Abstract
Transcriptional Intermediary Factor 2 (TIF2) is a key Androgen receptor (AR) coactivator that has been implicated in the development and progression of castration resistant prostate cancer (CRPC). This chapter describes the implementation of an AR-TIF2 protein-protein interaction (PPI) biosensor assay to screen for small molecules that can induce AR-TIF2 PPIs, inhibit the DHT-induced formation of AR-TIF2 PPIs, or disrupt pre-existing AR-TIF2 PPIs. The biosensor assay employs high content imaging and analysis to quantify AR-TIF2 PPIs and integrates physiologically relevant cell-based assays with the specificity of binding assays by incorporating structural information from AR and TIF2 functional domains along with intracellular targeting sequences using fluorescent protein reporters. Expression of the AR-Red Fluorescent Protein (RFP) “prey” and TIF2-Green Fluorescent Protein (GFP) “bait” components of the biosensor is directed by recombinant adenovirus (rAV) expression constructs that facilitated a simple co-infection protocol to produce homogeneous expression of both biosensors that is scalable for screening. In untreated cells, AR-RFP expression is localized predominantly to the cytoplasm and TIF2-GFP expression is localized only in the nucleoli of the nucleus. Exposure to DHT induces the co-localization of AR-RFP within the TIF2-GFP positive nucleoli of the nucleus. The AR-TIF2 biosensor assay therefore recapitulates the ligand-induced translocation of latent AR from the cytoplasm to the nucleus, and the PPIs between AR and TIF2 result in the colocalization of AR-RFP within TIF2-GFP expressing nucleoli. The AR-TIF2 PPI biosensor approach offers significant promise for identifying molecules with potential to modulate AR transcriptional activity in a cell-specific manner that may overcome the development of resistance and progression to CRPC.
Keywords: Protein-protein interaction biosensors, High content screening, Imaging, Image analysis
1. Introduction
Protein-protein interactions (PPIs) are integral to all cellular functions and therefore represent a large number of potential therapeutic targets for drug discovery [1–8]. Despite the critical importance of PPIs and the existence of many assay formats compatible with HTS/HCS, the dearth of approved PPI inhibitor/disruptor drugs suggests that the discovery of such molecules is not trivial [1–8]. Although PPI targets are often characterized as “undruggable” [9], the structural analysis of protein-protein complexes suggests that protein-binding interfaces contain discrete “hot spots” and that relatively small numbers of amino acids at the PPI interface contribute the majority of the binding energy [1–4, 6–8]. Furthermore, PPI contact surfaces exhibit some degree of flexibility with cavities, pockets, and grooves available for small molecule binding [1–4, 6–8]. Existing small molecule PPI inhibitors appear to bind to hotspots with much higher efficiencies and deeper within the target protein than do the contact atoms of the native protein partner [1–4, 6–8]. This chapter describes the implementation of a high content positional biosensor (PPIB) assay that we have developed to measure the PPIs between the Androgen Receptor (AR) and a key coactivator Transcriptional Intermediary Factor 2 (TIF2) (Fig. 1) [4]. The biosensor assay employs high content imaging and analysis to quantify AR-TIF2 PPIs and integrates physiologically relevant cell-based assays with the specificity of binding assays by incorporating structural information from AR and TIF2 functional domains along with intracellular targeting sequences and fluorescent reporters (Figs. 1, 2 and 3) [4].
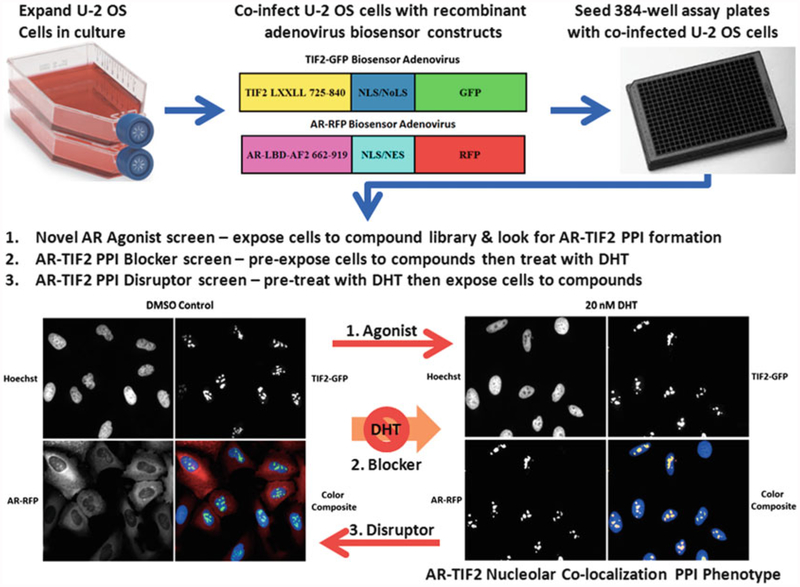
Fig. 1.
