Abstract
Background
Cognitive and language dysfunction is common among patients with glioma and has a significant impact on survival and health-related quality of life (HRQOL). Little is known about the factors that make individual patients more or less susceptible to the cognitive sequelae of the disease. A better understanding of the individual and population characteristics related to cognitive function in glioma patients is required to appropriately stratify patients, prognosticate, and develop more efficacious treatment regimens. There is evidence that allelic variation among genes involved in neurotransmission and synaptic plasticity are related to neurocognitive performance in states of health and neurologic disease.
Methods
We studied the association of single-nucleotide polymorphism variations in brain-derived neurotrophic factor (BDNF, rs6265), dopamine receptor 2 (DRD2, rs1076560), and catechol-O-methyltransferase (COMT, rs4680) with neurocognitive function and ability to return to work in glioma patients at diagnosis and at 3 months. We developed a functional score based on the number of high-performance alleles that correlates with the capacity for patients to return to work.
Results
Patients with higher-performing alleles have better scores on neurocognitive testing with the Repeatable Battery for the Assessment of Neuropsychological Status and Stroop test, but not the Trail Making Test.
Conclusions
A better understanding of the genetic contributors to neurocognitive performance in glioma patients and capacity for functional recovery is necessary to develop improved treatment strategies based on patient-specific factors.
Keywords: cognition, glioma, health-related quality of life, language, return to work
Approximately 688 000 adults are currently living with a brain tumor in the United States, and 77 600 new gliomas are diagnosed every year.1,2 Gliomas are associated with the highest estimated number of potential years of life lost relative to any cancer.3 While the survival of some may be short, many with low- and intermediate-grade histology and favorable tumor genetics will experience extended survival periods. Mainstay treatment options both for low- and high-grade glioma include surgery, radiation therapy, and chemotherapy. While much effort is dedicated to developing novel treatments to extend survival, relatively little is understood about how to mitigate the cognitive sequelae attributable to gliomas and oncological therapies.4 Glioma patients commonly experience language and nonlanguage cognitive dysfunction,5 which significantly affects health-related quality of life (HRQOL).6 A better understanding of the mechanisms underlying cognitive dysfunction is central to a comprehensive management approach for glioma patients. However, currently there is no way to predict patients expected to have poor functional outcomes and delayed recovery.
A component of cognitive susceptibility and treatment-related neurotoxicity in glioma patients may be based on genetic variations. Constitutive genetic traits may affect the degree of cognitive dysfunction as well as functional recovery that occurs following chemoradiation. There are several single-nucleotide polymorphisms (SNPs) associated with language and cognition that could be significant in glioma patients. Prior reports have published that polymorphisms of catechol-O-methyltransferase (COMT), brain-derived neurotrophic factor (BDNF), and dystrobrevin binding protein 1 (DNTPB) affect cognition in a diverse population of adult brain tumor patients.7 However, this study included a wide range of intra- and extraaxial supratentorial brain tumors including meningiomas, brain metastases, and gliomas. Intra- and extraaxial brain tumors have significantly different patterns of growth and parenchymal infiltration. Given the differing mechanisms of tumor invasion in a heterogeneous group of brain tumor patients, we sought to study genetic polymorphisms known to influence cortical and subcortical plasticity in the adult low- and high-grade glioma population. To investigate the possibility that cognitive outcomes in glioma patients are related to allele variations, we studied the following 3 polymorphisms associated with adult CNS plasticity and cognitive recovery in neurological diseases: BDNF, dopamine receptor D2 (DRD2), and COMT. Cognitive recovery can be quantified by objective neurocognitive task performance. However, assessment of return to work at full-time capacity allows for measurement of the functional impact of these cognitive changes, and thus offers additional insight into “real-world” patient performance. We therefore studied the ability to return to work in addition to neurocognitive task performance as a primary endpoint measurement for functional recovery.
BDNF is the most abundant trophic molecule in the adult brain.8BDNF plays a central role in CNS plasticity and is directly involved in synaptic regulation and axonal plasticity through its effects on proliferation and differentiation of oligodendrocyte progenitor cells and dopamine receptor expression.9–11BDNF has been shown to contribute to synaptogenesis, cortical reorganization, and motor recovery following acute cerebral ischemia.12–15 An allelic variant of BDNF may influence intracellular transport. A SNP at position 196 on exon 2 results in an amino acid switch from valine (Val) to methionine (Met) at codon 66 (rs6265, BDNF A/G).16,17 This polymorphism results in reduced BDNF secretion both locally and at the synaptic cleft, with implications on memory and cognitive impairment following systemic chemotherapy.18
DRD2 influences dopamine signaling by inhibiting adenylyl cyclase through G1-protein coupling.19 A SNP in intron 6 of DRD2 (rs10076560, DRD2 G/T) affects exon 6 splicing, resulting in decreased dopamine production.19 Aberrant dopamine signaling such as the DRD2 G/T polymorphism is also associated with poorer working memory and attention processing combined with reduced striatal and prefrontal cortical activity with functional MRI.19–21
Dopamine is metabolized by COMT, which is present in mature and developing oligodendrocytes.21 A common polymorphism of COMT is the Val (G) variant at position 158 (rs4680, COMT G/A), which leads to lower thermostability and thereby more dopamine degradation.21,22 Carriers of the Val (G) allele perform more poorly on tests of executive function where frontostriatal dopamine is depleted relative to Met (A) carriers.23,24
Previous work has shown that polymorphisms in the COMT, DRD2, and BDNF genes are associated with motor learning, cognition, and sensorimotor adaptation both in aging and disease states.22 Based on the known molecular function of these 3 genes as well as emerging clinical evidence describing an impact of allelic variation on cognitive function, BDNF, COMT, and DRD2 allele status may affect language and nonlanguage cognitive recovery in adult glioma patients. We created a polygene score based on the number of high-performance alleles (NHPA) for patients with WHO grade II-IV glioma to further investigate this relationship. The NHPA score is obtained by assigning a 1- (lowest performing) through 3- (highest performing) point system for each of the BDNF, COMT, and DRD2 genes based on a patient’s genotype. The NHPA score is the total number of points for all 3 genes. NHPA has been shown to be associated with language and cognitive recovery in patients with breast cancer, Alzheimer dementia, and glioma.13–15,21,25–27 The goal of this study is to test the hypothesis that the NHPA score based on common polymorphisms of BDNF, COMT, and DRD2 correlates with baseline and 3-month postdiagnosis cognitive recovery in addition to ability to return to work in patients with WHO II-IV glioma.
