Abstract
Background
Human chronic myelogenous leukemia (CML) is a hematopoietic stem cell disorder with high malignant and invasive activity. lncRNA SNHG5 has been reported to be upregulated in CML. However, whether it affects the proliferation, differentiation, and apoptosis in CML cells is still unknown. This study investigated the role of SNHG5 in CML and revealed the potential mechanism.
Material/Methods
K562 cells were transfected with shRNA, and the expression level of SNHG5 was assessd by quantitative RT-PCR. The proliferation ability was determined by CCK-8 assay. Western blot analysis was performed to detect protein expressions related to cell proliferation, differentiation, and apoptosis. Cell apoptosis rate was analyzed by flow cytometry. The DNA methylation level was determined by methylation-specific PCR (MSP).
Results
Our results show that inhibition of SNHG5 induced by RNA interference significantly inhibits K562 cells proliferation and induces cell differentiation with the increased expression of CD42b, CD11b, CD14, GATA-1, and β-globin. Flow cytometry analysis indicated that inhibition of SNHG5 significantly induced cell apoptosis with decreased expression of Bcl-2 and increased expression of Bax and cleaved capase-3. Additionally, Western blot and MSP analyses confirmed that inhibition of SNHG5 increased the expression of DR4 gene through suppressing its methylation.
Conclusions
Inhibition of SNHG5 suppressed K562 cell proliferation through inducing the differentiation and apoptosis by inhibiting methylation of DR4. Therefore, downregulated SNHG5 could play a key role in CML progression, and might provide a new strategy for the treatment of CML.
MeSH Keywords: Antigens, Differentiation; Apoptosis; Leukemia, Myelomonocytic, Chronic; Receptors, TNF-Related Apoptosis-Inducing Ligand; RNA, Long Noncoding
Background
Chronic myeloid leukemia (CML) is a malignant clonal disease, accounting for about 20% of adult leukemia, with a strong malignant and invasive ability [1]. CML is characterized by the occurrence of the Philadelphia chromosome, which has been regarded as the diagnostic marker for CML [2]. The Philadelphia chromosome results from aberrant rearrangements between chromosome 9 and 22, t(9; 22) (q34; q11), directly resulting in the fusion of the Abelson murine leukemia (ABL) gene on chromosome 9 and the breakpoint cluster region (BCR) gene in chromosome 22, followed by the formation of BCR-ABL1 oncogene, which encodes an oncoprotein [3]. This constitutively activated oncoprotein is closely involved in the proliferation, apoptosis, and differentiation of hematopoietic stem cells [4].
Hematopoietic transcription factor GATA-1, a member of the GATA family of transcription factors, is expressed in primitive and definitive erythroid cells, mast cells, eosinophils, megakaryocytes, and the Sertoli cells of the testis [5]. It is reported that GATA-1 is involved in hematopoietic specification control and differentation, and large studies have demonstrated that GATA-1 drives differentation of erythrocytes and megakaryocytes [6,7]. The K562 cell line is a human myelogenous leukemia cell line from a highly undifferentiated progenitor of the megakaryocytic and erythrocytic lineages, with potential for erythroid and megakaryocyte differentiation, and it is currently the major cell model used for research of cellular differentation and screening of medical treatment for CML [8,9]. Differentiation-inducing therapy seems to be a promising strategy for CML [10,11], and the discover of a potential differentiation inducer or molecule has become an important focus of CML treatment.
Long noncoding RNAs (lncRNAs), a class of single-stranded noncoding RNAs larger than 200 mucleotides in length and unable to be translated into protein [12], have attracted much attention in recent years for their abundant functions and complicated modulation mechanisms in many cellular processes, including erythroid modulation [13–15]. Small nucleolar RNA host gene 5 (SNHG5) is a well-studied lncRNA. Overexpression of SNHG5 has been reported in various human cancers, such as bladder cancer and colorectal cancer [16,17]. SNHG5 is upregulated in the bone marrow and plasma of acute myeloid leukemia patients, and could serve as a potential prognostic biomarker in acute myeloid leukemia [18]. SNHG5 promotes imatinib resistance in CML via acting as a competing endogenous RNA against miR-205-5p [19]. Thus, we hypothesized that SNHG5 is closely involved in myeloid leukemia. However, its exact biological function has not been investigated in previous studies. The aim of this study was to explore the role of SNHG5 in the incidence of CML and to identify the function of SNHG5 in cell proliferation, differentiation, and apoptosis during the progression of CML.