AR-TIF2 protein-protein interaction biosensor design, grayscale and color composite images of maximum and minimum plate controls, and potential screening formats. Recombinant adenovirus (rAV) AR and TIF2 biosensor constructs were created to co-infect and express the AR and TIF2 protein-protein interaction partners in cells. The AR-RFP “prey” protein interaction partner shuttles between the cytoplasm and the nucleus in a ligand-dependent manner and the rAV construct is composed of AR residues 662–919 encoding the AR-LBD and AF2 surface as a chimeric fusion protein with red fluorescent protein (RFP) and both nuclear localization and nuclear export sequences that are part of the chimera, and not specific to AR. The central region of TIF2 contains three α-helical LXXLL motifs that mediate the binding to ligand-bound AR, and a rAV construct was created to express TIF2 residues 725–840 as a chimeric fusion protein with green fluorescent protein (GFP) and a high affinity nuclear/nucleolar localization (NLS/NoLS) sequence derived from HIV Rev. The TIF2-GFP “bait” protein interaction partner is targeted to and anchored in the nucleoli within the cell nucleus. U-2 OS cells cultured in tissue culture flasks are harvested after exposure to trypsin, counted, co-infected with the AR-RFP and TIF2-GFP adenoviruses, seeded at 2500 cells per well in 384-well collagen-coated assay plates, cultured overnight at 37 °C, 5% CO2 and 95% humidity, and then treated for 30 min with0.5% DMSO or 20 nM DHT in 0.5% DMSO prior to formaldehyde fixation and Hoechst staining as described above. Individual gray-scale images of three fluorescent channels (Hoechst Ch1 blue, FITC Ch2 green, and Texas Red Ch3) were sequentially acquired on the IXU automated imaging platform using a 20×/0.45 NA objective, the 405 nm Ch1, 488 nm Ch2, and 561 nm Ch3 laser lines, and a Quad emission filter set as described above. Individual× 20 gray-scale and color composite images are presented; Ch1 Hoechst—blue, Ch2 TIF2-GFP—green, and Ch3 AR-RFP—red. In untreated U-2 OS cells expressing both biosensors, AR-RFP expression is localized predominantly to the cytoplasm and TIF2-GFP expression is localized only to nucleoli as indicated by the color composite images of cells with diffuse red cytoplasm and blue nuclei containing bright green TIF2-GFP puncta. After exposure to DHT for 30 min the AR-RFP colocalizes with the TIF2-GFP partner in nucleoli as indicated by the bright yellow AR-TIF2 puncta within the blue stained nuclei of color composite images. The AR-TIF2 biosensor therefore recapitulates the ligand-induced translocation of AR from the cytoplasm to the nucleus, and the PPIs between AR and TIF2 results in the colocalization of ARRFP and TIF2-GFP within the nucleolus. The AR-TIF2 PPIB HCS assay can be screened in three distinct formats: format 1, cells are exposed to compounds to identify novel AR agonists capable of inducing the formation of AR-TIF2 PPIs; format 2, cells are exposed to compounds prior to the addition of DHT to screen for compounds that block DHT-induced formation of AR-TIF2 PPIs; and format 3, cells are pretreated with DHT before compound addition to identify small molecules capable of disrupting pre-existing AR-TIF2 complexes
Fig. 2.