Methods
Patients
The study design is a retrospective review of a prospectively collected database. Patients were assessed as part of the Functional Wellness Initiative and Brain Tumor Program at the University of Michigan between 2015 and 2017, where the standard of care included neuropsychological and language assessments.28 A total of 128 adult patients with WHO II, III, and IV newly diagnosed glioma were included in the study population. Baseline assessments were made at the time of first clinic visit after diagnosis of a brain mass and before surgical resection. All patients were evaluated again at an approximate 3-month interval after this initial assessment. Study eligibility included age >18 years, newly diagnosed glioma, and no history of psychiatric or other neurological disorders via chart review as well as interview in the clinic setting. All participants were fluent in English and had a baseline Boston Diagnostic Aphasia Examination Severity Examination score ≥2 permitting them to participate in language and neuropsychological assessment. Patients receiving chemoradiation within 3 months of diagnosis were excluded from an analysis of ability to return to work because of the significant potential confounding influence of treatment on return to work. All patients with newly diagnosed WHO grade IV glioma and 16/50 patients with WHO grade II or III glioma received chemoradiation within 3 months of diagnosis, excluding them from this portion of the study and leaving 34/50 patients in the return-to-work analysis. Neurocognitive testing data were available for 38 patients. The University of Michigan Institutional Review Board approved the study protocol and written consent was obtained from all participants (HUM00092238).
Extent of Resection
Volumetric analysis was performed to assess for difference in extent of resection between experimental groups. Pre- and postoperative T1-weighted postcontrast MRI studies were segmented for volumetric analysis using Slicer 3D software.29 Preoperative imaging was within 1 month of surgery and postoperative imaging was within 48 hours of surgery. Percentage resection was calculated for each patient.
Plasma BDNF Measurement
All patients provided a blood sample for genotyping. Blood samples were collected with EDTA as the anticoagulant. Plasma was isolated from whole blood after it had been centrifuged at 1000 × g for 10 minutes at 4°C immediately. An additional centrifuge at 1000 × g for 10 minutes at 4°C was performed to remove platelets and then plasma was immediately stored at –80°C until it was thawed for assay. Plasma BDNF levels were quantified using ELISA. A Quantikine Human Cytokine Kit (R&D Systems, Minneapolis, MN, USA) and an ELISA reader were used to analyze plasma BDNF levels. All assays were performed in duplicate and expressed as pictogram per milliliter. To control for the inter- and intraassay variation that is inherent to immunoenzymatic techniques, we included a control human plasma in duplicate in every ELISA plate (Ramanakumar, J Clin Microbiol 2010).
Genotyping
Candidate SNPs (COMT Val158Met, DRD2 G>T, and BDNF Val66Met) were selected based on reports in the literature on association with cognitive functions in healthy and clinical populations and evidence from imaging and in vitro assays.7 Genomic DNA was extracted from blood using a DNase Blood & Tissue Kit in accordance with the manufacturer’s instructions (Qiagen, Germantown, MD, USA). Individual gene polymorphisms were examined using polymerase chain reaction (PCR). For PCR, 1 μL of human blood DNA was amplified using Plantinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA, USA) for 35 cycles at 94°C for 15 seconds, at 58°C for 25 seconds, and at 68°C for 20 seconds. The following primers were used: BDNF (Val66Met, rs6265), forward: 5’-CAGGTGAGAAGAGTGATGACCA-3’, reverse: 5’-ATGGACATGTTTGCAGCATC-3’, COMT (Val158 Met, rs4680), forward: 5’-CACAGGCAAGATCGTGGAC-3’, reverse: 5’-GCCTGGTGATAGTGGGTTTT-3’, and DRD2 (G/T, rs1076560), forward: 5’-GGGAGAGTCAGCTGGTGG-3’, reverse: 5’-GGCAGAACAGAAGTGGGGTA-3’. The PCR products were electrophoresed on a 1% agarose gel and extracted with a QIAquick Gel Extraction Kit (Qiagen) and then sent to the University of Michigan DNA sequencing core for DNA sequencing of gene polymorphism. The homozygous low performing allele (LPA) for each of the 3 gene loci identified included BDNF A/A, DRD2 T/T, and COMT G/G and received zero points. Heterozygotes include BDNF A/G, DRDT G/T, and COMT G/A and were given 1 point for each locus. The high-performing (HPA) homozygotes were given 2 points at each locus (BDNF G/G, DRD2 G/G, and COMT A/A). Patients with an NHPA score of 0-4 were assigned to the LPA group while those with an NHPA score of 5-6 were assigned to the HPA group. While not validated, this scoring system is being used as an exploratory method, as a first step for construction of a clinically useful polygene risk score in a glioma patient population.30
Neurocognitive Assessment
All patients completed a thorough neuropsychological evaluation. We assessed cognitive functioning for all participants using the Repeatable Battery for the Assessment of Neuropsychological Status (RBANS). The RBANS comprises 12 subtests that are used to calculate 5 age-adjusted index scores and a total score.31 The test battery was administered and scored by a neuropsychologist member of the study team or by a trained research assistant (E.M.B., N.G., A.B.).
Test indices included:
Immediate memory: list learning, story memory tasks
Visuospatial and constructional: Fig. copy and line orientation tasks
Language: picture naming and semantic fluency tasks
Attention: digit span and coding tasks
Delayed memory: list recall, story recall, Fig. recall, and list recognition tasks
Raw test scores were collected at baseline and at 3 months postdiagnosis. The Trail Making Test (TMT) and Stroop test were used as well to more thoroughly assess executive function.32,33 Return to work at full capacity was collected prospectively as a discrete yes/no value.
