Abstract
Posttraumatic stress disorder (PTSD) is associated with wide-spread immune dysregulation; however, little is known about the gene expression differences attributed to each PTSD symptom cluster. This is an important consideration when identifying diagnostic and treatment response markers in highly comorbid populations with mental and physical health conditions that share symptoms. To this aim, we utilized a transcriptome-wide analysis of differential gene expression in peripheral blood by comparing military service members: (1) with vs. without PTSD, (2) with high vs. low PTSD cluster symptom severity, and (3) with improved vs. not improved PTSD symptoms following 4 to 8 weeks of evidenced-based sleep treatment. Data were analyzed at a ± 2.0-fold change magnitude with subsequent gene ontology-based pathway analysis. In participants with PTSD (n=39), 89 differentially expressed genes were identified, and 94% were upregulated. In participants with high intrusion symptoms (n=22), 1,040 differentially expressed genes were identified, and 98% were upregulated. No differentially expressed genes were identified for the remaining two PTSD symptom clusters. Ten genes (C5orf24, RBAK, CREBZF, CD69, PMAIP1, AGL, ZNF644, ANKRD13C, ESCO1, and ZCCHC10) were upregulated in participants with PTSD and high intrusion symptoms at baseline and downregulated in participants with improved PTSD symptoms following treatment. Pathway analysis identified upregulated immune response systems and metabolic networks with a NF-kB hub, which were downregulated with symptom reduction. Molecular biomarkers implicated in intrusion symptoms and PTSD symptom improvement may inform the development of therapeutic targets for precise treatment of PTSD.
Keywords: posttraumatic stress disorder, military, cluster analysis, flashbacks, intrusion symptoms, transcriptome-wide, gene expression, inflammation, immune response, metabolic processes
1. Introduction
Posttraumatic stress disorder (PTSD) emerges in response to trauma exposure, with subsequent symptoms of reexperiencing the traumatic event in the form of intrusive recollections, flashbacks, or nightmares; persistent avoidance of stimuli associated with the traumatic event; negative alterations in cognition and mood; as well as a constant state of heightened alertness and increased arousal.[1] PTSD lifetime prevalence is estimated at 7.8% and doubles in military populations; it presents a significant public health concern due to the high rates of comorbid psychiatric disorders, sleep disorders, and physical health conditions.[2] It remains largely unknown why a subset of trauma exposed individuals develop PTSD, but likely involves changes in the molecular mechanisms that regulate stress susceptibility and resiliency.[3]
Transcriptome-wide analysis of differential gene expression allows for the unbiased identification of genes, pathways, and proteins implicated in the pathophysiology of PTSD. A recent mega-analysis synthesized extant data from five transcriptome-wide peripheral blood studies encompassing seven types of trauma in 229 PTSD participants and 311 controls.[4] Transcriptional dysregulation converged on the innate immune, cytokine, and type I interferon pathways; however, no discrete genes overlapped between all five studies and only two genes (IFI44L and GNG11) overlapped between the three combat exposure studies.[4] While these findings provide a broad view of immune dysregulation in PTSD, the results are inconclusive and little is known about the gene expression differences attributed to each PTSD symptom cluster. This is an important consideration when identifying diagnostic and treatment response markers in highly comorbid populations with mental and physical health conditions that share symptoms. Furthermore—with the exception of increased FKBP5 expression following cognitive behavioral therapy[5]—most PTSD research has investigated group differences at a single timepoint, leaving much to be understood about the genes and pathways implicated in treatment response and symptom reduction.
The current study aimed to address these limitations by analyzing gene expression differences between a homogeneous cohort of military service members with vs. without PTSD, as well as with high vs. low PTSD cluster symptom severity: intrusions, avoidance/numbing, and arousal. Additionally, follow-up data were used to examine gene expression changes associated with symptom reduction, following evidenced-based sleep treatment, to identify genes and pathways that may signify novel treatment targets. In doing so, we utilized a transcriptome-wide analysis of differential gene expression with subsequent gene ontology-based pathway analysis.
