Abstract
Genetic characterization of local breeds is essential to preserve their genomic variability, to advance conservation policies and to contribute to their promotion and sustainability. Genomic diversity of twenty European local pig breeds and a small sample of Spanish wild pigs was assessed using high density SNP chips. A total of 992 DNA samples were analyzed with the GeneSeek Genomic Profiler (GGP) 70 K HD porcine genotyping chip. Genotype data was employed to compute genetic diversity, population differentiation and structure, genetic distances, linkage disequilibrium and effective population size. Our results point out several breeds, such as Turopolje, Apulo Calabrese, Casertana, Mora Romagnola and Lithuanian indigenous wattle, having the lowest genetic diversity, supported by low heterozygosity and very small effective population size, demonstrating the need of enhanced conservation strategies. Principal components analysis showed the clustering of the individuals of the same breed, with few breeds being clearly isolated from the rest. Several breeds were partially overlapped, suggesting genetic closeness, which was particularly marked in the case of Iberian and Alentejana breeds. Spanish wild boar was also narrowly related to other western populations, in agreement with recurrent admixture between wild and domestic animals. We also searched across the genome for loci under diversifying selection based on FST outlier tests. Candidate genes that may underlie differences in adaptation to specific environments and productive systems and phenotypic traits were detected in potentially selected genomic regions.
Subject terms: Animal breeding, Genomics, Population genetics
Introduction
Pork is the most widely consumed meat worldwide1. Pig industry is mainly based on a limited number of cosmopolitan lean breeds which have been extensively used for breeding improvement. These highly selected breeds are raised in intensive production systems, focused on maximizing productivity, and supplying the market with fresh pork. Besides, many local, less performing breeds exist, although some of them are nowadays close to extinction. These traditional breeds are usually associated with local forms of pig husbandry and their meat is used for the production of high-quality and niche products. The Iberian pig, raised in the South-West of the Iberian Peninsula, is probably the most representative local pig breed, although many others are reared in European countries. Common characteristics of these breeds are a good environmental adaptation, rusticity, low muscle mass deposition and high adipogenic potential and, in many cases, superior meat quality traits2.
Conservation of small local breeds is mainly conditioned by their economic relevance but also depends on socio-cultural value, adaptation to local agro-climatic conditions, contribution to the development of local communities and marginal areas, and scientific importance. Efficient utilization of local breeds is needed and for this goal, technological advances, growing knowledge and innovative ideas founded on scientific research must be developed. The genetic characterization of these resources is a preliminary step for the development of conservation programs and to boost local breed promotion, and their sustainable use3.
Despite a few whole genome studies have been carried out for some European local pig breeds, such as the Iberian4,5 or Casertana6,7, many other autochthonous breeds have not been analyzed in detail yet and are considered untapped genetic resources. Recently, the diversity at several relevant candidate gene polymorphisms has been evaluated in twenty local European pig breeds8, providing information about the segregation of interesting markers for breeding or traceability9,10 purposes and giving some first insights into the genetic structure of these populations. Nevertheless, genetic characterization of animal breeds is usually addressed with the analysis of neutral markers like microsatellites or SNPs, as recommended by FAO11. High density SNP panels help to investigate genome wide diversity with a higher resolution. Dense SNP panels can be applied to a variety of genomics studies including inference on population history, structure and admixture12, estimation of effective population size4,5, QTL mapping strategies13 and whole genome association studies14 and genomic selection15. Also, comparative genomic diversity enables us to explore the degree of genomic variation and linkage disequilibrium (LD) among pig breeds. This as well helps to detect genomic regions that have been subject to selective sweeps in different pig populations16.
In this study, we analyzed genomic diversity of 20 European local pig breeds: Black Slavonian and Turopolje (Croatia), Basque and Gascon (France), Schwäbisch-Hällisches Schwein (Germany), Apulo-Calabrese, Casertana, Cinta Senese, Mora Romagnola, Nero Siciliano and Sarda (Italy), Lithuanian indigenous wattle and Lithuanian White old type (Lithuania), Alentejana and Bísara (Portugal), Moravka and Swallow-Bellied Mangalitsa (Serbia), Krškopolje pig (Slovenia) and Iberian and Majorcan Black (Spain). The genotyping information from a high density SNP chip has been employed to assess genomic diversity and structure and to identify selection signatures. This work is framed within the TREASURE project (https://treasure.kis.si), a multidisciplinary European Union funded project pointing toward the development of sustainable pork chains in several European local pig breeds.
Results and Discussion
A total of 985 pigs from 20 European autochthonous breeds and a small Spanish Wild Boar population (n = 7) were successfully genotyped with the GeneSeek ® Genomic Profiler (GGP) 70 K HD Porcine chip (Illumina Inc, USA). The sample sizes of each analyzed population are included in Table 1. Selection of animals for sampling was performed by local specialized personnel with a deep knowledge of each breed, in order to get representative samples within each analyzed population. A total of 60,451, out of 68,516 SNPs, remained after the removal of those with more than 10% missing genotypes or Minor Allele Frequency (MAF) lower than 0.01. The average within-breed MAF (Table 1, Fig. 1) ranged from 0.133 (Turopolje) to 0.294 (Sarda). In agreement with these results, Turopolje was the breed with the highest number of SNPs (29,740) with the lowest MAF values (ranging between 0.01 and 0.05) while Sarda had the highest number (15,350) of highly informative SNP markers with frequencies between 0.4 and 0.5 (Fig. 1 and Supplementary Table 1). The breeds showing the lowest number of informative SNPs were Alentejana, Basque, Iberian, Swallow-Bellied Mangalitsa, Mora Romagnola, Turopolje and Wild Boar, since more than 25% of their SNPs had MAF values lower than 0.05. On the contrary, the breeds showing the most informative genotyping results, with more than 20% of their SNPs with MAF higher than 0.40 were Sarda, Krškopolje, Schwäbisch-Hällisches Schwein, Bísara, Nero Siciliano, Old type Lithuanian White, Black Slavonian, Moravka and Lithuanian indigenous wattle.
Table 1.
Samples sizes (N), mean minor allele frequencies (MAF), observed (HO) and expected (HE) heterozigosities, inbreeding coefficient of an individual (I) relative to the subpopulation (S) (FIS) and Wright’s fixation index (FST), for each analyzed breed.
| Breed | N | MAF | HO | HE | FIS | FST |
|---|---|---|---|---|---|---|
| Alentejana | 48 | 0.193 | 0.248 | 0.259 | 0.041 | 0.116 |
| Apulo Calabrese | 53 | 0.228 | 0.258 | 0.305 | 0.138 | 0.118 |
| Basque | 39 | 0.169 | 0.240 | 0.233 | −0.026 | 0.147 |
| Bísara | 49 | 0.270 | 0.339 | 0.355 | 0.045 | 0.102 |
| Black Slavonian | 49 | 0.262 | 0.332 | 0.346 | 0.040 | 0.096 |
| Casertana | 54 | 0.246 | 0.291 | 0.327 | 0.095 | 0.110 |
| Cinta Senese | 54 | 0.220 | 0.300 | 0.300 | 0.011 | 0.101 |
| Gascon | 48 | 0.224 | 0.299 | 0.298 | −0.005 | 0.122 |
| Iberian | 49 | 0.202 | 0.251 | 0.270 | 0.077 | 0.110 |
| Krškopolje pig | 52 | 0.277 | 0.363 | 0.361 | −0.003 | 0.109 |
| Lithuanian indigenous wattle | 48 | 0.249 | 0.354 | 0.331 | −0.066 | 0.116 |
| Majorcan Black | 48 | 0.210 | 0.279 | 0.285 | 0.005 | 0.102 |
| Swallow-Bellied Mangalitsa | 50 | 0.192 | 0.257 | 0.259 | 0.006 | 0.125 |
| Mora Romagnola | 48 | 0.161 | 0.230 | 0.220 | −0.039 | 0.161 |
| Moravka | 50 | 0.267 | 0.348 | 0.353 | 0.014 | 0.101 |
| Nero Siciliano | 50 | 0.272 | 0.341 | 0.360 | 0.052 | 0.085 |
| Lithuanian White Old Type | 51 | 0.260 | 0.358 | 0.341 | −0.049 | 0.119 |
| Sarda | 48 | 0.294 | 0.358 | 0.382 | 0.060 | 0.092 |
| Schwäbisch-Hällisches Schwein | 49 | 0.264 | 0.349 | 0.342 | −0.016 | 0.110 |
| Turopolje | 50 | 0.133 | 0.195 | 0.187 | 0.046 | 0.159 |
| Wild Boar | 7 | 0.192 | 0.240 | 0.254 | 0.041 | 0.132 |
| Average | 47 | 0.228 (0.044) | 0.297 (0.053) | 0.303 (0.054) | 0.022 (0.050) | 0.115 (0.020) |
Figure 1.
