Abstract
Bacterial and fungal communities in biofilms are important components in driving biogeochemical processes in stream ecosystems. Previous studies have well documented the patterns of bacterial alpha diversity in stream biofilms in glacier-fed streams, where, however, beta diversity of the microbial communities has received much less attention especially considering both bacterial and fungal communities. A focus on beta diversity can provide insights into the mechanisms driving community changes associated to large environmental fluctuations and disturbances, such as in glacier-fed streams. Moreover, modularity of co-occurrence networks can reveal more ecological and evolutionary properties of microbial communities beyond taxonomic groups. Here, integrating beta diversity and co-occurrence approach, we explored the network topology and modularity of the bacterial and fungal communities with consideration of environmental variation in glacier-fed streams in Central Asia. Combining results from hydrological modeling and normalized difference of vegetation index, this study highlighted that hydrological variables and vegetation status are major variables determining the environmental heterogeneity of glacier-fed streams. Bacterial communities formed a more complex and connected network, while the fungal communities formed a more clustered network. Moreover, the strong interrelations among the taxonomic dissimilarities of bacterial community (BC) and modules suggest they had common processes in driving diversity and taxonomic compositions across the heterogeneous environment. In contrast, fungal community (FC) and modules generally showed distinct driving processes to each other. Moreover, bacterial and fungal communities also had different driving processes. Furthermore, the variation of BC and modules were strongly correlated with hydrological properties and vegetation status but not with nutrients, while FC and modules (except one module) were not associated with environmental variation. Our results suggest that bacterial and fungal communities had distinct mechanisms in structuring microbial networks, and environmental variation had strong influences on bacterial communities but not on fungal communities. The fungal communities have unique assembly mechanisms and physiological properties which might lead to their insensitive responses to environmental variations compared to bacterial communities. Overall, beyond alpha diversity in previous studies, these results add our knowledge that bacterial and fungal communities have contrasting assembly mechanisms and respond differently to environmental variation in glacier-fed streams.
Keywords: Biofilm, Co-occurrence, Hydrology, Dissimilarity, Modules, Microbial community
Introduction
Glaciers cover approximately 10% of the land surface on the Earth (Milner et al., 2017) and are important components of the hydrological cycle providing vital water resources (Barnett, Adam & Lettenmaier, 2005; Gardner et al., 2013; Zemp et al., 2015). However, glaciers are shrinking rapidly across the world due to accelerating global warming (Immerzeel, Van Beek & Bierkens, 2010; Sorg et al., 2012; Marzeion et al., 2014), and most of them are expected to disappear by 2050 (Zemp et al., 2006; IPCC, 2014). As a prominent component of the glacier forefront, glacier-fed streams have a highly heterogeneous environment due to longitudinal alterations of landcover, river hydrology and morphology, sediment transport, and biogeochemical processes (Hood & Scott, 2008; Laghari, 2013; Hotaling, Hood & Hamilton, 2017; Milner et al., 2017). For example, from glacier terminus to downstream, terrestrial vegetation increases (Zhang et al., 2013; Raynolds et al., 2015), stream channel lengthens (Milner, Brown & Hannah, 2009; Robinson, Thompson & Freestone, 2014), and water source compositions changes (Brown, Hannah & Milner, 2003).
Biofilms are hot spots of microbial diversity and activity in stream ecosystems (Geesey et al., 1978; Battin et al., 2016). Within stream biofilms, bacteria, fungi, and algae are the major components driving the bulk of metabolism and biogeochemical processes (Brittain & Milner, 2001; Battin et al., 2003; Von Schiller et al., 2007). The changing environment presents significant challenges for glacier-fed stream ecosystems. Previous studies have revealed that factors associated with glacier shrinkage have significant influences on the composition, diversity, and functional potential of bacterial communities in stream biofilms (Wilhelm et al., 2013, 2014; Ren, Gao & Elser, 2017; Ren et al., 2017). However, fungal communities in glacial systems are rarely studied (Edwards, 2015; Anesio et al., 2017). With the decrease in elevation, glacier coverage, and glacier source contribution to streamflow, as well as increase in distance to glacier terminus, bacterial communities showed increased alpha diversity as well as distinct taxonomic and functional compositions (Wilhelm et al., 2013, 2014; Ren, Gao & Elser, 2017; Ren et al., 2017). Biodiversity is important for generating and stabilizing ecosystem structure and functions (Loreau et al., 2001; Tilman, Isbell & Cowles, 2014). The positive effects of local species richness (alpha diversity) on ecosystem functioning have been widely confirmed by a growing number of studies (Chapin et al., 2000; Cardinale et al., 2012; Duffy, Godwin & Cardinale, 2017). However, comparing to alpha diversity, beta diversity is an underexplored facet of biodiversity (Mori, Isbell & Seidl, 2018), which accumulates from compositional variations among local assemblages and provides insights into the mechanisms underlining biodiversity changes and their ecological consequences (Anderson et al., 2011; Socolar et al., 2016). For ecological communities suffering intensive environmental fluctuations and disturbances, focusing on beta diversity is especially important (Mori, Isbell & Seidl, 2018). In addition, microorganisms in many environments often coexist in a complex network with positive and negative interactions among members, playing pivotal roles in community assembly (Fuhrman, 2009; Barberán et al., 2012; Shi et al., 2016). These interactions may imply biologically or biochemically meaningful relationships between microorganisms (Weiss et al., 2016). Microbial co-occurrence networks can reveal how taxa potentially interact with each other, how diverse taxa structure networks, and how networks are compartmentalized into modules of closely associated taxa, as well as how microbial communities responded to environmental variations (Newman, 2006; Fuhrman, 2009; De Menezes et al., 2015; Banerjee et al., 2016). In addition, modularity (the tendency of a network to contain sub-clusters of nodes) is an important ecological feature in many biological systems, providing opportunities to identify highly connected taxa and integrate high dimension data into predicted ecological modules (De Menezes et al., 2015; Shi et al., 2016). A module is defined as a group of densely connected operational taxonomic units (OTUs), which have less links with OTUs belonging to other modules (Shi et al., 2016), forming a clustered network topology (Barberán et al., 2012). Modules can help to reveal more ecological and evolutionary properties (Thompson, 2005; Olesen et al., 2007), which are easily overlooked when communities are studied as a whole or in taxonomic groups (Porter, Onnela & Mucha, 2009; Bissett et al., 2013; De Menezes et al., 2015). The relationships between microbial modules and environmental variables can improve our understanding of the influences of environmental variation on microbial community assembly (Lindström & Langenheder, 2012; De Menezes et al., 2015; Toju et al., 2016). However, previous studies in glacier-fed streams have only focused on the whole communities or certain taxonomic groups of bacteria and fungi (Robinson & Jolidon, 2005; Milner, Brown & Hannah, 2009; Wilhelm et al., 2013; Ren, Gao & Elser, 2017). The network and modularity features of bacterial and fungal communities in glacier-fed streams are remaining one of our knowledge gaps. Integrating beta diversity and network modularity can provide novel insights into assembly mechanisms of microbial communities in glacier-fed streams.
Glacier-fed streams in Tian Shan Mountains in Central Asia are particularly vulnerable to climate change, where glaciers contribute significantly to stream runoff (Aizen et al., 1997; Hagg et al., 2007; Sorg et al., 2012). Glacier shrinkage has been observed in the past decades (Farinotti et al., 2015) and will accelerate in the coming decades as temperature increases (Kraaijenbrink et al., 2017; Gao et al., 2018). Here, we investigated bacterial and fungal communities in two glacier-fed streams using high-throughput sequencing combined with hydrological modeling. We aimed to examine the microbial co-occurrence networks (considering both fungal and bacterial communities) and to assess their response patterns to environmental variation in these glacier-fed streams. In glacier foreland soil, bacterial and fungal communities had contrasting community structures and response patterns to environmental variables (Blaalid et al., 2012; Bradley, Singarayer & Anesio, 2014; Brown & Jumpponen, 2014). Glacier-fed streams have intimate connections with terrestrial ecosystems in the glacier foreland through multiple ways (Crump, Amaral-Zettler & Kling, 2012; Gutiérrez et al., 2015; Ruiz-González, Niño-García & Del Giorgio, 2015). Thus, we hypothesize that bacterial and fungal communities in glacier-fed streams have contrasting assembly mechanisms and respond differently to environmental variation.
