During the obligate vernalization response of Arabis alpina, gibberellin promotes flowering in cold and this is modulated by the PEP1 transcription factor.
Abstract
Regulation of flowering by endogenous and environmental signals ensures that reproduction occurs under optimal conditions to maximize reproductive success. Involvement of the growth regulator gibberellin (GA) in the control of flowering by environmental cues varies among species. Arabis alpina Pajares, a model perennial member of the Brassicaceae, only undergoes floral induction during vernalization, allowing definition of the role of GA specifically in this process. The transcription factor PERPETUAL FLOWERING1 (PEP1) represses flowering until its mRNA levels are reduced during vernalization. Genome-wide analyses of PEP1 targets identified genes involved in GA metabolism and signaling, and many of the binding sites in these genes were specific to the A. alpina lineage. Here, we show that the pep1 mutant exhibits an elongated-stem phenotype, similar to that caused by treatment with exogenous GA, consistent with PEP1 repressing GA responses. Moreover, in comparison with the wild type, the pep1 mutant contains higher GA4 levels and is more sensitive to GA prior to vernalization. Upon exposure to cold temperatures, GA levels fall to low levels in the pep1 mutant and in wild-type plants, but GA still promotes floral induction and the transcription of floral meristem identity genes during vernalization. Reducing GA levels strongly impairs flowering and inflorescence development in response to short vernalization treatments, but longer treatments overcome the requirement for GA. Thus, GA accelerates the floral transition during vernalization in A. alpina, the down-regulation of PEP1 likely increases GA sensitivity, and GA responses contribute to determining the length of vernalization required for flowering and reproduction.
Early physiological analysis of the floral transition proposed prominent roles for growth regulators. For example, in Arabidopsis (Arabidopsis thaliana), cytokinin was found to confer altered cell division patterns at the shoot meristem during floral induction and increased levels of GA were implicated in the flowering response to photoperiod (Vaughan, 1955; Langridge, 1957; Corbesier et al., 2003). More recently, molecular-genetic analyses have implicated GA as well as cytokinin, brassinosteroids, and auxin in aspects of the floral transition (Domagalska et al., 2007; Li et al., 2010; Bartrina et al., 2011; D’Aloia et al., 2011; Galvão et al., 2012; Porri et al., 2012; Yamaguchi et al., 2013). Nevertheless, how these responses are linked to the well-established transcriptional networks that control the floral transition in response to environmental cues is often not clear. Furthermore, how conserved these processes are among species is generally unknown, particularly as the role of GA in the control of flowering can vary from being floral promoting to floral repressive (Mutasa-Göttgens and Hedden, 2009). Here, we utilize Arabis alpina, an obligate vernalization-requiring perennial relative of Arabidopsis, to define the role of GA in the vernalization response.
Genetic analysis of the contribution of GA to the control of flowering is most developed in Arabidopsis. It was first described in the context of photoperiodic response, where applications of GA were found to overcome the delay in flowering observed under noninductive short days (SDs; Langridge, 1957). Similarly, mutations that strongly impair GA biosynthesis almost abolish flowering under SDs but have a less pronounced effect under inductive long days (LDs; Wilson et al., 1992). Nevertheless, overexpressing an enzyme in the shoot meristem that deactivates GA also significantly delayed flowering under LDs (Porri et al., 2012). Similarly, blocking GA signaling by combining mutations in each of the three genes encoding GIBBERELLIN INSENSITIVE DWARF1 (GID1) GA receptors strongly delayed flowering under LDs and SDs (Griffiths et al., 2006). Increased levels of GA4 at the shoot apex precede the floral transition under SDs, but no increase in expression of GA biosynthetic enzymes at the apex was detected (Eriksson et al., 2006). GA12, a precursor of active GAs, is systemically transported through the plant (Regnault et al., 2015), and an increase in transport might explain the increase in GA4 detected at the apex under SDs prior to the floral transition. Alternatively, the increased GA4 at the apex of older plants might be due to the additive effect of GA being transported from a higher number of leaves and thereby passing a threshold at the apex required to induce flowering. DELLA proteins are also important intermediates in GA signal transduction that influence flowering time. These proteins interact with and affect the activity of many transcription factors (Marín-de la Rosa et al., 2014), but when GID1 proteins bind GA they interact with DELLA proteins to promote their proteasomal degradation (Griffiths et al., 2006; Nakajima et al., 2006). Flowering is repressed by gain-of-function forms of DELLA that are not efficiently degraded in the presence of GA (Peng et al., 1997; Dill et al., 2001; Galvão et al., 2012). Several transcription factors that regulate flowering, notably in the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) class, interact with DELLAs (Yu et al., 2012). The promotion of transcription of APETALA1 by SPL9 during the early stages of floral primordium development is enhanced by interaction with DELLAs (Yamaguchi et al., 2014), whereas the transcription of FRUITFULL and MIR172b by SPL15 during floral induction under SDs is reduced by interaction with DELLA (Yu et al., 2012; Hyun et al., 2016). Thus, destabilization of DELLA by GA positively affects floral induction via SPL15 but negatively affects flower development via SPL9. Another example of floral-promoting transcription factors that are targeted and negatively regulated by DELLAs are the PHYTOCHROME INTERACTING FACTORs (PIFs; de Lucas et al., 2008; Feng et al., 2008). PIF4 and to some extent PIF3 promote flowering in response to high temperatures under SDs, although the significance of their involvement varies among experiments (Kumar et al., 2012; Galvão et al., 2015; Fernández et al., 2016). After floral induction, GA also affects other aspects of reproductive development in Arabidopsis, including flower development (Achard et al., 2004), inflorescence meristem size (Serrano-Mislata et al., 2017), and bolting of the inflorescence (Koornneef and van der Veen, 1980; Griffiths et al., 2006; Rieu et al., 2008).
In several species, GA biosynthesis is linked to the flowering response to vernalization. In lisianthus (Eustoma grandiflorum) and the Brassicaceae species field pennycress (Thlaspi arvense), precursors of active GAs are increased in abundance by vernalization, while in winter canola (Brassica napus), vernalization increased the levels of active GA (Hazebroek et al., 1993; Zanewich and Rood, 1995; Hisamatsu et al., 2004). In addition, in radish (Raphanus sativus) and T. arvense, application of exogenous GAs overcame the requirement of vernalization for flowering (Suge and Rappaport, 1968; Metzger, 1985). However, genetic analysis in Arabidopsis did not identify a role for GA in the vernalization response (Chandler et al., 2000). In this species, the vernalization response depends on the MADS box transcription factor FLOWERING LOCUS C (FLC), which is a floral repressor that blocks flowering until the plant is exposed to vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999). During vernalization, transcription of FLC is repressed, allowing flowering to proceed. Arabidopsis Columbia, the standard laboratory accession, does not express high levels of FLC mRNA and therefore flowers rapidly without vernalization. Therefore, Chandler et al. (2000) used fca mutants that contain high levels of FLC mRNA and showed a strong flowering response to vernalization. Combining fca with mutations that impair GA biosynthesis did not affect the vernalization response (Chandler et al., 2000), suggesting that GA is not required for flowering under these conditions. Nevertheless, analysis of the genome-wide binding sites (BSs) of FLC identified several genes involved in GA metabolism and signaling (Deng et al., 2011; Mateos et al., 2015), suggesting that FLC might inhibit flowering at least in part by repressing processes related to GA.
A. alpina is a perennial relative of the annual Arabidopsis, providing a comparative model system to study annual and perennial flowering patterns. A. alpina shows the typical perennial behavior of flowering repeatedly with alternating phases of vegetative and floral development. During the flowering phase, some shoots remain vegetative, ensuring the survival of the plant to the next year. Interestingly, the Spanish Pajares reference accession of A. alpina absolutely requires vernalization to flower (Wang et al., 2009). Vernalization causes transcriptional silencing of PERPETUAL FLOWERING1 (PEP1), but its expression increases again after vernalization, repressing the flowering of any shoots that did not flower in vernalization (Wang et al., 2009). The roles of PEP1 and FLC during vernalization are similar between their respective species, but comparison of PEP1 and FLC target genes in A. alpina and Arabidopsis, respectively, revealed that a low proportion of them are conserved (Mateos et al., 2017). Particularly, like FLC, PEP1 binds to GA-related genes, but many of the genes bound are not conserved between species (Mateos et al., 2017). Thus, in both of these species, repression of GA responses might contribute to the vernalization requirement, but the precise mechanism appears to differ between them. Nevertheless, many genes involved in the regulation of flowering are present in the conserved group, including the floral integrator genes SPL15 and SUPPRESSION OF OVEREXPRESSION OF CONSTANS1 and the floral development gene SEPALLATA3 (Mateos et al., 2017).
Here, we exploit the obligate vernalization response of A. alpina to investigate the interaction between vernalization and GA. We find that PEP1 negatively regulates GA signaling and reduces GA levels before vernalization. Also, we use paclobutrazol (PAC), an inhibitor of GA biosynthesis, and transgenic A. alpina overexpressing GA catabolic enzymes to show that GA promotes flowering during vernalization and that this is particularly important to determine flowering time and inflorescence development on exposure to short vernalization periods. We conclude that the GA pathway contributes to the flowering response of A. alpina to vernalization.