Material and Methods
Cell culture
K562 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and were cultured in RPMI1640 medium containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (all from Thermo Fisher Scientific, Inc., Waltham, MA, USA) and incubated in a humidified atmosphere of 5% CO2 at 37°C.
RNA interference
RNA interference for SNHG5 was done as follows. Cells were transfected with 50 nM shRNAs targeting SNHG5 or siGFP (GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen, CA) in accordance with the manufacturer’s instructions. After incubation for 72 h, the effect of shRNAs was conformed by qRT-PCR analysis.
Cell proliferation assay
After transfected with shRNA for 24, 48, and 72 h, the Cell Counting Kit-8 (CCK-8; Dojingdo Laboratories, Kumamoto, Japan) was used to detect cell proliferation ability. In brief, K562 cells were cultured in 96-well plates, and 24 h later, 10 μl CCK-8 regent was added to each well and incubated at 37°C for 3 h. Then, the absorbance was measured at 450 nm. Three separate experiments were performed for each assay.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from K562 cells using TRIzol Regent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. Reverse transcription was performed to synthesize cDNA. qPCR reactions were carried out using the ABI7500 system (Applied Biosystems, CA) and SYBR green dye (Takara Biotechnology, Dalian, China). Primers used are as followed:
SNHG5 forward primer (5′-CGCTTGGTTAAAACCTGACACT-3′)
and reverse primer (5′-CCAAGACAATCTGGCCTCTATC -3′);
DR4 forward primer (5′-GGAACACAGCATGTCAGTGCAA-3′)
and reverse primer (5′-TGTCACTCCAGGGCGTACAATC-3′),
and GAPDH forward primer (5′-GTGAGGAGGGGAGATTCAG-3′)
and reverse primer (5′-GCATCCTGGGCTACACTG-3′).
GAPDH was used as an internal control. Three separate experiments were performed for each assay.
Western blot analysis
The cultured K562 cells were harvested and lysed with protein extraction buffer. The concentration of protein was detected with a BCA kit (Beyotime, Shanghai, China), then equal amounts of cell extracts were separated on 12% SDS-PAGE gel and transferred into polyvinylidene fluoride (PVDF) membranes. Afterwards, the membranes were blocked with 5% nonfat milk and then incubated with primary antibodies against cyclin E1, CDK2, p27, CD42b, CD11b, CD14, GATA-1, Bcl-2, Bax, Cleaved Caspase-3, Caspase-3, DR4, GAPDH (Abcam, Cambridge, MA, USA), and β-globin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. Then, horseradish peroxidase-linked secondary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were added and incubated for 2 h at room temperature. The bands were visualized with ECL-PLUS/Kit (Beyotime, China) and quantified by Gel-Pro analysis software (Media Cybernrtics, Rockville, MD, USA). Three separate experiments were performed for each assay.
Cell apoptosis
Flow cytometry analysis was performed to analyze the apoptosis status of K562 cells using the Annexin V-FITC/PI apoptosis detection kit (Beijing Biosea Biotechnology, Beijing, China). Briefly, after being collected and washed with ice-cold PBS, cells were resuspended in binding buffer containing FITC-annexin V and PI. Then, cells were incubated in the dark for 15 min at room temperature and analyzed by use of a flow cytometer (Beckman Coulter, USA). Three separate experiments were performed for each assay.
Methylation-specific PCR (MSP)
The genomic DNA of cell was extracted according to the DNA extraction kit (Sangon Biotech Co., Shanghai, China). The methylation of DNA was performed using an EZ DNA Methylation Lighting Kit. The sequence of methylation primer (M) and non-methylation primer (U) of death receptor 4 (DR4) gene were: upstream (M): 5′-TTCGAATTTCGGGAGCGTAGC-3′, downstream (M): 5′-GTAATTCAATCCTCCCCGCGA-3′, and upstream (U): 5′-GTAGTGATTTTGAATTTTGGGAGTGTAGT-3′, downstream (U): 5′-CTCATAATTCAATCCCCACAA-3. PCR amplication was performed with an EpiTect MSP kit (Qiagen, Valencia, CA). The amplified products were analyzed by 40 g/L agarose gel electrophoresis (Beijing Liuyi Biotechnology Co., Beijing, China) at 100 V for 30 min, and gel imaging was observed and photographed under ultraviolet light.
