Short abstract
Objectives
To investigate the pathogenicity of Klebsiella pneumoniae (KPN) possessing rmpA or the aerobactin gene in infected mice.
Methods
BALB/c mice were divided into four groups (n = 10 per group) and infected with: string test-positive and rmpA-positive or aerobactin-positive KPN (group 1), string test-negative but rmpA-positive KPN (group 2), string test-negative but aerobactin-positive KPN (group 3), or string test- and rmpA/aerobactin-negative KPN (group 4). Mouse survival time was compared among groups, and the infection of livers, spleens, lungs, and kidneys and KPN growth were assessed in infected mice.
Results
Compared with the negative group (group 4), the survival rates of mice infected with rmpA- or aerobactin-positive KPN (groups 1–3) were significantly lower, their multi-organ injuries were significantly more severe, and bacterial enumeration was significantly higher.
Conclusions
Despite being string test-negative, aerobactin- or rmpA-positive KPN still exhibit high virulence and anti-immune effect activity. Therefore, the combination of the string test and gene detection of aerobactin and rmpA will be helpful in screening hypervirulent KPN.
Keywords: Hypervirulent, Klebsiella pneumoniae, rmpA, aerobactin, mice, pathogenicity
Introduction
Klebsiella pneumoniae (KPN) is the main pathogen of community-acquired and nosocomial infections, and leads to pneumonia, genitourinary infection, and sepsis.1,2 KPN is classified into two types: classical K. pneumoniae (cKP) primarily occurs in hospitals and long-term care facilities,3 while hypervirulent K. pneumoniae (hvKP) causes serious life threatening disease and organ failure in young, healthy individuals from the community.4–6 The strong virulence factors of hvKP are induced by iron carrier-related genes, including aerobactin, enterobacterin, samochelin, and yersiniabactin; the presence of aerobactin appears to be a defining hvKP trait.7 Virulence factor rmpA (regular mucoid phenotype) mediates capsule production and hypermucoviscosity, which is a critical virulence factor.8 Compared with cKP, a defining characteristic of hvKP is its capacity for metastatic spread from infection sites, with resulting devastating sequelae in the immune competent host. Although the string test is widely considered to be reliable for distinguishing hvKP, its sensitivity is not optimal. Indeed, some string test-positive KPN samples were previously shown to be cKP, while string test-negative K. pneumoniae strains were hvKP.8,9 However, the identification of hvKP still depends on string testing because no alternative rapid and reliable detection method exists. Therefore, this study aimed to assess whether rmpA and aerobactin contribute to the virulence of hvKP in infected mice and if they could be used to help screen hvKP in the clinical laboratory.
Methods
Bacterial strains
Sixty-nine consecutive K. pneumoniae culture-positive samples were isolated from the blood, urine, sputum, ascites, bile, and abscess fluid of patients with liver abscesses hospitalized at Linyi People’s Hospital from July 2015 to July 2016, identified as K. pneumoniae using an automated bacterial identification system (VITEK®2; bioMerieux, Hazelwood, MI, USA), and stored at –80°C. Clinical and laboratory data including age, sex, temperature, clinical features, and treatment received were collected from all patients and analyzed.
Ethics and consent
All experiments were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from the patients. The study design was reviewed and approved by the Ethics Committees of Shandong Medical College.
String test
A bacteriology inoculation loop was used to ‘stretch’ a colony away from the agar plate on which it was grown at 37°C overnight. The formation of a mucoid string >5 mm was defined as string test-positive.10
PCR detection of aerobactin, rmpA, and capsular serotype-specific genes
Genomic DNA was extracted from all K. pneumoniae strains using the thermal cracking method.1 Aerobactin, rmpA, and serotype-specific genes for K1, K2, K5, K20, K16, K57, and K54 capsular serotypes were amplified by PCR (ABI Veriti® Thermal Cycler; Applied Biosystems Asia Pte Ltd., Singapore) using primers that have been previously described.11,12 Amplification primers to detect capsular-type genes and virulence genes1,11,12 were synthesized by Shanghai Shenggong,1 Appendix.
