The NUCLEAR FACTOR Y, SUBUNIT B2 (NF-YB2) and NF-YB3 transcription factors have high sequence similarity but differentially regulate target genes in a drought and heat stress-specific manner.
Abstract
Functional diversification of transcription factors allows the precise regulation of transcriptomic changes under different environmental conditions. The NUCLEAR FACTOR Y (NF-Y) transcription factor comprises three subunits, NF-YA, NF-YB, and NF-YC, and is broadly diversified in plant species, whereas Humans (Homo sapiens) have one protein for each subunit. However, there remains much to be learned about the diversified functions of each subunit in plants. Here, we found that NF-YB2 and NF-YB3, which have the greatest sequence similarity to each other among NF-YB family proteins in Arabidopsis (Arabidopsis thaliana), are functionally diversified and specifically activate dehydration-inducible and heat-inducible genes, according to environmental conditions. Overexpression of NF-YB2 and NF-YB3 specifically enhanced drought and heat stress tolerance, respectively, and each single knockout mutant showed adverse stress-sensitive phenotypes. Transcriptomic analyses confirmed that overexpression of NF-YB2 and NF-YB3 largely affected the transcriptomic changes under dehydration and heat stress conditions, respectively. The DNA-binding profiles of each protein in planta also suggested that dehydration and heat stress increased the DNA-binding activity of NF-YB2 and NF-YB3 to dehydration-inducible and heat stress-inducible target genes, respectively. Moreover, phylogenetic analysis suggested that the NF-YB proteins of angiosperm plants belong to divergent NF-YB2 and NF-YB3 subgroups. These results demonstrate the functional diversification of NF-Y through evolutionary processes and how plants adapt to various abiotic stresses under fluctuating environments.
Abiotic stresses such as drought, high and low temperature, and high salinity are important factors that affect plant growth and reproduction (Mickelbart et al., 2015). Recent extreme weather events have damaged global food production and security (Lesk et al., 2016; Schauberger et al., 2017). Plants have developed various types of molecular strategies through evolution that are specifically induced according to environmental conditions. Transcriptomic analyses of plants treated with different abiotic stresses have revealed various stress-specific and common gene regulatory mechanisms (Rasmussen et al., 2013; Maruyama et al., 2017). For example, LATE EMBRYOGENESIS ABUNDANT (LEA) proteins and several sugar-biosynthetic enzymes are typical dehydration stress-inducible proteins that function as osmoprotectants and inducers of osmolytes, respectively (Hincha and Thalhammer, 2012; Keunen et al., 2013). Heat stress activates the expression of genes encoding the molecular chaperones HEAT SHOCK PROTEINs (HSPs; Jacob et al., 2017). These stress-specific transcriptomic changes are regulated by various transcription factors that are activated in response to specific abiotic stress (Song et al., 2016; Ohama et al., 2017).
Nuclear factor Y (NF-Y) is a transcription factor that is widely conserved among eukaryotes (Kumar et al., 2012; Li et al., 2016). A NF-Y trimer is composed of the NF-YA, NF-YB, and NF-YC subunits, and the trimer is known to bind to a CCAAT element on its target gene promoters to regulate their transcription (Myers and Holt, 2018). Humans (Homo sapiens) only have one family protein for each NF-Y subunit. However, subunits of NF-Y are broadly diversified in plant species; Arabidopsis (Arabidopsis thaliana) has 10 NF-YAs, 10 NF-YBs, and 10 NF-YCs (Petroni et al., 2012). The NF-YB and NF-YC proteins contain conserved H2B and H2A domains (which are related to histone H2B and H2A proteins), respectively, and these domains are necessary to form a dimer of NF-YB and NF-YC before trimerization. DNA POLYMERASE II SUBUNIT B3 (DPB3) and NEGATIVE COFACTOR 2α family proteins also have the H2A domain, and DPB4 and NEGATIVE COFACTOR 2β family proteins have the H2B domain (Petroni et al., 2012). These proteins are also suggested to be involved in transcriptional regulation (Kim et al., 2011; Yoshida et al., 2011), and previous studies suggested that these H2A-containing and H2B-containing proteins can interact with each other across protein families (Kim et al., 2011). Moreover, recent studies have suggested that dimers of NF-YB and NF-YC can interact with not only NF-YAs but also with other proteins containing a CO, CO-LIKE, and TOC1 domain and that DNA binding preference of the trimer is changed by the CO, CO-LIKE, and TOC1-containing subunit (Yoshida et al., 2011). Diversification of plant NF-Ys and their interacting proteins raises the possibility that different combinations of trimers have different molecular functions, and many studies have actually revealed the wide variations in NF-Y functions in plants, including flowering induction (Fujita et al., 2009; Yoshida et al., 2015), seed maturation (Nambara et al., 1998; Iuchi et al., 2001), root nodule formation (Zanetti et al., 2010), and responses to abiotic stresses such as drought, heat, and cold (Yoshida et al., 2011). However, detailed insight into the functional diversification of each NF-Y subunit is still limited.
We previously identified that Arabidopsis DPB3-1, also annotated as NF-YC10, interacted with the transcription factor DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A), and we suggested that DPB3-1 forms a trimer with NF-YA2 and NF-YB3 to regulate heat-stress–responsive genes together with DREB2A (Sato et al., 2014). DREB2A is a transcription factor that regulates both dehydration-inducible and heat-stress–inducible genes according to environmental conditions (Qin et al., 2008; Morimoto et al., 2017). The DREB2A protein is stabilized under these abiotic stress conditions (Morimoto et al., 2013; Mizoi et al., 2019), whereas it is easily degraded under nonstress conditions through a 26S proteasome pathway (Sakuma et al., 2006a). A DREB2A protein lacking its negative regulatory domain was constitutively stable even under nonstress conditions (DREB2A CA; DREB2A constitutively active form; Sakuma et al., 2006a, 2006b). DREB2A CA-overexpressing plants show enhanced expression of both dehydration-inducible and heat-inducible DREB2A target genes under nonstress conditions and drought-tolerant and heat stress-tolerant phenotypes (Sakuma et al., 2006a, 2006b). Our previous findings provided insights into the molecular functions of NF-YA2, NF-YB3, and DPB3-1 to control target selectivity by forming specific transcription complexes under specific environmental conditions (Sato et al., 2014). Meanwhile, we found that a trimer composed of NF-YA2, NF-YB2, and DPB3-1 also activated target promoters together with DREB2A in mesophyll protoplasts, but interestingly, expression patterns of NF-YB2 and NF-YB3 in plants the opposite of each other; NF-YB3 was induced by heat stress and suppressed by dehydration stress, and NF-YB2 was induced by dehydration stress and suppressed by heat stress (Sato et al., 2014). We therefore concluded that NF-YB3 is a candidate to form a transcriptional complex with NF-YA2, DPB3-1, and DREB2A during heat stress, although direct interaction between NF-YB3 and DREB2A was not detected in yeast cells (Sato et al., 2014). However, these results also suggested the functional diversification of NF-YB2 and NF-YB3, which have the greatest sequence similarity to each other among NF-YB family proteins (70% sequence identity). NF-YB2 and NF-YB3 are reported to have a redundant function: to induce flowering through activation of FLOWERING LOCUS T together with CONSTANS (CO; Siefers et al., 2009; Calvenzani et al., 2012; Sato et al., 2014). However, the functional diversification of NF-YB2 and NF-YB3 during abiotic stress is not well understood.
In this study, we revealed the functional diversification of NF-YB2 and NF-YB3 during dehydration and heat stress. Overexpressing plants and knockout mutants of NF-YB2 and NF-YB3 showed dehydration-specific and heat stress-specific phenotypes and gene expression patterns, respectively. Moreover, phylogenetic analysis revealed that proteins in divergent NF-YB2 and NF-YB3 subgroups are conserved among angiosperm plants but not bryophytes and lycophytes. These results provide important insight into the functional diversification of NF-Y proteins through evolution and provide a mechanistic understanding of the target selectivity of DREB2A under dehydration and heat stress conditions.
