Abstract
The combination of the specificity of antibodies and the programmability of DNA nanotechnology has provided the scientific community with a powerful tool to label and unambiguously distinguish a large number of subcellular targets using fluorescence-based read-out methods. Whereas primary antibodies are commercially available for a large class of targets, a general stoichiometric site-selective DNA labeling strategy for this affinity reagent is lacking. Here we present a universal, site-selective conjugation method using a small photo-cross-linkable protein G adaptor that allows labeling of antibodies of different host species with a controlled number of short oligonucleotides (ODNs). Importantly, we illustrate that this conjugation method can be directly performed on commercially available primary antibodies on a small scale and without cross-reactivity towards bovine serum albumin. In addition, we present a general benchtop-compatible strategy to purify DNA-labeled antibodies without a loss of function. The application of protein G-ODN-labeled primary antibodies is demonstrated by employing three well-known methods for detecting subcellular targets using fluorescence read-out, including flow cytometry, DNA-PAINT, and dSTORM. This work thus establishes a general and efficient platform for the synthesis of a library of unique ODN–antibody conjugates, facilitating the broader use of DNA-based programmable tags for multiplexed labeling to identify subcellular features with nanometer precision and improving our understanding of cellular structure and function.
Introduction
To unravel the structure, organization, and function of subcellular components in a crowded environment, specific orthogonal labeling of a large variety of biomolecules inside the cell is essential. Currently, antibodies are the preferred affinity reagents for the visualization of subcellular components because they offer exquisite control over specificity and are commercially available for a large class of targets. The predictability of DNA nanotechnology has provided a powerful tool for providing antibodies with unique, programmable labels that allow detection via various fluorescence-based read-out methods.1 In general, read-out methods based on DNA have the advantage that their coding capacity, in addition to a number of spectrally distinct fluorescent tags, relies on the complementarity of unique oligonucleotide (ODN) sequences, increasing the number of labels that can be simultaneously used.2,3 As a result, multiple fluorescence-based read-out methods are available that rely on the reversible binding of short imager strands,4,5 affinity-mediated signal amplification,6,7 and DNA strand displacement.8−10 In addition, advances in the field of DNA nanotechnology have provided a powerful tool for the design of well-defined nanostructures, the DNA origami technique.11−13 This development has allowed the design of nanostructures that facilitate control over optical properties (e.g., brightness, color) by the site-specific incorporation of fluorescently labeled ODNs.14,15 Combining the programmability of DNA nanotechnology with the specificity of antibody labeling therefore facilitates the design of programmable fluorescent tags that have the ability to label >100 subcellular targets and can be distinguished unambiguously.
Although primary antibodies are widely commercially available, a general stoichiometric site-selective ODN labeling strategy is missing. Traditionally, antibodies are functionalized with a modified ODN that targets chemical groups present in the native antibody (e.g., thiols and primary amines).16,17 However, this method lacks site-selectivity and stoichiometric control and can therefore result in antibodies with diminished binding capacity.18 Moreover, commercially available antibody solutions contain protein stabilizers, bovine serum albumin (BSA) in particular, which carry numerous functional groups that directly compete for reaction with the functionalized ODN. Several methods have been introduced to address these limitations, involving the introduction of noncanonical amino acids19 or specific labeling tags, including Snap-tags,20 HaloTags,21 and CLIP-tags.22 Additionally, coupling methods targeting specific regions on the antibody have been applied.23−26 However, these methods require genetic re-engineering of the antibody, are limited by the specific host species or subtype of the antibody, or are performed in the absence of stabilizing proteins.
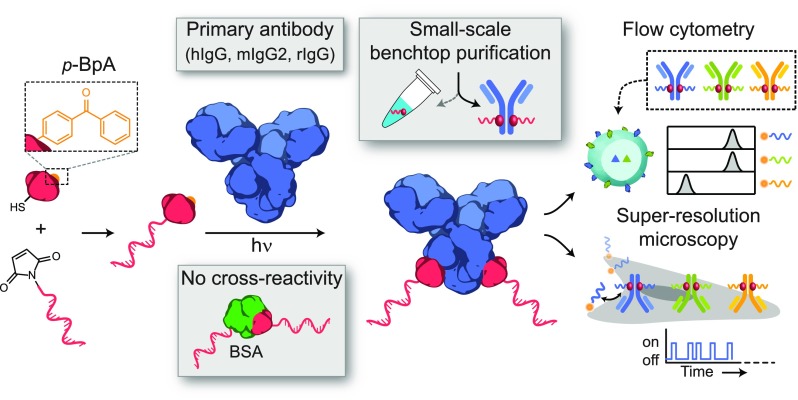
Here we present a general, benchtop-compatible strategy to site-selectively label and purify commercially available primary antibodies with short ODNs. Importantly, we confirm that this labeling method is selective for antibodies and shows no cross-reactivity toward BSA. This selectivity is achieved using an ODN-functionalized protein G adaptor27 (pG-ODN) that can be photo-cross-linked to the heavy-chain region of a native immunoglobulin G-type (IgG) antibody (Figure 1). Protein G is part of a larger class of proteins, among them protein A and protein L, which are able to selectively bind to a specific region of a native IgG antibody and therefore allow site-selective functionalization of antibodies.28−31 We previously developed, and successfully used, this strategy to decorate DNA nanostructures with antibodies and Fc-functionalized proteins.32 In this study, we optimized the ODN coupling efficiency to protein G, which allowed the direct conjugation of unpurified pG-ODN constructs to a native antibody, making multiplexed antibody labeling efficient and less time-consuming. We show that this strategy is compatible with human IgG1, mouse IgG2a, and rabbit IgG antibodies, which together cover ≥80% of the commercially available primary antibodies.33 In combination with a universal, benchtop-compatible purification method, we report on the successful labeling and purification of a subset of primary antibodies. To illustrate the universal applicability of the coupling strategy, cellular labeling with pG-ODN–antibody constructs is evaluated using flow cytometry and super-resolution microscopy techniques, including stochastic optical reconstruction microscopy (STORM)34 and DNA point accumulation for imaging in nanoscale topography (DNA-PAINT).2 Our results show successful cellular labeling using the pG-ODN–antibody conjugates and underline that covalent coupling of the pG-ODN construct to the antibody is required when multiplexed cellular labeling is performed.
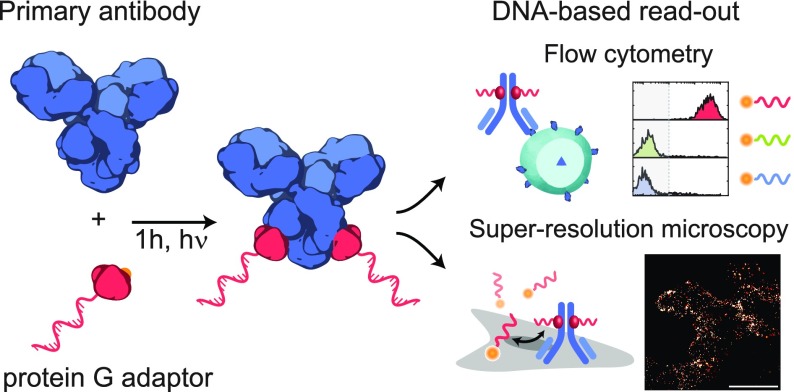
Figure 1.
