Abstract
The pleiotropic CCCTC-binding factor (CTCF) plays a role in homologous recombination (HR) repair of DNA double-strand breaks (DSBs). However, the precise mechanistic role of CTCF in HR remains largely unclear. Here, we show that CTCF engages in DNA end resection, which is the initial, crucial step in HR, through its interactions with MRE11 and CtIP. Depletion of CTCF profoundly impairs HR and attenuates CtIP recruitment at DSBs. CTCF physically interacts with MRE11 and CtIP and promotes CtIP recruitment to sites of DNA damage. Subsequently, CTCF facilitates DNA end resection to allow HR, in conjunction with MRE11–CtIP. Notably, the zinc finger domain of CTCF binds to both MRE11 and CtIP and enables proficient CtIP recruitment, DNA end resection and HR. The N-terminus of CTCF is able to bind to only MRE11 and its C-terminus is incapable of binding to MRE11 and CtIP, thereby resulting in compromised CtIP recruitment, DSB resection and HR. Overall, this suggests an important function of CTCF in DNA end resection through the recruitment of CtIP at DSBs. Collectively, our findings identify a critical role of CTCF at the first control point in selecting the HR repair pathway.
INTRODUCTION
DNA double-strand breaks (DSBs) represent the most damaging DNA injuries that can compromise genomic integrity and viability. DSBs can result from exogenous (UV, ionizing radiation and cytotoxic chemicals) and endogenous (cellular metabolites, reactive oxygen species and replication errors) insults. DSBs, if left unrepaired, can lead to fatal diseases, including cancer, growth and mental retardation, immune deficiency and developmental defects. To repair DSBs, eukaryotic cells employ mutually exclusive error-prone non-homologous end joining (NHEJ) or error-free homologous recombination (HR) repair. NHEJ involves ligation of the broken DNA ends and may create mutations, given that a homologous template is not available for repair. HR repair employs mostly homologous DNA in the sister chromatid as a template, which restores the correct DNA sequence. HR repair occurs predominantly during the S and G2 phases of the cell cycle, while NHEJ occurs throughout G1, S and G2.
The first control point for the DSB repair pathway occurs at the processing of the 5′ DNA end resection, which is catalyzed by MRE11 and CtIP (1,2). DNA end resection inversely influences the selection of the two major DSB repair pathways. Accordingly, extensive end resection suppresses NHEJ and permits HR repair (3). HR repair commences with the formation of extensive 3′-overhang single-stranded DNA (ssDNA), which requires the recruitment of MRE11 and CtIP at the DSB sites, facilitation of the nuclease activity of MRE11 controlled by CtIP, and involvement of the nucleases EXO1 and BLM/DNA2. Replication protein A (RPA) loads rapidly onto the resulting ssDNA and is simultaneously phosphorylated (3). Subsequently, the recombinase RAD51 displaces RPA in concert with BRCA1–BARD1, PALB2 and BRCA2 to form a helical nucleoprotein filament, thereby allowing homology search, strand invasion and sister chromatid exchange (4,5). Therefore, DNA end resection is a key step that controls the choice of the DSB repair pathway.
Although extensive studies have uncovered much about these critical steps in the regulation of DNA end resection and HR pathway choice between different DSB repair mechanisms, the process is complex and involves many additional proteins. Hence, how DSB repair proteins play a role in selecting the HR repair pathway within this exquisite network and how this process is controlled are largely underexplored.
Recently, a novel role of the multifunctional nuclear protein CCCTC-binding factor (CTCF) in HR-mediated DSB repair has been unveiled (6). CTCF is a transcription factor with 11 zinc finger (ZF) domains that function in many nuclear processes, including genomic organization, transcriptional regulation, insulator activity, VDJ recombination and HR-mediated repair. CTCF mutations in humans are linked to microcephaly and intellectual disability (7). In vivo evidence from CTCF knockout mice implicate CTCF as a haploinsufficient tumor suppressor (8), since heterozygous CTCF+/− mutations display greater susceptibility to irradiation-induced carcinogenesis, while homozygous CTCF−/− mutations result in embryonic lethality (8). Along with these mutation phenotypes, recent findings show that CTCF is recruited to damaged DNA sites and facilitates HR repair (9–11). Additionally, CTCF interacts with BRCA2 (10) and RAD51 (11), which are implicated in DSB repair by HR. Nevertheless, very little is known about the precise role of CTCF in HR and the key mechanism by which CTCF promotes HR.
In this study, we investigated the role of CTCF in HR-mediated DSB repair and its underlying mechanism. Via a proteomic approach, we identified MRE11 and CtIP as novel CTCF-interacting partners with a functional link to HR repair. We further corroborated that CTCF is recruited to DNA lesions in an MRE11-dependent manner, followed by CtIP recruitment, leading to DNA end resection at DSB sites and initiating HR. Consistent with these observations, CTCF depletion or truncation mutants are incapable of binding to CtIP sensitized cells to DNA damage. These data provide insights into a previously undescribed role of CTCF in promoting HR-mediated DNA repair by facilitating the formation of an initial HR complex for the onset of DNA end resection.
MATERIALS AND METHODS
Cells and reagents
HeLa, 293T and U2OS cell lines were purchased from ATCC, and these lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Welgene, Kyung-san, Republic of Korea) with 10% FBS (Hyclone, GE Healthcare, Chicago, IL, USA) and 1% penicillin/streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). U2OS-based DSB reporter cell lines, which stably express inducible AsiSI-ER (AsiSI-ER-U2OS; a generous gift from Dr. Gaëlle Legube, Universitéde Toulouse) (12) or inducible ER-mCherry-LacI-FokI-DD (FokI-U2OS; a generous gift from Dr. Roger Greenberg, University of Pennsylvania) (13) fusion protein, were grown in DMEM-containing puromycin. U2OS-based DR-GFP, EJ2-GFP, EJ5-GFP and SA-GFP cells (14) were kindly provided by Dr. Jeremy Stark (Beckman Research Institute of the City of Hope); they were maintained in DMEM without sodium pyruvate (Hyclone) containing 10% FBS and 1 μg/ml puromycin (Sigma-Aldrich, St. Louis, MO, USA). H1299.GC and H1299.EJ cell lines (15) were kindly provided by Dr. Hans Will and were cultured in DMEM-containing puromycin and G418, respectively. All cells were incubated at 37°C in a 5% CO2 incubator (BB15, Thermo Fisher Scientific).
Plasmid construction and transfection
GFP- and HA-tagged full-length and truncation mutants of CTCF and MRE11 were constructed by a classical PCR method using pEGFP-N1 (Addgene #6085-1) and pcDNA3-HA, respectively. The oligonucletoide sequences of the primers used for cloning are shown in Supplementary Table S1. All junction regions of CTCF and MRE11 constructs and the coding sequences of CTCF fragments were verified by DNA sequencing. Double-stranded short hairpin RNAs (shRNAs) targeting CTCF, MRE11 and CtIP were generated using pSUPER.retro.puro, an H1 promoter-driven RNA interference retroviral vector (Oligoengine, Seattle, WA). The sequences of the shRNAs are shown in Supplementary Table S2. Transfections were performed using Effectene Transfection Reagent (Qiagen, Carlsbad, CA) for expression in mammalian cells.
Antibodies
The CTCF antibodies were obtained from Abcam (ab128873, 1:2000 dilution for immunoblotting (IB) and 1:2000 for immunofluorescence (IF); ab70303, 2 μg for each chromatin immunoprecipitation (ChIP) sample), Cell Signaling Technology (#2899S, 1:1000 for immunoprecipitation (IP)) and Millipore (07-729, 2 μg for each ChIP sample). The other antibodies used for IB, IF, IP and ChIP analyses were as follows: HA (ab9110 from Abcam, 1:2000 for IB, ChIP, and IP), GFP (ab290 from Abcam, 1:2000 for IB and 1:1000 for IP), γH2AX (05-636 from Millipore, 1:2000 for IF and IB; ab2893 from Abcam, 2 μg for each ChIP sample), CtIP (61141 from Active Motif, 1:1000 for IF, IB, IP and ChIP), RPA (ab2175 from Abcam, 1:1000 for IF and 2 μg for ChIP), phospho-RPA (S4/S8) (A300-245A from Bethyl, 1:2000 for IB and IF), MRE11 (ab214 from Abcam, 1:2000 for IF, ChIP, IP and IB and 2 μg for ChIP; #4895S from Cell Signaling Technology for IP), NBS1 (ab32074 from Abcam, 1:2000 for IB), RAD50 (ab89 from Abcam, 1:2000 for IB and IP), Tubulin (05-829 from Millipore, 1:3000 for IB), RAD51 (ab176458 from Abcam, 2 μg for each ChIP sample), p53 (sc-126 from Santa Cruz, 1:2000 for IB), p21 (05-345 from Millipore, 1:2000 for IB), phospho-(serine/threonine) ATM/ATR substrate multi-monoclonal antibody (#6966S from Cell Signaling Technology, IB), BRCA1 (sc-642 from Santa Cruz, 1:1000 for IB), 53BP1 (NB100-305 from Novus, 1:2000 for IB) and β-actin (sc-47778 from Santa Cruz, 1:3000 for IB).
Tandem affinity purification (TAP) and mass spectrometry
The 293T cells transiently expressing S protein-Flag-Streptavidin binding peptide (SFB)-tagged CTCF (SFB-CTCF) were treated with or without γ-irradiation. The cells were lysed with TAP-NETN buffer (20 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, pH 8.0). After removal of cell debris by centrifugation, the crude lysates were incubated with Streptavidin-sepharose beads (GE Healthcare, Chicago, IL, USA). The bead-bound proteins were washed and eluted with biotin (Sigma Aldrich) in TAP-NETN buffer. The eluates were then incubated with S-protein agarose beads (Millipore, Burlington, MA, USA). The S-protein bead-bound proteins were washed, separated by SDS-PAGE and then visualized by staining with Instant Blue (Expedeon, Heidelberg, Germany). For LC-MS/MS analyses, the gel lanes were sliced into different bands and processed as follows. Briefly, the acetylated protein bands were divided into 10-mm sections and digested in the gel with trypsin. The tryptic digests were separated by online reversed-phase chromatography using a Thermo Scientific EASY-nLC 1200 UHPLC equipped with an autosampler using a reversed-phase peptide trap Acclaim PepMap™ 100 (75-μm inner diameter, 2-cm length) and a reversed-phase analytical column PepMap™ RSLC C18 (75-μm inner diameter, 15-cm length, 3-μm particle size), both from Thermo Scientific. This was followed by electrospray ionization at a flow rate of 300 nl·min−1. The chromatography system was coupled in line with an Orbitrap Fusion Lumos Mass Spectrometer. The obtained spectra were screened against the UniProt human database using Proteome Discoverer Sorcerer 2.1 software with a SEQUEST-based search algorithm. The comparative analysis of proteins identified in this study was performed using Scaffold 4 Q+S. All raw mass spectrometry proteomics data obtained in this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014441.
Laser micro-irradiation
U2OS cells were plated onto glass-bottomed culture dishes (SPL Life Science, Korea) and transfected with the indicated plasmids and siRNA. The cells were presensitized with 10 μM 5-bromo-2′-deoxyuridine (BrdU, Sigma) for 24–30 h and then subjected to laser micro-irradiation using an Eclipse T1 & A1 confocal system (Nikon Instruments Inc., Melville, NY, USA) at 405 nm for 3 s (32 lines/s) in a 37°C chamber containing 5% CO2. After the laser treatment, the cells were subjected to live-cell imaging or were processed for IF.
Immunofluorescence
After the indicated treatments (micro-irradiation, etoposide, 4′-hydroxytamoxifen [4-OHT], or γ-irradiation), the cells were fixed with 3.7% formaldehyde and permeabilized with 0.5% Triton X-100. After blocking with 1% BSA, the cells were incubated with the indicated primary and secondary antibodies. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma). Coverslips were mounted using Vectashield mounting medium (Vector Labs), and images were acquired using a confocal (LSM 710, Carl Zeiss, Oberkochen, Germany) or fluorescence microscope (Eclipse Ti-S, Nikon Instruments Inc.).