Materials and Methods
Study area
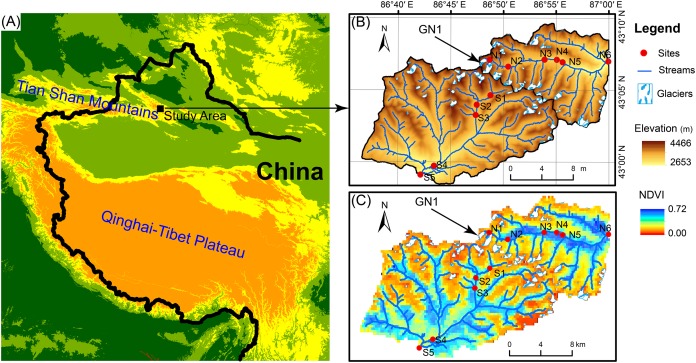
The Tian Shan mountains, known as the “water tower of Central Asia,” span a large area of Central Asia from northwestern China to southeastern Kazakhstan and from Kyrgyzstan to Uzbekistan (Fig. 1). In Tian Shan area, glaciers contribute considerably to water resources and play an important role in streamflow regimes (Sorg et al., 2012; Unger-Shayesteh et al., 2013). In China’s portion of the Tian Shan Mountains, there are 7,934 glaciers with a total area of 7,179 km2 and a total volume of 707 km3 (Guo et al., 2014). However, glaciers in the Tian Shan Mountains are extremely sensitive to global warming (Kraaijenbrink et al., 2017) and have been shrinking rapidly due to climate warming since the 1970s (Narama et al., 2010; Farinotti et al., 2015). For example, Urumqi Glacier No. 1 (GN1, 43°06′N, 86°49′E) is located in eastern Tian Shan Mountain at the headwater of the Urumqi River (Fig. 1) and retreated from an area of 1.95 km2 in 1962 to 1.65 km2 in 2010 (Zhang et al., 2014). In 2050, GN1 will likely lose up to 54% of the glacier area and 79% of the ice volume relative to 1980 (Gao et al., 2018).
Figure 1. Map of the study area.
(A) Location of the study area. (B) Elevation distribution of the study area. GN1 represents Urumqi Glacier No. 1. (C) The Normalized difference of vegetation index (NDVI) of the study area.
Field sampling
In June 2016, we investigated two glacier-fed streams in the Tian Shan Mountain. Water samples and benthic biofilm samples were collected from 11 sample sites in total spanning from the elevation of 3,828 to 2,646 m (Fig. 1). The sample sites were chosen along the streams in order to have heterogeneous environments, including different land vegetation and hydrological properties such as glacier contributions to stream flow. However, due to the constraint of accessibility, the sites were not spaced at equal intervals. At each sample site, six to nine submerged rocks were randomly sampled from the stream cross section below 10 cm. A sterilized nylon brush was used to remove the benthic biofilm from each stone in an area of 4.5 cm diameter on the upper surface. The slurry was rinsed with 500 mL sterile water. Approximately 10 mL of the mixed slurry was filtered through a 0.2-μm polycarbonate membrane filter (Whatman, Maidstone, UK) which was immediately frozen in liquid nitrogen in the field. After transported to the lab, the benthic biofilm samples were stored under −80 °C until DNA extraction. In addition, 500 mL water samples were collected for chemical analyses with three replications and stored under 4 °C.
Environmental factors
At each sample site, pH, conductivity (Cond), and elevation were measured in situ using a handheld pH meter (PHI 400 Series; Beckman Coulter, Brea, CA, USA), YSI meter (model 80, Yellow Springs, OH, USA), and GPS unit (Triton 500; Magellan, Santa Clara, CA, USA), respectively. Unfiltered water samples were directly used to measure total nitrogen (TN) and total phosphorus (TP). TN was analyzed by ion chromatography with prior persulfate oxidation (EPA 300.0). TP was analyzed using the ammonium molybdate method with prior oxidation (EPA 365.3). Filtered water samples (filtered with pre-combusted GF/F filters) were used to test nitrate (NO3−), ammonium (NH4+), soluble reactive phosphorus (SRP), and dissolved organic carbon (DOC). NO3− was analyzed using ion chromatography (EPA 300.0). NH4+ was analyzed using the indophenol colorimetric method (EPA 350.1). SRP was measured according to the ammonium molybdate method (EPA 365.3). DOC was measured using a total organic carbon (TOC) analyzer (TOC-VCPH; Shimadzu Scientific Instruments, Columbia, MD, USA). Water chemistry data was reported in our previous research (Ren, Gao & Elser, 2017).
In glacier-fed streams, both biotic and abiotic environments are tightly linked to the relative contributions of glacier melt and runoff to the stream flow (Milner et al., 2001; Hannah et al., 2007; Kuhn et al., 2011). According to the landscape-based hydrological model proposed by Gao et al. (2016, 2017), we classified the landscape into glaciated and non-glaciated. For each sample site, the proportion of glaciated area (GA) in its sub-catchment was calculated and the proportion of glacier source water (GS) in the total runoff was derived from the model (Gao et al., 2016, 2017). The hydrological distance to glacier terminus (GD) was measured according to the river channel network (Fig. 1). These hydrological parameters were also reported in our previous research (Ren, Gao & Elser, 2017).
In the study area, the vegetation (grassland) status was measured by the normalized difference vegetation index (NDVI) using the Terra moderate resolution imaging spectroradiometer Vegetation Indices (MOD13Q1) Version 6 data downloaded from USGS (https://earthexplorer.usgs.gov/) (Fig. 1). The MOD13Q1 product was generated on June 26, 2017 and has a resolution of 250 m. NDVI is calculated based on the absorption of red light and the reflection of infrared radiation by vegetation (Rouse et al., 1974). The equation is represented as NDVI = (NIR − RED)/(NIR + RED), where NIR is near infrared reflectance and RED is visible red reflectance. It has been demonstrated that NDVI exhibits close relationships with above-ground vegetation biomass and coverage (Carlson & Ripley, 1997; Eastwood et al., 1997; Ren et al., 2019). For each stream site, the average NDVI of its sub-catchment (the upstream area of the stream site) was calculated as the mean NDVI of each pixel in the sub-catchment.
DNA extraction, PCR, and sequencing
Bacterial 16S rRNA gene sequences and fungal 18S rRNA gene sequences were analyzed to determine the bacterial community (BC) and fungal community (FC), respectively. To determine the fungal community, the internal transcribed spacer regions and 18S rRNA genes are commonly used and provide similar results and congruent conclusions (Brown, Rigdon-Huss & Jumpponen, 2014). We used 18S rRNA gene sequencing to detect the fungal community in this study. DNA was extracted using the PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA, USA) following the manufacturer’s protocol. The V3–V4 regions of the 16S rRNA genes were amplified using the bacterium-specific forward and reverse primers 338F-ACTCCTACGGGAGGCAGCA and 806R-GGACTACHVGGGTWTCTAAT (Invitrogen, Vienna, Austria) (Huws et al., 2007; Masoud et al., 2011; Caporaso et al., 2012). The V4–V5 regions of the 18S rRNA genes were amplified using the fungus-specific forward and reverse primers 817F-TTAGCATGGAATAATRRAATAGGA and 1196R-TCTGGACCTGGTGAGTTTCC-3′ (Invitrogen, Vienna, Austria) (Borneman & Hartin, 2000). The forward primers were barcoded, and the barcodes were designed considering the balanced guanine–cytosine content, minimal homopolymer runs, and no self-complementarity of more than two bases to reduce internal hairpin propensity (Hawkins et al., 2018). PCR reaction systems were prepared using a Premix Taq Kit (Code No. RR902A; Takara, Kusatsu,, Japan) according to the manufacturer’s instructions. The total volume of each PCR reaction was 20 μL, containing 10 μL of 2×EX Premix Taq™ Polymerase, one μL of forward primer, one μL of reverse primer, one μL of NDA extraction, and seven μL of Nuclease-free water. The PCR reactions were conducted with a thermal cycler (ABI 2700, SeqGen, Torrance, CA, USA) with a temperature profile of 1-min hot start at 80 °C, followed by pre-denaturation at 94 °C for 5 min, 30 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 90 s), and a final extension at 72 °C for 10 min. The PCR amplicons were verified in 1.0% agarose with 1× TAE buffer using electrophoresis, purified using the Gel Extraction Kit (Qiagen, Hilden, Germany), and quantified by Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). One of the fungi sample (N2) was not successfully amplified. The purified and quantified DNA libraries were then pooled together according to their concentrations. The pooled library was sequenced on an Illumina MiSeq (PE300) sequencing platform.
Analyses
Raw sequence data of bacterial 16S rRNA (available at NCBI, PRJNA398147, SRP115356) and fungal 18S rRNA (available at NCBI, PRJNA542974, SRP198430) were processed using QIIME 1.9.0 (Caporaso et al., 2010). The forward and reverse reads were merged. The merged sequences were then assigned to samples based on the barcode. The barcode and primer sequence were cut off to truncate the sequences. The sequences with length >200 bp and mean quality score <20 were discarded. Using UCHIME algorithm (Edgar et al., 2011), the chimeric sequences were detected and removed. Finally, the effective sequences of 16S and 18S rRNA were grouped into OTUs against the Silva 132 database at 99% threshold.