The pathogenicity of KPN in infected mice
BALB/C female mice (18–22 g) were intraperitoneally injected with various titers of the bacterial strains. Four strains (forming four groups, n = 10 per group) were evaluated: string test-positive KPN with rmpA or aerobactin (ST+ group), string test-negative but rmpA-positive (rmpA+ group), string test-negative but aerobactin-positive (aerobactin+ group), and string test-negative plus rmpA/aerobactin negative (cKP group). Animals were followed for 7 days, with an in extremis state or death being used as the study end point. The LD50 of KPN was calculated using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) according to the number of mice that died. Average data of different titers of a given strain were presented as log titers. Hematoxylin and eosin (HE) staining of liver, spleen, lung, and kidney samples of infected mice was performed to evaluate morphological damage. Animal studies were approved by the Animal Care Committee of Shandong Medical College.
KPN colony counting
Mice (n = 5 per group) were intraperitoneally injected with 0.5 mL 2.3 × 108 colony forming units (CFU)/mL KPN. Bacterial counts in blood were measured 4 hours after the challenge. Briefly, blood samples were diluted continuously with physiological saline and applied to LB solid medium. The number of KPN colonies was calculated 4 hours later by the colony counting method.3
Statistical analyses
Data are presented as means ± standard errors. P < 0.05 was considered statistically significant based on the Bliss method.3 Findings of the rmpA- and aerobactin-positive group were compared with those of the negative group and string test-positive group. Paired t tests were used to compare quantitative data. All results were converted into log10 values, and log10-transformed values were used.
Results
Distribution of virulence factors and capsule types of KPN
String test-positive KPN was defined as high-mucous KPN, and 53.6% (37/69) KPN strains were string test-positive. Of these, 72.9% (34/37) were rmpA-positive and 100% (37/37) were aerobactin-positive. Among the other 32 strains, 9.4% (3/32) were rmpA-positive and 6.3% (2/32) were aerobactin-positive (Table 1). Among the 37 string test-positive strains, 54.1% (20/37) were K1 type, 29.7% (11/37) were K2 type, 8.1% (3/37) were K54 type, 5.4% (2/37) were K57 type, and 2.7% (1/37) were an unknown type. Among the 32 strains of non-mucous KPN, 3.1% (1/32) were K1 type and 12.5% (4/32) were K2 type (Table 1).
Table 1.
The 69 strains showing different KPN virulence genes and capsule type distribution.
| KPN type (n) | rmpA, n (%) | aerobactin, n (%) | K1, n (%) | K2, n (%) | K57, n (%) | K54, n (%) |
|---|---|---|---|---|---|---|
| High-mucous (37) | 34/37 (72.9%) | 37/37 (100%) | 20/37 (54.1%) | 11/37 (29.7%) | 2/37 (5.4%) | 3/37 (8.1%) |
| Non-mucous (32) | 3/32 (9.4%) | 2/32 (6.3%) | 1/32 (3.1%) | 4/32 (12.5%) | 0 (0) | 1/32 (3.1%) |
LD50 of KPN
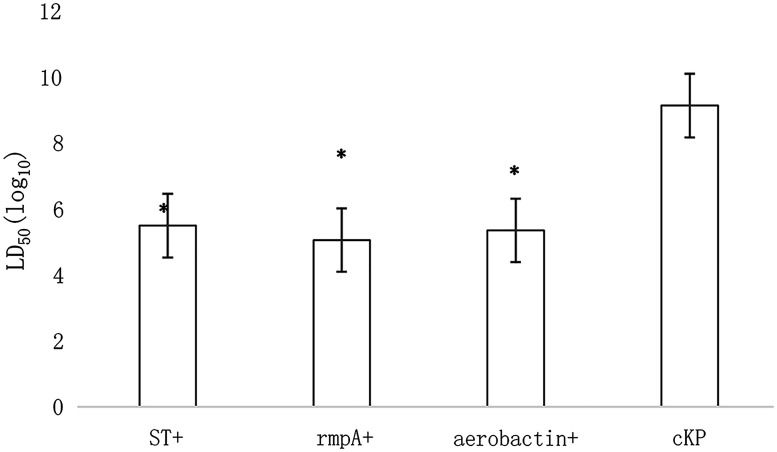
The LD50 of the high-mucous group was 3.12 × 105 CFU/mL, that of the rmpA-positive group was 1.13 × 105 CFU/mL, the aerobactin-positive group was 2.25 × 105 CFU/mL, and the cKP group was 1.36 × 109 CFU/mL. The LD50 between aerobactin- or rmpA-positive and string-positive KPN groups was comparable (Figure 1). The difference between aerobactin- or rmpA-positive KPN and cKP groups was statistically significant (P < 0.05).