Immunoprecipitation
The 293T cells that had been pretreated with or without 10 or 20 μM ATM inhibitor (KU55933 from Santa Cruz) for 1 h and then treated with 50 μM etoposide or vehicle (dimethyl sulfoxide (DMSO)) were lysed with NETN buffer (0.1 mM dithiothreitol (DTT), 1% NP-40, 150 mM NaCl, 40 mM Tris-HCl, pH 8.0) containing protease inhibitors on ice for 10 min. The cell lysates were clarified by centrifugation and incubated with nuclease digestion buffer (0.25 M sucrose, 1.5 mM Tris-HCl [pH 7.4], 80 mM NaCl, 3 mM KCl, 7.5 mM NaCl, 1 mM CaCl2, 0.1 mM DTT, 20 U/ml micrococcal nuclease) containing protease inhibitor with or without 150 U/ml Benzonase® (Sigma) or 0.1 μg/μl ethidium bromide (Qbiogene) for 15 min at 37°C. After centrifugation, the chromatin-bound proteins were collected and incubated with antibody against the indicated proteins for 12 h at 4°C, and then with protein A beads for 1 h at 4°C. The beads were then washed with NETN buffer three times and analyzed by IB. For tagged-protein IP, 293T cells treated with etoposide or vehicle were lysed in IP buffer (40 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM MgCl2, 0.2% Nonidet P-40, 0.4% Triton X-100) containing protease inhibitors on ice for 30 min. The cell lysates were clarified by centrifugation and incubated with the indicated antibody for 12 h at 4°C, followed by incubation with protein A beads for 1 h at 4°C. The precipitates were rinsed with wash buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.2% Triton X-100) three times, and the bound proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
ChIP assay
AsiSI-ER-U2OS cells (12) were treated with 300 nM 4-OHT (Sigma) for 4 h to induce AsiSI-ER nuclear localization and DSB generation by the AsiSI nuclease. FokI-U2OS reporter cells (13) expressing inducible ER-mCherry-LacI-FokI-DD were treated with 300 nM 4-OHT plus 1 μM Shield-I (Clontech) for 4 h to induce nuclear expression and stabilization of ER-mCherry-LacI-FokI-DD and DSB generation at the transgene-harboring Lac operator sequences by the FokI nuclease. Subsequently, ChIP was carried out using the EZ-ChIP Kit according to the manufacturer’s protocols (Millipore). The immunoprecipitated and input DNA were analyzed by quantitative PCR (qPCR) using the Rotor-Gene SYBR® Green PCR kit (Qiagen) on a Rotor-Gene Q system (Qiagen). The PCR conditions were an initial preincubation step at 95°C for 5 min followed by 45 cycles of 95°C for 5 s and 60°C for 30 s. The last amplification cycle was followed by a melting curve analysis to confirm the specificity of the PCR amplification. Relative IP values were calculated based on the threshold cycle (Ct) value using the 2−ΔΔCt method (16). The sequences of the primers used for qPCR are described in Supplementary Table S3.
HR, canonical and alternative NHEJ, and single-strand annealing (SSA) DNA repair assays
HR and NHEJ efficiencies were measured by using H1299 (human non-small cell lung carcinoma cell line)-based repair reporter cell lines (H1299.GC for HR and H1299.EJ for canonical NHEJ) (15) and U2OS-based repair reporter cell lines (DR-GFP for HR, EJ5-GFP for canonical NHEJ, EJ2-GFP for alternative NHEJ, and SA-GFP for single-strand annealing) (14), respectively. H1299.GC and H1299.EJ cells were transfected with CTCF-targeting shRNAs. U2OS DR-GFP, EJ2-GFP, EJ5-GFP and SA-GFP cells were transfected with siRNA targeting the 3′-UTR of CTCF. The following day, I-SceI and HA-CTCF constructs were transfected into each reporter cell line, and evaluated 48 h later for GFP-positive cells using the FACSAria III (BD Biosciences, San Jose, CA, USA).
Measurement of DNA end resection
Genomic DNA was extracted with a gDNA extraction kit protocol (Dneasy blood & Tissue Kit, Qiagen) from AsiSI-ER-U2OS cells transfected with the indicated siRNAs and HA-tagged CTCF constructs. The DNA was then digested with restriction enzymes (HindIII for a negative control and BamHI (chromosome1:89,458,296; CCBL2 gene) or BsgI (chromosome 1:110,319,090) for measuring resection). The level of ssDNAs generated by DNA end resection at the specific AsiSI sites (DSBs) was evaluated by qPCR using the primer sets shown in Supplementary Table S4. The percentage of ssDNA (ssDNA%) generated by resection at the DSBs was determined as previously described (17,18). Briefly, for each sample, a △Ct value was calculated by subtracting the Ct value of the mock-digested sample from the Ct value of the digested sample. The ssDNA% was calculated with the following equation: ssDNA% = 1/(2(△Ct − 1) + 0.5) × 100 (19).
siRNAs
siRNA duplexes with the following sequences were synthesized by Genolution, Inc. (Seoul, Korea): CTCF, 5′-GUAGAAGUCAGCAAAUUAAUU-3′; 3′-UTR of CTCF, 5′-GCUGUCUGAUGUUAGCAAAUU-3′; CtIP, 5′-GAAGGAUGAAGGACAGUUUUU-3′. Cells were transfected with 20 nM siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Immunoblotting
Immunoblotting experiments were performed as previously described (20). Representative results from at least three independent experiments are shown in the figures.
Cellular fractionation
Cellular fractionation was performed as described previously with slight modifications (21). Briefly, the cells were washed twice with ice-cold PBS and harvested. The cell pellets were resuspended for 15 min on ice in 200 μl extraction buffer [50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA] containing 0.1% Triton X-100 and supplemented with protease inhibitor cocktail and phosphatase inhibitors. Following centrifugation at 13,000 rpm (16,000 g) for 5 min, the supernatant was collected (S1 fraction). The pellets were further extracted for 15 min on ice using the same buffer and then collected. The pellets were further incubated in 200 μl of extraction buffer supplemented with 200 μg/ml RNase A but without Triton X-100 for 30 min at 25°C. Following centrifugation at 13,000 rpm (16,000 g) for 5 min, the supernatant was separated from the pellet. The pellets were resuspended in 1% SDS-containing PBS, boiled for 10 min and sonicated for 10 s (Ultrasonic Processor, Cole-Parmer, Canada) (P2 fraction). The cellular fractions were used for IB with antibodies against CTCF (1:5000), CtIP (1:1000), phospho-RPA (S4/S8) (1:1000), phospho-RPA (T21) (1:1000), γ-H2AX (S129) (1:2000) and α-tubulin (1:5000).
Clonogenic cell survival assay
HeLa cells were seeded into 6-well plates (2 × 104 cells/well) and allowed to adhere for 24 h prior to treatment. The cells were treated with 2 mM thymidine for 16 h, and then incubated in fresh media for 6 h, followed by treatment with 0, 2 or 5 μM etoposide for 1 h. After incubation for 24 h, the cells were seeded into 60-mm plates. After 10–14 days of incubation, the colonies were fixed with ice-cold methanol and stained with 0.4% Crystal Violet in 20% ethanol. Survival curves were plotted after normalizing for the plating efficiency mediated by CTCF silencing or overexpression. The clonogenic survival curves were constructed from at least three independent experiments.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated using the Hybrid-R Total RNA Purification Kit (GeneAll, Seoul, Korea). One microgram of total RNA was reverse-transcribed with PrimeScript RT Master Mix (Takara Bio, Shiga, Japan). After a 1:10 dilution, 2 μl of cDNA was used as a template in a 20 μl PCR mixture. The following primers were used for qPCR: CCBL2 (5′-GCCCAACAACACCAGCTCCT-3′ and 5′-CCTGGGCATCCTTGAGTTCC-3′); GAPDH (5′-AGCCACATCGCTCAGACAC-3′ and 5′-GCCCAATACGACCAAATCC-3′). Relative quantification of gene expression was carried out using the ΔΔCt method with GAPDH as a reference gene. The PCR program was as follows: initial denaturation at 95°C for 2 min, followed by 45 cycles of 95°C for 5 s and 60°C for 30 s. The final amplification cycle was followed by a melting curve analysis to confirm the specificity of the PCR amplification.
Statistical analysis
All data are expressed as means ± standard deviation (SD) of at least three replicates. The Student’s t-test was employed to analyze significant differences between datasets. Differences were considered statistically significant at P ≤ 0.05 (in figures: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
RESULTS
CTCF interacts with MRE11 upon DNA damage
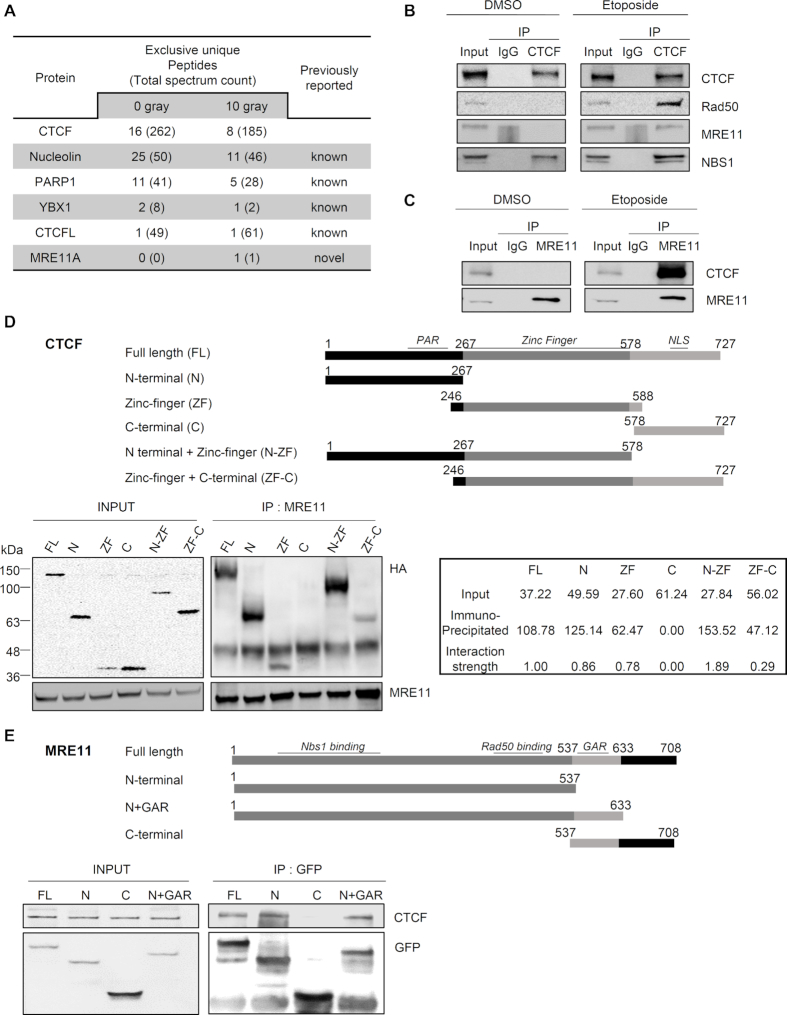
CTCF was recently shown to contribute to genomic integrity by facilitating HR (10,11). To clarify the role of CTCF in HR repair and its underlying mechanism, we analyzed proteins captured by co-immunoprecipitation (Co-IP) with SFB-tagged CTCF by a proteomics approach using 293T cells expressing SFB-tagged CTCF before and after γ-irradiation. Previously known CTCF-associated proteins, including PARP1 (22), YBX1 (23) and nucleolin (24,25), were co-immunoprecipitated either with or without γ-irradiation (Figure 1A). Interestingly, we also identified MRE11, a well-characterized component of the MRE11–RAD50–NBS1 (MRN) complex, as a CTCF-associated protein in a γ-irradiation-dependent manner. This association occurred even if the number of unique peptides or total spectrum count of CTCF in the presence of γ-irradiation was relatively lower than that in the absence of γ-irradiation (Figure 1A). Since only one unique peptide of MRE11A was observed, we needed to verify this DNA damage-responsive interaction. Thus, we performed forward and reciprocal Co-IP experiments with an antibody against CTCF or MRE11 using 293T cells with or without etoposide treatment. As shown in Figure 1B and C, endogenous CTCF and MRE11 interacted upon etoposide treatment, while they did not associate in the absence of treatment. This result suggests that the CTCF–MRE11 interaction occurred upon DNA damage, or was otherwise enhanced by DNA damage. Furthermore, an endogenous CTCF–MRE11 interaction was observed in chromatin fractions (Figure 1B and C; Supplementary Figure S1A), indicating that the CTCF–MRE11 interaction was likely to occur in chromatin-bound settings. We performed the Co-IP experiment in the presence of DNase or ethidium bromide, demonstrating that the CTCF–MRE11 interaction was not bridged by DNA and that the two proteins directly interacted (Supplementary Figure S1B). Taken together, the CTCF–MRE11 Co-IP results indicate that CTCF interacts with MRE11 in a chromatin-bound setting in response to DNA damage.
Figure 1.
CTCF interacts with MRE11 in response to DNA damage. (A) Selected protein list obtained from LC-MS/MS analysis after interactome tandem affinity purification of FLAG-SFB-tagged CTCF with or without γ-irradiation. The previously known and novel hits from MS results are shown. (B and C) Forward (B) and reciprocal (C) co-immunoprecipitation (co-IP) between endogenous CTCF and MRE11 in 293T cells, after the addition of DNase Benzonase, was performed with anti-CTCF (B) or anti-MRE11 (C) antibody, without (left, DMSO) and with (right, Etoposide) etoposide treatment. Immunoblot (IB) analysis was performed with the indicated antibodies. IgG immunoprecipitation (IP) was used as a negative control. See also Supplementary Figure S1. (D and E) Schematic representations of CTCF (D) and MRE11 (E) constructs used in this study (Top). The 293T cells were transfected with the indicated HA-tagged CTCF truncation constructs (D) or GFP-tagged MRE11 truncation constructs (E). Cell lysates were immunoprecipitated with anti-MRE11 (D) or anti-GFP (E) antibody, and immunoblot (IB) analyses were performed with the indicated antibodies. (D) Positions of molecular weight makers are denoted on the left of the INPUT SDS-PAGE gel. The interaction strengths between MRE11-HA and fusion CTCF proteins were normalized to the expression levels of HA fusion CTCF proteins, i.e. input. The interaction strength between MRE11-HA full-length CTCF was set as one, and the interaction strength of each HA fusion CTCF protein was calculated (bottom right panel); PAR, poly ADP-ribosylation; NLS, nuclear localization signal; N+GAR, N-terminus and glycine arginine rich motif.