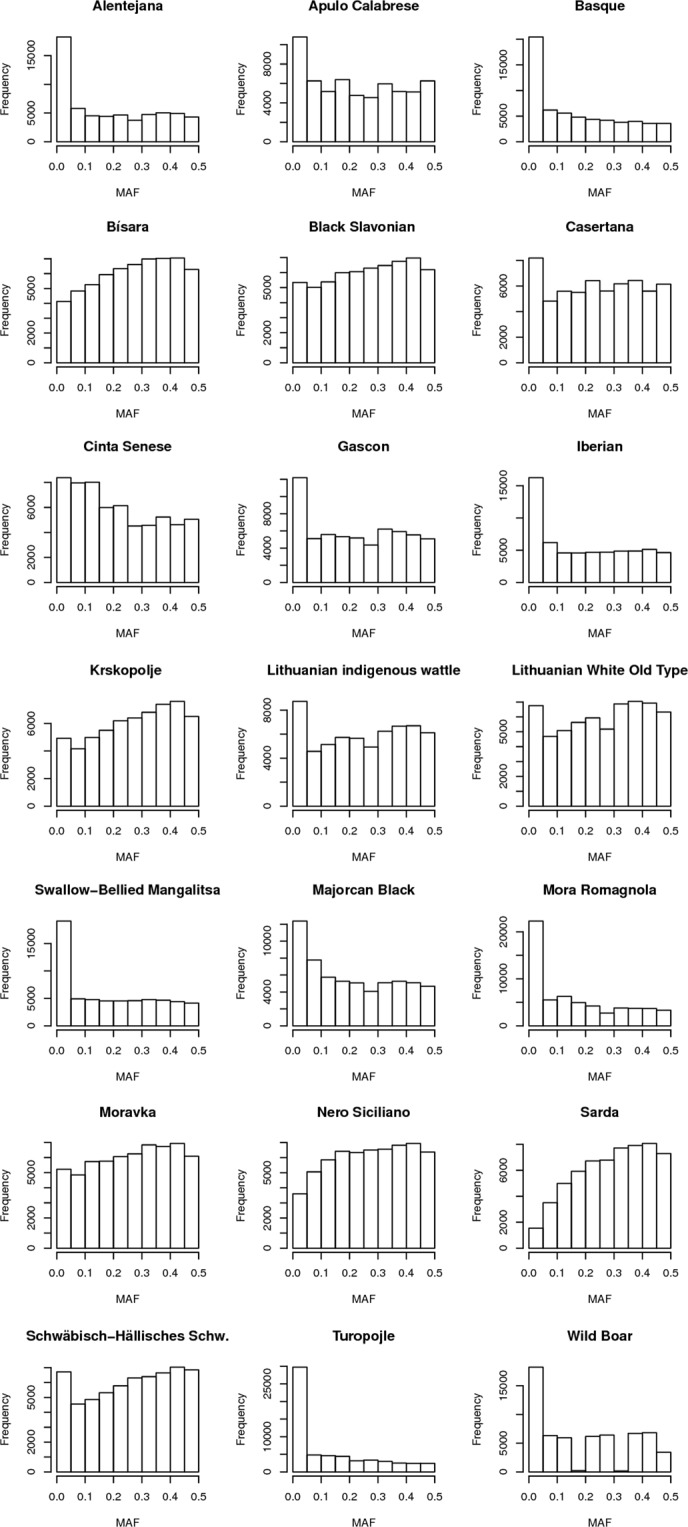
Frequency distribution of minor allele frequencies (MAF) in all the breeds.
In general, the informativity of the SNP chip was moderate in most of the studied breeds, which is not unexpected as these local breeds have not been considered for the design of commercial porcine chips17. The most widely used Illumina 60 K chip was designed and validated using samples from Berkshire, Duroc, Hampshire, Landrace, Large White, Meishan, Pietrain, Synthetic lines (Large White and Piétrain) and wild boar17. The GeneSeek ® GGP Porcine HD Genomic Profiler used in the present study is an improved version of the latter one, which was designed to include the most informative SNPs from the Porcine60K chip, according to findings in the same major commercial breeds, and that was complemented with new SNPs to improve the coverage of the chromosomes and the telomere regions, to better account for recombination18. In spite of the ascertainment bias implicit in the use of SNP chips, previous results have shown that these tools provide reliable estimates of genomic diversity for comparative studies between European populations, even in local breeds19.
Genetic diversity parameters and population structure
Within-breed genetic diversity
Genetic variability parameters for the analysed populations are presented in Table 1. Within-breed observed (HO) and expected (HE) heterozygosity ranged from 0.195 (Turopolje) to 0.363 (Krškopolje) and from 0.187 (Turopolje) to 0.382 (Sarda), respectively (Table 1). Turopolje, Mora Romagnola, Basque and Wild Boar exhibit the lowest HO and HE values and Krškopolje, Sarda and Old type Lithuanian White, the highest ones. Across-breeds averaged values for HO and HE were 0.297 (±0.053) and 0.303 (±0.054), respectively. These heterozygosity values are considerably lower than those reported previously for European cosmopolitan and Chinese pig breeds20–23, which ranged from 0.30–0.40 to 0.60–0.70 with an average of about 0.5, and similar to those reported for some local breeds19. The lowest heterozygosity values observed in Turopolje are in agreement with a previous study in which microsatellites were employed to assess the genetic diversity and population structure in eight populations corresponding to Balkan pig breeds24. In addition, Lithuanian indigenous wattle shows the smallest (−0.066) inbreeding coefficient of an individual relative to the subpopulation (FIS) while Apulo Calabrese showed the highest one (0.138) (Table 1). Negative FIS values were observed in Mora Romagnola, Schwäbisch-Hällisches Schwein, the two Lithuanian and the two French breeds (Gascon and Basque). Negative values indicate random mating among the individuals of the subpopulations but do not necessarily imply lower values of the total inbreeding coefficient which takes into account the accumulated inbreeding along the generations25.
Lack of selection programs and frequent or recurrent admixture, common to these untapped local pig breeds, should presumably lead to a higher degree of genetic diversity. However, the level of genetic variation in our local breeds is in general lower than that in cosmopolitan pig breeds16–20. The reason for this could be due to the small effective population size: in some of the breeds, only few founders were left at the beginning of the preservation programs2. This can also be the cause for high level of inbreeding observed in some of these breeds. For instance, the highest FIS values obtained in Apulo Calabrese and Casertana are in agreement with their endangered situation and small census26,27 and similar values were reported recently in an analysis based on candidate gene polymorphisms8.
Differentiation among breeds and genetic distances
The level of population differentiation can be quantified using the fixation indexes. The fixation Index (FST) for each breed (Table 1), estimated as the average of breed pairwise comparisons per SNP and then averaged by breed, shows the highest value for Mora Romagnola (0.161) and the lowest value for Sarda (0.092). The overall FST value from all the SNP markers was 0.115 (±0.020), indicating that most of the genetic variation occurred within populations rather than between breeds, as previously reported for pig populations28,29. The value obtained is concordant with previous works28, where the averaged FST value calculated between European breeds was 0.134, ranging from 0.021 to 0.209. The overall FST value obtained in the present work (analyzing data of 70K SNP chip) is considerably lower than the one observed in our previous study (0.27)8 performed with an array of selected SNPs. This difference is expected because in the latter study only 39 causal mutations and polymorphisms in candidate genes were used whereas, in the present work, SNP markers across the whole genome and mostly neutral were used6. In addition to this, the distribution of different genetic diversity parameters estimated for each SNP marker is shown in Supplementary Fig. 1.
Nei’s genetic distances30 between studied breeds range from the minimum value of 0.276 (observed between Alentejana and Iberian breeds) to the maximum value of 0.604 (observed between Apulo Calabrese and Mora Romagnola) (Supplementary Table 2), with an averaged genetic distance of 0.440 (±0.057). The Neighbor-Joining tree (NJ) constructed from these genetic distances (Fig. 2) is in general agreement with the geographical distribution of most of these breeds, namely the breeds geographically close cluster together, such as the two French breeds; Iberian, Alentejana and Wild Boar which come from the Iberian Peninsula; the two Lithuanian breeds (Lithuanian indigenous wattle and Lithuanian White old type), and the six Italian breeds (Apulo Calabrese, Casertana, Cinta Senese, Mora Romagnola, Nero Siciliano and Sarda) that are all placed in the middle of the unrooted tree (Fig. 2). These findings are expected considering that closely located breeds are more likely to share common ancestors. Besides this resemblance with the geographical distribution of the breeds, this tree cannot be used to infer any phylogenetic relationships, considering the complexity of events that might have contributed to construct the current genetic pools of the investigated breeds which share a common European origin.
Figure 2.