2. Methods and Materials
2.1. Study Design
This study was part of a larger study of US military service members presenting with sleep disturbance.[6] Participants underwent a sleep medicine and psychiatric evaluation and had blood drawn at baseline and at 12-week follow-up. At baseline, participants diagnosed with insomnia received 4 to 8 biweekly sessions of cognitive behavioral therapy for insomnia (CBT-i), and participants diagnosed with obstructive sleep apnea (OSA) received automatic positive airway pressure (APAP) therapy; participants with complex insomnia (i.e., comorbid insomnia and OSA) received CBT-i and APAP combination therapy. Our previous research found PTSD symptom reduction following treatment for sleep disturbance.[7] The Madigan Army Medical Center, institutional review board approved the study (#210090), and written informed consent was obtained prior to all study procedures.
2.2. Participant Characterization
The PTSD Checklist-Military version (PCL-M) was used to assess for posttraumatic symptom severity. This 17-item inventory includes one total score and 3 symptom cluster subscores. Total scores range from 17 (lowest severity) to 85 (highest severity). Participants with a baseline PCL-M score ≥50 formed the PTSD group (n=39) and participants with a baseline PCL-M score ≤25 formed the control group (n=27); these are the suggested cut-points for military prevalence.[8] The PTSD group was further divided into two groups: PTSD improved (baseline to follow-up PCL-M change score ≥5) and PTSD not-improved (baseline to follow-up PCL-M change score ≤0). A change score ≥5 is determined to be reliable and not due to chance.[9] (CONSORT Diagram, Supplementary Fig. 1).
2.3. Covariates
To build a parsimonious ANOVA model that would be generalizable, each gene expression analysis included dichotomous covariates of age (18-35 years/36-55 years), sex (male/female), race (white/non-white), mild traumatic brain injury (mTBI) (yes/no), and complex insomnia (yes/no). The Warrior Administered Retrospective Casualty Assessment Tool (WARCAT) was used to diagnose mTBI. A positive mTBI diagnosis was made if the participant indicated any loss of consciousness, loss of memory, and/or alteration in mental state due to the head being struck by or striking an object, or the brain undergoing an acceleration/deceleration. A complex insomnia diagnosis was made in accordance with the International Classification of Sleep Disorders, 2nd edition.[10]
2.4. Sample and Data Acquisition
Non-fasting venous blood was collected into PAXgene 2.5 mL tubes (PreAnalytiX Inc.; Hombrechtikon, Switzerland). Tubes were inverted 10 times, placed at room temperature for two hours, frozen at −20°C, and stored at −80°C until RNA extraction using the PAXgene Blood RNA Kit (PreAnalytiX Inc.; Hombrechtikon, Switzerland). Concentration and quality of extracted RNA were assessed using the NanoDrop DN-1000 (Thermo Fisher Scientific; Wilmington, DE, USA) and the 2100 Bioanalyzer (Agilent Technologies Inc.; Santa Clara, CA, USA); all samples met or exceeded an RNA Integrity Number of 7.0. Samples were reverse transcribed using the GeneChip 3’ IVT Expression Kit, converted to biotinylated cRNA, and hybridized at 45°C for 16 hours to Affymetrix HG-U133 Plus 2.0 microarrays (Affymetrix Inc.; Santa Clara, CA, USA). Microarrays were stained, washed, and fluorescent images were obtained using the GeneChip Scanner 3000 7G (Affymetrix Inc.; Santa Clara, CA, USA). Procedures were undertaken per manufacture protocols.
2.5. Statistical Analysis
Demographic, military, and clinical variables were analyzed with SPSS Statistics for Mac, version 23.0 (IBM Corp.; Armonk, NY, USA). Two-tailed t-tests were used for continuous variables and chi-square tests for categorical data; p-values ≤.05. PCL-M symptom cluster subgroups were created using equal 33.33 percentile cut-points, whereby participants who endorsed the highest symptoms (high 1/3) were compared with participants who endorsed the lowest symptoms (low 1/3).