RESULTS
NF-YB2 and NF-YB3 Gene Expression Patterns Correlate with DREB2A under Dehydration and Heat Stress Conditions
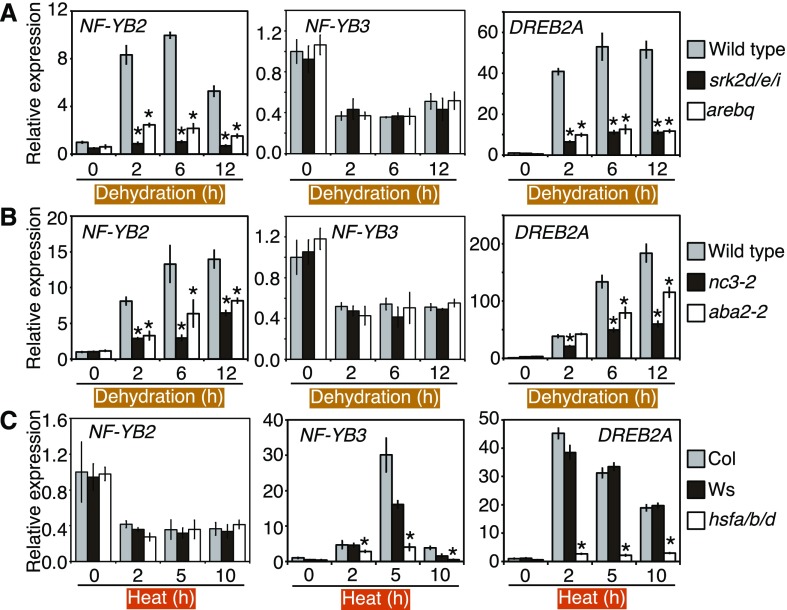
We previously reported that NF-YA2, NF-YB3, and DPB3-1 (NF-YC10) form a trimer and cooperatively function with DREB2A to activate their target genes specifically under heat stress conditions, and we suggested that NF-YB2, which has a highly similar sequence to NF-YB3 (70% sequence identity), has different functions than NF-YB3 under dehydration and heat stress conditions (Sato et al., 2014). First, gene expression patterns of NF-YB2 and NF-YB3 were examined. Induction of the NF-YB2 gene during dehydration stress was suppressed in the abscisic acid (ABA)-signaling mutants; triple mutants of the SNF1-RELATED PROTEIN KINASE2D (SRK2D), SRKD2E, and SRK2I (srk2d/e/i; Fujita et al., 2009); and quadruple mutants of the ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING PROTEIN1 (AREB1), AREB2, ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTOR1 (ABF1), and ABF3 (arebq; Yoshida et al., 2010; Fig. 1A). The ABA-deficient mutants; knockout mutants of NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (nc3-2) and ABA DEFICIENT2 (ABA2; aba2-2; Nambara et al., 1998; Iuchi et al., 2001) also showed decreased expression of NF-YB2 during dehydration stress (Fig. 1B). NF-YB2 induction was not completely abolished in these mutants (Fig. 1, A and B), suggesting that the NF-YB2 gene was induced through both ABA-dependent and ABA-independent mechanisms during dehydration stress as well as in a DREB2A-dependent manner (Kim et al., 2011). We also analyzed the expression levels of NF-YB2 in these mutants treated with ABA. The expression level of NF-YB2 was up-regulated by ABA treatment and was suppressed in the srk2d/e/i and arebq mutants (Supplemental Fig. S1A), confirming that the ABA-dependent pathway contributes to a part of NF-YB2 gene induction. In the ABA-deficient mutants, the suppression of NF-YB2 gene induction during ABA treatment was not observed (Supplemental Fig. S1B). These mutations had almost no effect on NF-YB3 expression during dehydration and ABA treatment (Fig. 1, A and B; Supplemental Fig. S1, A and B). DREB2A is also induced by HEAT SHOCK FACTOR A1s (HSFA1s) in response to heat stress (Yoshida et al., 2011). The hsfa1a/b/d mutants (hsfa1a hsfa1b hsfa1d triple mutant) showed decreased expression levels of NF-YB3 under heat stress conditions (Fig. 1C). This result showed that the expression of NF-YB3 in response to heat stress was HSFA1 dependent. NF-YB2 expression was not affected by the mutation of HSFA1s (Fig. 1C). AREB/ABFs and HSFA1s are known to bind to the ABA responsive element (ABRE) and heat shock element (HSE) on their target promoters, respectively (Kim et al., 2011; Yoshida et al., 2011). NF-YB2 and NF-YB3 also have ABRE and HSE sequences on their promoters, respectively (Supplemental Table S1), and expression of the NF-YB2 and NF-YB3 genes seems to be directly activated by AREB/ABFs and HSFA1s through these cis-elements, respectively. We also analyzed the conservation of these elements in the promoters of Arabidopsis NF-YB2 and NF-YB3 putatively orthologous genes across species and found that a DRE (GCCGAC) and an ABRE (ACGTGG) in the Arabidopsis NF-YB2 promoter and an HSE-like (GAAnTTC) and an ABRE (CCACGT) in the Arabidopsis NF-YB3 promoter are conserved among putatively orthologous promoters of Brassica sp. (Supplemental Fig. S2, A and B).
Figure 1.
Expression patterns of NF-YB2 and NF-YB3 in ABA signaling, ABA-deficient, and HSFA1 knockout mutants (hsfa1a/b/d) during dehydration and heat stress. A and B, Expression levels of NF-YB2, NF-YB3, and DREB2A during dehydration stress in the ABA-signaling mutants (A) and ABA-deficit mutants (B). The expression of NF-YB2, NF-YB3, and DREB2A genes was analyzed in the SRK2D triple mutants (srk2d/e/i) and ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING PROTEIN/FACTORs quadruple mutants (arebq; A) and NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (NCED3; nc3-2) and ABA DEFICIENT2 (ABA2; aba2-2) knockout mutants (B) by reverse transcription quantitative PCR (RT-qPCR. The 14-d-old seedlings grown on agar plates were treated with dehydration stress. The expression levels of NF-YB2, NF-YB3, and DREB2A in wild-type plants before stress treatment (0 h) were defined as 1.0. The error bars indicate SD (n = three experiments, involving 7 individual plants for each line). The asterisk indicates the time point at which the expression levels of each gene were significantly decreased compared with those in the wild-type plants (P < 0.05, Bonferroni-corrected Student’s t test). C, The expression levels of NF-YB2, NF-YB3, and DREB2A during heat stress in the hsfa1a/b/d mutants. The 14-d-old seedlings grown on agar plates were treated with heat stress (37°C). The expression levels of NF-YB2, NF-YB3, and DREB2A in the wild-type plants before stress treatment (0 h) were defined as 1.0. The error bars indicate SD (n = three experiments, involving 7 individual plants for each line). The asterisk indicates the time point at which the expression levels of each gene were significantly decreased compared with that of both wild-type plants (Col and Ws; P < 0.05, Bonferroni-corrected Student’s t test).
Overexpression of NF-YB2 and NF-YB3 Enhanced Drought and Heat Stress Tolerance, Respectively
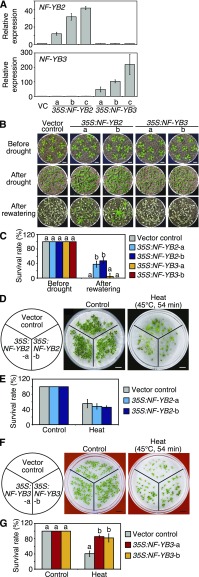
Next, NF-YB2-overexpressing and NF-YB3-overexpressing plants were generated to analyze their functions (35S:NF-YB2-a, b and c, and 35S:NF-YB3-a, b and c; Fig. 2A). These overexpressing plants showed early-flowering phenotypes (Supplemental Fig. S3, A and B) under long-day conditions (16-h photoperiod), as previously reported (Kumimoto et al., 2008). We therefore performed a drought stress tolerance test under short-day conditions (8-h photoperiod) to keep all the plant lines at the same developmental stages. After the drought stress treatment, the NF-YB2-overexpressing plants showed significantly higher survival rates, whereas the NF-YB3-overexpressing plants did not (Fig. 2, B and C). The overexpression of NF-YB2 did not affect the heat stress tolerance (Fig. 2, D and E), whereas overexpression of NF-YB3 significantly enhanced the heat stress tolerance (Fig. 2, F and G). Additionally, we analyzed water loss of detached leaves using overexpressors of NF-YB2, NF-YB3, and DREB2A CA. The results showed that overexpression of NF-YB2 and DREB2A CA suppressed water loss of leaves (Supplemental Fig. S3C). These results suggest that NF-YB2 and NF-YB3 are functionally differentiated and are positive regulators of drought and heat stress responses, respectively, despite the high protein sequence similarity.
Figure 2.
Drought and heat stress tolerance test in the NF-YB2-overexpressing and NF-YB3-overexpressing plants. A, Expression analysis of NF-YB2 and NF-YB3 in the NF-YB2-overexpressing and NF-YB3-overexpressing plants, respectively, by RT-qPCR. The expression levels of NF-YB2 or NF-YB3 in the VC plants were defined as 1.0. The error bars indicate SD (n = three experiments, involving 7 individual plants for each line). B, Drought stress tolerance of the NF-YB2-overexpressing and NF-YB3-overexpressing plants. Plants grown on agar plates for 12 d were transferred to soil and grown for 2 d. Water was withheld for 24 d. Scale bars = 1 cm. Photographs were taken before drought (top row), after drought (middle row), and 7 d after rewatering (bottom row). C, Percentages of surviving plants after rewatering. The error bars indicate the SD from five experiments, each involving 7 individual plants for each line. The letters above the bars indicate significant differences among plant lines before and after drought stress treatment (P < 0.05, according to Tukey’s multiple range test). D and F, Heat stress tolerance of the NF-YB2-overexpressing (D) and NF-YB3-overexpressing (F) plants. The 7-d-old seedlings were treated with or without 45°C for 54 min. Scale bars = 1 cm. The photographs were taken after a 7-d recovery period at 22°C with (right) or without (left) heat stress. E and G, Percentages of surviving plants after heat stress in the NF-YB2-overexpressing (E) and NF-YB3-overexpressing (G) plants. The error bars indicate the SD from three experiments, each involving 28 individual plants for each line. The survival rates of the NF-YB2-overexpressing plants after heat stress were evaluated using one-way ANOVA, but no significant differences were detected (P > 0.05). The letters above the bars in G indicate significant differences among plant lines with or without heat stress treatment (P < 0.05, according to Tukey’s multiple range test).