Schematic overview of the labeling strategy for the site-selective functionalization of primary antibodies with an oligonucleotide (ODN) via a small protein G adaptor without cross-reactivity toward stabilizing proteins (e.g., bovine serum albumin (BSA)). Protein G is functionalized and expressed with a cysteine coupled to a maleimide-functionalized ODN. Additionally, the protein G variant contains a non-natural amino acid, p-benzoylphenylalanine (p-Bpa), that covalently couples the protein G-ODN conjugate to the fragment crystallizable (Fc) region of a primary antibody, including human IgG (hIgG), mouse IgG2 (mIgG2), and rabbit IgG (rIgG), using long-wavelength UV light (365 nm). Antibody–ODN conjugates are purified via a benchtop-compatible method and can be directly used for multiplexed cellular labeling using either flow cytometry or super-resolution microscopy.
Results and Discussion
To facilitate the efficient synthesis of multiple unique pG-ODN constructs, we first focused on a labeling method to couple maleimide-functionalized ODNs to protein G with a high yield. Previous work showed that the N-terminal cysteine formed an unreactive thiazolidine adduct during pG expression, which limited the pG-ODN conjugation efficiency to ∼15%.32 In this previous work, it was hypothesized that the introduction of an additional amino acid before the N-terminal cysteine would resolve this problem and increase the conjugation efficiency. To this end, we site-specifically inserted a serine N-terminal to the cysteine and found that after pG expression, no adduct formation was observed (Figure S1).
As a result, the pG-ODN coupling efficiency was increased to >90% when pG was incubated with a five-fold molar excess of maleimide-functionalized ODN (Figure S2). Subsequently, we investigated if the pG-ODN reaction mixture could directly be used for antibody coupling without the initial purification of the pG-ODN construct. To this end, we incubated Cetuximab, a monoclonal IgG1 antibody, with different molar equivalents of pG-ODN. We compared the formation of pG-ODN–antibody conjugates using the unpurified pG-ODN reaction mixture and purified pG-ODN. SDS-PAGE analysis was used to monitor product formation, and gel band intensity analysis showed that >80% of the heavy chains were successfully labeled with pG-ODN using a 10-fold molar excess of unpurified pG-ODN (Figure 2A and Figure S3). In comparison, we observed >90% labeling efficiency when purified pG-ODN was used (Figure S4). Additionally, the lyophilization of the pG-ODN constructs did not decrease the reported conjugation efficiency, making these constructs ideal for long-term storage and shipping (Figure S5). We note that the antibody conjugation efficiency (∼80%) does not match the coupling efficiency observed after pG-ODN coupling (>90%); however, we hypothesize that pG–antibody binding is preferred over pG-ODN–antibody coupling due to the steric hindrance and electrostatic repulsion induced by the ODN. Nevertheless, on the basis of the observed heavy-chain labeling efficiency for unpurified pG-ODN, we expect that >95% of all antibodies (two heavy chains) contain at least one ODN sequence.
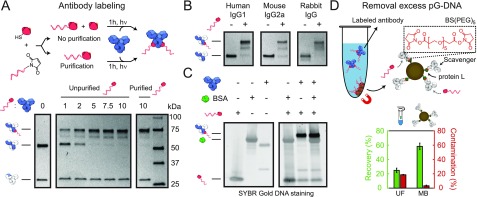
Figure 2.
Antibody–ODN labeling and purification. (A) Protein G (pG) was conjugated to a 20 nt 3′-maleimide-functionalized ODN. Human IgG1 (hIgG1) was incubated with varying molar equivalents of unpurified pG-ODN, and the labeling efficiency was analyzed and compared with purified pG-ODN under reducing SDS-PAGE conditions. Without pG-ODN purification, ∼20% of the heavy chains were coupled to pG instead of pG-ODN. (B) Reducing SDS-PAGE analysis of various IgG subclasses coupled to 10 equiv pG-ODN and a (C) hIgG1 antibody coupled to pG-ODN in the presence and absence of 0.07% (w/v) bovine serum albumin (BSA). (D) Excess of pG-ODN was removed using protein-L-functionalized magnetic beads covalently coupled to a scavenger antibody via a PEGylated bis(sulfosuccinimiyl)suberate linker (BS(PEG)5). The average recovery (green) and contamination (red) of the magnetic beads (MBs) were compared to ultrafiltration (UF). Error bars represent SD (n = 3).
To examine the general applicability of the pG-ODN labeling strategy, we evaluated the coupling efficiency for antibodies of different host species. Previously, it was shown that pG was also able to photo-cross-link to mouse IgG2a and Rabbit IgG.27 Rabbit IgGs, in particular, are an important class of antibodies because rabbits are the host species in which most primary antibodies are raised.33 Incubating both mouse IgG2a (mIgG2a) and rabbit IgG with a 10-fold excess of pG-ODN showed successful antibody coupling, emphasizing the universal applicability of pG-ODN labeling (Figure 2B).
Because commercially available primary antibodies are typically supplemented with protein stabilizers (e.g., BSA, glycerol, sodium azide), pG-ODN antibody labeling should be compatible with the presence of these additives. BSA, in particular, is known to interfere with more classic ODN-labeling methods targeting functional groups in the antibody (e.g., lysine, cysteine). Commercially available kits can be used to remove BSA; however, these protocols rely on extensive washing steps, resulting in the extensive loss of antibody. The exclusive selectivity of pG-ODN towards the heavy-chain region should overcome this problem, making the need for BSA-free antibody solutions redundant. To test this, we performed the coupling of an antibody to pG-ODN in the presence of 0.07% (w/v) BSA. As expected, the labeling of Cetuximab in the presence of BSA did not result in cross-reactivity, confirming the selectivity of pG-ODN constructs (Figure 2C). Additionally, we evaluated the coupling efficiency of pG-ODN in the presence of other common antibody additives, including TWEEN-20, glycerol, and sodium azide. Sodium azide is known to quench the reactive triplet state of p-Bpa and is therefore likely to inhibit the pG-ODN cross-linking.35 Whereas TWEEN-20 did not alter the coupling efficiency, both glycerol and sodium azide inhibited the formation of pG-ODN–antibody constructs (Figure S6). However, buffer exchange of the antibody using ultrafiltration to a glycerol/sodium azide free buffer did restore the coupling efficiency. From this, we conclude that the pG-ODN labeling strategy can be used in combination with commercially available antibodies.