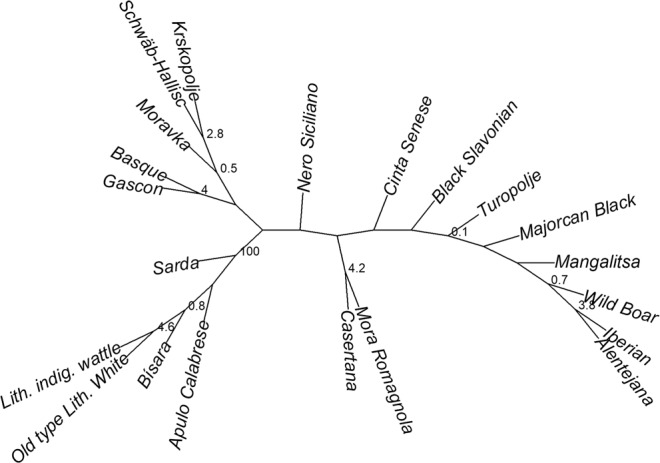
Neighbor-joining tree constructed with Nei’s distances. Numbers correspond to the percentage in which the node is recovered.
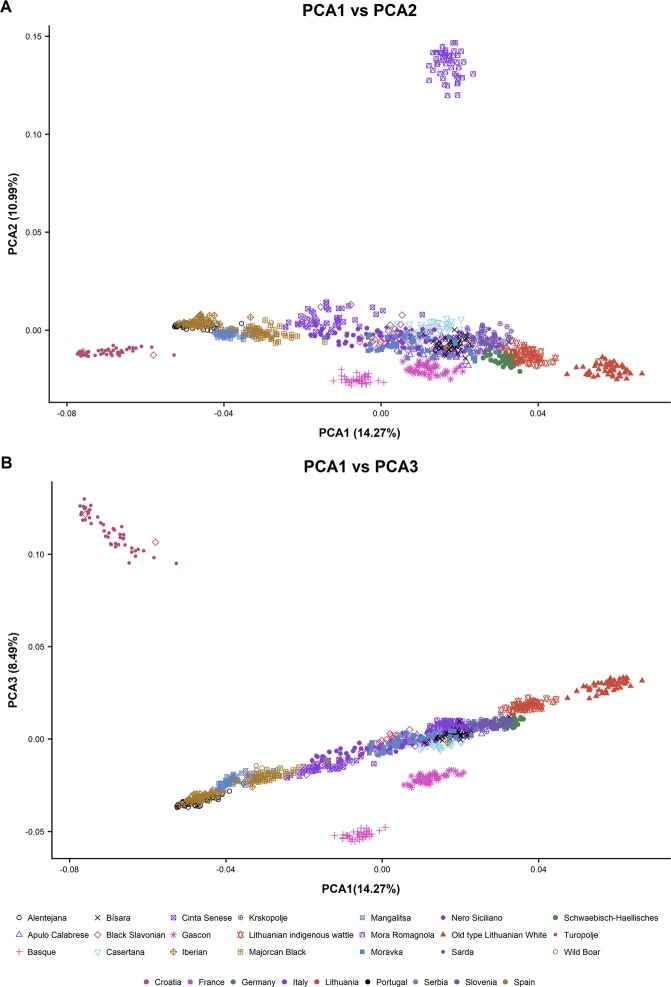
Principal component analysis (PCA) was employed to explore the clustering of individuals of different populations. The first three principal components explained 14.26%, 10.92% and 8.49% of the total variation. PCA allowed the visualization of groups formed by individuals belonging to the same breeds (Fig. 3). Moreover, clearly separated clusters were observed for Mora Romagnola, Turopolje, Gascon, Basque and Old Type Lithuanian White breeds. Some relationship of the clusters with the breeds’ geographical distribution could also be distinguished in few cases. For instance, Alentejana and Spanish populations group together as well as French and Lithuanian breeds, in agreement with the constructed NJ tree. On the other hand, the two Croatian breeds (Turopolje and Black Slavonian) plot very distant to each other, although two Black Slavonian pigs cluster with the isolated Turopolje group. The net differentiation between the two Croatian breeds is in agreement with previous results24 obtained after the study of several Balkan breeds with microsatellite markers, showing a clear distinction which is now confirmed at the genomic level and with a wider panel of breeds. The isolation of Turopolje breed is not surprising, as this breed is among the oldest ones in Europe, apparently being locally domesticated in the Middle Ages31, while Black Slavonian breed was formed at the end of the 19th century by crossing Mangalitsa with several imported breeds of pig. However, as both breeds were raised in the past in the same geographical area, the observed few exceptions in clustering may result from uncontrolled mating between them32. Moreover, previous works using microsatellite markers already showed higher genetic heterogeneity in the Black Slavonian population with one herd being clustered together with Turopolje pigs, suggesting moderate gene flow between the Black Slavonian breed and the Turopolje population, matching our results20.
Figure 3.
Genetic structure of the investigated 20 porcine breeds and Wild Boar population. Each point represents the eigenvalues of principal components 1 and 2 (A) and 2 and 3 (B). Points are colored according to the country and the shapes represent the different breeds.
Although the individuals are grouped by breeds, there is some overlapping among them, with a big cluster including many breeds. In the left end of this big cluster, Iberian and Alentejana breeds are completely overlapped, in agreement with their genetic closeness and common breeding history2,8. Next to the Iberian cluster, Spanish Wild Boar and Majorcan Black pigs are also closely grouped, all composing a nucleus of South-Western European populations. A close relatedness between the domestic breeds and the wild relative is observed. This finding is in agreement with the previously proposed recurrent admixture between wild and domesticated animals in Europe10,33, which might be especially intense in these local breeds, exploited in free-range systems, favoring a long history of genetic exchange with wild boars. The only breed coming from the Iberian Peninsula and being located far away from the south-western cluster is the Bísara breed, for which the separation from Iberian breed had been previously reported8,12. This is in agreement with its Celtic origin34. Interestingly (Swallow-bellied) Mangalitsa pigs are located in the middle of the western nucleus, quite far away from Moravka (the other Serbian breed), as already evidenced with the NJ tree. Genetic proximity between Hungarian Mangalitsa and Iberian pigs was reported previously by Herrero-Medrano et al.19. Next, four Italian breeds (all but Mora Romagnola) are clustered in the middle, together with Moravka and Black Slavonian pigs. The right end of the Italian cluster partially overlaps with Bísara and Krškopolje breeds, which are followed by Schwäbisch-Hällisches Schwein and Lithuanian Indigenous wattle, which is in the right end, close but separated from the other Lithuanian breed.
Interestingly, the breeds located in the right end of the PCA plots (Fig. 3A,B) are those with the highest heterozygosity values, which may be related to introgression or admixture with other breeds. In some cases Asian introgression, common in many European breeds, may be the explanation. In fact, for instance, the Old type Lithuanian White breed was developed by improving Lithuanian Indigenous wattle pigs with Large White, Middle White, Edelschwein, Berkshire and local Danish pigs35, which may explain its apparently high genetic diversity, despite several bottlenecks occurred since 2003, leading to a critical situation nowadays. The Schwäbisch-Hällisches Schwein breed was originated by crossing local pigs in Württemberg with Chinese pigs, and later by crossbreeding with pigs imported from England2. The creation of Black Slavonian breed, which was a dominant one on the territory of Croatia up to the mid-20th century, resulted from planned crossing between Mangalitsa, Berkshire, Poland China and Large Black (Cornwall) pig36,37. On the other hand, several breeds have been subjected to crossbreeding with cosmopolitan European breeds carrying Asiatic introgression, such as Krškopolje, which was likely crossed with German Landrace in times when the breed was cast out. Regarding the two Serbian breeds, Moravka was created as a result of unsystematic crossings of the old pig Šumadinka with Berkshire and possibly with Yorkshire38, while no Asian introgression is known for Swallow-Bellied Mangalitsa. This may explain the separation of both breeds observed in the NJ tree and PCA plots, as well as the proximity of Mangalitsa breed to the south-western populations, which are free of Asiatic introgression.
Linkage disequilibrium (LD) analyses
Differences in LD among populations result from the differences in population history and demography39,40 and detailed information on LD in domesticated animals is important because it is of high utility for fine mapping of genes41. Usually, a substantial extent of LD has been found in domestic species, which may be due to small effective population size in commercial populations. Our study provides an overview of LD patterns against physical distance in 20 European pig breeds and Iberian Wild Boar.