Statistics
Mean test scores were calculated for each RBANS test index and subset indices for patients at baseline and at 3 months postdiagnosis. The mean differences between the 2 time points were also calculated. Patients were separated into LPA and HPA groups based on their NHPA score. Patients were also stratified by LPA and HPA groups for the individual BDNF, COMT, and DRD2 genes. Two sample t-tests were performed to compare mean scores at baseline, at 3 months postdiagnosis, and the changes in mean scores between baseline and 3 months between NHPA groups (LPA 0-4 vs HPA 5-6). Across all RBANS indices and subtests, higher scores represent favorable performance. On the TMT test, the dependent variable measured was the ratio of the raw scores for TMT B to TMT A in seconds where higher scores equal more difficulty with executive function of set-shifting/multitasking. For the Stroop test, the raw interference T-score was taken where lower scores equate to more difficulty with response inhibition.34 A 1-sample proportion test was used to compare the allele distribution in our data to the study population. A Fisher exact test was used to compare the rate of returning to work at full capacity by 3 months postdiagnosis between the study groups given insufficient population sample size to support a chi-squared test. Given the exploratory nature of this study, we did not perform additional statistical tests to control for multiple comparisons. However, we lowered the P value threshold to .01 for statistical significance to address multiple comparisons. Statistical analysis was performed using SAS 9.4.
Results
Population Demographics
Relevant population characteristics are given in Table 1. Briefly, the median age of the study population was 51.5 years with 52 (41%) women and 76 (59%) men. Seventy-eight (61%) were diagnosed with a WHO IV glioma, 31 (24%) were diagnosed with a WHO III glioma, and 19 (15%) were diagnosed with a WHO II glioma. Thirty-nine participants (30%) completed college. The majority of gliomas (n = 66, 52%) were located within the frontal lobe. Seventy-six (59%) gliomas were located in the left hemisphere. Extent of resection and additional demographic variables such as age, gender, tumor grade, location, and size were similar in low- and high-NHPA groups with no significant difference based on NHPA score. Extent of resection in the LPA group was 87 ± 12.5% and 89 ± 7.2% in the HPA group. At the time of the 3-month interval assessment, 94 (73.4%) patients had received chemoradiation. This number increased to 93% by the time of last clinic follow-up.
Table 1.
Patient Demographics and Disease History
| Variable (%) | Glioma (n = 128) |
|---|---|
| Age at Study Entry, Years | |
| Mean (SD) | 53 |
| Median (range) | 51.5 (19-87) |
| Gender | |
| Female | 52 (41%) |
| Male | 76 (59%) |
| Education (%) | |
| Completed college | 39 (30%) |
| Did not complete college | 89 (70%) |
| Glioma Grade | |
| WHO II | 19 (15%) |
| WHO III | 31 (24%) |
| WHO IV | 78 (61%) |
| Treatment Type | |
| RT ± chemotherapy | 119 (93%) |
| Chemotherapy | 120 (94%) |
| Handedness | |
| Right-handed | 118 (92%) |
| Left-handed | 7 (6%) |
| Both | 3 (2%) |
| Smoking Status (%) | |
| Smoker | 8 (6%) |
| Nonsmoker | 120 (94%) |
| Tumor location (%) | |
| Frontal | 66 (52%) |
| Parietal | 26 (20%) |
| Temporal | 27(21%) |
| Occipital | 1 (1%) |
| Insular | 8 (6%) |
| Other (thalamus, brainstem) | 0 (%) |
| Tumor Side | |
| Left | 76 (59%) |
| Right | 52 (41%) |
| Antiepileptic Medications | |
| Yes | 105 (82%) |
| No | 23 (18%) |
Abbreviation: RT, radiation therapy.
Genotyping Results
We genotyped the widely reported COMT rs4680 (Val158Met), DRD2 rs1076560, and BDNF rs6265 (Val66Met) polymorphisms for all DNA samples. Table 2 shows that the distribution of alleles for each polymorphism within the low- and high-grade glioma cohorts did not follow expected allelic frequencies according to the Hardy–Weinberg equilibrium for the COMT and DRD2 genes. The observed allelic frequency for the BDNF gene was not statistically different relative to the general population. For each of the 3 genes, LPA and HPA have been described for which the protein function or production is affected by SNP variation. A composite NHPA score was generated for each patient based on the allelic status for the specified locus on the BDNF, DRD2, and COMT genes with points given according to allele status. The homozygous LPA for each of the 3 gene loci identified receives no points (BDNF A/A, DRD2 T/T, and COMT G/G). Heterozygotes are given 1 point for each locus (BDNF A/G, DRDT G/T, and COMT G/A). The high-performing homozygotes are given 2 points at each locus (BDNF G/G, DRD2 G/G, and COMT A/A). Therefore, the total number of HPA (ie, NHPA score) ranges from 0 to 6 (Table 3). There were no differences in population demographics between the groups with respect to mean age at presentation (LPA = 54.2, HPA = 53.6, P = .85), tumor side (LPA: 46% right-sided, HPA: 37% right-sided, P = .31), and extent of resection (LPA: 87%, HPA: 89%, P = .43).
Table 2.
Frequency of Allelic Distribution for COMT, DRD2, and BDNF genes
| Gene (RSID) |
Allele | % observed | % expected | P Value |
|---|---|---|---|---|
|
COMT
(rs4680) |
A/A | 34 (43/128) | 30 | .0001 |
| G/A | 55 (71/128) | 40 | ||
| G/G | 11 (14/128) | 30 | ||
|
DRD2
(rs1076560) |
G/G | 63 (80/128) | 75 | .0215 |
| G/T | 34 (44/128) | 23 | ||
| T/T | 3 (4/128) | 2 | ||
|
BDNF
(rs6265) |
G/G | 68 (87/128) | 62 | .3144 |
| A/G | 28 (36/128) | 35 | ||
| A/A | 4 (5/128) | 3 |
Abbreviations: BDNF, brain-derived neurotrophic factor; COMT, catechol-O-methyltransferase; DRD2, dopamine receptor 2; RSID, reference single-nucleotide polymorphism cluster ID;
The observed values represent the percentage of allelic distribution in our sample of adults with low- and high-grade glioma. The expected values represent the Hardy–Weinberg equilibrium for the percentage of allelic distribution in a mixed gender/age population for the BDNF allele though not for the COMT or DRD2 alleles.
Table 3.