Figure 1.
The LD50 of KPN.
KPN, Klebsiella pneumoniae; ST+, string test-positive; rmpA+, rmpA-positive; aerobactin+, aerobactin-positive. *P < 0.05.
The number of KPN in mice
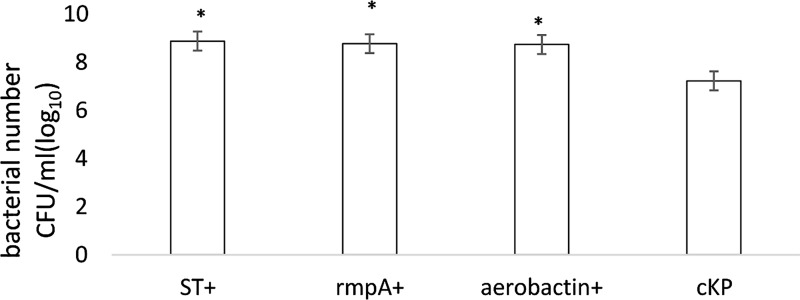
Medial numbers of string-positive, rmpA-positive, and aerobactin-positive KPN were 8.88 CFU/mL (log10), 8.74 CFU/mL (log10), and 8.77 CFU/mL (log10), respectively. These numbers were significantly higher than the cKP group (7.23 CFU/mL, P < 0.05; Figure 2).
Figure 2.
The anti-immune effect of KPN.
KPN, Klebsiella pneumoniae; ST+, string test-positive; rmpA+, rmpA-positive; aerobactin+, aerobactin-positive. *P < 0.05.
HE staining
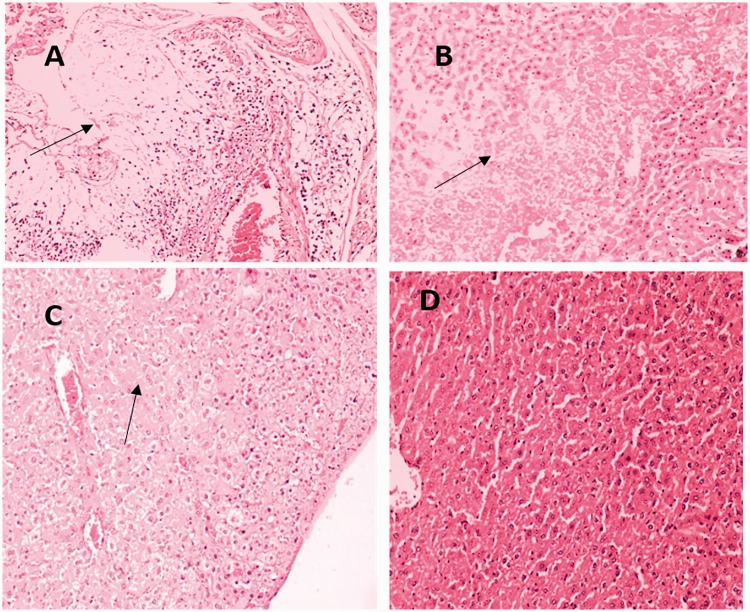
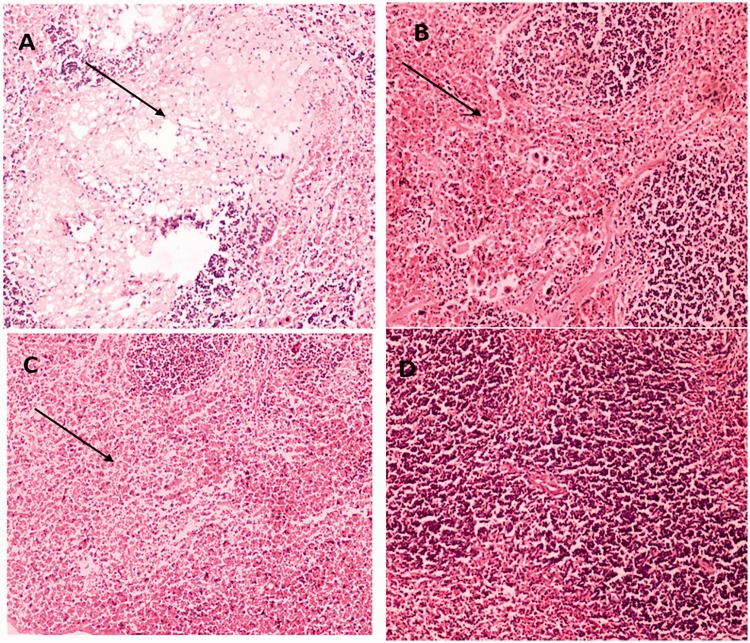
Compared with the cKP-injection group, more severe infiltration of inflammatory cells was observed in the liver, spleen, lung, and kidney of string-positive, rmpA-positive, and aerobactin-positive groups (Figures 3–6). Among the four tissues, inflammatory infiltration was most common and serious in the liver.