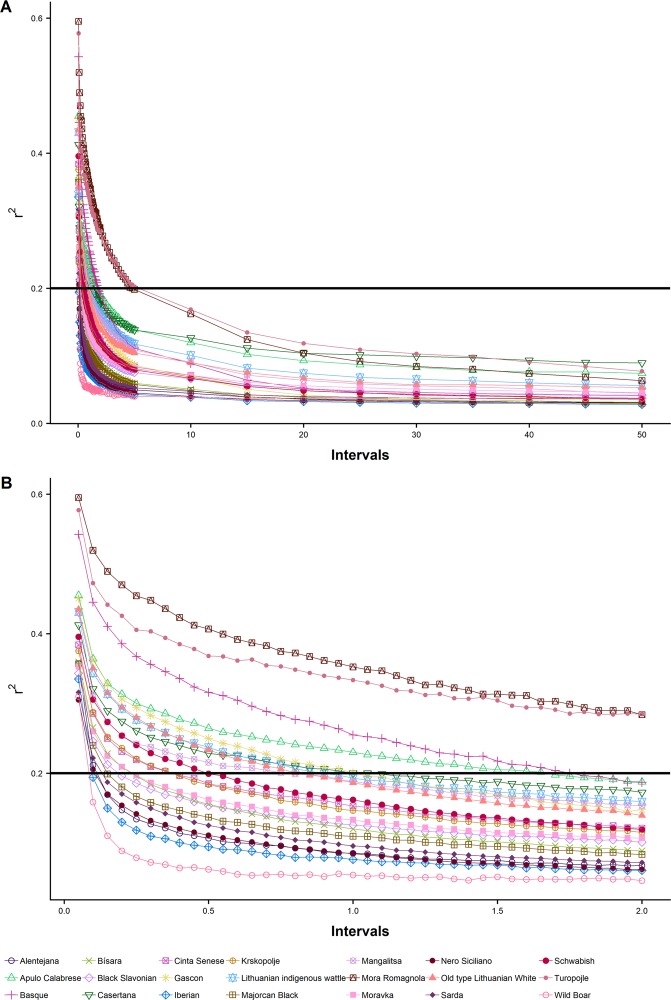
Different SNP marker sets defined by breed (Supplementary Table 3) were used to estimate LD for all SNP pairs in a distance lower than 50 Mb, dividing this window into three different categories: (a) 0 to 2 Mb, (b) 2 to 5 Mb, (c) 5 to 50 Mb; and averaging the r2 values in distance bins of 0.05, 0.20 and 5.0 Mb for classes a), b) and c), respectively (Supplementary Table 4). Overall LD across the genome between adjacent SNPs ranged from 0.289 in Wild Boar to 0.604 in Mora Romagnola.
Overall r2 values by breed were plotted against increasing distances (Fig. 4). As expected, most tightly linked SNP pairs have the highest r2 and average r2 rapidly decreases as distance increases, with a similar pattern to what has been observed in previous studies in pigs and in other species42–45. Values of r2 at short distances (0.00–0.05 Mb) ranged from 0.305 (Nero Siciliano) to 0.595 (Mora Romagnola); and at long distances (45–50 Mb) r2 values ranged from 0.028 (Iberian) to 0.089 (Casertana). The persistence and strength of LD varied among breeds. Focusing on the domestic breeds, while the LD of Iberian and Alentejana breeds decreased by the half at 0.15 Mb, showing the highest LD decay, the LD of Mora Romagnola and Turopolje decreased by the half at 1.8 and 1.75 Mb, respectively, showing the highest LD persistence. In addition to this, Fig. 4 reveals that all breeds showed r2 < 0.2 at distances lower than 2 Mb except Mora Romagnola and Turopolje, which showed r2 < 0.2 at distances lower than 5 Mb. This high level of long LD extent could point out that these breeds have experienced more unbalanced contributions (bottlenecks) or genetic drift compared with the other ones40. Similar r2 values for all the distances were observed for Iberian and Alentejana breeds, supporting the genetic closeness of these breeds already described above. Wild Boar showed the lowest extent of LD in agreement with previous findings28 and as expected for an outbred and non-admixed population. In a previous work44, European pig breeds showed high levels of significant differences in the extent of LD, in agreement with our results.
Figure 4.
Linkage disequilibrium decay. Average linkage disequilibrium plotted against distance between SNPs across the 18 autosomes for each breed.
Pairwise r2 estimates for short distances (0.05 Mb) were averaged by autosomes in all the breeds (Supplementary Table 5). These estimates revealed variation among chromosomes. While SSC1 was the chromosome with the highest LD in most breeds (Alentejana, Basque, Bísara, Casertana, Iberian, Krškopolje, Lithuanian indigenous wattle, Majorcan Black, Swallow-Bellied Mangalitsa, Moravka, Nero Siciliano, Old type Lithuanian White, Sarda, Schwäbisch-Hällisches Schwein, Wild Boar), SSC10 was the chromosome with the lowest LD observed in many breeds (Alentejana, Basque, Bísara, Black Slavonian, Cinta Senese, Iberian, Krškopolje, Swallow-Bellied Mangalitsa, Moravka, Nero Siciliano, Old type Lithuanian White, Schwäbisch-Hällisches Schwein, Turopolje, Wild Boar). Chromosomes with highest and lowest LD mean values were SSC3 (Mora Romagnola, r2 = 0.728) and SSC10 (Nero Siciliano, r2 = 0.256), respectively.
Effective population size across generations
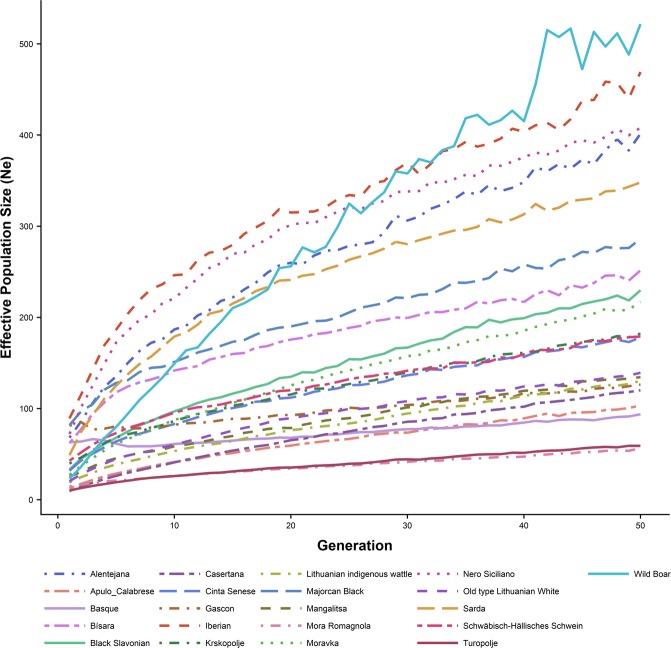
Synteny r2 estimates between all pairs within 50 Mb were computed to estimate Ne across 50 generations (Fig. 5, Supplementary Tables 6–26) using the recombination values for each chromosome showed in Supplementary Table 3. Wild Boar had the highest Ne 50 generations ago (521.68) and Mora Romagnola and Turopolje were the breeds with the lowest Ne values (56.63 and 59.08, respectively). Table 2 shows the current effective population size, with Iberian pig having the highest Ne (89.18), and Casertana the lowest value (9.44).
Figure 5.
Estimated effective population size (Ne) along 50 generations.
Table 2.
Current effective population size (Ne), standard deviation (SD) between brackets and sample size (N) by breed.
| Breed | Ne (SD) | N |
|---|---|---|
| Alentejana | 67.96 (0.10) | 48 |
| Apulo Calabrese | 12.38 (0.01) | 53 |
| Basque | 62.98 (0.12) | 39 |
| Bísara | 62.01 (0.06) | 49 |
| Black Slavonian | 33.11 (0.03) | 49 |
| Casertana | 9.44 (0.01) | 54 |
| Cinta Senese | 31.82 (0.03) | 54 |
| Gascon | 81.12 (0.12) | 48 |
| Iberian | 89.18 (0.16) | 49 |
| Krškopolje pig | 39.63 (0.03) | 52 |
| Lithuanian indigenous wattle | 18.81 (0.01) | 48 |
| Majorcan Black | 81.86 (0.12) | 48 |
| Swallow-Bellied Mangalitsa | 25.15 (0.02) | 50 |
| Mora Romagnola | 14.68 (0.01) | 48 |
| Moravka | 27.25 (0.02) | 50 |
| Nero Siciliano | 72.14 (0.08) | 50 |
| Old type Lithuanian White | 20.30 (0.01) | 51 |
| Sarda | 48.81 (0.04) | 48 |
| Schwäbisch-Hällisches Schwein | 42.74 (0.04) | 49 |
| Turopolje | 10.21 (0.01) | 50 |
| Wild Boar | 23.01 (0.06) | 7 |
Meuwissen46 recommended an effective population size of 100 in order to maintain the genetic diversity of a population, which is not accomplished in any of the populations analysed in the present work. Our findings further confirm the need for conservation strategies for many of the studied breeds. The most extreme cases are Casertana, Apulo Calabrese, Turopolje, Mora Romagnola and both Lithuanian pig breeds, for which conservation efforts are currently being undertaken27,47–49.
In fact, most local breeds are characterized by having small effective population sizes, which affects their diversity, and leads to high levels of LD and a high proportion of SNPs with fixed alleles. In general, breeds showing the highest levels of LD were those with the lowest effective population size and higher inbreeding (Apulo Calabrese, Casertana and Turopolje).
FST analyses
The estimation of FST index was used in order to detect genomic regions that could be involved in domestication, breed pattern establishment or selective breeding. A genome-wide scan of divergent genomic regions was carried out through the estimation of Wright’s FST at each marker as a measure of genetic differentiation. Candidate regions to diversifying selection were identified as those in the 99th percentile of the empirical distributions of sliding windows (Supplementary Figs 2–22). A total of 502 windows per breed were identified as outlier windows (Supplementary Tables 27–47) and when the outlier windows were adjacent, they were considered as the same genomic regions.