We next focused on the development of a universal purification approach to remove excess pG-ODN. To this extent, we systematically evaluated different purification methods that either focus on size-based separation or are specifically designed for antibody purification. First, we tested the separation of pG-ODN–antibody constructs and pG-ODN using size-exclusion chromatography (SEC). The relatively large difference in molecular weight between pG-ODN and labeled antibodies, 15 and 180 kDa, respectively, should provide enough separation to effectively purify the pG-ODN–antibody. To this end, we applied a high-performance liquid chromatography (HPLC) system with a size exclusion column that has a fractionation range between 5 × 103 and 1.25 × 106 Da. Using this approach, the pG-ODN–antibody conjugate was well-separated from unreacted pG-ODN (Figure S7). However, despite the excellent separation, we note that HPLC requires specialized equipment. Moreover, HPLC purification results in high sample dilution, making this method less appropriate for small sample volumes, which is typically the case when primary, more expensive antibodies are labeled. To address this limitation, we evaluated a second size-based purification technique using ultrafiltration. We performed a repetitive dilution concentration process using a regenerated cellulose membrane with a 100 kDa molecular weight cutoff (MWCO). In this process, the large pG-ODN–antibody should be retained by the membrane, whereas the smaller pG-ODN should flow through. In contrast with SEC, ultrafiltration resulted in limited separation of the pG-ODN–antibody and pG-ODN with a recovery yield of 25% and contamination of 19% (Figure 2D, bottom). The recovery yield and contamination percentage were calculated by gel analysis of the purified product and known concentrations of a reference antibody (Figure S8). To improve the separation of the pG-ODN–antibody and the free pG-ODN, we evaluated the performance of multiple (denaturing) washing buffers. Whereas the use of different washing buffers did improve the recovery yield (68%), the separation remained limited (Figure S9). Further optimization of this purification process, for example, the use of an acidic washing buffer, could therefore be performed to improve the limited separation.29−31
To design a universal benchtop-compatible purification method, we therefore decided to turn our attention to antibody-specific purification methods. In addition to protein G, several recombinant proteins are available that target specific regions of an antibody, including proteins A and L. Whereas protein A overlaps with the binding site of protein G, protein L specifically binds to kappa light chains of mouse and human IgG monoclonal antibodies.28 Using commercially available magnetic beads functionalized with protein L, we were able to successfully purify pG-ODN-functionalized Cetuximab (hIgG1) from pG-ODN (Figure S10). However, the limited compatibility of protein L with different subclasses or host species of antibodies prevents the general application of this purification method. Therefore, we developed a purification method that utilizes the protein L magnetic beads to target free pG-ODN, instead of the labeled antibody. We achieved this via the covalent attachment of a scavenging antibody, typically a secondary, less expensive antibody, to protein L beads by cross-linking the primary amines of both proteins using a bis-succinimide ester-activated PEG compound BS(PEG)5 (Figure 2D, top). Protein L ensures the correct orientation of the scavenger antibody, making the Fc region available for free pG-ODN binding. We evaluated multiple cross-linking conditions to increase the binding capacity of the scavenger beads and showed the successful capture of pG-ODN (Figure S11). Subsequently, we confirmed that this method can be used to purify pG-ODN-labeled antibodies, and after desalting of the purified mixture, a recovery of 58% and contamination of only 3% were achieved (Figure 2D, bottom; Figure S12). Whereas the relatively low recovery yield could be improved by further optimization of the washing steps, we primarily focused on the low contamination. Additionally, we note that the recovery yield is decreased to 30% when antibodies that have a high intrinsic affinity for protein L (hIgG, mIgG) are purified (Figure S13). Lastly, it was shown that this scavenging approach could be performed on a small scale (5 μg) and was compatible with Fc-fusion proteins (Figure S14). Protein-L-functionalized magnetic beads, in combination with a scavenging antibody, therefore provide an additional platform for the small-scale purification of antibodies from different host species and subtypes.
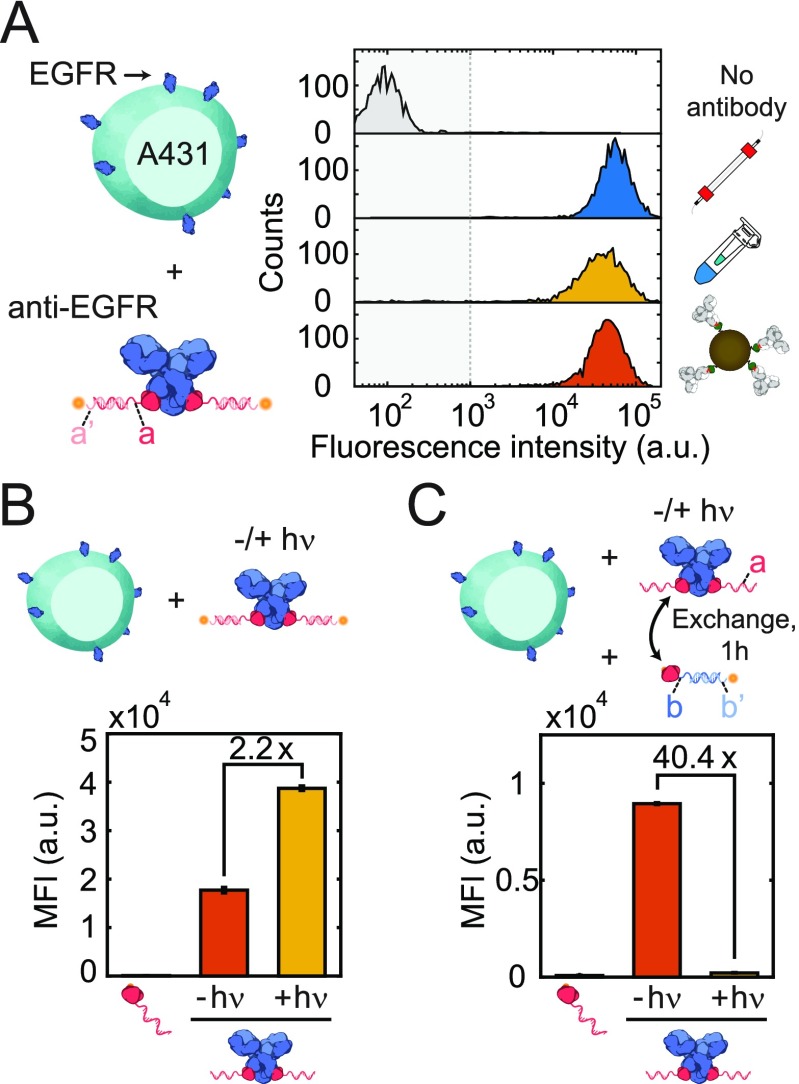
To test the activity of the pG-ODN functionalized antibodies after labeling and purification, we used fluorescence-activated cell sorting (FACS) analysis. To this end, we functionalized Cetuximab with a pG-ODN docking strand a. Subsequently, we preincubated pG-ODN-labeled Cetuximab that was purified using SEC, ultrafiltration, or magnetic scavenging beads with a CY5-functionalized ODN, a′, complementary to the pG-ODN docking strand. Eventually, we incubated A431 carcinoma cells, expressing high levels of epidermal growth factor receptor (EGFR), with pG-ODN–Cetuximab, hybridized to a CY5-labeled ODN, and subjected them to flow cytometry. In all cases, an increase in the fluorescence intensity was observed, indicating the binding of pG-ODN–Cetuximab to the EGFR receptor (Figure 3A and Figure S15).