RESULTS
PEP1 Binds to and Regulates Genes Involved in GA Metabolism and Signaling
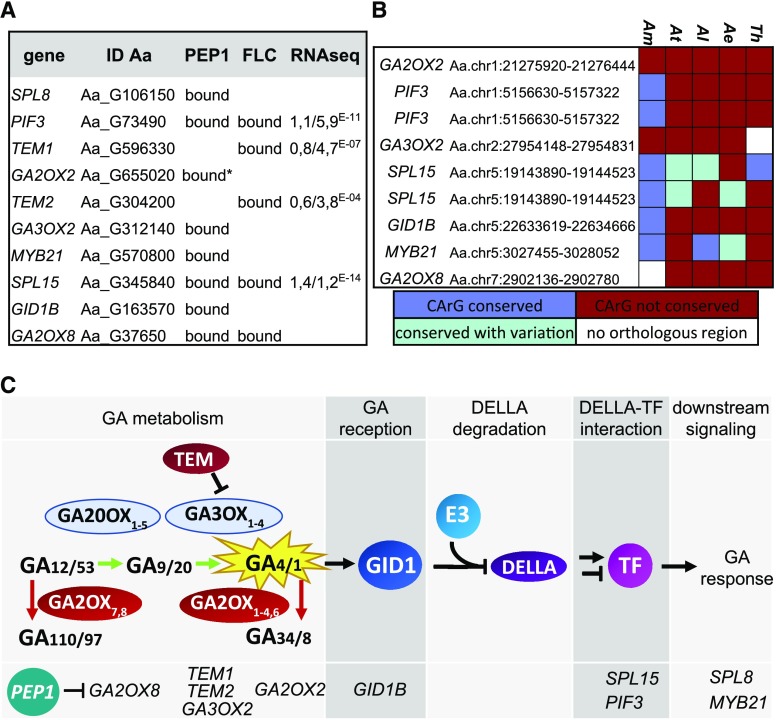
PEP1 target genes in A. alpina are enriched for genes involved in GA metabolism and hormone signaling (Mateos et al., 2017). Some of those genes with roles in GA metabolism and signaling were also differentially expressed in a genome-wide transcriptome analysis (RNA-sequencing) comparing wild-type and pep1 mutant plants prior to vernalization (Fig. 1A; Supplemental Fig. S1). Interestingly, most of the GA-related target genes were bound by PEP1 in A. alpina but not by FLC in Arabidopsis. To test whether PEP1 binding to GA-related genes is likely to be conserved in other closely related species, orthologous sequences of PEP1 BSs were extracted for several of these species, aligned to the A. alpina sequence, and the conservation of CArG-box motifs, which are bound by MADS-box transcription factors such as PEP1, was assessed. In Arabis montbretiana, the annual sister species of A. alpina, CArG-box motifs present in several GA-related genes were conserved (Fig. 1B), although those present in GA3OX2 and GA2OX2, genes encoding GA metabolic enzymes, were not conserved. Most CArG-boxes bound by PEP1 in A. alpina were lost in more distantly related Brassicaceae species such as Arabidopsis and its perennial sister species Arabidopsis lyrata or the basal Brassicaceae species Aethionema arabicum. Also, few of these sites were conserved in Tarenaya hassleriana, a member of the Cleomaceae family used as an outgroup. Although, we cannot exclude that orthologous genes in which the PEP1-bound CArG-box motif is not conserved contain another CArG box that is bound by the FLC ortholog of the second species, this was observed at only a low frequency in direct comparisons of FLC and PEP1 BSs in Arabidopsis and A. alpina, respectively (Mateos et al., 2017). Therefore, this pattern of conservation suggests that direct regulation of GA-related genes by PEP1 is not highly conserved in other Brassicaceae lineages. The BSs in SPL15 were exceptions; this gene contained two CArG-box motifs in A. alpina and at least one of them was conserved in each of the species tested (Fig. 1B). SPL15 is an important floral integrator that is posttranslationally regulated by GA (Hyun et al., 2016); thus, conservation of SPL15 BSs might be another indication that PEP1 and FLC have conserved functions in the regulation of flowering (Mateos et al., 2017).
Figure 1.
PEP1 binds and regulates genes involved in GA metabolism and signaling. A, List of GA-related genes that were bound or regulated by PEP1. This list includes all genes that were targeted by PEP1 (directly or indirectly as detected by Mateos et al. [2017]) and involved in GA metabolism or direct targets that are part of the Gene Ontology category GO:0009739: response to GA stimulus and have a confirmed function in GA signaling or have a published role in the response to GA. Binding information of FLC was gained in three previous studies (Deng et al., 2011; Mateos et al., 2015, 2017). For genes that were differentially regulated in apices of the pep1 mutant, the log2 (fold change) and P values are given. *, The chromatin immunoprecipitation (ChIP)-seq study detected weak binding of PEP1 to GA2OX2 but read enrichment was below the significance threshold. ChIP-qPCR results shown in Supplemental Figure S1, however, suggest that GA2OX2 is a significant target of PEP1. B, Heat map showing the conservation of CArG-boxes in different species. The heat map includes orthologous regions for A. alpina PEP1 BSs associated with GA-related genes in other species. The color code is as follows: dark blue, CArG-box is conserved; light blue, CArG-box is present but sequence varies from A. alpina; dark red, CArG-box is not conserved; and white, no orthologous region was identified. Species are as follows: Arabis montbretiana (Am), Arabidopsis (At), Arabidopsis lyrata (Al), Aethionema arabicum (Ae), and Tarenaya hassleriana (Th). C, Schematic overview of the different steps of the GA pathway. The middle row shows key enzymes involved in each step, and the bottom row lists genes that are targeted by PEP1 in each step.
To better understand the role of PEP1 in regulating GA-related processes, PEP1 targets were grouped according to their functions in the GA pathway. As illustrated in Figure 1C, PEP1 target genes are involved in different stages of the GA pathway from biosynthesis to GA signaling (Fig. 1C). Therefore, PEP1 appears to regulate GA metabolism as well as signaling in A. alpina.
The pep1-1 Mutation and GA Treatments Cause Similar Stem-Elongation Phenotypes
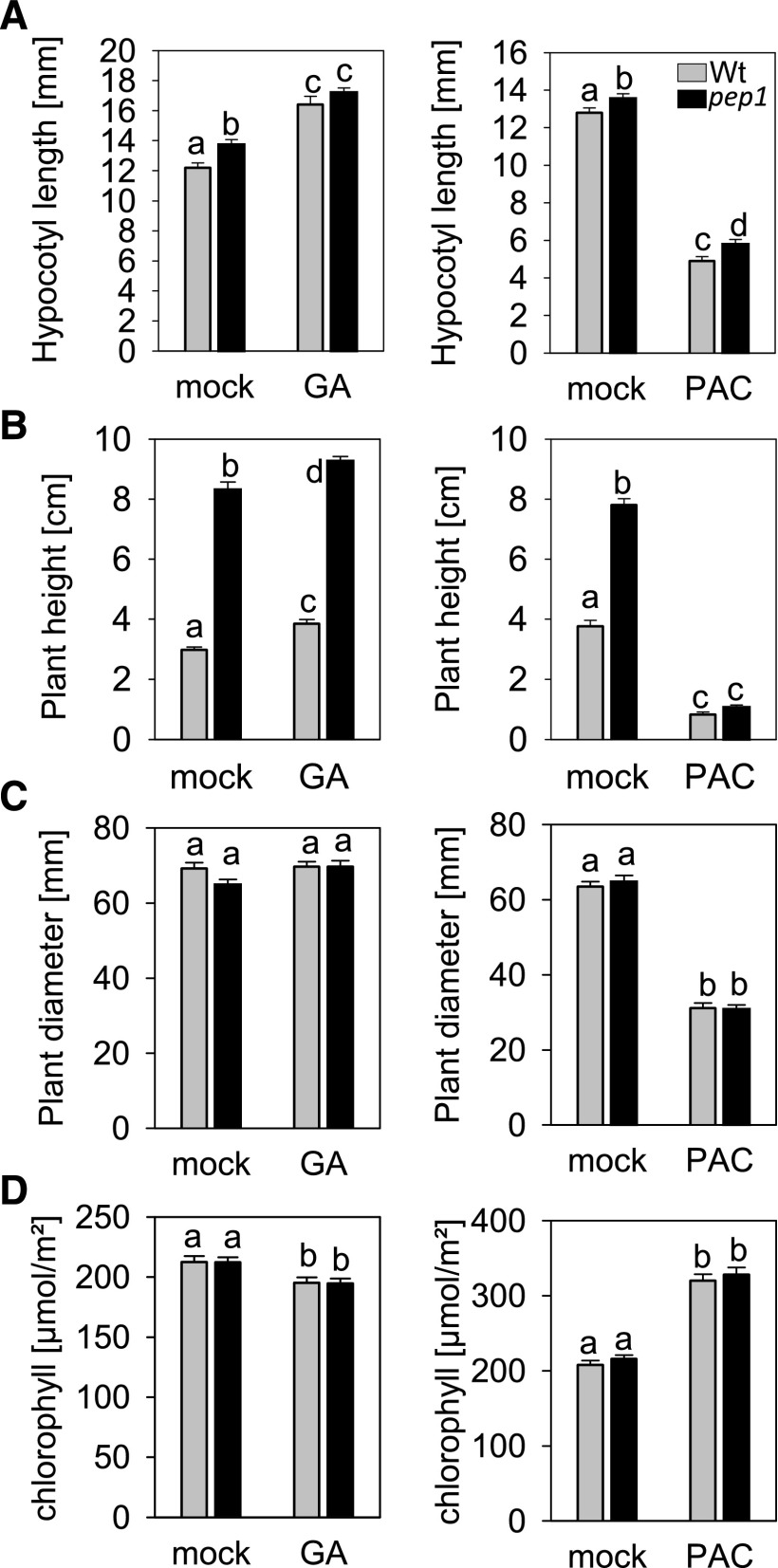
To test whether GA metabolism or signaling might be affected by the pep1 mutation, the mutant and the wild type were compared to identify differences in phenotypes associated with GA responses. The pep1 mutant had a longer hypocotyl and increased plant height relative to the wild type (Fig. 2, A and B). Inhibiting GA biosynthesis by application of PAC caused a strong reduction in the height of treated wild-type and pep1 mutant plants, and after PAC treatment the difference in height between the genotypes was no longer detected, indicating that GA biosynthesis is required for the taller phenotype of the pep1 mutant (Fig. 2B). No differences in plant diameter or chlorophyll content were detected between the genotypes, although these phenotypes are also regulated by GA, as they were affected by GA or PAC treatments (Fig. 2, C and D). Taken together, these results suggest that PEP1 negatively regulates GA metabolism or signaling, but this effect is specific to certain developmental programs.
