Abstract
Spontaneous post-transcriptional silencing of sense transgenes (S-PTGS) is established in each generation and is accompanied by DNA methylation, but the pathway of PTGS-dependent DNA methylation is unknown and so is its role. Here we show that CHH and CHG methylation coincides spatially and temporally with RDR6-dependent products derived from the central and 3′ regions of the coding sequence, and requires the components of the RNA-directed DNA methylation (RdDM) pathway NRPE1, DRD1 and DRM2, but not CLSY1, NRPD1, RDR2 or DCL3, suggesting that RDR6-dependent products, namely long dsRNAs and/or siRNAs, trigger PTGS-dependent DNA methylation. Nevertheless, none of these RdDM components are required to establish S-PTGS or produce a systemic silencing signal. Moreover, preventing de novo DNA methylation in non-silenced transgenic tissues grafted onto homologous silenced tissues does not inhibit the triggering of PTGS. Overall, these data indicate that gene body DNA methylation is a consequence, not a cause, of PTGS, and rule out the hypothesis that a PTGS-associated DNA methylation signal is transmitted independent of a PTGS signal.
INTRODUCTION
RNA-mediated gene silencing is a widely conserved mechanism in eukaryotes, and different classes of small RNAs with specialized roles in gene silencing have been identified. In plants, the majority of small RNAs derived from endogenous loci consist of short interfering RNAs (siRNAs) that are 24 nucleotides (nt) in length. These 24-nt siRNAs produced by DICER-LIKE 3 (DCL3) processing of double-stranded RNA (dsRNA) are loaded onto ARGONAUTE 3 (AGO3), AGO4, AGO6 or AGO9 to guide transcriptional gene silencing (TGS) and RNA-directed DNA methylation (RdDM) of homologous DNA, mostly at transposon and repetitive loci (1–3). The second largest class of plant endogenous small RNAs consists of microRNAs (miRNAs), which are produced by DCL1 and loaded onto AGO1, AGO2, AGO7 or AGO10 to guide the cleavage and/or translational repression of complementary mRNAs (4). The third class of endogenous small RNAs consists of 21-nt trans-acting siRNAs (ta-siRNAs), which are produced by DCL4 and loaded onto AGO1 to guide the cleavage of complementary mRNAs (4).
dsRNA derived from viruses and transgenes is also processed into 21-, 22- and 24-nt siRNAs by DCL4, DCL2 and DCL3, respectively. Like their endogenous counterparts, 24-nt exogenous siRNAs trigger RdDM and TGS of homologous promoter sequences, whereas 21- and 22-nt siRNAs trigger post-transcriptional gene silencing (PTGS) of homologous transcribed sequences (5). It is generally accepted that spontaneous, sense transgene-induced PTGS (S-PTGS) results from over-abundant transcription of aberrant mRNAs that lack a 5′ cap (6,7) or poly A tail (8). Likely, these aberrant RNAs are protected from degradation by SGS3 (9), and serve as substrates for endogenous RNA-dependent RNA polymerase 6 (RDR6). The resulting dsRNA is diced into primary siRNAs by DCL4 or DCL2, which are loaded onto AGO1 to guide the cleavage of homologous mRNAs. During transgene S-PTGS, the production of siRNAs is generally initiated at one end of the mRNA and progresses toward the other end (10), a phenomenon referred to as transitivity (11,12). Transitivity is also observed when targeting the 5′ or 3′ end of transgenes with siRNAs produced from a hairpin (IR-PTGS) or a virus (VIGS), and it occurs in both 5′-to-3′ and 3′-to-5′ directions (11,12). Remarkably, transitivity occurs on silenced transgenes, but generally not when endogenous genes are targeted by primary siRNAs produced from a hairpin or a virus (12). Therefore, it is unlikely that mRNA fragments produced by primary siRNA-guided cleavage of transcripts are substrates for RDR6 alone. More likely, transitivity results from recruitment of RDR6 to transgene RNAs in a complex with complementary primary siRNAs, in particular 22-nt siRNAs produced by DCL2 (10–11,13). In support of this hypothesis, ago1 mutants that can bind to small RNAs but not carry out small RNA-guided cleavage of target RNA, can nevertheless initiate transitive biogenesis of secondary siRNA using uncleaved target RNA as a template (14).
The formation of an aberrant RNA from a transgene locus does not necessarily result in S-PTGS, as these abnormal RNAs can be intercepted and degraded by RNA quality control (RQC) pathways. Indeed, S-PTGS and RQC compete for transgene-derived aberrant RNAs. Accordingly, genetic defects in RQC, including nonsense-mediated decay, deadenylation, decapping or XRN- or exosome-mediated degradation, allows the accumulation of aberrant RNAs and results in increased rates of spontaneous S-PTGS in Arabidopsis (6–7,15–19). Importantly, mutations in decapping components DPC1, DPC2 and VCS (VARICOSE), or dual impairment of XRN and exosome components, provokes the production of siRNAs from thousands of endogenous mRNA, ultimately causing the death of the plant, and this effect is suppressed by impairing DCL2, RDR6 or SGS3 (16,20). These reports clearly indicate that RQC is essential for eliminating endogenous aberrant RNAs that could otherwise induce PTGS.
Even before the discovery of siRNAs, it was demonstrated that a silencing signal was able to move and act throughout the plant during transgene S-PTGS (21,22). Using Nicotiana species, it was shown that a transgene-specific silencing signal could not only travel across a graft junction, but also through a segment of intervening non-transgenic stem to mediate graft-induced PTGS (G-PTGS) (21,22). Surprisingly, and adding further to the complexity of gene silencing pathways in plants, components of TGS have also been implicated in G-PTGS (23). RNA polymerase (Pol) IV and V are essential components of the RdDM/TGS pathway in plants. NRPD1 encodes the largest sub-unit of Pol IV, which in a complex with RDR2, is involved in the biogenesis of dsRNA and 24-nt siRNAs (24). NRPE1 is the largest sub-unit of Pol V, and nascent Pol V transcripts interact with complementary 24-nt siRNAs to recruit DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to methylate the associated DNA (25,26). The role of promoter DNA methylation in blocking transcription during TGS is well established (27). In contrast, the role of DNA methylation of the transcribed portion of the gene (i.e. the gene body), which is observed in transgene S-PTGS (28), remains elusive. Recently, it was shown that DNA methylation is also triggered at TAS loci producing 21–22-nt ta-siRNAs (29). Furthermore, the initial silencing of active transposons involves 21–22-nt siRNAs produced by RDR6, a component of the S-PTGS pathway, before DNA methylation and TGS become established and maintained by the canonical RdDM pathway (30–33). These results indicate that the establishment of TGS involves a cross-talk between S-PTGS and TGS via DNA methylation. However, whether DNA methylation induced by S-PTGS plays an active role in preventing, establishing, propagating or maintaining S-PTGS remains to be determined.
In the present work, we have deciphered the contribution of components of the RdDM pathway to PTGS-dependent DNA methylation, and whether DNA methylation is required or not for spontaneous and systemic PTGS in Arabidopsis. We used two transgenic lines carrying a 35S:GUS (β-glucuronidase) transgene: L1, which spontaneously triggers GUS S-PTGS, and 6b4, which constitutively expresses the GUS protein in a wild-type background but triggers S-PTGS when RQC is impaired, suggesting that, like L1, it produces aberrant RNAs but at a lower level (7,10,17,19). Previously, we showed that when 6b4 scions are grafted onto L1 rootstocks they undergo a form of PTGS hereafter referred to as G-PTGS (13). To elucidate how the silencing signal is transmitted or perceived/received, we grafted various mutants in both the 6b4 (expressing scions) and L1 (silenced rootstocks) genetic backgrounds. Further to the findings that RDR6 is required in the rootstock for systemic transmission of PTGS (13), we show that RDR6 is also required for establishing G-PTGS in scions. We also show that preventing de novo DNA methylation by impairing DRM2 in either the silenced rootstocks or expressing scions has no impact on S-PTGS or G-PTGS, indicating that DNA methylation is a consequence and not a cause of PTGS.