All the analyzed environmental variables, including GD, GA, GS, NDVI, elevation, pH, Cond, TN, NO3−, NH4+, TP, SRP, and DOC, were standardized using the “normalize” method in decostand function in vegan 2.5-3 package (Oksanen et al., 2007). Spearman correlation analyses (cor function in stats v3.6.0 package) were used to assess the pairwise relationships between environmental variables and visualized using corrplot function in corrplot 0.84 package. Mantel tests (mantel function in vegan 2.5-3 package) were used to assess the relationships among spatial dissimilarity (represented by geographic distance), overall environmental distance, hydrological dissimilarity, nutrient dissimilarity, and vegetation dissimilarity. Partial Mantel tests (mantel.partial function in vegan 2.5-3 package) were used to assess those relationships by controlling nutrient dissimilarity. The geographic distance was calculated based on the GPS coordinates of sample sites using distGeo function in geosphere 1.5-10 package. The environmental distance was represented by Euclidean distance based on all analyzed environmental variables. Hydrological dissimilarity was represented by Euclidean distance based on GD, GA, and GS. Nutrient dissimilarity was represented by Euclidean distance based on TN, NO3−, NH4+, TP, and SRP. Vegetation dissimilarity was represented by Euclidean distance based on NDVI. Euclidean distances were calculated using vegdist function in vegan 2.5-3 package (Oksanen et al., 2007). All the above statistical analyses were conducted in R 3.5.1 (R Development Core Team, 2017).
The co-occurrence networks of bacterial and fungal communities were assessed and visualized using Cytoscape (version 3.7.1) (Shannon et al., 2003). In network analyses, the pairwise correlations between OTUs (OTUs with a relative abundance >0.01%) were calculated using Spearman’s correlation based on the relative abundance of OTUs (data was transformed by Hellinger transformation using decostand function in vegan 2.5-3 package). Only strong (R > 0.7 OR R < −0.7) and significant (P < 0.01) correlations were considered in network analysis. ClusterMaker app (Morris et al., 2011) was used to analyze the modular structures of the co-occurrence networks. Modularity values greater than 0.4 suggest that the network has a modular structure (Newman, 2006). The group attributes layout algorithm was used to construct the networks based on modules. The basic topological metrics of networks were calculated, including number of nodes, number of edges, clustering coefficient, characteristic path length, network density, network heterogeneity, and modularity. The taxonomic dissimilarities (beta diversity) of the BC and FC were calculated based on Bray–Curtis distance in terms of the relative abundance of OTUs using R package vegan 2.5-3 (Oksanen et al., 2007). Moreover, the taxonomic dissimilarities of the major modules (modules have more than 15 nodes) were also calculated based on Bray–Curtis distance in terms of the relative abundance of OTUs in the module. BM1, BM2, BM3, and BM4 represent four major modules in the bacterial network. FM1, FM2, FM3, FM4, FM5, and FM6 represent six major modules in the fungal network. Mantel tests were used to assess the correlations between spatial and environmental dissimilarities and taxonomic dissimilarities of bacterial and fungal communities and modules. The relationships among different modules and communities for bacteria and fungi were also assessed using Mantel tests.
Results
Environmental variations and community taxonomic dissimilarities
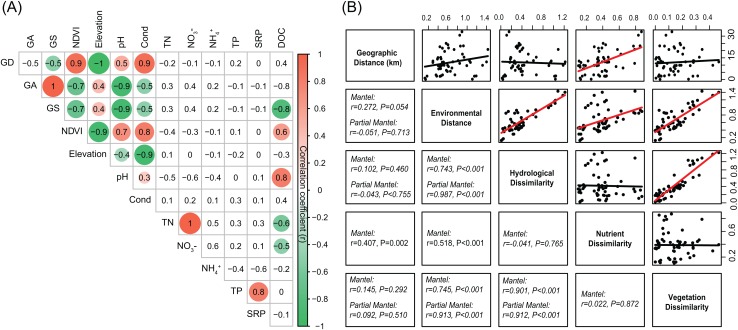
The environmental variables of the streams varied across the sampling sites and showed differing interrelationships (Fig. 2). GD, GA, GS, NDVI, elevation, pH, and conductivity were closely correlated with each other (Fig. 2A). DOC was negatively correlated with GS, TN, and NO3−, while positively correlated with NDVI and pH (Fig. 2A). Across the sampling sites, hydrological and vegetation dissimilarities had strong linear relationships with overall environmental dissimilarity, while nutrient dissimilarity only had a weak relationship (Fig. 2B). Moreover, hydrological and vegetation dissimilarities had a strong interrelationship with each other but without significant relationships to nutrient dissimilarity (Fig. 2B). Geographic distance only had a significant relationship with nutrient dissimilarity (Fig. 2B). In addition, Mantel tests showed that environmental, hydrological, and vegetation dissimilarities had significant positive relationships to taxonomic dissimilarities of BC but not to FC (Fig. 3). However, geographic distance and nutrient dissimilarity did not have significant relationships with taxonomic dissimilarities of both bacterial and fungal communities (Fig. 3).
Figure 2. (A) Pairwise-Spearman correlations between environmental variables.
The magnitude of correlation coefficient is only shown graphically with color scale when correlation is significant (P < 0.05). The color intensity and the size of the circle are proportional to the correlation coefficients. (B) Correlations between the different components of environmental dissimilarities. Spearman correlations (r) and associated P-values were calculated by Mantel test. Nutrient dissimilarity was controlled in Partial Mantel test.
Figure 3. Correlations between different components of environmental distance (environmental dissimilarity, hydrological dissimilarity, nutrient dissimilarity, and vegetation dissimilarity) and taxonomic dissimilarities of (A) bacterial and (B) fungal communities and modules.
Spearman correlations were calculated by Mantel test. The magnitude of correlation coefficient is only shown graphically with color scale when correlation is significant (P < 0.05). The color intensity and the size of the circle are proportional to the correlation coefficients.
Bacteria and fungi co-occurrence patterns and modular structures
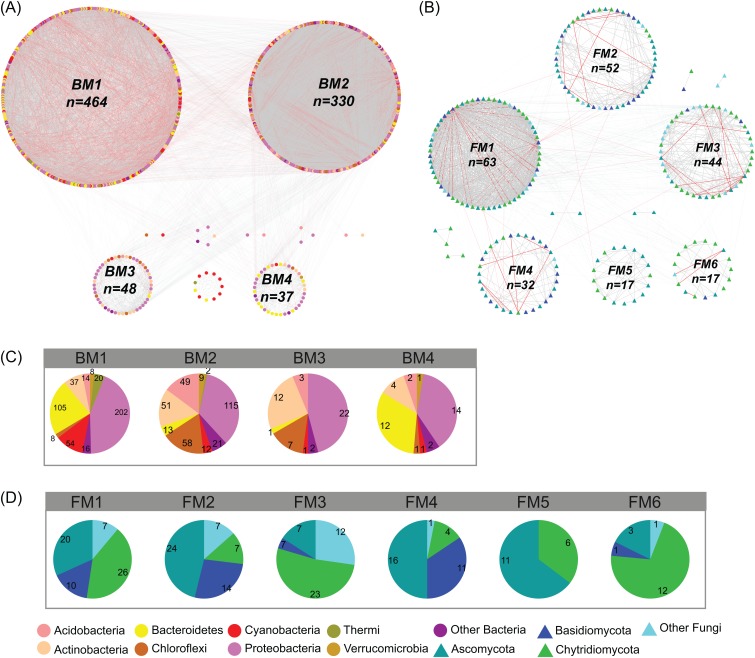
In the studied glacier-fed streams, bacteria and fungi formed complex co-occurrence networks (Fig. 4). The bacterial and fungal networks consisted of 904 and 238 nodes and 21,463 and 1,348 edges, respectively (Table 1). The bacterial network had a higher number of nodes and edges as well as a higher network density (Fig. 4; Table 1), indicating the bacterial network was more complex and connected than the fungal network. However, the fungal network exhibited a higher clustering coefficient, characteristic path length, network heterogeneity, and modularity, indicating that the fungal network had a more clustered topology than the bacterial network (Fig. 4; Table 1).
Figure 4. Co-occurrence network of (A) bacterial and (B) fungal communities.
Dot and triangle represent bacterial and fungal OTUs (OTUs with a relative abundance >0.01%), respectively. OTUs were colored according to major phylum (phylum with relative abundance >1%). Edges represent Spearman correlation relationships (P < 0.01). Gray lines indicate positive associations and pink lines indicate negative associations. The circles represent modules of the networks. The pie graphs in the below two panels show the composition of the modules in (C) bacterial and (D) fungal networks with the number representing the number of OTUs.