Figure 2.
GA treatment of wild-type (Wt) plants mimics the effects of the pep1-1 mutation. Phenotypes of wild-type versus pep1-1 mutant plants upon GA/PAC treatment are shown. A, Hypocotyl length of plants grown for 11 d in LD (three independent biological replicates; n ≥ 38). B, Height of plants grown for 5 weeks in LD. GA/mock, Three independent biological replicates, n ≥ 33; PAC/mock, two independent biological replicates; n ≥ 32. C, Plant diameter. Plants were grown for 3 weeks in LD in two independent biological replicates (n ≥ 21). D, Chlorophyll content. Plants were grown for 6 weeks in LD, and measurements were performed on the seventh true leaf (three independent biological replicates; n ≥ 38). All error bars represent se. For all phenotypes, n describes the total number of replicates after combining all biological replicates. Letters indicate significantly different groups determined by two-way ANOVA and multiple comparisons using the Bonferroni t test method that were performed within genotypes and within treatments. Groups were defined as significantly different at P ≤ 0.05.
PEP1 Reduces GA Sensitivity
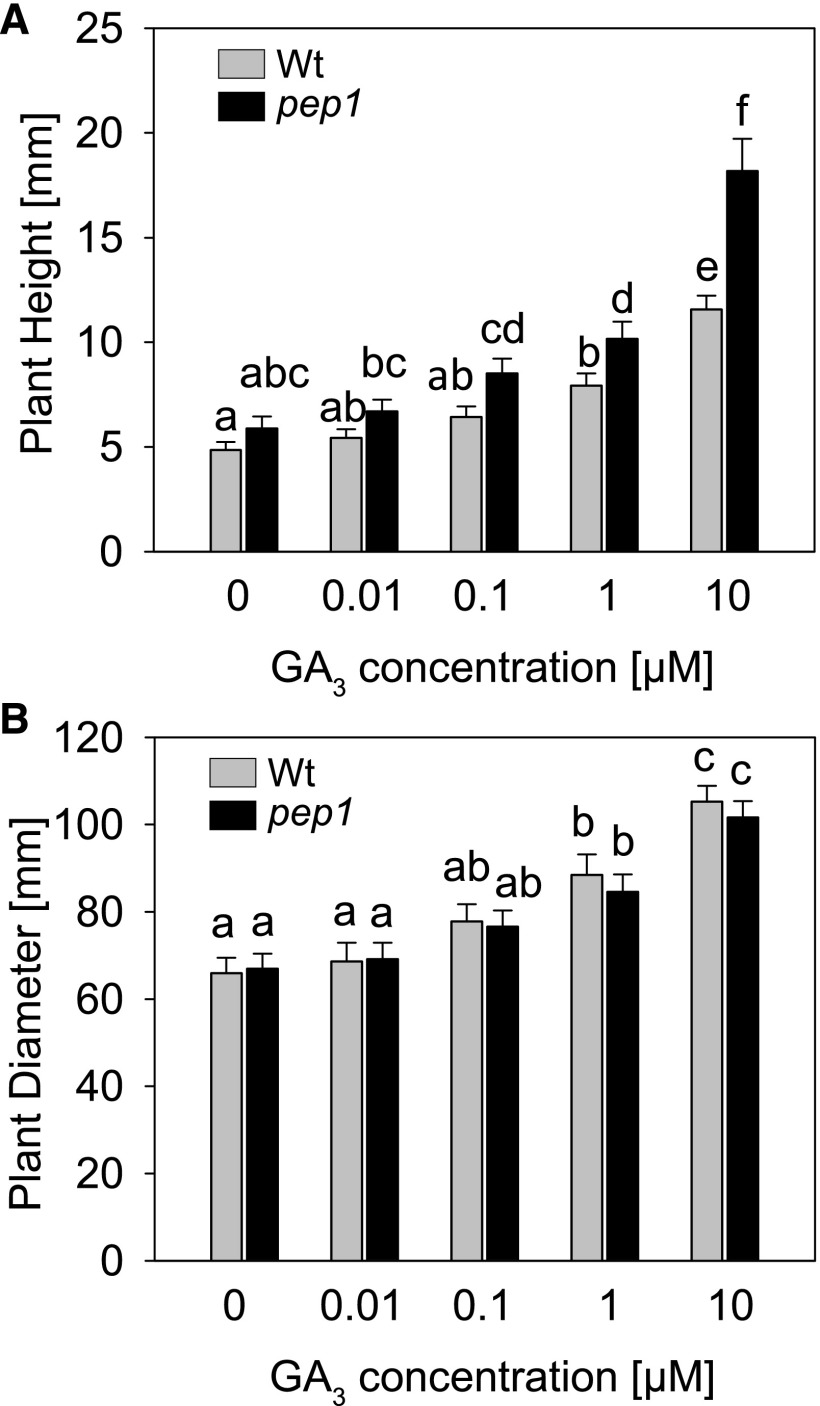
PEP1 was found to bind and regulate genes encoding GA signaling components (Fig. 1), suggesting that GA signaling might be more active in pep1 mutants. To compare GA signaling in the wild type and pep1 mutants, plants were treated with PAC to inhibit the synthesis of endogenous GA and the effects of application of defined concentrations of GA3 on GA-related phenotypes were scored. Plant height increased more rapidly with an increasing concentration of GA in the pep1 mutant compared with the wild type, and these differences between genotypes were significant for the higher concentrations (Fig. 3A). This suggests that the response to GA in the pep1 mutant is stronger and that GA signaling is more effective in the mutant. The increase of plant diameter with increasing GA concentration, however, was similar in both genotypes (Fig. 3B), indicating that the stronger GA response in the pep1 mutant is restricted to certain developmental programs, as observed previously for PAC treatments (Fig. 2).
Figure 3.
Effects of PEP1 on GA signaling. A, Plant height after 6.5 weeks in LD. B, Plant diameter after 6 weeks in LD. pep1-1 and wild-type (Wt) plants were treated simultaneously with PAC to inhibit the synthesis of endogenous GA, and different concentrations of GA3 were applied once per week to investigate the effect of the genotype on the response to GA. Five independent biological replicates were used (total number of replicates after combining all biological replicates; n ≥ 60). All error bars represent se. Letters indicate significantly different groups determined by two-way ANOVA and multiple comparisons using the Bonferroni t test method that were performed within genotypes and within treatments. Groups were defined as significantly different at P ≤ 0.05.
GA Promotes Floral Induction during Vernalization
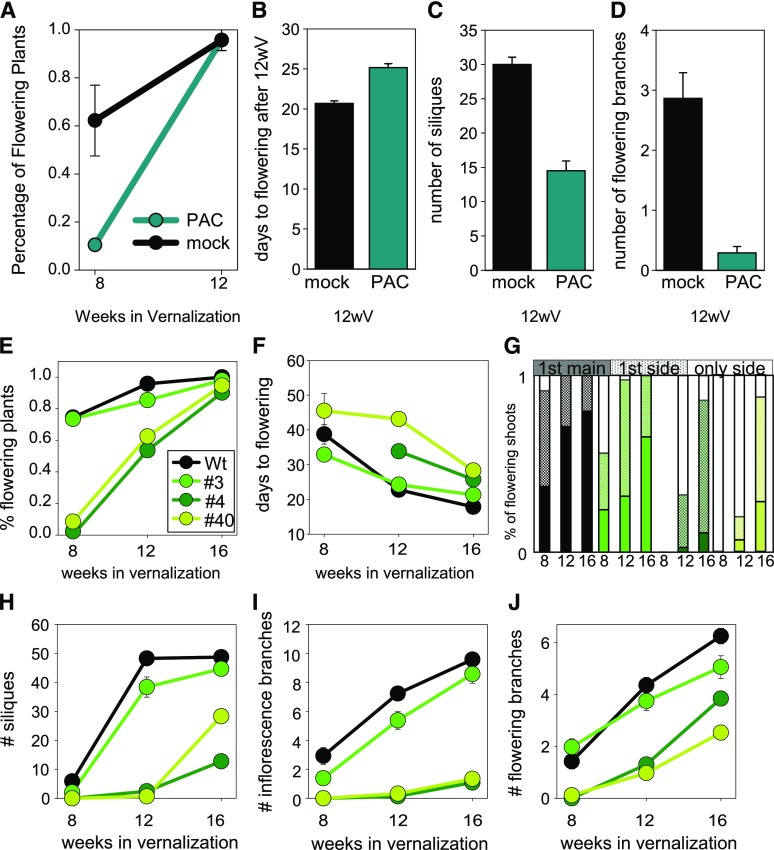
The main conserved function of PEP1 and other FLC orthologs is repression of flowering prior to vernalization, which is overcome by a progressive reduction in transcript levels during vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999; Wang et al., 2009). Considering that PEP1 represses GA metabolism and signaling, whether GA has a role during vernalization to promote flowering was tested. To achieve this, wild-type plants were repeatedly treated with PAC or the mock treatment (see “Materials and Methods”) during an 8-week vernalization period and flowering was scored. PAC treatments strongly reduced the proportion of plants that flowered after vernalization (Fig. 4A). Exposure of plants to 12 weeks of vernalization further enhances flowering and inflorescence development of wild-type plants (Wang et al., 2009; Lazaro et al., 2018). Treatment with PAC during a 12-week vernalization had no effect on the proportion of plants that flowered (Fig. 4A). However, after the 12-week vernalization, PAC treatment resulted in significantly delayed flowering, a reduced number of siliques on the main inflorescence, and a lower number of flowering branches (Fig. 4, B–D). To determine whether the role of GA is restricted to the vernalization period, PAC was applied only after vernalization. A reduction of GA levels after vernalization did not affect flowering time or the number of siliques and flowering branches (Supplemental Fig. S2), indicating that the role of GA in floral induction is restricted to the vernalization period. Complementary results were obtained by treatment of wild-type plants with exogenous GA. GA treatment was not sufficient to overcome the requirement for vernalization to induce flowering but increased the proportion of plants that flowered in response to relatively short vernalization periods (Supplemental Fig. S3). GA treatments had no effect on the proportion of plants that flowered after exposure to long durations of vernalization, as observed with PAC treatments. Taken together, these results indicate that GA promotes floral initiation and inflorescence development of A. alpina during vernalization.