MATERIALS AND METHODS
Plant material
35S:GUS lines 6b4 and L1 and mutants rdr6(sgs2–1), clsy1–6, nrpd1a-5, rdr2–5, dcl3–3, nrpe1(drd3–7), drd1–6, cmt3–7, drm2–3 and vcs-8 have been described previously (16,28,34–40). To generate transgene/mutant combination lines, 6b4 and L1 were crossed to the corresponding mutants, and F2 progenies were genotyped to identified plants homozygous for both the transgene and the mutation(s). To avoid potential effects of unlinked EMS mutations, two independent transgene/mutant combination lines were identified for each genotype (L1 rdr6, L1 nrpd1, L1 rdr2, L1 dcl3, L1 nrpe1, L1 drd1, L1 drm2, L1 drm2 cmt3, 6b4 rdr6, 6b4 drm2), and all experiments were done using the two independent batches of seeds, providing the two biological replicates analyzed in every figure and in Table 1.
Table 1.
Efficiency of transmission of S-PTGS from L1 rootstocks to 6b4 scions
| Number of silenced scions/total plants | |||
|---|---|---|---|
| Grafting combination | Exp 1 | Exp 2 | % of S-PTGS transmission |
| 6b4 // L1 | 35/35 | 24/24 | 100 |
| 6b4 // L1rdr6 | 0/13 | 0/8 | 0 |
| 6b4 // L1clsy1 | 6/6 | 5/5 | 100 |
| 6b4 // L1nrpd1 | 5/5 | 6/6 | 100 |
| 6b4 // L1rdr2 | 9/9 | 8/8 | 100 |
| 6b4 // L1dcl3 | 15/15 | 7/7 | 100 |
| 6b4 // L1nrpe1 | 12/12 | 16/16 | 100 |
| 6b4 // L1drd1 | 8/8 | 8/8 | 100 |
| 6b4 // L1drm2 | 7/7 | 6/6 | 100 |
| 6b4 // L1drm2 cmt3 | 6/6 | 16/16 | 100 |
| 6b4 rdr6 // L1 | 0/8 | 0/8 | 0 |
| 6b4 drm2 // L1 | 6/6 | 22/22 | 100 |
For each grafting combination, two biological replicates were analyzed. Shoots were considered silenced when exhibiting GUS activity < 20 fluorescent units per minute per microgram of total proteins, eight weeks after grafting. The efficiency of S-PTGS transmission is expressed as the percentage of silenced plants when summing the results of two replicates.
Growth conditions
Arabidopsis seeds were sown in vitro on a nutritive medium (1.3% S-medium Duchefa, 1% Phytoblend agar) and vernalized at 4°C for at least 2 days before being transferred to soil in culture chambers. Plants were grown at 23°C, 70% humidity, 120 μE.m−2 lighting and 16 h light/8 h dark (long-days) or 8 h light/16 h dark (short-days) photoperiod.
Grafting techniques
Grafting was performed as described in (41). Briefly, Arabidopsis seedlings are sown in vitro, and grown under long-day conditions for 6 days. Grafting is performed in 9 cm Petri dishes on Bouturage medium (Duchefa) added with a layer of nitrocellulose filter (Hybond) on the top. Seedlings were cut transverse with a fresh razor blade across the hypocotyl (90° butt graft). The scions and rootstocks of interest were then placed on the nitrocellulose filter. Under a binocular, scions and rootstocks hypocotyls were introduced into a silicon microtube (2 mm long) so that both attach tight to each other. Tube was used as collar to maintain the hypocotyls junction. The Petri dishes were incubated under short-day conditions (8 h light, 16 h dark) for 7–14 days. The grafts integrity was checked and grafted seedlings without adventitious roots were then transferred to soil and grown under short-day conditions.
GUS activity and GUS RNA analysis
GUS protein was extracted and GUS activity was quantified as described before (7) from plant leaves by monitoring the quantity of 4-methylumbelliferone products generated from the substrate 4-methylumbelliferyl-b-D-glucuronide (Duchefa) on a fluorometer (Thermo Scientific fluoroskan ascent).
RNA extraction and HMW or LMW RNA gel blot analyses were performed as described before (7). All RNA gel blot analyses were performed using 5–10 μg of total RNA. GUS, U6 and 25S probes have been described before (7).
DNA methylation analysis
DNA was extracted using the Nucleospin Plant II kit from Macheray-Nagel (REF740770.50). A total of 150 ng of DNA was digested with HaeIII, PagI, MspI or ScrFI DNA methylation-sensitive enzymes from ThermoFisher Scientific in 50 μl overnight at 37°C. Undigested DNA was used as control. DNA was diluted three times before quantitative polymerase chain reaction (qPCR). For qPCR DNA methylation assays, the fold change between digested and undigested DNA for the tested region is normalized to the fold change between digested and undigested DNA for a region that is not recognized by the enzymes. Results are expressed as (2∧-(Cq non digested DNA – Cq digested DNA) [sequence of the tested region] / 2∧-(Cq non digested DNA – Cq digested DNA) [sequence not cleaved by the enzymes]). All primers are listed in Supplementary Table S1.
For whole-genome bisulfite sequencing (WGBS), bisulfite treatment, library preparation and whole-genome sequencing (final depth of 20×) were performed by the BGI (China) using the HiSeq technology (Illumina) producing 100 bp paired-end reads. Reads were trimmed with Trim_Galore (Babraham Bioinformatics) and aligned to the Col-0 Arabidopsis thaliana TAIR10 reference genome and the T-DNA sequence with Bismark version 0.20.0 (Babraham Bioinformatics) using standard options (Bowtie2; 1 mismatch allowed). Identical pairs were collapsed.
RESULTS
The pattern of PTGS-induced DNA methylation coincides with RDR6 products
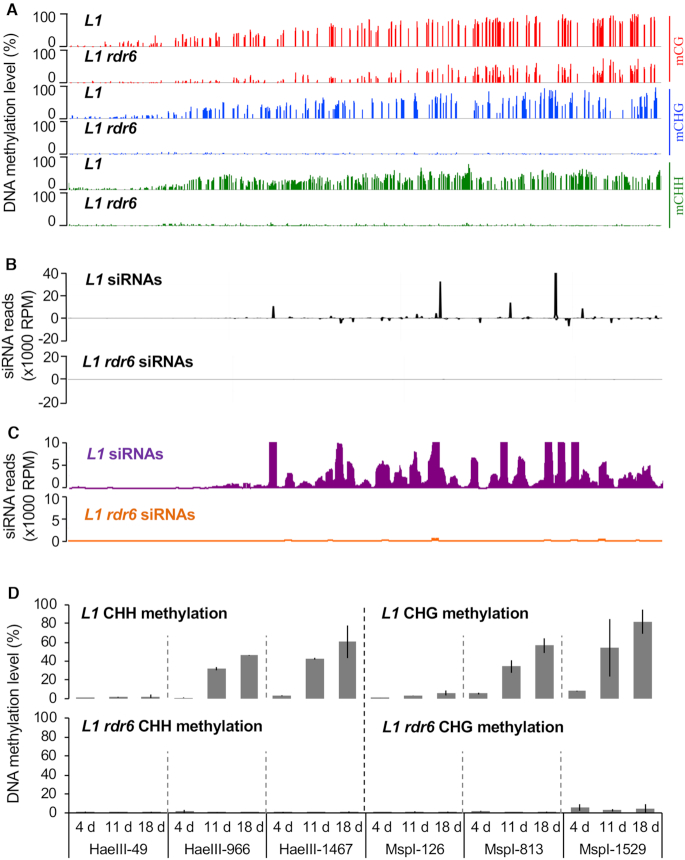
Using methylation-sensitive enzymes and Southern blot analyses, we previously reported that in adult L1 plants, S-PTGS correlates with CHH, CHG and CG methylation of the GUS coding sequence (28). However, the large probes used for these experiments did not allow determining the exact pattern of GUS DNA methylation. Moreover, sRNA-seq analyses revealed that RDR6-dependent siRNAs do not originate from the entire GUS sequence. Indeed, siRNAs were found in the central and 3′ part of GUS, but not in the 250 bases at its 5′ end (10,13). To determine if the pattern of GUS DNA methylation coincides with the pattern of GUS RDR6-dependent siRNA production, a whole-genome bisulfite sequencing (WGBS) was performed on silenced L1 and non-silenced L1 rdr6 adult plants. At first, the quality of our analysis was verified by looking at the DNA methylation profile of endogenous TAS genes, where DNA methylation occurs in regions producing RDR6-dependent siRNAs (29). Consistent with previous observations, CHH and CHG methylation was observed in TAS regions producing siRNAs in L1 wild-type plants, and was lost in L1 rdr6 mutants (Supplementary Figure S1). When looking at the GUS coding sequence, methylation was detected at CHH, CHG and CG sites in L1 plants with the exception of sites located in the first 250 bases at the 5′ end (Figure 1A), from which siRNAs are not produced (Figure 1B, C). L1 rdr6 plants, which lack GUS siRNAs, did not show CHH or CHG methylation in any part of the GUS coding sequence (Figure 1D). Nevertheless, CG methylation was observed in L1 rdr6 plants, although at a level lower than that in L1, confirming previous results obtained by southern blot analyses (28). Because CG methylation in coding sequences is found in both silenced transposons and actively transcribed genes, the level of CG methylation observed in the GUS coding sequence of L1 rdr6 plants reflects either the high level of transcription of the L1 locus or the fact that the GUS coding sequence has been targeted for DNA methylation in the past (i.e. in L1 plants prior to their exposure to EMS mutagenesis, which yielded L1 rdr6 mutants), and that CG methylation has been passively inherited. In any case, the results presented above strongly suggest that RDR6 products, either dsRNAs or siRNAs, guide DNA methylation in a homologous manner at CHH and CHG sites in the GUS coding sequence.