Table 1. Topological parameters of the network of bacterial and fungal communities.
| Topological parameters | Description | Bacterial | Fungal |
|---|---|---|---|
| Number of nodes | Number of OTUs in the network | 904 | 238 |
| Number of edges (in total) | Strong and significant correlations | 21,463 | 1,348 |
| Number of edges (positive) | Positive correlations | 18,419 | 1,253 |
| Number of edges (negative) | Negative correlations | 3,044 | 95 |
| Clustering coefficient | The fraction of observed vs. possible clusters for each node | 0.440 | 0.451 |
| Characteristic path length | The median of the means of the shortest path lengths connecting each vertex to all other vertices | 2.887 | 3.829 |
| Network density | The ratio of the number of edges and the number of possible edges | 0.053 | 0.048 |
| Network heterogeneity | Density distribution of connections between nodes | 0.929 | 1.107 |
| Modularity | Tendency of a network to contain sub-clusters of nodes | 0.52 | 0.599 |
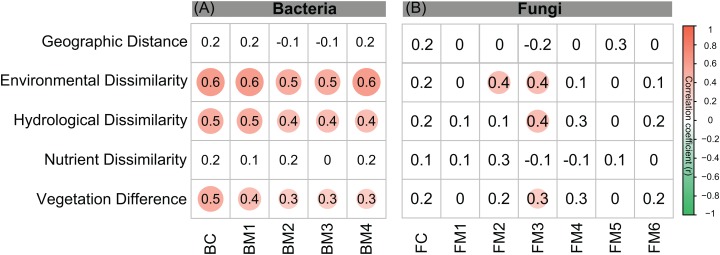
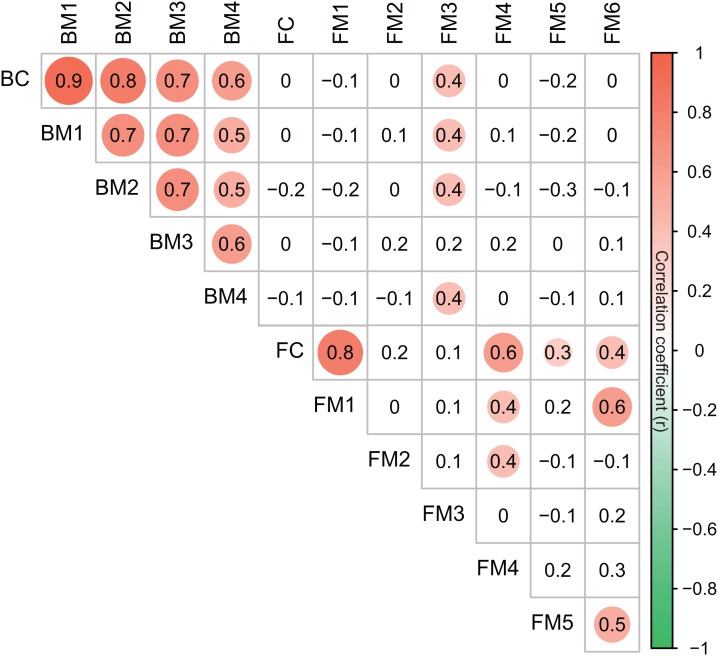
Co-occurrence networks can be compartmentalized into modules within which nodes are closely associated and are expected to share environmental preferences. We found that the OTUs in bacterial and fungal networks were grouped into four and six major modules (modules with more than 15 nodes), respectively (Figs. 4A and 4B). All the modules were formed by various microbial taxa (Figs. 4C and 4D), which shown differently across sample sites (Figs. S1 and S2). Modules had significantly different taxonomic compositions to each other (Tables S1 and S2). Mantel tests showed that the taxonomic dissimilarities of the BC and modules had strong interrelationships (Fig. 5). The taxonomic dissimilarities of FC and modules only had weak interrelationships. Between bacterial and fungal networks, only FM3 had strong relationships with BC and BMs. Mantel tests also revealed that all bacterial modules (BM1, BM2, BM3, and BM4) and one fungal module (FM3) were positively correlated with environmental, hydrological, and vegetation dissimilarities, but not with geographic distance and nutrient dissimilarity (Fig. 3).
Figure 5. Correlation matrix of the taxonomic dissimilarities for bacterial and fungal communities and modules.
The magnitude of correlation coefficient is only shown graphically with color scale when correlation is significant (P < 0.05). The color intensity and the size of the circle are proportional to Spearman correlation coefficients.
Discussion
Bacterial and fungal communities in stream biofilms are major components of glacier-fed stream ecosystems. The strong correlations between bacterial communities and environmental, hydrological, and vegetation characteristics suggest substantial influences of spatial heterogeneity and potential influences of glacier shrinkage on bacterial communities (Fig. 3). However, the variation of bacterial communities was not associated with stream nutrient variations and geographic distance. In our studied glacier-fed streams, the proportion of GA, the proportion of glacier source water (GS), the hydrological distance to glacier terminus (GD)and the NDVI were the environmental variables associated with longitudinal patterns of glacier-fed streams. Glacier-fed streams are fed by various sources, including ice-melt, snowmelt, and groundwater (Brown, Hannah & Milner, 2003). The contribution of different water sources varies longitudinally from the glacier terminus to downstream reaches and temporally with glacier shrinkage (Brittain & Milner, 2001; Milner, Brown & Hannah, 2009; Gao et al., 2016), resulting in distinct hydrology and physicochemical features which control the ecological structures and processes in glacier-fed streams (Brown, Hannah & Milner, 2003, 2007; Sertic Peric et al., 2015). Synchronizing with the hydrological changes in glacier-fed streams, vegetation growth and aboveground biomass in the catchment also has a clear elevation gradient (Carlyle, Fraser & Turkington, 2014) which will likely be amplified due to climate warming (Zhang et al., 2013; Raynolds et al., 2015). The changed landcover modifies terrestrial and aquatic biogeochemistry (Sadro, Nelson & Melack, 2012), and affects stream biofilms (Figueiredo et al., 2010; Nielsen et al., 2012). Thus, hydrological variables (GD, GA, and GS) and vegetation variable (NDVI) determine the environmental heterogeneity of glacier-fed streams and can potentially indicate glacier shrinkage. It has been widely demonstrated that glacier shrinkage alters watershed landcover and instream environments (Hood & Scott, 2008; Jacobsen & Dangles, 2012; Laghari, 2013; Raynolds et al., 2015; Milner et al., 2017), imposing impacts on bacterial communities (Nelson, Sadro & Melack, 2009; Hotaling, Hood & Hamilton, 2017). For example, the alpha diversity of biofilm bacteria decreased with the increases of elevation, the proportion of glacier area in the watershed, and the relative contribution of glacier sources to stream runoff (Wilhelm et al., 2013; Ren, Gao & Elser, 2017). Potential functions of bacterial communities are also significantly associated with hydrological factors (Ren et al., 2017). Our results further suggest strong influences of hydrological and vegetation characteristics on bacterial communities, leading to more different (biotic heterogenization) bacterial communities in glacier-fed streams.
In contrast to bacterial communities, environmental, hydrological, and vegetation dissimilarities were not significantly associated with the dissimilarity of fungal communities. Moreover, the variation fungal communities were also not associated to nutrient variations. The results suggest that FC variations were not affected by the environmental variation and might be insensitive to glacier shrinkage. We proposed that the unique responses of fungi may relate to the low temperature which can suppress the response of fungal communities to environmental variation. Fungi can survive and grow in harsh conditions with low temperatures such as glacier and snow by evolving various adaptive features (Hassan et al., 2016). These fungi are known as psychrophiles and psychrotrophs. Although they exist widely in cold environments, the optimum temperature for the growth of psychrophilic fungi is around 15 °C and for psychrotrophic fungi is 20 °C (Gounot, 1986; Robinson, 2001; Cavicchioli et al., 2002; Turchetti et al., 2008). In glacier-fed streams, water temperature is usually below 10 °C or even close to 0 °C in summer (Milner & Petts, 1994). More interestingly, the contrasting response patterns of bacterial and fungal communities were also found in glacier foreland soil ecosystems, where bacterial communities are strongly influenced by the presence of vegetation and environmental heterogeneity and show convergence (Brown & Jumpponen, 2014). In contrast to the bacterial communities in glacier foreland soil, fungal richness and diversity were more static and the community structure and distribution show a large extent of stochastic processes across the glacier foreland (Blaalid et al., 2012; Bradley, Singarayer & Anesio, 2014; Brown & Jumpponen, 2014). It has been revealed that bacteria and fungi in headwater streams are similar to communities in adjacent soil due to intimate associations between headwater streams and terrestrial ecosystems (Crump, Amaral-Zettler & Kling, 2012; Gutiérrez et al., 2015; Ruiz-González, Niño-García & Del Giorgio, 2015). The immigration and advection of allochthonous bacteria and fungi from terrestrial environments can influence bacterial and fungal communities in glacier-fed streams (Crump, Amaral-Zettler & Kling, 2012; Gutiérrez et al., 2015). The unique response of fungal communities in glacier-fed streams is congruent with the observations in the periglacial soils, suggesting differing trajectories of fungal and BC variations in glacier-fed streams.