Expression of DREB2A Target Genes was Enhanced in NF-YB2-Overexpressing and NF-YB3-Overexpressing Plants during Dehydration and Heat Stress, Respectively
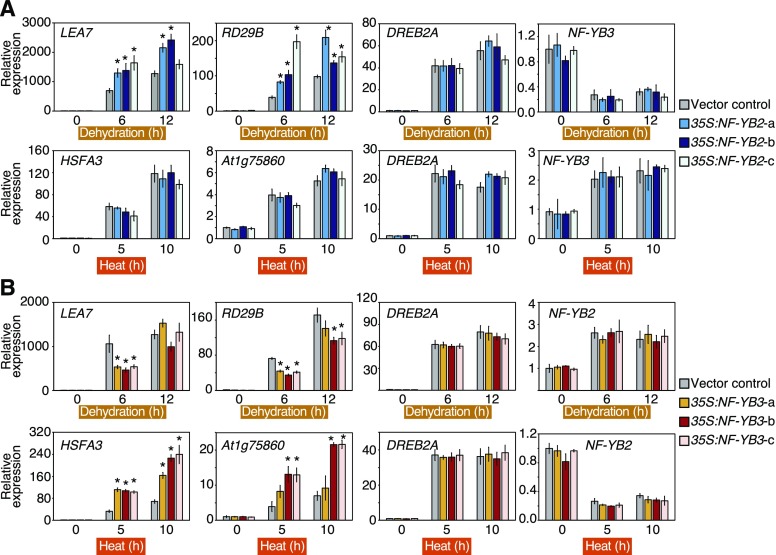
The expression levels of DREB2A target genes during dehydration and heat stress were analyzed in the NF-YB2-overexpressing and NF-YB3-overexpressing plants. Under dehydration stress conditions, dehydration stress-inducible DREB2A target genes LEA7 and RESPONSIVE TO DESICCATION29B (RD29B) were significantly up-regulated in NF-YB2-overexpressing plants, whereas the expression levels of DREB2A and NF-YB3 were not altered in these plants (Fig. 3A). We also found that the expression of these dehydration-inducible DREB2A target genes was suppressed in NF-YB3-overexpressing plants, while DREB2A and NF-YB2 expression levels were not altered (Fig. 3B). Under heat stress conditions, heat stress-inducible DREB2A target genes HSFA3 and At1g75860 were significantly up-regulated in NF-YB3-overexpressing plants, whereas no differences were observed in the expression of DREB2A and NF-YB2 (Fig. 3B). Overexpression of NF-YB2 did not affect the expression of these heat stress-inducible genes under heat stress conditions (Fig. 3A). Accumulation of the DREB2A protein was also not changed by the overexpression of NF-YB2 and NF-YB3 under dehydration and heat stress conditions (Supplemental Fig. S4, A and B). These results suggest that NF-YB2 and NF-YB3 are functionally diversified in response to dehydration and heat stress; that they cooperatively activate their target genes with DREB2A specifically under dehydration and heat stress conditions, respectively; and that NF-YB3 has negative effects on the expression of dehydration stress-inducible genes under dehydration stress conditions. To analyze the genetic interactions between the NF-YBs and DREB2A, we overexpressed NF-YB2 or NF-YB3 in the dreb2a-1 knockout mutants (Supplemental Fig. S4, C–F). As previously reported, gene induction of DREB2A-taget genes was suppressed during dehydration (LEA7 and RD29B) or heat stress (HSFA3 and At1g75860; Supplemental Fig. S4, G and H). The expression levels of the target genes were not affected by overexpression of NF-YB2 and NF-YB3 during dehydration and heat stress, respectively, in the dreb2a-1 mutants (Supplemental Fig. S4, G and H). These results suggested that NF-YB2 and NF-YB3 function to enhance the expression levels of these target genes in a DREB2A-dependent manner.
Figure 3.
Expression patterns of dehydration-inducible and heat-inducible DREB2A target genes during dehydration and heat stress. A and B, The expression levels of dehydration-inducible genes (LEA7, RD29B), heat-inducible genes (HSFA3 and At1g75860), DREB2A, NF-YB2, and NF-YB3 during dehydration and heat stress in the NF-YB2–overexpressing (A) and NF-YB3–overexpressing (B) plants. The expression levels were analyzed by RT-qPCR. The 12-d-old plants grown on agar plates were treated with dehydration or heat stress. The expression levels of each gene in the VC plants under the control condition (0 h) were defined as 1.0. The error bars indicate SD (n = three experiments, involving 7 individual plants for each line). Asterisks indicate significant differences from the VC plants at each time point (P < 0.05, Bonferroni-corrected Student’s t test), and the expression levels of each gene at other time points during dehydration and heat stress were evaluated using one-way ANOVA, but no significant differences were detected (P > 0.05).
Transcriptomic Analysis Confirmed Stress-Specific Functions of NF-YB2 and NF-YB3
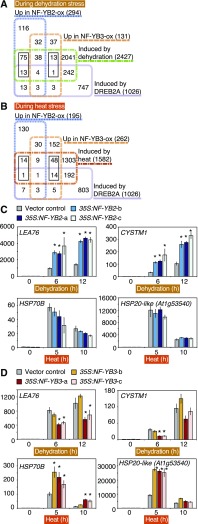
Transcriptomic analysis was performed in the NF-YB2-overexpressing and NF-YB3-overexpressing plants (35S:NF-YB2-b and 35S:NF-YB3-b) relative to the vector control (VC) plants under 6-h dehydration stress, 5-h heat stress, and nonstress conditions to reveal the stress-specific functions of NF-YB2 and NF-YB3 by RNA sequencing (Supplemental Data Sets S1–S6). We detected 2427 and 1582 genes up-regulated by dehydration and heat stress, respectively, reflecting a more than 3-fold increase in response to dehydration and heat stress in the VC plants (VC [control] vs VC [dry], and VC [control] vs VC [heat]; P < 0.05; Fig. 4, A and B). Previous studies revealed up-regulated genes by overexpression of DREB2A CA (Sakuma et al., 2006b; Mizoi et al., 2013). Under dehydration stress, overexpression of NF-YB2 and NF-YB3 resulted in up-regulation of 294 and 131 genes, respectively, more than 1.8-fold of the levels in the VC plants (VC [dry] vs NF-YB2ox [dry], and VC [dry] vs NF-YB3ox [dry]; P < 0.05; Fig. 4A). The proportion of dehydration-stress–inducible genes specifically up-regulated in the NF-YB2–overexpressing plants (88 genes; 30%) among the 294 genes up-regulated by NF-YB2 was significantly higher than that specifically up-regulated in the NF-YB3-overexpressing plants (14 gene; 11%) among the 131 genes up-regulated by NF-YB3 (P < 0.0001, Fisher’s exact test). Meanwhile, under heat stress, 195 and 262 up-regulated genes were detected in the NF-YB2-overexpressing and NF-YB3-overexpressing plants, respectively, reflecting more than 1.8-fold greater levels than those in the VC plants (VC [heat] vs NF-YB2ox [heat], and VC [heat] vs NF-YB3ox [heat]; P < 0.05; Fig. 4B). The proportion of heat-stress–inducible genes specifically up-regulated in the NF-YB3-overexpressing plants (62 genes; 24%) among the 262 genes up-regulated by NF-YB3 was significantly higher than that specifically up-regulated in the NF-YB2-overexpressing plants (15 gene; 8%) among the 195 genes up-regulated by NF-YB2 (P < 0.0001, Fisher’s exact test). These results suggest that NF-YB2 and NF-YB3 have specific functions under dehydration and heat stress, respectively, and that overexpression of these genes has greater effects on the transcriptomic changes under each stress. Moreover, the proportion of DREB2A CA-inducible genes among the 294 genes up-regulated in the NF-YB2-overexpressing plants during dehydration stress (33 genes; 11%) was significantly higher than that among Arabidopsis whole genes (1026 genes; 4%; P < 0.0001; Fisher’s exact test), whereas that among the 131 genes up-regulated in the NF-YB3-overexpressing plants during dehydration stress (11 genes; 8%) was not significantly higher (P > 0.01, Fisher’s exact test). During heat stress, the proportion of DREB2A CA-inducible genes among the 262 genes up-regulated in the NF-YB3-overexpressing plants (23 genes; 9%) was significantly higher than that among Arabidopsis whole genes (1026 genes; 4%; P < 0.0001; Fisher’s exact test), while that among the 195 genes up-regulated in the NF-YB2-overexpressing plants during heat stress (12 genes; 6%) was not significantly higher (P > 0.01, Fisher’s exact test). These results suggest that NF-YB2 and NF-YB3 share their downstream genes with DREB2A during dehydration and heat stress, respectively. Eighty-eight NF-YB2–specific up-regulated genes during dehydration stress were also dehydration-inducible genes, and among them, 13 genes were up-regulated in the DREB2A CA-overexpressing plants (Fig. 4A). Sixty-two NF-YB3–specific genes up-regulated during heat stress were also heat-inducible genes, and among them, 14 genes were up-regulated in the DREB2A CA-overexpressing plants (Fig. 4B). These genes are candidates for regulation by DREB2A together with NF-YB2 and NF-YB3 under dehydration and heat stress conditions, respectively (Supplemental Tables S2 and S3). Among them, several genes have DRE and CCAAT elements on their 1 kb-promoters, and expression of these genes was analyzed by qPCR because eukaryotic NF-Y trimers have been suggested to bind to CCAAT elements (Dolfini et al., 2012; Myers and Holt, 2018). The LEA76 and CYS-RICH TRANSMEMBRANE MODULE 1 (CYSTM1) genes were candidates regulated by NF-YB2 and DREB2A in the transcriptomic analysis (Supplemental Table S2) and were significantly up-regulated in NF-YB2–overexpressing plants during dehydration stress. The HSP70B and At1g53540 (HSP20-like) genes were candidates regulated by NF-YB3 and DREB2A (Supplemental Table S2) and were significantly up-regulated in the NF-YB3–overexpressing plants during heat stress (Fig. 4, C and D). Moreover, the overexpression of NF-YB3 suppressed the expression of LEA76 and CYSTM1 during dehydration stress (Fig. 4D).
Figure 4.