MRE11 is known to form the MRN complex with RAD50 and NBS1. By the Co-IP assay, we found that CTCF associated with RAD50 in the presence of etoposide, similar to that observed with MRE11 (Figure 1B and Supplementary Figure S1C). The CTCF–NBS1 interaction was augmented by etoposide treatment, although CTCF did bind to NBS1 without etoposide (Figure 1B). We next assessed whether ATM-mediated DNA damage signaling pathway is engaged in the DNA damage-induced interaction between CTCF and MRE11. By using an ATM inhibitor KU-55933, we found that etoposide-induced CTCF interaction with MRE11 (Figure 1A–C and Supplementary Figure S1) was abolished by the ATM inhibitor (Supplementary Figure S1D), suggesting that ATM-mediated DNA damage signaling pathway may regulate the CTCF–MRE11 interaction in response to DNA damage.
To identify the regions of CTCF that were responsible for the CTCF–MRE11 interaction, we generated truncated fragments of CTCF (Figure 1D). Anomalously, both N- (residues 1–267) and C-terminal (residues 578–727) fragments migrated slowly on SDS-PAGE (Figure 1D). In contrast, its ZF fragment (residues 246–588) appeared to migrate expectedly (Figure 1D). The N-terminus was observed at ∼64 kDa instead of 31 kDa as predicted; the C-terminus was observed at ∼37 kDa instead of 17 kDa as expected (Figure 1D and Supplementary Table S5). Consistent with this, similar aberrant mobility of bacterially purified N- and C-terminal fragments of CTCF on SDS-PAGE has been observed and reported previously (26), wherein they found that both terminal regions are extendend, monomeric and unstructured, and thereby migrate slowly. Accordingly, the anomalous mobility of N- and C-terminal fragments could be caused by their intrinsically disordered structure. Endogenous MRE11 pulled down full-length CTCF and fragments harboring either the N-terminus or ZF (Figure 1D). Considering the protein expression levels of the N-terminal and ZF fragments, the N-terminus alone was likely to be associated less with MRE11 than the ZF alone. By contrast, the C-terminus alone could not interact with MRE11. The N-terminal + Zinc finger (N-ZF) fragment associated more strongly with MRE11 than the N-terminal or ZF fragment alone (Figure 1D), indicating that the combined N-ZF fragment can associate additively or synergistically with MRE11. The Zn finger + C-terminal (ZF-C) fragment appeared to be associated less with MRE11 than with ZF (Figure 1D). Collectively, this result indicates that the N-terminal and ZF domain are marginally and robustly responsible, respectively, for the CTCF–MRE11 interaction, while the C-terminus is dispensable. Similarly, we generated three fragment mutants of MRE11 (Figure 1E). The CTCF-interacting region of MRE11 was mapped to the N-terminus of MRE11 (residues 1–537), which harbors its nuclease activity as well as the NBS1- and RAD50-interacting regions (27,28). However, the glycine-arginine rich (GAR) motif and C-terminus of MRE11 was not required for its CTCF interaction (Figure 1E).
Because endogenous CTCF associated with MRE11 in chromatin fractions (Figure 1B and C) through its N-terminal or ZF domain but not its C-terminus (Figure 1D), we next examined whether the N-terminal and ZF domains of CTCF bind to chromatin, and whether its C-terminal domain does not bind to chromatin, in the presence or absence of etoposide treatment. The C-terminus of CTCF was undetectable in chromatin fractions, its ZF domain was detectable in chromatin but not in soluble fractions, and its N-terminus was detectable in both fractions (Supplementary Figure S2). This indicates that the C-terminal domain fails to bind to chromatin regardless of etoposide treatment, contrary to the chromatin-binding capacity of the N-terminal and ZF domains. These observations suggest that CTCF bound to chromatin is capable of interacting with MRE11, due to the correlation between chromatin-binding abilities (Supplementary Figure S2) and MRE11–interaction strengths (Figure 1D) of the N-terminal and ZF domains of CTCF. Collectively, the CTCF–MRE11 interaction results support the hypothesis that CTCF associates with MRE11 in a chromatin-bound setting in response to DNA damage.
CTCF enrichment at sites of DNA damage depends on MRE11
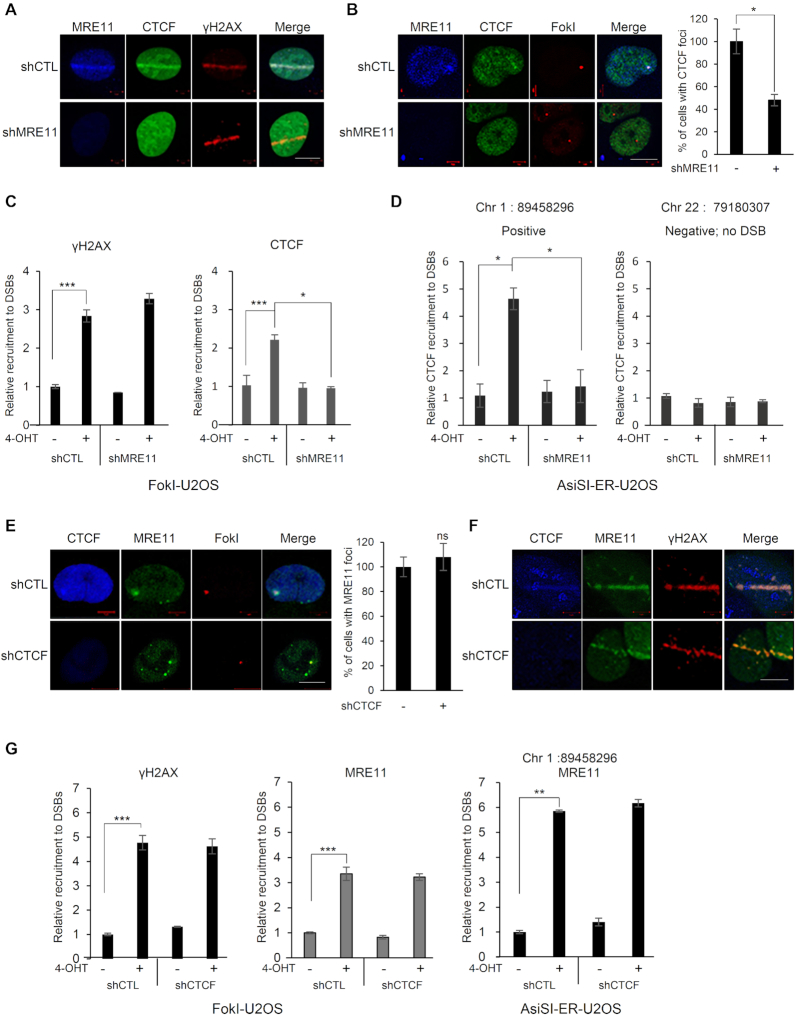
Most proteins involved in the DNA damage response, including MRE11, localize to DNA lesions and form repair foci. However, we did not observe CTCF foci by immunofluorescence staining following DNA damage. Using a live, laser micro-irradiation system, we observed that, similar to that of MRE11, CTCF was rapidly recruited to laser strips (Figure 2A). Furthermore, we tested the CTCF recruitment and its co-localization with MRE11 at DNA DSBs by using the previously described FokI–U2OS reporter system (a U2OS clone carrying repeats of the Lac operator in one locus of the genome), in which the mCherry-LacI-FokI endonuclease fusion protein introduces a single DSB in the genome (13). Consistent with the recruitment and MRE11-colocalization of CTCF onto laser strips, CTCF was recruited and co-localized with MRE11 at the FokI-induced DSB site (Figure 2B). As shown in Figure 2C, CTCF ChIP analysis revealed that CTCF was recruited to the FokI-induced DNA damage site, similar to the recruitment of γH2AX. In the FokI–U2OS reporter system, the mCherry-LacI-FokI nuclease results in a single DSB in the cell. Therefore, it is possible that CTCF accumulated at a single or a few local DSBs rather than multiple DSB sites. To rule out this possibility, we employed an additional AsiSI-ER-U2OS reporter system (12), in which the AsiSI nuclease cuts multiple AsiSI sites throughout the genome of a cell. Consistent with the recruitment of CTCF onto the FokI-induced DSB, CTCF was readily recruited to AsiSI-induced DSBs (Figure 2D). Altogether, these observations indicate that CTCF accumulates at DSBs and co-localizes with MRE11.
Figure 2.
MRE11-dependent recruitment of CTCF at sites of DNA damage. (A) MRE11-depleted (shMRE11) or control (shCTL) U2OS cells were presensitized with BrdU and subjected to laser micro-irradiation. Cells were fixed and stained with the indicated antibodies; scale bar: 10 μm. (B) Immunofluorescence was performed 4 h after induction of double-strand breaks by ER-mCherry-lacR-FokI-DD in the FokI-U2OS reporter cells transfected with shRNA targeting MRE11 (shMRE11) or control shRNA (shCTL); scale bar: 10 μm. The plot represents the percentage of cells positive for CTCF co-localized at mCherry-FokI foci. Data are the means ± SD of three independent experiments. More than 100 cells were counted in each experiment; *P≤ 0.05. (C) ChIP-qPCR was performed with an antibody to γ-H2AX or CTCF in the FokI-U2OS DSB reporter cells transfected with the indicated shRNA (shCTL as a control or shMRE11 targeting MRE11), with (+) or without (−) induction of DSBs by mCherry-LacI-FokI. The values of recruitment to DSBs were relative to those of cells without the induction of DSBs. All qPCR reactions were performed in triplicate, with the SEM values calculated from at least three independent experiments; *P≤ 0.05; ***P≤ 0.001. (D) AsiSI-ER-U2OS cells were transfected with the indicated shRNA, with (+) or without (−) induction of DSBs by AsiSI. CTCF and chromatin were immunoprecipitated with anti-CTCF antibody. The fold enrichment values were relative to those of cells without induction of DSBs. Primers on chromosome 22 (no DSB) were used as negative controls. Data are the means ± SD of at least three independent experiments, and all qPCR reactions were performed in triplicate. (E) Immunofluorescence was performed 4 h after induction of double-strand breaks (DSBs) by mCherry-LacI-FokI in the CTCF-depleted (shCTCF) or control (shCTL) FokI-U2OS cells; scale bar: 10 μm. Bar graph represents the percentage of cells positive for MRE11 co-localized at mCherry-LacI-FokI foci. Data are the means ± SD of three independent experiments. More than 100 cells were counted in each experiment. ns, not significant. (F) Recruitment of MRE11 (green) to DSBs induced by laser micro-irradiation in the CTCF-depleted (shCTCF) and control (shCTL) U2OS cells; scale bar: 10 μm. (G) ChIP-qPCR was performed with an antibody to γ-H2AX or MRE11 in FokI-U2OS cells (left and center), and AsiSI-ER-U2OS cells (right) transfected with the control (shCTL) or CTCF shRNA (shCTCF), with (+) or without (−) induction of DSBs by FokI (FokI-U2OS) or AsiSI (AsiSI-ER-U2OS). The fold enrichment values were relative to those of cells without induction of DSBs. Data are presented as means ± SD of three independent experiments, and all qPCR reactions were performed in triplicate; **P≤ 0.01; ***P≤ 0.001.
Given that CTCF interacted with MRE11 of the DNA damage sensor MRN complex upon DNA damage (Figure 1), and both CTCF and MRE11 were rapidly recruited to sites of damaged DNA (Figure 2), we investigated whether CTCF and MRE11 could affect each other’s recruitment to DNA damage sites. Depletion of MRE11 abolished CTCF recruitment to the laser-induced DNA damage sites, compared with that in control cells (Figure 2A). Consistent with the CTCF recruitment in the laser strips, in the cellular system carrying the mCherry-LacI-FokI reporter (FokI-U2OS), CTCF localization at DSBs was dramatically reduced upon MRE11 knockdown (Figure 2B). In a similar manner, CTCF ChIP results using FokI-U2OS (Figure 2C) and AsiSI-ER-U2OS (Figure 2D) reporter systems revealed that CTCF was recruited to the FokI- or AsiSI-induced DSBs significantly less in MRE11 knockdown cells than in control cells. However, depletion of CTCF had little effect on the recruitment of MRE11 to the FokI-induced DSBs (Figure 2E) as well as the laser strips (Figure 2F). In parallel, we found that CTCF depletion did not alter MRE11 accumulation at both mCherry-LacI-FokI- and AsiSI-induced DSBs by MRE11 ChIP analysis (Figure 2G). Together, these results indicate that the accumulation of CTCF to sites of damaged DNA requires MRE11, but not vice versa.