Figure 3.
Representative HE staining of liver tissues from mice injected with string test-positive KPN (a), rmpA-positive KPN (b), aerobactin-positive KPN (c), or string test-negative rmpA- and aerobactin-positive KPN (d).
(a) Liver abscess is indicated by arrows. (b) Severe steatosis inflammation and necrosis is indicated by arrows. (c) Inflammation effects are indicated by arrows. (d) No obvious lesions are seen.
HE, hematoxylin and eosin; KPN, Klebsiella pneumoniae.
Figure 4.
Representative HE staining of spleen tissues from mice injected with string test-positive KPN (a), rmpA-positive KPN (b), aerobactin-positive KPN (c), or string test-negative rmpA- and aerobactin-positive KPN (d).
(a) Spleen abscess is indicated by arrows. (b) Severe congestion is seen in the spleen. (c) Severe congestion in the spleen is indicated by arrows. (d) No obvious lesions are seen.
HE, hematoxylin and eosin; KPN, Klebsiella pneumoniae.
Figure 5.
Representative HE staining of lung tissues from mice injected with string test-positive KPN (a), rmpA-positive KPN (b), aerobactin-positive KPN (c), or string test-negative rmpA- and aerobactin-positive KPN (d).
(a) Pulmonary hemorrhage in the lung is indicated by arrows. (b) Congestion in the lung is indicated by arrows. (c) Congestion in the lung is seen. (d) No obvious lesions are seen.
HE, hematoxylin and eosin; KPN, Klebsiella pneumoniae.
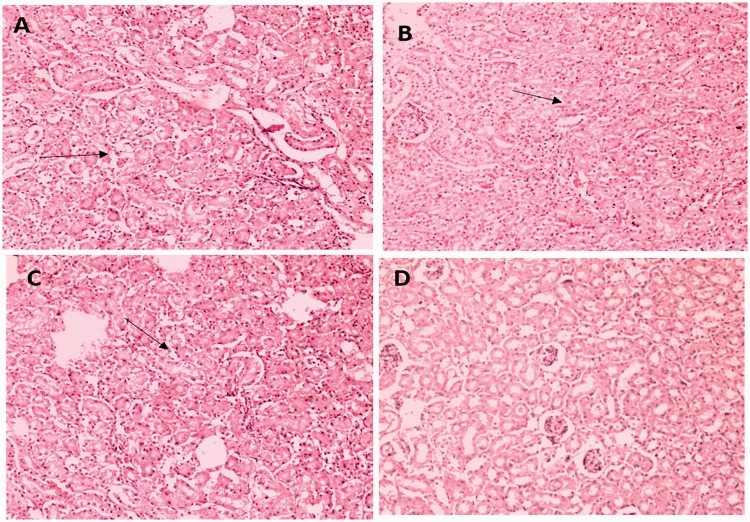
Figure 6.