Statistical analysis
All analyses were conducted using SPSS version 20.0 (IBM Co., Armonk, NY). Data are represented as means and SD of values obtained in at least 3 independent experiments. One-way ANOVA was used for statistical analyses. P<0.05 was regarded as statistically significant.
Results
Inhibition of SNHG5 suppressed the proliferation of K562 cells
To explore the inhibitory effect of SNHG5 on cell proliferation, we designed 2 different shRNAs against SNHG5 for transfection into K562 cells. The qPCR results showed that both SNHG5-shRNA1 and SNHG5-shRNA2 obviously decreased SNHG5 expression in K562 cells (Figure 1). As there was lower expression of SNHG5 when cells were transfected with SNHG5-shRNA2, we selected SNHG5-shRNA2 for use in subsequent experiments.
Figure 1.
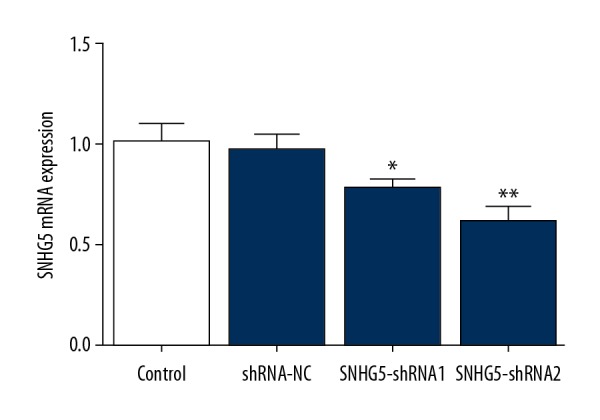
K562 cells were transfected with SNHG5-shRNA. The interference efficiencies in K562 cells were detected by RT-PCR. Data are expressed as the mean± SD of at least 3 experiments. * p<0.05, **p<0.01 vs. control.
As shown in Figure 2A, CCK-8 assay was performed to determine the effect of SNHG5 on cell proliferation. Downregulated SNHG5 significantly decreased the ability of cell proliferation, and the cell proliferation ability declined to nearly 50% when K562 cells were transfected with SNHG5-shRNA2 for 48 h. Western blot assay showed that downregulated SNHG5 significantly decreased the expression of cyclin-dependent kinase (CDK2) and cyclin E1, and increased the expression of p27, which is characterized as an anti-oncogene (Figure 2B, 2C). These results indicated that downregulation of SNHG5 dramatically suppressed the proliferation of K562 cells.
Figure 2.
Inhibition of SNHG5 significantly suppressed K562 cell growth. Cell proliferation ability of K562 cells was analyzed by CCK-8 assay at 24 h, 48 h, and 72 h (A). Western blot analysis was performed to examine the protein expression of cyclin E1 (B). Western blot analysis was performed to examine the protein expression cyclin-dependent kinase (CDK2) and p27 (C). Data are expressed as the mean ±SD of at least 3 experiments. * p<0.05, ** p<0.01, *** p<0.001 vs. control.
Inhibition of SNHG5 induced differentiation of K562 cells
To examine the potential role of SNHG5 in CML, we evaluated the effects of SNHG5 on cell differentiation. As shown in Figure 3A, the protein expression of CD42b, CD11b, and CD14 were increased with the transfection of SNHG5-shRNA2, indicating that K562 cells were induced to differentiate to mature granulocytes, monocytes, and megakaryocyte, respectively. GATA-1 is a vital nuclear transcription factor that regulates cell differentiation and erythroid cell development. β-globin was also considered as a marker of cell differentiation. The results showed that protein expressions of GATA-1 and β-globin were notably increased (Figure 3B), suggesting that downregulated SNHG5 induced K562 cells differentiation to erythrocytes. These results show that the inhibition of SNHG5 induced differentiation of K562 cells.
Figure 3.
Inhibition of SNHG5 induced the differentiation of K562 cells. Western blot analysis was performed to detect the protein expression of CD42b, CD11b, CD14 (A), GATA-1, and β-globin (B). Data are expressed as the mean ±SD of at least 3 experiments. ** p<0.01, *** p<0.001 vs. control.
