylated universal primer was annealed with the universal primer, both at a concentration of 40 μm in a volume of 5 μL, by incubation at 95°C for 10 min and then at 4°C overnight. Two μL of the annealed DNA mixture (100 pmol) was adjusted to a volume of 10 μL containing 1 × reaction mixture (66 mm Tris-HCl at pH 7.6, 10 mm MgCl2, 1 mm ATP, 1 mm DTT, 0.1 mg/mL bovine serum albumin, 0.2 mm dNTPs, and 5 mm T7 DNA polymerase [exo−]), incubated for 30 min at 37°C, and then diluted to one-tenth with one-fifth TE buffer. Unbiotinylated probes were prepared in the same manner using unbiotinylated universal primer instead of the biotinylated one. Biotinylation of the synthesized probes was confirmed by electrophoresis, and biotin was detected using streptavidin-conjugated alkaline phosphatase (PerkinElmer Japan) and western Blue Stabilized Substrate (Promega KK).
FLAG-tagged TF proteins were synthesized using an in vitro transcription/translation system (from NUProtein), according to the methods described in Matsuo et al. (2015). Template cDNAs for complementary RNA were amplified from corresponding cDNA clones in the Arabidopsis TF cDNA collection (Mitsuda et al., 2010) or, if not in the collection, prepared by RT-PCR using total RNA from H2O2-treated Arabidopsis seedlings. The amount and molecular weights of synthesized proteins were confirmed by immunoblotting analysis using anti-FLAG antibody (Wako Pure Chemical Industries). Unpurified translation products and biotinylated or unbiotinylated probes were mixed in the reaction buffer in a total volume of 8.5 μL containing 25 mm HEPES-KOH at pH 7.6, 40 mm KCl, 0.01% (v/v) TWEEN20, and 0.1% (w/v) bovine serum albumin in 96-well plates (FastGene 96-well PCR plates; NIPPON Genetics), and incubated for 30 min at room temperature (∼23°C). After the initial incubation, 2 μL of acceptor beads (AlphaScreen FLAG [M2] Detection Kit; PerkinElmer Japan) diluted to 1/40 with sterile distilled water were added to the reaction mixture and incubated again for 30 min in the dark at room temperature. Donor beads were diluted and added to the reaction mixture in the same way as the acceptor beads, then the reaction mixture was transferred to 384-well Optiwell microtiter plates (PerkinElmer) in dim light. After incubation in the dark for 3 h at 23°C ± 1°C, light emission was measured using an EnSpire reader (PerkinElmer Japan). Detected signals were normalized with those of the control samples with unbiotinylated probes. The data are based on triplicate experiments, with averages and ses.
Accession Numbers
H2O2 microarray data from this article can be found in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) under accession no. E-MTAB-4961. Supplemental Table S12 shows accession numbers of public microarray data used in this study. Identified H2O2-responsive genes with Arabidopsis Genome Initiative (AGI) codes are listed in Supplemental Tables S1 and S4. H2O2-responsive sORFs with Probe ID and other details are shown in Supplemental Table S2.
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Accumulation of H2O2 by H2O2 treatments.
Supplemental Figure S2. Subcellular localization of the H2O2-responsive genes.
Supplemental Figure S3. Two possible types of signal crosstalk at colocalized sites of promoter prediction.
Supplemental Figure S4. Heat map of colocalization analysis based on two kinds of time-course data of microarray analysis.
Supplemental Figure S5. Preparation of a 5′-biotinylated double-strand DNA probe.
Supplemental Figure S6. Possible secondary structure of probe and interaction with protein.
Supplemental Figure S7. Identified TF binding sequences by AlphaScreen assay with mutated probes.
Supplemental Table S1. Identified H2O2-responsive genes.
Supplemental Table S2. Expression profiles of H2O2-responsive sORFs.
Supplemental Table S3. Gene ontology analysis identified cell wall-associated genes in H2O2 response.
Supplemental Table S4. Identification of H2O2-responsive TFs by microarray and RT-qPCR.
Supplemental Table S5. Expression profiles of H2O2-responsive TFs in stress-related hormone responses.
Supplemental Table S6. Expression profiles of H2O2-responsive TFs under biotic stresses.
Supplemental Table S7. Expression profiles of H2O2-responsive TFs under abiotic stresses.
Supplemental Table S8. Expression profiles of H2O2-responsive TFs in overexpressor and knockdown mutants.
Supplemental Table S9. Probe sequences and results of protein–DNA binding assay.
Supplemental Table S10. Protein–DNA binding assay with mutated probes.