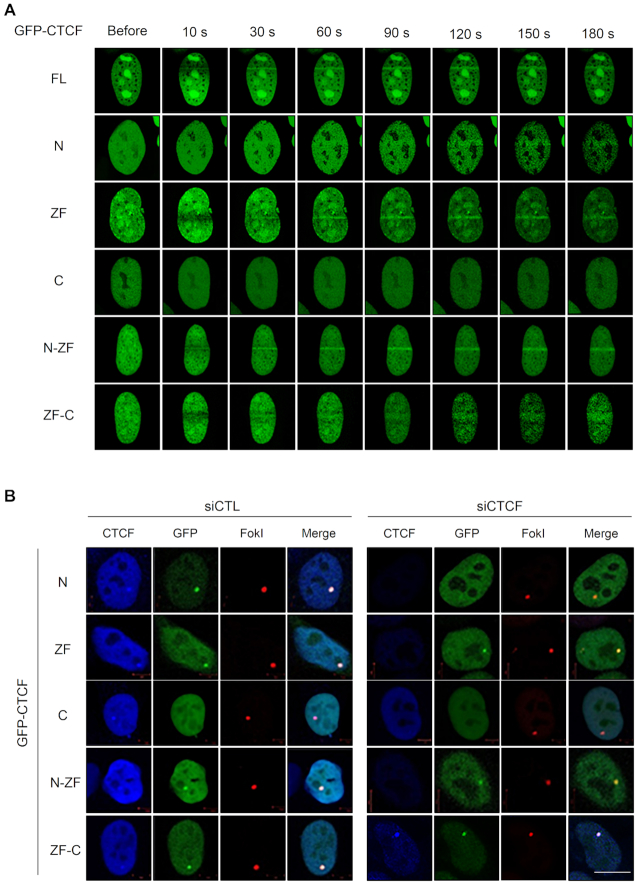
Because the recruitment of CTCF onto DNA damage sites was dependent on MRE11 (Figure 2), we investigated whether the MRE11–CTCF interaction was indeed implicated in the MRE11-mediated CTCF recruitment. Accordingly, we assessed the recruitment of ectopically overexpressed GFP-tagged full-length or truncated CTCF onto sites of DNA damage. As expected, we observed that GFP-tagged full-length CTCF was recruited to laser strips within seconds of the laser micro-irradiation (Figure 3A). GFP-tagged CTCF fragments harboring the N-terminal or ZF domain, which are able to interact with MRE11 with differing strengths (Figure 1D), were recruited in the laser strips (Figure 3A). The efficiency of CTCF fragment recruitment onto the laser strips was variable (Figure 3A) and appeared to correlate with the strength of interaction with MRE11 (Figure 1D). For example, the N-ZF and ZF fragments showed rapid and strong recruitment, while the N-terminal and ZF-C fragments showed weak but detectable recruitment. In contrast, the C-terminal fragment of CTCF, which did not interact with MRE11, failed to accumulate onto the laser strips (Figure 3A). This micro-irradiation result indicates that the CTCF–MRE11 interaction is required for CTCF recruitment to sites of DNA damage. To confirm this result, we monitored the recruitment of MRE11-interacting and non-interacting CTCF fragments in the FokI-U2OS reporter system by fluorescence microscopy. Consistent with the micro-irradiation result, the MRE11-interacting CTCF fragments (i.e. N-terminal, ZF, N-ZF and ZF-C) could be recruited at DSBs with different efficiencies, while recruitment of the C-terminal fragment did not occur (Figure 3B). The above results indicate that CTCF is recruited to sites of DNA damage in an MRE11-interaction-dependent manner.
Figure 3.
CTCF recruitment to DNA lesions requires its N-terminal or zinc-finger domain. (A) U2OS cells were transfected with GFP-tagged full-length CTCF and its truncation constructs (see Figure 1D, top), and were subjected to laser micro-irradiation for up to 180 s. (B) Recruitment of the indicated GFP-tagged CTCF proteins (green; see Figure 1D top) to double-strand breaks induced by mCherry-LacI-FokI (red) in the endogenous CTCF-depleted (siCTCF) or control (siCTL) FokI-U2OS cells. The ectopically complemented C-terminal domain of CTCF in CTCF knockdown cells (GFP-tagged C and ZF-C in siCTCF cells, right) is also recognized with an antibody to endogenous CTCF (blue); scale bar: 10 μm.
CTCF is required for CtIP localization to sites of DNA damage
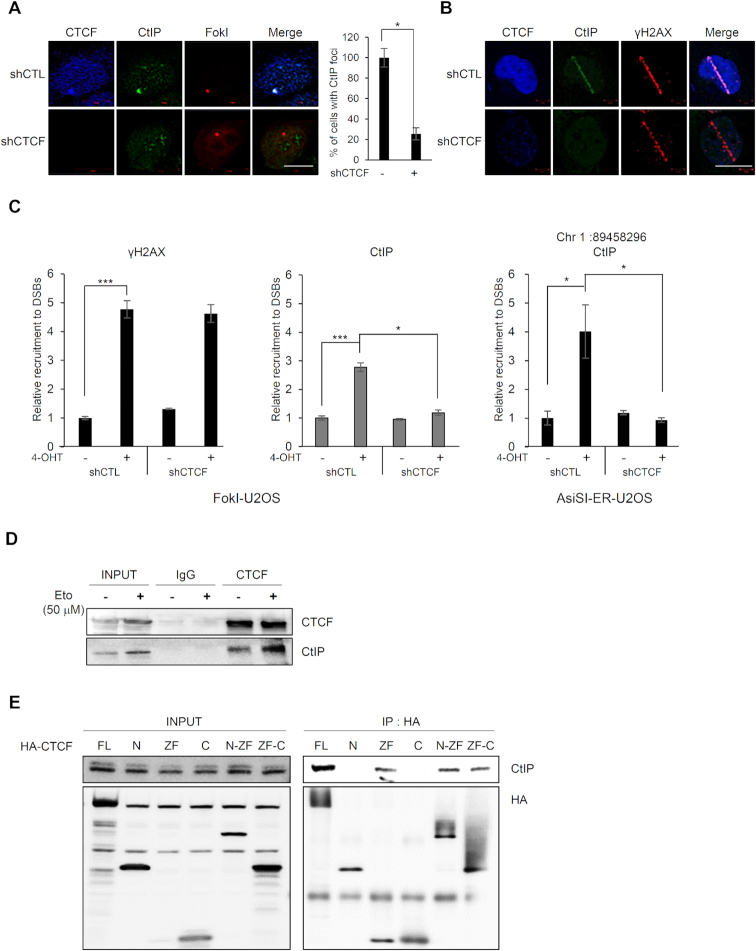
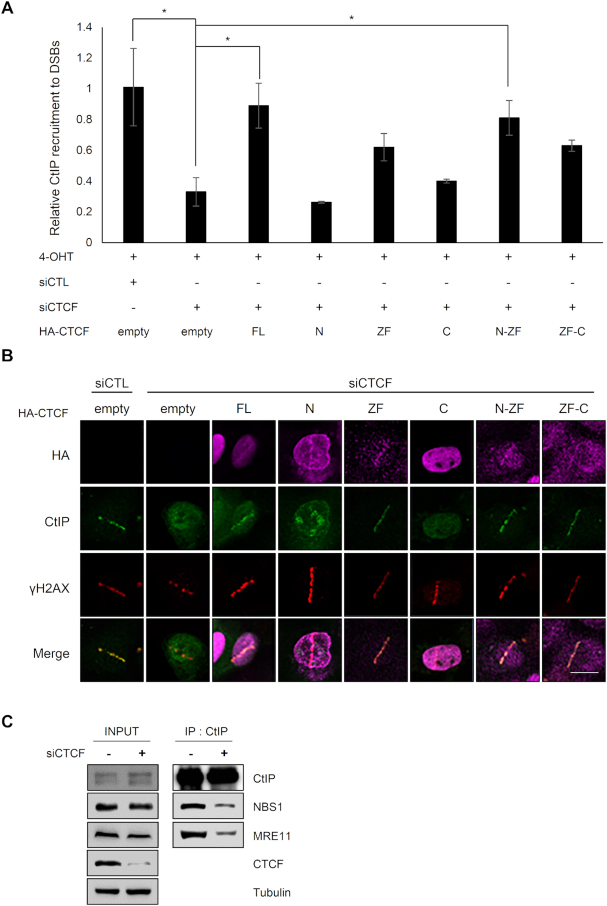
Since CTCF interacted with MRE11 (Figure 1), which cooperates with CtIP in HR (29), we hypothesized that CTCF would promote recruitment of CtIP at DNA lesions. Thus, we tested whether CTCF could affect CtIP recruitment to DNA lesions. We monitored CtIP recruitment at FokI-induced DSBs using the FokI–U2OS reporter system (Figure 4A) in CTCF-depleted cells. We found that CTCF depletion dramatically abrogated CtIP focus formation (Figure 4A), suggesting that CTCF is required for CtIP recruitment to sites of DNA damage. Next, we monitored CtIP recruitment in laser strips in the CTCF-knockdown cells. CTCF depletion profoundly attenuated CtIP recruitment in the laser strips (Figure 4B), similar to the defective CtIP recruitment with MRE11 depletion (Supplementary Figure S3A). To confirm this result, we examined whether CTCF depletion affected CtIP accumulation at DNA damage sites using a ChIP assay. In CTCF-depleted cells, CtIP accumulation onto FokI-induced or AsiSI-induced DSBs in the FokI–U2OS and AsiSI–ER-U2OS reporter cells, respectively, was dramatically reduced (Figure 4C), supporting the hypothesis that CtIP recruitment at DNA lesions depends on CTCF. In contrast, depletion of CtIP had little effect on CTCF or MRE11 recruitment to sites of DNA damage (Supplementary Figure S3B). These observations suggest that CtIP recruitment to DNA damage sites requires CTCF, but CtIP does not affect CTCF localization at these sites.
Figure 4.
CTCF is required for CtIP recruitment at DNA lesions. (A) Immunofluorescence was performed 4 h after induction of double-strand breaks (DSBs) by mCherry-LacI-FokI in CTCF-depleted (shCTCF) or control (shCTL) FokI-U2OS cells; scale bar: 10 μm. Bar graph represents the percentage of cells with CtIP (green) that co-localized at mCherry-FokI (red) foci. Data are the means ± SD of at least three independent experiments. More than 100 cells were counted in each experiment; *P≤ 0.05. (B) CTCF-depleted (shCTCF) or control (shCTL) U2OS cells were subjected to laser micro-irradiation. Cells were fixed and stained with the indicated antibodies. The scale bar represents 10 μm. (C) ChIP-qPCR was performed with an antibody to CtIP and γ-H2AX in the CTCF-depleted (shCTCF) or control (shCTL) FokI-U2OS cells (left) and AsiSI-ER-U2OS cells (right), with (+) or without (−) induction of DSBs. The fold enrichment values were relative to those of cells without induction of DSBs. Data are means ± SD of at least three independent experiments, and all qPCR reactions were performed in triplicate; *P≤ 0.05; ***P≤ 0.001. (D) CTCF interacts with CtIP. Co-immunoprecipitation assays were performed using control IgG and anti-CTCF with (+) or without (−) etoposide (Eto) treatment, and the immunoprecipitates were probed for the indicated proteins by western blotting (IB). (E) CTCF interacts with CtIP via its zinc finger domain. HA-tagged full-length CTCF and its truncated fragments (see Figure 1D, top) were expressed in 293T cells. HA-immunoprecipitates were resolved by SDS-PAGE, and blots were probed for HA (bottom) and CtIP (top).
Because CTCF was required for CtIP recruitment at DNA lesions (Figure 4A–C), we investigated whether CTCF could interact with CtIP. CTCF was capable of associating with CtIP in the presence or absence of DNA damage and the CTCF–CtIP interaction was enhanced in the presence of DNA damage (Figure 4D). We then mapped the regions of CTCF responsible for its interaction with CtIP. HA-tagged full-length CTCF and its fragments containing ZF domains (i.e. ZF, N-ZF and ZF-C) pulled down endogenous CtIP (Figure 4E). In contrast, CtIP was barely detected in the N- and C-terminal immunoprecipitates (Figure 4E). Therefore, we found that the ZF domain of CTCF was necessary for the CTCF–CtIP interaction.