Figure 1.
PTGS-induced DNA methylation is established de novo in each generation. (A) Average DNA methylation levels at CG, CHG and CHH sites of the GUS coding sequence in leaves of 8-week-old L1 and L1 rdr6plants grown under short day conditions. The methylation levels correspond to the ratios of methylated cytosines over the total number of cytosines based on WGBS. The screenshots were obtained with the Integrative Genome Browser (IGB). (B) Distribution of GUS siRNAs in L1 and L1 rdr6 plants. The graphic represents the normalized aligned reads per million, and is based on previously published data (10). (C) Close-up of data in (B), except only showing the distribution of siRNAs that were less abundant than 10000 RPM. (D) Time course of DNA methylation in L1 and L1 rdr6plants using representative CHH and CHG methylation-sensitive enzymes in the 5′, central and 3′ regions of the GUS coding sequence. Plants were sown in vitroand the aerial part was harvested 4, 11 or 18 DAG. Analyses were performed at the following sites: HaeIII-49, MspI-126, MspI-813, HaeIII-966, HaeIII-1467, MspI-1529 (see Supplementary Figure S2). The percentage of DNA methylation at each site is based on the difference in qPCR amplification between digested and mock template. Mean and standard deviation bars are based on two biological replicates.
PTGS-induced CHH and CHG DNA methylation is established de novo in each generation
To further address the relationship between CHH/CHG methylation and S-PTGS in L1 plants, we analyzed CHH and CHG methylation during the onset of S-PTGS at each generation and in various mutant backgrounds. To avoid using WGBS on a large scale, we assessed if a quantitative assay based on methylation-sensitive enzymes reflects the data obtained by WGBS. For this purpose, the level of DNA methylation at HaeIII and PagI sites (CHH) and MspI and ScrFI (CHG) sites distributed all over the GUS sequence (Supplementary Figure S2A) was monitored in silenced L1 and non-silenced L1 rdr6 adult plants. Methylation was not detected in L1 rdr6, but was detected at all tested CHH and CHG sites in L1 at a level similar to that observed in the methylome analysis (Supplementary Figure S2B), indicating that this simple method is a robust and reliable tool to monitor S-PTGS-induced DNA methylation.
We previously reported that GUS activity and GUS mRNA levels rapidly decrease in L1 during the first three weeks following germination, while GUS siRNA levels inversely increase (10,28). Whether DNA methylation is inherited through meiosis and precedes the establishment of S-PTGS during the development of the next generation, or is re-established in each generation concurrently with the establishment of S-PTGS has not been examined so far. To address this question, DNA methylation at representative HaeIII sites (CHH) and MspI sites (CHG) sites of the 5′, central and 3′ regions of the GUS coding sequence was monitored at 4, 11 and 18 days after germination (DAG) in silenced L1 and non-silenced L1 rdr6 plants of the fourth and fifth generations. CHH and CHG methylation were absent in L1 plants at 4 DAG, but progressively appeared in the 3′ and central regions of the GUS coding sequence with the onset of S-PTGS at about 10 DAG (Figure 1D). In contrast, CHH and CHG methylation remained absent in L1 rdr6 during this time course. These results therefore indicate that CHH methylation and CHG methylation are not inherited through meiosis in L1 plants but are re-established in each generation concurrently with the onset of S-PTGS.
S-PTGS-induced DNA methylation is locus-independent
To investigate whether the pattern of DNA methylation established during spontaneous L1 S-PTGS is locus independent or specific to the L1 locus, we assayed DNA methylation at another 35S:GUS locus called 6b4. In a wild-type background, the 6b4 does not trigger S-PTGS. Monitoring the level of DNA methylation at HaeIII and PagI sites (CHH) and MspI and ScrFI (CHG) sites distributed all over the GUS sequence (Supplementary Figure S2A) in 6b4 plants showed that, like non-silenced L1 rdr6 plants, 6b4 plants show no CHH methylation and very low levels of CHG methylation (Supplementary Figure S2B), confirming the correlation between S-PTGS and gene body CHH and CHG methylation.
The 6b4 locus triggers S-PTGS in RQC-deficient mutant backgrounds, for example, in the RNA decapping-defective vcs mutant (16). As shown by the analysis of GUS activity, GUS mRNA and GUS siRNA levels, 6b4 vcs plants trigger S-PTGS during the early development, similar to L1 plants (Figure 2A and B), indicating that the 6b4 locus actually produces aberrant RNAs, but at a rate insufficient to bypass RQC-mediated degradation and trigger S-PTGS in a wild-type background. DNA methylation analysis revealed that the CHH and CHG methylation profile of the GUS coding sequence in silenced 6b4 vcs plants is similar to that of spontaneously silenced L1 plants, i.e. at the 3′ but not 5′ end of the GUS coding sequence (Figure 2C), indicating that the pattern of S-PTGS-induced DNA methylation is locus-independent.
Figure 2.