A total of 19 genomic regions overlapped in five or more pig groups (breeds and/or wild boars) on chromosomes SSC1, SSC2, SSC6, SSC7, SSC8 and SSC13 (Table 3). In these regions, candidate genes related with reproduction (ADAD1, PRDM1 SPACA1 and SLCO4C1), lipid, carbohydrate and protein metabolism (PGD, UBE4B, RNF150, UBE2E1, CNR1, RBP7 and STARD4), growth and development (FER, IL2, IL15, IL21 and PRDM1), cellular homeostasis (ATG5), locomotor behavior (NOVA1, SOBP) and response to nutrient (BCKDHB) were identified. The region located on SSC8 (100.93–101.74) overlapped in seven breeds (Alentejana, Apulo Calabrese, Bísara, Cinta Senese, Gascon, Krškopolje, Moravka) and in Wild Boar and contained genes involved in growth and reproduction. The breeds with the highest number of overlapped regions (with five or more breeds) were Alentejana and Iberian in addition to Wild Boar (12 regions), whereas the breed with the lowest number was Mora Romagnola, in agreement with a specific genetic differentiation in this breed, also highlighted in the PCA analyses.
Table 3.
Genomic regions with outlier FST-windows shared among at least five breeds and genes annotated within these regions in Sscrofa11.1.
| Chr | Position (Mb) | Breeds | Genes | Function |
|---|---|---|---|---|
| SSC1 | 55.71–56.47 | AL, AC, CS,LI,MB | Sperm Acrosome Associated 1 (SPACA1) | Spermatogenesis |
| Cannabinoid Receptor 1 (CNR1) | Intramuscular fat deposition | |||
| SSC1 | 63.51–63.94 | AL, IB, KR, LI, MB, NS | — | — |
| SSC1 | 70.86–74.09 | AL, BI, IB, KR, LI,OW, SW | PR/SET Domain 1 (PRDM1) | Embrionic development |
| Autophagy Related 5 (ATG5) | Autophagy; cellular homeostasis | |||
| Sine Oculis Binding Protein Homolog (SOBP) | Locomotory Behaviour. Sensory perception | |||
| SSC1 | 84.00–85.65 | AL, BA, IB, MB, SW | Branched Chain Keto Acid Dehydrogenase E1 Subunit Beta (BCKDHB) | Response to nutrient |
| SSC2 | 107.46–108.09 | AL, CS, IB, SA, TU, WB | Solute Carrier Organic Anion Transporter Family Member 4C1 (SLCO4C1) | Establishment and maintenance of pregnancy |
| SSC2 | 111.79–112.28 | IB, MA, MV, TU, WB | — | — |
| SSC2 | 112.94–114.00 | AL, BI, IB, MA, WB | FER Tyrosine Kinase (FER) | Cell proliferation; Growth factor |
| SSC2 | 115.83–116.31 | AL, CA, LI, MA, WB | Star Related Lipid Transfer Domain Containing 4 (STARD4) | Cholesterol metabolism |
| SSC6 | 23.81–24.40 | AL, IB, MB, SA, WB | — | — |
| SSC6 | 65.52–66.05 | BI, BS, GA, KR, SA, WB | adherens junctions associated protein 1 (AJAP1) | Cell adhesion |
| SSC6 | 70.29–70.71 | AC, BA, BS, CA,KR, NS, WB |
Retinol Binding Protein 7 (RBP7) ubiquitination factor E4B (UBE4B) kinesin family member 1B (KIF1B) phosphogluconate dehydrogenase (PGD) |
Lipid binding Protein ubiquitination Vesicle transport. Development of nervous system Carbohydrate metabolism |
| SSC7 | 71.73–73.13 | CA, GA, MB, MV, OW | NOVA alternative splicing regulator 1 (NOVA1) | Locomotory behaviour |
| SSC8 | 57.90–60.06 | BA, CA, CS, GA, MB, NS | — | — |
| SSC8 | 85.68–86.25 | AL, BA, BI, GA, IB, MR, MV |
Interleukin 15 (IL15) Ring finger protein 150 (RNF150) |
Cell maturation Protein ubiquitination |
| SSC8 | 92.95–96.07 | AL, BA, CA, CS,GA, WB | Sodium channel and clathrin linker 1 (SCLT1) | Ciliogenesis |
| SSC8 | 99.32–99.73 | AL, AC, GA, MV, SA, WB | — | — |
| SSC8 | 100.93–101.74 | AL, AC, BI, CS, GA, KR, MV, WB | Interleukin 21 (IL21) | Growth factor activity |
| Interleukin 2 (IL2) | Growth factor activity | |||
| Adenosine Deaminase Domain Containing 1 (ADAD1) | Spermatid development | |||
| SSC13 | 10.52–10.63 | CS, IB, KR, MA, SA, WB | Ubiquitin conjugating enzyme E2 E1 (UBE2E1) | Protein ubiquitination |
| SSC13 | 14.11–15.16 | IB, MA, MB, NE, SA,TU, WB | Eomesodermin (EOMES) 5-azacytidine induced 2 (AZI2) | Embryonic development Immunity |
AL: Alentejana; AC: Apulo Calabrese; BA: Basque; BI: Bísara; BS: Black Slavonian; CA: Casertana; CS: Cinta Senese; GA: Gascon; IB: Iberian; KR: Krškopolje; LI: Lithuanian indigenous wattle; MA: Swallow-Bellied Mangalitsa; MB: Majorcan Black; MR: Mora Romagnola; MV: Moravka; NS: Nero Siciliano; OW: Old type Lithuanian White; SA: Sarda; SW: Schwäbisch-Hällisches Schwein;TU: Turopolje; WB: Wild Boar.
Besides, breed specific signatures were identified as those in the 99.9th percentile of the empirical distributions specifically detected in one breed but not in the remaining ones (Supplementary Tables 48–69). A total of 115 breed specific regions were detected in the 21 populations, with a mean of 5 ± 3 regions detected per breed. The number of specific signatures detected in each breed was quite variable. Several breeds showed a large number of specific regions, such as Turopolje (14 regions) or Mora Romagnola (12 regions), while the numbers were smaller in other ones, such as Iberian or Sarda with just one region detected. Chromosomes 1 and 13 were the ones harboring the highest’s numbers of breed specific regions (15 and 17, respectively). The higher differentiation observed on Turopolje and Mora Romagnola may suggest a higher genetic drift in those breeds due to their limited effective population size50. The identified regions contain interesting candidate genes involved in functions and pathways related to productive and behavioral traits. Although a detailed discussion of the genes identified in each of the quoted regions is beyond the scope of this work, some findings can be highlighted.
Several different olfactory receptor genes were detected within the potentially selected regions in Apulo Calabrese (SSC9), Gascon (SSC13), Iberian (SSC15), Lithuanian Indigenous wattle (SSC4), Swallow-Bellied Mangalitsa (SSC2) and Mora Romagnola (SSC1). This is in agreement with the large repertoire of functional olfactory receptor genes in pigs, the fast evolution of these genes33,51 and their relevant role in smell and food finding, especially in these extensively-reared breeds. The putative selection of different subfamilies in each breed could match with the specialization in the detection of specific odors, characteristic of each breeds’ environment, as there is a wide functional diversity among olfactory receptors subfamilies. In fact, different gene clusters have been potentially associated with the recognition of specific odorants42. Moreover, the relevance of these genes exceeds their role in olfaction as they are also implicated in reproductive and behavioral traits, which may influence fitness.
Also, candidate genes involved in relevant metabolic functions associated with the adipogenic phenotype of our breeds are identified within the putative selected regions. For instance, in the Basque breed, the genes ACOX1 and CPT1A, both involved in fatty acid metabolism are identified. NFKBIA and PPARGC1B which have both been associated with backfat thickness in pigs52,53 are detected in Krškopolje and Cinta Senese breeds, respectively. DECR1, a positional candidate gene for the first fatness QTL detected in pigs on SSC454,55, is observed in a potentially selected region in Gascon breed. DLK1, a gene with a fundamental role in muscle growth and fat deposition56, is detected in Majorcan Black. LPIN1, which has been associated with obese pig phenotypes57, is detected in Turopolje breed. Taste receptor genes play a fundamental role in survival through the identification of dietary nutrients or potentially toxic substances, being linked to eating behavior and adaptation to specific geographical locations and diets58 and potentially related to growth and fat deposition10,59. Among this gene family, the TAS2R16 gene was detected in a selection signature in the Turopolje breed. Genes involved in the endocrine regulation of growth and insulin signalling, are also observed: IRS1 is detected in Casertana; GAL and GALR2 in Basque, and members of the insulin like growth factor binding protein gene family are detected in Black Slavonian. These genes code for signalling molecules that integrate and coordinate numerous biologically key extracellular signals within the cell. Some of them are intermediate of the insulin signalling, with a key role in growth, fatness and energy homeostasis60,61.