Figure 3.
Flow cytometric analysis of EGFR-expressing A431 cells using 10 nM pG-ODN-functionalized Cetuximab, a, hybridized to a CY5-functionalized ODN, a′. (A) Antibody activity after purification using SEC, ultrafiltration, and magnetic beads. The fluorescence intensity of pG-ODN-Cetuximab-labeled A431 cells was compared with that of A431 cells incubated with only pG-ODN. (B) Labeling efficiency and (C) cross-contamination of pG-ODN noncovalently (−hν) or covalently (+hν) coupled to Cetuximab. pG-ODN-functionalized Cetuximab was incubated for 1 h with a 20 mol equiv of a competing pG-ODN sequence. MFI represents the median fluorescence intensity, and error bars represent the SD (n = 3).
To emphasize the importance of irreversible coupling, we compared the labeling efficiency and cross-contamination of antibodies labeled covalently (+hν) and noncovalently (−hν) with pG-ODN. To this end, Cetuximab was incubated with pG-ODN in the presence and absence of UV illumination. Subsequently, A431 cells were incubated with pG-ODN–Cetuximab to label the EGFR receptor. After labeling, a CY5-functionalized ODN, complementary to the pG-ODN sequence, was introduced, and the cells were analyzed using flow cytometry. The 2.2-fold difference in median fluorescence shows a clear increase in labeling efficiency when the antibody is labeled covalently (Figure 3B), which underlines the importance of irreversible labeling.
Because cellular labeling procedures are typically performed with multiple antibodies, traces of pG-ODN present after purification could bind other antibodies, resulting in unwanted cross-contamination. To evaluate the degree of cross-contamination, we incubated noncovalently and covalently labeled pG-ODN–Cetuximab with a 20-fold molar excess of a competing pG-ODN docking sequence, b, for 1 h. Subsequently, A431 cells were incubated with the reaction mixture, and after labeling of the EGFR receptor, a CY5-functionalized ODN, b′, complementary to the competing pG-ODN sequence, was added. The observed median fluorescence for the cells that were labeled with noncovalent pG-ODN–Cetuximab showed pG-ODN exchange resulting in cross-contamination (Figure 3C). In contrast, when pG-ODN was coupled covalently to Cetuximab via photoinduced coupling, no cross-contamination was observed. Additionally, it was shown that free Fc binding sites could be blocked during cellular labeling using a five-fold molar excess of pG compared to the competing pG-ODN sequence, nullifying cross-contamination (Figure S16). These results show that multiple pG-ODN-functionalized antibodies can be used simultaneously, even when the pG-ODN antibody coupling is not quantitative and free Fc sites are still available.
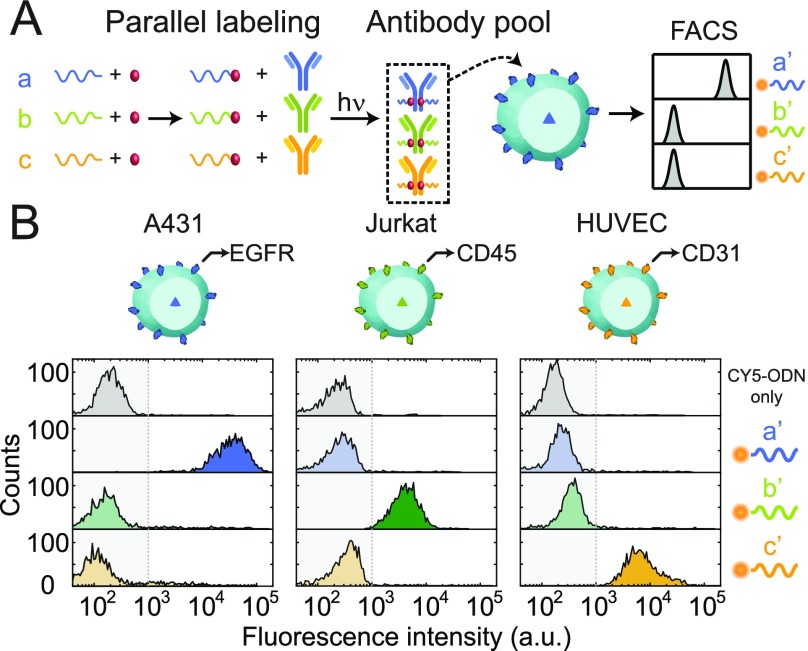
Thus far, we have shown the synthesis and purification of pG-ODN–antibody constructs and successfully used them for cellular labeling. Next, we tested the multiplex applicability of the pG-ODN labeling strategy by coupling three primary antibodies, Cetuximab, mouse IgG2a anti-CD45, and mouse IgG2a anti-CD31, to a unique pG-ODN docking sequence, a, b, and c, respectively. Subsequently, we performed the simultaneous labeling of three cell types with all of the pG-ODN functionalized antibodies, including A431 carcinoma cells, Jurkat T cells, and human umbilical vein endothelial cells (HUVECs) expressing EGFR, CD45, and CD31, respectively. Because quantitative pG-ODN labeling was not observed for all antibodies, we introduced a five-fold molar excess of pG with respect to pG-ODN to block any remaining free Fc sites. After cellular labeling, we introduced CY5-functionalized ODNs, a′, b′, and c′, respectively, complementary to a specific pG-ODN docking sequence, and analyzed the cells using flow cytometry (Figure 4A). The presence of only one distinct fluorescent population per CY5-functionalized strand showed the successful labeling of targets without the exchange of pG-ODN sequences, verifying that pG-ODN–antibody conjugates are suitable for multiplexed cellular labeling and imaging (Figure 4B).
Figure 4.
Characterization of protein G-ODN cross-talk. (A) Cetuximab, anti-CD45, and anti-CD31 are labeled in parallel with a unique pG-ODN construct, containing ODN sequences a, b, and c, respectively. Subsequently, the pG-ODN-labeled antibodies are pooled, and cells expressing a specific target protein are incubated with the antibody pool. Eventually, CY5-labeled ODNs, a′, b′, and c′, respectively, complementary to the unique pG-ODN sequence, are used to detect the presence of each antibody on the cell surface. (B) Flow cytometric analysis of three cell lines (A431, Jurkat T cells, and HUVECs) expressing EGFR, CD45, and CD31, respectively. Cells were incubated with 10 nM of the antibody pool containing pG-ODN-labeled Cetuximab, mouse IgG2a anti-CD45, and mouse IgG2a anti-CD31 and subsequently labeled with 100 nM of a CY5-functionalized ODN.