The different patterns of bacterial and fungal communities in glacier-fed streams were further supported by network analysis, which showed that the bacterial communities formed a more complex and connected network, while the fungal communities formed a more clustered network (Fig. 4; Table 1). Microbial communities are complex assemblages comprised by highly interactive taxa (Fuhrman, 2009; Jones, Hambright & Caron, 2018). This study is the first to explore the organization of the bacterial and fungal communities in glacier-fed streams using a co-occurrence approach with consideration of the driving forces. In general, communities with tight co-occurrence interactions and high complexity have a lower stability and are more susceptible to disturbance (Montoya, Pimm & Sole, 2006; Saavedra et al., 2011). The highly connected and complex bacterial network suggests that bacterial communities in the glacier-fed streams were more sensitive to environmental variations, especially to instream hydrological properties and land vegetation. Moreover, in a complex network, the highly interconnected species are grouped into a module (Barabási & Oltvai, 2004; Newman, 2006). The strong interrelations among the taxonomic dissimilarities of BC and modules suggest that they had common processes in driving diversity and composition across various environments (Delgado-Baquerizo et al., 2018). In contrast, FC and modules generally had distinct driving processes to each other. Moreover, the fungal and bacterial communities also had different driving processes (except FM3 and bacterial communities and modules). Consistent with these findings, the variation of all bacterial modules and FM3 were strongly associated with environmental variation except nutrient dissimilarity, while fungal modules (except FM3) did not respond to environmental variation. Thus, our results suggest that, in our studied area, environmental variation had strong influences on bacterial communities and their assembly mechanisms but not on fungal communities (except one module) in biofilms of glacier-fed streams. The fungal communities may have unique assembly mechanisms which lead to their insensitive responses to environmental variations.
Conclusion
Glacier shrinkage imposes significant influences on glacier-fed streams. Integrating beta diversity, our study provides the first co-occurrence network analyses of bacterial and fungal communities in glacier-fed streams. Firstly, this study highlighted that hydrological variables and vegetation status are important components in determining environment heterogeneity of glacier-fed streams and are indicator variables of glacier shrinkage. Then we identified co-occurrence properties of the microbial communities and their responses to environmental variations. Bacterial communities formed a more complex and connected network, while the fungal communities formed a more clustered network. Nutrients were insignificant to the assemblies of both bacterial and fungal communities in these glacier-fed streams. However, hydrological properties and vegetation status impose significant influences on assemblies of the BC but not on the FC. The results suggest the influences of glacier shrinkage on bacterial communities. However, fungi communities might be insensitive to glacier shrinkage. The results would add our knowledge of microbial community assembly mechanisms and the responses of microbial communities to environmental variations caused by glacier shrinkage.
Supplemental Information
Figure S1 Distributions of OTUs across sample sites for bacterial modules.
Figure S2 Distributions of OTUs across sample sites for fungal modules.
Table S1 Pairwise dissimilarity tests of taxonomic composition among bacterial modules using PERMANOVA (adonis function in vegan package 2.5-3). R2 and P-values (in bracket) are shown.
Table S2 Pairwise dissimilarity tests of taxonomic composition among fungal modules using PERMANOVA (adonis function in vegan package 2.5-3). R2 and P-values (in bracket) are shown.
Acknowledgments
We are grateful to anonymous reviewers for the suggestions of writing and analysis, to Zhao QD, Han TD, and Ren Y for assistance in the field.
Funding Statement
This work was supported by the National Natural Science Foundation (41801036), National Key R&D Program of China (2017YFE0100700), the Key Program of National Natural Science Foundation of China (41730646), the project from the Key Laboratory for Mountain Hazards and Earth Surface Process, Institute of Mountain Hazards and Environment, Chinese Academy of Sciences (KLMHESP-17-02), and the project from the State Key Laboratory of Cryospheric Sciences, Cold and Arid Regions Environment and Engineering Research Institute, Chinese Academy of Sciences (SKLCS-OP-2016-04). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare that they have no competing interests.
Author Contributions
Ze Ren conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Hongkai Gao conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
References
- Aizen et al. (1997).Aizen VB, Aizen EM, Melack JM, Dozier J. Climatic and hydrologic changes in the Tien Shan, Central Asia. Journal of Climate. 1997;10:1393–1404. doi: 10.1175/1520-0442(1997)010<1393:CAHCIT>2.0.CO;2. [DOI] [Google Scholar]
- Anderson et al. (2011).Anderson MJ, Crist TO, Chase JM, Vellend M, Inouye BD, Freestone AL, Sanders NJ, Cornell HV, Comita LS, Davies KF, Harrison SP, Kraft NJB, Stegen JC, Swenson NG. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecology Letters. 2011;14(1):19–28. doi: 10.1111/j.1461-0248.2010.01552.x. [DOI] [PubMed] [Google Scholar]
- Anesio et al. (2017).Anesio AM, Lutz S, Chrismas NAM, Benning LG. The microbiome of glaciers and ice sheets. NPJ Biofilms Microbiomes. 2017;3(1):1–10. doi: 10.1038/s41522-017-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee et al. (2016).Banerjee S, Baah-Acheamfour M, Carlyle CN, Bissett A, Richardson AE, Siddique T, Bork EW, Chang SX. Determinants of bacterial communities in Canadian agroforestry systems. Environmental Microbiology. 2016;18(6):1805–1816. doi: 10.1111/1462-2920.12986. [DOI] [PubMed] [Google Scholar]
- Barabási & Oltvai (2004).Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nature Reviews Genetics. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Barberán et al. (2012).Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME Journal. 2012;6(2):343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett, Adam & Lettenmaier (2005).Barnett TP, Adam JC, Lettenmaier DP. Potential impacts of a warming climate on water availability in snow-dominated regions. Nature. 2005;438(7066):303–309. doi: 10.1038/nature04141. [DOI] [PubMed] [Google Scholar]
- Battin et al. (2016).Battin TJ, Besemer K, Bengtsson MM, Romani AM, Packmann AI. The ecology and biogeochemistry of stream biofilms. Nature Reviews Microbiology. 2016;14(4):251–263. doi: 10.1038/nrmicro.2016.15. [DOI] [PubMed] [Google Scholar]
- Battin et al. (2003).Battin TJ, Kaplan LA, Denis Newbold J, Hansen CME. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature. 2003;426(6965):439–442. doi: 10.1038/nature02152. [DOI] [PubMed] [Google Scholar]
- Bissett et al. (2013).Bissett A, Brown MV, Siciliano SD, Thrall PH. Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecology Letters. 2013;161:128–139. doi: 10.1111/ele.12109. [DOI] [PubMed] [Google Scholar]
- Blaalid et al. (2012).Blaalid R, Carlsen T, Kumar S, Halvorsen R, Ugland KI, Fontana G, Kauserud H. Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Molecular Ecology. 2012;21(8):1897–1908. doi: 10.1111/j.1365-294X.2011.05214.x. [DOI] [PubMed] [Google Scholar]