Different and abundant genes involved in proteolysis were also found, such as HEDTC2 and IDE in Alentejana; USP54 in Casertana; CAPN10, RNPEPL1 in Iberian; PSMA6 in Krškopolje, CTSV in Turopolje, or UBE4B, RNF150 and UBE2E1 detected in 5 or more breeds simultaneously (Table 3). Increased protein turnover has been proposed as a potential determinant for the limited muscle growth usually observed in local pigs2,62,63. A signal of diversifying selection was reported by Wilkinson et al.64 and Ai et al.65 in European pig breeds, close to the EDNRB gene, which is implicated in coat color pattern in mammals. This same region has been detected in the present work in Apulo Calabrese breed. Nevertheless, signals of selection were not detected near the two main known coat color genes, KIT and MC1R, for which allelic variation is associated with many of the coat color variants in pigs66,67. This could be due to incomplete coverage or informativity of the SNP chip in these particular regions.
Regarding Wild Boar-specific selection signatures, only three regions were detected. Two of them are located in SSC7, an autosome that has been repeatedly shown to be associated with domestication and behavior-related traits in QTL and GWAS studies68. Interestingly, the TECTB gene is potentially included in a selected genome region in Wild Boar. This gene is expressed in the inner ear and has a main role in hearing, which may be associated with survival in wild environmental conditions. One of the regions detected in SSC7 (33.55–33.59 cM) is located very close to PPARD gene (31.22–31.29 cM), related to ear morphology, fat deposition and growth, and detected previously as being located in a differentiated genomic region between European breeds and Wild Boar64.
In previous works analyzing different commercial and traditional pig populations, a number of regions showing between-breed signatures of selection has been identified16,64. In these studies, as well as in the present one, genes mapped to these regions can be considered as candidates under selection in pig breeds. Some common biological functions have been detected in different works, such as olfaction, growth or muscle development69,70. Nevertheless, when comparing different works, variable regions and genes have been observed probably due to differences in the breeds analyzed, statistical methods, SNP density or sample size. Especially, domestic pigs under different evolution and production conditions show different selection signatures and in our case all tested populations are locally produced breeds, which have not been selected for lean meat content, muscularity or enhanced reproduction. Thus, in a differentiation analysis among those breeds, expected signatures may be weaker than those observed in commercial and highly selected genotypes, or more related to domestication and breed standards establishment than actual artificial selection processes.
Conclusions
The obtained results were useful for the characterization of the genomic diversity of autochthonous European pig breeds. Results highlighted the genetic closeness among several of these domesticated breeds, and with their wild ancestor, as well as clear differentiation of some other ones and confirm the need of conservation programs to protect their genetic pools. Linkage disequilibrium patterns and extent have been determined at the genome level for a wide repertoire of European traditional breeds, showing potential effects of admixture and inbreeding. Putative signals of selection were detected for regions containing genes involved in growth, muscle development, reproduction, metabolism, behavior and sensory perception. Our findings improve the knowledge about the genome biology of European local pig breeds, and provide candidate genes for relevant traits, as well as useful information for future conservation, association or selection approaches.
Methods
Animals and sampling
Experts of each country, including personnel from breeding organizations and herd books, selected the animals to be included in the analyses in order to get a representative sampling of each breed. Selection of individuals for sampling was performed by avoiding highly related animals (no full- or half-sibs), balancing between sexes and prioritizing adult individuals or at least animals with adult morphology. Blood samples were obtained from 39 to 54 individuals from each one of the 20 local pig breeds included in the study: Black Slavonian and Turopolje (Croatia), Basque and Gascon (France), Schwäbisch-Hällisches Schwein (Germany), Apulo-Calabrese, Casertana, Cinta Senese, Mora Romagnola, Nero Siciliano and Sarda (Italy), Lithuanian indigenous wattle and Lithuanian White old type (Lithuania), Alentejana and Bísara (Portugal), Moravka and Swallow-Bellied Mangalitsa (Serbia), Krškopolje pig (Slovenia) and Iberian and Majorcan Black (Spain). Besides, seven European wild boars were employed as outgroup. Specialized professionals from each institution that provided animal material obtained blood samples following standard routine monitoring procedures and guidelines, at farm or at slaughter. No procedures with animals were performed that would demand ethical protocols according to Directive 2010/63/EU (2010) and blood samples were obtained as a general breeding procedure or previously collected DNA only reused here. A total of 992 DNA samples were genotyped.
The genomic DNA was extracted from leukocytes present in 8–15 mL of peripheral blood, collected in Vacutainer tubes containing 10% 0.5 M EDTA (ethylenediaminetetraacetic acid, disodium dihydrate salt) at pH 8.0. The extraction was performed using either a standardized phenol-chloroform, high-salt method or a commercial kit71.
Genotyping
Samples were genotyped with the GeneSeek ® GGP Porcine HD Genomic Profiler v1 (Illumina Inc, USA), which includes 68,516 SNPs evenly distributed with a median of 25 kb gap spacing and sharing 42,372 markers with Illumina porcine SNP60 chip. The average genotyping call rate was 0.94.
Genotype quality control (QC) and data filtering were performed using PLINK72. SNPs with MAF lower than 0.01 or more than 10% missing genotypes were excluded from the analyses in a preliminary filter to inspect the distribution of MAF across the genotyped SNPs. In addition, an individual call rate threshold was set to 95% and ten samples (two Bísara, two Casertana, one Cinta Senese, one Moravka two Nero Siciliano and two Schwäbisch-Hällisches) were removed for further analyses.
Data analyses
A total of 51,219 SNPs mapped on the 18 autosomes on Sus Scrofa 11.1 were used to compute, for the 20 studied breeds, the following indicators of genetic diversity: observed (HO) and expected heterozygosity (HE), inbreeding coefficient of an individual (I) relative to the subpopulation (S) (FIS), fixation index (FST) and inbreeding coefficient of an individual (I) relative to the total (T) population (FIT), heterozygosity index based on observed heterozygosity in individuals within breeds (HI) based on expected heterozygosity in subpopulation (HS) and based on expected heterozygosity for overall breeds (HT). Calculations were made with VCFtools software73. Nei’s genetic distances12 were calculated in R74 environment. Pairwise distances were used to build a NJ tree with the nj function belonging to the ape75 library in R. In addition, population structure was inspected trough PCA analyses performed with DISSECT software tool76.
Linkage disequilibrium (LD) analyses
Markers with MAF lower than 0.05, with more than a 10% of missing values, significantly deviating from Hardy-Weinberg equilibrium (P < 8.27 × 10−7) and unmapped or mapped in sexual chromosomes were excluded from LD analysis. This QC filtering was carried out independently for each breed and the number of SNP used is showed in Supplementary Table 3. The LD coefficient r2 was estimated for all marker pairs less than 50 Mb for each population and autosome independently using PLINK. To plot LD vs physical distance between markers, following Saura et al.4, we divided SNP pairs into three distance classes, (a) 0 to 2 Mb, (b) 2 to 5 Mb and (c) 5 to 50 Mb. Distance bins of 0.05, 0.20 and 5.0 Mb were used for classes (a), (b) and (c), respectively, and average r2 values for each bin were plotted against physical distance. Sample sizes were similar for all the breeds except for Wild Boar. As sample size can have an influence on LD estimation, a correction for sampling size was used to estimate r2 in Wild Boar as follows: (r2 − 1/N) (1 − 1/N)77.
Effective population size across generations
Estimates of effective population size (Ne) for each population were computed using the relationship between LD and Ne according to the following equation39,78:
where c is the distance between SNPs (Morgans), Ne is the sample size and absence of mutation was assumed () and N is equal to the number of diploid individuals sampled in the analyses. r2 estimates were computed between all pairs within 50 Mb windows within chromosome. Physical distances were converted to genetic distances in Morgans taking into account the specific recombination rate estimated for each chromosome by Muñoz et al.79. To estimate population size per generation, r2 between SNP pairs at a determined specific genetic distance corresponding to t = 1/2c80, where t is the generation, was considered. Finally, Ne at each generation was estimated through a non-linear least square approach based on the equation mentioned above. Ne estimates were calculated back for a period of 50 generations based on the same set of genotypes used in the linkage disequilibrium analyses (Supplementary Table 3).
FST analyses
Hardy-Weinberg equilibrium was inspected individually in all breeds. SNPs significantly deviating from Hardy-Weinberg equilibrium (P < 8.27 × 10−7) in at least one of the studied breeds were removed. A total of 828 SNPs was extracted from the subset of 51,219 SNPs used to compute genetic diversity parameters. Pairwise Wright’s FST81 were estimated as a measure of genetic differentiation according to the method described in Wilkinson et al.64. For each breed, a total of 21 breed-pairwise comparisons at each SNP were obtained and they were averaged to get overall FST for each SNP per breed. FST values were averaged in sliding windows of 13 SNPs centred at the 7th SNP which determined the genomic location of the window. Regions in the 99th percentile of the empirical distributions of windows per breed were defined as candidate regions to genetic differentiation. Genes were annotated with Biomart tool82.
