Abstract
Background
We previously demonstrated that etonogestrel concentrations were 82% lower in women using etonogestrel contraceptive implants plus efavirenz-based ART compared with women not receiving ART.
Objectives
To investigate the genetic contribution to this previously observed drug–drug interaction through studying SNPs in genes known to be involved in efavirenz, nevirapine or etonogestrel metabolism in the same group of women.
Patients and methods
Here, we present a secondary analysis evaluating SNPs involved in efavirenz, nevirapine and etonogestrel metabolism and associated etonogestrel pharmacokinetics among 57 women, 19 not receiving ART (control group), 19 receiving efavirenz- (600 mg daily) based ART and 19 receiving nevirapine- (200 mg twice daily) based ART. Associations between patient genotype and etonogestrel pharmacokinetic parameters were determined through univariate and multivariate linear regression. This study was registered at clinicaltrials.gov (NCT02082652).
Results
Within the control group, CYP2B6 983 T>C was associated with 27% higher etonogestrel Cmax and 28% higher AUC0–24weeks. In the efavirenz group CYP2B6 516 G>T was associated with 43% lower etonogestrel Cmin and 34% lower AUC0–24weeks. For participants receiving nevirapine, NR1I2 63396 C>T was associated with 39% lower etonogestrel Cmin and 37% lower AUC0–24weeks.
Conclusions
This study demonstrates the influence of pharmacogenetics on the extent of drug–drug interactions between etonogestrel and efavirenz- or nevirapine-based ART. Efavirenz plus the etonogestrel contraceptive implant results in a detrimental drug–drug interaction irrespective of patient genetics, which is worsened in women possessing variant alleles for these CYP2B6 SNPs.
Introduction
Within sub-Saharan Africa, 80% of new HIV cases in adolescents are among girls.1 More highly effective contraceptive options are needed to support the needs of this growing demographic and to help reduce the incidence of mother to child transmission. The etonogestrel subdermal implant is an effective contraceptive method recommended by the WHO.2 The antiretroviral drug efavirenz is a first-line HIV medication also recommended by the WHO; however, concomitant use of efavirenz and the etonogestrel implant results in a significant drug–drug interaction resulting in reduced etonogestrel exposure and unintended pregnancies.3–6
We previously demonstrated etonogestrel concentrations to be 82% lower in Ugandan women receiving efavirenz-based ART compared with women not receiving ART, while nevirapine-based ART did not result in a significant drug–drug interaction with etonogestrel.6 Additionally our group has previously reported an association between CYP2B6 SNPs with alterations in the pharmacokinetics of levonorgestrel released from a subdermal implant when prescribed concomitantly with efavirenz or nevirapine.7 Etonogestrel and levonorgestrel are both approved for use as progestin-only contraceptive implants and have similar metabolism pathways, both being primarily metabolized by CYP3A4.8,9 We sought to investigate potential associations between SNPs involved in efavirenz, nevirapine and etonogestrel metabolism with etonogestrel pharmacokinetics in the same group of women, including SNPs within the CYP2B6, NR1I2, CYP3A4 and ABCB1 genes.
NR1I2 encodes the pregnane X receptor (PXR) responsible for regulation of expression of multiple enzymes including CYP3A4.8,10ABCB1 SNPs have previously been associated with alterations in efavirenz plasma concentrations.11CYP2B6 SNPs have been linked with alterations in efavirenz and nevirapine pharmacokinetics in a multitude of studies within patients of different ethnicities.12–20 Efavirenz is an inducer of CYP3A4 activity, resulting in enhanced systemic clearance of co-administered CYP3A4 substrates.21–23 Furthermore, efavirenz activates PXR, which is responsible for transcriptional regulation of CYP3A4, in a dose-dependent manner in vitro.23 We hypothesize that alterations in efavirenz or nevirapine concentrations, caused by SNPs within associated genes, would have a secondary effect of altering etonogestrel metabolism, through the antiretroviral drug altering the activity of enzymes involved in the metabolism of etonogestrel.21–23
Patients and methods
Ethical approval
All study procedures occurred at the Infectious Disease Institute (IDI) in Kampala, Uganda and were approved by the University of Pittsburgh (PRO14010195), the Joint Clinical Research Centre and Uganda National Council of Science and Technology (HS 1618). This study followed the Declaration of Helsinki and was registered at clinicaltrials.gov (NCT02082652).
Study design and cohort
Full information on the study design and participants has been described previously by Chappell et al.6 In brief, this pharmacogenetics substudy included 57 of the 60 Ugandan women enrolled into the parent study, 19 receiving nevirapine- (200 mg twice daily) and 19 receiving efavirenz- (600 mg daily) based ART for HIV treatment. Statistical analysis was also completed for the 19 participants within the antiretroviral-naive (control) arm of the study to assess the influence of pharmacogenetics in the absence of concomitant ART. Exclusionary criteria included, but were not limited to, HIV RNA >400 copies/mL in participants receiving ART, CD4+ cell count <350 cells/mm3 in the antiretroviral-naive group and coadministration of medication contraindicated for use with etonogestrel, efavirenz or nevirapine within the respective groups. In light of the growing number of cases of observed pregnancies in women receiving efavirenz who have a contraceptive implant, participants in the efavirenz group had a copper intrauterine device inserted prior to study initiation to minimize risk of unintended pregnancy in the event of etonogestrel contraceptive failure.