Frequency of NHPA Score in Our Mixed Sample of Adults with WHO II, III, and IV Gliomas.
| NHPA | Frequency (%) WHO II and III | Frequency (%) WHO IV | Total | 3-Month RTW 42% (10/24) |
|---|---|---|---|---|
| 0 | 0 | 0 | 0 | |
| 1 | 0 | 0 | 0 | |
| 2 | 2(1/50) | 3 (2/78) | 2 (3/128) | |
| 3 | 16 (8/50) | 15 (12/78) | 16 (20/128) | 18.2% (2/11) |
| 4 | 24 (12/50) | 29 (23/78) | 27 (35/128) | |
| 5 | 48 (24/50) | 40 (31/78) | 43 (55/128) | 61.5% (8/13)* |
| 6 | 10(5/50) | 13 (10/78) | 12 (15/128) |
Abbreviations: NHPA, number of high-performance alleles; RTW, return to work.
The frequency of NHPA scores in patients by WHO grade. Forty-two percent of patients returned to work. The frequency of return to work was higher in patients with an NHPA score ≥5 and was significantly higher than in patients with an NHPA score <5. (P = .047).
Return-to-Work Results by NHPA Score
Analysis of ability to return to work at full capacity was limited only to individuals with WHO II and III gliomas for whom the decision was made to not administer chemoradiation during the first 3 months after diagnosis. A greater percentage of HPA patients were able to return to work at full capacity 3 months after diagnosis compared with those in the LPA group (LPA: 18.2%, HPA: 61.5%, P = .047) (n = 24) (Table 3).
Association of SNPs, NHPA Scores and Plasma BDNF
Similarly, patients in the HPA group had higher circulating plasma BDNF levels compared with the LPA group (HPA: 1114.21, LPA: 742.56 pg/mL, P = .047) (Table 4). However, this latter finding was significant only in patients with WHO IV gliomas; no statistically significant difference in circulating plasma BDNF was seen in patients with grade II and III gliomas regardless of NHPA score. This pattern was also observed when examining plasma BDNF based on WHO grade and genotype (Table 5). Plasma BDNF for patients with WHO IV glioma is significantly lower in the A/G cohort relative to those who are G/G homozygous (344.52 vs 1354.59 pg/mL, P = .047) (Table 5).
Table 4.
Plasma BDNF by WHO grade and NHPA Score
| Glioma Grade | Plasma BDNF(pg/ml) | ||
|---|---|---|---|
| NHPA (≤4) | NHPA (5-6) | P Value | |
| WHO II and III | 885.28 (9/47) | 785.39 (38/47) | .1508 |
| WHO IV | 742.56 (14/75) | 1114.21 (61/75) | .0473 |
Abbreviations: BDNF, brain-derived neurotrophic factor; NHPA, number of high-performance alleles.
Plasma BDNF levels are significantly higher in the high-performing NHPA 5-6 patients relative to the low-performing NHPA ≤4 group; however, this difference is observed only in WHO IV glioma patients rather than in patients with WHO II-III lower-grade glioma.
Table 5.
Plasma BDNF by WHO Grade and BDNF Genotype
| Glioma Grade | Plasma BDNF (pg/ml) | ||
|---|---|---|---|
| BDNF Val/Val | BDNF Val/Met | P Value | |
| WHO II and III | 693.36 (31/47) | 1122.03 (16/47) | .1366 |
| WHO IV | 1354.59 (52/75) | 344.52 (23/75) | .0471 |
Abbreviations: BDNF, brain-derived neurotrophic factor; Met, methionine; Val, valine.
Plasma BDNF levels are significantly higher in the BDNF Val/Met patients relative to the BDNF Val/Val patients; however, this difference is observed only in WHO IV glioma patients rather than in patients with WHO II-III lower-grade glioma.
Association of SNPs, NHPA Scores and Neurocognitive Outcomes
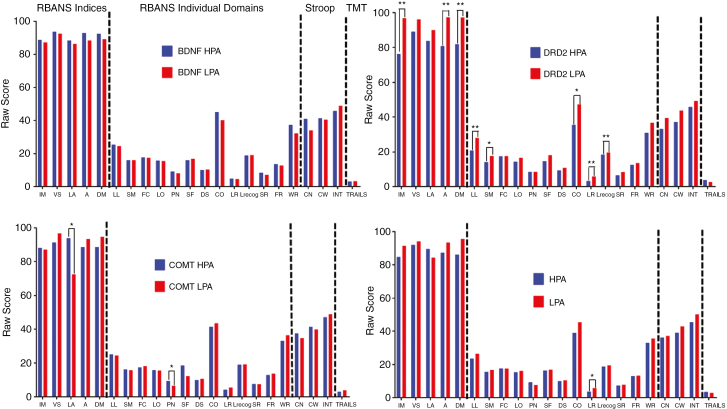
We assessed neurocognitive function in patients at baseline and 3 months postdiagnosis using RBANS, Stroop, and TMT tests. The total number of patients with complete genotyping and neuropsychological test data at baseline and 3 months was 32 and 25, respectively. Patients in the HPA group performed better on the RBANS list recall test at baseline, though otherwise there were no differences in baseline performance by NHPA group (Fig. 1). Interestingly, when comparing patients with DRD2 LPA to those with HPA, patients with LPA performed better on RBANS immediate memory, delayed memory, and attention tasks at baseline (Fig. 1). Patients with COMT HPA performed better on picture-naming language testing (Fig. 1). There were no differences in RBANS performance by BDNF genotype at baseline.
Fig. 1.
Performance at Baseline by Allele and NHPA Score.
Mean scores for RBANS indices, RBANS individual domains, Stroop, and TMT tests (from left to right) at baseline by individual low- (LPA) and high- (HPA) performing BDNF (top left), DRD2 (top right), and COMT (bottom left) alleles along with total number of high-performing alleles (NHPA) (LPA = NHPA 0 to 4, HPA = NHPA 5 to 6) (bottom right). RBANS data are presented as mean raw score. Stroop data are presented as mean interference T score. TMT data are presented as mean ratio of the score on Part B to Part A (Trails B/A). A indicates attention; CN, color naming; CO, coding; CW, color-word condition; DM, delayed memory; DS, digit span; FC, figure copy; FR, figure recall; IM, immediate memory; LA, language; LL, list learning; LO, line orientation; LR, list recall; Lrecog, list recognition; INT, interference; PN, picture naming; RBANS, Repeatable Battery for the Assessment of Neuropsychological Status; SF, semantic fluency; SM, story memory; SR, story recall; TMT, Trail Making Test; VS, visuospatial; WR, word recognition. *P < .05; **P < .01.