2.6. Gene Expression Analysis
Expression values were computed from raw Affymetrix CEL files using Partek Genomics Suite 6.6 (Partek Inc.; St. Louis, MO, USA). Robust Multi-array Average normalization was used, which includes background correction, quantile normalization, median polish summarization, and batch effect control. ANOVA was used to compare baseline expression data between groups and paired t-tests were used to compared baseline to follow-up gene expression changes within groups. Gene lists were generated based on a criterion of ± 2.0-fold change magnitude using a false discovery rate (FDR) ≤.05. This criterion was determined by a power analysis indicating that the given ANOVA at a ± 2.0-fold change magnitude, would require at least 35 samples to achieve a 90% confidence level.[11] All gene lists were uploaded into the Ingenuity Pathway Analysis, Core Analysis version June 11, 2018 (IPA, QIAGEN; Redwood City, CA, USA). The Human Genome U133A 2.0 Array reference set was used and considered both direct and indirect relationships when confidence was experimentally observed. P-values were calculated using the right-tailed Fisher’s exact test. All microarray data are Minimum Information about a Microarray Experiment compliant and accessible through NCBI’s Gene Expression Omnibus Series accession number GSE81761.
3. Results
3.1. Baseline Gene Expression Differences Between PTSD vs. Controls
There were no significant differences in demographics, body mass index (BMI), military characteristics, and blood withdrawal times between the PTSD (n=39) and control (n=27) groups. Prevalence of complex insomnia and mTBI were greater in the PTSD group compared with controls x2=9.031; p=0.003 and x2=32.125; p=<0.001 respectively. The PTSD severity mean (SD) was 61.3 (7.7) in the PTSD group and 21.7 (2.7) in the control group t=−25.552; p=<0.001. (Demographic Table, Supplementary Table 1). Expression analysis revealed 98 probesets, representing 89 distinct genes, and 4 uncharacterized genes, differentially expressed between participants with PTSD and controls (Supplementary Table 2). Most probesets (94%) were upregulated in the PTSD group, including RBAK and CENPK, which had a ≥2.5-fold change magnitude; FDR ≤.05.
3.2. Baseline Gene Expression Differences Between High vs. Low Intrusion Symptoms
Expression analysis revealed 1,327 probesets, representing 1,040 distinct genes, and 45 uncharacterized genes, differentially expressed between participants with high intrusion (n=22) vs. low intrusion (n=24) symptoms (Supplementary Table 3). Most probesets (98%) were upregulated in the high intrusion group, including RSL24D1, CETN3, ZNF117, SCOC, NDUFA5, TFEC, ZCCHC10, COMMD6, CD69, and CENPK, which had a ≥3.5-fold change magnitude; FDR ≤.05. There were no differentially expressed probesets between participants with high avoidance/numbing (n=20) vs. low avoidance/numbing (n=22) symptoms, or high arousal (n=15) vs. low arousal (n=23) symptoms. All between group demographics, BMI, military characteristics, and blood withdrawal times were non-significant. All participants in the high intrusion group were also in the PTSD group, and all participants in the low intrusion group were also in the control group; however, the middle third percentile consisted of both PTSD participants and controls.
3.3. Gene Expression Changes Associated with PTSD Symptom Improvement
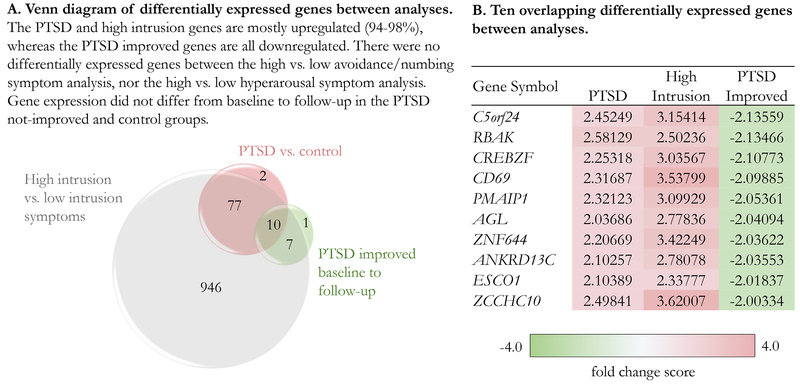
Expression analysis revealed 20 probesets, representing 18 distinct genes, and 1 uncharacterized gene, differentially expressed in the PTSD improved group (n=12) from baseline to follow-up (Supplementary Table 4). All probesets were downregulated at follow-up and 10 of the probesets overlapped with the PTSD vs. control gene list and the high intrusion vs. low intrusion gene list, but with an inverse relationship (Fig. 1). There were no differentially expressed probesets in the PTSD not-improved group (n=11) or the control group (n=20) from baseline to follow-up. There were no baseline gene expression differences between the PTSD improved and PTSD not-improved groups using a less conservative ANOVA with a ± 1.0-fold change magnitude; FDR ≤ .05. Between group demographics, BMI, military characteristics, and blood withdrawal times were non-significant.