Inhibition of SNHG5 induced cell apoptosis
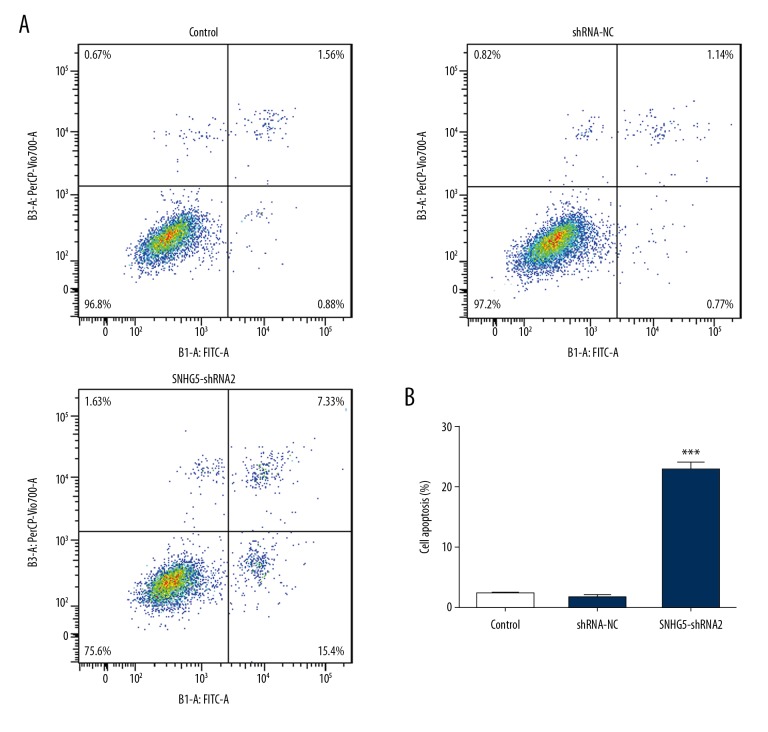
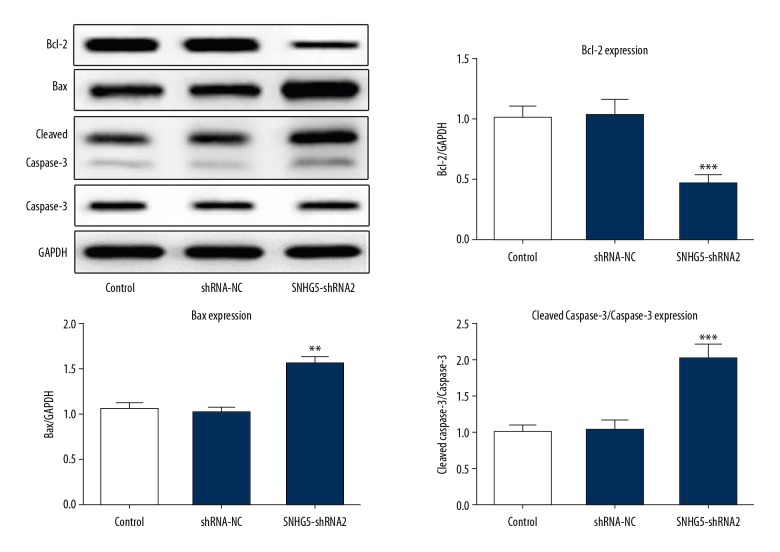
We explored whether SNHG5 depletion inhibited cell growth through promoting the apoptosis of K562 cells. Annexin V-FITC/PI flow cytometry results showed that the apoptosis rate was notably increased in SNHG5-shRNA2-transfected cells (Figure 4A, 4B). The activity of apoptosis marker caspase-3 and the Bcl-2 family proteins, Bcl-2, and Bax, were estimated using Western blot analysis (Figure 5). We detected a decrease in Bcl-2 and an increase in Bax and caspase-3 following SNHG5 downregulation. Taken together, these results suggest that inhibition of SNHG5 induced cell apoptosis of K562 cells.
Figure 4.
Inhibition of SNHG5 induced cell apoptosis as shown by flow cytometry analysis (A) and a quantification of cell apoptosis proportion was generated (B). Data are expressed as the mean ±SD of at least 3 experiments. *** p<0.001 vs. control.