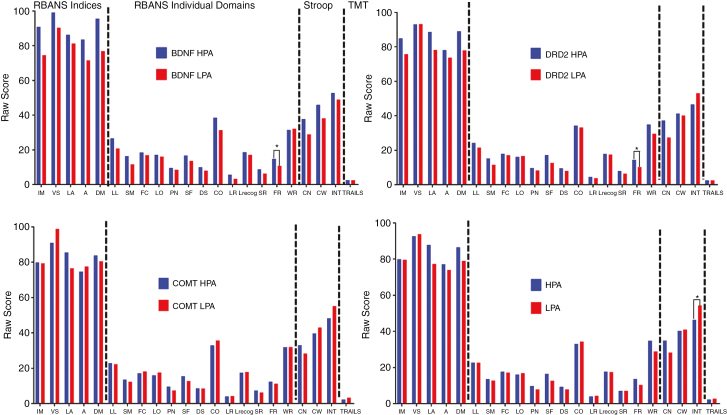
Three months after diagnosis, patients with DRD2 and BDNF HPA performed better than those with LPA on the Fig. recall task (Fig. 2). Counterintuitively, patients in the LPA group performed better than those in the HPA group on the Stroop interference test at 3 months postdiagnosis (Fig. 2).
Fig. 2.
Performance at 3 Months by Allele and NHPA Score.
Mean scores for RBANS indices, RBANS individual domains, Stroop, and TMT tests (from left to right) at 3-months postdiagnosis by individual low- (LPA) and high- (HPA) performing BDNF (top left), DRD2 (top right), and COMT (bottom left) alleles along with total number of high-performing alleles (NHPA) (LPA = NHPA 0 to 4, HPA = NHPA 5 to 6) (bottom right). RBANS data are presented as mean raw score. Stroop data are presented as mean interference T score. TMT data are presented as mean ratio of the score on Part B to Part A (Trail B/A). A indicates attention; CN, color naming; CO, coding; CW, color-word condition; DM, delayed memory; DS, digit span; FC, Fig. copy; FR, Fig. recall; IM, immediate memory; LA, language; LL, list learning; LO, line orientation; LR, list recall; Lrecog, list recognition; INT, interference; PN, picture naming; RBANS, Repeatable Battery for the Assessment of Neuropsychological Status; SF, semantic fluency; SM, story memory; SR, story recall; TMT, Trail Making Test; VS, visuospatial; WR, word recognition. *P < .05; **P < .01.
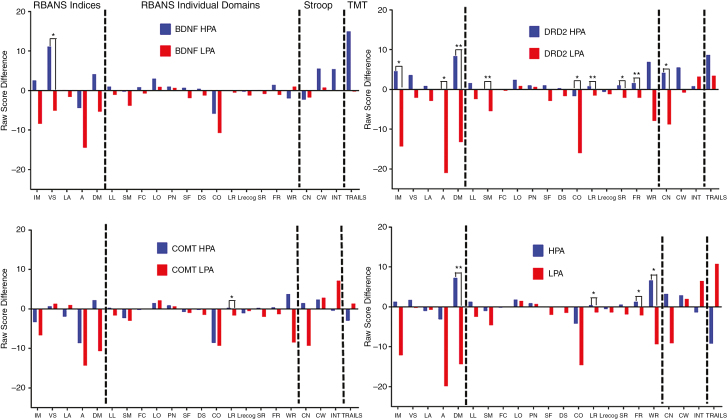
Next, we assessed the mean change in score across RBANS indices, subtests, Stroop test, and TMT between baseline and 3 months postdiagnosis to better understand neurocognitive recovery within this time period. Task performance was better in the HPA group in the delayed memory, list recognition, and figure recall domains (Fig. 3). HPA patients also performed better on the Stroop word recognition task. By individual gene allele status, patients with DRD2 HPA overwhelmingly performed better on RBANS and Stroop tests across a number of RBANS subtests (Fig. 3). Furthermore, patients with DRD2 HPA had improved neurocognitive test scores across all RBANS and Stroop test subsections on average 3 months after diagnosis (Fig. 3). No significant difference in performance on the TMT test was observed at baseline or at 3 months regardless of BDNF, DRD2, or COMT allele or NHPA group.
Fig. 3.
Three-Month Change in Performance by Allele and NHPA Score.
Mean difference in scores for RBANS, Stroop, and TMT tests from baseline to 3 months postdiagnosis by individual low- (LPA) and high- (HPA) performing BDNF, COMT, and DRD2 alleles along with total number of high-performing alleles (NHPA) (LPA = NHPA 0 to 4, HPA = NHPA 5 to 6). A indicates attention; CN, color naming; CO, coding; CW, color-word condition; DM, delayed memory; DS, digit span; FC, figure copy; FR, figure recall; IM, immediate memory; LA, language; LL, list learning; LO, line orientation; LR, list recall; Lrecog, list recognition; INT, interference; PN, picture naming; RBANS, Repeatable Battery for the Assessment of Neuropsychological Status; SF, semantic fluency; SM, story memory; SR, story recall; TMT, Trail Making Test; VS, visuospatial; WR, word recognition. *P < .05; **P < .01.
Discussion
Polymorphisms in the DRD2, COMT, and BDNF genes are known to affect protein function on the molecular level and also correlate with functional recovery in neurologic illness. Therefore, a polygenic risk score may be clinically relevant for patients with glioma. In our patient population, polymorphisms in the DRD2, COMT, and BDNF genes have a significant impact on neurocognitive performance. Furthermore, an NHPA score for all 3 alleles reveals that patients with a higher score return to work on a more frequent basis 3 months postdiagnosis. This may be due to neurocognitive recovery across multiple domains including delayed memory, list recall, figure recall, and word recall (Table 3). However, there is no linear relationship between NHPA and return to work in this series. It may be that a more granular assessment of return-to-work status is required to detect differences between patients based on NHPA scores rather than a more dichotomized assessment separating patients into HPA and LPA cohorts as was performed here. Also, it may be that effect on return-to-work status appears on a different time scale, as we have chosen to analyze at 3 months. Additional efforts will be made to explore these relationships.