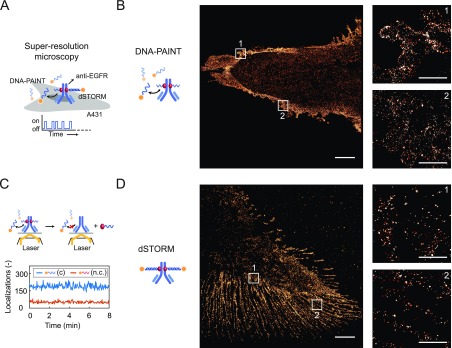
Although flow cytometry is an excellent tool to study cell populations, it is unable to provide a detailed view of the structure and organization of subcellular components. Super-resolution techniques, however, are able to visualize cellular targets with nanometer precision and therefore have the potential to increase our understanding of cell structure and function. DNA-PAINT and, in particular, exchange-PAINT, rely on the use of antibodies functionalized with a unique ODN to achieve multiplexed super-resolution imaging with a spatial resolution down to ∼5 nm.2,36 To illustrate that pG-ODN-functionalized primary antibodies could be directly used for DNA-PAINT, we demonstrated in situ imaging in a fixed A431 carcinoma cell using pG-ODN-functionalized Cetuximab that was purified using SEC (Figure 5A). We obtained super-resolution images of the EGFR receptor and observed little nonspecific binding when a noncomplementary imager strand was used (Figure 5B and Figure S17). Recently, Jungmann and coworkers also successfully obtained super-resolution images when using ODN-functionalized protein A and G variants as secondary, noncovalent labeling reagents for DNA-PAINT.37 Because pG-ODN is covalently coupled to the antibody using UV light, we additionally investigated whether the pG-ODN antibody coupling is reversed when the construct is exposed to high laser intensity, as those employed during DNA-PAINT. To validate the stability of the pG-ODN–antibody construct, we quantified the number of localizations during image acquisition over the course of 8 min (Figure 5C). The observed number of localizations remained constant over time, confirming that the photoconjugated pG-ODN–antibody construct is stable when exposed to high laser intensity. This result also allowed the use of pG-ODN–antibody constructs for cellular staining in combination with direct STORM (dSTORM), which requires high illumination power. Successful dSTORM images of the EGFR receptor on A431 cells were acquired using a complementary CY5-functionalized imager strand, hybridized to a pG-ODN–Cetuximab construct (Figure 5D and Figure S18). Taken together, these results show the compatibility of the pG-ODN labeling approach with super-resolution techniques, including DNA-PAINT and dSTORM.
Figure 5.
Protein G-ODN–antibody conjugates for applications in super-resolution microscopy. (A) A431 carcinoma cells are labeled with a pG-ODN–Cetuximab construct containing either a short (11 nt) or a long (20 nt) docking strand. Subsequently, the labeled cells are fixated to a glass slide, and super-resolution images are obtained using the transient binding of a short imager strand (DNA-PAINT) or using a long irreversible binder in combination with a photoswitchable dye (dSTORM). (B) DNA-PAINT super-resolution image obtained using ATTO647N-functionalized imager strands (20 000 frames, 20 Hz frame rate). A zoom-in on two defined areas shows the distribution of EGFR receptors. (C) Number of localizations observed over the course of 8 min during a DNA-PAINT acquisition using complementary (c) or non-complementary (n.c.) imager strands. (D) Super-resolution image obtained with dSTORM using a CY5-functionalized imager strand (20 000 frames, 65.5 Hz frame rate), including a zoom-in on two defined areas that show the distribution of EGFR receptors. Scale bars: 5 and 1 μm for the zoom-in images.
Conclusions
In this work, we have developed a generally applicable ODN–antibody coupling method using a small protein G adaptor that site-selectively targets the heavy chain of an IgG antibody. We successfully demonstrated pG-ODN labeling of antibodies from different host species without cross-reactivity towards BSA. Importantly, we showed that the pG-ODN labeling did not affect the native function of the antibody. In combination with the universal, benchtop-compatible purification strategy using magnetic beads, this ODN labeling method is directly applicable to commercially available primary antibodies. Because multiple pG-ODN conjugates can be constructed in parallel and lyophilized without a loss of function, the potential of the pG-ODN labeling strategy lies in the synthesis of a library of pG-ODN constructs that can directly be used for antibody labeling. This could eventually facilitate the implementation of the multiplexing abilities of DNA-based read-out methods for the detection of a large variety of subcellular components and make these methods accessible for a broader scientific community. Additionally, we envision the use of pG-ODN–antibody constructs in more quantitative imaging applications, owing to the unique ability of pG-ODN conjugates to selectively label an antibody with a controlled number of ODNs.
Materials and Methods
Recombinant Protein Cloning, Expression, and Purification
A pET28a(+) vector encoding the pG gene as reported previously32 was site-specifically mutated by the insertion of an N-terminal serine using the QuikChange Lightning multi site-directed mutagenesis kit (Agilent) according to the manufacturer’s instructions using the following forward and reverse primers (ser codon underlined), 5′-TTTAAGAAGGAGATATAACATGAGTTGCTGGTCCCATCCG-3′ and 5′-CGGATGGGACCAGCAACTCATGTTATATCTCCTTCTTAAAG-3′, respectively. Insertion was confirmed by a DNA sequencing service provided by StarSEQ (Mainz, Germany). Plasmid DNA of the pG gene was cotransformed with the pEVOL-pBpF vector, encoding for the orthogonal aminoacyl tRNA synthetase/tRNA pair (kindly provided by Peter Schultz), into E. coli BL21(DE3) (Novagen) for protein expression. A single colony of freshly transformed cells was cultured at 37 °C in 500 mL of 2xYT medium supplemented with 50 μg/mL kanamycin (Merck) and 25 μg/mL chloramphenicol (Sigma-Aldrich). When the OD600 of the culture reached ∼0.6, protein expression was induced by the addition of β-d-1-thiogalactopyranoside (IPTG, Applichem), arabinose (Sigma-Aldrich), and the unnatural amino acid p-BpA (Bachem) in a final concentration of 1 mM, 0.02% (w/v), and 1 mM, respectively. The induced protein expression was carried out for ∼18 h at 25 °C, and subsequently, the cells were harvested by centrifugation at 10 000g for 10 min at 4 °C. The cell pellet was resuspended in BugBuster (5 mL/g pellet, Merck) supplemented with benzonase (5 μL/g pellet, Merck) and incubated for 45 min on a shaking table. The suspension was centrifugated at 40 000g for 30 min at 4 °C, and the supernatant was subjected to Ni-NTA affinity chromatography on a gravity column. His-tagged pG was loaded on the column and washed with washing buffer (1× PBS, 370 mM NaCl, 10% (v/v) glycerol, 20 mM imidazole, pH 7.4) before elution with his-elution buffer (1× PBS, 370 mM NaCl, 10% (v/v) glycerol, 250 mM imidazole, pH 7.4). Subsequently, the Ni-NTA elution fraction was loaded on a Strep-Tactin column, washed with washing buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 8.0), and eluted using wash buffer supplemented with 2.5 mM desthiobiotin (IBA Life Sciences). Proteins were stored at −80 °C in 1 mL aliquots of 50 μM at −80 °C in a buffer containing 100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 2 mM TCEP at pH 8.0. The concentration of pG was calculated on the basis of the absorption at 280 nm (ND-1000, Thermo Scientific), assuming an extinction coefficient of 15 470 M–1 cm–1, and the purity of pG was assessed on reducing SDS-PAGE and liquid chromatography quadrupole time-of-flight mass spectrometry (Q-Tof).