Sample and data collection
Study visits occurred at 1, 4, 12 and 24 weeks after implant placement. Blood samples were taken in order to determine the etonogestrel concentration at each study visit. For efavirenz and nevirapine, a single timed blood sample was taken twice before implant insertion and 4, 12 and 24 weeks after implant insertion. For nevirapine sampling, blood was drawn 11–13 h after the participant’s last nevirapine dose. For efavirenz, sampling was completed 12–14 h after the last efavirenz dose. Etonogestrel concentrations were quantified from plasma through week 24 after etonogestrel implant placement, using HPLC-MS.24 For nevirapine and efavirenz quantification, HPLC was performed utilizing validated methods.25,26 The pharmacokinetic parameters included in this study were AUC from entry to week 24 (AUC0–24weeks), Cmax, Tmax and Cmin. Cmax and Cmin represent the highest and lowest concentrations observed over the entire study period. AUC was calculated using the trapezoidal rule (Phoenix WinNonlin, Certara®).
Genotyping
Patient DNA was extracted from whole blood through use of the manufacturer’s protocol (E.Z.N.A Blood DNA Mini Kit; Omega Bio-tek, Norcross, GA, USA). Genotyping was completed using a real-time allelic discrimination PCR assay on a DNA Engine Chromo4 system (Bio-Rad Laboratories, Hercules, CA, USA). The PCR protocol involved denaturation at 95°C for 10 min, followed by 50 cycles of amplification at 92°C for 15 s and annealing at 60°C for 1 min 30 s. Samples were genotyped for the following SNPs utilizing Taqman assays: CYP2B6 516 G>T (rs3745274), 983 T>C (rs28399499) and 15582 C>T (rs4803419), NR1I2 63396 C>T (rs2472677), CYP3A4 392 G>A (rs2740574), ABCB1 4036 A>G (rs3842) and 3435 C>T (rs1045642) using Taqman Genotyping Master mix and corresponding Taqman Genotyping assays purchased from Thermo Fisher Scientific (Wilmington, DE, USA). Opticon Monitor v.3.1 software (Bio-Rad Laboratories) was used to obtain allelic discrimination plots and identify genotypes.
Statistical analysis
Compliance for each SNP with Hardy–Weinberg equilibrium was tested through previously outlined methods.27 Genotypes were coded for regression analyses as 0 = homozygous common allele, 1 = heterozygous and 2 = homozygous variant allele. Categorical variables were described using relative frequencies; continuous variables were described using the median and IQR. The Shapiro–Wilk test was used to test for normality, with P ≤ 0.05 considered as statistically significant. Associations between patient genotype and etonogestrel pharmacokinetic parameters were determined through univariate and multivariate linear regression. A univariate analysis through linear enter regression was carried out in order to identify independent variables associated with etonogestrel pharmacokinetic parameters within each study group. Variables with P ≤ 0.2 for the univariate analysis were carried through to a linear backwards multivariate analysis, with P ≤ 0.05 considered statistically significant. All statistical analyses were carried out using IBM SPSS Statistics v.24 (IBM Armonk, NY, USA). All charts were produced using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA).
Results
Etonogestrel, efavirenz and nevirapine pharmacokinetics
In total, 57 women living with HIV were included in the analysis, 19 receiving efavirenz, 19 receiving nevirapine and 19 not receiving ART (control group). All genotypes and patient characteristics are summarized in Table 1. The median (IQR) age and weight of all participants was 28 years (25–34 years) and 57 kg (50–69 kg). All SNPs were in Hardy–Weinberg equilibrium, with the exception of ABCB1 4036 A>G, which compromises interpretation of this SNP. Statistically significant univariate and multivariate regression analysis results of each group are presented in Table 2. Full regression analysis results are shown in Table S1 (available as Supplementary data at JAC Online).
Table 1.
Characteristics and genotype frequencies of the study participants at entry
| Total (n = 57) | Control group (n = 19) | Efavirenz group (n = 19) | Nevirapine group (n = 19) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | ||||||||||||
| age (years) | 28 (25–34) | 27 (24–30) | 29 (23–35) | 32 (28–35) | ||||||||
| height (cm) | 160 (155–163) | 160 (154–165) | 157 (150–165) | 161 (155–164) | ||||||||
| weight (kg) | 57 (50–69) | 62 (49–78) | 56 (48–64) | 56 (51–82) | ||||||||
| CD4 count (cells/mm3) | 624 (441–1050) | 832 (624–1483) | 449 (274–1072) | 544 (428–853) | ||||||||
| Genotype frequencies | ||||||||||||
| CYP2B6 516G>T (rs3745274) (%) | GG | GT | TT | GG | GT | TT | GG | GT | TT | GG | GT | TT |
| 40 | 53 | 7 | 57 | 37 | 6 | 32 | 58 | 11 | 32 | 63 | 5 | |
| CYP2B6 983T>C (rs28399499) (%) | TT | CT | CC | TT | CT | CC | TT | CT | CC | TT | CT | CC |
| 82 | 18 | 0 | 84 | 16 | 0 | 84 | 16 | 0 | 79 | 21 | 0 | |
| CYP2B6 15582C>T (rs4803419) (%) | CC | CT | TT | CC | CT | TT | CC | CT | TT | CC | CT | TT |
| 89 | 11 | 0 | 89 | 11 | 0 | 95 | 5 | 0 | 84 | 16 | 0 | |
| NR1I2 63396C>T (rs2472677) (%) | CC | CT | TT | CC | CT | TT | CC | CT | TT | CC | CT | TT |
| 39 | 44 | 17 | 37 | 42 | 21 | 37 | 47 | 16 | 47 | 42 | 11 | |
| CYP3A4 392G>A (rs2740574) (%) | GG | AG | AA | GG | AG | AA | GG | AG | AA | GG | AG | AA |
| 47 | 44 | 9 | 47 | 37 | 16 | 42 | 58 | 0 | 53 | 37 | 10 | |
| ABCB1 4036A>G (rs3842) (%) | AA | AG | GG | AA | AG | GG | AA | AG | GG | AA | AG | GG |
| 70 | 14 | 16 | 84 | 11 | 5 | 58 | 21 | 21 | 68 | 11 | 21 | |
| ABCB1 3435C>T (rs1045642) (%) | CC | CT | TT | CC | CT | TT | CC | CT | TT | CC | CT | TT |
| 74 | 26 | 0 | 68 | 32 | 0 | 68 | 32 | 0 | 84 | 16 | 0 | |
Values are shown as median (IQR) and percentage of population.