Our results so far have established that CTCF interacts with CtIP through its ZF domain, and depletion of CTCF abrogates CtIP recruitment at sites of damaged DNA. Next, we tested whether the CTCF–CtIP interaction was required for the CTCF-mediated CtIP recruitment to DSBs. We co-transfected cells with siRNA targeting the 3′-UTR of endogenous CTCF and with full-length CTCF or its truncated mutants. Using the FokI-U2OS DSB reporter system with fluorescence microscopy (Supplementary Figure S4A) and the ChIP assay (Figure 5A), CtIP was found to be recruited to FokI-induced DSBs in endogenous CTCF-depleted cells expressing exogenous, full-length CTCF or its fragments containing the ZF region (Supplementary Figure S4A). We found that the ZF, N-ZF and ZF-C domains, which are capable of associating with CtIP, restored the impaired CtIP recruitment into DSBs, indicating that the ZF domain of CTCF is sufficient for CtIP recruitment to DSBs. By contrast, the N- and C-terminal fragments did not rescue CTCF-depleted cells from impaired CtIP recruitment to FokI-induced DSBs. Consistently, ChIP analysis showed that CTCF fragments carrying the ZF domain restored CtIP recruitment to DSBs in CTCF-depleted cells, comparably to full-length CTCF (Figure 5A). In contrast, the N- and C-terminal fragments did not rescue CTCF-depleted cells from impaired CtIP recruitment to DSBs. These results suggest that the CTCF–CtIP interaction is necessary for CtIP recruitment to DSBs.
Figure 5.
CtIP recruitment to DNA damage requires interactions with the zinc finger domain of CTCF. (A) Recruitment of CtIP to mCherry-LacI-FokI-induced DSBs was evaluated by ChIP–qPCR in the CTCF-depleted FokI–U2OS cells complemented by the indicated HA fusion CTCF proteins (see Figure 1D, top). CtIP and chromatin were immunoprecipitated with an anti-CtIP antibody. qPCR was performed for the quantitative analysis of ChIP samples. Fold recruitment values were relative to those of cells without induction of DSBs. Data are means ± SD of at least three independent experiments, and all qPCR reactions were performed in triplicate; *P≤ 0.05. (B) GFP–CtIP co-localization (green) at γH2AX foci (red) in CTCF-depleted U2OS cells complemented by the indicated HA fusion CTCF proteins (see Figure 1D, top). CTCF-depleted (siCTCF) and control (siCTL) U2OS cells were subjected to laser micro-irradiation. Cells were fixed and stained with the antibodies indicated on the left. The scale bar represents 10 μm. (C) Co-immunoprecipitation of endogenous CtIP and MRE11 in CTCF-depleted (+) or control (−) 293T cells was performed with anti-CtIP antibody. Immunoblot (IB) analysis was performed with the antibodies indicated on the right.
To confirm this result further, we determined whether the zinc finger domain of CTCF was required and sufficient for CtIP recruitment in the laser strips. CTCF depletion abolished CtIP recruitment to the γ-H2AX laser line, indicative of DNA lesions, although formation of the γ-H2AX strip was not affected by CTCF deficiency (Figure 5B). Consistent with the results using the U2OS-DSB-reporter system with FokI (Figure 5A and Supplementary Figure S4A), full-length CTCF or its fragments harboring the ZF domain restored CtIP localization at the γ-H2AX strips in the CTCF-depleted cells (Figure 5B). By contrast, the N- and C-terminal fragments did not rescue the localization of CtIP at the γ-H2AX strips (Figure 5B). That CtIP was recruited to the γ-H2AX strip in the presence of its full-length or ZF-fragment form verified the significance of CTCF and its ZF (Figure 5B). These results indicate that the CtIP-interacting ZF domain of CTCF is necessary and sufficient for CtIP recruitment at DSBs, in agreement with the U2OS–DSB reporter results.
Considering that the MRN complex interacts with CtIP, we questioned why CtIP required CTCF for its enrichment at DNA lesions, and how CTCF contributed to CtIP accumulation at DSBs. To address this, we first investigated whether CTCF contributes to CtIP interaction with the MRN complex. Accordingly, we examined the interaction of CtIP proteins with the MRN by immunoprecipitating CtIP with or without CTCF depletion, and we found that less MRN co-immunoprecipitated with CtIP from extracts of CTCF-depleted cells (Figure 5C). This result suggests that CTCF assists CtIP interaction with the MRN complex, which in turn results in enhanced recruitment of CtIP along with CTCF at DNA lesions. Based on the findings that CTCF is dependent on MRE11 for its DSB recruitment (Figure 2A–D) and is required for CtIP enrichment at DNA lesions (Figure 4A–C) through DNA damage-promoted CTCF–MRN/CTCF–CtIP interactions (Figures 1 and 4D) and the CTCF-assisted CtIP–MRN interaction (Figure 5C), CtIP recruitment kinetics must correlate with those of CTCF and its fragments if the CTCF-dependent CtIP enrichment at DSBs is attributable to CtIP recruitment. Otherwise, if the CTCF dependency is attributable to CtIP retention, full-length CTCF and its fragments must affect the stability (such as length or extent) of CtIP residence at DNA lesions, rather than the timing of CtIP recruitment. To test this, we conducted time-lapse imaging analyses of CtIP recruitment into the laser strips in CTCF-depleted cells complemented with or without full-length CTCF or its fragments. As shown in Figure 3A, the CTCF recruitment to laser lines could be detected as early as 10 s after micro-irradiation. In contrast, the CtIP recruitment to laser lines was significantly delayed up to 60 s compared with that of CTCF. In CTCF-knockdown cells, CtIP failed to translocate to laser lines; however, reintroduction of full-length CTCF rescued the CtIP recruitment by 60 s, which became more clearly defined over time (Supplementary Figure S4B). The difference between the time points of CTCF (10 s, earlier) and CtIP (60 s, later) recruitment onto the laser tracks, together with the findings that CtIP depletion did not detectably affect CTCF accumulation at sites of damaged DNA (Supplementary Figure S3B) but CTCF depletion, conversely, hampered CtIP recruitment (Figure 4A–C), raised the possibility that CTCF acts upstream of CtIP in DNA end resection. Complementation of the N-terminal CTCF fragment, which was weakly recruited at 10 s (Figure 3A) and was not able to interact with CtIP (Figure 4E), failed to recruit CtIP (Supplementary Figure S4B) because the N-terminus was not able to interact with CtIP, resulting in no CtIP recruitment. In contrast, all CTCF fragments harboring the ZF (i.e. ZF, N-ZF and ZF-C), which interacted with both MRE11 and CtIP (Figures 1D and 4E) and were recruited by 10 s following micro-irradiation (Figure 3A), caused CtIP recruitment by 60 s and persistence until 240 s, similar to that in the presence of full-length CTCF (Supplementary Figure S4B). The C-terminal CTCF, which is incapable of DSB recruitment and CtIP interaction, failed to recruit CtIP (Supplementary Figure S4B). These kinetic analyses of CtIP recruitment onto laser strips (Supplementary Figure S4B) reveal that CTCF promoted CtIP recruitment possibly in a CTCF–CtIP interaction-dependent manner. However, these kinetic results do not exclude the possibility that CTCF also stabilizes CtIP retention instantly after its recruitment to DNA lesions, resulting in elongated and/or intensive retention in addition to CTCF-dependent CtIP recruitment. This is presumably caused by CTCF intensifying the CtIP–MRN interaction (Figure 5C) and interacting more strongly with MRN and CtIP in response to DNA damage (Figures 1 and 4D).
CTCF enhances CtIP-mediated DNA end resection
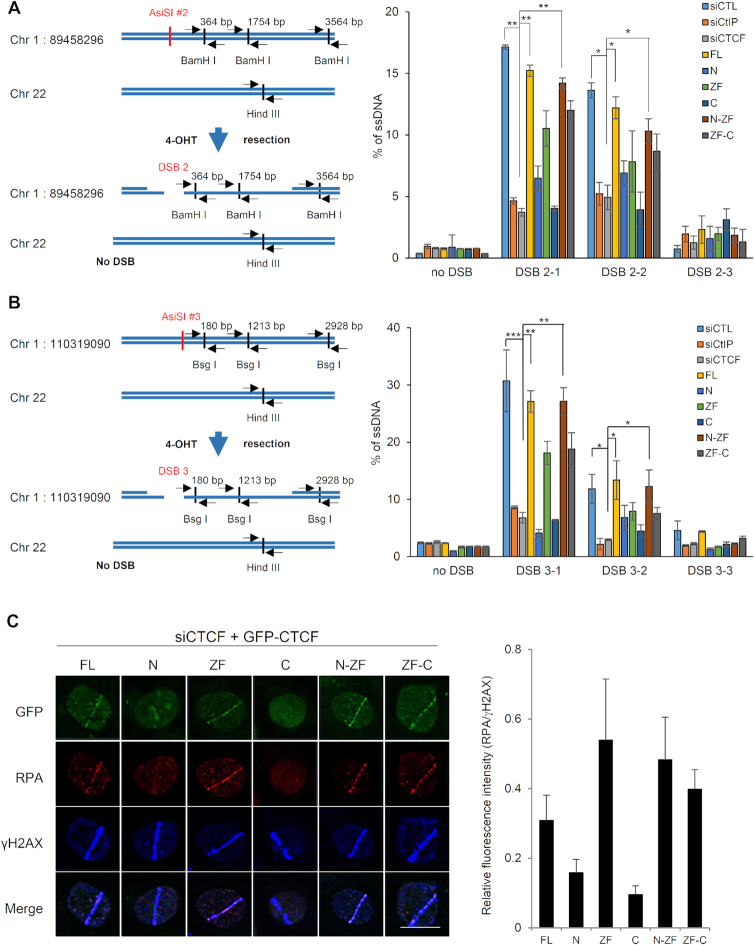
MRE11 cooperates with CtIP in 5′-to-3′ DSB resection, an early step in DNA repair that controls HR initiation (29). As our results revealed that CTCF interacts with MRE11 (Figure 1) and CtIP (Figure 4D and E), and that MRE11 is required for CTCF recruitment (Figure 2) and CTCF is then required for CtIP recruitment to damaged DNA sites (Figure 5), we hypothesized that CTCF would enhance MRE11–CtIP-mediated DNA end resection via CtIP recruitment to DSBs. To test this hypothesis, we first conducted a DNA end resection assay in the absence of CTCF by using the AsiSI–ER–U2OS reporter system. We found that CTCF knockdown remarkably reduced DNA end resection at the AsiSI-induced DSB on chromosome 1, position 89,458,296 to a level similar to that achieved by depleting CtIP, the key HR cofactor for the essential catalytic enzyme MRE11, compared with that in cells expressing control siRNA (Figure 6A). The efficiency of DNA end resection at DSBs was similar in CtIP- and CTCF-depleted cells, confirming the requirement of CTCF for DNA end resection. This reduction in DNA end resection was recovered by reintroduction of the siRNA-resistant full-length CTCF or its fragments containing the ZF domain (Figure 6A), indicating that the resection is not compromised unless the fragment containing the ZF is absent. The ZF fragment by itself showed a modest resection recovery, compared with the robust resection conferred by N-ZF, but the expression level of the ZF fragment was significantly lower than that of N-ZF and ZF-C, possibly explaining the lower DNA resection recovery by the isolated fragment. However, the expression levels of N-ZF and ZF-C did not correspond proportionally to their resection recovery efficiencies. This suggests that the N-terminus is able to interact with MRE11 (Figure 1D) but not CtIP (Figure 4E) and plays a structural role in coordinating MRE11 and CtIP in concert with the ZF domain, which is able to concomitantly interact with both MRE11 and CtIP. However, the C-terminus is unable to interact with both MRE11 and CtIP (Figures 1D and 4E) and appears to be dispensable for DNA end resection. In contrast, the N- and C-terminal fragments of CTCF without the ZF domain failed to reverse the DNA end resection defect in the CTCF-knockdown cells. The DNA end resection efficiencies of the CTCF fragments correlated with the ability of each fragment to recruit CtIP into DSBs, indicating that CTCF facilitates CtIP recruitment at DNA lesions and regulates the resulting DNA end resection. Next, to examine whether CTCF cooperates with CtIP in DNA end resection during DSB repair, resection efficiencies were assessed in CtIP and CTCF double-knockdown cells. The co-depletion of CTCF and CtIP reduced end resection at the AsiSI-induced DSBs to similar levels as observed with either single depletion (Supplementary Figure S5A), suggesting an epistatic relationship between CTCF and CtIP in end resection at DSBs. Therefore, these results suggest an important role for CTCF in regulating DNA end resection by recruiting CtIP at DSBs.
Figure 6.
CTCF depletion impairs DNA end resection. (A) Quantification of ssDNA generated by 5′-end resection at three sites (left, 2-1, 2-2, and 2–3 are 364, 1754 and 3564 bp from DSB, respectively and the paired primers across BamH 1 restriction sites are indicated as black arrow pairs) around the AsiSI-induced DSB (red) on chromosome 1:89,458,296 position in AsiSI–ER–U2OS cells transfected with the indicated shRNAs and HA-tagged CTCF constructs (right plot). The primers (black arrow pair) on chromosome 22 (no DSB) across a Hind III restriction site were used for negative control (left). Data are means ± SD of at least three independent experiments, and all qPCR reactions were performed in triplicate; *P≤ 0.05, **P≤ 0.01 (see Supplementary Figure S13A indicating levels of depleted CTCF or CtIP and complemented HA-tagged CTCF proteins.). (B) Design of qPCR primers and probes for measurement of resection at sites, 3-1 (180 bp), 3-2 (1213 bp) and 3-3 (2928 bp) near an AsiSI-induced DSB (red) on chromosome 1:110,319,090 position (left). Quantification of ssDNA generated from resection at three sites (left, 3-1, 3-2 and 3-3 located at Bsg I restriction sites) as in part (A) (right plot). Data represent means ± SEM of three independent experiments (see Supplementary Figure S13B indicating levels of silenced and complemented proteins.). (C) Accumulation of RPA onto the laser strips in CTCF-depleted U2OS cells complemented with the indicated GFP-tagged full-length or truncated CTCF proteins (left, see Figure 1A, above). Relative fluorescent intensities of RPA to those of γ-H2AX are presented in the right plot as the means ± SD of at least three independent experiments. More than 30 cells were counted in each experiment.