Transcriptomic analysis of NF-YB2-overexpressing and NF-YB3-overexpressing plants during dehydration and heat stress. A, Venn diagram comparing the dehydration-inducible genes, DREB2A CA (DREB2A constitutively active form)-inducing genes, and the up-regulated genes in the NF-YB2–overexpressing and NF-YB3–overexpressing plants during dehydration stress. The sums of numbers in black squares represent the NF-YB2 (75+13) –specific or NF-YB3 (13+1) –specific up-regulated genes with dehydration inducibility. The total numbers of each set of genes are presented in parentheses. B, Venn diagram comparing the heat-inducible genes, DREB2A CA-inducing genes, and the up-regulated genes in the NF-YB2–overexpressing and NF-YB3–overexpressing plants during heat stress. The sums of numbers in black squares represent the NF-YB2 (14+1) –specific or NF-YB3 (48+14) –specific up-regulated genes with heat inducibility. The total numbers of each set of genes are presented in parentheses. C and D, The expression analysis of several dehydration-inducible or heat-inducible genes during dehydration or heat stress in the NF-YB2–overexpressing (C) and NF-YB3–overexpressing (D) plants. The expression levels of candidate target genes of DREB2A with dehydration-inducibility or heat-inducibility that were specifically up-regulated in the NF-YB2–overexpressing {LEA76 and CYSTM1) or NF-YB3–overexpressing (HSP70B and At1g53540) plants were analyzed by RT-qPCR. The expression levels of each gene in the VC plants were under the control condition (0 h) defined as 1.0. The error bars indicate SD (n = three experiments, involving 7 individual plants for each line). Asterisks indicate significant differences from the VC plants at each time point (P < 0.05, Bonferroni-corrected Student’s t test).
Knockout Mutants of NF-YB2 and NF-YB3 Showed Dehydration and Heat Stress-Sensitive Phenotypes, Respectively
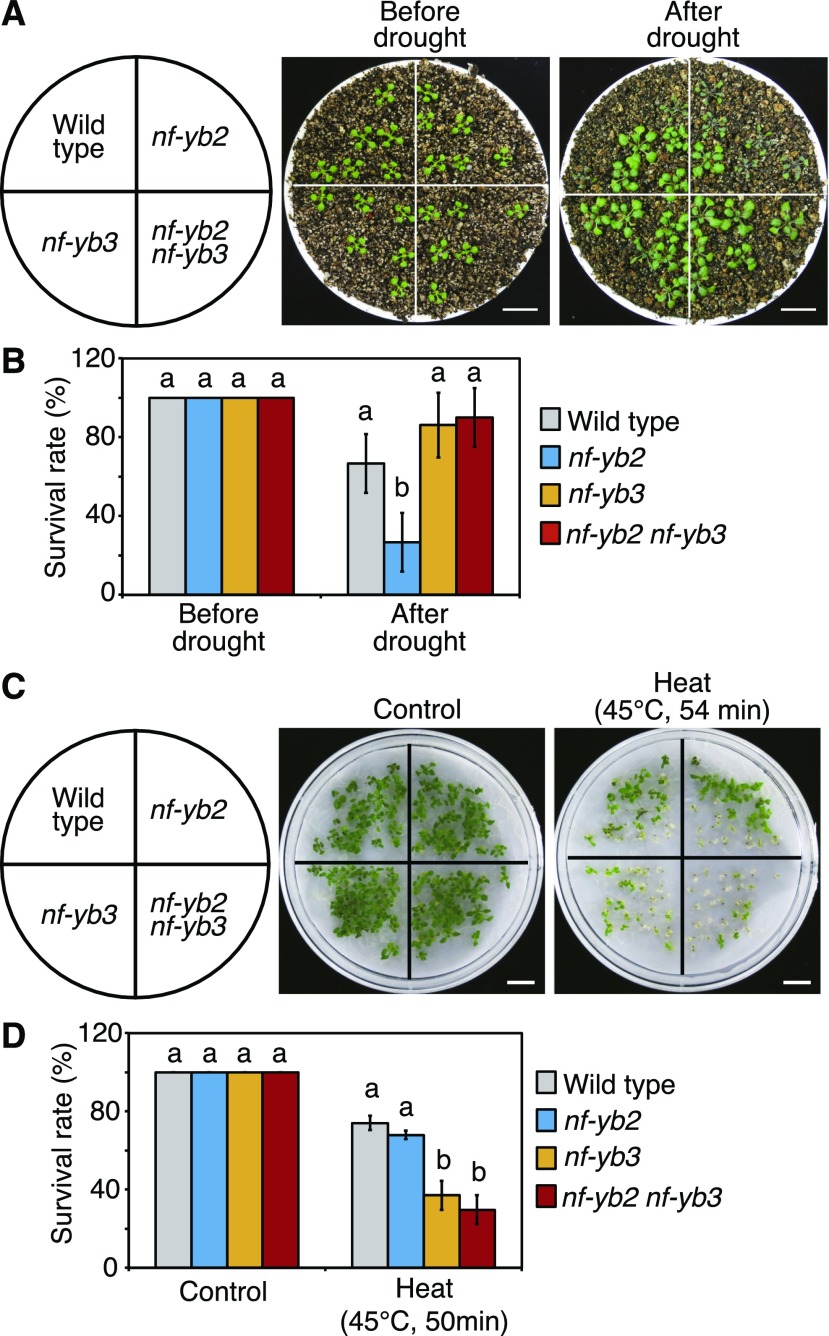
To analyze the contribution of NF-YB2 and NF-YB3 to stress responses under dehydration and heat stress conditions, single-knockout mutants of NF-YB2 (nf-yb2; salk_025666) and NF-YB3 (nf-yb3; salk_062245) and a double-knockout nf-yb2 nf-yb3 mutant were isolated (Supplemental Fig. S5, A and B). As previously reported (Kumimoto et al., 2008), these knockout mutants showed late-flowering phenotypes under long-day conditions (Supplemental Fig. S5C). Drought stress tolerance tests were therefore performed under short-day conditions to keep all lines at the same developmental stages. The results revealed that the nf-yb2 plants showed drought stress sensitivity. Meanwhile, the nf-yb3 and nf-yb2 nf-yb3 plants showed no significant changes in drought stress tolerance compared with that of the wild-type plants (Fig. 5, A and B). A previous paper showed that nf-yb3 knockout mutants were sensitive to drought stress (Zhang et al., 2015). These differences probably resulted from the experimental conditions; we focused on phenotypic differences between the nf-yb2 and nf-yb3 mutants. A heat stress tolerance test revealed that the nf-yb3 and nf-yb2 nf-yb3 plants were sensitive to heat stress and that the nf-yb2 plants did not show significant changes relative to the wild-type plants (Fig. 5, C and D). These results indicated that NF-YB2 and NF-YB3 play different roles and are important factors for survival under drought and heat stress conditions, respectively. In water loss tests of detached leaves, the nf-yb3 and nf-yb2 nf-yb3 plants showed significant suppression of water loss, while the nf-yb2 plants did not show phenotypic differences (Supplemental Fig. S5D). The phenotype of nf-yb3 coincides with the suppressive effect of NF-YB3 on dehydration stress-inducible genes (Fig. 3B).
Figure 5.
Drought and heat stress tolerance in the nf-yb2, nf-yb3, and nf-yb2 nf-yb3 knockout mutants. A, Drought stress tolerance of the nf-yb2, nf-yb3, and nf-yb2 nf-yb3 knockout mutants. Plants grown on agar plates for 10 d were transferred to soil and grown for 2 d. Water was withheld for 22 d. Scale bars = 1 cm. Photographs were taken before drought (left) and 7 d after rewatering (right). B, Percentages of surviving plants after rewatering. The error bars indicate the SD from six experiments, each involving 6 individual plants for each line. The letters above the bars indicate significant differences among plant lines before or after drought stress treatment (P < 0.05, according to Tukey’s multiple range test). C, Heat stress tolerance of the nf-yb2, nf-yb3, and nf-yb2 nf-yb3 mutants. The 7-d-old seedlings were treated with or without 45°C for 54 min. Scale bars = 1 cm. The photographs were taken after a 7-d recovery period at 22°C with (right) or without (left) heat stress. D, Percentages of surviving plants after heat stress. The error bars indicate the SD from three experiments, each involving 28 individual plants for each line. The letters above the bars indicate significant differences among plant lines with or without heat stress treatment (P < 0.05, according to Tukey’s multiple range test).