- Borneman & Hartin (2000).Borneman J, Hartin RJ. PCR primers that amplify fungal rRNA genes from environmental samples. Applied and Environmental Microbiology. 2000;66(10):4356–4360. doi: 10.1128/AEM.66.10.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, Singarayer & Anesio (2014).Bradley JA, Singarayer JS, Anesio AM. Microbial community dynamics in the forefield of glaciers. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1795):e20140882. doi: 10.1098/rspb.2014.0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittain & Milner (2001).Brittain JE, Milner AM. Ecology of glacier-fed rivers: current status and concepts. Freshwater Biology. 2001;46(12):1571–1578. doi: 10.1046/j.1365-2427.2001.00845.x. [DOI] [Google Scholar]
- Brown, Hannah & Milner (2003).Brown LE, Hannah DM, Milner AM. Alpine stream habitat classification: An alternative approach incorporating the role of dynamic water source contributions. Arctic, Antarctic, and Alpine Research. 2003;35(3):313–322. doi: 10.1657/1523-0430(2003)035[0313:ASHCAA]2.0.CO;2. [DOI] [Google Scholar]
- Brown, Hannah & Milner (2007).Brown LE, Hannah DM, Milner AM. Vulnerability of alpine stream biodiversity to shrinking glaciers and snowpacks. Global Change Biology. 2007;13(5):958–966. doi: 10.1111/j.1365-2486.2007.01341.x. [DOI] [Google Scholar]
- Brown & Jumpponen (2014).Brown SP, Jumpponen A. Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Molecular Ecology. 2014;23(2):481–497. doi: 10.1111/mec.12487. [DOI] [PubMed] [Google Scholar]
- Brown, Rigdon-Huss & Jumpponen (2014).Brown SP, Rigdon-Huss AR, Jumpponen A. Analyses of ITS and LSU gene regions provide congruent results on fungal community responses. Fungal Ecology. 2014;9:65–68. doi: 10.1016/j.funeco.2014.02.002. [DOI] [Google Scholar]
- Caporaso et al. (2010).Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, Mcdonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso et al. (2012).Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale et al. (2012).Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava DS, Naeem S. Biodiversity loss and its impact on humanity. Nature. 2012;486(7401):59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- Carlson & Ripley (1997).Carlson TN, Ripley DA. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sensing of Environment. 1997;62(3):241–252. doi: 10.1016/S0034-4257(97)00104-1. [DOI] [Google Scholar]
- Carlyle, Fraser & Turkington (2014).Carlyle CN, Fraser LH, Turkington R. Response of grassland biomass production to simulated climate change and clipping along an elevation gradient. Oecologia. 2014;174(3):1065–1073. doi: 10.1007/s00442-013-2833-2. [DOI] [PubMed] [Google Scholar]
- Cavicchioli et al. (2002).Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR. Low-temperature extremophiles and their applications. Current Opinion in Biotechnology. 2002;13(3):253–261. doi: 10.1016/S0958-1669(02)00317-8. [DOI] [PubMed] [Google Scholar]
- Chapin et al. (2000).Chapin FS, III, Zavaleta ES, Eviner VT, Naylor RL, Vitousek PM, Reynolds HL, Hooper DU, Lavorel S, Sala OE, Hobbie SE, Mack MC, Díaz S. Consequences of changing biodiversity. Nature. 2000;405(6783):234–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- Crump, Amaral-Zettler & Kling (2012).Crump BC, Amaral-Zettler LA, Kling GW. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME Journal. 2012;6(9):1629–1639. doi: 10.1038/ismej.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Menezes et al. (2015).De Menezes AB, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environmental Microbiology. 2015;17(8):2677–2689. doi: 10.1111/1462-2920.12559. [DOI] [PubMed] [Google Scholar]
- Delgado-Baquerizo et al. (2018).Delgado-Baquerizo M, Reith F, Dennis PG, Hamonts K, Powell JR, Young A, Singh BK, Bissett A. Ecological drivers of soil microbial diversity and soil biological networks in the Southern Hemisphere. Ecology. 2018;99(3):583–596. doi: 10.1002/ecy.2137. [DOI] [PubMed] [Google Scholar]
- Duffy, Godwin & Cardinale (2017).Duffy JE, Godwin CM, Cardinale BJ. Biodiversity effects in the wild are common and as strong as key drivers of productivity. Nature. 2017;549(7671):261–264. doi: 10.1038/nature23886. [DOI] [PubMed] [Google Scholar]
- Eastwood et al. (1997).Eastwood JA, Yates MG, Thomson AG, Fuller RM. The reliability of vegetation indices for monitoring saltmarsh vegetation cover. International Journal of Remote Sensing. 1997;18(18):3901–3907. doi: 10.1080/014311697216739. [DOI] [Google Scholar]
- Edgar et al. (2011).Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards (2015).Edwards A. Coming in from the cold: potential microbial threats from the terrestrial cryosphere. Frontiers in Earth Science. 2015;3:e12. doi: 10.3389/feart.2015.00012. [DOI] [Google Scholar]
- Farinotti et al. (2015).Farinotti D, Longuevergne L, Moholdt G, Duethmann D, Mölg T, Bolch T, Vorogushyn S, Güntner A. Substantial glacier mass loss in the Tien Shan over the past 50 years. Nature Geoscience. 2015;8(9):716–722. doi: 10.1038/ngeo2513. [DOI] [Google Scholar]
- Figueiredo et al. (2010).Figueiredo RO, Markewitz D, Davidson EA, Schuler AE, Watrin ODS, De Souza Silva P. Land-use effects on the chemical attributes of low-order streams in the eastern Amazon. Journal of Geophysical Research. 2010;115:1–14. doi: 10.1029/2009JG001200. [DOI] [Google Scholar]
- Fuhrman (2009).Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459(7244):193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Gao et al. (2017).Gao H, Ding Y, Zhao Q, Hrachowitz M, Savenije HHG. The importance of aspect for modelling the hydrological response in a glacier catchment in Central Asia. Hydrological Processes. 2017;31(16):2842–2859. doi: 10.1002/hyp.11224. [DOI] [Google Scholar]
- Gao et al. (2016).Gao H, Han T, Liu Y, Zhao Q. Use of auxiliary data of topography, snow and ice to improve model performance in a glacier-dominated catchment in Central Asia. Hydrology Research. 2016;48(5):1418–1437. doi: 10.2166/nh.2016.242. [DOI] [Google Scholar]
- Gao et al. (2018).Gao H, Li H, Duan Z, Ren Z, Meng X, Pan X. Modelling glacier variation and its impact on water resource in the Urumqi Glacier No. 1 in Central Asia. Science of the Total Environment. 2018;644:1160–1170. doi: 10.1016/j.scitotenv.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Gardner et al. (2013).Gardner AS, Moholdt G, Cogley JG, Wouters B, Arendt AA, Wahr J, Berthier E, Hock R, Pfeffer WT, Kaser G, Ligtenberg SRM, Bolch T, Sharp MJ, Hagen JO, Van Den Broeke MR, Paul F. A reconciled estimate of glacier contributions to sea level rise: 2003 to 2009. Science. 2013;340(6134):852–857. doi: 10.1126/science.1234532. [DOI] [PubMed] [Google Scholar]
- Geesey et al. (1978).Geesey GG, Mutch R, Costerton JW, Green RB. Sessile bacteria: an important component of the microbial population in small mountain streams. Limnology and Oceanography. 1978;23(6):1214–1223. doi: 10.4319/lo.1978.23.6.1214. [DOI] [Google Scholar]
- Gounot (1986).Gounot AM. Psychrophilic and psychrotrophic microorganisms. Experientia. 1986;42(11–12):1192–1197. doi: 10.1007/BF01946390. [DOI] [PubMed] [Google Scholar]
- Guo et al. (2014).Guo WQ, Liu SY, Yao XJ, Xu JL, Shangguan DH, Wu LZ, Zhao JD, Liu Q, Jiang ZL, Wei JF, Bao WJ, Yu PC, Ding LF, Li G, Li P, Ge CM, Wang Y. The Second Glacier Inventory Dataset of China (Version 1.0). Cold and Arid Regions Science Data Center at Lanzhou. 2014. http://card.westgis.ac.cn/data/f92a4346-a33f-497d-9470-2b357ccb4246 http://card.westgis.ac.cn/data/f92a4346-a33f-497d-9470-2b357ccb4246
- Gutiérrez et al. (2015).Gutiérrez MH, Galand PE, Moffat C, Pantoja S. Melting glacier impacts community structure of Bacteria, Archaea and Fungi in a Chilean Patagonia fjord. Environmental Microbiology. 2015;17(10):3882–3897. doi: 10.1111/1462-2920.12872. [DOI] [PubMed] [Google Scholar]
- Hagg et al. (2007).Hagg W, Braun LN, Kuhn M, Nesgaard TI. Modelling of hydrological response to climate change in glacierized Central Asian catchments. Journal of Hydrology. 2007;332(1–2):40–53. doi: 10.1016/j.jhydrol.2006.06.021. [DOI] [Google Scholar]
- Hannah et al. (2007).Hannah DM, Brown LE, Milner AM, Gurnell AM, McGregor GR, Petts GE, Smith BPG, Snook DL. Integrating climate–hydrology–ecology for alpine river systems. Aquatic Conservation: Marine and Freshwater Ecosystems. 2007;17(6):636–656. doi: 10.1002/aqc.800. [DOI] [Google Scholar]