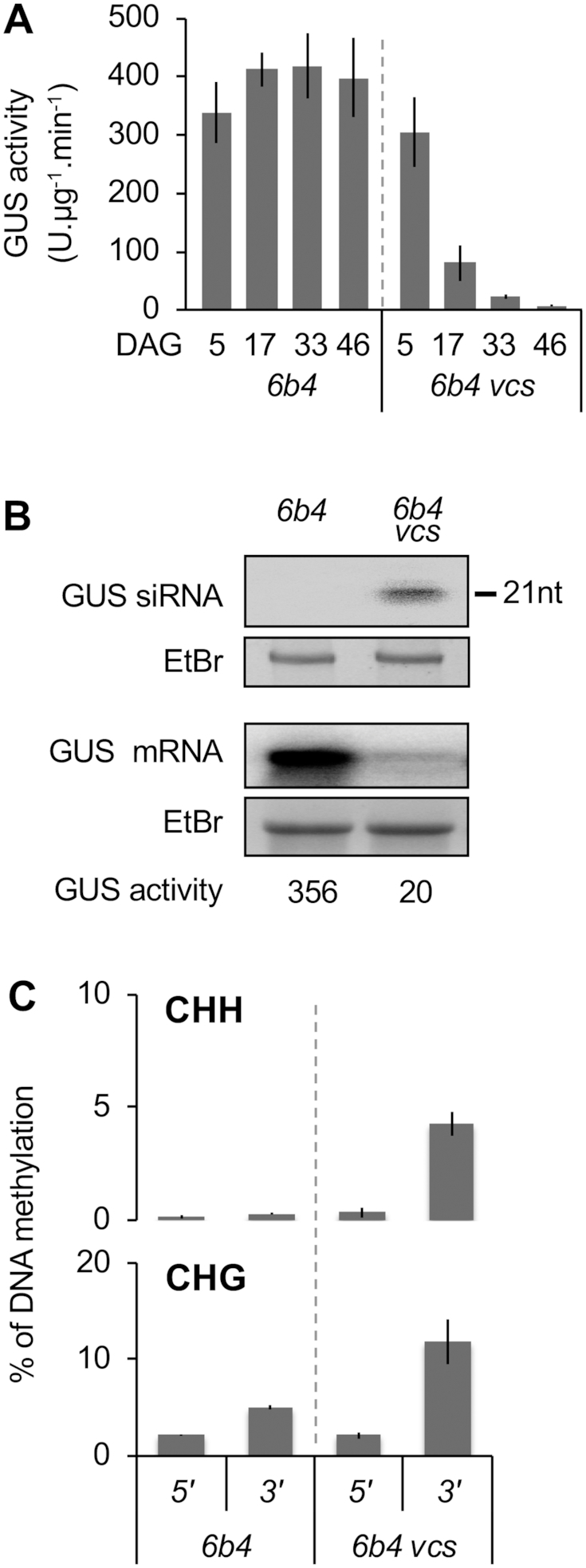
The pattern of PTGS-induced DNA methylation is locus-independent. (A) Time course of S-PTGS in plants of the indicated genotypes grown under long day conditions. GUS activity was measured in leaves of 16 plants, and is expressed in fluorescent units per minute per microgram of total proteins. Error bars: standard deviation. (B) GUS siRNA and mRNA accumulation in 17-day-old plants of the indicated genotypes. The ethidium bromide signal is shown as loading control. (C) Percentage of DNA methylation at representative CHH and CHG methylation-sensitive enzymes of the 5′ and 3′ regions of the GUS coding sequence in leaves of 17-day-old plants of the indicated genotypes. Analyses were performed at HaeIII-49, MspI-126, HaeIII-1467 and MspI-1529 (see Supplementary Figure S2). The percentage of DNA methylation was calculated as described in Figure 1D. Mean and standard deviation bars are based on two biological replicates.
PTGS-induced DNA methylation requires RDR6, NRPE1, DRD1, DRM2 and CMT3
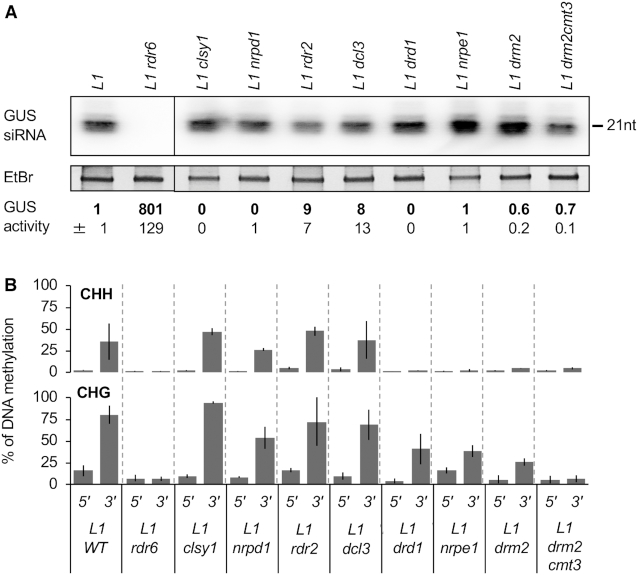
Next, we investigated the effect of RdDM mutations on S-PTGS triggering and PTGS-induced DNA methylation in L1 plants. Unlike L1 rdr6 plants, which exhibited high GUS activity and lacked GUS siRNAs, L1 clsy1, L1 nrpd1, L1 rdr2, L1 dcl3, L1 nrpe1, L1 drd1 and L1 drm2 plants lacked GUS activity and accumulated GUS siRNAs, similar to L1 control plants (Figure 3A), indicating that RdDM mutations do not compromise the establishment or maintenance of S-PTGS in L1. These results are consistent with the fact that none of these mutations were recovered in the extensive genetic screen for S-PTGS-deficient mutants in the L1 genetic background (28,38–39,42–45).
Figure 3.
Specific components of the RdDM pathway are required for PTGS-induced DNA methylation but not PTGS establishment. (A) GUS siRNA accumulation and GUS activity in leaves of 8-week-old plants of the indicated genotypes grown under short day conditions. RNA was extracted from a bulk of four plants for small RNA gel blots, and ethidium bromide staining is shown as loading control. Distinct boxes indicate that the samples were separated by tracks that are not relevant for this work. The original blot is presented in Supplementary Figure S3. GUS activity is expressed as fluorescent units per minute per microgram of total proteins quantified by Bradford. Averages and standard deviations correspond to 16 plants. (B) Percentage of DNA methylation at representative CHH and CHG methylation-sensitive enzymes of the 5′ and 3′ region of the GUS coding sequence in leaves of plants of the indicated genotypes grown under short day conditions. Analyses were performed at HaeIII-49, MspI-126, HaeIII-1467 and MspI-1529 (see Supplementary Figure S2). The percentage of DNA methylation was calculated as described in Figure 1D. Mean and standard deviation bars are based on two biological replicates.
Although none of the RdDM mutations compromised spontaneous S-PTGS in L1, analysis of CHH and CHG methylation within the GUS coding sequence of L1 revealed that the nrpe1, drd1 and drm2 mutations abolish CHH methylation, and for most of these mutations, also decrease the level of CHG methylation (Figure 3B). By contrast, CHH and CHG methylation was not greatly affected in clsy1, nrpd1, rdr2 or dcl3 mutants (Figure 3B). Thus, PTGS-induced DNA methylation at CHH and CHG sites requires NRPE1, DRD1 and DRM2, but not CLSY1, NRPD1, RDR2 and DCL3. Considering that: i) 97.5% of the 21-to-24-nt GUS siRNA population in L1 consists in 21- and 22-nt long molecules (10,13), and ii) CHH and CHG methylation is abolished in L1 rdr6 (Figure 1A), the PTGS-induced DNA methylation observed at the L1 GUS locus appears reminiscent of DNA methylation induced at endogenous TAS loci and at actively transcribed transposon loci producing RDR6-dependent 21- and 22-nt siRNAs (30–33). Most likely, the CLSY1-NRPD1-RDR2-DCL3 steps of the RdDM pathway are dispensable because PTGS-related DNA methylation relies on RDR6 products.
Of note, CHH methylation is equally reduced in rdr6, nrpe1, drd1 and drm2 mutants, whereas CHG methylation is less reduced in nrpe1, drd1 and drm2 compared with rdr6. This suggests that the RDR6-NRPE1-DRD1-DRM2 pathway contributes to the establishment and maintenance of CHH methylation, and only to the establishment of the CHG methylation. CMT3 is most likely responsible for the maintenance of CHG methylation induced by DRM2 during S-PTGS. Supporting this hypothesis, analysis of a L1 drm2 cmt3 double mutant revealed a reduction in CHG methylation similar to that observed in L1 rdr6 (Figure 3B), but still without abolishing L1 S-PTGS (Figure 3A).
Impairing CHH and CHG methylation does not affect the production of a systemic PTGS signal
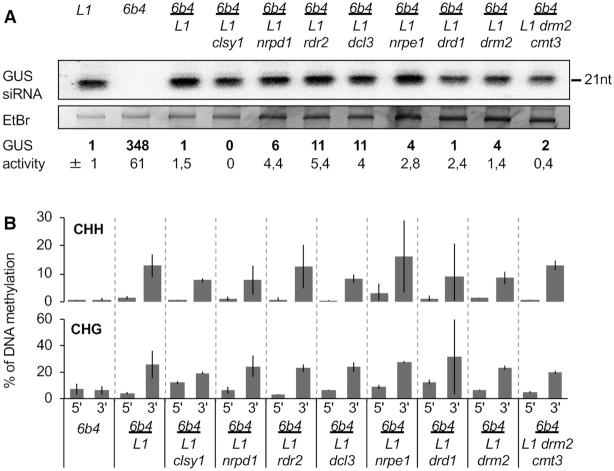
We previously reported that PTGS is transmitted from p35S:GUS tobacco rootstocks undergoing S-PTGS to non-silenced p35S:GUS scions (21). The same phenomenon is observed in Arabidopsis when grafting non-silenced scions of line 6b4 onto silenced rootstocks of line L1 (13). Indeed, seven weeks after grafting, all 6b4 scions grafted onto L1 rootstocks show low GUS activity and high GUS siRNA accumulation (Figure 4), indicating that they had become silenced by a type of PTGS hereafter referred to as G-PTGS. Self-grafted L1 (L1//L1) controls remained silenced, indicating that grafting per se does not compromise L1 S-PTGS. In addition, self-grafted 6b4 (6b4//6b4) controls remained non-silenced, indicating that grafting per se does not trigger 6b4 G-PTGS.