Fig. 1. PTSD associated differential gene expression.
A. Venn diagram of differentially expressed genes between analyses. The PTSD and high intrusion associated genes were mostly upregulated (94% and 98%), whereas the PTSD improved associated genes were all downregulated. B. Ten overlapping differentially expressed genes between analyses with fold change score.
3.4. Ingenuity Pathway Analysis
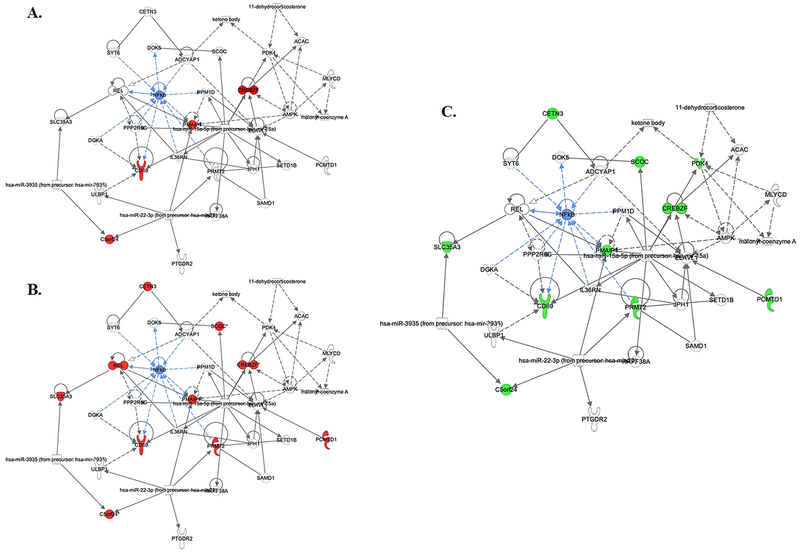
All three gene lists were uploaded into Ingenuity Pathway Analysis to compute the top physiological systems and networks associated with each comparison (Table 1). Briefly, immune response and immune cell trafficking systems were upregulated in the PTSD and high intrusion groups at baseline, and downregulated in the PTSD improved group at follow-up. Networks of lipid metabolism, nucleic acid metabolism, and small molecule biochemistry, with a nuclear factor kappa-B (NF-kB) hub were linked to PTSD symptom improvement (Fig. 2.).
Table 1. Ingenuity Pathway Analysis.
Legend for color-coding of physiological systems and networks: red text = comprised solely of upregulated genes, green text = comprised solely of downregulated genes, black text = comprised of both upregulated and downregulated genes.
| PTSD vs. Control (92 upregulated and 6 downregulated probesets) |
High Intrusion vs. Low Intrusion (1307 upregulated and 20 downregulated probesets) |
PTSD Improved (baseline to follow-up) (0 upregulated and 20 downregulated probesets) |
|||
|---|---|---|---|---|---|
| Top Physiological Systems (p-value range) | |||||
| Cell-mediated Immune Response | 4.75E-03-4.75E-03 | Tissue Development | 4.90E-02-6.79E-04 | Cell-mediated Immune Response | 2.61E-02-1.06E-03 |
| Embryonic Development | 4.43E-02-4.75E-03 | Immune Cell Trafficking | 4.30E-02-2.38E-03 | Hematological System Function | 4.95E-02-1.06E-03 |
| Hematological System Function | 3.74E-02-4.75E-03 | Humoral Immune Response | 4.30E-02-2.58E-03 | Hematopoiesis | 2.61E-02-1.06E-03 |
| Hematopoiesis | 2.35E-02-4.75E-03 | Hematological System Function | 4.90E-02-3.12E-03 | Immune Cell Trafficking | 1.92E-02-1.06E-03 |
| Immune Cell Trafficking | 2.35E-02-4.75E-03 | Digestive System Function | 3.16E-03-3.16E-03 | Lymphoid Structure | 2.61E-02-1.06E-03 |
| Top Networks (score) | |||||
| 1. Cardiovascular Disease, Developmental Disorder, Endocrine System Disorders | 43 | 1. RNA Post-Transcriptional Modification, Cellular Assembly, Developmental Disorder | 46 | 1. Lipid Metabolism, Nucleic Acid Metabolism, Small Molecule Biochemistry | 25 |
| 2. Cell Death and Survival, Behavior, Cell Cycle | 32 | 2. RNA Damage and Repair, Cellular Development | 46 | 2. Cell Cycle, Connective Tissue Development, Cancer | 19 |
| 3. Cellular Growth and Proliferation, Endocrine System Disorders, Gastrointestinal Disease | 24 | 3. Protein Synthesis, RNA Post-Transcriptional Modification, Hematological Disease | 43 | ||
Fig. 2. The lipid metabolism, nucleic acid metabolism, and small molecule biochemistry network with NF-kB hub.