Preparation of Reaction ODNs
In a typical reaction, to a solution of 10 nmol ODN in water (10 μL) was added 1× PBS, pH 7.2 (30 μL) and 100 nmol Sulfo-SMCC (Thermo Scientific) in DMSO (40 μL). The reaction was incubated at 850 rpm for 2 h at 20 °C. Excess Sulfo-SMCC was removed using two rounds of ethanol precipitation. SMCC-labeled ODNs were precipitated by the addition of 10% (v/v) 5 M NaCl and 300% (v/v) ice-cold EtOH and incubating for 75 min at −30 °C. The reaction mixture was centrifuged at 19 000g for 30 min at 4 °C, the pellet was reconstituted in 1× PBS (pH 7.2), and the precipitation was repeated. After centrifugation, the pellet was washed in 95% (v/v, in water) ice-cold EtOH, centrifuged at 19 000g for 15 min, and lyophilized.
General Procedure for the Conjugation of ODN to pG
For the conjugation of pG to a SMCC-functionalized ODN, an aliquot of pG was buffer-exchanged to (100 mM sodium phosphate, 25 μM TCEP, pH 7) using a PD10 desalting column (GE Healthcare). Subsequently, desalted pG was concentrated to a final concentration of 50 μM using Amicon 3 kDa MWCO centrifugal filters (Merck Millipore). Ten nmol lyophilized SMCC-functionalized ODN was reconstituted in 40 μL of 50 μM pG (2 nmol), resulting in a five times excess of maleimide–ODN. The reaction was shaken at 850 rpm for 3 h at 20 °C. The coupling efficiency was assessed using SDS-PAGE under nonreducing conditions. The purification, if applicable, of pG-ODN was performed using fast protein liquid chromatography (FPLC, ÄKTA Prime, GE Healthcare) with an anion-exchange HiTrap Q HP column (1 mL, GE Healthcare) using a salt gradient with a start and end concentration of 100 and 500 mM NaCl in 50 mM Tris-HCl (pH 7.5), respectively. Elution fractions were collected and analyzed by measuring online absorption at 280 nm and SDS-PAGE under nonreducing conditions. pG-ODN conjugates were aliquoted and stored at −80 °C.
General Procedure for pG-ODN Antibody Labeling
Before the conjugation of the antibody to the pG-ODN, all antibodies were buffer-exchanged to 1× PBS (pH 7.4) using Amicon 10 kDa MWCO centrifugal filters (Merck Millipore).
In the absence of BSA: In a typical reaction for SDS-PAGE gel analysis, a 10 μL aliquot containing 0.4 μM of antibody and 4 μM of pG-ODN was exposed for 1 h to UV light (λ = 365 nm) at 4 °C.
In the presence of BSA: For the conjugation of the antibody to pG-ODN in the presence of BSA, the pG-ODN reaction mixture was first incubated with 15 nmol of DTT and shaken at 850 rpm for 30 min at 20 °C to deactivate remaining maleimide–ODNs. Subsequently, pG-ODN was buffer-exchanged using a Zeba spin desalting column, 7000 MWCO, 0.5 mL (Thermo Scientific).
General Procedure for the Antibody Functionalization of Protein L Magnetic Beads
The scavenging antibody Cetuximab (Erbitux, Merck) was buffer-exchanged to conjugation buffer (100 mM sodium phosphate, 70 mM NaCl, 0.05% TWEEN-20 (v/v), pH 8.0) using a Zeba spin desalting column (7000 MWCO, 0.5 mL, Thermo Scientific) according to the manufacturer’s instructions. In a typical reaction, 25 μL of Pierce protein L magnetic beads was added to 75 μL of conjugation buffer. The beads were washed twice in 200 μL of conjugation buffer, after which the magnetic beads were incubated with 50 μg of scavenging antibody in a final volume of 250 μL. The tube was rotated for 1 h at room temperature. To cross-link the antibody to the beads, 0.5 μL of 250 mM BS(PEG)5 (Thermo Scientific) (dissolved in dry DMSO) was added to the reaction mixture. Cross-linking was performed for 30 min in a rotating wheel at room temperature, after which the reaction was deactivated by the addition of 25 μL of 1 M Tris-HCl (pH 7.5). Deactivation was performed for 15 min in a rotating wheel at room temperature. Subsequently, the magnetic beads were collected with a magnetic stand, and the supernatant was discarded. The scavenging beads were incubated two times for 5 min with 200 μL of washing buffer (100 mM Tris-HCl, 1 M NaSCN, pH 7.5), subsequently washed two times with 200 μL of storage buffer (1× PBS, 0.05% TWEEN-20 (v/v), pH 7.4), and stored at 4 °C.
General Procedure for the Purification of pG-ODN Functionalized Antibody
This procedure is optimized to purify 12.5 μL of 4 μM antibody–pG-ODN from 40 μM uncoupled pG-ODN using 25 μL of magnetic scavenging beads. Twenty-five μL of scavenging beads was washed two times in binding buffer (1× PBS, 870 mM NaCl, 0.05% TWEEN-20 (v/v), pH 7.4). The beads were redissolved in 37.5 μL of binding buffer, and 12.5 μL of the antibody–pG-ODN reaction mixture was added and incubated for 15 min in a rotating wheel at room temperature. The scavenging beads were collected with a magnetic stand, and the supernatant was discarded. Subsequently, the beads were incubated two times for 5 min with 200 μL of washing buffer (100 mM Tris-HCl, 1 M NaSCN, pH 7.5). The whole process was repeated two more times to remove all pG-ODN. Amicon 50 kDa MWCO centrifugal filters were used to purify the antibody–pG-ODN conjugates from unreacted maleimide–ODN conjugates and buffer-exchange the purified conjugates to 1× PBS, pH 7.4. The removal of the pG-ODN conjugate and the recovery of the antibody-coupled pG-ODN were assessed based on the gel band intensity analysis using SDS-PAGE under reducing conditions.