NF-YB2 and NF-YB3 Play Important Roles in the Full Expression of Dehydration-Inducible and Heat Stress-Inducible DREB2A Target Genes, Respectively
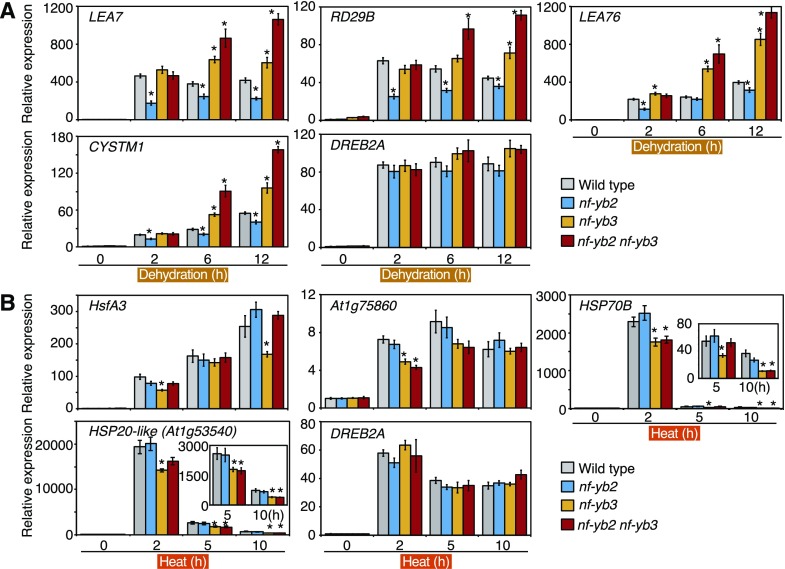
The expression levels of dehydration-inducible and heat stress-inducible genes were analyzed in the nf-yb2, nf-yb3, and nf-yb2 nf-yb3 knockout mutants. During dehydration stress, induction of the dehydration stress-inducible DREB2A target genes was significantly suppressed in the nf-yb2 mutants (Fig. 6A). Meanwhile, induction of the heat-stress-inducible DREB2A target genes was significantly suppressed in the nf-yb3 mutants (Fig. 6B). The expression levels (Fig. 6) and protein accumulation (Supplemental Fig. S6, C and D) of DREB2A were not different among the wild-type and knockout mutants. These results suggest that NF-YB2 and NF-YB3 are necessary for the full expression of these target genes specifically under dehydration and heat stress conditions, respectively. The nf-yb3 mutants showed enhanced expression of dehydration stress-inducible genes during dehydration stress (Fig. 6A), suggesting that NF-YB3 has negative effects on the induction of these dehydration stress-inducible genes. However, knockout of NF-YB2 did not affect the expression of heat stress-inducible genes (Fig. 6B). These data also indicated the functional diversity of NF-YB2 and NF-YB3 in terms of the repressive effects on their target genes under stress conditions. In the nf-yb2 nf-yb3 mutants, some heat-inducible genes such as At1g75860 and HSP70B were down-regulated, and some dehydration-inducible genes such as LEA7 and RD29B were up-regulated (Fig. 6). These data suggested that knockout of NF-YB3 had greater effects than did knockout of NF-YB2 on the regulation of dehydration-inducible genes during dehydration stress.
Figure 6.
Expression patterns of dehydration-inducible and heat-inducible DREB2A target genes during dehydration and heat stress. A and B, Expression analysis of dehydration-inducible (A) or heat-inducible (B) DREB2A target genes during dehydration and heat stress in the nf-yb2, nf-yb3, and nf-yb2 nf-yb3 knockout plants. The expression levels of DREB2A and dehydration-inducible (LEA7, RD29B, LEA76, and CYSTM1) and heat-inducible (HSFA3, At1g75860, HSP70B, and At1g53540) DREB2A target genes during dehydration and heat stress were analyzed by RT-qPCR. The 14-d-old plants grown on agar plates were treated with dehydration or heat stress. The expression levels of each gene in the VC plants under the control condition (0 h) were defined as 1.0. The error bars indicate SD (n = three experiments, involving 7 individual plants for each line). Asterisks indicate significant differences from the VC plants at each time point (P < 0.05, Bonferroni-corrected Student’s t test). Small values of HSP70B and At1g53540 (HSP20-like) gene expression under 5- and 10-h heat stress conditions were enlarged. The expression levels of DREB2A at each time point during dehydration and heat stress were evaluated using one-way ANOVA, but no significant differences were detected (P > 0.05).
NF-YB2 and NF-YB3 Show Stress-Specific DNA-Binding Profiles to the Promoter of DREB2A Target Genes
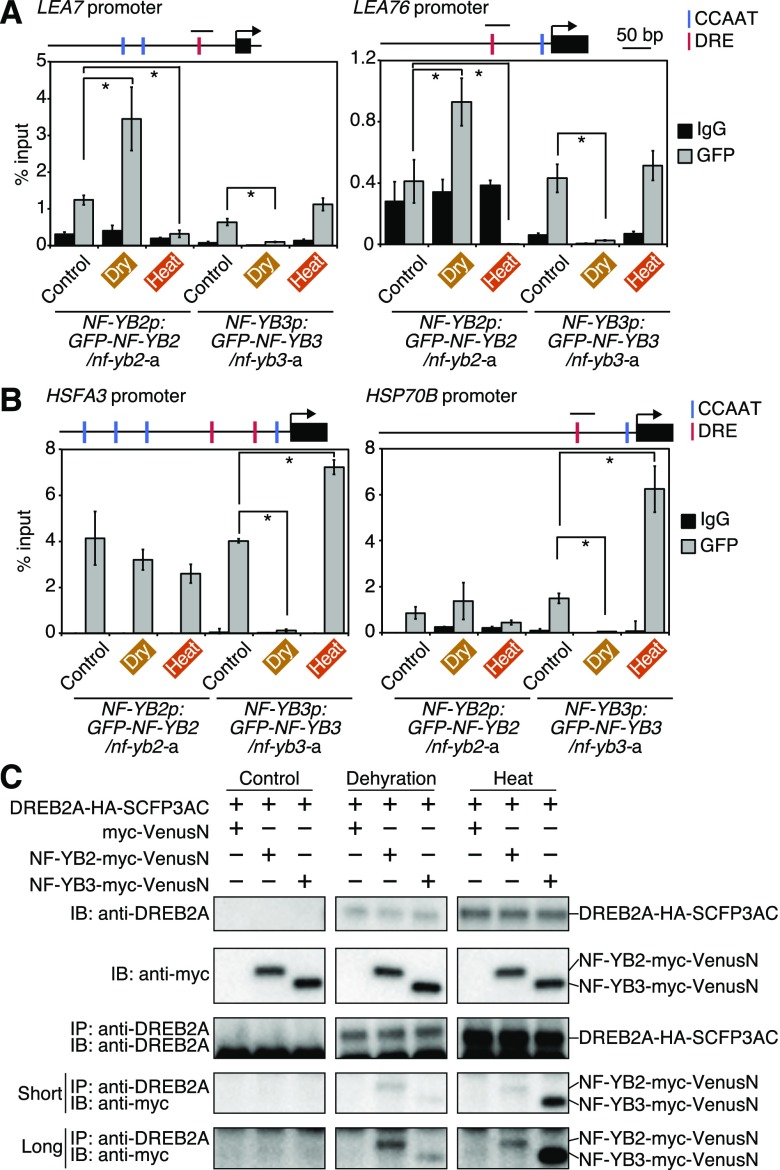
To analyze the DNA-binding profiles of NF-YB2 and NF-YB3 to target gene promoters during stress, transgenic nf-yb2 or nf-yb3 plants expressing NF-YB2 or NF-YB3 fused to GFP driven by its native promoter were generated (NF-YB2p:GFP-NF-YB2/nf-yb2-a, b and NF-YB3p:GFP-NF-YB3:nf-yb3-a, b; Supplemental Fig. S7, A and B). The complementation of GFP-NF-YB2 and GFP-NF-YB3 recovered the drought-sensitive phenotypes of nf-yb2 (Supplemental Fig. S7, C and D) and heat-sensitive phenotypes of nf-yb3 (Supplemental Fig. S7, E and F), respectively, indicating that NF-YB2 and NF-YB3 fused to GFP were functional. The suppressed expression levels of LEA7 and HSFA3 during dehydration and heat stress were also recovered by complementation of NF-YB2 and NF-YB3, respectively (Supplemental Fig. S7, G and H). LEA7 and LEA76 are candidate target genes with dehydration stress inducibility, and HSFA3 and HSP70B have heat stress inducibility (Fig. 6). These genes have DRE and CCAAT sequences on their promoters within 1 kb upstream of translational start sites (Fig. 7, A and B). Chromatin immunoprecipitation (ChIP) assays revealed that dehydration stress increased the DNA binding of NF-YB2 to the LEA7 and LEA76 promoters (Fig. 7A), while heat stress increased the DNA binding of NF-YB3 to the HSFA3 and HSP70B promoters (Fig. 7B). These results suggest the possibility that these dehydration-inducible and heat-inducible genes are direct targets of NF-YB2 and NF-YB3 under dehydration and heat stress conditions, respectively. Moreover, heat stress decreased the DNA binding of NF-YB2 to the promoters of the dehydration-inducible genes (Fig. 7A), and dehydration stress decreased that of NF-YB3 to those of heat-inducible genes (Fig. 7B). It is suggested that the transcriptional complexes, including NF-YB2 and NF-YB3, were dissociated from the promoters of target genes that were not induced during specific stress conditions. We also found differences in the DNA-binding profiles of NF-YB2 and NF-YB3; heat stress did not affect the DNA-binding of NF-YB2 to the promoters of the heat-inducible genes (Fig. 7B), while dehydration stress clearly suppressed that of NF-YB3 to those of the dehydration-inducible genes (Fig. 7A). These data suggested differences in the molecular functions of NF-YB2 and NF-YB3 toward heat-inducible and dehydration-inducible genes, respectively. In our previous study, direct interactions between NF-YBs (NF-YB2 and NF-YB3) and DREB2A were not detected in yeast cells (Sato et al., 2014). To analyze whether they form a transcriptional complex in plants, transgenic plants overexpressing one fusion protein consisting of DREB2A and the C-terminal half of SCFP3A with a hemagglutinin (HA) tag (DREB2A-HA-SCFP3AC) and one fusion protein consisting of NF-YBs (NF-YB2 or NF-YB3) and the N-terminal half of Venus with a myc tag (NF-YB2-myc-VenusN or NF-YB3-myc-VenusN) were generated. In bimolecular fluorescence complementation (BiFC) assays, fluorescent signals that indicated the interaction between DREB2A and NF-YB3 were detected in nuclei during heat stress (37°C, 5 h; Supplemental Fig. S7I). This suggested that DREB2A and NF-YB3 form a transcriptional complex during heat stress. Signals indicating the interaction between DREB2A and NF-YB2 could not be detected even under dehydration stress conditions (6 h), probably because the accumulated levels of the DREB2A protein were too low to be detected during drought conditions, or water loss from cells decreased signal intensity. We therefore performed coimmunoprecipitation (IP) assays using the same transgenic lines after crosslinking to identify the proteins in transcriptional complexes. After IP of DREB2A-HA-SCFP3AC using an anti-DREBB2A antibody after dehydration stress (6 h), the NF-YB2-myc-VenusN protein as well as the NF-YB3-myc-VenusN protein were detected after heat stress (37°C, 5 h; Fig. 7C). The differences in the protein levels of NF-YB2 and NF-YB3 immunoprecipitated during dehydration and heat stress suggested the stress-specific formation of transcriptional complexes with DREB2A.