- Hassan et al. (2016).Hassan N, Rafiq M, Hayat M, Shah AA, Hasan F. Psychrophilic and psychrotrophic fungi: a comprehensive review. Reviews in Environmental Science and Bio/Technology. 2016;15(2):147–172. doi: 10.1007/s11157-016-9395-9. [DOI] [Google Scholar]
- Hawkins et al. (2018).Hawkins JA, Jones SJ, Finkelstein IJ, Press WH. Indel-correcting DNA barcodes for high-throughput sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(27):E6217–E6226. doi: 10.1073/pnas.1802640115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood & Scott (2008).Hood E, Scott D. Riverine organic matter and nutrients in southeast Alaska affected by glacial coverage. Nature Geoscience. 2008;1(9):583–587. doi: 10.1038/ngeo280. [DOI] [Google Scholar]
- Hotaling, Hood & Hamilton (2017).Hotaling S, Hood E, Hamilton TL. Microbial ecology of mountain glacier ecosystems: biodiversity, ecological connections, and implications of a warming climate. Environmental Microbiology. 2017;19(8):2935–2948. doi: 10.1111/1462-2920.13766. [DOI] [PubMed] [Google Scholar]
- Huws et al. (2007).Huws SA, Edwards JE, Kim EJ, Scollan ND. Specificity and sensitivity of eubacterial primers utilized for molecular profiling of bacteria within complex microbial ecosystems. Journal of Microbiological Methods. 2007;70(3):565–569. doi: 10.1016/j.mimet.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Immerzeel, Van Beek & Bierkens (2010).Immerzeel WW, Van Beek LPH, Bierkens MFP. Climate change will affect the Asian water towers. Science. 2010;328(5984):1382–1385. doi: 10.1126/science.1183188. [DOI] [PubMed] [Google Scholar]
- IPCC (2014).IPCC . Climate change 2014: synthesis report. Geneva: Intergovernmental Panel on Climate Change; 2014. [Google Scholar]
- Jacobsen & Dangles (2012).Jacobsen D, Dangles O. Environmental harshness and global richness patterns in glacier-fed streams. Global Ecology and Biogeography. 2012;21(6):647–656. doi: 10.1111/j.1466-8238.2011.00699.x. [DOI] [Google Scholar]
- Jones, Hambright & Caron (2018).Jones AC, Hambright KD, Caron DA. Ecological patterns among bacteria and microbial Eukaryotes derived from network analyses in a low-salinity lake. Microbial Ecology. 2018;75(4):917–929. doi: 10.1007/s00248-017-1087-7. [DOI] [PubMed] [Google Scholar]
- Kraaijenbrink et al. (2017).Kraaijenbrink PDA, Bierkens MFP, Lutz AF, Immerzeel WW. Impact of a global temperature rise of 1.5 degrees Celsius on Asia’s glaciers. Nature. 2017;549(7671):257–260. doi: 10.1038/nature23878. [DOI] [PubMed] [Google Scholar]
- Kuhn et al. (2011).Kuhn J, Andino P, Calvez R, Espinosa R, Hamerlik L, Vie S, Dangles O, Jacobsen D. Spatial variability in macroinvertebrate assemblages along and among neighbouring equatorial glacier-fed streams. Freshwater Biology. 2011;56(11):2226–2244. doi: 10.1111/j.1365-2427.2011.02648.x. [DOI] [Google Scholar]
- Laghari (2013).Laghari JR. Melting glaciers bring energy uncertainty. Nature. 2013;502(7473):617–618. doi: 10.1038/502617a. [DOI] [PubMed] [Google Scholar]
- Lindström & Langenheder (2012).Lindström ES, Langenheder S. Local and regional factors influencing bacterial community assembly. Environmental Microbiology Reports. 2012;4(1):1–9. doi: 10.1111/j.1758-2229.2011.00257.x. [DOI] [PubMed] [Google Scholar]
- Loreau et al. (2001).Loreau M, Naeem S, Inchausti P, Bengtsson J, Grime JP, Hector A, Hooper DU, Huston MA, Raffaelli D, Schmid B, Tilman D, Wardle DA. Biodiversity and ecosystem functioning: Current knowledge and future challenges. Science. 2001;294(5543):804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- Marzeion et al. (2014).Marzeion B, Cogley JG, Richter K, Parkes D. Attribution of global glacier mass loss to anthropogenic and natural causes. Science. 2014;345(6199):919–921. doi: 10.1126/science.1254702. [DOI] [PubMed] [Google Scholar]
- Masoud et al. (2011).Masoud W, Takamiya M, Vogensen FK, Lillevang S, Al-Soud WA, Sørensen SJ, Jakobsen M. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. International Dairy Journal. 2011;21(3):142–148. doi: 10.1016/j.idairyj.2010.10.007. [DOI] [Google Scholar]
- Milner et al. (2001).Milner AM, Brittain JE, Castella E, Petts GE. Trends of macroinvertebrate community structure in glacier-fed rivers in relation to environmental conditions: a synthesis. Freshwater Biology. 2001;46(12):1833–1847. doi: 10.1046/j.1365-2427.2001.00861.x. [DOI] [Google Scholar]
- Milner, Brown & Hannah (2009).Milner AM, Brown LE, Hannah DM. Hydroecological response of river systems to shrinking glaciers. Hydrological Processes. 2009;23(1):62–77. doi: 10.1002/hyp.7197. [DOI] [Google Scholar]
- Milner et al. (2017).Milner AM, Khamis K, Battin TJ, Brittain JE, Barrand NE, Fuereder L, Cauvy-Fraunie S, Gislason GM, Jacobsen D, Hannah DM, Hodson AJ, Hood E, Lencioni V, Olafsson JS, Robinson CT, Tranter M, Brown LE. Glacier shrinkage driving global changes in downstream systems. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(37):9770–9778. doi: 10.1073/pnas.1619807114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner & Petts (1994).Milner AM, Petts GE. Glacial rivers: physical habitat and ecology. Freshwater Biology. 1994;32(2):295–307. doi: 10.1111/j.1365-2427.1994.tb01127.x. [DOI] [Google Scholar]
- Montoya, Pimm & Sole (2006).Montoya JM, Pimm SL, Sole RV. Ecological networks and their fragility. Nature. 2006;442(7100):259–264. doi: 10.1038/nature04927. [DOI] [PubMed] [Google Scholar]
- Mori, Isbell & Seidl (2018).Mori AS, Isbell F, Seidl R. β-Diversity, Community assembly, and Ecosystem functioning. Trends in Ecology & Evolution. 2018;33(7):549–564. doi: 10.1016/j.tree.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris et al. (2011).Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, Bader GD, Ferrin TE. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. 2011;12(1):e436. doi: 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narama et al. (2010).Narama C, Kääb A, Duishonakunov M, Abdrakhmatov K. Spatial variability of recent glacier area changes in the Tien Shan Mountains, Central Asia, using Corona (~1970), Landsat (~2000), and ALOS (~2007) satellite data. Global and Planetary Change. 2010;71(1–2):42–54. doi: 10.1016/j.gloplacha.2009.08.002. [DOI] [Google Scholar]
- Nelson, Sadro & Melack (2009).Nelson CE, Sadro S, Melack JM. Contrasting the influences of stream inputs and landscape position on bacterioplankton community structure and dissolved organic matter composition in high-elevation lake chains. Limnology and Oceanography. 2009;54(4):1292–1305. doi: 10.4319/lo.2009.54.4.1292. [DOI] [Google Scholar]
- Newman (2006).Newman MEJ. Modularity and community structure in networks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen et al. (2012).Nielsen A, Trolle D, Søndergaard M, Lauridsen TL, Bjerring R, Olesen JE, Jeppesen E. Watershed land use effects on lake water quality in Denmark. Ecological Applications. 2012;22(4):1187–1200. doi: 10.1890/11-1831.1. [DOI] [PubMed] [Google Scholar]
- Oksanen et al. (2007).Oksanen J, Kindt R, Legendre PO, Hara B, Stevens MHH, Oksanen MJ, Suggests MASS. The vegan package. Community Ecology Package. 2007;10:631–637. [Google Scholar]
- Olesen et al. (2007).Olesen JM, Bascompte J, Dupont YL, Jordano P. The modularity of pollination networks. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(50):19891–19896. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, Onnela & Mucha (2009).Porter MA, Onnela JP, Mucha PJ. Communities in networks. Notices of the AMS. 2009;56:1082–1097. [Google Scholar]
- R Development Core Team (2017).R Development Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2017. [Google Scholar]
- Raynolds et al. (2015).Raynolds M, Magnusson B, Metusalemsson S, Magnusson SH. Warming, sheep and volcanoes: land cover changes in Iceland evident in satellite NDVI trends. Remote Sensing. 2015;7(8):9492–9506. doi: 10.3390/rs70809492. [DOI] [Google Scholar]