Representative HE staining of kidney tissues from mice injected with string test-positive KPN (a), rmpA-positive KPN (b), aerobactin-positive KPN (c), or string test-negative rmpA- and aerobactin-positive KPN (d). (a) Kidney tubular degeneration is indicated by arrows. (b) Vacuolar degeneration of kidney is indicated by arrows. (c) Renal tubule epithelial cell swelling is indicated by arrows. (d) No obvious lesions are seen.
HE, hematoxylin and eosin; KPN, Klebsiella pneumoniae.
Discussion
Minimizing the incidence of metastasis and sequelae consequences associated with KPN infection and providing optimal therapy requires the prompt recognition of hvKP. These needs are becoming even more urgent with recent reports about the acquisition of hvKP multidrug resistance.10 Although progress has been made in understanding various virulence factors leading to the clinical syndrome, rapid diagnostic tests have not yet been developed and an increased understanding of the epidemiology of hvKP has not been addressed.
Capsule K1 is the most common type of hvKP,13 and 21 of the 69 KPN strains were K1 type in this study. We also found that 95.2% (20/21) of K1 type strains were string test-positive and rmpA- or aerobactin-positive, while 4.8% (1/21) were string test-negative but aerobactin-positive. The K1 type has been shown to be highly associated with virulence, but the detection of non-K1 hvKP strains cannot be ignored. Therefore, the pathogenicity of string test-negative K1 type carrying virulence genes should be further validated.
rmpA is a plasmid-located virulence factor that regulates the hvKP high-mucus phenotype. Strains carrying rmpA have been significantly associated with the hypermucoviscosity phenotype, and a significant correlation was detected with purulent tissue infections such as abscesses of the liver, lung, neck, psoas muscle, and other foci.14 Among 37 string test-positive KPN strains in the present study, 72.9% were rmpA-positive, while 9.4% of 32 string test-negative KPN strains were also rmpA-positive.
Iron plays an important role in the growth and reproduction of bacteria, and aerobactin functions in bacterial iron acquisition, growth, and/or virulence, so is a critical virulence factor for hvKP. The contribution of aerobactin to virulence is dependent on both its innate biologic activity and the level of protein expression, which is a defining trait of hvKP strains. Previously, aerobactin was shown to be expressed at much higher levels in hvKP than in cKP.14,15 The high-pathogenic strain NTUH-K2044 carries a full-length aerobactin gene that is not found in traditional KPN.7,16 In the present study, 100% of high-mucous strains were found to carry aerobactin, as well as 6.3% of non-mucus strains, while 72.9% of high-mucous KPN carried rmpA compared with 9.4% of non-mucus strains. Aerobactin has been shown to have high sensitivity to a diagnosis of hvKP, while rmpA has high specificity. Our detection of KPN is consistent with these findings.
Although string test-positive KPN is usually considered to be hvKP, a small portion of string test-positive KPN in the present study did not show severe pathogenicity as hvKP, yet some string test-negative KPN showed severe pathogenicity. Thus our results indicate that combining rmpA/aerobactin detection with string testing will be more efficient to select true-positive hvKP than string testing alone.
In the infected host, serum KPN is resistant to the bactericidal effect of the immune system, while hvKP has a stronger anti-immune effect than cKP.17 Compared with cKP strains, the LD50 of KPN carrying rmpA or the aerobactin gene but string test-negative was similar to that of string test-positive KPN, reflecting greater anti-immune effects.
In summary, combining the string test with genetic detection of rmpA and the aerobactin gene appears to be more beneficial in improving the detection rate of hvKP than the string test alone. Further study should evaluate the feasibility and efficiency of this combination method.
Acknowledgments
Zhentao Ma assisted with HE staining and provided pathological information about the tissues.