Figure 4.
The role of GA in the induction of flowering during vernalization. A to D, Effects of PAC treatment on flowering. Wild-type plants were grown for 5 weeks in LD and then vernalized for 8 or 12 weeks. During vernalization, plants were treated once per week with PAC or mock (see “Materials and Methods”), and flowering-related traits were scored. A, Percentage of flowering plants after 8 or 12 weeks of vernalization (n ≥ 45 grown in two independent biological replicates). B, Time after a 12-week vernalization treatment until the first flower opened (n ≥ 79 grown in four independent biological replicates). C, Number of individual siliques formed on the main inflorescence after a 12-week vernalization (does not include siliques on inflorescence branches; n ≥ 55 grown in three independent biological replicates). D, Number of flowering side branches after a 12-week vernalization (n ≥ 38 grown in two independent biological replicates). All error bars represent se. E to J, Effects of the KNAT1::GA2OX7 transgene on flowering. Data are shown for three independent transformants compared with wild-type (Wt) Pajares. Plants were grown for 8 weeks in LD and then vernalized for 8, 12, or 16 weeks. Different flowering-related traits were measured. E, Percentage of flowering plants after different vernalization times. F, Time after vernalization until the first flower opened. G, Shoots that contained open flowers. Data are shown for each genotype after 8, 12, or 16 weeks of vernalization. Genotypes are shown using the same color code as for E. Solid portion of each column, Plants that flowered first on the main shoot; stippled portion of each column, plants that flowered first on a side branch and later on the main shoot; unfilled portion of each column, plants that only flowered on side branches and not on the main shoot. H, Number of individual siliques on the main inflorescence (does not include siliques on inflorescence branches). I, Number of flowering branches on the main inflorescence. J, Number of flowering side branches. Experiments were performed in at least two biological replicates (total n = 33). All error bars represent se.
The role of GA during vernalization was further explored in a genetic experiment. To reduce the level of GA, GA2OX7, which encodes a GA-inactivation enzyme, was expressed in A. alpina from the KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1) promoter, which is specifically expressed in meristems in Arabidopsis, and this pattern is likely to be conserved in A. alpina but has not been directly tested (Lincoln et al., 1994; Porri et al., 2012). Three independent transgenic lines (#3, #4, and #40) expressing the transgene were selected (Fig. 4E; Supplemental Fig. S4). Analysis of line #3 was performed with plants in a segregating population that were homozygous or heterozygous for the transgene, because plants homozygous for the transgene were not self-fertile. For line #4, homozygous plants were fertile but produced relatively few seeds, so that experiments were performed using heterozygous and homozygous plants. For line #40, all experiments were performed with homozygous plants. For lines #4 and #40, the proportion of plants flowering after 8 weeks of vernalization was strongly reduced, and even after 12 weeks of vernalization only around 60% of the plants flowered (Fig. 4E; Supplemental Fig. S4C). After an extended vernalization period of 16 weeks, however, almost all plants flowered (Fig. 4E; Supplemental Fig. S4C). Line #3 responded in a similar way, but the effect was weaker (Fig. 4E; Supplemental Fig. S4C). Although the phenotype of line #3 was the weakest, it contained the highest amount of GA2OX7 mRNA (Supplemental Fig. S4B). The explanation for the discrepancy between the severity of phenotype and GA2OX7 mRNA level is not clear, but it might be due to spatial differences in expression among lines, which could also explain why some lines were infertile as homozygotes while others were not. The transgenic lines also showed delayed opening of the first flower, a reduced number of inflorescence branches, fewer siliques on the main inflorescence, and a reduced number of flowering branches (Fig. 4, F–J). As in the wild type treated with PAC, these traits became less severe with longer vernalization times, but even after 16 weeks of vernalization, inflorescence development was strongly compromised in the transgenic plants (Fig. 4G). These results confirm those obtained with PAC treatments that reduction in GA levels delays the floral transition during vernalization.
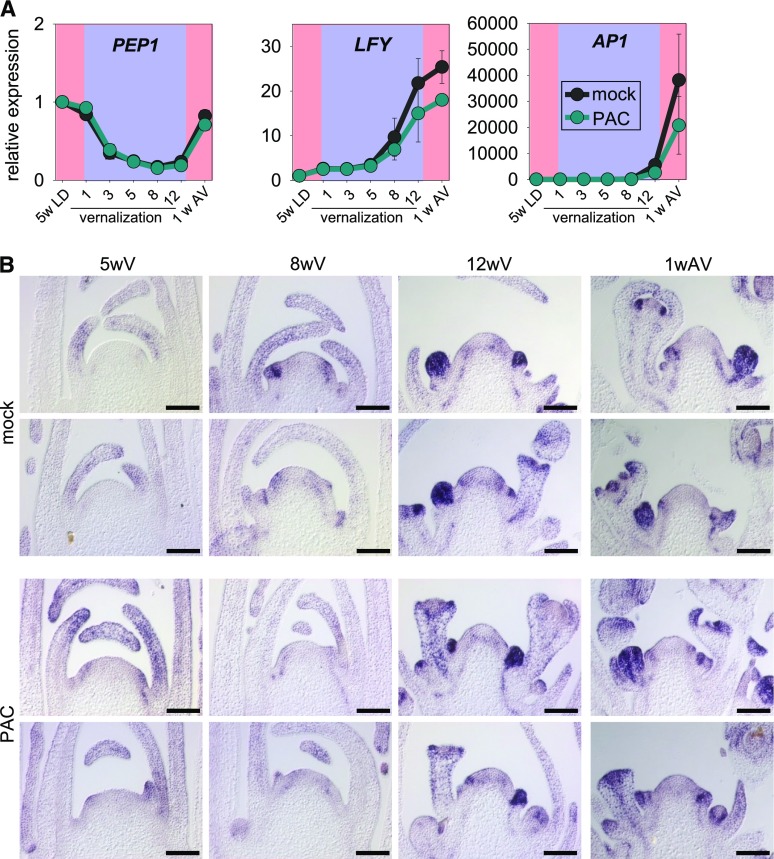
To understand how GA affects floral induction during vernalization, the expression of floral marker genes was measured by reverse transcription quantitative PCR (RT-qPCR). Whereas the PAC treatment during vernalization did not affect PEP1 mRNA levels, induction of the floral meristem identity genes LEAFY (LFY) and APETALA1 (AP1) was reduced (Fig. 5A). Similar to the mock-treated samples, in PAC-treated samples LFY and AP1 mRNAs increased after 8 or 12 weeks of vernalization, respectively, but the increment was slower in the treated samples, and even after vernalization these mRNAs remained at lower levels in PAC-treated plants (Fig. 5A). These results indicate that PAC treatment reduces the expression levels of LFY and AP1 either by delaying its induction and lowering the number of primordia expressing the genes or reducing their levels in the same number of developing primordia.
Figure 5.
Effects of GA on the expression of floral marker genes during vernalization. A, RT-qPCR analysis of floral marker gene expression during vernalization in wild-type plants. Plants were grown as described for Figure 4A. Data are shown as means ± se (n = 2 biological replicates). Expression was normalized to PP2A and to expression in the wild type at the start of the experiment. On the x axis, 5LD indicates plants grown for 5 weeks in LD before vernalization, numbers represent weeks in vernalization, and 1AV represents plants grown for 1 week after vernalization. B, In situ hybridization of LFY mRNA in wild-type plants treated with PAC or mock during vernalization. Wild-type plants were grown for 5 weeks in LD and then transferred to vernalization conditions. Apices were harvested for in situ hybridization at 5, 8, or 12 weeks during vernalization. After 12 weeks of vernalization, plants were transferred out of vernalization, and apices were again harvested 1 week after the end of vernalization (1wAV). During vernalization, plants were treated weekly with PAC or mock and flowering-related traits were measured. Bars = 100 µm.
To understand the effects of PAC treatments on the spatial distribution of LFY mRNA, in situ hybridizations were performed in apices that were treated with PAC or mock during a vernalization time course (Fig. 5B). After 5 weeks of vernalization, meristems of nontreated plants were domed, indicating that floral transition was occurring. By contrast, meristems of PAC-treated plants were still flat after 5 weeks of vernalization, suggesting that floral transition was delayed. This delay was also visible after 8 weeks of vernalization, where flower buds in meristems of mock-treated plants were more advanced and LFY expression in the buds was more extensive compared with PAC-treated plants. After 12 weeks of vernalization, apices of treated and nontreated plants contained similarly advanced flowers.
In summary, these findings suggest that GA acts during vernalization to promote floral transition and flower development.
PEP1 Affects the Induction of GA-Related Genes during Vernalization
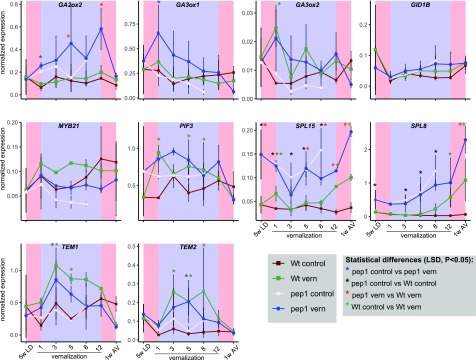
To further explore the interaction between PEP1 and GA in flowering, the expression patterns of PEP1 targets that are involved in the GA pathway were analyzed during vernalization. Transcript levels were tested comparing the wild type and pep1 mutant during vernalization and without exposure to vernalization. In both genotypes, transcript levels of PEP1 targets encoding the GA biosynthesis enzymes GA3OX1 and GA3OX2 were increased in abundance during the first weeks of vernalization and then fell again toward the end of vernalization (Fig. 6). PIF3, whose protein product is regulated by GA (de Lucas et al., 2008; Feng et al., 2008), showed a similar pattern. GA2OX2 mRNA, which encodes a GA inactivation enzyme, also increased during vernalization treatment in the pep1 mutant but not in the wild type, suggesting that in the wild type PEP1 reduces the transcriptional induction of GA2OX2. SPL8 and SPL15 showed expression patterns complementary to PEP1: their expression levels increased during vernalization in wild-type and pep1 plants as well as in nonvernalized pep1 plants but not in nonvernalized wild-type plants (Fig. 6). This pattern suggests that they are increased in expression during floral induction or inflorescence development. TEMPRANILLO1 (TEM1) and TEM2 showed a different pattern. These genes were induced during vernalization, but their expression levels decreased again after vernalization. Unlike the majority of PEP1 targets, these genes reached higher levels in the wild type (Fig. 6), indicating that PEP1 activity increases their expression. In the case of TEM1 and TEM2, this does not appear to be a direct effect, because they were not detected as bound by PEP1.