Figure 4.
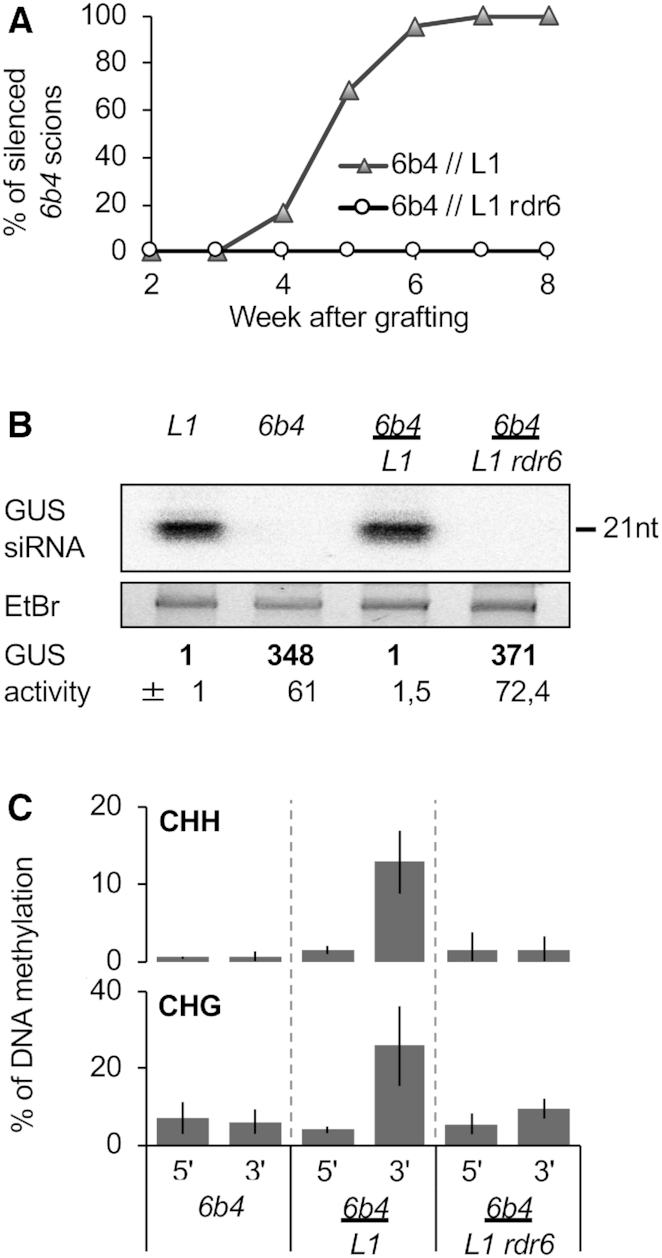
PTGS transmission from L1 rootstocks to 6b4 scions requires siRNA amplification in L1 and triggers PTGS-induced DNA methylation in 6b4. (A) Kinetics of S-PTGS establishment in leaves of 6b4 shoots grafted onto L1 and L1 rdr6 roots and grown under short day conditions. GUS activity was measured in scion leaves every week. The graph represents the average of two independent experiments involving at least eight plants each (see Table 1). S-PTGS transmission efficiency is expressed as the percentage of silenced scions, i.e. scions exhibiting GUS activity <20 fluorescent units per minute per microgram of total proteins, whereas control 6b4 plants exhibits GUS activity ∼350 fluorescent units per minute per microgram of total proteins. (B) GUS activity and GUS siRNA accumulation in scion leaves of plants of the indicated genotypes grown under short day conditions for eight weeks after grafting. GUS activity is expressed as fluorescent units per minute per microgram of total proteins quantified by Bradford. Averages and standard deviations correspond to the number of plants indicated in Table 1. RNA was extracted from a bulk of four plants. Ethidium bromide staining is shown as loading control. (C) Percentage of DNA methylation at representative CHH and CHG sites in the 5′ and 3′ regions of the GUS coding sequence in scion leaves of plants of the indicated genotypes grown under short day conditions for eight weeks after grafting. Analyses were performed at HaeIII-49, MspI-126, HaeIII-1467 and MspI-1529 restriction sites (see Supplementary Figure S2). The percentage of DNA methylation was calculated as described in Figure 1D. Mean and standard deviation bars are based on two biological replicates.
To determine if, like during L1 and 6b4 vcs S-PTGS, G-PTGS induction in 6b4 scions grafted onto L1 rootstocks induces CHH and CHG methylation of the 3′ but not 5′ of GUS coding sequence, we assessed CHH and CHG methylation at 5′ and 3′ regions of the GUS coding sequence in 6b4 controls and 6b4 scions grafted onto L1 rootstocks. Results indicate that G-PTGS induction in 6b4 scions grafted onto L1 rootstocks induces a CHH and CHG methylation pattern similar to that of spontaneously silenced L1 plants (Figure 4C versus Figure 3B), confirming that PTGS-induced DNA methylation occurs at the 3′ region, but not the 5′ region of GUS coding sequence, independent of the locus considered (L1 and 6b4) and independent of the way PTGS is triggered (S-PTGS and G-PTGS).
Because spontaneous triggering of S-PTGS and associated DNA methylation is impaired in L1 rdr6 plants (Figure 3), we tested if the production or amplification of the silencing signal was also impaired. None of the 6b4 scions grafted onto L1 rdr6 roots became silenced (Table 1 and Figure 4A), indicating that rdr6 prevents the production of the systemic silencing signal. CHH and CHG methylation were not observed in 6b4 scions grafted onto L1 rdr6 rootstocks (Figure 4C), indicating that grafting per se does not induce DNA methylation in the scions. Rather, DNA methylation in 6b4 scions grafted onto L1 rootstocks likely results directly from the triggering of G-PTGS in grafted 6b4 scions.
In contrast to grafts involving L1 rdr6 rootstocks, G-PTGS was triggered in all 6b4 scions grafted onto L1 clsy1, L1 nrpd1, L1 rdr2, L1 dcl3, L1 nrpe1, L1 drd1 and L1 drm2 rootstocks (Table 1 and Figure 5A), indicating that these mutations do not prevent the production of the mobile silencing signal in the root. Moreover, 6b4 scions grafted onto L1 drm2 cmt3 rootstocks also triggered G-PTGS (Figure 5A), establishing that PTGS-induced DNA methylation is not necessary for the production of the systemic silencing signal. Notably, CHH and CHG methylation were observed in 6b4 scions grafted onto L1 clsy1, L1 nrpd1, L1 rdr2, L1 dcl3, L1 nrpe1, L1 drd1, L1 drm2 and L1 drm2 cmt3 roots (Figure 5B), ruling out that the induction of CHH and CHG methylation in 6b4 scions grafted onto L1 rootstocks is due to the transmission of a systemic DNA methylation signal from the rootstock. Rather, DNA methylation is most likely induced in 6b4 scions grafted onto L1 rootstocks as a consequence of the triggering of G-PTGS in response to the signal transmitted from L1 rootstocks.
Figure 5.