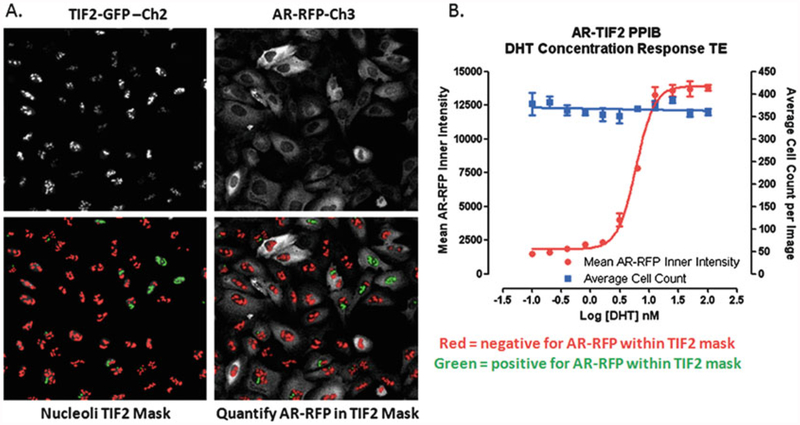
Translocation Enhanced Image Analysis Module. (a) Image segmentation-derived TIF2-GFP positive nucleoli masks in the FITC and Texas Red channels. Enlarged and cropped gray-scale images for presentation purposes of TIF2-GFP (Ch2) and AR-RFP (Ch3) from U-2 OS cells co-infected with both biosensor adenoviruses and then cultured overnight without further treatment. The translocation enhanced (TE) image analysis module utilizes the TIF2-GFP biosensor component in Ch2 to create a mask of the nucleoli. The bright fluorescent puncta in Ch2 with TIF2-GFP fluorescent intensities >750 gray levels over background, an approximate width of 4.0 μm, a minimum area of 5.0 μm2, and a maximum area < 150 μm2 are classified by the image segmentation as TIF2-GFP positive nucleoli and used to create translocation masks. AR-RFP images from Ch3 are segmented into nucleoli regions using the masks derived from the detected TIF2-GFP positive nucleoli in Ch2. The red or green color of the nucleoli masks indicates whether the correlation coefficient for colocalization of the AR-RFP signal within the TIF2-GFP positive nucleoli was below (red) or above (green) a preset threshold (typically 0.25). (b) Quantitative data extracted by the Translocation Enhanced image analysis module: average inner fluorescent intensity of the Ch3 AR-RFP signal within Ch2-derived masks of TIF2-GFP positive nucleoli; average cell count per image determined from the number of Hoechst stained nuclei quantified in Ch1 images. U-2 OS cells were co-infected with the AR-RFP and TIF2-GFP rAV biosensors, 2500 cells were seeded into the wells of 384-well assay plates, cultured overnight at 37 °C, 5% CO2 and 95% humidity, and then treated with the indicated concentrations of DHT for 30 min. Cells were then fixed and stained with Hoechst, 20× images in three fluorescent channels were acquired on the IXU automated imaging platform, and the extent of DHT-induced AR-TIF2 PPIs was quantified using the TE image analysis module using the average inner intensity of AR-RFP within TIF2-GFP positive nucleoli parameter. To control for differences in cell numbers, the average number of Hoechst stained nuclei per image was also quantified by the TE image analysis module. The mean ± sd (n = 3) average inner intensity of AR-RFP within the TIF2-GFP positive nucleoli (●) and cell counts per image (■) at DHT concentrations ranging between 0.001 and 100 nM are presented
Fig. 3.