Figure 7.
DNA-binding profiles of NF-YB2 and NF-YB3 to the target gene promoters, and protein interactions of NF-YB2 and NF-YB3 with DREB2A during dehydration and heat stress. A and B, ChIP-RT-qPCR assays of the NF-YB2 and NF-YB3 proteins under nonstress (control), dehydration (dry,) and heat stress conditions to the promoters of the dehydration-inducible (A) and heat-inducible (B) DREB2A target genes in the NF-YB2p:GFP-NF-YB2/nf-yb2 and NF-YB3p:GFP-NF-YB3/nf-yb3 plants. A schematic diagram displays the models of each target gene and amplified regions by RT-qPCR. The coding regions and CCAAT and DRE sequences are indicated as closed boxes and blue and red lines, respectively. The 14-d-old plants grown on agar plates were treated with or without dehydration or heat stress. IP was performed using an anti-GFP antibody or normal mouse igG (IgG; negative control). The error bars indicate the SD from six experiments. Asterisks indicate significant differences between two values (P < 0.05, Student’s t test). C, Co-IP of NF-YB2-myc-VenusN and NF-YB3-myc-VenusN with DREB2A-HA-SCFP3AC in the transgenic plants under nonstress, dehydration, and heat stress conditions. The 14-d-old plants grown on agar plates were treated with or without dehydration or heat stress. Protein extracts before and after IP were analyzed by immunoblotting using anti-DREB2A or antimyc antibodies. Short and long exposures of the immunoblots using the antimyc antibody after IP are shown.
Phylogenetic Analysis Suggests the Evolutionary Diversification of NF-YB2 and NF-YB3 Subgroups
Our current studies indicated that Arabidopsis NF-YB2 and NF-YB3 have dehydration-specific and heat-stress–specific functions. To speculate on the evolutionary conservation of NF-YB2–specific and NF-YB3–specific functions in various plant species, phylogenetic analysis was performed. The phylogenetic tree of NF-YB indicated that all analyzed angiosperm plants have NF-YB family proteins that belong to divergent NF-YB2 and NF-YB3 subgroups (Supplemental Fig. S8). Another subgroup to which Arabidopsis NF-YB1, NF-YB8, and NF-YB10 belong also contains NF-YB proteins of all angiosperms (Supplemental Fig. S8). Meanwhile, green algae (Chlamydomonas reinhardtii) has only one NF-YB protein among these three subgroups (Supplemental Fig. S8). Bryophytes (Physcomitrella patens) and lycophytes (Selaginella moellendorffii) have NF-YB proteins that are closely related to the subgroup of Arabidopsis NF-YB1, NF-YB8, and NF-YB10; however, they do not have diversified NF-YB proteins that differentially belong to the NF-YB2 and NF-YB3 subgroups (Supplemental Fig. S8). These data suggest that functional diversification between NF-YB2 and NF-YB3 is conserved among angiosperms.
DISCUSSION
The NF-Y family proteins in plant species are more widely diversified compared with those in Homo sapiens (Petroni et al., 2012). The NF-Y transcription factors form a trimer composed of NF-YA, NF-YB, and NF-YC subunits when they work together, and wide diversification of plant NF-Y proteins raises the possibility that different combinations of these three subunits result in trimers with different molecular functions (Siefers et al., 2009; Myers and Holt, 2018). Previous studies have revealed the functions of NF-Y subunits involved in several biological phenomena such as flowering, embryo development, and abiotic stress responses (Nelson et al., 2007; Kumimoto et al., 2008; Yamamoto et al., 2009). Our current study revealed functional diversification between NF-YB2 and NF-YB3, which have the greatest sequence similarity (70% sequence identity) among all NF-YB proteins in Arabidopsis. Overexpression of NF-YB2 and NF-YB3 resulted in enhanced drought and heat stress tolerance, respectively (Fig. 2), and increased expression levels of several stress-specific inducible genes under each stress condition (Fig. 3). Moreover, the nf-yb2 and nf-yb3 knockout mutants were drought sensitive and heat stress sensitive, respectively (Fig. 5), and showed significant decreases in the expression of several stress-inducible genes during dehydration and heat stress (Fig. 6). RNA-sequencing also confirmed that NF-YB2 and NF-YB3 regulated their specific target genes during dehydration and heat stress, and overexpression of these genes had greater effects on transcriptomic changes in dehydration and heat stress responses, respectively (Fig. 4). These results clearly indicated that NF-YB2 and NF-YB3 were functionally diversified and activated target genes specifically in response to dehydration and heat stress. The DNA binding profiles also confirmed the stress-specific dynamics of NF-YB2 and NF-YB3 on the target gene promoters. Dehydration stress increased the DNA binding of NF-YB2 to the dehydration-inducible gene promoters (Fig. 7A) but did not affect that to the heat-inducible gene promoters (Fig. 7B). In contrast, heat stress increased NF-YB3 binding to the heat-inducible gene promoters (Fig. 7B) but did not affect that to the dehydration-inducible gene promoters (Fig. 7A). The gene expression patterns of NF-YB2 and NF-YB3 also coincide with the stress-specific functions of NF-YB2 and NF-YB3 (Fig. 1).
Moreover, unexpected functions of NF-YB3 during dehydration stress were indicated. The nf-yb3 knockout mutants showed enhanced expression of dehydration-inducible genes during dehydration stress (Fig. 5A), and overexpression of NF-YB3 resulted in decreased expression of those genes (Fig. 3). Meanwhile, knockout and overexpression of NF-YB2 did not affect the expression of heat-inducible genes (Figs. 3 and 5B). These data suggested that NF-YB3 had negative effects on the induction of dehydration-inducible genes during dehydration stress, but NF-YB2 did not have negative effects on heat-inducible genes during heat stress. In the ChIP assays, heat stress did not affect NF-YB2 binding to the heat-inducible gene promoters (Fig. 7B), whereas dehydration stress decreased the binding of NF-YB3 to the dehydration-inducible promoters (Fig. 7A). These data also suggested the negative functions of NF-YB3 in dehydration-inducible genes, but NF-YB2 did not show such negative functions in heat-inducible genes. It is still possible that these repressive effects on the expression of the dehydration-inducible genes may be caused indirectly by NF-YB3 knockout. Further studies will be necessary to reveal the more detailed mechanisms by which NF-YB3 exerts negative effects on dehydration-inducible genes and what causes differences between NF-YB2 and NF-YB3 functions toward their target genes.
The possibility of functional conservation among plant species opens the door to the application of NF-YB2 and NF-YB3 to improve drought and heat stress tolerance in agriculturally important crops. We previously reported that Arabidopsis DPB3-1 (NF-YC10) and rice (Oryza sativa) OsDPB3-2 had conserved functions and that overexpression of Arabidopsis DPB3-1 enhanced heat stress tolerance in both Arabidopsis and rice (Sato et al., 2014, 2016). Other studies have provided evidence that NF-Y could be a useful target for improving agricultural traits in response to abiotic stress: Arabidopsis NF-YB1 conferred drought stress tolerance in maize (Zea mays; Nelson et al., 2007), overexpression of rice NF-YA7 increased drought tolerance in rice (Lee et al., 2015), and so on. Arabidopsis NF-YB2 and NF-YB3 or other NF-YB proteins with conserved functions in other plant species might be important factors for manipulating drought and heat stress tolerance in crops.
Our current study also suggested the mechanisms underlying the target selectivity of DREB2A, which regulated both dehydration-inducible and heat stress-inducible genes in response to environmental conditions. The genes that were differentially expressed in NF-YB2-overexpressing and NF-YB3-overexpressing or knockout plants during dehydration and heat stress, respectively, are also known to be target genes of DREB2A (Sakuma et al., 2006b; Figs. 3 and 6). Genetic interaction and physical protein interaction assays suggested that NF-YB2 and NF-YB3 cooperatively activated their target genes together with DREB2A specifically during dehydration and heat stress, respectively (Fig. 7C; Supplemental Fig. S4, G and H). Moreover, transcriptomic data showed that NF-YB2 and NF-YB3 shared a significant proportion of their downstream genes with DREB2A CA during dehydration and heat stress, respectively (Fig. 4, A and B). These results indicated that there is an overlap between stress-specific target genes of DREB2A and NF-YBs (NF-YB2 or NF-YB3). Moreover, our transcriptomic analysis revealed that overexpression of NF-YB2 and NF-YB3 enhanced the expression levels of dehydration-inducible and heat-inducible genes, respectively, that were not downstream of DREB2A (Fig. 4, A and B), suggesting that NF-YB2 and NF-YB3 are involved in DREB2A-independent pathways, for example, ABA-dependent pathways during dehydration stress (Yoshida et al., 2014) or HSFA2 pathways during heat stress (Ohama et al., 2017). Future studies are needed to reveal what factors other than DREB2A form transcriptional complexes with NF-YB2 and NF-YB3, and enhance drought and heat stress responses, respectively.