PTGS transmission from L1 roots to 6b4 scions and PTGS-induced DNA methylation in 6b4 do not require any RdDM component in L1. (A) GUS siRNA accumulation and GUS activity in scion leaves of plants of the indicated genotypes grown under short day conditions for eight weeks after grafting. RNA was extracted from a bulk of four plants for small RNA gel blots, and ethidium bromide staining is shown as loading control. GUS activity is expressed as fluorescent units per minute per microgram of total proteins quantified by Bradford. Averages and standard deviations correspond to the number of plants indicated in Table 1. (B) Percentage of DNA methylation at representative CHH and CHG sites in the 5′ and 3′ regions of the GUS coding sequence in scion leaves of plants of the indicated genotypes grown under short day conditions for 8 weeks after grafting. Analyses were performed at HaeIII-49, MspI-126, HaeIII-1467 and MspI-1529 restriction sites (see Supplementary Figure S2). The percentage of DNA methylation was calculated as described in Figure 1D. Mean and standard deviation bars are based on two biological replicates.
Impairing DNA methylation in recipient tissues does not affect the triggering of PTGS upon reception of a systemic PTGS signal
To test further the above hypothesis, G-PTGS triggering and PTGS-induced methylation were examined in 6b4 rdr6 scions grafted onto L1 rootstocks. Indeed, 6b4 rdr6 scions cannot produce RDR6-dependent dsRNAs or siRNAs and only receive those produced by the L1 rootstocks. None of the 6b4 rdr6 scions grafted onto L1 rootstocks became silenced (Table 1). Moreover, no GUS siRNAs were detected by northern analysis in 6b4 rdr6 scions grafted onto L1 rootstocks (Figure 6A). This result indicates that if GUS dsRNAs or siRNAs are transmitted from silenced L1 rootstocks to 6b4 rdr6 scions, they are not sufficiently abundant to be detected by northern blots, and clearly not present in sufficient amounts to degrade a significant amount of the GUS mRNA in 6b4 rdr6 scions. Therefore, it is likely that, upon reception of the silencing signal, G-PTGS needs to be initiated via RDR6 in scions to ensure the production of siRNAs in amounts sufficient for degrading all of the GUS mRNA and the triggering GUS DNA methylation.
Figure 6.
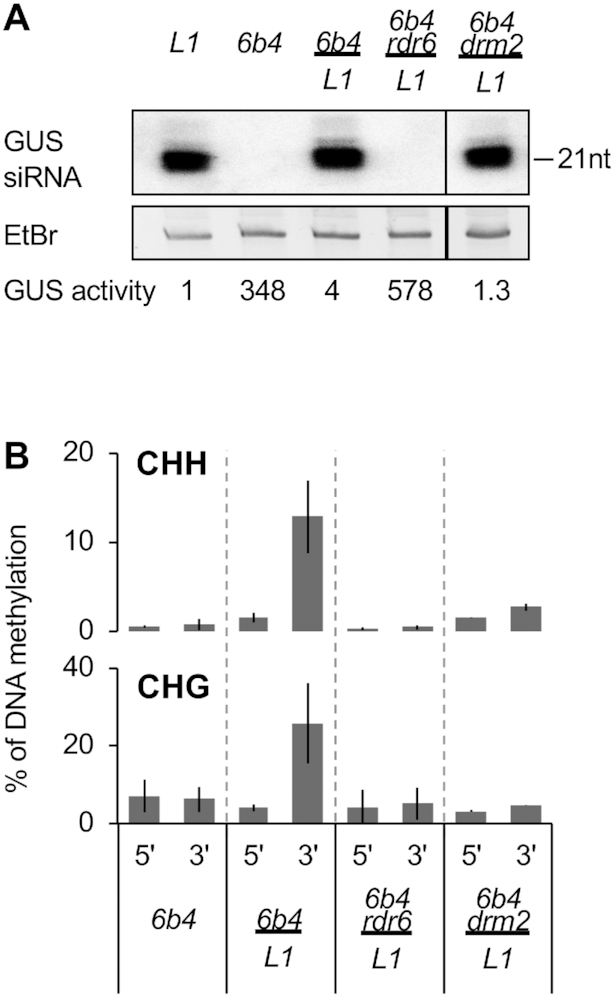
PTGS transmission from L1 rootstocks to 6b4 scions requires RDR6 but not DRM2 in 6b4. (A) GUS activity and GUS siRNA accumulation in scion leaves of plants of the indicated genotypes grown under short day conditions for eight weeks after grafting. GUS activity is expressed as fluorescent units per minute per microgram of total proteins quantified by Bradford. RNA was extracted from a bulk of four plants. Ethidium bromide staining is shown as loading control. Distinct boxes indicate that the samples were separated by tracks that are not relevant for this work. The original blot is presented in Supplementary Figure S4. (B) Percentage of DNA methylation at representative CHH and CHG methylation-sensitive enzymes in the 5′ and 3′ regions of the GUS coding sequence in scion leaves of plants of the indicated genotypes grown under short day conditions for eight weeks after grafting. Analyses were performed at HaeIII-49, MspI-126, HaeIII-1467 and MspI-1529 (see Supplementary Figure S2). The percentage of DNA methylation was calculated as described in Figure 1D. Mean and standard deviation bars are based on two biological replicates.
To determine if, upon reception of the systemic signal produced by L1 rootstocks, G-PTGS initiation in 6b4 scions requires first the induction of GUS DNA methylation by GUS dsRNAs or siRNAs transmitted from the L1 rootstocks, 6b4 drm2 scions were grafted onto L1 rootstocks. If G-PTGS initiation required the establishment of GUS DNA methylation, one would expect 6b4 drm2 to not trigger G-PTGS when grafted onto L1 rootstocks. However, 6b4 drm2 scions grafted onto L1 rootstocks triggered G-PTGS as efficiently as 6b4 scions grafted onto L1 rootstocks (Table 1 and Figure 6A) and produced similarly high levels of GUS siRNAs (Figure 6A), indicating that DNA methylation is not required for the G-PTGS initiation in grafted scions. Lastly, unlike in 6b4 scions grafted onto L1 rootstocks, G-PTGS triggering in 6b4 drm2 scions grafted onto L1 rootstocks was not accompanied by CHH and CHG methylation of the GUS coding sequence (Figure 6B), an effect stronger than that observed in L1 drm2 (Figure 3B), confirming that DRM2-mediated de novo DNA methylation is a dispensable consequence of the establishment of PTGS.
DISCUSSION
DNA methylation is instrumental for TGS in plants (27). It is maintained by the RdDM pathway, which uses CLSY1/2/3/4-NRPD1-RDR2-DCL3-dependent 24-nt siRNAs to target homologous DNA via the action of AGO4/6/9, NRPE1, DRD1 and DRM2. DNA methylation is also observed during PTGS, but the way it is triggered and its role have long remained mysterious. Here we show that during PTGS of a sense transgene, CHH and CHG methylation is established de novo in the central and 3′ portions of the transcribed region in each generation, following the same profile of siRNA production along the transcript (Figure 1). CLSY1, NRPD1, RDR2 and DCL3 are dispensable, whereas NRPE1, DRD1 and DRM2 are required for PTGS-induced DNA methylation (Figure 3). Because methylation in the transcribed portion of the transgene also requires RDR6 (Figure 3), these results suggest that PTGS-induced DNA methylation depends on RDR6 products, long dsRNAs and/or 21–22-nt siRNAs produced by DCL2 and DCL4, and not on 24-nt siRNAs, thus explaining why CLSY1, NRPD1, RDR2 and DCL3 are dispensable. This result is consistent with the fact that very low levels of 24-nt siRNAs are produced during S-PTGS (10,13). Therefore, it is likely that during S-PTGS, RDR6 products play two roles: i) RDR6-derived long dsRNAs are processed into 21- and 22-nt siRNAs by DCL4 and DCL2, which are loaded onto AGO1 to guide mRNA cleavage (44), and ii) RDR6 products, long dsRNAs and/or 21–22-nt siRNAs, guide the methylation of homologous DNA in an NRPE1-, DRD1- and DRM2-dependent manner. Such a situation resembles that of endogenous TAS genes, which produce 21–22-nt ta-siRNAs that are loaded onto AGO1 to regulate their targets by PTGS (46) and which undergo DNA methylation in a RDR6-AGO4-AGO6-NRPE1-dependent but NRPD1-RDR2-DCL3-independent manner (29). Very few 21–22-nt ta-siRNAs were found loaded onto AGO4 and AGO6, compared to AGO1. Nevertheless, it has been proposed that AGO4 and AGO6 guide DNA methylation of TAS loci, mostly because DNA methylation is reduced in ago4 and ago6 mutants (29). Adding to the complexity of the system, de novo DNA methylation of the endogenous FWA promoter triggered by VIGS, which involves predominantly 21- and 22-nt siRNAs, was shown to be NRPE1-DRM2-dependent but NRPD1-RDR2-DCL3-AGO4-independent (47), but this may be due to the redundancy between AGO4 and AGO6. Lastly, WGBS analysis revealed that the quadruple dcl1 dcl2 dcl3 dcl4 mutant still exhibit de novo DNA methylation in Arabidopsis (48), suggesting that dsRNAs that are longer than siRNAs can trigger DNA methylation (49). Therefore, whether RDR6-derived long dsRNAs guide the methylation of homologous DNA independently of AGO4/6 proteins or after processing into 21–22-nt siRNAs that are loaded onto AGO4/6 remains an open question.