Table 2.
Statistically significant results from univariate and multivariate linear regression analysis within each study group
| Univariate linear regression |
Multivariate linear regression |
|||||
|---|---|---|---|---|---|---|
| P | β (95% CI) | r 2 | P | β (95% CI) | r 2 | |
| Efavirenz group | ||||||
| log10 ENG Cmax | ||||||
| CYP2B6 516G>T (rs3745274) | 0.135 | −0.085 (−0.2, 0.0) | 0.126 | |||
| CYP2B6 983T>C (rs28399499) | 0.014 | −0.222 (−0.4,−0.5) | 0.307 | 0.003 | −0.237 (−0.4, 0.1) | 0.518 |
| CYP2B6 15582C>T (rs4803419) | 0.070 | −0.277 (−0.6, 0.3) | 0.180 | |||
| ABCB1 4036A>G (rs3842) | 0.110 | 0.068 (0.0, 0.2) | 0.144 | |||
| ENG Tmax | ||||||
| log10 weight (log10 kg) | 0.199 | 3.005 (−1.7, 7.8) | 0.095 | |||
| CYP2B6 516G>T (rs3745274) | 0.045 | 0.507 (0.0, 1.0) | 0.216 | 0.045 | 0.507 (0.0, 1.0) | 0.216 |
| log10 ENG Cmin | ||||||
| CYP2B6 516G>T (rs3745274) | 0.003 | −0.102 (−0.2, 0.0) | 0.423 | 0.003 | −0.102 (−0.2, 0.0) | 0.423 |
| log10 ENG AUC0–24weeks | ||||||
| CYP2B6 516G>T (rs3745274) | 0.028 | −0.098 (−0.2, 0.0) | 0.255 | 0.008 | −0.106 (−0.2, 0.0) | 0.487 |
| CYP2B6 983T>C (rs28399499) | 0.062 | −0.142 (−0.3, 0.0) | 0.190 | 0.016 | −0.158 (−0.3, 0.0) | 0.487 |
| Nevirapine group | ||||||
| log10 ENG Cmax | ||||||
| CYP2B6 983T>C (rs28399499) | 0.013 | 0.187 (0.0, 0.3) | 0.313 | 0.013 | 0.187 (0.0, 0.3) | 0.313 |
| NR1I2 63396C>T (rs2472677) | 0.058 | −0.091 (−0.2, 0.0) | 0.196 | |||
| log10 ENG Cmin | ||||||
| CYP2B6 983T>C (rs28399499) | 0.062 | 0.114 (0.0, 0.2) | 0.190 | |||
| NR1I2 63396C>T (rs2472677) | 0.010 | −0.091 (−0.2, 0.0) | 0.329 | 0.010 | −0.091 (−0.2, 0.0) | 0.028 |
| log10 ENG AUC0–24weeks | ||||||
| CYP2B6 983T>C (rs28399499) | 0.080 | 0.125 (0.0, 0.3) | 0.170 | |||
| CYP3A4 392G>A (rs2740574) | 0.154 | 0.063 (−0.2, 0.0) | 0.116 | 0.004 | 0.096 (−0.2, 0.0) | 0.643 |
| NR1I2 63396C>T (rs2472677) | 0.004 | −0.116 (−0.2, 0.0) | 0.388 | <0.001 | −0.139 (−0.2,−0.1) | 0.643 |
| Control group | ||||||
| log10 ENG Cmax | ||||||
| CYP2B6 983T>C (rs28399499) | 0.053 | 0.159 (0.0, 0.3) | 0.203 | 0.013 | 0.193 (0.0, 0.3) | 0.416 |
| CYP3A4 392G>A (rs2740574) | 0.133 | 0.063 (0.0, 0.1) | 0.128 | 0.028 | 0.083 (0.0, 0.2) | 0.416 |
| log10 ENG AUC0–24weeks | ||||||
| CYP2B6 983T>C (rs28399499) | 0.043 | 0.156 (0.0, 0.3) | 0.219 | 0.011 | 0.188 (0.0, 0.3) | 0.415 |
| CYP3A4 392G>A (rs2740574) | 0.160 | 0.056 (0.0, 0.1) | 0.113 | 0.034 | 0.076 (0.0, 0.1) | 0.415 |
ENG, etonogestrel.
Univariate linear regression (P ≤0.2) completed, all statistically significant results then carried through to multivariate linear regression analysis (P ≤0.05). All statistically significant variables from multivariate linear regression are shown in bold.