Supplemental Table S11. Primer sequences used in this study.
Supplemental Table S12. Public microarray data used in this study.
Acknowledgments
The authors appreciate Dr. Frank Van Breusegem and Dr. Inge De Clercq of Ghent University for critical reading of the article. We thank Dr. Masanori Okamoto of Utsunomiya University and Dr. Takashi Hirayama of Okayama University for kindly providing Arabidopsis mutant seeds, and also Dr. Mutsutomo Tokizawa and Mr. Daichi Obata in our laboratory for technical assistance with protein–DNA binding assays.
Footnotes
This work was supported in part by the Japan Science and Technology Agency, Advanced Low Carbon Technology Research and Development Program (to Y.Y.Y.), Grant-in-Aid for Scientific Research on Innovative Areas “Plant Perception” (grant no. 23120511 to Y.Y.Y.), and the Japan Advanced Plant Science Network.
Articles can be viewed without a subscription.
References
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer P-J, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Banti V, Mafessoni F, Loreti E, Alpi A, Perata P (2010) The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 152: 1471–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MD, Czarnecka E, Gurley WB (1992) Mutational analysis of a plant heat shock element. Plant Mol Biol 19: 665–675 [DOI] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N (2014) ROS as key players in plant stress signalling. J Exp Bot 65: 1229–1240 [DOI] [PubMed] [Google Scholar]
- Benn G, Wang CQ, Hicks DR, Stein J, Guthrie C, Dehesh K (2014) A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J 80: 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN (2002) Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol Microbiol 44: 73–88 [DOI] [PubMed] [Google Scholar]
- Choudhury FK, Rivero RM, Blumwald E, Mittler R (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90: 856–867 [DOI] [PubMed] [Google Scholar]
- Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debener T, Lehnackers H, Arnold M, Dangl JL (1991) Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J 1: 289–302 [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8: 192–200 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR (2003) Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J 33: 691–705 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S (1992) Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol 99: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y, Nave R, Boyce JM, Nachmias D, Knight MR, Fromm H (2008) Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett 582: 943–948 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Fryer MJ, Lawson T, Slattery K, Truman W, Smirnoff N, Asami T, Davies WJ, Jones AM, Baker NR, et al. (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govrin EM, Levine A (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10: 751–757 [DOI] [PubMed] [Google Scholar]
- Govrin EM, Rachmilevitch S, Tiwari BS, Solomon M, Levine A (2006) An elicitor from Botrytis cinerea induces the hypersensitive response in Arabidopsis thaliana and other plants and promotes the gray mold disease. Phytopathology 96: 299–307 [DOI] [PubMed] [Google Scholar]
- Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y, Wang Y, Wang J (2008) Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J Genet Genomics 35: 105–118 [DOI] [PubMed] [Google Scholar]
- Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. (2013) Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci USA 110: 2395–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A (1998) Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem 273: 26857–26861 [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hayami N, Sakai Y, Kimura M, Saito T, Tokizawa M, Iuchi S, Kurihara Y, Matsui M, Nomoto M, Tada Y, et al. (2015) The responses of Arabidopsis ELIP2 to UV-B, high light, and cold stress are regulated by a transcriptional regulatory unit composed of two elements. Plant Physiol 169: 840–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Vásquez A, Salinas P, Holuigue L (2015) Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci 6: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieno A, Naznin HA, Sawaki K, Koyama H, Sakai Y, Ishino H, Hyakumachi M, Yamamoto YY (2013) Analysis of environmental stress in plants with the aid of marker genes for H2O2 responses. Methods Enzymol 527: 221–237 [DOI] [PubMed] [Google Scholar]
- Huitema E, Vleeshouwers VG, Francis DM, Kamoun S (2003) Active defence responses associated with non-host resistance of Arabidopsis thaliana to the oomycete pathogen Phytophthora infestans. Mol Plant Pathol 4: 487–500 [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J (2003) Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15: 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Keil C, Leach RW, Faizaan SM, Bezawada S, Parsons L, Baryshnikova A (2016) Treeview 3.0 (beta 1)—Visualization and analysis of large data matrices. https://zenodo.org/record/160573#.XPE77xZKhpg [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA 109: 17129–17134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun N, Liu M, Liu J, Du B, Wang X, Qi X (2013) An autoregulatory loop controlling Arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol 162: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P (2001) Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J Biol Chem 276: 19937–19944 [DOI] [PubMed] [Google Scholar]