Appendix
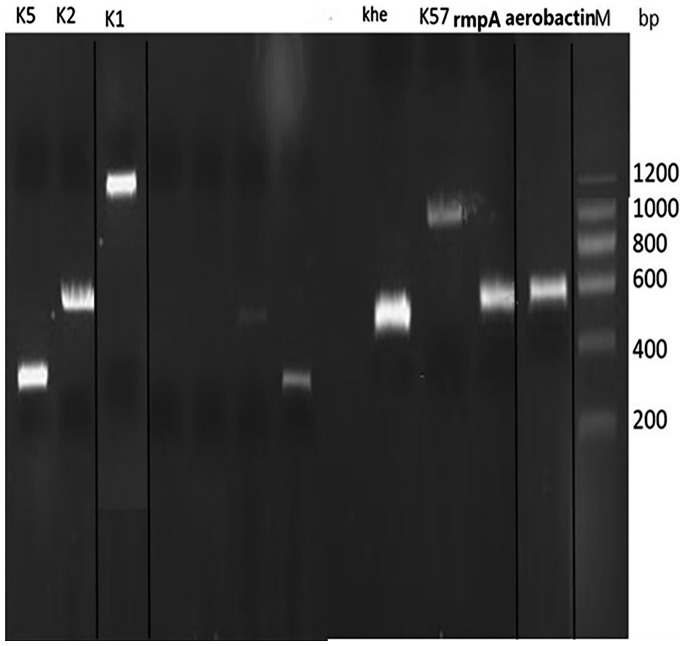
Figure. Multiplex PCR gel image of KPN isolates. KPN, Klebsiella pneumoniae; M, DNA marker; bp, base pair.
Khe: 428 bp; K1: 1283 bp; K2: 641 bp; K5: 280 bp; K57: 1037 bp; rmpA: 536 bp; aerobactin: 556 bp.
Author contribution
Yunfang Sun designed the research, took part in all experiments, and wrote the main manuscript. Guili Li collected the bacteria and prepared figures. Shuhong Sun and Zhongyuan Zhao performed KPN identification experiments.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on a reasonable request basis.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This study was sponsored by a Project of Health and Family Planning Commission of Shandong Province Program (2015WS0201), Shandong, China.
References
- 1.Sun Y, Wu H, Shen D. Clinical and molecular analysis of Klebsiella pneumoniae causing liver abscess in China. J Mol Microbiol Biotechnol 2016; 7: 669–671. [DOI] [PubMed] [Google Scholar]
- 2.Lin YT, Wang YP, Wang FD, et al. Community-onset Klebsiella pneumoniae in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front Microbiol 2015; 9: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 2013; 4: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moellering RC., Jr. NDM-1—a cause for worldwide concern. N Engl J Med 2010; 363: 2377–2379. [DOI] [PubMed] [Google Scholar]
- 5.Snitkin ES, Zelazny AM, Thomas PJ, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Transl Med 2012; 4: 148ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomakova DK, Hsiao CB, Beanan JM, et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumoniae: an emerging and under recognized pathogenic variant. Eur J Clin Microbiol Infect 2012; 31: 981–989. [DOI] [PubMed] [Google Scholar]
- 7.Russo TA, Olson R, MacDonald U. Aerobactin, but not Yersiniabactin, Salmochelin, or Enterobactin, enables the growth/survival of hypervirulent Klebsiella pneumoniae ex vivo and in vivo. Infect Immun 2015; 83: 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadasy KA, Domiati-Saad R, Tribble MA. Invasive Klebsiella pneumoniae in North America. Clin Infect Dis 2007; 45: e25–e28. [DOI] [PubMed] [Google Scholar]
- 9.Patel PK, Russo TA, Karchmer AW. Hypervirulent Klebsiella pneumoniae. Open Forum Infect Dis 2014; 1: ofu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis 2014; 58: 225–232. [DOI] [PubMed] [Google Scholar]
- 11.Compain F, Babosan A, Brisse S, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol 2014; 52: 4377–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turton JF, Perry C, Elgohari S, et al. PCR characterization and typing of K. Pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol 2010; 59: 541–547. [DOI] [PubMed] [Google Scholar]
- 13.Yeh KM, Kurup A, Siu LK, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol 2007; 45: 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng HY, Chen YS, Wu CY, et al. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 2010; 192: 3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung SW, Chae HJ, Park YJ, et al. Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol Infect 2013; 141: 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo TA, Shon AS, Beanan JM, et al. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One 2011; 6: e26734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soto E, Marchi S, Beierschmitt A, et al. Interaction of non-human primate complement and antibodies with hypermucoviscous Klebsiella pneumoniae. Vet Res 2016; 47: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on a reasonable request basis.