- Ren, Gao & Elser (2017).Ren Z, Gao H, Elser JJ. Longitudinal variation of microbial communities in benthic biofilms and association with hydrological and physicochemical conditions in glacier-fed streams. Freshwater Science. 2017;36(3):479–490. doi: 10.1086/693133. [DOI] [Google Scholar]
- Ren et al. (2017).Ren Z, Gao H, Elser JJ, Zhao Q. Microbial functional genes elucidate environmental drivers of biofilm metabolism in glacier-fed streams. Scientific Reports. 2017;7(1):e12668. doi: 10.1038/s41598-017-13086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren et al. (2019).Ren Z, Niu D, Ma P, Wang Y, Fu H, Elser JJ. Cascading influences of grassland degradation on nutrient limitation in a high mountain lake and its inflow streams. Ecology. 2019;100:e02755. doi: 10.1002/ecy.2755. [DOI] [PubMed] [Google Scholar]
- Robinson (2001).Robinson CH. Cold adaptation in Arctic and Antarctic fungi. New Phytologist. 2001;151(2):341–353. doi: 10.1046/j.1469-8137.2001.00177.x. [DOI] [Google Scholar]
- Robinson & Jolidon (2005).Robinson CT, Jolidon C. Leaf breakdown and the ecosystem functioning of alpine streams. Journal of the North American Benthological Society. 2005;24(3):495–507. doi: 10.1899/04-100.1. [DOI] [Google Scholar]
- Robinson, Thompson & Freestone (2014).Robinson CT, Thompson C, Freestone M. Ecosystem development of streams lengthened by rapid glacial recession. Fundamental and Applied Limnology. 2014;185(3):235–246. doi: 10.1127/fal/2014/0667. [DOI] [Google Scholar]
- Rouse et al. (1974).Rouse JW, Haas RH, Schell JA, Deering DW. Monitoring vegetation systems in the Great Plains with ERTS. Third Earth Resources Technology Satelite-1 Symposium; Washington, D.C., USA. 1974. pp. 309–317. [Google Scholar]
- Ruiz-González, Niño-García & Del Giorgio (2015).Ruiz-González C, Niño-García JP, Del Giorgio PA. Terrestrial origin of bacterial communities in complex boreal freshwater networks. Ecology Letters. 2015;18(11):1198–1206. doi: 10.1111/ele.12499. [DOI] [PubMed] [Google Scholar]
- Saavedra et al. (2011).Saavedra S, Stouffer DB, Uzzi B, Bascompte J. Strong contributors to network persistence are the most vulnerable to extinction. Nature. 2011;478(7368):233–235. doi: 10.1038/nature10433. [DOI] [PubMed] [Google Scholar]
- Sadro, Nelson & Melack (2012).Sadro S, Nelson CE, Melack JM. The influence of landscape position and catchment characteristics on aquatic biogeochemistry in high-elevation lake-chains. Ecosystems. 2012;15(3):363–386. doi: 10.1007/s10021-011-9515-x. [DOI] [Google Scholar]
- Sertic Peric et al. (2015).Sertic Peric M, Jolidon C, Uehlinger U, Robinson CT. Long-term ecological patterns of alpine streams: an imprint of glacial legacies. Limnology and Oceanography. 2015;60(3):992–1007. doi: 10.1002/lno.10069. [DOI] [Google Scholar]
- Shannon et al. (2003).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi et al. (2016).Shi S, Nuccio EE, Shi ZJ, He Z, Zhou J, Firestone MK. The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecology Letters. 2016;19(8):926–936. doi: 10.1111/ele.12630. [DOI] [PubMed] [Google Scholar]
- Socolar et al. (2016).Socolar JB, Gilroy JJ, Kunin WE, Edwards DP. How should beta-diversity inform biodiversity conservation? Trends in Ecology & Evolution. 2016;31(1):67–80. doi: 10.1016/j.tree.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Sorg et al. (2012).Sorg A, Bolch T, Stoffel M, Solomina O, Beniston M. Climate change impacts on glaciers and runoff in Tien Shan (Central Asia) Nature Climate Change. 2012;2(10):725–731. doi: 10.1038/nclimate1592. [DOI] [Google Scholar]
- Thompson (2005).Thompson JN. The geographic mosaic of coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- Tilman, Isbell & Cowles (2014).Tilman D, Isbell F, Cowles JM. Biodiversity and ecosystem functioning. Annual Review of Ecology, Evolution, and Systematics. 2014;45(1):471–493. doi: 10.1146/annurev-ecolsys-120213-091917. [DOI] [Google Scholar]
- Toju et al. (2016).Toju H, Kishida O, Katayama N, Takagi K. Networks depicting the fine-scale co-occurrences of fungi in soil horizons. PLOS ONE. 2016;11(11):e0165987. doi: 10.1371/journal.pone.0165987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetti et al. (2008).Turchetti B, Buzzini P, Goretti M, Branda E, Diolaiuti G, D’Agata C, Smiraglia C, Vaughan-Martini A. Psychrophilic yeasts in glacial environments of Alpine glaciers. FEMS Microbiology Ecology. 2008;63(1):73–83. doi: 10.1111/j.1574-6941.2007.00409.x. [DOI] [PubMed] [Google Scholar]
- Unger-Shayesteh et al. (2013).Unger-Shayesteh K, Vorogushyn S, Farinotti D, Gafurov A, Duethmann D, Mandychev A, Merz B. What do we know about past changes in the water cycle of Central Asian headwaters? A review. Global and Planetary Change. 2013;110:4–25. doi: 10.1016/j.gloplacha.2013.02.004. [DOI] [Google Scholar]
- Von Schiller et al. (2007).Von Schiller D, Marti E, Riera JL, Sabater F. Effects of nutrients and light on periphyton biomass and nitrogen uptake in Mediterranean streams with contrasting land uses. Freshwater Biology. 2007;52(5):891–906. doi: 10.1111/j.1365-2427.2007.01742.x. [DOI] [Google Scholar]
- Weiss et al. (2016).Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, Xia LC, Xu ZZ, Ursell L, Alm EJ, Birmingham A, Cram JA, Fuhrman JA, Raes J, Sun F, Zhou J, Knight R. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME Journal. 2016;10(7):1669–1681. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm et al. (2014).Wilhelm L, Besemer K, Fasching C, Urich T, Singer GA, Quince C, Battin TJ. Rare but active taxa contribute to community dynamics of benthic biofilms in glacier-fed streams. Environmental Microbiology. 2014;16(8):2514–2524. doi: 10.1111/1462-2920.12392. [DOI] [PubMed] [Google Scholar]
- Wilhelm et al. (2013).Wilhelm L, Singer GA, Fasching C, Battin TJ, Besemer K. Microbial biodiversity in glacier-fed streams. ISME Journal. 2013;7(8):1651–1660. doi: 10.1038/ismej.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp et al. (2015).Zemp M, Frey H, Gärtner-Roer I, Nussbaumer SU, Hoelzle M, Paul F, Haeberli W, Denzinger F, Ahlstrøm AP, Anderson B, Bajracharya S, Baroni C, Braun LN, Cáceres BE, Casassa G, Cobos G, Dávila LR, Delgado Granados H, Demuth MN, Espizua L, Fischer A, Fujita K, Gadek B, Ghazanfar A, Ove Hagen J, Holmlund P, Karimi N, Li Z, Pelto M, Pitte P, Popovnin VV, Portocarrero CA, Prinz R, Sangewar CV, Severskiy I, Sigurđsson O, Soruco A, Usubaliev R, Vincent C. Historically unprecedented global glacier decline in the early 21st century. Journal of Glaciology. 2015;61(228):745–762. doi: 10.3189/2015JoG15J017. [DOI] [Google Scholar]
- Zemp et al. (2006).Zemp M, Haeberli W, Hoelzle M, Paul F. Alpine glaciers to disappear within decades? Geophysical Research Letters. 2006;33(13):L1350413. doi: 10.1029/2006GL026319. [DOI] [Google Scholar]
- Zhang et al. (2013).Zhang Y, Gao J, Liu L, Wang Z, Ding M, Yang X. NDVI-based vegetation changes and their responses to climate change from 1982 to 2011: a case study in the Koshi River Basin in the middle Himalayas. Global and Planetary Change. 2013;108:139–148. doi: 10.1016/j.gloplacha.2013.06.012. [DOI] [Google Scholar]
- Zhang et al. (2014).Zhang G, Li Z, Wang W, Wang W. Rapid decrease of observed mass balance in the Urumqi Glacier No. 1, Tianshan Mountains, central Asia. Quaternary International. 2014;349:135–141. doi: 10.1016/j.quaint.2013.08.035. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Distributions of OTUs across sample sites for bacterial modules.
Figure S2 Distributions of OTUs across sample sites for fungal modules.
Table S1 Pairwise dissimilarity tests of taxonomic composition among bacterial modules using PERMANOVA (adonis function in vegan package 2.5-3). R2 and P-values (in bracket) are shown.
Table S2 Pairwise dissimilarity tests of taxonomic composition among fungal modules using PERMANOVA (adonis function in vegan package 2.5-3). R2 and P-values (in bracket) are shown.
Data Availability Statement
The following information was supplied regarding data availability: