Abstract
Purpose:
The genomic landscape of gliomas has been characterized and now contributes to disease classification, yet the relationship between molecular profile and disease progression and treatment response remain poorly understood.
Experimental Design:
We integrated prospective clinical sequencing of 1,004 primary and recurrent tumors from 923 glioma patients with clinical and treatment phenotypes.
Results:
13% of glioma patients harbored a pathogenic germline variant, including a subset associated with heritable genetic syndromes and variants mediating DNA repair dysfunctions (29% of the total) that were associated with somatic biallelic inactivation and mechanism-specific somatic phenotypes. In astrocytomas, genomic alterations in effectors of cell-cycle progression correlated with aggressive disease independent of IDH-mutation status, arose preferentially in enhancing tumors (44% vs. 8%, P<0.001), were associated with rapid disease progression following tumor recurrence (HR=2.6, P=0.02), and likely preceded the acquisition of alkylating therapy-associated somatic hypermutation. Thirty-two percent of patients harbored a potentially therapeutically actionable lesion, of whom 11% received targeted therapies. In BRAF-mutant gliomas, response to agents targeting the RAF/MEK/ERK signaling axis was influenced by the type of mutation, its clonality, and its cellular and genomic context.
Conclusions:
These data reveal genomic correlates of disease progression and treatment response in diverse types of glioma and highlight the potential utility of incorporating genomic information into the clinical decision-making for patients with glioma.
Keywords: neuro-oncology, genomics, precision medicine, glioma, glioblastoma
Translational Relevance
Gliomas comprise a group of primary brain tumors of considerable molecular and clinical heterogeneity. We hypothesized that prospective molecular characterization in the clinical care of gliomas can reveal correlates of key clinical disease states and therapeutic response. The genomic correlates of malignant progression and response to molecularly targeted therapies was context-specific. In lower-grade gliomas, genetic alterations in genes related to cell-cycle control were associated with tumor progression, contrast enhancement on brain imaging, and poor prognosis at recurrence. The susceptibility of tumors to therapy-induced hypermutation varied by glioma subtype and was highest in IDH-mutant tumors. Clinical responses to molecularly targeted treatments, typified by those targeting mutant BRAF, correlated with mutant allele type, clonality, and glioma subtype. Collectively, integrative clinico-genomic characterization of recurrent gliomas can begin to unravel the clinical and therapeutic complexity of glioma.
Introduction
Gliomas are a clinically diverse collection of primary brain tumors. Molecular characterization of previously untreated gliomas has established the landscape of genomic alterations (1–5). Mutations in the metabolic genes Isocitrate dehydrogenase (IDH) 1 and 2 and co-deletion of chromosome arms 1p and 19q stratify prognostically distinct disease subgroups and have therefore recently been incorporated into the World Health Organization (WHO) classification of diffuse glioma (2,6). IDH-wildtype (IDH-WT) gliomas follow a distinct molecular pathogenesis from IDH-mutant gliomas and are more aggressive (7). However, significant clinical heterogeneity exists within all molecular subgroups of adult diffuse glioma.
To date, the molecular attributes of recurrent gliomas remain less well understood. Initial studies have begun to unravel temporal and evolutionary dynamics of this process, and heterogeneity exists in the mutational evolution of both IDH-mutant and IDH-WT gliomas (8,9). Moreover, exogenous pressures such as therapy can drive unique evolutionary trajectories (10). Nevertheless, the specific aberrations in key genes and pathways that drive aggressive clinical phenotypes and treatment response remain largely unexplored in glioma patients.
We prospectively characterized the molecular alterations of 923 patients with glioma, many of which were acquired after disease recurrence or initiation of treatment. We integrated these genomic data with clinical and treatment phenotypes to identify genomic aberrations that were associated with clinical behavior, genomic evolution on therapy, and response to therapy.
Materials and Methods
Patient cohort and clinical details
1,004 tumor specimens and their matched normal controls were acquired from 923 adult patients that underwent prospective genomic profiling as part of their routine care during the time period July 2013 to July 2017. This study was approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board (IRB) and was conducted in accordance with International Ethical Guidelines for Biomedical Research Involving Human Subjects, Good Clinical Practice guidelines, the Declaration of Helsinki, and local laws. All patients provided written informed consent for sequencing.
Gliomas were classified using the WHO 2016 Classification of Tumors of the Central Nervous System (6). An integrated diagnosis was established by incorporating histologic features of the tumor and class-defining molecular features from the sequenced resection. Ten tumors defied classification and received a descriptive diagnosis. Clinical annotation was obtained for all patients and included 1) date of initial diagnosis; 2) date of patient death and/or date of last contact with hospital; 3) surgical interventions; 4) lines of systemic treatment and radiotherapy; and 5) date of first progression as determined by RANO criteria for patients with sequenced primary specimens (Tables S1–3).
For 392/447(88%) sequenced specimens from patients with WHO grade II and III oligodendrogliomas, WHO grade II and III astrocytomas, or IDH-mutant glioblastomas (GBM), the presurgical MRI was available and was reviewed whether it included a nodular area of contrast enhancement. Radiographic responses were also determined by a neuro-radiologist (R.J.Y.) without knowledge of the genomic information or clinical history. Volumetric analysis was performed using iNtuition 4.4.13 (TeraRecon, Foster City, CA). Contrast enhanced T1-weighted images were examined and tumors were contoured slice-by-slice to construct the tumor volumes. Response criteria were defined as progressive disease at more than 25% tumor volume increase summed across all lesions; stable disease at less than 25% increase; partial response at 25% to 85% decrease; and near-complete response if the tumor volume decreased by at least 85%. Progression-free survival was calculated from the time of surgery of the sequenced resection using RANO criteria (11).
Genomic sequencing and analysis
Tumor DNA was extracted from formalin-fixed paraffin embedded (FFPE) specimens. Normal DNA was extracted from mononuclear cells from patient-matched peripheral blood. Specimens underwent sequencing in CLIA-certified environments using either MSK-IMPACT (n=837, paired tumor-normal sequencing) or FoundationOne (n=167, unmatched tumor sequencing) sequencing platforms (Table S2). Both platforms cover protein-coding exons of cancer-associated genes, as previously described (12–14), and detect somatic single-nucleotide variants, insertions, deletions, copy-number alterations, and certain gene fusions and structural variants. All variants identified by MSK-IMPACT were manually reviewed. Genes included on each version of the platforms are listed in Table S4.
For detection of co-deletion of chromosome arms 1p and 19q in samples sequenced on MSK-IMPACT, allele-specific DNA copy number analysis was performed using FACETS (www.github.com/mskcc/facets, version 0.3.9). The co-deletion was called if at least 75% of both chromosome arms were lost in a sample. Similarly, for detecting concurrent chromosome 7 gain and chromosome 10 loss, any sample with at least 90% of each chromosome affected was considered altered, and all determinations were manually reviewed for accuracy. Co-deletion of 1p19q in samples sequenced on the FoundationOne assay was detected using a read-coverage based method for copy number detection as previously described. Briefly, in these samples copy-number was inferred at the regions of arms 1p, 1q, 19p, and 19q commonly bound in FISH 1p19q co-deletion assays (1p chr1:3402506–3775797; 1q chr1:178816670–179434670; 19p chr19:12380625–12779591; 19q chr19:48109453–48348586). Samples harbored 1p19q co-deletion if the inferred copy number at the 1p and 19q regions was less than or equal to half of the copy number at 1q and 19p regions. The clonality of BRAF mutations was estimated from MSK-IMPACT samples as previously described (15). Briefly, allele-specific copy number was used to estimate local BRAF-spanning copy number and tumor purity, with which the most likely fraction of cancer cells harboring the mutation was derived.
Germline analysis
Pathogenic and likely pathogenic (P/LP) germline variants were identified in all patients sequenced with MSK-IMPACT. The pipeline for germline variant detection utilized here was as previously described (16). All patients were anonymized prior to analysis except for 32 patients who had consented for identified germline analysis, whose pathogenic or likely pathogenic variants were signed out in their electronic medical record by clinical geneticists and referred to genetic counseling. Germline variants identified in all patients were subsequently classified as P/LP if rare in the general population (<0.0003 minor allele frequency across subpopulations in the non-TCGA subset of ExAC, nor more than 5 observations with the cohort) and fulfilling one of the following criteria: 1) signed out as P/LP in identified germline analysis in any other patient and cancer type within our institutional initiative; 2) considered pathogenic by ClinVar with no conflicting curation; 3) predicted to result in the truncation of a tumor-suppressor gene; or 4) occurring at a somatic hotspot residue in an oncogene. Tumor-suppressor genes and oncogenes were defined by OncoKB (17) as of March 2018. Those variants presumed attributable to clonal hematopoiesis (CH) were identified by the following criteria and excluded from pathogenicity assessment: 1) mutations occurring in non-CH gene with a mutant allele fraction in the normal sample <0.25 or <0.35 for indel and single-nucleotide variants, respectively; 2) mutations occurring in CH-associated genes ATM, DNMT3A, JAK2, SF3B1, SRFS2, STAG2, or TET2 with a somatic mutant allele fraction in the tumor less than the expected minimum ploidy-corrected allele frequency (corrected by a factor of 0.35). Finally, the CH-associated variant JAK2 V617F was removed in all cases it was observed (18). For P/LP germline variants in an established tumor-suppressor gene, biallelic loss was determined in the corresponding tumor specimen by determining the somatic zygosity of the locus and/or presence of a likely oncogenic somatic mutation in the same gene.
Somatic variant classification
Genomic variants were classified as oncogenic or likely oncogenic by the following criteria: 1) annotation with OncoKB (www.github.com/oncokb/oncokb-annotator, accession Nov, 2017); and 2) analysis of recurrent mutations across a collection of a total of 3,130 glioma samples (see External data sources) (19,20). Unless otherwise stated, all analyses described herein utilize only variants considered oncogenic or likely oncogenic according to these criteria. Levels of clinical actionability were similarly annotated using OncoKB. Only those actionable alterations of levels 1–3 evidence were considered, which include FDA-approved and standard-of-care biomarkers for drugs approved in the treatment of gliomas or other indications, as well as those with compelling clinical evidence predicting response in any indication. Alterations with only biological evidence supporting efficacy (OncoKB level 4), which includes EGFR amplifications, were excluded from consideration at this time. After the data freeze for analysis, IDH1 and IDH2 mutations were upgraded to OncoKB level 2B for glioma due to the approval of IDH-directed therapy (Ivosidenib) in acute myeloid leukemias, but to ensure analytical consistency, these remain level 4 for the purposes of present analysis.
Alkylator-induced hypermutation
Mutational signatures were inferred using single-nucleotide mutations for any sample with greater than or equal to five somatic mutations using a method that assigns the fraction of mutations attributable to known mutational signatures (21) using a basin-hopping algorithm (www.github.com/mskcc/mutation-signatures). For each sample, the mutation burden was determined as the number of mutations per Mb of capture targets. Samples for which at least 50% of mutations were attributable to signature 11 (temozolomide-associated signature) and the mutation burden exceeded 1.5 median absolute deviations from the median were called hypermutated due to alkylator exposure. The latter criterion was applied separately for samples sequenced on MSK-IMPACT and FoundationOne, respectively.
MGMT promoter methylation
MGMT promoter methylation was assessed on the primary or recurrent surgical specimen per standard clinical testing procedures in CLIA-certified environments. The majority of samples underwent local assessment at Memorial Sloan Kettering Cancer Center either by pyrosequencing or methylation-specific real-time PCR after sodium bisulfite conversion. For patient-level methylation status (available for 636 patients), borderline methylation of the promoter was deemed negative, and patients with discordant results from two time points were deemed methylation positive.
External data sources
Publicly available glioma mutational data were acquired from AACR GENIE (22) (498 samples, www.cbioportal.org/genie), The Cancer Genome Atlas (1,2) and Foundation Medicine (894 and 739 samples, respectively, both www.portal.gdc.cancer.gov). For standardization and cross-platform comparisons, all somatic mutations were re-annotated with VEP (version 88) using vcf2maf (www.github.com/mskcc/vcf2maf, version 1.6.14).
Statistical analyses
All statistical analyses were carried out using the R statistical programming environment, using base functions for statistical testing and the survival package for outcome analyses. Differences in proportions were tested using two-sided Fisher’s exact test. P-values adjusted for multiple hypotheses are indicated throughout the text. The log-rank test was used for assessing outcome differences across defined groups, and Cox proportional hazards modeling was used to determine the significance of predictors in multivariable analyses. Any comparisons of gene- or variant-level counts were carried out on a per-patient basis to account for patients for whom multiple sequenced specimens exist.
Data availability
The prospective somatic mutational and clinical data have been deposited for visualization and analysis in the cBioPortal for Cancer Genomics (www.cbioportal.org/study?id=glioma_mskcc_2019).
Results
Somatic and germline genomic landscape of clinically sequenced gliomas
To identify genomic alterations underlying aggressive and recurrent disease, we performed prospective sequencing of 1,004 tumors from 923 glioma patients (Fig. 1A, Tables 1, S1). The cohort comprised 355 IDH-mutant (IDH1 R132, IDH2 R140/R172) and 649 IDH-wildtype (WT) tumors classified based on histology and molecular features according to WHO 2016 guidelines (6) (Table 1, S1). We also curated clinical information, outcomes, and treatment data for all patients to identify treatment-associated alterations and explore tumor evolution on systemic therapy (Fig. 1, Tables 1, S1–3).
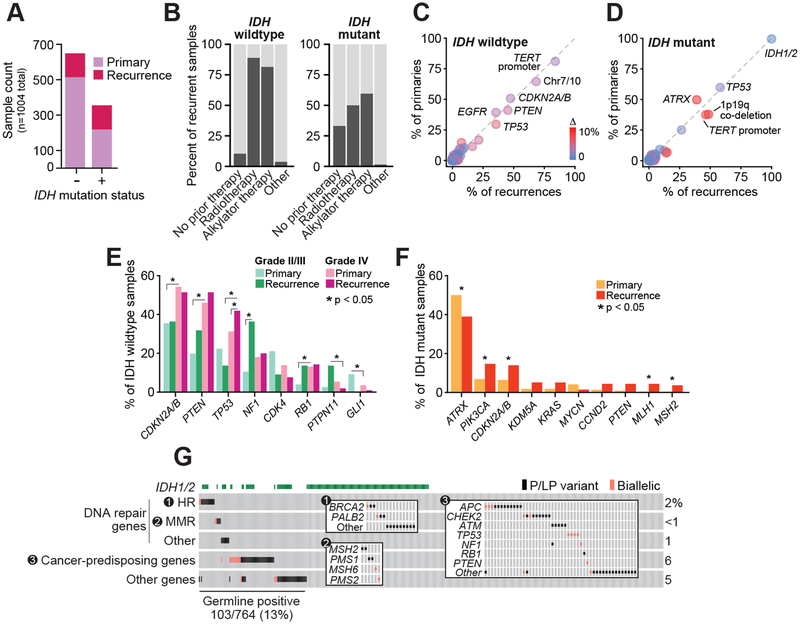
Figure 1. Clinical and genomic characteristics of prospectively characterized gliomas.
A) The number of primary (n=733; 219 IDH-mutant and 514 IDH WT) and recurrent tumor specimens (n=271; 136 IDH-mutant and 135 IDH WT) characterized in the study cohort are shown by IDH-mutation status. B) The percentage of recurrent specimens acquired at any point after one or more lines of the indicated therapeutic modality (91 of 136 of IDH-mutant and 121 of 135 IDH WT recurrences acquired after at least one line of treatment). C-D) The frequency of key glioma-associated mutations in primary and recurrent IDH-WT or IDH-mutant tumors indicate glioma subtype-defining lesions are largely stable over time and therapy. Cardinal genomic lesions are labeled, circles are colored by absolute difference in frequency between primary and recurrent tumors (IDH wildtype: n=514 and 135 primary and recurrent tumors, respectively; IDH-mutant: n=219 and 136 primary and recurrent tumors, respectively). E-F) The frequency of aberrations in those genes with differing frequencies (P<.05 or a 2.5 fold-change) in IDH WT and IDH-mutant tumors (panels E and F, respectively) across grade or sample type (primary versus recurrence). IDH WT: 80, 420 and 14 primary tumors of grades II/III, IV or mixed/indeterminate grade, respectively, and 12, 106, and 17 recurrent tumors of grades II/III, IV or mixed/indeterminate grade; IDH-mutant: 218 and 136 primary and recurrent tumors, respectively). G) The frequency and pattern of pathogenic and likely pathogenic abnormalities in the germline of patients with IDH-WT or -mutant disease is shown. Inset panels highlight specific subgroups genes by pathway of interest. Somatic biallelic loss following a germline lesion is indicated by pink squares. DNA repair genes labeled as “Other” are ATR, ERCC2, ERCC5 and MUTYH.
Table 1. Cohort characteristics.
Major clinical and diagnostic characteristics of the study cohort.
| All patients (n=923) | |
|---|---|
| Age at diagnosis (years) | |
| Median | 52 |
| Range | 11–90 |
| Sex (%) | |
| Male | 557 (60) |
| Female | 366 (40) |
| WHO Class at diagnosis (%) [a] | |
| IDH-WT | 596 |
| Glioblastoma | 468 (47) |
| Anaplastic astrocytoma | 65 (7) |
| Diffuse astrocytoma | 23 (2) |
| Gliosarcoma | 15 (2) |
| Diffuse midline glioma, H3 K27M-mutant | 5 (<1) |
| Other | 20 (2) |
| IDH-mutant | 327 |
| Oligodendroglioma | 93 (9) |
| Diffuse astrocytoma | 86 (9) |
| Anaplastic astrocytoma | 80 (8) |
| Anaplastic oligodendroglioma | 43 (4) |
| Glioblastoma | 24 (2) |
| Other | 1 (<1) |
| Prior chemotherapy (specimens)a (%) | |
| Yes | 196 (20) |
| No | 808 (80) |
| Prior radiotherapy (specimens)a (%) | |
| Yes | 191 (19) |
| No | 813 (81) |
Total tumor specimens, n=1,004.
Broadly, the cohort consisted of the full spectrum of disease including IDH-mutant grade II and III oligodendrogliomas with co-deletion of chromosomes 1p and 19q (hereafter referred to as 1p19q), IDH-mutant and IDH-WT grade II and III astrocytomas with intact 1p19q, grade IV IDH-WT and IDH-mutant GBMs, and other rare glioma subtypes. In contrast to previous studies of primary untreated gliomas (1–4), our cohort consisted of both primary and recurrent tumor samples (n=733 and 271, 73 and 27%, respectively), the latter of which were acquired after a median of one line (range 0–8) of treatment (Fig. 1B, Fig. S1). We also molecularly characterized matched pre- and post-treatment samples from 57 patients (Fig. S2).
The prevalence and co-occurrence of alterations in core glioma genes and loci (IDH1/2, CDKN2A/B, EGFR, PTEN, NF1, TP53, TERT promoter, CIC, FUBP1, ATRX, 1p19q, chromosomes 7 and 10) in this cohort of patients with advanced disease was consistent with established associations to glioma subtypes inferred from primary untreated disease (Fig. S3, Tables S5–6). Tumors from 21 patients harbored gene fusions, including 16 patients with FGFR3-TACC3 fusions and five patients with rare EGFR or BRAF kinase domain-retaining fusion events. The primary tumors in this cohort were characterized by the presence of these alterations at frequencies comparable with contemporary cohorts, such as The Cancer Genome Atlas (Fig. S4). Moreover, these genomic lesions associated with glioma subtypes largely occurred with similar frequencies in recurrent gliomas (Fig. 1C–D).
Exploring grade-specific differences among IDH-WT tumors (80 and 420 primary grade II/III and IV tumors; 12 and 106 recurrent grade II/III and IV tumors, respectively), alterations in CDKN2A/B, PTEN, and TP53 were more common in higher-grade (grade IV) disease independent of whether the affected tumor was a primary or recurrence. By contrast, alterations in NF1 and PTPN11 were more common in the recurrences of patients that started as lower grade disease (grades II and III, P<0.05; Fig. 1E). Among IDH-mutant tumors, significant differences occurred predominantly among less frequently altered genes (less than 15% of cases in either subset; Fig. 1F).
To increase our statistical power to identify novel mutational hotspots in known glioma genes such as EGFR and PDGFRA, we combined our prospective cohort with retrospectively sequenced glioma cohorts (n=3,130 sequenced gliomas) (2,3,22). This analysis identified 34 previously uncharacterized hotspot mutations in EGFR and PDGFRA as well as novel candidate driver mutations in MAP3K1 and SF3B1, two genes not previously associated with glioma (Table S7).
We next explored the incidence of pathogenic germline alleles in this prospectively characterized cohort. In total, 13% (103/764) of evaluable patients harbored at least one deleterious or likely deleterious alteration in the germline of which 54% were of high, moderate, or low penetrance. (Fig. 1G, Table S8). Notably, 28% (29/103) of all such pathogenic alleles targeted diverse effectors of DNA repair. Three patients harbored pathogenic or likely pathogenic germline BRCA2 mutations. Six patients had germline defects in mismatch repair (MMR) genes (two MSH2, two PMS1, and one each MSH6 and PMS2). Two of these six patients had tumors with somatic biallelic inactivation of the germline pathogenic allele, loss of protein expression by immunohistochemistry, and a clinical presentation consistent with Lynch syndrome. Moreover, both tumors were hypermutated with a mutational signature consistent with microsatellite instability (MSI). The other four germline MMR cases retained heterozygosity of the pathogenic germline MMR defect in their tumors and were microsatellite stable (MSS). Pathogenic germline alleles were also observed in genes involved in high penetrance heritable genetic syndromes previously reported in patients with glioma (NF1, TP53, PTEN, and RB1). There was no association between the presence of a pathogenic germline alteration and age at diagnosis nor was there an association with pathological grade at diagnosis (25/167, 15/153, and 61/432 of grade II, III and IV tumors, respectively, P=0.3) or IDH status (38/265 versus 65/499 of IDH-mutant and -wildtype tumors, respectively, P=0.7).
Genomic correlates of tumor MRI contrast enhancement and progression
Given the long natural history of lower-grade gliomas, we determined which genomic alterations were associated with malignant progression, using contrast enhancement as a surrogate for this common event in the natural history of this disease (23,24). We reviewed the pre-operative magnetic resonance imaging (MRI) associated with each resection from which the specimen was sequenced and then performed a pathway-level enrichment analysis of genetic alterations found in the corresponding tumor specimen (Table S9). In IDH-mutant tumors, genomic alterations in key effectors of the cell cycle and both the RTK/RAS and PI3K/AKT signaling pathways arose significantly more frequently in tumors with MRI contrast enhancement [n=150 non-enhancing (111 and 39 primaries and recurrences, all grade II/III) and 148 enhancing (77 and 71 primaries and recurrences, 110 grade II/III and 38 grade IV), respectively; Fig. 2A]. Aberrant effectors of cell-cycle control were the most significantly associated with enhancing disease (Bonferroni corrected P<.001). These alterations occurred almost exclusively in 1p19q-intact tumors at a frequency that could not be otherwise explained by the difference in grade (44% of 8% enhancing and non-enhancing 1p19q-intact tumors, respectively; 3% and 4% respectively in 1p19q co-deleted tumors; Fig. 2B). By contrast, PI3K/AKT and RTK-RAS alterations were found in enhancing gliomas irrespective of 1p19q status but arose preferentially in enhancing tumors with 1p19q co-deletion (P=.002; Fig. S5).
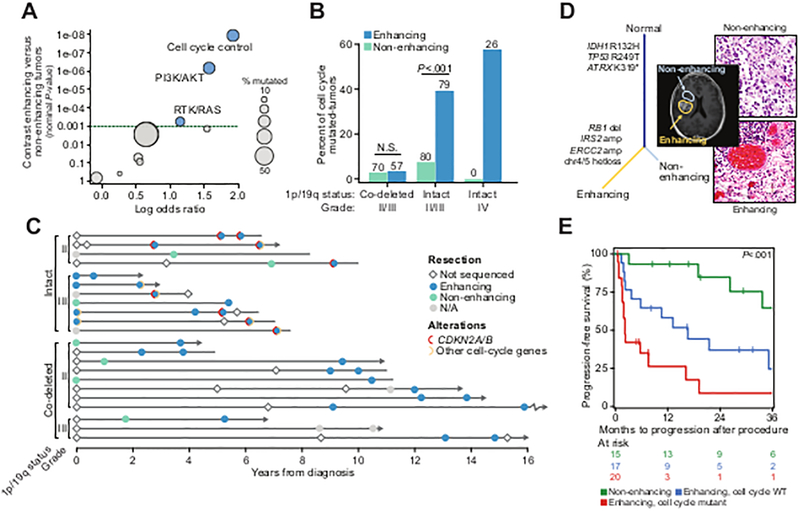
Figure 2. Genomic correlates of contrast-enhancement on brain imaging in IDH-mutant tumors.
A) Genomic alterations in core molecular and signaling pathways are associated with enhancing disease in IDH-mutant gliomas [n=150 and 148 non-enhancing and enhancing primary and recurrent tumors (111 and 39 versus 77 and 71, respectively) and recurrent tumors, respectively; n=42 and 270 sequenced grade IV and grade II/III specimens, respectively). All but 24 tumors were grade II-III at diagnosis. B) The rate of alterations in effectors of the cell cycle across IDH-mutant glioma of different grade and 1p19q status (N cases indicated above each bar), and by presence of enhancement on presurgical MRI. P-values indicate significance, Fisher’s exact test. C) The molecular and clinical evolution in 22 patients with IDH-mutant disease [n=11 each of 1p19q-intact (four and seven grade II and III) and co-deleted (eight and three grade II and III), respectively] in the cohort with multiple specimens sequenced over time, grouped by grade and 1p19q status, indicates the temporal association of enhancing disease and the acquisition of a lesion in a cell-cycle effector. Arrows indicate patients still alive at last follow-up, lines without arrows indicate deceased patients. D) In a representative de novo IDH-mutant secondary GBM, two spatially distinct components of the primary tumor were profiled, one non-enhancing lower grade focus and one grade IV enhancing lesion. While clonally related (as represented by sample tree showing truncal mutations in IDH1, TP53 and ATRX), only the enhancing component possessed the cell cycle lesion (RB1 homozygous deletion). E) In IDH-mutant 1p19q intact tumors (n=47 and 18 grade II and III at diagnosis), progression-free survival from the time of recurrent surgery is significantly shorter in patients whose recurrence exhibit pre-surgical MRI enhancement and whose tumors harbored a cell-cycle alteration. Multivariate statistics from Cox proportional hazards model are presented in Fig. S6B.
We next assessed the temporal relationship between contrast enhancement and genetic alterations in patients with IDH-mutant gliomas for whom we had sequenced two or more tumor specimens throughout the course of their disease (n=22). Eleven of these patients had 1p19q-intact tumors (four and seven were grade II and III at diagnosis, respectively) and 11 had 1p19q co-deleted tumors (eight and three were grade II and III at diagnosis, respectively). Integrating imaging data into these longitudinal genomic profiles, we found that cell-cycle aberrations were associated with the acquisition of contrast enhancement only in astrocytomas (6/11), but not 1p19q co-deleted tumors (0/11, P=0.01) (Fig. 2C). Furthermore, patients whose tumor had acquired CDKN2A/B deletions at recurrence progressed rapidly. Two patients with less common cell-cycle alterations (CDK4 amplification and RB1 mutation, respectively) in their primary tumors remained progression-free for 5 and 6 years, respectively, before another tumor recurrence with CDKN2A/B mutation, followed by disease acceleration and death within 15 and 11 months, respectively. Taken together, these data suggest that loss of the cyclin-dependent kinase inhibitor CDKN2A gene, which is common in GBM and associated with more aggressive tumor biology in mouse models of glioma (25), may have a greater effect on tumor progression than alterations in other cell-cycle pathway members.
To further explore the association between cell-cycle alterations and contrast-enhancing disease, we sequenced multiple adjacent regions of a de novo IDH-mutant GBM that included both lower-grade non-enhancing and grade IV enhancing components (Fig. 2D). While canonical, truncal lesions existed in both regions, an RB1 deletion was detected only in the enhancing component of the tumor.
While contrast enhancement is an established marker of malignant progression in lower-grade glioma (26), among recurrent grade II and III IDH-mutant 1p19q intact tumors with enhancement (47 and 18 were grade II and III at diagnosis, respectively), cell-cycle alterations were independently associated with shorter progression-free survival (HR=2.6, 95% CI 1.6–5.7, log-rank P=0.02; Fig. 2E, Fig. S6). It was notable that cell-cycle alterations also occurred more frequently in recurrent tumor specimens (Fig. 1F). Among the rare subset of IDH-WT lower-grade astrocytomas, cell-cycle alterations were less frequent in grade II tumors, increased in frequency with grade III lesions (4/24 vs. 51/71, P<0.001), and were associated with worse progression-free survival (Fig. S7–8). Collectively, these results indicate that alterations in effectors of cell-cycle progression are associated with poor prognosis in 1p19q-intact lower grade gliomas independent of IDH status and are associated with molecular progression and transformation in the IDH-mutant subset.
Treatment-induced hypermutation across glioma subtypes
In addition to core pathway alterations associated with poor outcomes, we explored the relationship between DNA hypermutation and prior systemic therapy with alkylating chemotherapeutics, in particular temozolomide (TMZ) (8,10). We first determined the prevalence of alkylating therapy-induced hypermutation in TMZ-exposed tumors by integrating the overall somatic tumor mutational burden with mutational signature analysis. This analysis distinguished hypermutation due to therapy from hypermutation due to underlying defects in MMR associated with microsatellite instability (Fig. 3A). Thirty-eight tumors were hypermutated (n=14 IDH WT, 8 IDH-mutant 1p19q-intact, and 16 IDH-mutant 1p19q co-deleted), and therapy-induced hypermutation was significantly more common in IDH-mutant than in IDH-WT gliomas (24/82 vs. 14/114; 29% and 12% of alkylator-exposed tumors, respectively, P=0.004) and in patients with MGMT methylation (17/57 vs. 8/79; 30% and 10% of patients with available MGMT status, P=0.006). Among the IDH-mutant gliomas, therapy-associated hypermutation arose more commonly in the subset harboring 1p19q co-deletion (16/41 versus 8/38, 39 and 21% respectively, P=.09) (Fig. S9). We did not observe therapy-induced hypermutation in any tumor not previously exposed to alkylating therapy.
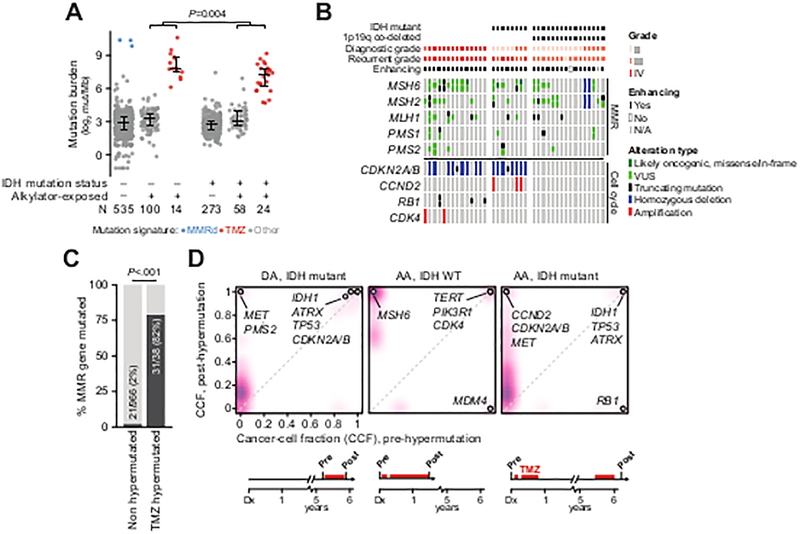
Figure 3. Genomic context of somatic hypermutation in glioma.
A) Somatic mutation rates in IDH-wildtype and IDH-mutant samples acquired prior to or after exposure to alkylator-based chemotherapy. Samples with elevated mutation burden were attributable to either mismatch repair (MMR) deficiency or prior alkylator exposure as reflected in their mutational signatures. B) TMZ-hypermutated gliomas (n=14 IDH WT, 8 IDH-mutant 1p19q-intact, and 16 IDH-mutant 1p19q co-deleted; grade at diagnosis and recurrence as indicated) have nearly obligate lesions in effectors of MMR independent of the status of IDH, 1p19q, or diagnostic and recurrent grade, whereas cell-cycle lesions (predominantly copy-number alterations and not mutations) arose only in 1p19q intact tumors independent of IDH status and diagnostic grade. C) Mutations in MMR genes occur infrequently in gliomas that do not develop alkylator-mediated hypermutation as compared to those with therapy-induced hypermutation. D) In three representative patients with matched pre-treatment and post-TMZ specimens, clonal evolution of a TMZ-hypermutated clone was evident (purple is proportional to the density of somatic mutations arising at a given cancer cell fraction). In all cases, a cell-cycle abnormality precedes therapy (labeled). The abbreviated clinical course for each patient is shown at bottom. DA: diffuse astrocytoma, AA: anaplastic astrocytoma.
Phenotypically, alkylator-induced hypermutation was associated with contrast enhancement on brain MRI in all but one patient, regardless of glioma subtype [among grade II-III tumors: 23 enhancing tumors of 24 hypermutators (16 and 8 grade II and III at diagnosis) versus 156 of 342 in non-enhancing tumors (177 and 165 grade II and III at diagnosis); P<.001]. Of the 38 total hypermutated recurrent tumors, 31 (82%) harbored mutations in effectors of MMR (27) independent of their IDH or 1p19q status (Fig. 3B–C). In total, 67% (16/24) of IDH-mutant tumors with therapy-associated hypermutation also underwent malignant progression independent of 1p19q status (Fig. 3B). Moreover, all tumors of astrocytic origin that relapsed with therapy-induced hypermutation (n=22), regardless of IDH status, harbored a cell-cycle alteration. Most of these alterations (15/26) were focal deletions of CDKN2A/B that could not otherwise be ascribed to either the MMR defect or the mechanism of hypermutation in these patients (Fig. 3B). We also identified 16 TMZ-hypermutated IDH-mutant 1p19q co-deleted tumors. These tumors appear to have a similar frequency of MMR defects as the TMZ-hypermutated 1p19q-intact tumors, consistent with their treatment resistance, but generally lack clear cell-cycle progression defects.
To determine the timing of cell-cycle alterations in the 1p19q-intact tumors relative to therapy, we sequenced a matched tumor specimen acquired prior to therapy in a subset of patients (n=3) with TMZ-hypermutated recurrences. In all such cases, a cell-cycle alteration preceded therapy (Fig. 3D). In two cases, this alteration was retained upon hypermutation. In one case, convergent evolution driven by an ongoing selective pressure was evident, in which the pre-treatment RB1 aberration was lost, but a different cell-cycle alteration targeting CDKN2A/B emerged as the incumbent clone. In all cases, these cell-cycle aberrations appeared clonal in both the pre-treatment and post-therapy tumors.
Actionable alterations and response to targeted treatment in gliomas
Beyond alkylating chemotherapy, patients received a diversity of other therapeutic modalities. Aside from IDH mutations, whose sensitivity to investigational IDH inhibitors is currently being evaluated in clinical trials, 65 of 327 (20%) and 230 of 596 (39%) IDH-WT and -mutant tumors respectively (P<.001) harbored a genomic alteration with investigational therapeutic significance, including standard-of-care biomarkers predictive of response to FDA-approved therapies in other cancer types (Fig. S10). Of these, 32 patients received a molecularly targeted therapy approved, or considered standard-of-care, for the treatment of either gliomas or other cancer types (levels of evidence 1–3, OncoKB; see Methods). An additional 32 patients received a targeted agent on another basis, such as preclinical evidence for efficacy. Collectively, the patients receiving molecularly targeted therapy were predominantly IDH-WT GBMs (60%). Many of these therapies targeted mutant effectors of aberrant RTK/MAPK signaling including RAF and MEK inhibitors now used routinely to treat patients with diverse cancer types with activating BRAF mutations (28–30).
Oncogenic BRAF mutations were present in 29 samples (2% of study cohort), and we investigated the genomics and clinical response of these patients in further detail. The frequency and distribution of BRAF-activating mutations in our cohort were largely consistent with other contemporary cohorts (Fig. S11). Both monomer-signaling V600E mutations (30) and less common activating non-V600 hypoactive BRAF mutations (31) were identified, but arose in a histology-dependent manner. BRAF V600 mutations arose clonally and nearly exclusively in atypical IDH-WT GBMs with epithelioid histological features, as recently reported (32), or high-grade tumors that arose from lower-grade lesions (17 of 17 such tumors, Fig. 4A–B). By contrast, prototypical IDH-WT GBMs had a mix of largely subclonal V600 and non-V600 oncogenic BRAF mutations (G466, G469, and L597; n=5 of 10) (33), which in this subtype typically associated with co-occurring chromosome 7 gain and 10 loss (4 of 6 versus 2 of 13 patients among non-conventional GBMs, P=0.05) (Fig. 4A–B).
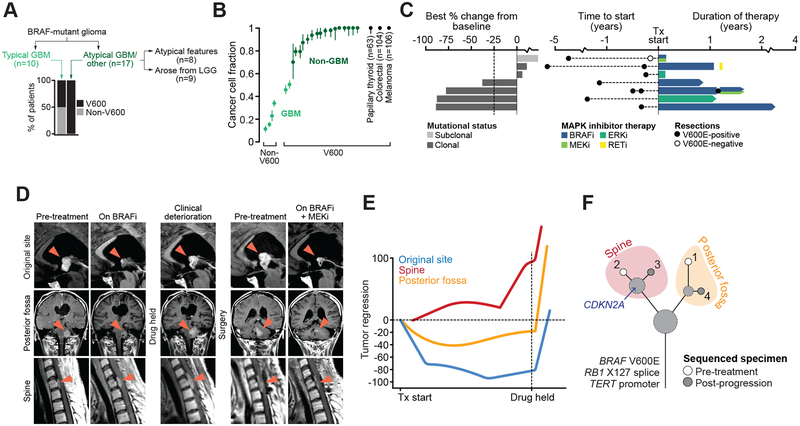
Figure 4. BRAF mutations and response to RAF/MEK/ERK-directed therapy.
A) The distribution of different BRAF mutations broken down by glioma type in the study cohort. B) The fraction of cancer cells harboring either V600E or non-V600 activating mutations in BRAF indicates GBMs harbor largely subclonal non-V600 BRAF mutations while non-GBM histologies possess predominantly clonal V600E mutations. The median and interquartile range for cancer cell fractions of BRAF V600 mutations in other BRAF-mutant cancer types are shown for context. C) On left, the best response in BRAF V600-mutated patients receiving MAPK-directed targeted therapy as measured by change in the sum of diameters across all measured lesions from initial scan. At right is the duration of therapy and date of resection and BRAF status. D) The clinical course of a single BRAF-mutant patient that progressed from low- to high-grade disease, and then experienced a durable 87% tumor regression on MAPK targeted therapy (bottommost patient in panel c). Imaging is shown for three lesions (original site of disease, posterior fossa, and spinal metastasis). RAF monotherapy was ultimately held after initial response for consolidative radiotherapy, upon which the patient clinically deteriorated and was subsequently treated with combination RAF+MEK therapy, to which they re-responded with similar lesion-specific differences in the degree of response. E) The degree of tumor regression over time is shown in the three disease sites. F) The evolutionary relationship among multiple sequenced lesions in the same patient in panels (D-E) indicated little additional genetic heterogeneity among sites except for a CDKN2A deletion specific to the non-responding spine lesion.
Seven of the BRAF V600E-mutant gliomas were treated with diverse MAPK-directed therapy (RAF, MEK, ERK, or RET inhibitor therapy). Of these, four patients had partial or near-complete responses (ongoing at 9 to 41 months), whereas two had stable disease as best response (Fig. 4C). The single patient with a BRAF V600 mutation who progressed immediately had the only treated tumor with a subclonal BRAF V600E (Fig. S12). Indeed, this subclonal BRAF mutation was identified in a diagnostic specimen acquired years prior to treatment and was not detected in a second specimen acquired just prior to, but sequenced after, the start of combined RAF+MEK inhibitor treatment.
Despite the variability of responses to MAPK-directed therapy, clinical benefit was nevertheless apparent in patients with stable disease, as one such patient remained on RAF inhibitor monotherapy for greater than one year. Glioma patients who responded to RAF inhibitor therapy typically harbored quiescent disease that underwent radiographic transformation after years of surveillance (8 to 26) prior to surgical resection. In other responders, there was lesion-to-lesion heterogeneity in the depth and durability of response. One such BRAF V600E-mutant patient achieved a durable 78% tumor regression overall, with the original site of disease undergoing near-complete regression whereas a lesion in the posterior fossa responded partially, and another spine lesion achieved only stable disease (Fig. 4D–E). To explore the basis for this mixed treatment response, we sequenced four spatiotemporally distinct lesions from this patient and found that the variability in drug sensitivity could not be attributed to BRAF V600 heterogeneity, as the sensitizing mutation arose early and was clonal in all lesions. Moreover, sequencing revealed only modest genetic heterogeneity pre-treatment that did not change with RAF inhibitor monotherapy. Notably, however, the spinal metastasis that did not respond to RAF inhibition possessed a homozygous deletion of CDKN2A that was absent from the responding lesions (Fig. 4F). CDKN2A alterations have previously been associated with reduced response to RAF inhibition in BRAF V600-mutant melanomas (34). Collectively, these data indicate that the spectrum of BRAF activating alleles varies by glioma subtype, that BRAF mutations can arise subclonally limiting drug response to RAF inhibitor therapy, but when clonal may engender responses that are conditioned in part by their co-mutational context.
Discussion
Diffuse glioma is a group of diseases with considerable molecular and clinical heterogeneity that has confounded new therapeutic approaches. We performed an integrated clinico-genomic analysis of prospectively sequenced gliomas to identify associations between somatic and germline alterations and key clinical phenotypes. In total, 13% of patients had pathogenic or likely pathogenic variants in their germline, including rare high penetrance alleles that we could, through mutational signature analysis, establish as relevant to the pathogenesis of their disease (n=2, Lynch syndrome). Though familial brain tumors have been reported, defined genetic syndromes are thought to account for only a small proportion of these cases and the genetic determinants of inheritance remains poorly understood (35,36). Notably, many of the germline alterations identified in our cohort targeted effectors of DNA repair, a subset of which were associated with somatic hypermutation in primary untreated tumors due to a mismatch-repair deficiency. Collectively, our data indicate that germline pathogenicity may play a greater role in gliomagenesis than previously appreciated and warrants further investigation.
Our data suggests that acquired mutations in effectors of cell-cycle progression may underlie malignant progression in lower-grade astrocytomas. These alterations were unique to astrocytomas, arose independent of IDH status, and were absent in 1p19q co-deleted tumors. These data from our prospectively sequenced cohort support the findings from recent retrospective studies which suggested that loss of the CDKN2A gene is associated with shorter overall survival in astrocytomas (37–39). Our data further suggest that the acquisition of cell-cycle alterations in recurrent astrocytomas is associated with the development of specific phenotypes including MRI contrast enhancement. Moreover, the acquisition of these alterations appears to be associated with the transition from indolent to malignant disease and conferred a poor prognosis independent of the grade of that recurrent tumor.
The alkylating agents TMZ and BCNU/CCNU are the most widely used drugs in the treatment of adult diffuse glioma. We identified alkylator therapy-associated hypermutation in 22% of recurrent tumors sequenced after alkylating therapy exposure (8,10,40) in both low- and high-grade predominantly enhancing tumors with and without IDH mutations. In 1p19q-intact tumors, TMZ hypermutation always arose in tumors with a cell-cycle alteration that could not be attributable to and likely preceded the MMR dysfunction that ultimately led to TMZ resistance. In 1p19q-co-deleted tumors, TMZ hypermutation arose in the absence of any alteration in the cell-cycle machinery, indicating that the mechanism of susceptibility to therapy-induced hypermutation may vary by molecular subtype. Collectively, these cohort-level and longitudinal analyses, despite limited sample size, suggest that cell-cycle aberrations may represent a pre-treatment biomarker of susceptibility to therapy-induced hypermutation in certain gliomas.
Integrating treatment phenotypes with mutational data, we identified two subtypes of BRAF-mutant gliomas. Conventional GBMs displayed subclonal BRAF mutations including both monomer-signaling V600 and known oncogenic non-V600 hypoactivating mutations. By contrast, BRAF mutations in atypical GBMs of epithelioid histology or those arising as malignant progression from lower-grade tumors were always clonal BRAF V600 mutations. While exploratory, responses to MAPK-directed therapy in these patients appeared limited to those with clonal V600 mutations, but also varied as a function of their co-mutational context. Overall, the differences in the type and clonality of oncogenic BRAF mutations in glioma indicate they may play a varying role in the pathogenesis of such tumors and reflect distinct therapeutic sensitivities. More broadly, these findings suggest that the selection of genotype-directed therapy may require a deeper understanding of how clinical response is influenced not only by the presence or absence of a sensitizing alteration, but also its allele-specific biology, clonality, and co-mutational context.
The genomic landscape of primary gliomas has been characterized extensively and an increasing number of studies are exploring how these tumors evolve spatially and temporally. Nevertheless, the use of prospective molecular characterization in the clinical care of gliomas is limited by multiple factors. Principle among these is the absence of a large-scale dataset of molecular data integrated with curated clinical data and treatment phenotypes to prompt studies of which molecular alterations may underlie clinical disease states and inform more rational clinical trial design. Our analysis begins to reveal such associations and we hope, through subsequent functional studies coupled to cooperative data sharing efforts (22,41), will catalyze future efforts to unravel the clinical and therapeutic complexity of glioma.
Supplementary Material
Acknowledgements
We thank members of the Marie-Josée and Henry R. Kravis Center for Molecular Oncology for useful discussions. This work was supported by National Institutes of Health awards P30 CA008748, U54 OD202355, R35 NS105109 (I.K.M.), R01 CA207244 (D.M.H., B.S.T.), R01 CA204749 (B.S.T.); National Brain Tumor Society Defeat GBM Initiative (I.K.M.); the American Cancer Society (B.S.T.), the Sontag Foundation, and the Josie Robertson Foundation (B.S.T.).
Conflicts of interest: A.L.L. reports research funding from Nantomics and is an equity holder in Sanofi; R.J.Y. received advisory board honoraria from Agios Pharmaceuticals and Puma Biotechnology; D.H.H. received consulting fees or advisory honoraria from Atara Biotherapeutics, Chugai Pharma, CytomX Therapeutics, Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, Debiopharm Group, ArQule, Genentech and reports research funding from AstraZeneca, Puma Biotechnology, Loxo Oncology; B.T.L. received consulting fees from Genentech, Thermo Fisher Scientific, Guardant Health, Mersana Therapeutics, Hengrui Therapeutics, and Biosceptre Australia; M.F.B. received research funding from Illumina and consulting fees from Roche; D.B.S. is a member of the scientific advisory boards of Pfizer and Loxo Oncology, an equity holder in Loxo Oncology, a clinical advisory board member of Intezyne, and advisory board honoraria from Illumina; Z.K.S. spouse received consulting fees from Genentech/Roche, Novartis, Regeneron, Biomarin, and Fortress Bio; M.E.G. was an employee of and equity holder in Foundation Medicine. J.C. was an employee of and equity holder in Foundation Medicine. T.J.K. received research funding from Eli Lilly, Merck, and Ludwig; C.G. receives research support from Pharmacyclics and Bayer and consulting fees from BTG; K.B. is an equity holder in MMT; E.P. received advisory board honoraria from AstraZeneca; A.O. received consulting fees or was an advisory board member for Stemline, Merck, Novocure, AstraZeneca, Bristol-Myers Squibb, Inovio, and Alexion; L.M.D. received scientific advisory board honoraria from Sapience Therapeutics, Tocagen, BTG International, Roche, Syndax; A.B. receives research funding from Arix Bioscience; V.S.T. is a co-founding investigator and consultant for BlueRock Therapeutics. I.K.M. reports research funding from General Electric, Amgen, and Lilly; advisory roles with Agios, Puma Biotechnology, and Debiopharm Group; and honoraria from Roche. No other disclosures are reported.
References
- 1.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155(2):462–77 doi 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372(26):2481–98 doi 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17(1):98–110 doi 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455(7216):1061–8 doi 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med 2015;372(26):2499–508 doi 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131(6):803–20 doi 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Reuss DE, Kratz A, Sahm F, Capper D, Schrimpf D, Koelsche C, et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 2015;130(3):407–17 doi 10.1007/s00401-015-1454-8. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, Zairis S, et al. Clonal evolution of glioblastoma under therapy. Nat Genet 2016;48(7):768–76 doi 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai H, Harmanci AS, Erson-Omay EZ, Li J, Coskun S, Simon M, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet 2016;48(1):59–66 doi 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014;343(6167):189–93 doi 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen PY, Chang SM, Van den Bent MJ, Vogelbaum MA, Macdonald DR, Lee EQ. Response Assessment in Neuro-Oncology Clinical Trials. J Clin Oncol 2017;35(21):2439–49 doi 10.1200/JCO.2017.72.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17(3):251–64 doi 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31(11):1023–31 doi 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang MT, Penson A, Desai NB, Socci ND, Shen R, Seshan VE, et al. Small-Cell Carcinomas of the Bladder and Lung Are Characterized by a Convergent but Distinct Pathogenesis. Clin Cancer Res 2018;24(8):1965–73 doi 10.1158/1078-0432.CCR-17-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandelker D, Zhang L, Kemel Y, Stadler ZK, Joseph V, Zehir A, et al. Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA 2017;318(9):825–35 doi 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017 doi 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 2017;21(3):374–82 e4 doi 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 2016;34(2):155–63 doi 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov 2018;8(2):174–83 doi 10.1158/2159-8290.CD-17-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47(12):1402–7 doi 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium APG. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7(8):818–31 doi 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierallini A, Bonamini M, Bozzao A, Pantano P, Stefano DD, Ferone E, et al. Supratentorial diffuse astrocytic tumours: proposal of an MRI classification. Eur Radiol 1997;7(3):395–9 doi 10.1007/s003300050173. [DOI] [PubMed] [Google Scholar]
- 24.Pallud J, Capelle L, Taillandier L, Fontaine D, Mandonnet E, Guillevin R, et al. Prognostic significance of imaging contrast enhancement for WHO grade II gliomas. Neuro Oncol 2009;11(2):176–82 doi 10.1215/15228517-2008-066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchougounova E, Kastemar M, Bråsäter D, Holland EC, Westermark B, Uhrbom L. Loss of Arf causes tumor progression of PDGFB-induced oligodendroglioma. Oncogene 2007;26(43):6289–96 doi 10.1038/sj.onc.1210455. [DOI] [PubMed] [Google Scholar]
- 26.van den Bent MJ, Wefel JS, Schiff D, Taphoorn MJ, Jaeckle K, Junck L, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol 2011;12(6):583–93 doi 10.1016/S1470-2045(11)70057-2. [DOI] [PubMed] [Google Scholar]
- 27.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res 2006;66(8):3987–91 doi 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373(8):726–36 doi 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364(26):2507–16 doi 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480(7377):387–90 doi 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548(7666):234–8 doi 10.1038/nature23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korshunov A, Chavez L, Sharma T, Ryzhova M, Schrimpf D, Stichel D, et al. Epithelioid glioblastomas stratify into established diagnostic subsets upon integrated molecular analysis. Brain Pathol 2018;28(5):656–62 doi 10.1111/bpa.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korshunov A, Chavez L, Sharma T, Ryzhova M, Schrimpf D, Stichel D, et al. Epithelioid glioblastomas stratify into established diagnostic subsets upon integrated molecular analysis. Brain Pathol 2017. doi 10.1111/bpa.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nathanson KL, Martin AM, Wubbenhorst B, Greshock J, Letrero R, D’Andrea K, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436). Clin Cancer Res 2013;19(17):4868–78 doi 10.1158/1078-0432.CCR-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blumenthal DT, Cannon-Albright LA. Familiality in brain tumors. Neurology 2008;71(13):1015–20 doi 10.1212/01.wnl.0000326597.60605.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheurer ME, Etzel CJ, Liu M, Barnholtz-Sloan J, Wiklund F, Tavelin B, et al. Familial aggregation of glioma: a pooled analysis. Am J Epidemiol 2010;172(10):1099–107 doi 10.1093/aje/kwq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirahata M, Ono T, Stichel D, Schrimpf D, Reuss DE, Sahm F, et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol 2018. doi 10.1007/s00401-018-1849-4. [DOI] [PubMed] [Google Scholar]
- 38.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016;164(3):550–63 doi 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reis GF, Pekmezci M, Hansen HM, Rice T, Marshall RE, Molinaro AM, et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II-III) astrocytomas. J Neuropathol Exp Neurol 2015;74(5):442–52 doi 10.1097/NEN.0000000000000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 2015;28(3):318–28 doi 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Consortium G Glioma through the looking GLASS: molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium. Neuro Oncol 2018;20(7):873–84 doi 10.1093/neuonc/noy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The prospective somatic mutational and clinical data have been deposited for visualization and analysis in the cBioPortal for Cancer Genomics (www.cbioportal.org/study?id=glioma_mskcc_2019).