Figure 5.
Inhibition of SNHG5 significantly upregulated the expression of Bax and cleaved caspase-3, but downregulated the expression of Bcl-2 compared to the control in K562 cells. Data are expressed as the mean ±SD of at least 3 experiments. ** p<0.01, *** p<0.001 vs. control.
Inhibition of SNHG5 suppressed methylation of DR4
To further investigate the mechanism and effect of SNHG5 on apoptosis of K562 cells, we assessed the expression of DR4. It has been reported that aberrant methylation of the DR4 gene promoter region in cancers results in decreased expression of DR4, and then the occurrence and development of tumors [20]. The results showed that inhibition of SNHG5 obviously increased the protein expression and mRNA level of DR4 (Figure 6A, 6B). Additionally, MSP analysis provided evidence that inhibition of SNHG5 suppressed the methylation of DR4 CpG islands (Figure 6C). Taken together, these results indicate that SNHG5 regulates the expression of DR4 through inducing the methylation of its CpG island, thus regulating cell apoptosis in CML.
Figure 6.
Inhibition of SNHG5 changed the expression and methylation of DR4. The protein expression of DR4 was determined by Western blot (A) and the mRNA level of DR4 gene was analyzed by qRT-PCR (B). The methylation of DR4 CpG sites was analyzed by methylation-specific PCR (MSP) analysis in K562 cells (C). Data are expressed as the mean ±SD of at least 3 experiments. *** p<0.001 vs. control.
Discussion
CML is a hematopoietic stem cell disorder characterized as the arrest of leukocyte differentiation and inhibition of apoptosis at the promyelocyte stage. It has been reported that some anti-cancer drugs can inhibit the growth of leukemia cells by inducing cell apoptosis or differentiation [21]. Thus, it is necessary to seek new therapeutic treatments and potential mechanisms to improve the prognosis for CML patients.
Apoptosis is one of the main regulatory mechanisms of CML K562 cells. In the present study, we evaluated the anti-proliferative and apoptosis-inducing effect of downregulated SNHG5 on CML K562 cells. The results of CCK-8 assay showed that the inhibition of SNHG5 inhibited proliferation and growth of K562 cells, and flow cytometry analysis results indicated that inhibition of SNHG5 significantly induced cell apoptosis. Apoptosis is an important cell death mechanism induced by stimulation via regulating the balance between Bcl-2 and Bax, which play a key role in determining whether cells undergo apoptosis [22]. Bcl-2 and Bax belong to the Bcl-2 family, and Bcl-2 acts as an anti-apoptotic protein while Bax acts as a pro-apoptotic protein [23,24]. Therefore, we assessed the effect of activation of the pro-apoptotic factor Bax, together with suppression of anti-apoptotic factor Bcl-2. The results indicated that inhibition of SNHG5 increased the expression of Bax while downregulating the expression of Bcl-2. Cleaved caspase-3, which has a key role in apoptosis, is closely associated with cell growth. Our results showed that the level of cleaved caspase-3 was increased, accelerating the apoptosis of K562 cells.
Differentiation is another main regulatory mechanism for CML K562 cells. In the process of differentiation and maturation, the antigenic markers on the surface of hematopoietic cells change constantly. Different antigens can be markers of different stages for cell differentiation. CD11b is a marker of differentiation in mature granulocytes, expressed in neutrophils, alkaline granulocytes, acidic granulocytes, and monocytes [25]. CD14 is the marker antigen of monocytes, and is mainly expressed in monocytes [26]. CD42b is the megakaryocyte differentiation antigen [10]. In the present study, the results showed that downregulation of SNHG5 significantly increased the expression of CD11b, CD14, and CD42b, indicating that K562 cells tended to differentiate into mature granulocytes, monocytes, and megakaryocyte. Hemoglobin synthesis, the specific marker for erythroid differentiation, was determined by the expression of globin genes and heme synthesis. GATA-1 is involved in transcriptional activation of globin genes and binds to the β-globin locus to regulate the expression of β-globin [27]. Our results showed that downregulation of SNHG5 significantly increased the expression of GATA-1 and β-globin, as expected, indicating enhanced globin genes-induced hemoglobin synthesis and erythroid differentiation of K562 cells.