Figure 6.
Effects of PEP1 and vernalization on the expression of GA-related genes. RT-qPCR expression analysis is shown for genes involved in the GA pathway during vernalization in pep1-1 versus wild-type (Wt) plants. Plants were grown for 5 weeks in LD, then transferred to 4°C for 12 weeks or kept under control conditions (SD; 21°C), and then shifted back to LD. Apical samples were taken at Zeitgeber time 8 (ZT8). Data are shown as means ± se (n = 2 biological replicates). Expression was normalized to PP2A. Significance was tested using ANOVA. On the x axis, 5LD indicates plants grown for 5 weeks in LD before vernalization, numbers represent weeks in vernalization, and 1AV represents plants grown for 1 week after vernalization.
In summary, most GA-related genes tested increased in expression rapidly in response to cold temperatures before down-regulation of PEP1, and their expression increased further in the pep1 mutant, suggesting that PEP1 modulates their response to cold.
PEP1 Affects GA Levels before But Not during Vernalization
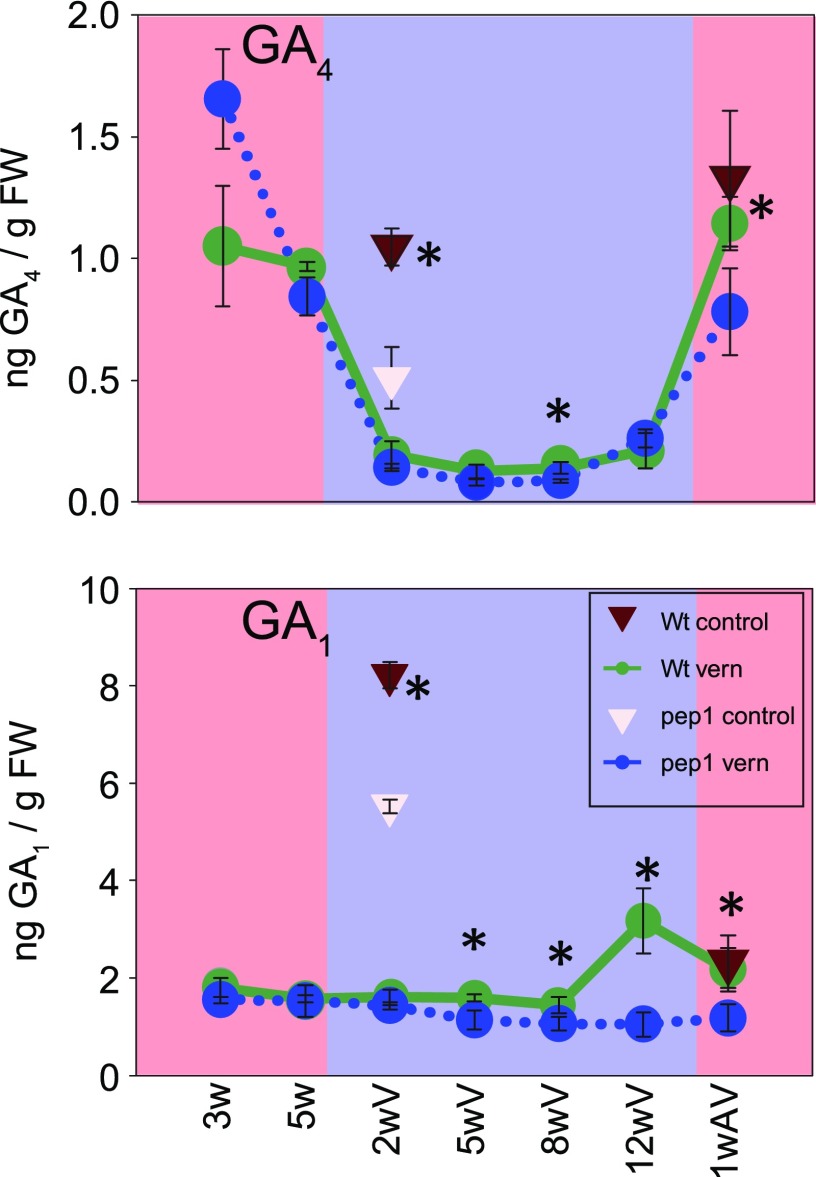
The findings that GA promotes floral induction during vernalization (Figs. 2 and 3; Supplemental Fig. S3) and that genes encoding GA biosynthetic enzymes were induced during the cold treatment (Fig. 6) suggested that GA levels might increase during vernalization. To test this hypothesis, levels of active GA were measured in apical samples of wild-type and pep1 mutant plants during vernalization and in control samples of nonvernalized plants at selected time points. The pep1 mutant contained significantly higher levels of GA4 when grown for 3 weeks prior to vernalization (Fig. 7). This difference likely contributes to the increased height of pep1 mutants compared with wild-type plants detected 5 weeks after germination (Fig. 2B). After 5 weeks, however, no difference in GA4 levels was detected between wild-type and pep1 plants (Fig. 7). Surprisingly, during vernalization, GA4 levels strongly decreased and reached similar trough levels in both genotypes (Fig. 7). An increase of GA4 levels occurred after plants were shifted back to warm temperatures, so that they reached levels similar to those present prior to cold exposure. The increase in GA4 did not coincide with the timing of floral induction in A. alpina, which occurs during vernalization, but rather with the outgrowth of the inflorescence. GA1, which is synthesized by a parallel pathway to GA4 through early 13-hydroxylation and shows lower biological activity (Cowling et al., 1998; Magome et al., 2013), did not strongly change between conditions or genotypes and accumulated to higher levels than GA4 (Fig. 7). Also, precursors or degradation products of active GAs did not show patterns that might suggest higher GA levels during vernalization or in the pep1 mutant at later stages, although in PEP1 plants GA12 and GA34 seemed to fall progressively in abundance during vernalization, similar to the pattern of PEP1 mRNA repression (Supplemental Fig. S5).
Figure 7.
Effects of PEP1 and vernalization on levels of active GA. Levels of active GAs in apices of pep1-1 and wild-type (Wt) plants are shown. Plants were grown for 5 weeks in LD, then transferred to 4°C for 12 weeks or kept under control conditions (SD; 21°C), and then shifted back to LD. Apical samples were taken at ZT8. Data are shown as means ± sd (n = 3 biological replicates, except the wild type and pep1-1 at 5 weeks of LD, where n = 2). Asterisks indicate significant differences between genotypes at the same time points (*, P ≤ 0.05, Student’s t test). FW, Fresh weight.
Lower GA levels during vernalization correspond to increased expression of GA2OX2 in cold temperatures (Fig. 6); however, two genes encoding GA biosynthetic enzymes, GA3OX1 and GA3OX2, were also induced during vernalization. To test whether this induction might be due to feedback regulation of the decreased GA levels, expression levels were tested in response to GA and PAC treatment. Indeed, expression levels of GA3OX1 and GA3OX2 were reduced by GA treatment and increased by PAC treatment, respectively, suggesting that they are susceptible to negative feedback regulation (Supplemental Fig. S6). In contrast, GA2OX2 expression responded positively to GA levels (Supplemental Fig. S6). Several genes involved in GA signaling showed a similar pattern to the genes involved in GA biosynthesis (Supplemental Fig. S6), suggesting that low GA levels also positively affect their expression.
In conclusion, PEP1 appears to negatively affect GA levels before vernalization to regulate plant height, but during vernalization GA levels were strongly decreased and not affected by PEP1, suggesting that PEP1 regulation of GA levels does not contribute to the control of floral induction during vernalization.
DISCUSSION
We investigated the involvement of GAs in the flowering response of A. alpina to vernalization. PEP1, a transcription factor that confers the vernalization requirement, bound to and regulated genes encoding enzymes for GA metabolism or components of the signaling pathway prior to and during vernalization. The pep1 mutant contained more GA4 than wild-type plants before vernalization and was more sensitive to treatments with bioactive GA. During vernalization, GA4 levels strongly decreased upon exposure to cold, reaching similar levels in wild-type and pep1 mutant plants, but nevertheless PAC treatments and analysis of transgenic plants overexpressing GA 2-oxidase demonstrated that GA was required to promote floral induction during short vernalization periods. As the GA-related target genes for PEP1 are different from those of FLC, and its BSs in these genes are not conserved across the Brassicaceae, novel linkages between the vernalization response and the GA pathway appear to have evolved recently in the Arabis lineage.