CTCF was robustly translocated to laser lines (Figures 2A, F and 3A) and efficiently recruited CtIP (Figures 4B and 5B; Supplementary Figure S4B) to the DSB lines, which were induced by laser micro-irradiation that randomly introduces DNA damage to the whole genome. These observations raise the possibility that CTCF is localized to DSBs irrespective of DNA sequence, transcriptional activity or chromatin status. Nonetheless, current evidence from genome-wide approaches suggests that actively transcribed regions are fragile and DSBs within actively transcribed regions are favorable for HR rather than NHEJ by preferentially accumulating HR factors (30–32). Accordingly, the transcription factor CTCF might be localized through its ZF DNA-binding domain to transcribed chromatin, in which DSBs are generated and inherently HR-prone. To test these two possibilities, we first confirmed whether there are CTCF-binding cis elements near the AsiSI-induced DSB site at position 89,458,296 on chromosome 1. The CCBL2 gene locus is mapped to this position and, therefore, active chromatin is possibly formed. This CCBL2 gene (chromosome1:89,458,296) is not a CTCF target gene, to exclude any possibility that CTCF binds to this active chromatin site in advance of cleavages generated by the AsiSI-ER nuclease (already present before DNA damage induction), without regard to DNA damage. Through ChIP-qPCR on the site, we confirmed the absence of CTCF under normal conditions before DNA damage (Figure 2D). We further evaluated the impact of CTCF depletion on transcription of the CCBL2 gene and found that this gene was robustly transcribed and its transcription was barely altered by CTCF depletion (Supplementary Figure S6). This suggests that CTCF hardly participates in the transcription of CCBL2 and that CTCF is indeed recruited into DSBs around active chromatin regions of this gene in response to DNA damage. However, this does not exclude the possibility that CTCF engages in DSB repair at active chromatin regions but not at inactive chromatin regions, because HR rather than NHEJ is chosen as the predominant repair pathway in the context of active chromatin (30–32). Thus, CTCF in combination with other HR components can be recruited to these transcribed sites.
Hence, we employed one additional AsiSI cut site (chromosome1:110,319,090), which is not mapped to any gene locus, in addition to the AsiSI cut site resident at the CCBL2 gene (chromosome1:89,458,296), previously described above. The site was chosen in an unbiased manner and because of the presence of restriction sites nearby for subsequent resection experiments. We first assessed DSB induced by the AsiSI-ER fusion nuclease on this inactive chromatin belonging to no gene (chromosome1:110,319,090) by the enrichment of γH2AX, an indicator of DNA damage. We found that γ-H2AX accumulated at this inactive chromatin region (chromosome1:110,319,090) following induction of the AsiSI-ER nuclease. This indicates that the inducible AsiSI-ER robustly introduces DSBs into both this inactive chromatin site (Supplementary Figure S7) and the active chromatin site, CCBL2 (chromosome1:89,458,296). Then, we asked whether CTCF could be recruited on the AsiSI-induced DSBs located at this inactive chromatin and whether the existence of CTCF on the DSBs affected the level of CtIP and DNA end resection. Similarly, CTCF was enriched at DSBs located in the inactive chromatin (Supplementary Figure S7), strongly suggesting that CTCF is recruited into DSBs at both active and inactive chromatin. Next, we examined whether CTCF depletion influences CtIP recruitment and consecutive DNA end resection at DSBs around inactive chromatin regions. Similar to that of DSBs in active chromatin regions, CTCF depletion reduced CtIP accumulation (Supplementary Figure S7) and DNA end resection efficiency at inactive chromatin regions (Figure 6B). As expected, knockdown of CtIP with or without CTCF knockdown reduced DNA end resection efficiency in the inactive chromatin regions to a similar level as in the single depletion of either CTCF or CtIP (Supplementary Figure S5B). This is consistent with the effects in active regions (Supplementary Figure S5A), supporting that CTCF functions in the same resection process, as does CtIP. Together, these results from the active and inactive chromatin experiments suggest that CTCF has a critical role in CtIP recruitment and DNA end resection without regard to chromatin status.
The cell-cycle-dependent regulatory mechanism for the choice between canonical NHEJ (cNHEJ) and HR is important. In S/G2 phases, when sister chromatids are available for HR, MRE11–CtIP is responsible for DSB resection, which involves nucleolytic processing of DSBs to produce ssDNA tails that are required for HR (33). We therefore examined whether CTCF depletion affects cell cycle distribution, and found that cell cycle distribution profiles were comparable in control and CTCF-depleted cells (Supplementary Figure S8), consistent with the findings of a previous report (11). This result suggests that CTCF-promoted HR is not caused by the regulation of cell cycle progression and that CTCF promotes HR through a different mechanism.
During the process of resection, RPA coats ssDNA overhangs and is then phosphorylated (29). To further determine whether CTCF deficiency affects resection, we treated control and CTCF-depleted cells with etoposide and assessed RPA phosphorylation and chromatin binding. In asynchronous cells, the levels of chromatin-bound phospho-RPA and CtIP had changed little in CTCF-depleted cells (Supplementary Figure S9A). Thus, we evaluated the effects of CTCF deficiency on RPA phosphorylation and chromatin binding of CtIP in S/G2 phases. H2AX phosphorylation (γ-H2AX), which occurred independently of DSB resection, was similar in etoposide-treated control and CTCF-depleted cells in S/G2 phases (Supplementary Figure S9B). However, the levels of RPA phosphorylation and CtIP bound to chromatin were lower in CTCF-depleted cells in S/G2 phases compared to that in control cells (Supplementary Figure S9B). In addition, RAD51 assembly at DSBs was profoundly reduced in CTCF-depleted cells in comparison to that in control cells (Supplementary Figure S9C), suggesting that CTCF promoted DSB resection, and thus leading to ssDNA generation. Thus, CTCF is able to promote ssDNA formation competent for RPA phosphorylation and the chromatin binding of phospho-RPA.
To further ascertain the role of CTCF in DNA end resection, we tested whether CTCF depletion would affect DNA damage-induced RPA focus formation, since decreased RPA phosphorylation in CTCF-depleted cells (Supplementary Figure S9D and E) correlated with decreased DSB resection (Figure 6A and B). Knockdown of endogenous CTCF diminished RPA focus formation, either in the mCherry-LacI-FokI reporter system (Supplementary Figure S9D) or after γ-irradiation (Supplementary Figure S9E). The ChIP assay using the AsiSI–ER–U2OS reporter system confirmed that γ-H2AX was enriched at DSBs in the untranscribed (on chromosome1:110,319,090; inactive chromatin) regions similar to that in the transcribed (CCBL2 gene on chromosome1:89,458,296, active chromatin) regions (Supplementary Figure S7), but RPA enrichment at DSBs near both these transcribed and untranscribed chromatin regions was remarkably impaired in cells depleted of endogenous CTCF (Supplementary Figure S9F). These results show that the RPA coating of ssDNA resulting from DNA end resection at DSBs was defective in cells lacking endogenous CTCF despite the fact that γ-H2AX focus formation was not affected. Using verified, CTCF-targeted siRNA and siRNA-resistant CTCF constructs, we further evaluated the RPA coating achieved by full-length CTCF or its fragments in cells depleted of endogenous CTCF. In cells lacking endogenous CTCF, either full-length CTCF or its fragments harboring the ZF domains were able to reverse the RPA recruitment defect (Figure 6C). By contrast, the N- and C-terminal fragments that failed to reverse the CtIP recruitment defect (Figure 5) did not restore the impaired RPA recruitment in cells depleted of endogenous CTCF (Figure 6C). Together, our results suggest an important role of CTCF in the DNA end resection step of HR by enhancing CtIP recruitment to DSBs.
Cellular role of CTCF in DNA-damage response
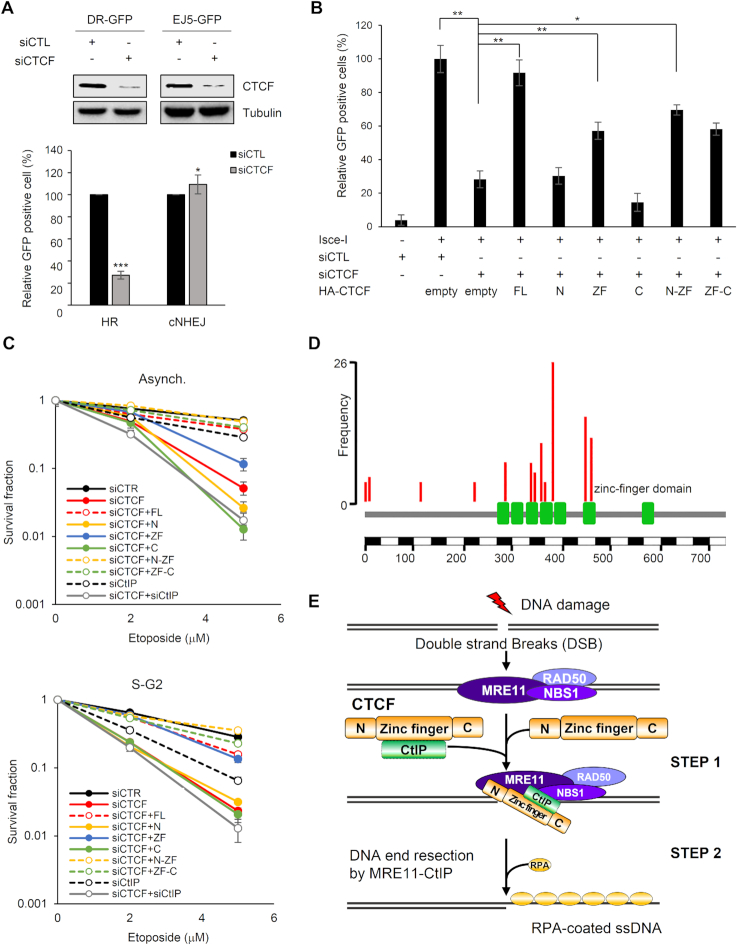
Our results reveal that CTCF was recruited to DNA lesions in an MRE11-dependent fashion (Figures 1 and 2), and that CTCF promoted CtIP recruitment, leading to DNA end resection at DSBs (Figures 4–6). DNA end resection catalyzed by the key HR proteins MRE11 and CtIP is essential for initiation of the HR repair pathway (34). Thus, we surmised that CTCF-mediated CtIP recruitment and DNA end resection are required for efficient HR. To test this supposition, we first assessed the DNA repair efficiency of CTCF by H1299-based HR/cNHEJ repair analysis (15). Using verified CTCF-targeted shRNA, we measured the repair efficiency achieved by CTCF and found that CTCF depletion alleviated HR but did not alter cNHEJ, revealing that CTCF is critical for HR but does not affect cNHEJ (Supplementary Figure S10A). To confirm this in the other well-established U2OS-based reporter systems, we evaluated HR and cNHEJ repair efficiency. CTCF-knockdown in these systems significantly lessened HR and marginally increased cNHEJ (Figure 7A, <10%), which is consistent with the effects of CTCF depletion on HR but inconsistent with the lack of detectable effects on cNHEJ in the H1299-based reporter systems. This suggests that CTCF indeed promotes HR, while CTCF seems to have no effects (or only subtle effects) on cNHEJ in a cell-type or cell context-dependent manner.
Figure 7.