Cellular Labeling for Flow Cytometry
A431, Jurkat T, and HUVECs were cultured in a 175 cm2 flask. Cells were harvested at a confluency of ∼80%, washed in labeling buffer (1× PBS, 0.1% BSA (w/v), pH 7.4), and diluted to a final concentration of 3.5 × 106 cells/mL in labeling buffer. Subsequently, 12.5 μL of the cell suspension was incubated in a final volume of 250 μL of labeling buffer containing 10 nM of pG-ODN-labeled antibody. The reaction mixture was shaken at 400 rpm for 30 min at room temperature. Subsequently, the labeled cells were centrifuged for 5 min at 1500g, and the supernatant was removed. The pelleted cells were redissolved in labeling buffer containing 100 nM of the complementary CY5-labeled ODN. The reaction mixture was incubated and centrifuged as previously described and subsequently analyzed using flow cytometry. For the multiplexed experiment (Figure 4), the labeling buffer was supplemented with 1.5 μM pG.
Super-Resolution Sample Preparation and Imaging
A431 cells (ATCCCRL-1555) were seeded on an eight-well glass-bottomed μ-slide (ibidi, Germany) overnight. Live-cell immunolabeling started with 5 min of acclimation of the cells at room temperature, followed by 5 min of acclimation at 4 °C. Cells were incubated for 45 min at 4 °C in DMEM/3% BSA with 1 μg/mL of pG-ODN–Cetuximab. Then, three 5 min washes with PBS at 4 °C were followed by formalin 5% and glutaraldehyde 0.25% fixation for 10 min. Ultimately, cells were washed three times for 5 min with PBS and stored in the fridge before imaging. Super-resolution images were acquired using a Nikon N-STORM microscope in total internal reflection fluorescence (TIRF) mode. The system was equipped with a Nikon 100×, 1.49 NA oil immersion objective and an Andor iXON3 camera. Images were acquired onto a 256 × 256 pixel region (40.96 × 40.96 μm) and analyzed with NIS Element Nikon software. Samples were illuminated with a 647 nm laser at 140 mW, 16 ms, and 20 000 frames for STORM or 70 mW, 50 ms, and 20 000 frames for DNA-PAINT. To perform STORM imaging, we used a specific buffer to induce dye photoswitching: 5% w/v glucose, 100 mM cysteamine, 0.5 mg/mL glucose oxidase, and 40 μg/mL catalase in PBS. For DNA-PAINT imaging, we diluted DNA imager ATTO647N to 1 nM in PBS (500 mM NaCl).
Acknowledgments
We thank L. A. Tiemeijer and C. M. Sahlgren for providing and culturing the HUVECs. Sinem K. Saka from the Wyss Institute for Biologically Inspired Engineering is gratefully acknowledged for valuable insights and fruitful discussions. This work was supported by the European Research Council (ERC) (project no. 677313 BioCircuit), an NWO-VIDI grant from The Netherlands Organization for Scientific Research (NWO, 723.016.003), and funding from the Ministry of Education, Culture and Science (Gravity programs, 024.001.035 & 024.003.013)
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.9b00490.
Extensive experimental methods and supplemental figures of purification strategies and characterization experiments (PDF)
Author Contributions
⊥ G.A.O.C. and B.J.H.M.R. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Chen Y.-J.; Groves B.; Muscat R. A.; Seelig G. (2015) DNA Nanotechnology from the Test Tube to the Cell. Nat. Nanotechnol. 10 (9), 748–760. 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- Jungmann R.; Avendaño M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. (2014) Multiplexed 3D Cellular Super-Resolution Imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11 (3), 313–318. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade O. K.; Woehrstein J. B.; Nickels P. C.; Strauss S.; Stehr F.; Stein J.; Schueder F.; Strauss M. T.; Ganji M.; Schnitzbauer J.; et al. (2019) 124-Color Super-Resolution Imaging by Engineering DNA-PAINT Blinking Kinetics. Nano Lett. 19, 2641. 10.1021/acs.nanolett.9b00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Steinhauer C.; Scheible M.; Kuzyk A.; Tinnefeld P.; Simmel F. C. (2010) Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami. Nano Lett. 10 (11), 4756–4761. 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- Chen K. H.; Boettiger A. N.; Moffitt J. R.; Wang S.; Zhuang X. (2015) Spatially Resolved, Highly Multiplexed RNA Profiling in Single Cells. Science 348 (6233), aaa6090–aaa6090. 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson S.; Gullberg M.; Jarvius J.; Olsson C.; Pietras K.; Gústafsdóttir S. M.; Östman A.; Landegren U. (2002) Protein Detection Using Proximity-Dependent DNA Ligation Assays. Nat. Biotechnol. 20 (5), 473–477. 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Sano T.; Smith C. L.; Cantor C. R. (1992) Immuno-PCR: Very Sensitive Antigen Detection by Means of Specific Antibody- DNA Conjugates. Science 258 (5079), 120–122. 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- Bailey R. C.; Kwong G. A.; Radu C. G.; Witte O. N.; Heath J. R. (2007) DNA-Encoded Antibody Libraries: A Unified Platform for Multiplexed Cell Sorting and Detection of Genes and Proteins. J. Am. Chem. Soc. 129 (7), 1959–1967. 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahotre S. N.; Chang Y. M.; Wieland A.; Stammen S. R.; Kwong G. A. (2018) Individually Addressable and Dynamic DNA Gates for Multiplexed Cell Sorting. Proc. Natl. Acad. Sci. U. S. A. 115 (17), 4357–4362. 10.1073/pnas.1714820115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J.; Cordier G. A.; Borbely J. S.; Sandoval Álvarez Á.; Lakadamyali M. (2014) Cross-Talk-Free Multi-Color STORM Imaging Using a Single Fluorophore. PLoS One 9 (7), e101772. 10.1371/journal.pone.0101772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund P. W. K. (2006) Folding DNA to Create Nanoscale Shapes and Patterns. Nature 440 (7082), 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Dietz H.; Douglas S. M.; Shih W. M. (2009) Folding DNA into Twisted and Curved Nanoscale Shapes. Science 325 (5941), 725–730. 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S. M.; Dietz H.; Liedl T.; Högberg B.; Graf F.; Shih W. M. (2009) Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature 459 (7245), 414–418. 