A. Four upregulated genes associated with PTSD (vs. control). B. Ten upregulated genes associated with high intrusion (vs. low intrusion) symptoms. C. Ten downregulated genes associated with PTSD symptom improvement from baseline to follow-up.
4. Discussion
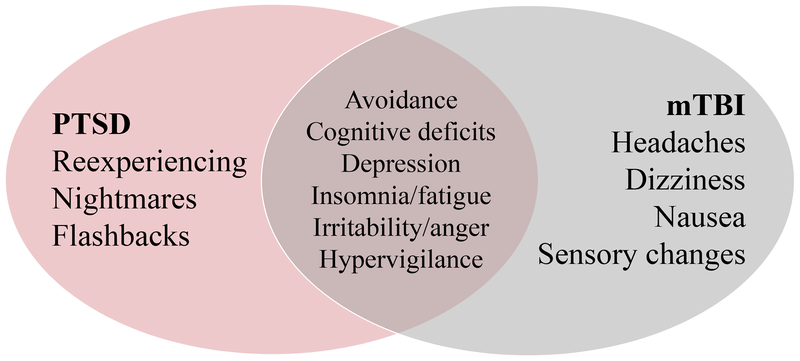
We found that between group (PTSD vs. control) gene expression differences were almost entirely attributed to the intrusion symptom cluster (98%), and there were no gene expression differences attributed to the other symptom clusters. One possible explanation for this finding is that intrusion symptoms (e.g., reexperiencing, nightmares, and flashbacks) are hallmark symptoms of PTSD, whereas pathological symptoms of avoidance, negative alterations in cognition and mood and arousal overlap with other conditions prevalent in military cohorts, most notably mTBI (Fig. 3). If participants endorse PTSD-like symptoms that are attributed to comorbid conditions—for example, cognitive deficits sustained from a mTBI—gene expression profiles may be represented by different genes and pathways and therefore mask group differences.
Fig. 3. Venn diagram of symptom overlap between posttraumatic stress disorder (PTSD) and mild traumatic brain injury (mTBI).
Intrusion symptoms (e.g., reexperiencing, nightmares, and flashbacks) are specific to PTSD. Whereas avoidance/numbing symptoms (e.g., avoidance of people, places, and things that are trauma reminders and cognitive and mood alterations) and arousal symptoms (e.g., insomnia, irritability, and increased startle) overlap with other conditions.
Another possible explanation for these findings is that trauma exposure incites gene expression changes implicated in intrusion symptoms, and the other symptom clusters develop subsequently in response. For example, physiological reactivity to trauma cues (i.e., reexperiencing) could cause secondary avoidance, insomnia, irritability, and hyperarousal symptoms. We also found that grouping based on high vs. low intrusion symptoms yielded a more robust gene expression profile than grouping based on PTSD vs. control. It’s possible that the PTSD group included false positive cases—participants who met the cut-off score by endorsing PTSD-like symptoms attributed to comorbid conditions. We previously reported that brain volume alterations were unrelated to overall PTSD severity, but inversely related to intrusion symptom scores.[12] Taken together, these findings suggest that intrusion symptoms are important to consider when investigating biomarkers in highly comorbid populations with symptom overlap.
We identified ten genes upregulated in participants with PTSD and high intrusion symptoms at baseline, which were downregulated in participants with improved PTSD symptoms following treatment. Of note, RBAK, a DNA transcription regulator, was the top upregulated gene associated with PTSD and second to top downregulated gene linked to symptom improvement. Decreased expression levels of RBAK were identified in the hippocampus of ‘resilient’ mice, which may point to its role in memory processes.[13] CD69, a gene involved in lymphocyte proliferation and cellular signal transmission, was remarkably upregulated in participants with high intrusion symptoms—an analysis we employed to control for overlapping mTBI symptoms. A transcriptome mega-analysis identified upregulation of CD69 in men with interpersonal trauma, but not combat trauma.[4] These discrepancies may reflect different cohort characteristics (e.g., type of trauma, comorbidities) that may influence gene expression profiles. Lastly, C5orf24 was the most downregulated gene associated with symptom improvement. Prior research has linked DNA methylation levels of C5orf24 to negative affect scores in drug addicts.[14]
The IPA converged on immune response as the most reliably upregulated gene-ontology system characterizing a PTSD diagnosis and high intrusion symptoms. These findings are in line with our previous transcriptome work[15] and those of others.[4] However, our current findings extend these results by showing that PTSD symptom reduction results in a downregulation of these same immune response systems, in addition to lipid metabolism networks. This reflects our prior results indicating normalized levels of inflammatory and metabolic biomarkers in women who recovered from PTSD.[16] PTSD symptom severity was also related to networks with a NF-kB hub. NF-kB is a linchpin in the immune system, a reported requisite for memory reconsolidation,[17] and is positively correlated with PTSD symptom severity in women with childhood abuse trauma.[18]
This study has a number of limitations including the small sample size; however, efforts were made to match participants with (vs. without) PTSD on important potentially confounding variables. Still, replication in larger, more diverse samples, including civilian cohorts is needed. Moreover, microarray is limited in dynamic range, specificity and sensitivity, novel transcript detection, and low-abundance transcript detection. Findings can potentially be confounded by a lack of epigenetic assays to address causative relationships of the differentially expressed genes.
5. Conclusions
Although prior research has identified putative risk factors that are associated with PTSD, the identification of discrete diagnostic biomarkers and therapeutic targets remains elusive. This study reported significant differentially expressed genes in individuals with high intrusion symptoms, which highlights the potential utility of this type of analysis in highly comorbid populations. Further classification of gene expression changes underlying treatment response and symptom reduction could lead to precision care initiatives and the development of new and effective therapeutics.
Supplementary Material
Fig. 1. CONSORT Diagram. This figure depicts the sample size starting at screening and continuing through baseline, follow-up, and analysis; the number of participants excluded at each stage of the study is provided.
Table 1: Baseline demographics, military, and clinical characteristics.
Table 2: Summary of 98 probesets, representing 89 distinct genes, and 4 uncharacterized genes that were differentially expressed in PTSD (n=39) participants compared with controls (n=27) at baseline.
Table 3: Summary of 1327 probesets, representing 1040 distinct genes, and 45 uncharacterized genes that were differentially expressed in participants with high intrusion (n=22) compared with low intrusion (n=24) symptoms at baseline
Table 4: Summary of 20 probesets, representing 18 distinct genes, and 1 uncharacterized gene that were differentially expressed in participants with improved PTSD symptoms from baseline to follow-up (n=12)
Highlights.
Gene expression differences were almost entirely attributed to the intrusion symptom cluster.
There were no gene expression differences attributed to the other PTSD symptom clusters.
PTSD-like symptoms that are due to comorbid conditions may be represented by different genes
Immune response systems and metabolic networks with a NF-kB hub were upregulated in PTSD and downregulated with symptom reduction.
7. Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The work was performed at the National Institutes of Health (NIH), Bethesda, MD, USA and received publication clearance by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest: none
References
- 1.Association, American Psychiatric. Diagnostic and statistical manual of mental disorders (American Psychiatric Publishing: Arlington, VA: ). [Google Scholar]
- 2.Kessler RC, et al. , Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry, 1995. 52(12): p. 1048–60. [DOI] [PubMed] [Google Scholar]
- 3.Girgenti MJ, et al. , Molecular and Cellular Effects of Traumatic Stress: Implications for PTSD. Curr Psychiatry Rep, 2017. 19(11): p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breen MS, et al. , PTSD Blood Transcriptome Mega-Analysis: Shared Inflammatory Pathways across Biological Sex and Modes of Trauma. Neuropsychopharmacology, 2018. 43(3): p. 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabo C, Kelemen O, and Keri S, Changes in FKBP5 expression and memory functions during cognitive-behavioral therapy in posttraumatic stress disorder: a preliminary study. Neurosci Lett, 2014. 569: p. 116–20. [DOI] [PubMed] [Google Scholar]
- 6.Mysliwiec V, et al. , Sleep disorders in US military personnel: a high rate of comorbid insomnia and obstructive sleep apnea. Chest, 2013. 144(2): p. 549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rusch HL, et al. , Improved sleep quality is associated with reductions in depression and PTSD arousal symptoms and increases in IGF-1 concentrations. J Clin Sleep Med, 2015. 11(6): p. 615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.“Using the PTSD Checklist (PCL).” In. 2012. VA National Center for PTSD, 1–3. [Google Scholar]
- 9.Monson CM, et al. , Change in Posttraumatic Stress Disorder Symptoms: Do Clinicians and Patients Agree? Psychological AssessmeT, 2008. 20(2): p. 131–138. [DOI] [PubMed] [Google Scholar]
- 10.Medicine, American Academy of Sleep. 2005. International Classification of Sleep Disorders: Diagnostic and Coding Manual. Second edition (American Academy of Sleep Medicine: Westchester, IL: ). [Google Scholar]
- 11.Muller KE, et al. , Power Calculations for General Linear Multivariate Models Including Repeated Measures Applications. J Am Stat Assoc, 1992. 87(420): p. 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroes MC, et al. , Association between flashbacks and structural brain abnormalities in posttraumatic stress disorder. Eur Psychiatry, 2011. 26(8): p. 525–31. [DOI] [PubMed] [Google Scholar]
- 13.Mozhui K, et al. , Strain differences in stress responsivity are associated with divergent amygdala gene expression and glutamate-mediated neuronal excitability. J Neurosci, 2010. 30(15): p. 5357–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lax E, et al. , A DNA Methylation Signature of Addiction in T Cells and Its Reversal With DHEA Intervention. Front Mol Neurosci, 2018. 11: p. 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guardado P, et al. , Altered gene expression of the innate immune, neuroendocrine, and nuclear factor-kappa B (NF-kappaB) systems is associated with posttraumatic stress disorder in military personnel. J Anxiety Disord, 2016. 38: p. 9–20. [DOI] [PubMed] [Google Scholar]
- 16.Gill JM, et al. , Women in recovery from PTSD have similar inflammation and quality of life as non-traumatized controls. J Psychosom Res, 2013. 74(4): p. 301–6. [DOI] [PubMed] [Google Scholar]
- 17.Si J, et al. , Activation of NF-kappaB in basolateral amygdala is required for memory reconsolidation in auditory fear conditioning. PLoS One, 2012. 7(9): p. e43973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pace TW, et al. , Increased peripheral NF-kappaB pathway activity in women with childhood abuse-related posttraumatic stress disorder. Brain Behav Immun, 2012. 26(1): p. 13–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. 1. CONSORT Diagram. This figure depicts the sample size starting at screening and continuing through baseline, follow-up, and analysis; the number of participants excluded at each stage of the study is provided.
Table 1: Baseline demographics, military, and clinical characteristics.
Table 2: Summary of 98 probesets, representing 89 distinct genes, and 4 uncharacterized genes that were differentially expressed in PTSD (n=39) participants compared with controls (n=27) at baseline.
Table 3: Summary of 1327 probesets, representing 1040 distinct genes, and 45 uncharacterized genes that were differentially expressed in participants with high intrusion (n=22) compared with low intrusion (n=24) symptoms at baseline
Table 4: Summary of 20 probesets, representing 18 distinct genes, and 1 uncharacterized gene that were differentially expressed in participants with improved PTSD symptoms from baseline to follow-up (n=12)