Control group
Within the control group, CYP2B6 983 T>C was significantly associated with higher log10 etonogestrel Cmax (P = 0.013, β = 0.193) and higher log10 etonogestrel AUC0–24weeks (P = 0.011, β = 0.188); equivalent to 10% higher etonogestrel Cmax and 76% higher etonogestrel AUC0–24weeks in participants heterozygous CT compared with those homozygous TT. CYP3A4 392 G>A was also significantly associated with higher log10 etonogestrel Cmax (P = 0.028, β = 0.083) and higher log10 etonogestrel AUC0–24weeks (P = 0.034, β = 0.076); equivalent to 64% higher etonogestrel Cmax and 63% higher etonogestrel AUC0–24weeks in participants homozygous G compared with those homozygous A (Tables 2 and 3).
Table 3.
Etonogestrel, efavirenz and nevirapine pharmacokinetic parameters shown as median (IQR), summarized by associated CYP2B6, NR1I2 or CYP3A4 genotype
|
CYP2B6 516G>T (rs3745274) |
CYP2B6 983T>C (rs28399499) |
NR1I2 63396C>T (rs2472677) |
CYP3A4 392G>A (rs2740574) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GT | TT | TT | CT | CC | CC | CT | TT | GG | AG | AA | |
| Allele frequency | ||||||||||||
| EFV group | 6 | 11 | 2 | 16 | 3 | 0 | 7 | 9 | 3 | 8 | 11 | 0 |
| NVP group | 6 | 12 | 1 | 15 | 4 | 0 | 9 | 8 | 2 | 10 | 7 | 2 |
| control group | 11 | 7 | 1 | 16 | 3 | 0 | 7 | 8 | 4 | 9 | 7 | 3 |
| ENG Cmax (pg/mL) | ||||||||||||
| EFV group | 160 (158–185) | 133 (102–207) | 97 (85–109) | 148 (109–207) | 93 (75–102) | – | 102 (101–207) | 136 (85–220) | 148 (114–178) | 148 (108–213) | 114 (101–185) | – |
| NVP group | 585 (533–895) | 514 (489–781) | 693 | 533 (498–705) | 913 (701– 1124) | – | 701 (498– 1124) | 585 (514–705) | 480 (460–500) | 502 (489–701) | 650 (585–895) | 674 (500–847) |
| control group | 840 (756–959) | 868 (685–974) | 527 | 840 (650–971) | 1157 (959– 1196 | – | 949 (922– 1022) | 840 (756–971) | 667.5 (650– 685) | 756 (527–949) | 959 (868–972) | 922 (840–974) |
| ENG Tmax (week) | ||||||||||||
| EFV group | 1 (1–1) | 1 (1–1) | 2.5 (1–4) | 1 (1–1) | 1 (1–1) | – | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) | – |
| NVP group | 1 (1–1) | 1 (1–1) | 1 | 1 (1–1) | 1 (1–1) | – | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) |
| control group | 1 (1–1) | 1 (1–1) | 1 | 1 (1–1) | 1 (1–1) | – | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) | 1 (1–1) |
| ENG Cmin (pg/mL) | ||||||||||||
| EFV group | 81 (63–84) | 65 (57–76) | 46 (40–52) | 67 (53–81) | 60 (57–62) | – | 57 (53–71) | 158 (133–185) | 57 (53–104) | 60 (53–85) | 67 (57–80) | – |
| NVP group | 302 (269–461) | 514 (489–781) | 368 | 343 (280–375) | 438 (369–507) | – | 369 (324–461) | 349 (302–394) | 222 (174–269) | 333 (280–404) | 354 (302–461) | 322 (269–375) |
| control group | 321 (281–427) | 393 (249–513) | 297 | 300 (249–420) | 480 (407–509) | – | 268 (249–427) | 374 (297–513) | 318 (243–393) | 297 (209–480) | 407 (374–513) | 300 (249–427) |
| ENG AUC0–24weeks (pg·week/mL) | ||||||||||||
| EFV group | 2052 (1679– 2669) | 1664 (1537– 2146) | 1364 (968– 1760) | 1760 (1587– 2405) | 1405 (1142– 1597) | – | 1537 (1405– 2405) | 1978 (1679– 2146) | 1587 (1571– 2669) | 1679 (1587– 2669) | 1664 (1405– 2148) | – |
| NVP group | 9048 (8492– 16217) | 9902 (8095– 11299) | 13420 | 9805 (8095– 11299) | 13185 (11540– 14829) | – | 11540 (9902– 15145) | 8492 (8088– 11299) | 7179 (5311– 9048) | 9805 (7778– 11540) | 10978 (8492– 16217) | 12096 (9048– 15145) |
| control group | 10492.5 (9753.5– 14300) | 10765 (10024.5– 14740.5) | 7855 | 10332 (8636– 11794.5) | 14300 (13448.5– 16205.5) | – | 10765 (10024.5– 14300) | 10492.5 (9753.5– 15821.5) | 9484 (8636– 10332) | 10185.5 (6112– 13448.5) | 11794.5 (10332– 14740.5) | 10765 (9753.5– 14478) |
| EFV C12–14h (mg/L) | 2.1 (2.0–2.7) | 3.2 (2.9–6.6) | 8.9 (8.1–9.7) | 2.9 (2.5–4.3) | 9.3 (7.05–11.4) | – | 3.0 (2.9–6.6) | 2.9 (2.0–4.9) | 3.3 (2.7–6.6) | 2.7 (2.1–4.9) | 3.2 (2.7–9.3) | – |
| NVP C11–13h (mg/L) | 5.9 (5.6–7.1) | 6.4 (4.8–7.9) | 11.0 | 6.2 (4.7–7.1) | 7.6 (7.4–7.8) | – | 6.5 (4.7–7.9) | 6.2 (5.9–11.0) | 6.0 (4.8–7.1) | 5.6 (4.0–7.9) | 6.3 (5.9–11.0) | 6.2 (7.3–5.2) |
ENG, etonogestrel; EFV, efavirenz; NVP, nevirapine.