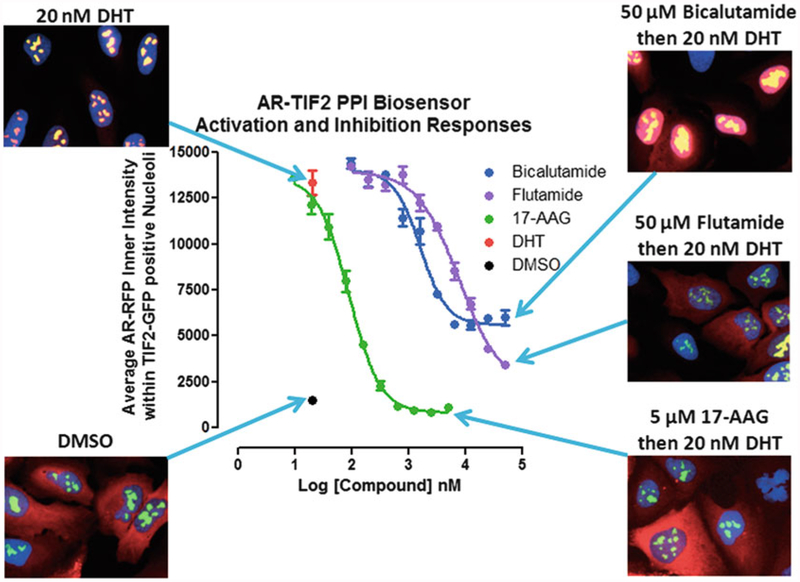
Inhibition of DHT-induced AR-TIF2 PPI responses and corresponding AR-TIF2 distribution phenotypes. U-2 OS cells were co-infected with the AR-RFP and TIF2-GFP rAV biosensors, 2500 cells were seeded into the wells of 384-well assay plates, cultured overnight at 37 °C, 5% CO2 and 95% humidity, and then exposed to compounds at the indicated concentrations for 1 h. Cells were then treated with 20 nM DHT for 30 min, fixed and stained with Hoechst, 20× images in three fluorescent channels were acquired on the IXU automated imaging platform, and the AR-TIF2 PPIs were quantified using the TE image analysis module as described above. The mean ± sd (n = 3) average inner intensity of AR-RFP within the TIF2-GFP positive nucleoli in cells exposed to the indicated concentrations of Bicalutamide (●), Flutamide (●), or 17-AAG (●), for 1 h and then treated with 20 nM DHT (●) or 0.5% DMSO (●) are presented. Experimental data from one of five independent experiments are presented. Representative 40× color composite images of the AR-TIF2 biosensor phenotypes of co-infected U-2 OS cells pre-exposed to 0.5% DMSO, 50 μM bicalutamide in 0.5% DMSO, 50 μM flutamide in 0.5% DMSO, or 5 μM 17-AAG in 0.5% DMSO for 1 h prior to 30 min treatment ± 20 nM DHT are shown
The subcellular localization of macromolecules in specific cellular compartments is a tightly regulated process [10–12]. For example, although molecules <40 kDa passively diffuse through the nuclear pore complexes (NPCs) in the nuclear envelope, cargos 40 kDa require an active transport process facilitated by specific receptor proteins to enter the nucleus from the cytoplasm [10–12]. Protein cargos ≥40 kDa bearing a suitable nuclear localization sequence (NLS) bind to an importin-α adaptor receptor that recognizes the NLS and forms a complex with an importin-β transport receptor that facilitates docking interactions with the nucleoporins that line the NPC [10–12]. Protein export from the nucleus is also mediated by the assembly of a complex between exportin-1 (CRM-1), Ran-GTP, and protein cargos bearing a leucine-rich nuclear export sequence (NES) [10–12]. The steady-state localization of a protein that moves between the cytoplasm and the nucleus is a function of the balance between the operational strengths and/or accessibility of its NLS and NES sequences [10–12]. Since the nucleus and the cytoplasm can be readily identified, separated, and quantified independently by image analysis methods, the regulated nucleo-cytoplasmic localization of proteins is frequently used in HCS assays as a surrogate for signaling pathway activation [13–18]. Additionally, the specific targeting sequences that direct proteins to specific subcellular sites or compartments have also been exploited to design positional biosensors to quantify PPIs [2–4, 11, 12, 19, 20]. Positional PPI biosensors typically consist of two parts, a “bait” biosensor that is targeted and anchored to a specific cellular location and a “prey” biosensor designed to shuttle between distinct subcellular compartments [2–4, 11, 12, 19, 20]. Colocalization of both the biosensors to the same site in the cell indicates the formation of productive PPI complexes. Stauber and colleagues pioneered the design of positional PPI biosensors that target “bait” PPI partners to the nucleolus using expression constructs that incorporate a NES deficient HIV-1 Rev. sequence and a fluorescent reporter protein [11, 12, 19, 20]. The matching “prey” PPI bio-sensor partners are designed to shuttle between the cytoplasm and the nucleus by integrating both the NLS and NES sequences with the fluorescent reporter protein [11, 12, 19, 20].
Prostate cancer (CaP) is the most common solid tumor and second leading cause of cancer death among men in Western countries [21–25]. Existing frontline androgen ablation therapies (AAT) either target androgen production or function. Although the initial responses to these therapies are generally favorable, approximately 20% of patient’s progress to castration-resistant metastatic CaP (CRPC) for which there currently is no cure [21–25]. The AR is a nuclear hormone receptor (NR) that is a ligand-dependent and DNA-sequence specific transcriptional regulator involved in prostate growth, terminal differentiation, and function [21–28]. Latent AR in the cytoplasm exists in a complex with heat-shock proteins (Hsp) 90 and 70 that maintain the NR in a stable, partially unfolded state primed for high affinity interactions with androgens [21–24]. Agonist binding induces AR homo-dimerization, trafficking to the nucleus, binding to specific DNA response element sequences in the promoter/enhancer regions of AR target genes, recruitment of coactivators, assembly of the core transcriptional machinery, and activation of transcription [21–24]. Coactivators recruited by ligand activated AR amplify the assembly of the transcription complex and regulate tissue specific spatiotemporal gene expression [27, 28]. Elevated coactivator levels reduce ligand concentration requirements and elicit a more rapid transcriptional response [27, 28]. Overexpression of AR and/or its co-activators, or shifts in the balance between coactivators and corepressors, are thought to play a role in the emergence of resistance in CRPC [21–24, 29–31]. TIF2 (SRC-2) is a member of the steroid receptor coactivator SRC/p160 family that has been implicated in the development and progression of CRPC [21, 22, 25, 27, 28, 31–36]. TIF2 stabilizes the AR and AR-ligand binding, facilitates AR N/C interactions, promotes chromatin remodeling enzyme recruitment, and AR transcriptional activation [21, 22, 27, 28]. There is a significant correlation between tumor TIF2 expression and CaP aggressiveness [25, 32–34]. Transient TIF2 overexpression increased AR responses to adrenal androgens and non-AR ligands, while TIF2 antisense oligo or siRNA knockdowns reduced AR target gene expression and slowed the proliferation of androgen-dependent and independent CaP cells [25, 32]. Prolonged AR localization and TIF2 recruitment to AR target gene promoters has been associated with the development of CRPC, and it was suggested that small molecules that block AR-TIF2 PPIs might have therapeutic value [25, 29–32, 35].
Agonist binding to AR induces a conformational change in the AR ligand-binding domain (AR-LBD) to form the Activation Function 2 (AF2) surface that binds the LXXLL motifs of SRC/ p160 coactivators including TIF2 [27–30, 36]. The chimeric AR and TIF2 biosensor components were cloned into separate recombinant adenovirus (rAV) expression constructs (Fig. 1), a high efficiency co-expression system that we have exploited previously for other PPIB HCS assays [2–4]. The AR “prey” biosensor was created to express AR residues 662–919 that encompass the ARLBD as a chimeric fusion protein with red fluorescent protein (RFP) and included both an NLS and an NES sequence (Fig. 1) [4]. The TIF2 “bait” biosensor was created to express residues 725–840 of TIF2 that contains three α-helical LXXLL motifs as a chimeric fusion protein with GFP and a high affinity nuclear/nucleolar localization (NLS/NoLS) sequence derived from HIV Rev (Fig. 1) [4]. Anchoring the “bait” biosensor in the nucleolus facilitates both the image acquisition process and the subsequent quantification of the colocalization of the “prey” biosensor by image analysis (Figs. 1 and 2) [4]. The use of the recombinant adenovirus expression vectors enables the development of a relatively simple co-infection protocol to produce homogeneous expression of both biosensor components that is scalable for screening (Fig. 1) [4]. The U-2 OS osteosarcoma cell line was acquired from American Type Culture Collection and was maintained in McCoy’s 5A medium with 2 mM L-glutamine (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA), and 100 U/mL penicillin and streptomycin (Invitrogen, Carlsbad, CA) in a humidified incubator at 37 °C, 5% CO2 and 95% humidity. In untreated U-2 OS cells co-infected with both rAVs, AR-RFP expression is localized predominantly to the cytoplasm and TIF2-GFP expression is localized only in the nucleoli of the nucleus, as indicated by the diffusely red cytoplasm and blue nuclei containing bright green TIF2-GFP puncta of the corresponding composite images (Fig. 1) [4].