While most basic, translational, and clinical research focuses on disease etiology, treatment, and prognosis, less attention has been given to functional outcomes. Outside of performance on individual cognitive tasks, 1 important measure of functional outcome is an individual’s ability to return to work during or after medical treatment for a serious illness. Return to work has been extensively studied in patients with cardiac disease following acute coronary events and after systemic cancer treatments. However, less attention has been given to return-to-work assessment in patients with neurologic illness such as traumatic brain injury, stroke, and brain tumors. For patients with glioma, functional metrics such as ability to return to work is influenced by the timing of oncological therapies. For this reason, return-to-work analysis for this study includes only individuals not receiving oncological therapies during the study period as fatigue is a common finding among glioma patients receiving chemoradiation and influences return-to-work analysis. Furthermore, we included only individuals who returned to work at full capacity. Here, we demonstrate that genetics may play a significant role in recovery after brain tumor surgery and may influence a patient’s ability to return to work.
Neurocognitive recovery in adult glioma patients is poorly understood. It is commonly believed that the majority of patients recover to some extent without interventions such as cognitive rehabilitation. Questions remain, however, regarding the timing of recovery and how to best identify vulnerable patient populations. Here, we evaluated neurocognitive performance by SNP variation at baseline and 3 months postdiagnosis. While both LPA and HPA cohorts experienced neurocognitive improvements across a number of domains, change in performance was superior for HPA vs LPA patients. This indicates that patients with a higher NHPA score may have an increased capacity for recovery. While better performance was seen in the visuospatial and list recall domains by BDNF HPA and COMPT HPA, respectively, these findings are likely driven by the robust differences seen across many test domains by DRD2 genotype. DRD2 genotype may, therefore, be particularly indicative of a patient’s capacity for recovery. Interestingly, those with DRD2 LPA perform better on neurocognitive testing at baseline. These patients also have less capacity for recovery as indicated by the performance regression they display at 3 months while those with the HPA make small but significant gains. These data may have implications when selecting glioma patients to participate in outpatient cognitive rehabilitation.
The primary functional domain of change with respect to COMT, BDNF, and DRD2 SNP heterozygotes was memory (delayed memory, list recall, figure recall, and word recall specifically). This is no surprise given that 59% of the study population had tumors within the dominant hemisphere, thereby in close proximity to mesial temporal structures. The influence of the rs4680 SNP in the COMT gene has previously been reported both in healthy individuals as well as those with psychiatric disorders. A Met/Val substitution at codon 158 defines this SNP. Those with a Val substitution perform more poorly on tests of executive function when compared with those who are Met carriers. Such a substitution is associated with higher enzymatic activity and more rapid dopamine degradation leading to dopamine depletion in the prefrontal cortex as a leading hypothesis for the cognitive effects observed in patients with this substitution. The effect of this gene variant has also been demonstrated in breast cancer patients, in whom Val carriers treated with chemotherapy had worse cognitive dysfunction compared with healthy untreated carriers, implicating a cancer treatment-related effect modification. Correa et al explored the relationship between this SNP and cognitive function in brain tumor patients. This study used a heterogeneous group of tumor patients regardless of infiltrative characteristics, histology, disease status, or WHO grade. Using a battery of 8 cognitive assessments, performance on 1 test (Hopkins Verbal Learning Test–Delay) was found to be reduced in patients with the rare and unfavorable Val/Val homozygous genotype.7 However, they observed no difference in task performance between COMT allele status and cognitive performance at 3 months. We observed a statistically significant difference in response to inhibition scores between LPA and HPA groups where the LPA group performed more poorly on the Stroop interference test. This is additional evidence implicating an interaction between NHPA and cognitive and executive functioning in glioma patients.
In the current patient population, those who have the BDNF A/G genotype do indeed have significantly lower circulating plasma BDNF levels. An important distinction should be made with respect to BDNF production and secretion at the postsynaptic cleft and circulating plasma BDNF as assessed in this study. There are several publications focused on circulating plasma BDNF in neurodegenerative disorders and psychiatric illness.9,13,17 Many believe that BDNF synthesis is mediated within the CNS. Evidence suggests that the brain is the major, but not sole, contributor to circulating BDNF plasma levels in humans.35,36 If it is therefore assumed the brain is the primary source of circulating BDNF, then it also stands to reason that changes in systemic circulating BDNF neurophysiology due to the presence of a glioma may indeed influence circulating plasma levels. In this analysis, plasma BDNF among BDNF LPA was decreased only in patients with WHO IV gliomas. This finding is likely based on the integrity of the blood-brain barrier in low-grade vs high-grade gliomas. High-grade gliomas cause histopathologic and cytoarchitecture changes that compromise the blood-brain barrier. This is also apparent on MRI, which demonstrates 90% of WHO IV gliomas exhibit contrast enhancement with gadolinium due to breakdown of the blood-brain barrier.37 Changes in peripheral BDNF due to altered BDNF neurophysiology in glioma patients may mask changes in basal levels due to somatic germline variations in the BDNF gene. The blood-brain barrier is also relatively more intact in patients with WHO II and III glioma, perhaps an explanation for the lack of statistical significance in peripheral plasma BDNF in these patients.
The data also demonstrate that allelic frequencies for the DRD2 and COMT genes were significantly different in the study population relative to the expected distribution. There are fewer patients with the lowest-performing COMT G/G allele in the glioma study population. With respect to the DRD2 gene, there are fewer patients with the highest-performing G/G allele relative to expected frequencies. There are many potential explanations for these findings, the most intriguing of which is that the presence of a particular COMT or DRD2 polymorphism may have an association with gliomagenesis. The biological mechanisms involved in tumor initiation and progression as a factor of COMT or DRD2 function by allelic variation is outside the scope of the current study. However, these associations should be further investigated. At present, there are limited basic science and clinical data that connect DRD2 or COMT function to cancer. For instance, the Cancer Genome Atlas dataset for 1133 glioma patients contains information for only 1 patient with a COMT gene mutation and 5 for DRD2. Additional research is required to better understand why the COMT rs4680 and DRD2 rs1076560 polymorphism frequency distributions are different in glioma patients.