Efavirenz C12–14h (mg/L) and nevirapine C11–13h (mg/L) determined from individual participant’s geometric mean value calculated from concentration measured at study entry and weeks 1, 4, 12, 24 and 48 summarized for the group as median (IQR).
Efavirenz group
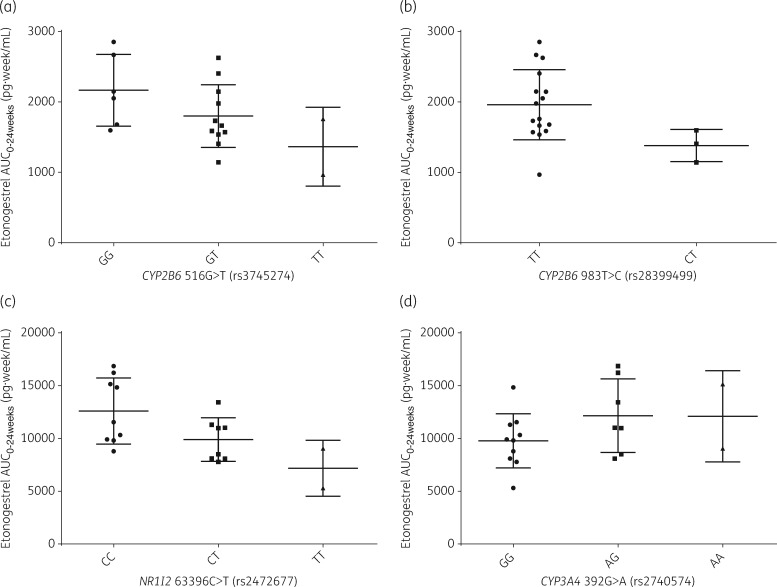
As shown in Table 2 and Figure 1, CYP2B6 516 G>T was associated with a lower log10 etonogestrel Cmin (P = 0.003, β=−0.102) and lower log10 etonogestrel AUC0–24weeks (P = 0.008, β=−0.106) for participants receiving efavirenz. This equates to a 43% difference in etonogestrel Cmin and a 34% difference in etonogestrel AUC0–24weeks between participants with homozygous G and homozygous T genotypes for CYP2B6 516 G>T, respectively (see Table 3).
Figure 1.
Etonogestrel pharmacokinetics compared by statistically significant genotype within the efavirenz (a and b) and nevirapine (c and d) groups. Data are represented by mean (SD) and compared by genotype for each of the SNPs significantly associated with etonogestrel AUC0–24weeks found through multivariate analysis (P = 0.05) within the efavirenz group (a and b) and the nevirapine group (c and d).
CYP2B6 983 T>C was associated with lower log10 etonogestrel Cmax (P = 0.003, β = −0.237) and lower log10 etonogestrel AUC0–24weeks (P = 0.016, β = −0.158), which equates to a 37% difference in etonogestrel Cmax and a 20% difference in etonogestrel AUC0–24weeks between participants who were homozygous T and heterozygous CT for CYP2B6 983 T>C when prescribed efavirenz alongside the etonogestrel contraceptive implant (see Tables 2, 3 and Figure 1).
Based on prior data, an etonogestrel concentration of 90 pg/mL is the minimum concentration required to suppress ovulation.6,28 In the context of the two SNPs associated with changes in etonogestrel exposure in the efavirenz group, we observed that the median etonogestrel concentration in all participants, regardless of genotype, fell below this concentration at all visits after the week 4 visit (Table 4). Further, participants who were homozygous (TT) or heterozygous (GT) for CYP2B6 516 G>T and those heterozygous CT for CYP2B6 983 T>C had a median concentration below 90 pg/mL by the week 4 visit.
Table 4.
Etonogestrel concentration per week of study summarized by significant CYP2B6 SNP genotype within the efavirenz group
| Etonogestrel concentration (pg/mL) |
||||
|---|---|---|---|---|
| week 1 | week 4 | week 12 | week 24 | |
| CYP2B6 516G>T (rs3745274) | ||||
| GG (n = 6) | 160 (158–185) | 92 (79–107) | 74 (68–110) | 81 (63–84) |
| GT (n = 11) | 114 (101–220) | 78 (58–135) | 53 (48–83) | 62 (53–80) |
| TT (n = 2) | 92 (85–99) | 72.5 (36–109) | 50.5 (36–65) | 46 (40–52) |
| CYP2B6 983T>C (rs28399499) | ||||
| TT (n = 16) | 148 (108–207) | 92 (78–121) | 64 (53–83) | 67 (53–81) |
| CT (n = 3) | 94 (75–102) | 58 (48–74) | 54 (32–68) | 60 (57–62) |
| CC (n = 0) | – | – | – | – |
Values are shown as median (IQR)
As anticipated, efavirenz plasma concentration (C12–14h) was 76% higher in participants homozygous T for CYP2B6 516 G>T and 69% higher in participants heterozygous CT for CYP2B6 983 T>C compared with participants who were homozygous T (Table 3).