CTCF–CtIP interaction contributes to CTCF-mediated homologous recombination (HR) and survival upon exposure to DNA damage. (A) U2OS-based homologous recombination- (DR-GFP) or canonical non-homologous end joining- (EJ5-GFP) reporter cell lines were co-transfected with siRNA (control, siCTL, or endogenous CTCF-specific, siCTCF) and I-SceI as indicated. Homologous recombination (HR) and canonical non-homologous end joining (cNHEJ) efficiencies were analyzed as in part (A). The plotted values are means ± SEM from at least three independent experiments; *P≤ 0.05; ***P≤ 0.001. (B) HR reporter cells (DR-GFP) were transfected with the indicated siRNAs and HA-tagged CTCF constructs together with I-SceI and subjected to an HR assay as in part (A) (see Supplementary Figure S13C indicating knockdown of endogenous CTCF and supplementation with the full-length or truncated constructs of CTCF.) The plotted values are means ± SEM from at least three independent experiments; *P≤ 0.05; **P≤ 0.01. (C) Effect of ectopic expression of the indicated HA-tagged CTCF proteins on survival of CTCF-depleted HeLa cells after treatment with etoposide. HeLa cells were transfected with the indicated siRNAs and HA-tagged CTCF constructs. The cells in asynchronous or S/G2 phase were treated with etoposide, and survival fractions were confirmed by a clonogenic survival assay. The intensities and areas of the colonies were measured using ImageJ software (see Supplementary Figure S13D showing depletion of CTCF or CtIP and supplemented CTCF protein levels.). (D) Somatic point mutations (nonsense and missense) of CTCF from Catalogue of Somatic Mutations in Cancer (COSMIC) as of 13 October, 2018. Among them, 14 substitution mutations were identified in ≥4 samples across tumors in the COSMIC database. Eight out of the 14 mutations were located on the zinc finger (ZF) domain, which was defined as Pfam (ID P49711). As the frequency of mutations in the ZF domain of CTCF was higher than that in its N-terminal domain, the somatic mutations were significantly enriched within the ZF domain in cancer. (E) Model for the role of CTCF in DNA end resection of HR-mediated repair via CtIP recruitment. The MRN complex recognizes and binds to double-strand breaks (DSBs) and recruits CTCF to sites of DNA damage. In turn, CTCF promotes the recruitment of CtIP to DSBs and enhances DNA end resection.
Since CTCF, whose recruitment depends on MRE11 (Figure 2), was found to play a role in DNA end resection by recruiting CtIP to DSBs (Figure 6) and CtIP-mediated DSB resection is required for HR initiation (29), we wondered whether the ZF domain of CTCF, which is able to recruit CtIP to DSBs (Figure 5) and achieve significant DSB resection (Figure 6A and B), supports HR repair by establishing functional HR links between MRE11, CTCF and CtIP. To do this, we evaluated DSB repair efficiency in CTCF-knockdown cells expressing siRNA-resistant full-length CTCF or its fragments, using U2OS-based reporter assays for HR and cNHEJ repair (14). As shown in Figure 7B, full-length CTCF and its fragments harboring ZF domains efficiently restored HR, consistent with the requirement of CTCF in CtIP recruitment and DSB resection, which is a key prerequisite for HR. The efficiencies of HR rescue by the CTCF constructs were correlated with those of the rescued CtIP recruitment and DNA resection in CTCF-depleted cells. Conversely, cNHEJ repair efficiency was not affected by CTCF depletion and reintroduction of the CTCF constructs in the U2OS-based (Supplementary Figures S10B and S11) and H1299-based (Supplementary Figure S10C) knockdown cells. These results indicate that CTCF-mediated CtIP recruitment at DSBs is critical for the function of CTCF in HR. Moreover, in agreement with previous findings (34,35), we found that depletion of CtIP resulted in a significant reduction in HR repair efficiency (Supplementary Figures S10B and S11), indicating that CTCF is required to promote HR, similar to that of CtIP. Additionally, depletion of both CTCF and CtIP attenuated HR to a level that was similar to that of either single depletion (Supplementary Figure S10B). This further supports the supposition that CTCF functions in the same genetic pathway as CtIP during DSB repair, consistent with the results of the DNA end resection assay. Taken together, these results raise the possibility that CTCF and CtIP facilitate HR, whose initiation depends on DSB resection, but they are not engaged in cNHEJ. This differs from the key HR player, BRCA1, which promotes HR and concomitantly antagonizes cNHEJ.
Unlike cNHEJ, alternative NHEJ (aNHEJ) and SSA repair depend on several HR proteins, including MRN and CtIP, like HR, these repair pathways are initiated by DSB resection, but they do not involve homologous sister chromatids (36,37). Thus, we examined whether CTCF-assisted resection at DSBs can promote aNHEJ and SSA repair. To do this, we conducted U2OS-based aNHEJ and SSA repair assays (14) in CTCF-depleted cells. CTCF depletion led to >50% reductions in aNHEJ and SSA efficiencies (Supplementary Figures S10B and S11), contrary to its subtle effect on cNHEJ (Figure 7A and Supplementary Figure S10A,B). Reintroduction of full-length CTCF or its ZF fragments into cells depleted of endogenous CTCF rescued defective SSA and aNHEJ, analogous to the effects on HR. These results indicate that reduced SSA and aNHEJ levels by CTCF depletion could be caused by impaired activity of DSB resection. Furthermore, the efficiencies of all tested DSB repair pathways (HR, SSA, aNHEJ and cNHEJ) in CTCF-depleted cells were comparable to those in CtIP-depleted cells (Supplementary Figure S10B), indicating a functional similarity between CTCF and CtIP in DNA repair. Collectively, these repair assay results suggest that CTCF facilitates HR, SSA and aNHEJ repair by recruiting CtIP and subsequently promoting MRE11/CtIP-mediated resection.
To corroborate these findings, we assessed the relevance of the ZF domain of CTCF, which is the CtIP-interacting region, for CTCF-driven HR in DNA damage sensitivity. To test this, cells depleted of endogenous CTCF with siRNA were compared with cells expressing control siRNA for their sensitivity to etoposide. In clonogenic cell survival assays, exposure of the cells to etoposide reduced the viability of the CTCF-depleted cells (Figure 7C). Consistent with the HR rescue efficiency of either the full-length CTCF or its ZF domain fragments in CTCF-depleted cells (Figure 7B and Supplementary Figure S10B), the cells exhibited partial complementation for survival (Figure 7C). While the CTCF-depleted and CtIP-depleted cells were equivalent with regard to HR and other repair pathways (Figure 7B and Supplementary Figure S10B) as well as DNA end resection efficiency (Figure 6A and B), there were clear differences between the CTCF-depleted and CtIP-depleted cells with regard to etoposide sensitivity (Figure 7C). The CTCF-depleted cells were hypersensitive to etoposide compared to that of the CtIP-depleted cells (Figure 7C). These differential etoposide sensitivities conferred by either CTCF or CtIP depletion indicate that CTCF likely contributes to DNA damage responses in addition to DNA repair, in which CTCF functions in concert with CtIP. This was also supported by the etoposide sensitivity of CTCF- and CtIP-codepleted cells, which was similar to that of CTCF-depleted cells in asynchronized status and marginally increased in S/G2 phases (Figure 7C). Together, our results illustrate the biological importance of the interaction between MRE11–CTCF–CtIP in DNA damage response via HR priming, and provide mechanistic evidence for a role of CTCF in the DNA end resection step of HR by facilitating CtIP recruitment to DSBs.
DISCUSSION
CTCF is the master genome organizer (38) identified lately to contribute to genome integrity by participating in HR (10, 11). Despite this importance, a definite role of CTCF in HR and its underpinning mechanism have been largely overlooked. This oversight has been due, in part, to a seemingly unrelated HR function distinct from the numerous, reported genome regulatory functions, such as transcriptional activation/repression, insulation, imprinting, X-chromosome inactivation and affecting 3D chromatin architecture. Here, we report that CTCF facilitates HR through DNA end resection by recruiting CtIP to DSBs (Figure 7E). This assertion is based on our findings that depletion of CTCF causes a loss of CtIP localization at DNA lesions (Figure 4A–C), impaired DSB resection (Figure 6A and B; Supplementary Figure S5A and B) and compromised HR (Figure 7A and Supplementary Figure S10A).
Our analysis also establishes a mechanistic basis for CTCF–MRE11–CtIP cooperation (Figure 7E). Ectopic expression of fragments harboring the ZF region of CTCF rescued MRE11-dependent CTCF recruitment (Figure 3), CtIP recruitment (Figure 5A and B; Supplementary Figure S4A), DSB end resection (Figure 6A and B) and HR (Figure 7B and Supplementary Figure S10B) defects in CTCF-depleted cells. Thus, our findings suggest that the ZF region of CTCF is necessary for the role of CTCF in HR. CTCF interacted with CtIP through its ZF region (Figure 4E). The importance of the CTCF–CtIP interaction for CTCF-mediated HR was also supported by the correlations between the rescue efficiencies and CtIP interaction strengths of each fragment harboring the ZF region. The N-terminus of CTCF assisted ZF-mediated HR processes, including DNA end resection (Figure 6A and B) and CtIP stabilization, but not recruitment (Figure 5A), considering our finding that the N-terminal domain did not interact with CtIP (Figure 4E and Supplementary Figure S4B). What might be possible explanations for this auxiliary role of its N-terminus that is not necessarily essential for the CTCF–CtIP interaction but contributes to DNA end resection? An appealing clue exists in the functional importance of NBS1 in connecting CtIP to S/G2 cyclin-dependent kinase (CDK) and ATM for CtIP phosphorylation (39) and then linking this phosphorylated CtIP to the MRE11/RAD50 nuclease for resection (40,41). This suggests that NBS has a structural role in bridging CtIP to stimulate the MRE11 nuclease. Likewise, the N-terminus of CTCF interacts with MRE11 and thereby seems likely to have a structural role in coordinating MRE11 with CtIP, which is bound to and presented by the ZF domain. This consequently leads to more efficient resection than with ZF alone, albeit the functionally minimal region of CTCF for HR is indeed the ZF domain. Interestingly, according to the Catalogue of Somatic Mutations in Cancer, the most frequently mutated CTCF nucleotides in human cancers are enriched within the ZF domain, suggesting that its ZF domain is likely to be essential for error-free DNA repair and genome integrity (Figure 7D). Taken together, reversal of these CTCF-depletion defects by its ZF region is, therefore, dependent on the interaction between CtIP and the ZF region of CTCF, which likely leads to CtIP recruitment at DSBs, end resection and HR initiation.
The CTCF-assisted end resection at DSBs seems to apply not only to HR (Figure 7A, and Supplementary Figure S10A) but also to the aNHEJ and SSA pathways (Supplementary Figure S10B), which are initiated by end resection at DSBs and share the DSB resection with HR (36,37). When assessed using aNHEJ/SSA repair reporters, knockdown of CTCF suppressed aNHEJ and SSA and this diminished aNHEJ/SSA repair efficiency was equivalent to that of CtIP-depleted cells (Supplementary Figure S10B), supporting that CTCF contributes to end resection at DSBs and resection-dependent repair pathways. Moreover, additional CTCF depletion had minimal impact on defective DNA end resection and impaired HR/aNHEJ/SSA in CtIP-depleted cells and vice versa, consistently and strongly suggesting that CTCF and CtIP function in the same pathway during DSB repair. The finding that CTCF and CtIP did not engage or marginally engaged in cNHEJ (Figure 7A and Supplementary Figure S10A–C), consistent with previously reported results that CtIP depletion does not detectably increase cNHEJ in U2OS cells (34,35), is reminiscent of their functional connection to the resection and resection-dependent repair pathways. The epistatic nature of CTCF and CtIP, both single- and co-depletion, on end resection and DSB repair pathways reinforces this notion. By contrast, an earlier study reported that depletion of CtIP from 293T cells modestly increased cNHEJ (1.3- to 1.4-fold) (42), suggesting that CtIP suppresses cNHEJ in 293T cells. Together, these contradictory effects of CtIP on cNHEJ suggest that cell type or cell context influences the involvement of HR proteins in cNHEJ, and, if involved, their role in cNHEJ. The major distinction between the previous controversial results is the different cell lines used for the studies. Thus, in our study, we employed two U2OS- and H1299-based repair reporter systems, which have wild-type and homozygous partial deletion mutant p53, respectively, and regulate the cell cycle checkpoint at the G1–S transition by directly targeting p21 transcription, the Ink41/Cip1 inhibitor, to arrest cells at the NHEJ-predominant G1 phase. We therefore asked whether DNA damage-induced p21 expression is impaired in H1299 cells. Induction of p21 following etoposide treatment was detected in all tested repair reporter cell lines except for H1299 (Supplementary Figure S12), suggesting that the induced p21 activates the G1-phase checkpoint and enhances cNHEJ efficiency in U2OS-based repair reporter cell lines. In contrast, the defective p21 induction by p53 gene deletion in H1299-based cell lines may not contribute to increased cNHEJ efficiency. Accordingly, this disparity between the U2OS and H1299 cell lines may cause differences in cell-cycle arrest at G1 between them, which might be advantageous (U2OS) and disadvantageous (H1299) for cNHEJ. We can speculate that small but differential effects of CTCF depletion and ectopic expression of its full-length gene or truncated fragments on cNHEJ in H1299 (no effect) or U2OS (marginal effect, <10%) cells may be mirrored by this differential cell context. Notwithstanding the small marginal effect of CTCF depletion on cNHEJ in U2OS cells, CTCF can promote HR largely without interfering with cNHEJ. Our results further reveal a functional correlation between CTCF and CtIP in end resection and DSB repair, as well as a functional distinction between CTCF–CtIP and BRCA1 in antagonizing cNHEJ. This, in turn, allows us to hypothesize that CTCF promotes HR by a mechanism distinct from BRCA1′s molecular role in counteracting 53BP1 by promoting 53BP1 dephosphorylation and releasing its essential partners, RIF1 and PTIP, which are required for end protection, from resection (43–46). Rather, CTCF would promote HR by facilitating early steps in the HR pathway, which is essential for HR-mediated repair, HR choice or both. Consistent with our hypothesis, we identified the MRN complex and CtIP as novel CTCF-interacting partners (Figures 1 and 3), both of which control the decision to repair DSB by HR through the regulation of DSB end resection (47). Prompted by these findings, we further revealed that CTCF enhanced DSB end resection by recruiting CtIP through the CTCF–CtIP interaction (Figures 5 and 6).