There are several limitations to the present study. As a study with a retrospective design, the influence of preexisting cognitive dysfunction in carriers of certain HPA or LPA cannot be controlled. We excluded individuals with known trauma or psychiatric history. The number of patients also limits the power to detect small or moderate effects and interactions among SNPs and cognitive outcome metrics as well as tumor grade, type, location, size, and multimodal treatment modalities. By isolating 3 potentially important alleles of 3 genes, we aimed to increase the power to detect associations with any 1 or more genetic signatures. With a narrow focus and a limited sample size, the ability to establish relationships across a broader spectrum of genes and polymorphisms that may be associated with functional outcomes in adult glioma patients is limited. It is also possible that variants may strongly influence task performance based on a biological mechanism that more heavily affects cognitive function than the candidate SNPs chosen for this study. With decreasing cost and increasing efficiency of whole-exome sequencing methods, an appropriate next step will be to continue this study through genomewide association methods to evaluate high-risk foci for functional impairment. These areas represent opportunities for future research.
Selection of the optimal neurocognitive task can be challenging as each battery of tasks requires consideration with respect to interpretation of results. Although our data analysis was retrospective, we prospectively decided to use the RBANS as an efficient measure that has multiple forms and covers several domains of neurocognitive functions often overlooked in oncology patient populations. Whereas most brief oncology clinical trial batteries consist only of measures of verbal memory, phonemic fluency, and processing speed/executive function, the RBANS also allows for measurement of visuospatial abilities (perception, reproduction/organization, and recall) in addition to verbal learning and memory, processing speed, language, and attention. The RBANS also represents the co-norming of several widely used test formats, aiding clinical interpretation. The sensitivity of the RBANS to detect mild cognitive impairment in elderly patients has been found to be low compared with normal controls on some RBANS subtests. We addressed shortcomings of the RBANS test to address executive functioning by including both the Stroop and TMT. We further elected to use the ratio of the TMT Part B to Part A as a validated method for increasing the sensitivity for executive function further.38 We found few impaired cognitive domains at 3 months postdiagnosis compared to baseline, and it is possible that our measures were unable to detect subtle though still potentially meaningful persisting cognitive deficits. Notably, however, we were able to demonstrate improvements in cognitive functioning between baseline and 3 months, consistent with the literature on trajectory of cognitive changes over the course of treatment in glioma patients.
Despite the mentioned limitations, this study identifies genetic factors that may be important in risk stratification, clinical decision making, and as a target for the development of future novel therapeutic strategies for brain tumor patients to improve cognitive outcomes and QOL. The NHPA model has not been proposed or studied previously. It is the first step toward validating such a polygene score method for a glioma patient population. Its value in this study is as a discovery tool, though our study is significantly limited by the fact that this method has not yet been validated. A large, prospective, longitudinal study is needed to validate the NHPA score model presented here and to further explore the role of relevant genes and polymorphisms that may contribute to improved outcomes for brain tumor patients. All these efforts will lead to a greater understanding of which patients recover and which continue to have functional impairments throughout oncological therapies.
Conclusions
Recent molecular classification has allowed for the identification of individuals with “favorable” genetics resulting in extended disease-free survival periods.39 However, throughout this survival period, more than 70% of patients experience language, motor, or cognitive disabilities, which impairs personal and professional relationships, with major implications on QOL.6 Little is known about the mechanisms by which gliomas affect cognition and cognitive recovery. We demonstrate a relationship between allelic variations in 3 genes associated with cognition and performance function at baseline and at 3 months posttreatment in patients with WHO grade II-IV glioma. We demonstrate that patients with a higher NHPA polygene index score return to work on a significantly more frequent basis 3 months after diagnosis compared with those with lower scores. Studies such as this provide an enhanced understanding of factors that might affect patient outcomes and will permit strategies appropriately focused on enhancing survival while maximizing QOL.
Funding
This work was supported by grants from the University of California San Francisco Specialized Program of Research Excellence, the Robert Wood Johnson Foundation, the Alfred A. Taubman Foundation, and the Mark Trauner Brain Research Fund to Senior Author S.L.H.J.
Conflict of interest statement. None declared
References
- 1. Ostrom QT, Gittleman H, Xu J, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009-2013. Neuro Oncol. 2016;18(suppl 5):v1–v75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang AS, Ostrom QT, Kruchko C, et al. Complete prevalence of malignant primary brain tumors registry data in the United States compared with other common cancers, 2010. Neuro Oncol. 2017;19(5):726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rouse C, Gittleman H, Ostrom QT, et al. Years of potential life lost for brain and CNS tumors relative to other cancers in adults in the United States, 2010. Neuro Oncol. 2016;18(1):70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Béhin A, Delattre J-Y. Neurologic sequelae of radiotherapy on the nervous system. In: Schiff D, Wen PY, eds. Cancer Neurology in Clinical Practice. New York: Humana Press; 2003:173–191. [Google Scholar]
- 5. Correa DD. Neurocognitive function in brain tumors. Curr Neurol Neurosci Rep. 2010;10(3):232–239. [DOI] [PubMed] [Google Scholar]
- 6. Taphoorn MJ, Klein M. Cognitive deficits in adult patients with brain tumours. Lancet Neurol. 2004;3(3):159–168. [DOI] [PubMed] [Google Scholar]