Nevirapine group
For participants on nevirapine treatment, NRI12 63396 C>T was associated with lower log10 etonogestrel Cmin (P = 0.010, β = −0.091) and lower log10 etonogestrel AUC0–24weeks (P <0.001, β = −0.013); equivalent to 39% lower etonogestrel Cmin and 37% lower etonogestrel AUC0–24weeks in participants homozygous TT compared with those homozygous CC. CYP2B6 983 T>C was associated with higher log10 etonogestrel Cmax (P = 0.013, β = 0.187), which equates to a etonogestrel Cmax difference of 41% between homozygous T and heterozygous CT participants. CYP3A4 392 G>A was associated with higher log10 etonogestrel AUC0–24weeks (P = 0.004, β = 0.096), which equates to an 18% difference in log10 etonogestrel AUC0–24weeks between homozygous G and homozygous A participants (Tables 2 and 3).
Nevirapine median plasma concentration (C12–14h) was 7% lower in participants homozygous T for NRI12 63396 C>T compared with participants homozygous C, and 18% higher in participants heterozygous for CYP2B6 983 T>C compared with participants homozygous T. Furthermore, for participants homozygous A for CYP3A4 392 G>A, nevirapine plasma concentration (C12–14h) was 10% higher than in participants homozygous G (Table 3).
Discussion
This study demonstrates associations between genetic variations in CYP2B6 516 G>T and 983 T>C with multiple pharmacokinetic parameters of etonogestrel in women treated with efavirenz using etonogestrel contraceptive implants. Our group has previously described a genetic association between SNPs in CYP2B6 and lower pharmacokinetics of levonorgestrel given as a subdermal implant in women receiving efavirenz.7 Here we describe 33% lower etonogestrel AUC0–24weeks within homozygous T participants compared with homozygous G participants for CYP2B6 516 G>T. For levonorgestrel AUC0–24weeks, 64% lower results were observed for homozygous T participants compared with homozygous G for CYP2B6 516 G>T.7 Furthermore, 20% lower etonogestrel AUC0–24weeks was seen between homozygous C and heterozygous CT participants for CYP2B6 983 T>C,7 similar to the 23% lower levonorgestrel AUC0–24weeks observed between these genotypes in our previous study. Greater reductions were seen in etonogestrel pharmacokinetic exposure in the presence of CYP2B6 SNPs associated with reduced efavirenz metabolism. This finding may be explained by higher concentrations of efavirenz resulting in increased CYP3A4 activity and expression that is known to enhance elimination of etonogestrel. This is supported by a previous study of the effect of varying concentrations of efavirenz on CYP3A4 activity that demonstrated a dose-dependent induction of CYP3A4 by efavirenz.22 Furthermore, CYP2B6 516 G>T and 983 T>C have been shown to result in reduced CYP2B6 expression.18,29
In previous work, we reported an association between NRI12 63396 C>T and higher levonorgestrel Tmax. Also, we observed an association between CYP2B6 516 G>T and higher levonorgestrel Cmin and Cmax.7 The consistent findings of these two studies strengthen the evidence base in support of a genetic contribution to the drug–drug interaction between contraceptive hormonal treatments and efavirenz- or nevirapine-based ART. Taken together, these studies imply that greater risk of contraceptive failure exists in women with variant alleles for CYP2B6 SNPs who receive efavirenz and levonorgestrel- or etonogestrel-based contraceptive implants.
Within the nevirapine group, NRI12 63396 C>T, CYP3A4 392 G>A and CYP2B6 983 T>C were associated with alterations in etonogestrel pharmacokinetics. The association of CYP3A4 392 G>A with higher log10 etonogestrel AUC0–24weeks is a novel finding in this study. CYP3A4 392 G>A is found in the promoter region of CYP3A4.30 The presence of this SNP alters the transcription binding site of the promoter region, where it is hypothesized to effect protein binding and thus reduce gene expression.30 This mechanism of action may explain the observed relationship, as reduced expression of CYP3A4 results in lesser metabolism of etonogestrel, irrespective of the presence of nevirapine, as demonstrated within HIV-positive women using a etonogestrel contraceptive implant without ART in the control group (Tables 2 and 3), where CYP3A4 392 G>A was associated with higher log10 etonogestrel AUC0–24weeks. The relationship between CYP2B6 983 T>C and higher etonogestrel Cmax contradicts that observed within the efavirenz group and is surprising given that nevirapine is an inducer of CYP3A4.31 However, this result mirrors the findings within the control group, where CYP2B6 983 T>C was associated with a 27% higher etonogestrel Cmax between TT and CT genotype patients. Additionally these findings mirror that observed within our levonorgestrel study, where CYP2B6 516 G>T was significantly associated with higher levonorgestrel Cmin and Cmax within the nevirapine group.7 While these consistent findings support the legitimacy of an association, a biological mechanism for this interaction is yet to be elucidated. The contradictory nature of the relationship between nevirapine pharmacokinetics and CYP2B6 983 T>C has been discussed previously, and a larger cohort study would be required to confirm the strength of the observations within our two studies.32
Notably, due to the extent of the interaction between efavirenz and etonogestrel observed (82% lower etonogestrel exposure), the median concentration of etonogestrel for all participants, irrespective of CYP2B6 genotype, fell below the concentration desired to suppress ovulation after week 4. Clinical studies are currently under way to determine the suitability of a dose alteration of etonogestrel or levonorgestrel to overcome this observed drug–drug interaction in patients receiving efavirenz. These studies are in the form of patients receiving either two etonogestrel (132 mg) or two levonorgestrel (300 mg) implants at once: clinical trials.gov registration numbers NCT03282799 and NCT02722421, respectively.33,34 The findings of these studies will be useful in determining if this approach can mitigate the interaction observed between efavirenz and progestin-based implants.