The PTGS-induced DNA methylation pathway also resembles the mechanism involved in the re-silencing of reactivated transposons (30–33). However, in the case of transposons, PTGS-dependent DNA methylation of the transposon coding sequences or body appears as a first step toward the establishment of TGS through de novo methylation of the transposon promoter sequences, which is subsequently maintained by the canonical RdDM pathway and its associated 24-nt siRNAs. This contrasts transgene PTGS-induced DNA methylation, which does not transform into TGS, at least at the L1 locus. Remarkably, during the establishment of L1 S-PTGS, siRNA production starts from the 3′ end of the GUS transgene, followed by methylation of the corresponding DNA sequences, and progresses in the 5′ direction. However, siRNA and DNA methylation corresponding to the 5′ end of the GUS coding sequence were not observed (Figure 1). This suggests either a mechanism limiting transitivity or the fact that only aberrant RNAs are used to produce siRNAs via RDR6. Indeed, if the 5′ end of the GUS coding sequence is not part of the aberrant RNAs produced by the transgene locus, it is normal to not find siRNAs corresponding to this region. In any case, the absence of RDR6 products at the 5′ end of the GUS coding sequence may prevent the spreading of DNA methylation into the promoter and the subsequent transformation of PTGS-induced DNA methylation into TGS at the L1 locus. Although this hypothesis remains to be proven, the fact that TAS genes, which also produce siRNAs from the 3′ and central, but not the 5′ parts of their transcribed regions, do not undergo TGS and do not exhibit DNA methylation in their promoter sequences makes it a reasonable hypothesis.
The grafting experiments performed in this study also address whether a systemic signal for DNA methylation migrates independently of a systemic PTGS signal. Indeed, it was previously shown that both 21-, 22- and 24-nt siRNAs move through graft-unions (50), but only mobile 24-nt siRNAs are assumed to trigger DNA methylation at distance (51). The GUS siRNAs produced by the L1 locus consist in 97.5% of 21–22-nt siRNAs and only 2,5% of 24-nt siRNAs (10,13). Because DNA methylation is unchanged in L1 dcl3 compared to L1 (Figure 3), it is likely that RDR6 products, long dsRNAs and/or 21–22-nt siRNAs produced by DCL2 and DCL4 trigger GUS DNA methylation, and that 24-nt siRNAs play a very minor role in this process. However, it remained possible that these 24-nt siRNAs migrate at long distance and participate in the systemic triggering of DNA methylation. To test this hypothesis, 6b4 scions were grafted onto L1 or L1 dcl3 rootstocks. GUS DNA methylation was similarly triggered in both cases (Figure 5), suggesting that DNA methylation is triggered in 6b4 scions grafted onto L1 rootstocks as a consequence of the triggering of G-PTGS in 6b4 scions and not as the consequence of the movement of a DCL3-dependent DNA methylation signal produced by L1 rootstocks. Supporting this hypothesis, GUS DNA methylation was not observed in 6b4 rdr6 scions grafted onto L1 rootstocks (Figure 6), likely because the 24-nt siRNAs transmitted from the L1 rootstock are insufficient to trigger GUS DNA methylation in the scion, just like the 21–22-nt siRNAs transmitted from the L1 rootstock are insufficient to trigger the degradation of GUS mRNA in the scion. Moreover, the fact that the drm2 mutation prevents PTGS-induced DNA methylation but not G-PTGS initiation in 6b4 drm2 scions grafted onto L1 rootstocks (Table 1 and Figure 6), confirms that DNA methylation is not required for PTGS to occur.
To conclude, our results strongly suggest that in line L1, RDR6 dsRNA and/or siRNA products are sufficient to trigger GUS S-PTGS and GUS DNA methylation using the RdDM components NRPE1, DRD1 and DRM2 (Figure 7). GUS RDR6 products also are able to migrate through graft unions to elicit G-PTGS and GUS DNA methylation in grafted 6b4 scions. Nevertheless, DNA methylation appears dispensable for PTGS to occur either spontaneously or upon grafting, indicating that gene body DNA methylation is a consequence and not a cause of PTGS.
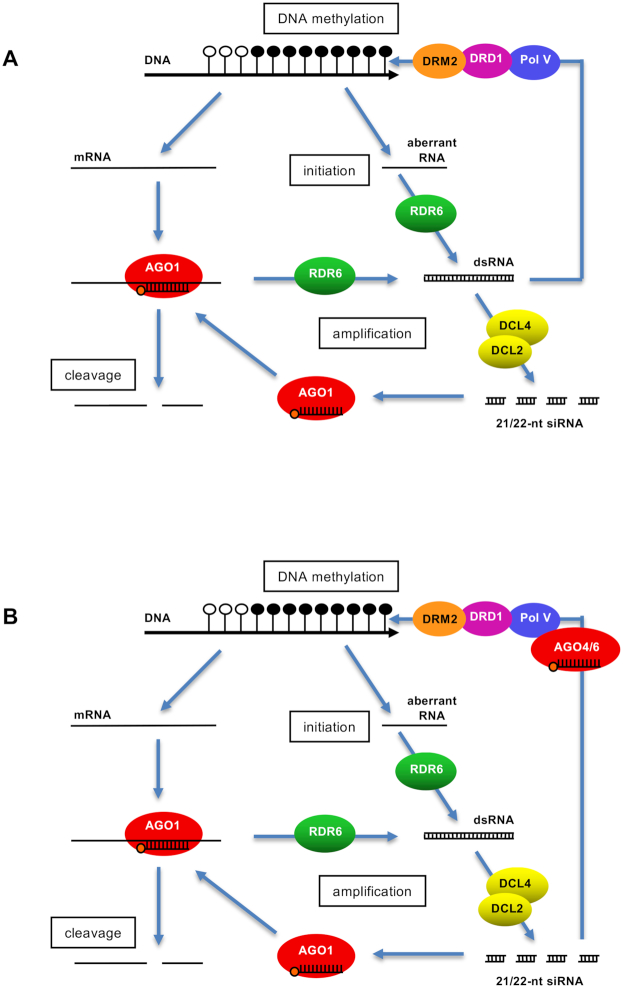
Figure 7.
Alternative models for PTGS-induced DNA methylation. S-PTGS is induced owing to the production of transgene aberrant RNAs that escape complete degradation by RQC pathways and are transformed into dsRNAs by RDR6, processed into primary 21–22-nt siRNAs by DCL2/DCL4, methylated by HEN1 and loaded onto AGO1 to guide the cleavage of regular mRNA. The binding of 22-nt siRNAs produced by DCL2 to mRNA may favor the recruitment of RDR6, leading to the production of secondary siRNAs and to amplifying the degradation process. To explain the pattern of DNA methylation in the transgene body, two hypotheses can be evoked: (A) RDR6-derived long dsRNAs directly trigger transgene DNA methylation through Pol V (NRPE1), DRD1 and DRM2, or (B) Part of the PTGS-derived siRNAs are loaded onto AGO4 and/or AGO6 and trigger transgene DNA methylation through Pol V (NRPE1), DRD1 and DRM2.
DATA AVAILABILITY
BS-seq data are available from the ENA database under the accession number PRJEB31038.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marjori Matzke, David Baulcombe and Steve Jacobsen for providing mutant seeds, and Maud Rivard for assistance in genotyping.