Exposure to DHT induces the colocalization of AR-RFP within the TIF2-GFP positive nucleoli of the nucleus, as indicated by the bright yellow of the AR-TIF2 puncta within the blue stained nuclei of the corresponding composite images (Fig. 1) [4]. The AR-TIF2 biosensor assay therefore recapitulates the ligand-induced translocation of latent AR from the cytoplasm to the nucleus, and the PPIs between AR and TIF2 result in the colocalization of ARRFP within TIF2-GFP expressing nucleoli [4]. Variations in AR coregulator expression levels between normal tissues and CaP cell lines contribute to their altered androgen responsiveness [37, 38]. NRs selectively recruit coactivator complexes, and diverse ligands preferentially recruit different coregulator cohorts [27, 28, 39]. Some NR ligands only activate a subset of target genes, and it is believed that such gene selectivity is cell or tissue specific and reflects the ratio of coactivators to corepressors which determines whether an on or off signal is processed [36, 39]. Coregulators may also exhibit different functions depending upon the specific promoter context [26, 27, 36]. NR coactivator recruitment profiles therefore influence the tissue-specific spatiotemporal gene expression responses to NR ligands [27, 36, 37].
To analyze and quantify AR-TIF2 PPIs in digital images acquired on either the ImageXpress Ultra (IXU) confocal or ImageXpress Micro (IXM) wide-field automated imaging platforms, we utilized the translocation enhanced (TE) image analysis module (Fig. 2) [4]. The bright fluorescent TIF2-GFP puncta apparent in the Ch2 images were used to define a translocation mask of the nucleoli within the Hoechst nuclei (Ch1 images, not shown) of U-2 OS cells (Fig. 2a). The TIF2-GFP puncta objects in Ch2 that had fluorescent intensities above a background threshold with suitable morphologic characteristics (width, length, and area) were classified by the image segmentation as nucleoli and used to generate a TIF2 mask (Fig. 2a, red and green masks). For images acquired on the IXU platform, these settings were generally applicable [4]; TIF2-GFP positive nucleoli were defined as objects with fluorescent intensities >750 gray levels over background, an approximate width of 4.0 μm, a minimum area of 5.0 μm2, and a maximum area ≤150 μm2. AR-RFP images in Ch3 were then segmented into an “Inner” nucleolus region using the masks generated from the detected TIF2-GFP positive nucleoli in Ch2 (Fig. 2a). The color of the nucleoli masks correspond to whether the correlation coefficient for colocalization of the AR-RFP signal within the TIF2-GFP masks was below (red) or above (green) a preset threshold (typically ≥0.25) (Fig. 2a). The TE image analysis module outputs quantitative data including: the selected TIF2-GFP positive nucleoli count in Ch2; the average fluorescent intensities of the TIF2-GFP positive nucleoli in Ch2; and the average and integrated fluorescent intensities of AR-RFP signals in Ch3 within TIF2-GFP positive masks (Fig. 2b). In U-2 OS cells expressing both biosensors, 30 min exposure to DHT at the indicated concentrations induced a concentration-dependent increase in the average inner intensity of the AR-RFP signal colocalized within TIF-2-GFP positive nucleoli (Fig. 2b). In the same cells, DHT exposure did not alter the average cell count per image determined from the number of Hoechst stained nuclei quantified in Ch1 images (Fig. 2b). In four independent concentration response experiments, DHT exhibited an EC50 of 5.33 ± 1.0 nM for the induction of AR-TIF2 PPIs, values that correlate closely with previously published DHT EC50 values determined in other AR cell-based assay formats [40].