According to an attractive hypothesis, the chromatin status where DSBs occur is another regulatory factor for the choice between the cNHEJ and HR pathways; indeed, actively transcribed regions favor HR associated with transcription by a DNA–RNA hybrid-dependent recruitment of RAD52, XPG and BRCA1, followed by recruitment of RAD51 (30,32). CTCF is a transcription factor and we found that it recruited CtIP and sequentially promoted end resection at two AsiSI cut sites, which reside within the CCBL2 gene (chromosome 1:89,458,296). We extended the CTCF relevance to another AsiSI cut site (chromosome 1:110,319,090), which does not belong to any gene, in an inactive chromatin region. We revealed that CTCF functions in CtIP recruitment, end resection and HR independently from transcription per se and, thus, CTCF can contend with DSBs at both active and inactive chromatin sites. However, this does not exclude the possibility that CTCF participates in transcription-associated HR (32).
BRCA1 also promotes CtIP-mediated DNA end resection via its interaction with CtIP (48). However, a previous study using a mouse model also showed that loss of the CtIP–BRCA1 interaction has little effect on resection and genomic integrity (49). Rather, there are additional routes for the initiation of HR via enhancing DSB end resection, wherein p75 (50), USP4 (51) or AUNIP (52) interacts with CtIP, promoting CtIP accumulation at DSBs for efficient end resection and HR. CTCF was required for CtIP accumulation at DSBs and enhanced DSB end resection and HR (Figures 4–7). Mechanistically, CTCF functioned with MRE11 and CtIP, critical nucleases for DSB end resection (Figures 5 and 6), acting downstream of MRE11 (Figure 2) and upstream of CtIP in the resection (Figures 4–6). CTCF is required for this attribute, where CTCF-mediated CtIP recruitment facilitates DSB end resection by the MRE11–CtIP nuclease. Then, CTCF may recruit RAD51 via its interaction to promote the formation of the RAD51 presynaptic filament onto a long ssDNA stretch generated from DNA end resection in HR, as shown by our results as well as those of a previous report (11). More importantly, CTCF interacted with MRE11, especially in the presence of DNA damage (Figure 1). Thus, this DNA damage-responsive association between CTCF and MRE11 enabled CTCF to engage in the DNA damage response. Furthermore, the ability of CTCF to recruit CtIP at DNA lesions (Figure 5) contributed to the formation of the HR initiation complex, which conferred DNA end resection activity (Figure 6A and B).
The earlier findings showed that CTCF is required for the recruitment of BRCA2 (10), which targets RAD51 on RPA-coated ssDNA, and the C-terminal domain of CTCF interacts with RAD51 (11). Therefore, these CTCF–BRCA2 and CTCF–RAD51 interactions likely contribute to the RAD51 presynaptic/synaptic complex assembly. In addition, we herein found that CTCF acts at DSB resection, a step upstream of RAD51 filament assembly in cooperation with BRCA2 (10,11). Taken together, our results, as well as previously published results, reveal the multifaceted role of CTCF in HR-mediated DSB repair. In addition, CTCF-depleted cells succumbed to etoposide, which was significantly more hypersensitive than that of CtIP-depleted cells (Figure 7C). Furthermore, the etoposide sensitivity of double CTCF- and CtIP-depleted cells was not dissimilar from that of CTCF-depleted cells in asynchronized status, but was subtly increased in S/G2 phases (Figure 7C), indicating that, although CTCF and CtIP likely function in the same pathways during DNA repair, CTCF may have additional roles in DNA damage responses. As a transcription factor or chromatin organizer, CTCF would further manage DNA damage in addition to HR.
Our results also showed a novel attribute and role of CTCF in the first control point of DSB repair at the DNA end resection step of HR-mediated repair (Figure 7B and E; Supplementary Figure S10A). Thus, the findings from our study extend the understanding of how CTCF contributes to DNA damage repair and tumor suppression functions. Elucidating the mechanisms for HR has important implications in understanding tumor pathogenesis and therapy. Tumors depleted in BRCA1 and BRCA2 are severely impaired in HR, and are therefore sensitive to therapies that utilize PARP inhibitors. Based on our study, depletion of CTCF renders wild-type BRCA1/2 cancer cells HR-incompetent. These results could be used to develop new therapeutic strategies for HR-competent tumor therapy with PARP inhibitors.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Roger A. Greenberg for the ER-mCherry-LacI-FokI-DD-U2OS reporter (FokI-U2OS), Dr Gaëlle Legube for the AsiSI-ER-U2OS reporter, Dr Jeremy M. Stark for the GFP-reporter cell lines used for the HR/cNHEJ/aNHEJ/SSA repair assays and Dr Sunyoung Chae for technical help in micro-irradiation.
Notes
Present address: Joo Seok Han, Neuracle Genetics Inc., Seoul 02841, Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF), Korean Government [2017M2A2A7A01021034, 2017R1A2B4010146, 2017R1D1A1B03031171, IBS-R022-D1]. Funding for open access charge: National Research Foundation of Korea (NRF), Korean Government [2017R1A2B4010146].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jimeno S., Femandez-Avila M.J., Cruz-Garcia A., Cepeda-Garcia C., Gomez-Cabello D., Huertas P.. Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res. 2015; 43:987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yuan J., Chen J.. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J. Biol. Chem. 2009; 284:31746–31752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010; 17:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Forget A.L., Kowalczykowski S.C.. Single-molecule imaging brings Rad51 nucleoprotein filaments into focus. Trends Cell Biol. 2010; 20:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 6. Pena-Hemandez R., Marques M., Hilmi K., Zhao T., Saad A., Alaoui-Jamali M.A., del Rincon S.V., Ashworth T., Roy A.L., Emerson B.M. et al.. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-1. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E677–E686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gregor A., Oti M., Kouwenhoven E.N., Hoyer J., Sticht H., Ekici A.B., Kjaergaard S., Rauch A., Stunnenberg H.G., Uebe S. et al.. De novo mutations in the genome organizer CTCF cause intellectual disability. Am. J. Hum. Genet. 2013; 93:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kemp C.J., Moore J.M., Moser R., Bernard B., Teater M., Smith L.E., Rabaia N.A., Gurley K.E., Guinney J., Busch S.E. et al.. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014; 7:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Izhar L., Adamson B., Ciccia A., Lewis J., Pontano-Vaites L., Leng Y., Liang A.C., Westbrook T.F., Harper J.W., Elledge S.J.. A systemic analysis of factors localized to damaged chromatin reveals PARP-dependent recruitment of transcription factors. Cell Rep. 2015; 11:1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hilmi K., Jangal M., Marques M., Zhao T., Saad A., Zhang C., Luo V.M., Syme A., Rejon C., Yu Z. et al.. CTCF facilitates DNA double-strand break repair by enhancing homologous recombination repair. Sci. Adv. 2017; 3:e1601898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lang F., Li X., Zheng W., Li Z., Lu D., Chen G., Gong D., Yang L., Fu J., Shi P. et al.. CTCF prevents genomic instability by promoting homologous recombination-directed DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:10912–10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G.. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010; 29:1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., Greenberg R.A.. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013; 20:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gunn A., Stark J.M.. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods. Mol. Biol. 2012; 920:379–391. [DOI] [PubMed] [Google Scholar]
- 15. Mund A., Schubert T., Staege H., Kinkley S., Reumann K., Fritsch L., Battisti V., Ail-Si-Ali S., Hoffbeck A.S. et al.. SPOC1 modulates DNA repair by regulating key determinants of chromatin compaction and DNA damage response. Nucleic Acids Res. 2012; 40:11363–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 17. Nicolette M.L., Lee K., Guo Z., Rani M., Chow J.M., Lee S.E., Paull T.T.. MRE11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2010; 17:1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Y., Caron P., Leqube G., Paull T.T.. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014; 42:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zierhut C., Diffley J.F.. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008; 27:1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nguyen N.H., Park H.J., Yang S.S, Choi K.S., Lee J.S.. Anti-cancer efficacy of nonthermal plasma dissolved in a liquid, liquid plasma in heterogeneous cancer cells. Sci. Rep. 2016; 6:29020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Andegeko Y., Moyal L., Mittelman L., Tsarfaty I., Shiloh Y., Rotman G.. Nuclear retention of ATM at sites of DNA double-strand breaks. J. Biol. Chem. 2001; 276:38224–38230. [DOI] [PubMed] [Google Scholar]
- 22. Zampieri M., Guastafierro T., Calabrese R., Ciccarone F., Bacalini M.G., Reale A., Perillli M., Passananti C., Caiafa P.. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem. J. 2012; 441:645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chemukhin I.V., Shamsuddin S., Robinson A.F., Carne A.F., Paul A., El-Kady A.I., Lobanenkov V.V., Klenova E.M.. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 2000; 275:29915–29921. [DOI] [PubMed] [Google Scholar]
- 24. Witcher M., Emerson B.M.. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009; 34:271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zlatanova J., Caiafa P.. CTCF and its protein partners: divide and rule. J. Cell Sci. 2009; 122:1275–1284. [DOI] [PubMed] [Google Scholar]
- 26. Martinez S.R., Miranda J.L.. CTCF terminal segments are unstructured. Protein Sci. 2010; 19:1110–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim J.E., Minter-Dykhouse K., Chen J.. Signaling networks controlled by the MRN complex and MCD1 during early DNA damage responses. Mol. Carcinog. 2006; 45:403–408. [DOI] [PubMed] [Google Scholar]
- 28. Williams R.S., Williams J.S., Tainer J.A.. MRE11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break singnaling, and the chromatin template. Biochem. Cell Biol. 2007; 85:509–520. [DOI] [PubMed] [Google Scholar]
- 29. Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P.. Ctp1 is a cell-cycle-regulated protein that functions with MRE11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007; 28:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburin V., Miller K.M., Jackson S.P. et al.. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014; 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marnef A. Cohen, Legube G.. Transcription-coupled DNA double-strand break repair: active genes need special care. J. Mol. Biol. 2017; 429:1277–1288. [DOI] [PubMed] [Google Scholar]
- 32. Yasuhara T., Kato R., Hagiwara Y., Shiotani B., Yamauchi M., Nakada S., Shibata A., Miyagawa K.. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell. 2018; 175:558–570. [DOI] [PubMed] [Google Scholar]
- 33. Chen H., Lisby M., Symington L.S.. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell. 2013; 50:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shamanna R.A., Lu H., de Freitas J.K., Tian J., Croteau D.L., Bohr V.A.. WRN regulates pathway choice between classical and alternative non-homologous end joining. Nat. Commun. 2016; 7:13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu Y., Ayrapetov M.K., Xu C., Gursoy-Yuzugullu O., Hu Y., Price B.D.. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012; 48:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sallmyr A., Tomikinson A.E.. Repair of DNA double-strand breaks by mammalian alternative end-joining pathway. J. Biol. Chem. 2018; 293:10536–10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ahrabi S., Sarkar S., Pfister S.X., Pirovano G., Higgins G.S., Porter A.C., Humphrey T.C.. A role for human homologous recombination factors in suppressing microhomology-mediated end joining. Nucleic Acids Res. 2016; 44:5743–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Phillips J.E., Corces V.G.. CTCF: master weaver of the genome. Cell. 2009; 137:1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W. et al.. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013; 9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 41. Anand R., Jasrotia A., Bundschuh D., Howard S.M., Ranjha L., Stucki M., Cejka P.. NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 2019; 38:e101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkac J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. et al.. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 44. Champman J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., Boulton S.J.. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 2013; 49:858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feng L., Li N., Li Y., Wang J., Gao M., Wang W., Chen J.. Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discov. 2015; 1:15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Isono M., Niimi A., Oike T., Hagiwara Y., Sato H., Sekine R., Yoshida Y., Isobe S.Y., Obuse C., Nishi R. et al.. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 2017; 18:520–532. [DOI] [PubMed] [Google Scholar]
- 47. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 48. Daley J.M., Sung P.. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell Biol. 2014; 34:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cruz-Garcia A., Lopez-Saavedra A., Huertas P.. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014; 23:451–459. [DOI] [PubMed] [Google Scholar]
- 50. Reczek C.R., Szabolcs M., Stark J.M., Ludwig T., Baer R.. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 2013; 201:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Daugaard M., Baude A., Fugger K., Povlsen L.K., Beck H., Sorensen C.S., Petersen N.H., Sorensen P.H., Lukas C., Bartek J. et al.. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012; 19:803–810. [DOI] [PubMed] [Google Scholar]
- 52. Lou J., Chen H., Han J., He H., Huen M.S.Y., Feng X.H., Liu T., Huang J.. AUNIP/C1orf135 directs DNA double-strand breaks towards the homologous recombination repair pathway. Nat. Commun. 2017; 8:985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.