Integration of the GA Pathway and the Obligate Vernalization Response of A. alpina
Floral induction of A. alpina Pajares only occurs in the cold during vernalization (Wang et al., 2009, 2011). This response occurs because PEP1 expression prior to vernalization fully blocks flowering, while during vernalization PEP1 transcription is reduced, allowing floral induction to proceed, and then after vernalization its transcription is rapidly reactivated, preventing flowering of any meristems that did not undergo floral induction during vernalization. Thus, flowering of this perennial species relies upon pathways that act in the cold. Recently, we showed that under these environmental conditions, a pathway including the transcription factor SPL15 is essential for a successful flowering response and that mutations in SPL15 prevent the flowering of A. alpina in response to vernalization (Hyun et al., 2019). In Arabidopsis, SPL15 activates the transcription of MIR172b, which negatively regulates the expression of APETALA2-like transcription factors (Hyun et al., 2016). A screen for mutants of A. alpina Pajares that flower independently of vernalization identified mutations in the AP2 ortholog, PEP2, suggesting that this floral repressor plays a critical role in blocking flowering of A. alpina prior to vernalization (Bergonzi et al., 2013; Lazaro et al., 2019). As PEP1 directly binds to SPL15 to repress its transcription and AaSPL15 transcription rises in vernalization (Mateos et al., 2017; Hyun et al., 2019), these data suggest that upon vernalization of A. alpina increased expression of SPL15 activates MIR172b transcription that in turn allows the repression of PEP2 and flowering. The effectiveness of this pathway may be dependent on GA, as DELLA proteins interact with SPL15 to reduce its transcriptional activity at the posttranslational level (Hyun et al., 2016). Thus, the presence of GA in the shoot meristem may be a requirement for SPL15 to activate its targets and for an efficient flowering response to short vernalization periods. Our observation that many plants with reduced GA levels in the apex do not flower in response to short vernalization periods, but that all flower in response to longer vernalization periods (Fig. 3A), suggests that the repression of SPL15 activity by DELLA can eventually be overcome, perhaps because SPL15 levels increase during vernalization or because DELLA interaction does not completely abolish SPL15 activity.
The relationship between GA and the vernalization response was tested genetically in Arabidopsis (Chandler et al., 2000). The vernalization response of fca-1 mutants was unaffected in fca-1 ga-1 double mutants, and this may be explained by floral induction of these plants occurring after vernalization rather than during the cold exposure, as occurs in A. alpina. Stable repression of FLC transcription by vernalization in Arabidopsis allows flowering to occur after the return to warm LDs through the photoperiodic pathway (Hyun et al., 2019). Under these LD conditions, the effects of spl15 and GA biosynthetic mutations on flowering are much reduced (Wilson et al., 1992; Hyun et al., 2016). Previous work indicated that in Arabidopsis, GA is most strongly required for flowering under noninductive SDs (Wilson et al., 1992; Chandler et al., 2000), which supports the importance of the interaction between DELLA and SPL15, as spl15 mutants in the Columbia accession also exhibit their strongest delay on flowering under SDs (Hyun et al., 2016). It remains to be tested whether GA plays a role in the vernalization response of Arabidopsis under conditions in which flowering occurs in the cold through the SPL15 pathway.
In addition to delaying the earliest stages of floral induction, such as meristem doming, reduction of GA levels in vernalization by PAC treatment or introduction of the KNAT1::GA2ox7 transgene also impaired inflorescence development, so that plants produced fewer flowers (Figs. 4 and 5). This reduction was probably due to a slower transition to flowering during vernalization in the presence of lower GA levels, so that the inflorescence meristem was not completely transitioned when plants were returned to warm temperatures and PEP1 transcription was reactivated. The reduction in GA would therefore effectively extend the time plants need to be exposed to cold temperatures for a full vernalization response. In support of this conclusion, the reduction of GA levels with PAC applications after vernalization did not affect inflorescence development (Supplemental Fig. S2). Thus, depletion of GA levels during vernalization indirectly affected outgrowth and development of the inflorescence after vernalization.
Regulation of GA Metabolism and Signaling by PEP1 and Related MADS Box Transcription Factors
Unexpectedly, the levels of GA4 fell in apices of A. alpina during vernalization when flowering occurred but increased again after vernalization. By contrast, the levels of GA1 were unchanged during or after vernalization and appeared higher overall than those of GA4. These patterns appeared to differ from those described in apices of Arabidopsis during floral induction under SDs, when GA4 levels rose sharply prior to flowering and were higher than those of GA1 (Eriksson et al., 2006). Thus, in Arabidopsis, GA4 is believed to be the active GA in the promotion of flowering, and GA1 is less bioactive than GA4 in Arabidopsis and rice (Oryza sativa; Cowling et al., 1998; Magome et al., 2013). During vernalization of A. alpina, local changes in GA4 levels, for example only in some meristematic cells, might promote floral induction and not be detected by the sampling methods used here. A clear relationship between GA levels and flowering in vernalization has been described in other species. For example, in T. arvense, vernalization induced the production of precursors of active GA causing bolting (Hazebroek et al., 1993), while in winter canola, GA levels increased toward the end of vernalization (Zanewich and Rood, 1995). In R. sativus and T. arvense, application of exogenous GA overcame the requirement for vernalization (Suge and Rappaport, 1968; Metzger, 1985), whereas in A. alpina, it did not overcome the vernalization requirement but reduced the duration of vernalization required for a full flowering response. Furthermore, GA4 levels increased again after vernalization in A. alpina but GA levels in vernalized apices did not exceed those in nonvernalized apices, and this moderate increase was substantially lower than was detected in apices of Arabidopsis during the floral transition in SDs (Eriksson et al., 2006). Similar to A. alpina, a moderate increase of active GAs at the end of vernalization was associated with bolting in winter canola (Zanewich and Rood, 1995). In other species, levels of precursors of active GAs increased during vernalization (Hazebroek et al., 1993; Hisamatsu et al., 2004), although levels of several GA precursors were also not elevated in A. alpina in response to vernalization (Supplemental Fig. S5).
PEP1 binds to and regulates genes that encode proteins in the GA pathway from biosynthesis to downstream signaling (Fig. 1C). The transcript levels of some of those target genes are up-regulated in the pep1 mutant prior to vernalization, such as GA3OX1, SPL15, and SPL8 (Fig. 6). Furthermore, the pep1 mutant exhibits elevated GA levels and increased sensitivity to GA prior to vernalization that contribute to the elongated internodes of the mutant. The Arabidopsis ortholog of PEP1, FLC, is closely related to the MADS box transcription factor SHORT VEGETATIVE PHASE (SVP), which was previously shown to repress transcription of the GA20OX2 gene, which encodes an enzyme involved in GA biosynthesis (Andrés et al., 2014). The svp mutant also contains elevated levels of GA in the shoot apex. Thus, the repression of GA levels appears to be a shared function of this class of MADS box transcription factors. In addition, pep1 showed higher levels of several GA signaling genes that might cause the elongated internodes and promote flowering. Two SPL genes that are known to affect flowering in Arabidopsis showed higher expression levels in the pep1 mutant. SPL8 is involved in flower development (Zhang et al., 2007) and SPL15 is a floral integrator gene at the shoot meristem (Hyun et al., 2016). The GID1B gene encodes a GA receptor that most likely causes a general increase in GA signaling that could cause different phenotypes (Griffiths et al., 2006; Nakajima et al., 2006). PIF3 was shown to be involved in hypocotyl elongation (Feng et al., 2008) and to promote flowering in the ambient temperature pathway (Kumar et al., 2012; Galvão et al., 2015). Deregulation of GA-related processes in the pep1 mutant is associated with elongated internodes and increased height of the plant, so in wild-type A. alpina PEP1 confers a compact phenotype that might be advantageous in the cold alpine environments in which it grows (Bliss, 1962).
During vernalization, additional GA-related genes are expressed at higher levels in the pep1 mutant than in the wild type, such as GA2OX2, while those expressed at higher levels prior to vernalization remain differentially expressed. Thus, several GA-related genes are negatively regulated by PEP1 during vernalization (Fig. 6), as shown previously for many cold stress-related genes (Mateos et al., 2017). Also, although PEP1 acts almost exclusively as a transcriptional repressor (Mateos et al., 2017), the GA-related gene MYB21 is bound by PEP1 and down-regulated in the pep1 mutant during vernalization (Fig. 6). This effect might be an indirect consequence of PEP1 also regulating genes upstream of MYB21. In Arabidopsis, MYB21 is induced by GAs during stamen development and a triple myb mutant exhibited short stamens and was sterile (Cheng et al., 2009). TEM1 and TEM2 are also down-regulated in the pep1 mutant during vernalization. TEM genes encode negative regulators of floral induction that directly repress GA3OX1 and GA3OX2 (Castillejo and Pelaz, 2008; Osnato et al., 2012). Thus, they might be responsible for the up-regulation of GA3OX1, which is not a direct PEP1 target, in the pep1 mutant.
The observation that GA4 levels are increased in the pep1 mutant compared with wild-type plants prior to vernalization initially led us to propose that the repression of PEP1 transcription during vernalization might cause GA levels to increase and thereby promote flowering. However, in cold temperatures, GA levels fell to similarly low levels in the wild type and pep1 mutants. Similarly, at the time PEP1 mRNA levels were repressed in cold temperatures, there was no corresponding increase in GA levels. The reduction in GA levels in cold temperatures might be due to reduced activity of biosynthetic enzymes at low temperature. Strikingly, genes encoding for GA biosynthetic enzymes and not GA degradation enzymes are up-regulated during vernalization in A. alpina. Expression levels of these genes are complementary to GA levels and not PEP1 expression levels, and PAC treatment caused similar changes in gene expression. Thus, these gene expression changes are most likely caused by feedback regulation of low GA levels and not by vernalization through PEP1. Low GA levels were also described to positively feedback on expression levels of genes encoding GA biosynthetic enzymes in Arabidopsis (Chiang et al., 1995; Phillips et al., 1995; Mitchum et al., 2006; Achard et al., 2008; Rieu et al., 2008), and increased expression of those genes was observed during vernalization in E. grandiflorum and T. arvense (Hazebroek and Metzger, 1990; Hazebroek et al., 1993; Mino et al., 2003).