Pre-exposure of biosensor expressing U-2 OS cells to the indicated concentrations of anti-androgens (flutamide and bicalutamide) or a Hsp 90 inhibitor (17-N-allylamino-17-demethoxygeldanamycin, 17-AAG) for 1 h prior to treatment with 20 nM DHT inhibited the colocalization of AR-RFP within TIF2-GFP positive nucleoli in a concentration-dependent manner (Fig. 3). 17-AAG completely inhibited the DHT-induced formation of AR-TIF2 PPIs, while flutamide and bicalutamide were only partial inhibitors. 17-AAG exhibited an IC50 of 88.5 ± 12.5 nM and flutamide exhibited an IC50 of 7.6 ± 2.4 μM for DHT-induced AR-TIF2 PPIs. In high-resolution 40× images acquired on the IXM, both 17-AAG and flutamide produced a cytoplasmic AR-RFP distribution phenotype similar to DMSO controls indicating that both of these compounds blocked DHT-induced AR-RFP nuclear translocation (Fig. 3). Bicalutamide exhibited an IC50 of 1.6 ± 0.4 μM for DHT-induced AR-TIF2 PPIs, but produced a diffuse AR-RFP nuclear distribution phenotype indicating that it prevented AR-RFP recruitment into TIF2-GFP positive nucleoli, presumably by inhibiting the PPIs between AR and TIF2. The AR-TIF2 PPIB assay was able to identify and quantify the concentration-dependent inhibitory effects of two FDA-approved anti-androgen CaP drugs, and an Hsp 90 inhibitor that prevents AR from assuming a folded state primed for high affinity interactions with androgenic ligands. High-resolution images of the AR-RFP distribution phenotype allowed us to distinguish between compounds that block AR translocation and those that block AR-TIF2 PPIs. The EC50 values for DHT and the IC50 values for 17-AAG, flutamide, and bicalutamide in the AR-TIF2 PPIB assay correlate closely with published values from other assay formats [40–42], indicating that the rAV expression system does not significantly alter the concentration responses of known AR modulators, and that the AR and TIF2 subdomains of the biosensors faithfully recapitulate the responses of the full length proteins.
The AR-TIF2 PPIB HCS assay can be configured to conduct screening campaigns in three distinct formats [4]: format 1, cells are exposed to compounds to identify novel AR agonists capable of inducing the formation of AR-TIF2 PPIs; format 2, cells are exposed to compounds prior to the addition of DHT to screen for compounds that block DHT-induced formation of AR-TIF2 PPIs; and format 3, cells are pretreated with DHT before compound addition to identify small molecules capable of disrupting pre-existing AR-TIF2 complexes (Fig. 1). In our experience, compound exposures of 3 h have typically been sufficient for us to measure compound effects.
2. Materials
AR agonist dihydrotestosterone (DHT).
Anti-androgen inhibitor test compounds, e.g., flutamide and bicalutamide.
Hsp 90 inhibitor, 17-N-allylamino-17-demethoxygeldanamycin (17-AAG).
Formaldehyde solution, 37%.
Dimethyl sulfoxide (DMSO) (99.9% high performance liquid chromatography-grade, under argon).
Hoechst 33342.
U-2 OS osteosarcoma cell line.
Culture medium: McCoy’s 5A medium, 2 mM L-glutamine, 10% fetal bovine serum, and 100 U/mL penicillin and streptomycin.
Humidified incubator at 37 °C, 5% CO2, and 95% humidity.
Dulbecco’s Mg2+ and Ca2+ free phosphate-buffered saline (PBS).
Trypsin 0.25%, 1 g/L EDTA solution.
Recombinant adenovirus TIF2-GFP and AR-RFP Biosensors: Recombinant adenovirus expression constructs bearing the individual TIF2-GFP (TagGFP, Evrogen, Inc.) and AR-RFP (Tag RFP, Evrogen, Inc.) protein-protein interaction partners were obtained from Cyprotex.
384-well collagen-I-coated barcoded assay microplates.