Our present study also raised several questions about the molecular functions of NF-YB2 and NF-YB3. First, it is important to identify dehydration-specific and heat stress-specific trimers including NF-YB2 or NF-YB3. Our previous work suggested that a heat stress–specific trimer composed of NF-YA2, NF-YB3, and DPB3-1 (NF-YC10) worked in cotyledons during heat stress (Sato et al., 2014); however, NF-YB3 is also expressed in true leaves (Siefers et al., 2009; Sato et al., 2014), and the expression patterns of several DREB2A target genes were altered in 14-d-old nf-yb3 mutants (Fig. 5B). These results imply that another NF-YC protein forms a transcriptional complex with NF-YB3 in true leaves during heat stress. In conclusion, we found that the NF-YB2 and NF-YB3 proteins, which had high sequence similarity to each other, had different functions in response to environmental conditions (Supplemental Fig. S9). These results provide insights into the evolution and diversification of NF-Y proteins in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Heynh. ecotype Columbia and Wassilewskija plants were grown on germination medium (GM) agar plates at 22°C under a 16-h or 8-h photoperiod at a photon flux density of 40 μm-2s−1 as described previously in Sakuma et al. (2006a, 2006b). The 16-d-old plants were transferred to perlite-containing soil (Dio Chemicals) and grown under similar conditions. For analysis of flowering time, five plants were transferred to soil in a plastic pot with a diameter of 7.5 cm. The T-DNA insertion lines for NF-YB2 and NF-YB3 were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus; nf-yb2; salk_025666, nf-yb3; salk_062245). The T-DNA insertions were detected by amplifying the insert-flanking region of the genomic DNA using the primers shown in Supplemental Table S4. The double mutant nf-yb2 nf-yb3 was constructed by genetic crosses between nf-yb2 and nf-yb3. The triple-knockout mutant srk2d srk2e srk2i (srk2d/e/i), quadruple mutant areb1 areb2 abf1 abf3 (arebq), triple-knockout mutant hsfa1a hsfa1b hsfa1d (hsfa1a/b/d), nc3-2, aba2-2 and dreb2a-1 mutants, and 35S:DREB2A CA transgenic plants were previously described (Nambara et al., 1998; Iuchi et al., 2001; Fujita et al., 2009; Yoshida et al., 2011, 2015). Transgenic plants were generated by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101 (pMP90) cells.
Stress Treatment
The dehydration, heat stress, and ABA treatments were performed as described previously (Liu et al., 1998; Sato et al., 2014) with minor modifications. For dehydration stress, the plants grown on GM agar plates were transferred to petri dishes covered with Parafilm M. For heat stress, the GM agar plates on which plants were grown were transferred to a 37°C incubator (LPH-350SP; NK system). The heat and drought stress tolerance tests were performed as previously described in Sakuma et al. (2006a, 2006b), with minor modifications. For the drought stress tolerance test, the plants grown on GM agar plates under an 8-h photoperiod were transferred to soil in plastic pots and grown for 2 d under the same conditions. Water supply was withheld until plants withered. The survival rates of plants were calculated 7 d after rewatering under control conditions. For the heat stress tolerance test, plants were grown on two layers of filter paper containing 4 ml of liquid GM medium in 90-mm plastic Petri dishes under control conditions with supplying 2 ml of sterilized water every 3 d. The 7-d-old plants were subjected to heat stress (45°C, 54 min) in a paper box (width 13 mm × depth 130 mm × height 50 mm) in an incubator (SLC-25A; Mitsubishi Electric). For ABA treatment, 14-d-old plants were transferred to a Petri dish filled with distilled water containing 100 μm ABA (Sigma-Aldrich). For measurement of water loss, rosette leaves of 14-d-old plants were detached and placed on a petri dish. The loss of fresh weight was monitored at the indicated time.
RNA Preparation and RT-qPCR
The total RNA from seedlings was isolated with RNAiso plus (TaKaRa) according to the supplier’s instructions. The complementary DNA (cDNA) was synthesized using SuperScript IV (Thermo Fisher), and RT-qPCR was performed using an Applied Biosystems 7500 Fast Real-Time PCR system and THUNDERBIRD SYBR qPCR Mix (TOYOBO) according to the supplier’s instructions. Triplicate measurements were made for each cDNA sample, and the values were normalized according to the amounts of 18S rRNA. The primers used for RT-qPCR are shown in Supplemental Table S4.
Protein Immunoblot Analysis
Protein immunoblot analysis was performed as previously described (Morimoto et al., 2013) with minor modifications. The total proteins were extracted from seedlings using protein extraction buffer (8 m urea, 5 mm [v/v] 2-mercaptoethanol, 2 mm EDTA, 1% [w/v] SDS, 50 mm Tris-HCl [pH 6.8]), and mixtures were centrifuged at 20,000g for 10 min at room temperature. The supernatant was boiled at 95°C for 3 min. The resultant extracts, which corresponded to a fresh weight of 4 mg seedling, were separated by SDS-PAGE. The separated proteins were blotted to an immune-blot polyvinylidene difluoride membrane (BIO RAD) using a Trans-Blot Turbo Transfer System (BIO RAD). For immunoblotting, a polyclonal anti-DREB2A antibody (1: 2500 dilution; Morimoto et al., 2013) or antimyc-antibody (1: 2000 dilution; Millipore) was used as a primary antibody, and goat antirabbit or antimouse IgG peroxidase-conjugate (Thermo Fisher) was used as a secondary antibody. The signals were developed by SuperSignal West Dura Extended Duration Substrate (Thermo Fisher) according to the manufacturer’s protocol and detected with an image analyzer (ChemiDoc MP, Bio-Rad). The Rubisco large subunit (rbcL) was detected by Ponceau S (APRO SCIENCE) according to the manufacturer’s instructions.
RNA Sequencing and Data Analysis
cDNA libraries were constructed using TruSeq RNA Sample Preparation Kit v2 (Illumina), and the libraries were sequenced by NextSeq 500 (Illumina). The produced bcl files were converted to fastq files by bcl2fastq (Illumina). The reads were analyzed as previously described (Notaguchi et al., 2015). The data were deposited to the DNA Data Bank of Japan (DRA007270). The genes up-regulated by DREB2A CA were reported in previous works (Sakuma et al., 2006a; Mizoi et al., 2013).
ChIP Assay
ChIP assays were performed using an EpiQuik Plant ChIP kit (Epigenetek) according to the user guide. Plant samples (one gram fresh weight) with or without stress treatment were collected. The polyclonal anti-GFP antibody (Tanaka et al., 2012) was used for IP of the DNA-protein complex.
BiFC and Co-IP Assays
GFP fluorescence was observed by using a confocal laser scanning microscope (Olympus FLUOVIEWV FV 1000) as described previously (Sato et al., 2018). For co-IP assays, 2 g of plant materials crosslinked by 1% (w/v) formaldehyde were ground in liquid nitrogen and dissolved in an extraction buffer containing 10 mm Tris-HCl (pH 8.0), 0.4 m Suc, 5 mm 2-mercaptoethanol, and a protease inhibitor mixture (cOmplete, EDTA-free; Roche Applied Science). The mixture was filtered through two layers of Miracloth (Merck) and centrifuged at 3,000g for 20 min at 4°C. The nuclear pellet was resuspended with 1 ml of a lysis buffer containing 10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 0.2 m NaCl, 1% (v/v) Triton X-100, and a protease inhibitor mixture (cOmplete, EDTA-free; Roche Applied Science) and placed on ice for 5 min. The solution was centrifuged at 3,000g for 10 min at 4°C. The pellet was resuspended with 500 μL of a sonication buffer containing 10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 0.15 m NaCl, 0.1% (w/v) SDS, 100 μm MG132 (Calbiochem), and a protease inhibitor mixture (cOmplete, EDTA-free; Roche Applied Science), sonicated, and centrifuged at 12,000g for 10 min at 4°C. The supernatant was incubated with 3 μg of an anti-DREB2A antibody (Morimoto et al., 2013) for 1 h at 4°C with rotation, and 50 μL of Protein G Sepharose 4 Fast Flow (GE Healthcare) was added and incubated for 1 h at 4°C with rotation. The resin was collected after centrifugation at 12,000g for 20 s at 4°C and washed with 3 mL of sonication buffer. Proteins were eluted with 50 μL of an elution buffer containing 50 mm Tris-HCl (pH 7.5), 1% (w/v) SDS, and 100 mm dithiothreitol at 95°C for 3 min and subjected to immunoblot analysis.
Sequence Alignment and Phylogenetic Analysis
The peptide and promoter sequences of the NF-YB family were obtained through Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html) using PANTHER ID (PTHR11064). The family proteins were aligned using the MUSCLE program by SnapGene software (https://www.snapgene.com/). The resultant alignments were manually adjusted and visualized through the BoxShade program (https://embnet.vital-it.ch/software/BOX_form.html). The neighbor-joining phylogenetic tree was constructed using MEGA6 software (http://www.megasoftware.net/) with the Poisson model, 10,000 bootstrap replicates, and pairwise deletion for gap/missing data treatment using conserved regions of the NF-YB proteins. The accession numbers of NF-YB family proteins used to construct the phylogenetic tree are shown in Supplemental Table S5.