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.; Jungmann R.; Leifer A. M.; Li C.; Levner D.; Church G. M.; Shih W. M.; Yin P. (2012) Submicrometre Geometrically Encoded Fluorescent Barcodes Self-Assembled from DNA. Nat. Chem. 4 (10), 832–839. 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehrstein J. B.; Strauss M. T.; Ong L. L.; Wei B.; Zhang D. Y.; Jungmann R.; Yin P. (2017) Sub–100-Nm Metafluorophores with Digitally Tunable Optical Properties Self-Assembled from DNA. Sci. Adv. 3 (6), e1602128. 10.1126/sciadv.1602128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agasti S. S.; Wang Y.; Schueder F.; Sukumar A.; Jungmann R.; Yin P. (2017) DNA-Barcoded Labeling Probes for Highly Multiplexed Exchange-PAINT Imaging. Chem. Sci. 8 (4), 3080–3091. 10.1039/C6SC05420J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trads J. B.; Tørring T.; Gothelf K. V. (2017) Site-Selective Conjugation of Native Proteins with DNA. Acc. Chem. Res. 50 (6), 1367–1374. 10.1021/acs.accounts.6b00618. [DOI] [PubMed] [Google Scholar]
- Szabó Á.; Szendi-Szatmári T.; Ujlaky-Nagy L.; Rádi I.; Vereb G.; Szöllősi J.; Nagy P. (2018) The Effect of Fluorophore Conjugation on Antibody Affinity and the Photophysical Properties of Dyes. Biophys. J. 114 (3), 688–700. 10.1016/j.bpj.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazane S. A.; Sok D.; Cho E. H.; Uson M. L.; Kuhn P.; Schultz P. G.; Smider V. V. (2012) Site-Specific DNA-Antibody Conjugates for Specific and Sensitive Immuno-PCR. Proc. Natl. Acad. Sci. U. S. A. 109 (10), 3731–3736. 10.1073/pnas.1120682109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A.; Gendreizig S.; Gronemeyer T.; Pick H.; Vogel H.; Johnsson K. (2003) A General Method for the Covalent Labeling of Fusion Proteins with Small Molecules in Vivo. Nat. Biotechnol. 21 (1), 86–89. 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Los G. V.; Encell L. P.; McDougall M. G.; Hartzell D. D.; Karassina N.; Zimprich C.; Wood M. G.; Learish R.; Ohana R. F.; Urh M.; et al. (2008) HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 3 (6), 373–382. 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Gautier A.; Juillerat A.; Heinis C.; Corrêa I. R.; Kindermann M.; Beaufils F.; Johnsson K. (2008) An Engineered Protein Tag for Multiprotein Labeling in Living Cells. Chem. Biol. 15 (2), 128–136. 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Rosen C. B.; Kodal A. L. B.; Nielsen J. S.; Schaffert D. H.; Scavenius C.; Okholm A. H.; Voigt N. V.; Enghild J. J.; Kjems J.; Tørring T.; et al. (2014) Template-Directed Covalent Conjugation of DNA to Native Antibodies, Transferrin and Other Metal-Binding Proteins. Nat. Chem. 6 (9), 804–809. 10.1038/nchem.2003. [DOI] [PubMed] [Google Scholar]
- Yu C.; Tang J.; Loredo A.; Chen Y.; Jung S. Y.; Jain A.; Gordon A.; Xiao H. (2018) Proximity-Induced Site-Specific Antibody Conjugation. Bioconjugate Chem. 29 (11), 3522–3526. 10.1021/acs.bioconjchem.8b00680. [DOI] [PubMed] [Google Scholar]
- Yamada K.; Shikida N.; Shimbo K.; Ito Y.; Khedri Z.; Matsuda Y.; Mendelsohn B. A. (2019) AJICAP: Affinity Peptide Mediated Regiodivergent Functionalization of Native Antibodies. Angew. Chem., Int. Ed. 58 (17), 5592–5597. 10.1002/anie.201814215. [DOI] [PubMed] [Google Scholar]
- Nielsen T. B.; Thomsen R. P.; Mortensen M. R.; Kjems J.; Nielsen P. F.; Nielsen T. E.; Kodal A. L. B.; Cló E.; Gothelf K. V. (2019) Peptide-Directed DNA-Templated Protein Labelling for The Assembly of a Pseudo-IgM. Angew. Chem., Int. Ed. 58 (27), 9068–9072. 10.1002/anie.201903134. [DOI] [PubMed] [Google Scholar]
- Hui J. Z.; Tamsen S.; Song Y.; Tsourkas A. (2015) LASIC: Light Activated Site-Specific Conjugation of Native IgGs. Bioconjugate Chem. 26 (8), 1456–1460. 10.1021/acs.bioconjchem.5b00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björck L. (1988) Protein L. A Novel Bacterial Cell Wall Protein with Affinity for Ig L Chains. J. Immunol. 140 (4), 1194–1197. [PubMed] [Google Scholar]
- Konrad A.; Eriksson Karlström A.; Hober S. (2011) Covalent Immunoglobulin Labeling through a Photoactivable Synthetic Z Domain. Bioconjugate Chem. 22 (12), 2395–2403. 10.1021/bc200052h. [DOI] [PubMed] [Google Scholar]
- Kanje S.; Hober S. (2015) In Vivo Biotinylation and Incorporation of a Photo-Inducible Unnatural Amino Acid to an Antibody-Binding Domain Improve Site-Specific Labeling of Antibodies. Biotechnol. J. 10 (4), 564–574. 10.1002/biot.201400808. [DOI] [PubMed] [Google Scholar]
- Kanje S.; von Witting E.; Chiang S. C. C.; Bryceson Y. T.; Hober S. (2016) Site-Specific Photolabeling of the IgG Fab Fragment Using a Small Protein G Derived Domain. Bioconjugate Chem. 27, 2095. 10.1021/acs.bioconjchem.6b00346. [DOI] [PubMed] [Google Scholar]
- Rosier B. J. H. M.; Cremers G. A. O.; Engelen W.; Merkx M.; Brunsveld L.; de Greef T. F. A. (2017) Incorporation of Native Antibodies and Fc-Fusion Proteins on DNA Nanostructures via a Modular Conjugation Strategy. Chem. Commun. 53 (53), 7393–7396. 10.1039/C7CC04178K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Results for “Primary Antibodies” | abcam: Antibodies, Proteins, Kits. https://www.abcam.com/products?sortOptions=Relevance&keywords=primary+antibodies&selected.classification=Primary+antibodies (accessed August 16, 2019).
- Rust M. J.; Bates M.; Zhuang X. (2006) Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 3 (10), 793–796. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnam S.; Hamblett I.; Tonnesen H. H. (2000) Photoreactivity of Biologically Active Compounds. XVI. Formation and Reactivity of Free Radicals in Mefloquine. J. Photochem. Photobiol., B 56 (1), 25–38. 10.1016/S1011-1344(00)00056-7. [DOI] [PubMed] [Google Scholar]
- Schnitzbauer J.; Strauss M. T.; Schlichthaerle T.; Schueder F.; Jungmann R. (2017) Super-Resolution Microscopy with DNA-PAINT. Nat. Protoc. 12 (6), 1198–1228. 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- Schlichthaerle T.; Ganji M.; Auer A.; Kimbu Wade O.; Jungmann R. (2019) Bacterial-Derived Antibody Binders as Small Adapters for DNA-PAINT Microscopy. ChemBioChem 20 (8), 1032–1038. 10.1002/cbic.201800743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.