- Mackerness AH, Surplus SL, Blake P, John CF, Buchanan-Wollaston V, Jordan BR, Thomas B (1999) Ultraviolet-B-induced stress and changes in gene expression in Arabidopsis thaliana: Role of signalling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ 18: 105–109 [Google Scholar]
- Matsuo M, Johnson JM, Hieno A, Tokizawa M, Nomoto M, Tada Y, Godfrey R, Obokata J, Sherameti I, Yamamoto YY, et al. (2015) High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol Plant 8: 1253–1273 [DOI] [PubMed] [Google Scholar]
- Mikami K, Mori IC, Matsuura T, Ikeda Y, Kojima M, Sakakibara H, Hirayama T (2016) Comprehensive quantification and genome survey reveal the presence of novel phytohormone action modes in red seaweeds. J Appl Phycol 28: 2539–2548 [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, Fujita M, Shinozaki K, Matsui M, Ohme-Takagi M (2010) Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol 51: 2145–2151 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: The new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H (2007) ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas M, Ryan CA (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96: 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N, Ranjan A, Pant P, Tripathi RK, Ateek F, Pandey HP, Patre UV, Sawant SV (2013) CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Roccaro M, Schön M, Logemann E, Somssich IE (2010) Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 64: 912–923 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu Z-Q, Cao Z, Mei C, Xin Q, Wu F-Q, Wang X-F, Du S-Y, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Ye T, Chen F, Deng J, Yang P, Zhang Y, Chan Z (2014) The cysteine2/histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiol 165: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C (1997) Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A, Nie S, Xing D (2012) Nitric oxide-mediated maintenance of redox homeostasis contributes to NPR1-dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiol 160: 1081–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokizawa M, Kobayashi Y, Saito T, Kobayashi M, Iuchi S, Nomoto M, Tada Y, Yamamoto YY, Koyama H (2015) SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and other transcription factors are involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 expression. Plant Physiol 167: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokizawa M, Kusunoki K, Koyama H, Kurotani A, Sakurai T, Suzuki Y, Sakamoto T, Kurata T, Yamamoto YY (2017) Identification of Arabidopsis genic and non-genic promoters by paired-end sequencing of TSS tags. Plant J 90: 587–605 [DOI] [PubMed] [Google Scholar]
- van Buer J, Cvetkovic J, Baier M (2016) Cold regulation of plastid ascorbate peroxidases serves as a priming hub controlling ROS signaling in Arabidopsis thaliana. BMC Plant Biol 16: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inzé D, et al. (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Vermeirssen V, De Clercq I, Van Parys T, Van Breusegem F, Van de Peer Y (2014) Arabidopsis ensemble reverse-engineered gene regulatory network discloses interconnected transcription factors in oxidative stress. Plant Cell 26: 4656–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Panchuk II, Mullineaux PM, Schöffl F (2006) Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol Biol 61: 733–746 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports 1: 639–647 [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Shimada Y, Kimura M, Manabe K, Sekine Y, Matsui M, Ryuto H, Fukunishi N, Abe T, Yoshida S (2004) Global classification of transcriptional responses to light stress in Arabidopsis thaliana. Endocytobiosis Cell Res 15: 438–452 [Google Scholar]
- Yamamoto YY, Yoshioka Y, Hyakumachi M, Maruyama K, Yamaguchi-Shinozaki K, Tokizawa M, Koyama H (2011) Prediction of transcriptional regulatory elements for plant hormone responses based on microarray data. BMC Plant Biol 11: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Hieno A, Obata D, Tokizawa M, Nomoto M, Tada Y, Kusunoki K, Koyama H, Hayami N (2017) Prediction of bipartite transcriptional regulatory elements using transcriptome data of Arabidopsis. DNA Res 24: 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
- Yu GL, Katagiri F, Ausubel FM (1993) Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol Plant Microbe Interact 6: 434–443 [DOI] [PubMed] [Google Scholar]
- Zhang L, Li Y, Xing D, Gao C (2009) Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J Exp Bot 60: 2073–2091 [DOI] [PubMed] [Google Scholar]
- Zhao CR, Ikka T, Sawaki Y, Kobayashi Y, Suzuki Y, Hibino T, Sato S, Sakurai N, Shibata D, Koyama H (2009) Comparative transcriptomic characterization of aluminum, sodium chloride, cadmium and copper rhizotoxicities in Arabidopsis thaliana. BMC Plant Biol 9: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HH, McClung CR (1996) The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol Gen Genet 251: 196–203 [DOI] [PubMed] [Google Scholar]
- Zhong HH, Young JC, Pease EA, Hangarter RP, McClung CR (1994) Interactions between light and the circadian clock in the regulation of CAT2 expression in Arabidopsis. Plant Physiol 104: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]