Plasmid Constructions
To generate the 35S:NF-YB2 and NF-YB3 constructs, the NF-YB2 and NF-YB3 coding sequences were inserted into the XbaI and XhoI sites of the pGKX or pGHX vector (Qin et al., 2008). To generate the NF-YB2p:GFP-NF-YB2 and NF-YB3p:GFP-NF-YB3 constructs, the NF-YB2 and NF-YB3 coding sequences were initially inserted into the XbaI and XhoI sites of the pGKX-NsGFP vector (Qin et al., 2008). The GFP-NF-YB2-Nos terminator (NosT) or GFP-NF-YB3-NosT sequences and 1-kb upstream promoter sequences from translational start sites of NF-YB2 and NF-YB3 were inserted into the SalI and KpnI sites and SacI and PstI sites of the pGreen0129 vectors (Hellens et al., 2000), respectively. To generate the constructs for BiFC and co-IP, NF-YBs (NF-YB2 and NF-YB3) and DREB2A coding sequences were inserted into the XbaI and XhoI sites or SpeI and ClaI sites of the pVYNE and pSCYCE vector (Waadt et al., 2008), respectively.
Statistical Analysis
The significant differences between the control plants and transgenic plants, control conditions, and stress conditions were analyzed by Bonferroni-corrected Student’s t test, Student’s t test, one-way ANOVA, Tukey’s multiple range test, or Fisher’s exact test. P values < 0.01 were considered to indicate statistical significance in Fisher’s exact test, and P values < 0.05 were considered to indicate statistical significance in other tests.
Accession Numbers
The sequence data of genes in this article can be obtained in Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html) under the following accession numbers: NF-YB2 (At5g47640), NF-YB3 (At4g14540), DREB2A (At5g05410), LEA7 (At1g52690), RD29B (At5g52300), RD17 (At1g20440), HSFA3 (At5g03720), At1g75860, LEA76 (At3g15670), CYSTM1 (At1g05340), HSP79B (At1g16030), and At1g53540.
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Expression levels of NF-YB2, NF-YB3 and DREB2A in the ABA-signaling and ABA-deficient mutants during ABA treatment.
Supplemental Figure S2. Alignments of promoters of NF-YB family genes among Brassica species.
Supplemental Figure S3. Early-flowering phenotypes under long-day conditions and water loss of the NF-YB2-overexpressing and NF-YB3-overexpressing plants.
Supplemental Figure S4. Protein accumulation of DREB2A in the NF-YB2-overexpressing and NF-YB3-overexpressing plants during dehydration and heat stress, and genetic interactions between NF-YBs (NF-YB2 and NF-YB3) and DREB2A.
Supplemental Figure S5. Isolation of nf-yb2 and nf-yb3 knockout mutants and late-flowering phenotypes under long-day conditions and water loss of the nf-yb2, nf-yb3 and nf-yb2 nf-yb3 mutants.
Supplemental Figure S6. Protein accumulation of DREB2A in the nf-yb2, nf-yb3 and nf-yb2 nf-yb3 mutants during dehydration and heat stress.
Supplemental Figure S7. Recovery of drought and heat stress tolerance by the complementation of GFP-NF-YB2 in nf-yb2 and GFP-NF-YB3 in nf-yb3, respectively, and protein interaction between DREB2A and NF-YB2 or NF-YB2 during dehydration and heat stress.
Supplemental Figure S8. A phylogenetic tree of NF-YB family proteins across various plant species.
Supplemental Figure S9. A current model of how NF-YB2 and NF-YB3 regulate their target genes together with of DREB2A under drought and heat stress conditions.
Supplemental Table S1. Numbers and positions of cis-acting elements related to abiotic stress on promoters of NF-YB2 and NF-YB3.
Supplemental Table S2. Candidate target genes by DREB2A and NF-YB2 during dehydration stress.
Supplemental Table S3. Candidate target genes by DREB2A and NF-YB3 during heat stress.
Supplemental Table S4. Primers used in this study.
Supplemental Table S5. NF-YB family proteins across various plant species belonging to the same subgroups of Arabidopsis thaliana NF-YB2, NF-YB3, NF-YB1, NF-YB8 and NF-YB10.
Supplemental Dataset S1. Dehydration-stress-inducible genes in the control plants.
Supplemental Dataset S2. Heat-stress-inducible genes in the control plants.
Supplemental Dataset S3. Upregulated genes in the NF-YB2-overexpressing plants during dehydration stress.
Supplemental Dataset S4. Upregulated genes in the NF-YB3-overexpressing plants during dehydration stress.
Supplemental Dataset S5. Upregulated genes in the NF-YB2-overexpressing plants during heat stress.
Supplemental Dataset S6. Upregulated genes in the NF-YB3-overexpressing plants during heat stress.
Acknowledgments
We thank Fuyuko Shimoda, Saho Mizukado, Saya Kikuchi, Manami Masuda, Hiroko Kobayashi, Kumiko Matsuo, Michie Etoh, Yuriko Tanaka, Sayuri Murasaki, Ayami Furuta, and Tomomi Shinagawa for excellent technical assistance and Etsuko Toma for skillful editorial assistance.
Footnotes
This work was supported by Japan Society for the Promotion of Science (Grant-in-Aid for JSPS Fellows 25-4185 to H.S.; Grant-in-Aid for Scientific Research for Young Scientists [B] 16K21626 to H.S.; and Grant-in-Aid for Scientific Research on Innovative Areas JP16H01475 and JP18H04792 to F.T. and 15H05960 to K.Y.-S.).
Articles can be viewed without a subscription.
References
- Calvenzani V, Testoni B, Gusmaroli G, Lorenzo M, Gnesutta N, Petroni K, Mantovani R, Tonelli C (2012) Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dolfini D, Gatta R, Mantovani R (2012) NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47: 29–49 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hincha DK, Thalhammer A (2012) LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans 40: 1000–1003 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Jacob P, Hirt H, Bendahmane A (2017) The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol J 15: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keunen E, Peshev D, Vangronsveld J, Van Den Ende W, Cuypers A (2013) Plant sugars are crucial players in the oxidative challenge during abiotic stress: extending the traditional concept. Plant Cell Environ 36: 1242–1255 [DOI] [PubMed] [Google Scholar]
- Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, Yamaguchi-Shinozaki K (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52: 2136–2146 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ (2008) The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723 [DOI] [PubMed] [Google Scholar]
- Lee DK, Kim HI, Jang G, Chung PJ, Jeong JS, Kim YS, Bang SW, Jung H, Choi YD, Kim JK (2015) The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci 241: 199–210 [DOI] [PubMed] [Google Scholar]
- Lesk C, Rowhani P, Ramankutty N (2016) Influence of extreme weather disasters on global crop production. Nature 529: 84–87 [DOI] [PubMed] [Google Scholar]
- Li K, Yu R, Fan LM, Wei N, Chen H, Deng XW (2016) DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat Commun 7: 11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Ogata T, Kanamori N, Yoshiwara K, Goto S, Yamamoto YY, Tokoro Y, Noda C, Takaki Y, Urawa H, et al. (2017) Design of an optimal promoter involved in the heat-induced transcriptional pathway in Arabidopsis, soybean, rice and maize. Plant J 89: 671–680 [DOI] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet 16: 237–251 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Ohori T, Moriwaki T, Kidokoro S, Todaka D, Maruyama K, Kusakabe K, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2013) GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol 161: 346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Kanazawa N, Kidokoro S, Takahashi F, Qin F, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K (2019) Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J Biol Chem 294: 902–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, Mizoi J, Qin F, Kim J-S, Sato H, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2013) Stabilization of Arabidopsis DREB2A is required but not sufficient for the induction of target genes under conditions of stress. PLoS One 8: e80457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, Ohama N, Kidokoro S, Mizoi J, Takahashi F, Todaka D, Mogami J, Sato H, Qin F, Kim JS, et al. (2017) BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc Natl Acad Sci USA 114: E8528–E8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers ZA, Holt BF III (2018) NUCLEAR FACTOR-Y: Still complex after all these years? Curr Opin Plant Biol 45(Pt A): 96–102 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, et al. (2007) Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA 104: 16450–16455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M, Higashiyama T, Suzuki T (2015) Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol 56: 311–321 [DOI] [PubMed] [Google Scholar]
- Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22: 53–65 [DOI] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF III, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. (2008) Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J (2013) Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol 161: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006a) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2006b) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Mizoi J, Tanaka H, Maruyama K, Qin F, Osakabe Y, Morimoto K, Ohori T, Kusakabe K, Nagata M, Shinozaki K, Yamaguchi-Shinozaki K (2014) Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 26: 4954–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Todaka D, Kudo M, Mizoi J, Kidokoro S, Zhao Y, Shinozaki K, Yamaguchi-Shinozaki K (2016) The Arabidopsis transcriptional regulator DPB3-1 enhances heat stress tolerance without growth retardation in rice. Plant Biotechnol J 14: 1756–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Takasaki H, Takahashi F, Suzuki T, Iuchi S, Mitsuda N, Ohme-Takagi M, Ikeda M, Seo M, Yamaguchi-Shinozaki K, Shinozaki K (2018) Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc Natl Acad Sci USA 115: E11178–E11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauberger B, Archontoulis S, Arneth A, Balkovic J, Ciais P, Deryng D, Elliott J, Folberth C, Khabarov N, Müller C, et al. (2017) Consistent negative response of US crops to high temperatures in observations and crop models. Nat Commun 8: 13931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE IV, Tayrose G, Holt BF III (2009) Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol 149: 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58: 843–856 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim JM, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21: 133–139 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Maruyama K, Mogami J, Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ 38: 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Blanco FA, Beker MP, Battaglia M, Aguilar OM (2010) A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 22: 4142–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhang D, Liu Y, Luo C, Zhou Y, Zhang L (2015) Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol Biochem 94: 153–164 [DOI] [PubMed] [Google Scholar]