Supplementary information
Acknowledgements
This work has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 634476 for the project with acronym TREASURE. The content of this article reflects only the authors’ view and the European Union Agency is not responsible for any use that may be made of the information it contains.
Author Contributions
C.O., L.F., M.C.P. and A.I.F. conceived and designed the study and obtained funding. Y.N. and F.G. performed the wet lab work. M.M. conducted bioinformatics analyses. M.S., R.C., J.M.M., R.Q., J.T., V.M., I.D., M.J.M., J.R., C.Z., V.R., J.P.A., C.R., R.S., D.K. and M.G. provided samples. C.O., M.M., R.B., J.G.C., A.R., O.F., G.S., S.B., V.J.U., R.C., J.M.M., R.Q., I.D., M.J.M., J.E., D.K., L.F., M.C.P. and A.I.F. contributed to data interpretation. C.O. and M.M. wrote the paper. All authors read and approved the submitted version.
Data Availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The raw genetic datasets generated during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49830-6.
References
- 1.FAO. Sources of Meat. Food and Agriculture Organization. Available at, http://www.fao.org/ag/againfo/themes/en/meat/backgr_sources.html, accessed 6/9/2018 (2018).
- 2.Čandek-Potokar, M., Nieto, R. & Project, T. Intech book European Local Pig Breeds - Diversity and Performance A study of project TREASURE (2019).
- 3.Weitzman ML. What to preserve? An application of diversity theory to crane conservation. Q. J. Econ. 1993;108:157–183. doi: 10.2307/2118499. [DOI] [Google Scholar]
- 4.Saura M, Tenesa A, Woolliams JA, Fernández A, Villanueva B. Evaluation of the linkage-disequilibrium method for the estimation of effective population size when generations overlap: An empirical case. BMC Genom. 2015;16:922. doi: 10.1186/s12864-015-2167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silió L, Barragán C, Fernández AI, García-Casco J, Rodríguez MC. Assessing effective population size, coancestry and inbreeding effects on litter size using the pedigree and SNP data in closed lines of the Iberian pig breed. J. Anim. Breed. Genet. 2016;133:145–154. doi: 10.1111/jbg.12168. [DOI] [PubMed] [Google Scholar]
- 6.Bertolini F, et al. Exploiting phenotype diversity in a local animal genetic resource: Identification of a single nucleotide polymorphism associated with the tail shape phenotype in the autochthonous Casertana pig breed. Livest. Sci. 2018;216:148–152. doi: 10.1016/j.livsci.2018.08.007. [DOI] [Google Scholar]
- 7.Schiavo G, et al. Taking advantage from phenotype variability in a local animal genetic resource: identification of genomic regions associated with the hairless phenotype in Casertana pigs. Anim. Genet. 2018;49:321–325. doi: 10.1111/age.12665. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz M, et al. Diversity across major and candidate genes in European local pig breeds. PLoS One. 2018;13:e0207475. doi: 10.1371/journal.pone.0207475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontanesi, L., Scotti, E., Gallo, M., Nanni Costa, L. & Dall’Olio, S. Authentication of ‘mono-breed’ pork products: Identification of a coat colour gene marker in Cinta Senese pigs useful to this purpose. Livest. Sci. 184, 71–77 (2016).
- 10.Ribani A, et al. Signatures of de-domestication in autochthonous pig breeds and of domestication in wild boar populations from MC1R and NR6A1 allele distribution. Anim. Genet. 2019;50:166–171. doi: 10.1111/age.12771. [DOI] [PubMed] [Google Scholar]
- 11.Groeneveld LF, et al. Genetic diversity in farm animals - A review. Anim. Genet. 2010;41:6–31. doi: 10.1111/j.1365-2052.2010.02038.x. [DOI] [PubMed] [Google Scholar]
- 12.Herrero-Medrano JM, et al. Conservation genomic analysis of domestic and wild pig populations from the Iberian Peninsula. BMC Genet. 2013;14:106. doi: 10.1186/1471-2156-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muñoz M, et al. Survey of SSC12 regions affecting fatty acid composition of intramuscular fat using high-density SNP data. Front. Genet. 2012;2:101. doi: 10.3389/fgene.2011.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontanesi L, Schiavo G, Galimberti G, Calò DG, Russo V. A genomewide association study for average daily gain in Italian large white pigs. J. Anim. Sci. 2014;92:1385–1394. doi: 10.2527/jas.2013-7059. [DOI] [PubMed] [Google Scholar]
- 15.Knol EF, Nielsen B, Knap PW. Genomic selection in commercial pig breeding. Anim. Front. 2016;6:15–22. doi: 10.2527/af.2016-0003. [DOI] [Google Scholar]
- 16.Rubin C-J, et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos AM, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS One. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samorè AB, Fontanesi L. Genomic selection in pigs: State of the art and perspectives. Ital. Journ. Anim. Sci. 2016;15:211–232. doi: 10.1080/1828051X.2016.1172034. [DOI] [Google Scholar]
- 19.Herrero-Medrano JM, et al. Whole-genome sequence analysis reveals differences in population management and selection of European low-input pig breeds. BMC Genom. 2014;15:601. doi: 10.1186/1471-2164-15-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laval G, et al. Genetic diversity of eleven European pig breeds. Genet. Sel. Evol. 2000;32:187. doi: 10.1186/1297-9686-32-2-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan B, et al. Genetic variation analysis within and among Chinese indigenous swine populations using microsatellite markers. Anim. Genet. 2002;33:422–427. doi: 10.1046/j.1365-2052.2002.00898.x. [DOI] [PubMed] [Google Scholar]
- 22.Luetkemeier ES, Sodhi M, Schook LB, Malhi RS. Multiple Asian pig origins revealed through genomic analyses. Mol. Phylogenet. Evol. 2010;54:680–686. doi: 10.1016/j.ympev.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Blackburn H, Faria DA, Wilson C, Paiva SR. P4066 Genetic diversity of pig populations from the US mainland, Pacific islands and China: Autosomal SNP evaluation. J. Anim. Sci. 2016;94:112. doi: 10.2527/jas2016.94supplement4112x. [DOI] [Google Scholar]
- 24.Druml T, et al. Genetic diversity, population structure and subdivision of local Balkan pig breeds in Austria, Croatia, Serbia and Bosnia-Herzegovina and its practical value in conservation programs. Genet. Sel. Evol. 2012;44:5. doi: 10.1186/1297-9686-44-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caballero, A. Genética Cuantitativa. (Síntesis, 2017).
- 26.ANAS. Libro Genealogico. Available at, http://www.anas.it/ (2019).