Use of physiologically based pharmacokinetic modelling to examine the effect of a reduction in efavirenz dose (600 to 400 mg) on the previously observed interaction between the 150 mg levonorgestrel subdermal implant and efavirenz predicted that efavirenz dose reduction would not fully mitigate the effect of efavirenz co-administration.35,36 A similar investigation would be of utility for etonogestrel, given the greater degree of variation in etonogestrel concentrations observed between week 1 and week 24 when prescribed concomitantly with efavirenz (geometric mean at week 24 = 66 pg/mL: a 51% reduction in etonogestrel concentration from study week 1) compared with that seen for levonorgestrel prescribed alongside efavirenz at study week 24 (geometric mean at week 24 = 280 pg/mL: 31% reduction in levonorgestrel concentration from study week 1).6,35
Our study included only Ugandan women of African ancestry, with the significant SNPs in the efavirenz group found predominantly in African patients.32 Further pharmacogenetics studies in women of different ethnicities would be necessary to understand if women of particular ethnicities are at higher risk of contraceptive implant failure compared with others. Future studies would benefit from recruitment of a larger sample size, given the limited number of patients within the statistically significant populations and that ABCB1 4036 A>G was not in Hardy–Weinberg equilibrium.
Overall, drug–drug interactions between hormonal contraceptive implants and antiretroviral drugs may significantly compromise contraceptive efficacy in HIV-positive women and limit clinical treatment options in resource-constrained settings. In our participants receiving efavirenz, a cumulative effect of the CYP2B6 SNP variant alleles on etonogestrel concentrations was observed throughout the study even though CYP2B6 is not involved in etonogestrel metabolism. This study demonstrates the influence of patient genetics on the pharmacokinetic exposure of contraceptive hormones mediated via a drug–drug interaction.
Supplementary Material
Acknowledgements
We thank the patients, staff and administration of the Infectious Diseases Institute, Makerere University, Kampala, Uganda, for their collaboration and support.
Funding
This work was supported by the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number 1R01HD085887 (Principal Investigator K. K. S.) and the Society of Family Planning Research Fund grant #SFPRF14-12.
Transparency declarations
C. A. C. is receiving research funding from Gilead Sciences and Merck though the Magee-Womens Research Institute and has served as a consultant for Gilead Sciences. B. A. C. was on a Merck contraceptive advisory board and has received research funding through Medicines360 and Merck, all managed by the Magee-Womens Research Institute. M. S. reports grants from ViiV and Janssen, outside the submitted work. A. O. has received research funding from Merck, AstraZeneca, Pfizer, ViiV and Janssen, has consulted for Merck and ViiV, and is also a coinventor of patents relating to nanotechnology-based drug delivery systems. M. L. has received research funding from ViiV and Janssen. All other authors: none to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Society of Family Planning Research Fund.
References
- 1.The Global Fund. Results Report 2017 2017; 50 https://data.unicef.org/topic/hivaids/global-regional-trends/.
- 2.WHO. WHO Statement on Progestogen-Only Implants.2015. https://www.who.int/reproductivehealth/publications/family_planning/statement-progestogen-implants/en/.
- 3. Vieira CS, Bahamondes MV, de Souza RM. et al. Effect of antiretroviral therapy including lopinavir/ritonavir or efavirenz on etonogestrel-releasing implant pharmacokinetics in HIV-positive women. J Acquir Immune Defic Syndr 2014; 66: 378–85. [DOI] [PubMed] [Google Scholar]
- 4. Leticee N, Viard J-P, Yamgnane A. et al. Contraceptive failure of etonogestrel implant in patients treated with antiretrovirals including efavirenz. Contraception 2012; 85: 425–7. [DOI] [PubMed] [Google Scholar]
- 5. McCarty E, Keane H, Quinn K. et al. Implanon® failure in an HIV-positive woman on antiretroviral therapy resulting in two ectopic pregnancies. Int J STD AIDS 2011; 22: 413–4. [DOI] [PubMed] [Google Scholar]
- 6. Chappell CA, Lamorde M, Nakalema S. et al. Efavirenz decreases etonogestrel exposure: a pharmacokinetic evaluation of implantable contraception with antiretroviral therapy. AIDS 2017; 31: 1965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neary M, Lamorde M, Olagunju A. et al. The effect of gene variants on levonorgestrel pharmacokinetics when combined with antiretroviral therapy containing efavirenz or nevirapine. Clin Pharmacol Ther 2017; 102: 529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maddox DD, Rahman Z.. Etonogestrel (Implanon), another treatment option for contraception. PT 2008; 33: 337. [Google Scholar]
- 9. Moreno I, Quiñones L, Catalán J. et al. Influence of CYP3A4/5 polymorphisms in the pharmacokinetics of levonorgestrel: a pilot study. Biomedica 2012; 32: 570–7. [DOI] [PubMed] [Google Scholar]
- 10. Istrate MA, Nussler AK, Eichelbaum M. et al. Regulation of CYP3A4 by pregnane X receptor: the role of nuclear receptors competing for response element binding. Biochem Biophys Res Commun 2010; 393: 688–93. [DOI] [PubMed] [Google Scholar]