- 7. Correa DD, Satagopan J, Cheung K, et al. COMT, BDNF, and DTNBP1 polymorphisms and cognitive functions in patients with brain tumors. Neuro Oncol. 2016;18(10):1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cramer SC. A window into the molecular basis of human brain plasticity. J Physiol. 2008;586(23):5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. [DOI] [PubMed] [Google Scholar]
- 10. Hyman C, Hofer M, Barde YA, et al. BDNF is a neurotrophic factor for dopaminergic neurons of the substantia nigra. Nature. 1991;350(6315):230–232. [DOI] [PubMed] [Google Scholar]
- 11. Ramos-Cejudo J, Gutiérrez-Fernández M, Otero-Ortega L, et al. Brain-derived neurotrophic factor administration mediated oligodendrocyte differentiation and myelin formation in subcortical ischemic stroke. Stroke. 2015;46(1):221–228. [DOI] [PubMed] [Google Scholar]
- 12. Schäbitz WR, Berger C, Kollmar R, et al. Effect of brain-derived neurotrophic factor treatment and forced arm use on functional motor recovery after small cortical ischemia. Stroke. 2004;35(4):992–997. [DOI] [PubMed] [Google Scholar]
- 13. Shishkova VN, Zotova LI, Maljukova NG, et al. An assessment of cerebrolysin effect on BDNF level in patients with post stroke aphasia depending on carbohydrate metabolism disorders [article in Russian]. Zh Nevrol Psikhiatr Im S S Korsakova. 2015;115(5):57–63. [DOI] [PubMed] [Google Scholar]
- 14. Stanne TM, Tjärnlund-Wolf A, Olsson S, Jood K, Blomstrand C, Jern C. Genetic variation at the BDNF locus: evidence for association with long-term outcome after ischemic stroke. PloS One. 2014;9(12):e114156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uchida K, Baba H, Maezawa Y, et al. Increased expression of neurotrophins and their receptors in the mechanically compressed spinal cord of the spinal hyperostotic mouse (twy/twy). Acta Neuropathol. 2003;106(1):29–36. [DOI] [PubMed] [Google Scholar]
- 16. Pearson-Fuhrhop KM, Cramer SC. Genetic influences on neural plasticity. PM R. 2010;2(12 suppl 2):S227–S240. [DOI] [PubMed] [Google Scholar]
- 17. Hünnerkopf R, Strobel A, Gutknecht L, Brocke B,Lesch KP. Interaction between BDNF Val66Met and dopamine transporter gene variation influences anxiety-related traits. Neuropsychopharmacology. 2007;32(12):2552–2560. [DOI] [PubMed] [Google Scholar]
- 18. Small BJ, Rawson KS, Walsh E, et al. Catechol-O-methyltransferase genotype modulates cancer treatment-related cognitive deficits in breast cancer survivors. Cancer. 2011;117(7):1369–1376. [DOI] [PubMed] [Google Scholar]
- 19. Luykx JJ, Broersen JL, de Leeuw M. The DRD2 rs1076560 polymorphism and schizophrenia-related intermediate phenotypes: a systematic review and meta-analysis. Neurosci Biobehav Rev. 2017;74(Pt A):214–224. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Bertolino A, Fazio L, et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci U S A. 2007;104(51):20552–20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mamiya PC, Richards TL, Coe BP, Eichler EE, Kuhl PK. Brain white matter structure and COMT gene are linked to second-language learning in adults. Proc Natl Acad Sci U S A. 2016;113(26):7249–7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noohi F, Boyden NB, Kwak Y, et al. Interactive effects of age and multi-gene profile on motor learning and sensorimotor adaptation. Neuropsychologia. 2016;84:222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Frias CM, Annerbrink K, Westberg L, Eriksson E,Adolfsson R,Nilsson LG. COMT gene polymorphism is associated with declarative memory in adulthood and old age. Behav Genet. 2004;34(5):533–539. [DOI] [PubMed] [Google Scholar]
- 24. Stuart K, Summers MJ, Valenzuela MJ, Vickers JC. BDNF and COMT polymorphisms have a limited association with episodic memory performance or engagement in complex cognitive activity in healthy older adults. Neurobiol Learn Mem. 2014;110:1–7. [DOI] [PubMed] [Google Scholar]
- 25. Lessmann V. Neurotrophin-dependent modulation of glutamatergic synaptic transmission in the mammalian CNS. Gen Pharmacol. 1998;31(5):667–674. [DOI] [PubMed] [Google Scholar]
- 26. Matzilevich DA, Rall JM, Moore AN, Grill RJ,Dash PK. High-density microarray analysis of hippocampal gene expression following experimental brain injury. J Neurosci Res. 2002;67(5):646–663. [DOI] [PubMed] [Google Scholar]
- 27. Vaughn KA, Ramos Nuñez AI, Greene MR, Munson BA,Grigorenko EL,Hernandez AE. Individual differences in the bilingual brain: the role of language background and DRD2 genotype in verbal and non-verbal cognitive control. J Neurolinguistics. 2016;40:112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kitchen Andren KA, Gabel NM, Stelmokas J, Rich AM, Bieliauskas LA. Population base rates and disease course of common psychiatric and neurodegenerative disorders. Neuropsychol Rev. 2017;27(3):284–301. [DOI] [PubMed] [Google Scholar]
- 29. Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30(9):1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17(7):392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Randolph C, Tierney MC, Mohr E, Chase TN. The Repeatable Battery for the Assessment of Neuropsychological Status (RBANS): preliminary clinical validity. J Clin Exp Neuropsychol. 1998;20(3):310–319. [DOI] [PubMed] [Google Scholar]
- 32. Bench CJ, Frith CD, Grasby PM, et al. Investigations of the functional anatomy of attention using the Stroop test. Neuropsychologia. 1993;31(9):907–922. [DOI] [PubMed] [Google Scholar]
- 33. Tombaugh TN. Trail Making Test A and B: normative data stratified by age and education. Arch Clin Neuropsychol. 2004;19(2):203–214. [DOI] [PubMed] [Google Scholar]
- 34. Chafetz M, Matthews LH. A new interference score for the Stroop test. Arch Clin Neuropsychol. 2004;19(4):555–567. [DOI] [PubMed] [Google Scholar]
- 35. Matthews VB, Aström MB, Chan MH, et al. Brain-derived neurotrophic factor is produced by skeletal muscle cells in response to contraction and enhances fat oxidation via activation of AMP-activated protein kinase. Diabetologia. 2009;52(7):1409–1418. [DOI] [PubMed] [Google Scholar]
- 36. Krabbe KS, Nielsen AR, Krogh-Madsen R, et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia. 2007;50(2):431–438. [DOI] [PubMed] [Google Scholar]
- 37. Scott JN, Brasher PM, Sevick RJ, Rewcastle NB,Forsyth PA. How often are nonenhancing supratentorial gliomas malignant? A population study. Neurology. 2002;59(6):947–949. [DOI] [PubMed] [Google Scholar]
- 38. Arbuthnott K, Frank J. Trail Making Test, Part B as a measure of executive control: validation using a set-switching paradigm. J Clin Exp Neuropsychol. 2000;22(4):518–528. [DOI] [PubMed] [Google Scholar]
- 39. Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108(1): 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]