- 27.Peretti V, Ciotola F, Ianuzzi L. Characterization, conservation and sustainability of endangered animal breeds in Campania (Southern Italy) Nat. Sci. 2013;5:1–9. [Google Scholar]
- 28.Zhang C, Plastow G. Genomic Diversity in Pig (Sus scrofa) and its Comparison with Human and other Livestock. Curr. Genomics. 2011;12:138–146. doi: 10.2174/138920211795564386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edea Z, Kim S-W, Lee K-T, Kim TH, Kim K-S. Genetic Structure of and evidence for admixture between Western and Korean native pig breeds revealed by single nucleotide polymorphisms. Asian-Australasian J. Anim. Sci. 2014;27:1263. doi: 10.5713/ajas.2014.14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei M. Genetic Distance between Populations. Am. Nat. 1972;106:283–292. doi: 10.1086/282771. [DOI] [Google Scholar]
- 31.Karolyi Danijel, Luković Zoran, Salajpal Krešimir, Škorput Dubravko, Vnučec Ivan, Mahnet Željko, Klišanić Vedran, Batorek-Lukač Nina. European Local Pig Breeds - Diversity and Performance. A study of project TREASURE. 2019. Turopolje Pig (Turopoljska svinja) [Google Scholar]
- 32.Gvozdanović K, et al. Genetic diversity of autochthonous pig breeds analyzed by microsatellite markers and mitochondrial DNA D-loop sequence polymorphism. Anim. Biotechnol. 2019;30:242–251. doi: 10.1080/10495398.2018.1478847. [DOI] [PubMed] [Google Scholar]
- 33.Groenen MAM, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491(7424):393. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Megens HJ, et al. Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: Differences in microsatellite variation between two areas of domestication. Genet. Sel. Evol. 2008;40:103–128. doi: 10.1186/1297-9686-40-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razmaitė Violeta, Šveistienė Rūta, Jatkauskienė Virginija, Juška Remigijus, Leikus Raimondas, Batorek-Lukač Nina. European Local Pig Breeds - Diversity and Performance. A study of project TREASURE. 2019. Lietuvos Vietinė (Lithuanian Indigenous Wattle) Pig. [Google Scholar]
- 36.Bradić M, et al. Microsatellite analysis of the genetic diversity in the Black Slavonian pig. Acta Vet.-Beogr. 2007;57:209–215. doi: 10.2298/AVB0703209B. [DOI] [Google Scholar]
- 37.Margeta Vladimir, Gvozdanović Kristina, Kušec Goran, Djurkin Kušec Ivona, Batorek-Lukač Nina. European Local Pig Breeds - Diversity and Performance. A study of project TREASURE. 2019. Black Slavonian (Crna slavonska) Pig. [Google Scholar]
- 38.Savić Radomir, Radović Čedomir, Petrović Milica, Gogić Marija, Radojković Dragan, Batorek-Lukač Nina. European Local Pig Breeds - Diversity and Performance. A study of project TREASURE. 2019. Moravka Pig. [Google Scholar]
- 39.Tenesa A, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002;3:299. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 41.Nordborg M, Tavaré S. Linkage disequilibrium: What history has to tell us. Trends Genet. 2002;182:83–90. doi: 10.1016/S0168-9525(02)02557-X. [DOI] [PubMed] [Google Scholar]
- 42.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 43.Farnir F, et al. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 2000;10:220–227. doi: 10.1101/gr.10.2.220. [DOI] [PubMed] [Google Scholar]
- 44.Amaral AJ, Megens HJ, Crooijmans RPMA, Heuven HCM, Groenen MAM. Linkage disequilibrium decay and haplotype block structure in the pig. Genetics. 2008;179:569–579. doi: 10.1534/genetics.107.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stratz P, Wimmers K, Meuwissen THE, Bennewitz J. Investigations on the pattern of linkage disequilibrium and selection signatures in the genomes of German Piétrain pigs. J. Anim. Breed. Genet. 2014;131:473–482. doi: 10.1111/jbg.12107. [DOI] [PubMed] [Google Scholar]
- 46.Meuwissen TH. Accuracy of breeding values of ‘unrelated’ individuals predicted genotyping. Genet. Sel. Evol. 2009;41:35. doi: 10.1186/1297-9686-41-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grunenfelder H-P. Saving the Turopolje pig. An International Pilot Project in Croatia in collaboration with EURONATUR. Stočarstvo Časopis za Unapr. stočarstva. 1994;48:361–364. [Google Scholar]
- 48.Razmaite, V. Lithuanian pig genetic resources and their conservation. In 59th Annual Meeting of the European Association for Animal Production (2008).
- 49.Crovetti, A., Sirtori, F., Pugliese, C., Franci, O. & Bozzi, R. Pedigree analysis of Cinta Senese and Mora Romagnola breeds. In 8th International Symposium on the Mediterranean Pig, Slovenia, Ljubljana, October 10th−12th, 2013. 41–44 (2013).
- 50.Kullo IJ, Ding K. Patterns of population differentiation of candidate genes for cardiovascular disease. BMC Genet. 2007;8:48. doi: 10.1186/1471-2156-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen DT, et al. The complete swine olfactory subgenome: expansion of the olfactory gene repertoire in the pig genome. BMC Genom. 2012;13:584. doi: 10.1186/1471-2164-13-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flisar T, Kunej T, Kovac M, Dovc P. Effect of PPARGC-1 gene on backfat thickness in pigs. Acta Agric. Slov. 2006;88:11–18. [Google Scholar]
- 53.Fontanesi L, et al. A genome wide association study for backfat thickness in Italian Large White pigs highlights new regions affecting fat deposition including neuronal genes. BMC Genom. 2012;13:583. doi: 10.1186/1471-2164-13-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersson L, et al. Genetic mapping of quantitative trait loci for growth and fatness in pigs. Science. 1994;263:1771–1774. doi: 10.1126/science.8134840. [DOI] [PubMed] [Google Scholar]
- 55.Clop A, et al. Detection of QTL affecting fatty acid composition in the pig. Mamm. Genome. 2003;14:650–656. doi: 10.1007/s00335-002-2210-7. [DOI] [PubMed] [Google Scholar]
- 56.Kim KS, Kim JJ, Dekkers JCM, Rothschild MF. Polar overdominant inheritance of a DLK1 polymorphism is associated with growth and fatness in pigs. Mamm. Genome. 2004;15:552–559. doi: 10.1007/s00335-004-2341-0. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, et al. The mRNA of lipin1 and its isoforms are differently expressed in the longissimus dorsi muscle of obese and lean pigs. Mol. Biol. Rep. 2011;38:319–325. doi: 10.1007/s11033-010-0110-6. [DOI] [PubMed] [Google Scholar]
- 58.da Silva EC, et al. Characterization of the porcine nutrient and taste receptor gene repertoire in domestic and wild populations across the globe. BMC Genom. 2014;15:1057. doi: 10.1186/1471-2164-15-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clop A, et al. Identification of protein-damaging mutations in 10 swine taste receptors and 191 appetite-reward genes. BMC Genomics. 2016;17:685. doi: 10.1186/s12864-016-2972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lindskog S, et al. Galanin of the homologous species inhibits insulin secretion in the rat and in the pig. Acta Physiol. Scand. 1990;139:591–596. doi: 10.1111/j.1748-1716.1990.tb08963.x. [DOI] [PubMed] [Google Scholar]
- 61.Gual P, Le Marchand-Brustel Y, Tanti JF. Positive and negative regulation of insulin signaling through IRS-1 phosphorylation. Biochimie. 2005;87:99–109. doi: 10.1016/j.biochi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 62.Rivera-Ferre MG, Aguilera JF, Nieto R. Differences in whole-body protein turnover between Iberian and Landrace pigs fed adequate or lysine-deficient diets. J. Anim. Sci. 2006;84:3346–55. doi: 10.2527/jas.2005-405. [DOI] [PubMed] [Google Scholar]
- 63.Óvilo C, et al. Longissimus dorsi transcriptome analysis of purebred and crossbred Iberian pigs differing in muscle characteristics. BMC Genomics. 2014;15:413. doi: 10.1186/1471-2164-15-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilkinson S, et al. Signatures of Diversifying Selection in European Pig Breeds. PLoS Genet. 2013;9:e1003453. doi: 10.1371/journal.pgen.1003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ai, H., Huang, L. & Ren, J. Genetic Diversity, Linkage Disequilibrium and Selection Signatures in Chinese and Western Pigs Revealed by Genome-Wide SNP Markers. PLoS One8 (2013). [DOI] [PMC free article] [PubMed]
- 66.Fang, M., Larson, G., Ribeiro, H. S., Li, N. & Andersson, L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5 (2009). [DOI] [PMC free article] [PubMed]
- 67.Fontanesi L, Russo V. Molecular Genetics of coat Colour in Pigs. Acta Agric Slov. 2013;4:16. [Google Scholar]
- 68.Ramos-Onsins SE, Burgos-Paz W, Manunza A, Amills M. Mining the pig genome to investigate the domestication process. Heredity (Edinb). 2014;113:471. doi: 10.1038/hdy.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amaral AJ, et al. Genome-wide footprints of pig domestication and selection revealed through massive parallel sequencing of pooled DNA. PLoS One. 2011;6:e14782. doi: 10.1371/journal.pone.0014782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li M, et al. Detecting mitochondrial signatures of selection in wild Tibetan pigs and domesticated pigs. Mitochondrial DNA. 2016;27:747–752. doi: 10.3109/19401736.2014.913169. [DOI] [PubMed] [Google Scholar]
- 71.Montgomery GW, Sise JA. Extraction of dna from sheep white blood cells. New Zeal. J. Agric. Res. 1990;33:437–441. doi: 10.1080/00288233.1990.10428440. [DOI] [Google Scholar]
- 72.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fehske H., Schneider R., Weiße A., editors. Computational Many-Particle Physics. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008. [Google Scholar]
- 75.Paradis E, Claude J, Strimmer K. Bioinformatics. 2004. APE: Analyses of phylogenetics and evolution in R language; pp. 289–290. [DOI] [PubMed] [Google Scholar]
- 76.Canela-Xandri O, Law A, Gray A, Woolliams JA, Tenesa A. A new tool called DISSECT for analysing large genomic data sets using a Big Data approach. Nat. Commun. 2015;6:10162. doi: 10.1038/ncomms10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 78.Sved JA. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 1971;2:125–141. doi: 10.1016/0040-5809(71)90011-6. [DOI] [PubMed] [Google Scholar]
- 79.Muñoz M, et al. Recombination rates across porcine autosomes inferred from high-density linkage maps. Anim. Genet. 2012;43:620–623. doi: 10.1111/j.1365-2052.2011.02301.x. [DOI] [PubMed] [Google Scholar]
- 80.Hayes BJ, Visscher PM, McPartlan HC, Goddard ME. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 2003;13:635–643. doi: 10.1101/gr.387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wright S. Evolution in mendelian populations. Genetics. 1931;16:97. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smedley D, et al. BioMart - Biological queries made easy. BMC Genom. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The raw genetic datasets generated during the current study are available from the corresponding author on reasonable request.