Studies have demonstrated that tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) exerts effective antitumor activity, and TRAIL-induced apoptosis of carcinoma cells mainly results from combining with its death receptors, such as DR4, activating and conducting apoptotic signals through the death domain, and initiating a cascade reaction, leading to cell apoptosis [28–30]. Therefore, high expression of DR4 can promote the TRAIL-induced apoptosis of carcinoma cells. DR4 plays the dominant role in death signal transduction in human B chronic lymphatic leukemia, and also plays a crucial role in the occurrence, development, and prognosis of acute myeloid leukemia [30,31]. Aberrant methylation of the DR4 gene promoter region has been reported to cause decreased expression of DR4 genes, and then the occurrence, development, and invasion of cancers [20,32]. It has been demonstrated that Decitabine terminates the methylation level of DR4 gene promoters and restores the mRNA expression of DR4, thus inhibiting the proliferation of K562 cells [30]. In this study, we performed qRT-PCR and Western blot analyses to examine the level of DR4 protein and mRNA, respectively, and conducted MSP to determine DR4 gene promoter methylation status in CML K562 cells. With the transfection of SNHG5-shRNA2, the mRNA and protein expression of DR4 was higher, and DR4 gene promoter region CpG islands of K562 cells exhibited lower methylation levels. These findings suggest that downregulation of SNHG5 inhibits DR4 gene promoter methylation, which upregulates the expression level of the DR4 gene, partly resulting in an increased apoptosis of K562 cells.
Conclusions
Collectively, downregulating SNHG5 effectively inhibited the proliferation and induced apoptosis of K562 cells, which was likely due to terminating the methylation of DR4 gene promoters and preserving the mRNA and protein expression of DR4. Thus, the findings of this study may provide new experimental data for the investigation of the underlying mechanism of CML and provide a novel strategy for the treatment of CML.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Quintas-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113(8):1619–30. doi: 10.1182/blood-2008-03-144790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaleem B, Shahab S, Ahmed N, Shamsi TS. Chronic myeloid leukemia – prognostic value of mutations. Asian Pac J Cancer Prev. 2015;16(17):7415–23. doi: 10.7314/apjcp.2015.16.17.7415. [DOI] [PubMed] [Google Scholar]
- 3.Usmani SZ, Yunus SA, Jamal Y. Overview of chronic myeloid leukemia patients in Pakistan in the pre-imatanib era. Asian Pac J Cancer Prev. 2009;10(6):1039–40. [PubMed] [Google Scholar]
- 4.Kantarjian H, O’Brien S, Jabbour E, et al. Improved survival in chronic myeloid leukemia since the introduction of imatinib therapy: A single-institution historical experience. Blood. 2012;119(9):1981–87. doi: 10.1182/blood-2011-08-358135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005;25(4):1215–27. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsumura KR, DeVilbiss AW, Pope NJ, et al. Transcriptional mechanisms underlying hemoglobin synthesis. Cold Spring Harb Perspect Med. 2013;3(9):a015412. doi: 10.1101/cshperspect.a015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16(1):137–47. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Rowley PT, Ohlsson-Wilhelm BM, Farley BA, LaBella S. Inducers of erythroid differentiation in K562 human leukemia cells. Exp Hematol. 1981;9(1):32–37. [PubMed] [Google Scholar]
- 9.Gahmberg CG, Andersson LC. K562 – a human leukemia cell line with erythroid features. Semin Hematol. 1981;18(1):72–77. [PubMed] [Google Scholar]
- 10.Han SH, Kim J, Her Y, et al. Phytosphingosine promotes megakaryocytic differentiation of myeloid leukemia cells. BMB Rep. 2015;48(12):691–95. doi: 10.5483/BMBRep.2015.48.12.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vieira Torquato HF, Ribeiro-Filho AC, Buri MV, et al. Canthin-6-one induces cell death, cell cycle arrest and differentiation in human myeloid leukemia cells. Biochim Biophys Acta Gen Subj. 2017;1861(4):958–67. doi: 10.1016/j.bbagen.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernando TR, Contreras JR, Zampini M, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16(1):126. doi: 10.1186/s12943-017-0692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lammens T, Durinck K, Wallaert A, et al. Long non-coding RNAs in leukemia: Biology and clinical impact. Curr Opin Hematol. 2017;24(4):353–58. doi: 10.1097/MOH.0000000000000354. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Y, Jiao C, Lin Y, et al. lncRNA UCA1 contributes to imatinib resistance by acting as a ceRNA against miR-16 in chronic myeloid leukemia cells. DNA Cell Biol. 2017;36(1):18–25. doi: 10.1089/dna.2016.3533. [DOI] [PubMed] [Google Scholar]