Authors’ contributions: C.T., B.J.C. and H.V. designed the research. C.T., A.Y., N.B., T.E. and U.W. performed the research. C.T., A.Y., N.B., T.E. and H.V. analyzed the data. C.T., B.J.C. and H.V. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Agence Nationale pour la Recherche [ANR-10-LABX-40, ANR-11-BSV6-007 to H.V.]; Fondation Louis D. de l’Institut de France [to H.V.]; Australian Research Council [DP120103966, DP150104048 to B.J.C.]; INRA Post-doctoral Fellowship (to C.T.). Funding for open access charge: ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Havecker E.R., Wallbridge L.M., Hardcastle T.J., Bush M.S., Kelly K.A., Dunn R.M., Schwach F., Doonan J.H., Baulcombe D.C.. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010; 22:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C.. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004; 2:642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Z., Liu X., Guo X., Wang X.J., Zhang X.. Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat Plants. 2016; 2:16049. [DOI] [PubMed] [Google Scholar]
- 4. Bologna N.G., Voinnet O.. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014; 65:473–503. [DOI] [PubMed] [Google Scholar]
- 5. Martinez de Alba A.E., Elvira-Matelot E., Vaucheret H.. Gene silencing in plants: a diversity of pathways. Biochim. Biophys. Acta. 2013; 1829:1300–1308. [DOI] [PubMed] [Google Scholar]
- 6. Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R.. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004; 306:1046–1048. [DOI] [PubMed] [Google Scholar]
- 7. Gy I., Gasciolli V., Lauressergues D., Morel J.B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C.. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007; 19:3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo Z., Chen Z.. Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell. 2007; 19:943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukunaga R., Doudna J.A.. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009; 28:545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parent J.S., Jauvion V., Bouche N., Beclin C., Hachet M., Zytnicki M., Vaucheret H.. Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res. 2015; 43:8464–8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mlotshwa S., Pruss G.J., Peragine A., Endres M.W., Li J., Chen X., Poethig R.S., Bowman L.H., Vance V.. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One. 2008; 3:e1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaistij F.E., Jones L., Baulcombe D.C.. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002; 14:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taochy C., Gursanscky N.R., Cao J., Fletcher S.J., Dressel U., Mitter N., Tucker M.R., Koltunow A.M.G., Bowman J.L., Vaucheret H. et al.. A genetic screen for impaired systemic RNAi highlights the crucial role of DICER-LIKE 2. Plant Physiol. 2017; 175:1424–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arribas-Hernandez L., Marchais A., Poulsen C., Haase B., Hauptmann J., Benes V., Meister G., Brodersen P.. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell. 2016; 28:1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R.. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008; 14:854–866. [DOI] [PubMed] [Google Scholar]
- 16. Martinez de Alba A.E., Moreno A.B., Gabriel M., Mallory A.C., Christ A., Bounon R., Balzergue S., Aubourg S., Gautheret D., Crespi M.D. et al.. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015; 43:2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moreno A.B., Martinez de Alba A.E., Bardou F., Crespi M.D., Vaucheret H., Maizel A., Mallory A.C.. Cytoplasmic and nuclear quality control and turnover of single-stranded RNA modulate post-transcriptional gene silencing in plants. Nucleic Acids Res. 2013; 41:4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thran M., Link K., Sonnewald U.. The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 2012; 72:368–377. [DOI] [PubMed] [Google Scholar]
- 19. Yu A., Saudemont B., Bouteiller N., Elvira-Matelot E., Lepere G., Parent J.S., Morel J.B., Cao J., Elmayan T., Vaucheret H.. Second-Site Mutagenesis of a Hypomorphic argonaute1 Allele Identifies SUPERKILLER3 as an Endogenous Suppressor of Transgene Posttranscriptional Gene Silencing. Plant Physiol. 2015; 169:1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X., Zhu Y., Liu X., Hong X., Xu Y., Zhu P., Shen Y., Wu H., Ji Y., Wen X. et al.. Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015; 348:120–123. [DOI] [PubMed] [Google Scholar]
- 21. Palauqui J.C., Elmayan T., Pollien J.M., Vaucheret H.. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997; 16:4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Voinnet O., Vain P., Angell S., Baulcombe D.C.. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998; 95:177–187. [DOI] [PubMed] [Google Scholar]
- 23. Brosnan C.A., Mitter N., Christie M., Smith N.A., Waterhouse P.M., Carroll B.J.. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:14741–14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haag J.R., Ream T.S., Marasco M., Nicora C.D., Norbeck A.D., Pasa-Tolic L., Pikaard C.S.. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell. 2012; 48:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson L.M., Du J., Hale C.J., Bischof S., Feng S., Chodavarapu R.K., Zhong X., Marson G., Pellegrini M., Segal D.J. et al.. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014; 507:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S.. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009; 41:630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Law J.A., Jacobsen S.E.. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010; 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elmayan T., Balzergue S., Beon F., Bourdon V., Daubremet J., Guenet Y., Mourrain P., Palauqui J.C., Vernhettes S., Vialle T. et al.. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998; 10:1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu L., Mao L., Qi Y.. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 2012; 160:990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mari-Ordonez A., Marchais A., Etcheverry M., Martin A., Colot V., Voinnet O.. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 2013; 45:1029–1039. [DOI] [PubMed] [Google Scholar]
- 31. McCue A.D., Nuthikattu S., Slotkin R.K.. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 2013; 10:1379–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCue A.D., Panda K., Nuthikattu S., Choudury S.G., Thomas E.N., Slotkin R.K.. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. EMBO J. 2015; 34:20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K.. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant Physiol. 2013; 162:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beclin C., Boutet S., Waterhouse P., Vaucheret H.. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 2002; 12:684–688. [DOI] [PubMed] [Google Scholar]
- 35. Dunoyer P., Himber C., Voinnet O.. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005; 37:1356–1360. [DOI] [PubMed] [Google Scholar]
- 36. Greenberg M.V., Ausin I., Chan S.W., Cokus S.J., Cuperus J.T., Feng S., Law J.A., Chu C., Pellegrini M., Carrington J.C. et al.. Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics. 2011; 6:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J.. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005; 37:761–765. [DOI] [PubMed] [Google Scholar]
- 38. Le Masson I., Jauvion V., Bouteiller N., Rivard M., Elmayan T., Vaucheret H.. Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell. 2012; 24:3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parent J.S., Bouteiller N., Elmayan T., Vaucheret H.. Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 2015; 81:223–232. [DOI] [PubMed] [Google Scholar]
- 40. Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C.. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007; 19:1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Turnbull C.G., Booker J.P., Leyser H.M.. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 2002; 32:255–262. [DOI] [PubMed] [Google Scholar]
- 42. Boutet S., Vazquez F., Liu J., Beclin C., Fagard M., Gratias A., Morel J.B., Crete P., Chen X., Vaucheret H.. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003; 13:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jauvion V., Elmayan T., Vaucheret H.. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell. 2010; 22:2697–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morel J.B., Godon C., Mourrain P., Beclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H.. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002; 14:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mourrain P., Beclin C., Elmayan T., Feuerbach F., Godon C., Morel J.B., Jouette D., Lacombe A.M., Nikic S., Picault N. et al.. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000; 101:533–542. [DOI] [PubMed] [Google Scholar]
- 46. Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P.. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004; 16:69–79. [DOI] [PubMed] [Google Scholar]
- 47. Bond D.M., Baulcombe D.C.. Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:917–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang D.L., Zhang G., Tang K., Li J., Yang L., Huang H., Zhang H., Zhu J.K.. Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res. 2016; 26:66–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dalakouras A., Dadami E., Wassenegger M., Krczal G., Wassenegger M.. RNA-directed DNA methylation efficiency depends on trigger and target sequence identity. Plant J. 2016; 87:202–214. [DOI] [PubMed] [Google Scholar]
- 50. Molnar A., Melnyk C.W., Bassett A., Hardcastle T.J., Dunn R., Baulcombe D.C.. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010; 328:872–875. [DOI] [PubMed] [Google Scholar]
- 51. Lewsey M.G., Hardcastle T.J., Melnyk C.W., Molnar A., Valli A., Urich M.A., Nery J.R., Baulcombe D.C., Ecker J.R.. Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E801–E810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BS-seq data are available from the ENA database under the accession number PRJEB31038.