- 11. Swart M, Ren Y, Smith P. et al. ABCB1 4036A>G and 1236C>T polymorphisms affect plasma efavirenz levels in South African HIV/AIDS patients. Front Genet 2012; 3: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haas DW, Ribaudo HJ, Kim RB. et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS 2004; 18: 2391–400. [PubMed] [Google Scholar]
- 13. Rotger M, Colombo S, Furrer H. et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics 2005; 15: 1–5. [DOI] [PubMed] [Google Scholar]
- 14. Wyen C, Hendra H, Vogel M. et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemother 2008; 61: 914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schipani A, Wyen C, Mahungu T. et al. Integration of population pharmacokinetics and pharmacogenetics: an aid to optimal nevirapine dose selection in HIV-infected individuals. J Antimicrob Chemother 2011; 66: 1332–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hui K, Lee S, Lam T.. Dose optimization of efavirenz based on individual CYP2B6 polymorphisms in Chinese patients positive for HIV. CPT Pharmacometrics Syst Pharmacol 2016; 5: 182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nightingale S, Chau TT, Fisher M. et al. Efavirenz and metabolites in cerebrospinal fluid: relationship with CYP2B6 c.516G→T genotype and perturbed blood–brain barrier due to tuberculous meningitis. Antimicrob Agents Chemother 2016; 60: 4511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Solus JF, Arietta BJ, Harris JR. et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics 2004; 5: 895–931. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, Sönnerborg A, Rane A. et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genomics 2006; 16: 191–8. [DOI] [PubMed] [Google Scholar]
- 20. Haas DW, Gebretsadik T, Mayo G. et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis 2009; 199: 872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marzolini C, Rajoli R, Battegay M. et al. Physiologically based pharmacokinetic modeling to predict drug–drug interactions with efavirenz involving simultaneous inducing and inhibitory effects on cytochromes. Clin Pharmacokinet 2017; 56: 409–20. [DOI] [PubMed] [Google Scholar]
- 22. Mouly S, Lown KS, Kornhauser D. et al. Hepatic but not intestinal CYP3A4 displays dose-dependent induction by efavirenz in humans. Clin Pharmacol Ther 2002; 72: 1–9. [DOI] [PubMed] [Google Scholar]
- 23. Hariparsad N, Nallani SC, Sane RS. et al. Induction of CYP3A4 by efavirenz in primary human hepatocytes: comparison with rifampin and phenobarbital. J Clin Pharmacol 2004; 44: 1273–81. [DOI] [PubMed] [Google Scholar]
- 24. Moser C, Zoderer D, Luef G. et al. Simultaneous online SPE-LC-MS/MS quantification of six widely used synthetic progestins in human plasma. Anal Bioanal Chem 2012; 403: 961–72. [DOI] [PubMed] [Google Scholar]
- 25. Almond LM, Hoggard PG, Edirisinghe D. et al. Intracellular and plasma pharmacokinetics of efavirenz in HIV-infected individuals. J Antimicrob Chemother 2005; 56: 738–44. [DOI] [PubMed] [Google Scholar]
- 26. Almond LM, Edirisinghe D, Dalton M. et al. Intracellular and plasma pharmacokinetics of nevirapine in human immunodeficiency virus-infected individuals. Clin Pharmacol Ther 2005; 78: 132–42. [DOI] [PubMed] [Google Scholar]
- 27. Rodriguez S, Gaunt TR, Day IN.. Hardy–Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 2009; 169: 505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Diaz S, Pavez M, Moo-Young A. et al. Clinical trial with 3-keto-desogestrel subdermal implants. Contraception 1991; 44: 393–408. [DOI] [PubMed] [Google Scholar]
- 29. Hofmann MH, Blievernicht JK, Klein K. et al. Aberrant splicing caused by single nucleotide polymorphism c. 516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther 2008; 325: 284–92. [DOI] [PubMed] [Google Scholar]
- 30. Jin T, Yang H, Zhang J. et al. Polymorphisms and phenotypic analysis of cytochrome P450 3A4 in the Uygur population in northwest China. Int J Clin Exp Pathol 2015; 8: 7083.. [PMC free article] [PubMed] [Google Scholar]
- 31. Riska P, Lamson M, MacGregor T. et al. Disposition and biotransformation of the antiretroviral drug nevirapine in humans. Drug Metab Dispos 1999; 27: 895–901. [PubMed] [Google Scholar]
- 32. Neary M, Owen A.. Pharmacogenetic considerations for HIV treatment in different ethnicities: an update. Expert Opin Drug Metab Toxicol 2017; 13: 1169–81. [DOI] [PubMed] [Google Scholar]
- 33. Kimberly K, Scarsi LC, Nakalema S. et al. Double-dose levonorgestrel does not fully overcome interaction with efavirenz. In: Conference on Retroviruses and Opportunistic Infections, Seattle, WA, USA,2019. Oral Abstract 51.
- 34.Clinicaltrials.gov. Pharmacologic Strategies for the Etonogestrel Implant in HIV-Infected Women ( NCT03282799) https://clinicaltrials.gov/ct2/show/NCT03282799.
- 35. Scarsi KK, Darin KM, Nakalema S. et al. Unintended pregnancies observed with combined use of the levonorgestrel contraceptive implant and efavirenz-based antiretroviral therapy: a three-arm pharmacokinetic evaluation over 48 weeks. Clin Infect Dis 2016; 62: 675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts O, Rajoli RK, Back DJ. et al. Physiologically based pharmacokinetic modelling prediction of the effects of dose adjustment in drug–drug interactions between levonorgestrel contraceptive implants and efavirenz-based ART. J Antimicrob Chemother 2018; 73: 1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.