- 16.Ma Z, Xue S, Zeng B, Qiu D. lncRNA SNHG5 is associated with poor prognosis of bladder cancer and promotes bladder cancer cell proliferation through targeting p27. Oncol Lett. 2018;15(2):1924–30. doi: 10.3892/ol.2017.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damas ND, Marcatti M, Come C, et al. SNHG5 promotes colorectal cancer cell survival by counteracting STAU1-mediated mRNA destabilization. Nat Commun. 2016;7:13875. doi: 10.1038/ncomms13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Sun CK. Long noncoding RNA SNHG5 is up-regulated and serves as a potential prognostic biomarker in acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2018;22(11):3342–47. doi: 10.26355/eurrev_201806_15154. [DOI] [PubMed] [Google Scholar]
- 19.He B, Bai Y, Kang W, et al. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res. 2017;7(8):1704–13. [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KH, Lim SW, Kim HG, et al. Lack of death receptor 4 (DR4) expression through gene promoter methylation in gastric carcinoma. Langenbecks Arch Surg. 2009;394(4):661–70. doi: 10.1007/s00423-009-0484-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Wang L, Pan XJ, Zhang H. Monocytic differentiation of K562 cells induced by proanthocyanidins from grape seeds. Arch Pharm Res. 2012;35(1):129–35. doi: 10.1007/s12272-012-0114-y. [DOI] [PubMed] [Google Scholar]
- 22.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science. 1997;275(5303):1132–36. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 23.Del Gaizo Moore V, Schlis KD, Sallan SE, et al. BCL-2 dependence and ABT-737 sensitivity in acute lymphoblastic leukemia. Blood. 2008;111(4):2300–9. doi: 10.1182/blood-2007-06-098012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nechushtan A, Smith CL, Hsu YT, Youle RJ. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 1999;18(9):2330–41. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason KD, Juneja SK, Szer J. The immunophenotype of acute myeloid leukemia: Is there a relationship with prognosis? Blood Rev. 2006;20(2):71–82. doi: 10.1016/j.blre.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Ziegler-Heitbrock HW, Ulevitch RJ. CD14: Cell surface receptor and differentiation marker. Immunol Today. 1993;14(3):121–25. doi: 10.1016/0167-5699(93)90212-4. [DOI] [PubMed] [Google Scholar]
- 27.Yu CH, Cui NX, Wang Y, et al. Changes in DNA methylation of erythroid-specific genes in K562 cells exposed to catechol in long term. Toxicol In Vitro. 2017;43:21–28. doi: 10.1016/j.tiv.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 28.Maldonado ME, Bousserouel S, Gosse F, et al. Implication of NF-kappaB and p53 in the expression of TRAIL-death receptors and apoptosis by apple procyanidins in human metastatic SW620 cells. Biomedica. 2010;30(4):577–86. [PubMed] [Google Scholar]
- 29.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: Decisions between life and death. Int J Biochem Cell Biol. 2007;39(7–8):1462–75. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Chen Y, Pei X, et al. Effects of Decitabine on the proliferation of K562 cells and the expression of DR4 gene. Saudi J Biol Sci. 2018;25(2):242–47. doi: 10.1016/j.sjbs.2017.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacFarlane M, Inoue S, Kohlhaas SL, et al. Chronic lymphocytic leukemic cells exhibit apoptotic signaling via TRAIL-R1. Cell Death Differ. 2005;12(7):773–82. doi: 10.1038/sj.cdd.4401649. [DOI] [PubMed] [Google Scholar]
- 32.Horak P, Pils D, Kaider A, et al. Perturbation of the tumor necrosis factor – related apoptosis-inducing ligand cascade in ovarian cancer: Overexpression of FLIPL and deregulation of the functional receptors DR4 and DR5. Clin Cancer Res. 2005;11(24 Pt 1):8585–91. doi: 10.1158/1078-0432.CCR-05-1276. [DOI] [PubMed] [Google Scholar]