A reduction of GA content in response to prolonged cold treatment was previously detected in R. sativus (Nakayama et al., 1995). In Arabidopsis, a short cold treatment was found to induce the expression of GA2OX genes and thereby decrease GA levels, which caused repression of growth (Achard et al., 2008). Similarly, the decrease in PEP1 expression during vernalization does not seem to cause an increase in GA levels. GA biosynthesis might be required for flowering to proceed in vernalization but may not increase during vernalization, so that GA has a permissive rather than a regulated role in the process. Another possibility is that the increased GA sensitivity detected prior to vernalization in pep1 mutants might also occur during vernalization to trigger flowering after PEP1 repression. Several GA signaling genes that were associated with the promotion of flowering in Arabidopsis (Griffiths et al., 2006; Nakajima et al., 2006; Zhang et al., 2007; Suzuki et al., 2009; Galvão et al., 2015; Hyun et al., 2016) were induced during vernalization in A. alpina. Thus, the effect of GA on floral induction during vernalization might also be caused by changes in the expression of GA signaling genes, and their roles could be tested by reverse genetic approaches based, for example, on CRISPR-Cas9. Alternatively, small changes in GA levels in some meristematic cells during floral induction might not be detectable by the method we used in this study, but they might cause the phenotypic changes in flowering time and flower development. Cellular markers for GA levels that can be detected in vivo may more sensitively detect changes in GA levels during vernalization (Rizza et al., 2017). Application of such approaches will extend our understanding of how GA contributes to the flowering pathway that promotes floral induction in the cold during vernalization.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabis alpina Pajares accession that was used as the wild type and the pep1-1 mutant were described previously in Wang et al. (2009). The KNAT1::GA2OX7 transgene was described to promote meristem-specific expression of Arabidopsis (Arabidopsis thaliana) GA2OX7, which encodes a GA inactivation enzyme. In Arabidopsis, the transgene caused strongly decreased GA levels and associated phenotypes (Lincoln et al., 1994; Schomburg et al., 2003; Porri et al., 2012). Here, this construct was transformed into the pep1-1 mutant by floral dip (Clough and Bent, 1998). T1 plants were crossed to Pajares wild-type plants, and F3 plants homozygous for wild-type PEP1 were identified by genotyping using a cleaved-amplified polymorphic sequence marker. PCR using the primers HY88 and HY89 was followed by a restriction digest with the enzyme AseI (New England Biolabs) for gel electrophoresis analysis. The presence of the KNAT1::GA2OX7 transgene was identified based on the strong GA-deficient phenotype (dwarf and dark green).
Seeds were stratified on moist soil at 4°C in darkness for 3 d. Plants were grown at a light intensity of about 200 μmol m−2 s−1 under LD conditions (16 h of light/8 h of dark) at 21°C and 60% to 70% humidity. For vernalization, adult plants (5–8 weeks old) were shifted to 4°C under SD conditions (8 h of light at 17 μmol m−2 s−1/16 h of dark). For all experiments, trays where shifted weekly to avoid positional artifacts.
ChIP
For ChIP experiments, wild-type and pep1-1 mutant plants were grown for 2 weeks in LD and 2 g of aboveground tissue was harvested at ZT8. One microliter of PEP1 antiserum (Albani et al., 2012) was used for immunoprecipitation. ChIP was performed as previously described in Gendrel et al. (2002). For ChIP-qPCR, three independent biological replicates were performed, and samples were purified using the PCR Clean-Up Gel purification kit (Macherey-Nagel) and eluted in 20 μL of water. Samples were diluted 1:10, 3 μL was used for qPCR with the SYBR Green master mix (Bio-Rad), and primers are listed in Supplemental Table S1. qPCR was performed in three technical replicates for each biological replicate. Data are represented as fold change of immunoprecipitate (IP) divided by input [2(−IP/input)]. Significance of the differences between genotypes was defined as P ≤ 0.05 after Student’s t test. ChIP-seq data were obtained from previous publications (Deng et al., 2011; Mateos et al., 2017).
Analysis of Gene Expression
For gene expression analysis, samples were collected at ZT8. For apical samples, apex-enriched tissue of 16 plants was combined. For leaf samples of juvenile plants, leaves of five plants were pooled. RNA was extracted using the RNAeasy Plant Mini Kit (Qiagen), and samples were treated with RNase-free DNase (Ambion) according to the manufacturer’s instructions. Five microliters of RNA was used for complementary DNA synthesis using the SuperScriptII DNA polymerase (Invitrogen). Three microliters of complementary DNA (diluted 1:10) was used for qPCR with my-budget Taq-DNA-Polymerase and EvaGreen Dye (Biotium) for detection in a 10-μL reaction volume. Primers are listed in Supplemental Table S1. qPCR was performed in three technical replicates for each biological replicate in a CFX384 Touch Real-Time PCR System (Bio-Rad) using PP2A as the housekeeping gene. At least two biological replicates were performed, and mean and se were calculated for normalized data (for details on individual experiments, see figure legends). Genome-wide expression data were obtained from Mateos et al. (2017).
Analysis of CArG-box Conservation in Related Species
To identify orthologous sequences in other species with varying evolutionary distances (Koch et al., 2001; Clauss and Koch, 2006; Couvreur et al., 2010; Hu et al., 2011; Karl et al., 2012; Willing et al., 2015), BSs were aligned to orthologous regions. Therefore, orthologs of genes associated with the BSs were identified by reciprocal BLAST, and these genes, including 5 kb upstream of the transcriptional start site and 3 kb downstream of the transcriptional end site, were extracted as orthologous loci. Finally, the best alignment of the BS in the orthologous locus was obtained by local Smith and Waterman alignments (Smith and Waterman, 1981), and the percentage identity between the BS and the orthologous sequence was calculated. Genome assemblies and annotation files of Arabidopsis lyrata (A. lyrata v1.0; Hu et al., 2011) were downloaded from Phytozome v11.0. Assemblies and annotations of the genomes of Aethionema arabicum (Haudry et al., 2013) and Tarenaya hassleriana (Cheng et al., 2013) were downloaded from https://genomevolution.org/coge/. The Arabis montbretiana genome assembly and annotation were kindly provided by Wen-Biao Jiao and Korbinian Schneeberger prior to publication.
CArG-box motifs in the BSs of PEP1 target genes were identified with the MEME software (Bailey and Elkan, 1994; Bailey, 2011) using the zoops (motif occurrence zero or one per sequence) model. Parameters were set to identify the 10 most significantly enriched motifs of a length between five and 20 nucleotides. In addition, a manual search for the sequence MYHWAWWWRGWWW was performed. Alignments of PEP1 BSs with orthologous sequences were performed with the LAGAN program in mvista (Frazer et al., 2004). These alignments were then checked for conservation of the CArG-box motifs identified in the A. alpina sequence.
Phenotypic Analysis
Plant height and diameter were measured using a ruler. Chlorophyll content was measured using the SPAD-502 leaf chlorophyll meter (Markwell et al., 1995). For each data point (one plant of one biological replicate), the average of three technical repetitions of measurements on the same leaf was created. Measurements were performed on the seventh true leaf. Flowering time was determined by the number of days until the first flower opened. Furthermore, flowering was scored by counting the number of individual siliques at the main shoot without including siliques at side branches and by counting the number of flowering inflorescence branches and flowering side branches. All experiments were performed in independent biological replicates. The numbers of technical and biological replicates for each experiment are indicated in the figure legends. All data points are represented as means ± se. ANOVA was performed using the SigmaStat 3.5 software.
Application of Exogenous GA and PAC
Plants were sprayed weekly with 20 μm GA4 (Sigma-Aldrich; stock solution: 100 mm in ethanol and 0.1% [v/v] Silwet L-77 [Loveland Industries]) or mock (0.1% [v/v] ethanol and 0.1% [v/v] Silwet). The GA biosynthesis inhibitor PAC (Sigma-Aldrich) was dissolved in dimethyl sulfoxide (stock concentration, 100 mg mL−1), and plants were sprayed weekly with 1 mg mL−1 PAC, 0.1% (v/v) Silwet, or 1% (v/v) dimethyl sulfoxide, with 0.1% (v/v) Silwet as the mock treatment. Unless indicated otherwise, treatment was started after germination and continued throughout the experiment. For the GA sensitivity test, plants were sprayed weekly with PAC as described above to inhibit the biosynthesis of endogenous GA. In addition, different concentrations of GA3 (Sigma-Aldrich) were directly added to the soil with water once per week. Therefore, 200 μL of GA3 solution (different concentrations dissolved in ethanol) was added to 1 L of water to yield final concentrations of 0, 0.01, 0.1, 1, and 10 μm GA3. The experiment was performed in two biological replicates. Data of five biological replicates were combined to yield a total n of at least 60.
In Situ Hybridization
LFY mRNA in situ hybridization was performed on apical samples as described previously in Wang et al. (2009).
Quantification of GA
Plants were grown in LDs for 5 weeks, then vernalized for 12 weeks, and finally transferred back to LDs. Between 100 and 200 mg fresh weight was harvested in liquid nitrogen per sample. Samples were harvested at ZT8 in three biological replicates, and GAs were quantified at IBMCP as described previously in Seo et al. (2011). The thoroughly ground tissue was suspended in 80% (v/v) methanol and 1% (v/v) acetic acid containing internal standards and mixed by shaking for 1 h at 4°C. The extract was kept at −20°C overnight and then centrifuged, and the supernatant was dried in a vacuum evaporator. The dry residue was dissolved in 1% (v/v) acetic acid and passed consecutively through a reverse-phase column Oasis HLB (30 mg; Waters) and a cationic exchange Oasis MCX eluted with methanol, as described in Seo et al. (2011). The final residues were dried and dissolved in 5% (v/v) acetonitrile and 1% (v/v) acetic acid, and the hormones were separated by UHPLC with a reverse Accucore C18 column (2.6 µm, 100 mm length; Thermo Fisher Scientific) with a 2% to 55% (v/v) acetonitrile gradient containing 0.05% (v/v) acetic acid at 400 µL min−1 over 21 min. The hormones were analyzed with a Q-Exactive mass spectrometer (Orbitrap detector; Thermo Fisher Scientific) by targeted selected ion monitoring (capillary temperature, 300°C; S-lens RF level, 70; resolution, 70,000) and electrospray ionization (spray voltage, 3 kV; heater temperature, 150°C; sheath gas flow rate, 40 µL min−1; auxiliary gas flow rate, 10 µL min−1) in negative mode. The concentrations of GAs in the extracts were determined using embedded calibration curves and the Xcalibur 2.2 SP1 build 48 and TraceFinder. The internal standards for quantification of each of the different GAs were the deuterium-labeled [17,17-2H]GAs (purchased from OlChemim). Data are represented as means of three biological replicates ± sd, and Student’s t test was employed to test for significant differences between genotypes for all time points.
Accession Numbers
The sequences of the A. alpina genes used in this study are available at www.arabis-alpina.org in version 4 of the A. alpina genome sequence using the gene identifiers shown in Figure 1A and the coordinates shown in Figure 1B.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. ChIP-qPCR to detect PEP1 binding to GA2OX2.
Supplemental Figure S2. The role of GA in flower development after vernalization.
Supplemental Figure S3. Exogenous GA application during vernalization accelerates flowering.
Supplemental Figure S4. Transgenic plants expressing KNAT::GA2OX7 in the Pajares background.
Supplemental Figure S5. Effect of PEP1 and vernalization on levels of precursors and degradation products of active GA.
Supplemental Figure S6. Feedback of GA levels on the expression of GA-related genes.
Supplemental Table S1. Primers used in this study.
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft via the Cluster of Excellence on Plant Science (CEPLAS, EXC 1028 to V.T.), by the Alexander von Humboldt-Stiftung (postdoctoral fellowship) and Argentinean National Council of Sciences (CONICET to J.L.M.), by the European Research Council (N′339113 HyLife to G.C.), and by a Core Grant of the Max Planck Society to G.C.
Articles can be viewed without a subscription.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani MC, Castaings L, Wötzel S, Mateos JL, Wunder J, Wang R, Reymond M, Coupland G (2012) PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet 8: e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Porri A, Torti S, Mateos J, Romera-Branchat M, García-Martínez JL, Fornara F, Gregis V, Kater MM, Coupland G (2014) SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA 111: E2760–E2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. (2011) DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S, Albani MC, Ver Loren van Themaat E, Nordström KJV, Wang R, Schneeberger K, Moerland PD, Coupland G (2013) Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340: 1094–1097 [DOI] [PubMed] [Google Scholar]
- Bliss LC. (1962) Adaptations of Arctic and Alpine plants to environmental conditions. Arctic 15: 117–144 [Google Scholar]
- Castillejo C, Pelaz S (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Chandler J, Martinez-Zapater JM, Dean C (2000) Mutations causing defects in the biosynthesis and response to gibberellins, abscisic acid and phytochrome B do not inhibit vernalization in Arabidopsis fca-1. Planta 210: 677–682 [DOI] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, van den Bergh E, Zeng P, Zhong X, Xu J, Liu X, Hofberger J, de Bruijn S, Bhide AS, Kuelahoglu C, et al. (2013) The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25: 2813–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss MJ, Koch MA (2006) Poorly known relatives of Arabidopsis thaliana. Trends Plant Sci 11: 449–459 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Prinsen E, Jacqmard A, Lejeune P, Van Onckelen H, Périlleux C, Bernier G (2003) Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J Exp Bot 54: 2511–2517 [DOI] [PubMed] [Google Scholar]
- Couvreur TLP, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K (2010) Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol 27: 55–71 [DOI] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol 117: 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aloia M, Bonhomme D, Bouché F, Tamseddak K, Ormenese S, Torti S, Coupland G, Périlleux C (2011) Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J 65: 972–979 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108: 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Jung HS, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ (2007) Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Böhlenius H, Moritz T, Nilsson O (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V, Takahashi Y, Le Gourrierec J, Coupland G (2016) Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J 86: 426–440 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: Computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Horrer D, Küttner F, Schmid M (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Galvão VC, Collani S, Horrer D, Schmid M (2015) Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J 84: 949–962 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA (2002) Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297: 1871–1873 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- Hazebroek JP, Metzger JD (1990) Thermoinductive regulation of gibberellin metabolism in Thlaspi arvense L. I. Metabolism of [2H]ent-kaurenoic acid and [14C]gibberellin A12-aldehyde. Plant Physiol 94: 157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazebroek JP, Metzger JD, Mansager ER (1993) Thermoinductive regulation of gibberellin metabolism in Thlaspi arvense L. II. Cold induction of enzymes in gibberellin biosynthesis. Plant Physiol 102: 547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T, Koshioka M, Mander LN (2004) Regulation of gibberellin biosynthesis and stem elongation by low temperature in Eustoma grandiflorum. J Hortic Sci Biotechnol 79: 354–359 [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, et al. (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G (2016) Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell 37: 254–266 [DOI] [PubMed] [Google Scholar]
- Hyun Y, Vincent C, Tilmes V, Bergonzi S, Kiefer C, Richter R, Martinez-Gallegos R, Severing E, Coupland G (2019) A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363: 409–412 [DOI] [PubMed] [Google Scholar]
- Karl R, Kiefer C, Ansell SW, Koch MA (2012) Systematics and evolution of Arctic-Alpine Arabis alpina (Brassicaceae) and its closest relatives in the eastern Mediterranean. Am J Bot 99: 778–794 [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T (2001) Molecular systematics of the Brassicaceae: Evidence from coding plastidic matK and nuclear Chs sequences. Am J Bot 88: 534–544 [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge J. (1957) Effect of day-length and gibberellic acid on the flowering of Arabidopsis. Nature 180: 36–37 [Google Scholar]
- Lazaro A, Obeng-Hinneh E, Albani MC (2018) Extended vernalization regulates inflorescence fate in Arabis alpina by stably silencing PERPETUAL FLOWERING1. Plant Physiol 176: 2819–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Zhou Y, Giesguth M, Nawaz K, Bergonzi S, Pecinka A, Coupland G, Albani MC (2019) PERPETUAL FLOWERING2 coordinates the vernalization response and perennial flowering in Arabis alpina. J Exp Bot 70: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Y, Chen S, An L (2010) Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J Exp Bot 61: 4221–4230 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S (2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA 110: 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñate-Sánchez L, Holdsworth MJ, Bhalerao R, Alabadí D, et al. (2014) Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol 166: 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwell J, Osterman JC, Mitchell JL (1995) Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynth Res 46: 467–472 [DOI] [PubMed] [Google Scholar]
- Mateos JL, Madrigal P, Tsuda K, Rawat V, Richter R, Romera-Branchat M, Fornara F, Schneeberger K, Krajewski P, Coupland G (2015) Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol 16: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, Krajewski P, Coupland G (2017) Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc Natl Acad Sci USA 114: E11037–E11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger JD. (1985) Role of gibberellins in the environmental control of stem growth in Thlaspi arvense L. Plant Physiol 78: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino M, Oka M, Tasaka Y, Iwabuchi M (2003) Thermoinduction of genes encoding the enzymes of gibberellin biosynthesis and a putative negative regulator of gibberellin signal transduction in Eustoma grandiflorum. Plant Cell Rep 22: 159–165 [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E, Hedden P (2009) Gibberellin as a factor in floral regulatory networks. J Exp Bot 60: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Nakayama M, Yamane H, Nojiri H, Yokota T, Yamaguchi I, Murofushi N, Takahashi N, Nishijima T, Koshioka M, Katsura N, et al. (1995) Qualitative and quantitative analysis of endogenous gibberellins in Raphanus sativus L. during cold treatment and the subsequent growth. Biosci Biotechnol Biochem 59: 1121–1125 [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S (2012) TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun 3: 808. [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NE, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Regnault T, Davière JM, Wild M, Sakvarelidze-Achard L, Heintz D, Carrera Bergua E, Lopez Diaz I, Gong F, Hedden P, Achard P (2015) The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat Plants 1: 15073. [DOI] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG, et al. (2008) The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J 53: 488–504 [DOI] [PubMed] [Google Scholar]
- Rizza A, Walia A, Lanquar V, Frommer WB, Jones AM (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants 3: 803–813 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM (2003) Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Jikumaru Y, Kamiya Y (2011) Profiling of hormones and related metabolites in seed dormancy and germination studies. Methods Mol Biol 773: 99–111 [DOI] [PubMed] [Google Scholar]
- Serrano-Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S, Sablowski R (2017) DELLA genes restrict inflorescence meristem function independently of plant height. Nat Plants 3: 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147: 195–197 [DOI] [PubMed] [Google Scholar]
- Suge H, Rappaport L (1968) Role of gibberellins in stem elongation and flowering in radish. Plant Physiol 43: 1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Park SH, Okubo K, Kitamura J, Ueguchi-Tanaka M, Iuchi S, Katoh E, Kobayashi M, Yamaguchi I, Matsuoka M, et al. (2009) Differential expression and affinities of Arabidopsis gibberellin receptors can explain variation in phenotypes of multiple knock-out mutants. Plant J 60: 48–55 [DOI] [PubMed] [Google Scholar]
- Vaughan JG. (1955) The morphology and growth of the vegetative and reproductive apices of Arabidopsis thaliana (L.) Heynh., Capsella bursa-pastoris (L.) Medic. and Anagallis arvensis L. Bot J Linn Soc 55: 279–301 [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC (2009) PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427 [DOI] [PubMed] [Google Scholar]
- Wang R, Albani MC, Vincent C, Bergonzi S, Luan M, Bai Y, Kiefer C, Castillo R, Coupland G (2011) Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23: 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing EM, Rawat V, Mandáková T, Maumus F, James GV, Nordström KJV, Becker C, Warthmann N, Chica C, Szarzynska B, et al. (2015) Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants 1: 14023. [DOI] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D (2013) A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell 24: 271–282 [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D (2014) Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641 [DOI] [PubMed] [Google Scholar]
- Yu S, Galvão VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW (2012) Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 24: 3320–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanewich KP, Rood SB (1995) Vernalization and gibberellin physiology of winter canola: Endogenous gibberellin (GA) content and metabolism of [3H]GA1 and [3H]GA20. Plant Physiol 108: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schwarz S, Saedler H, Huijser P (2007) SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol 63: 429–439 [DOI] [PubMed] [Google Scholar]