3. Methods
Aspirate spent tissue culture medium from U-2 OS cells in tissue culture flasks that are <70% confluent (see Note 1), wash cell monolayers 1× with PBS, and expose cells to trypsin-EDTA until they detached from the surface of the tissue culture flasks. Add serum containing tissue culture medium to neutralize the trypsin. Transfer the cell suspension to a 50 mL capped sterile centrifuge tube and centrifuge at 500 × g for 5 min to pellet the cells. Resuspend cells in serum containing tissue culture medium and count the number of trypan blue excluding viable cells using a hemocytometer.
Co-infect 1 × 107U-2 OS cells with TIF2-GFP and AR-RFP adenovirus by incubating cells with the manufacturer’s recommended volume of virus (see Note 2), typically 5 μL/106 cells, in 1.0 mL of culture medium for 1 h at 37 °C, 5% CO2 in a humidified incubator with periodic inversion (every 10 min) to maintain cells in suspension.
Dilute co-infected cells to 6.25 × 104 cells/mL in culture media and 40 μL (2500 cells) were seeded in each well of a 384-well collagen-I-coated barcoded microplate using an automated bulk liquid handler (see Note 3).
Incubate assay plates overnight at 37 °C, 5% CO2 in a humidified incubator (see Note 4).
As described above, the AR-TIF2 PPIB HCS assay can be configured to conduct screening campaigns in three distinct formats. Format 1: To identify novel AR agonists capable of inducing the formation of AR-TIF2 PPIs, use an automated liquid-handling device to transfer 5 μL of diluted compounds or plate controls (DHT 20 nM final, or DMSO 0.2% final) to appropriate wells for a final screening concentration of 20 μM and incubate plates at 37 °C, 5% CO2 in a humidified incubator for 30 min (see Notes 5–7). Format 2: To identify compounds that block DHT-induced formation of AR-TIF2 PPIs, use an automated liquid-handling device to transfer 5 μL of diluted compounds to appropriate wells for a final screening concentration of 20 μM, or 5 μL of diluted DMSO (0.2% final) to plate controls, and incubate plates at 37 °C, 5% CO2 in a humidified incubator for 1–3 h. After the appropriate compound exposure period use an automated liquid-handling device to transfer 5 μL of DHT (20 nM final) to compound treated wells and maximum plate controls, minimum plate controls receive media alone, and incubate assay plates at 37 °C, 5% CO2 in a humidified incubator for 30 min (see Notes 5–7). Format 3: To identify small molecules capable of disrupting pre-existing ARTIF2 complexes, use an automated liquid-handling device to transfer 5 μL of DHT (20 nM final) to compound treated wells and maximum plate controls, minimum plate controls receive media alone, and incubate assay plates at 37 °C, 5% CO2 in a humidified incubator for 30 min. After 30 min use an automated liquid-handling device to transfer 5 μL of diluted compounds to appropriate wells for a final screening concentration of 20 μM or 5 μL of diluted DMSO (0.2% final) to plate controls and incubate plates at 37 °C, 5% CO2 in a humidified incubator for 1–3 h (see Notes 5–7).
Fix and stain the nuclei of cells by the adding 50 μL of pre-warmed (37 °C) 7.4% formaldehyde (final is 3.7%) and 2 μg/mL Hoechst 33342 in PBS using a liquid handler and incubate at room temperature for 30 min (see Note 8).
Aspirate the liquid and wash the plates twice with 75 μL of PBS using a liquid handler (see Note 9). Seal with adhesive aluminum plate seals with the last 75 μL wash of PBS in place.
Acquire fluorescent images in three independent channels on an automated imaging platform (see Note 10) (e.g., ImageX-press Ultra or Micro automated imaging platforms) (Figs. 1, 2 and 3). The ImageXpress Ultra (IXU) platform (Molecular Devices LLC, Sunnyvale, CA) is a fully integrated point-scanning confocal automated imaging platform configured with four independent solid-state lasers providing four excitation wavelengths of 405, 488, 561, and 635 nm. The IXU was equipped with a Quad filter cube providing emission ranges of 417–477 nm, 496–580 nm, 553–613 nm, and 645–725 nm and four independent photomultiplier tubes (PMTs) each dedicated to a single detection wavelength. The IXU utilizes a dedicated high-speed infra-red laser auto-focus system, has a 4-position automated objective changer with air objectives (10×, 20×, 40×, and 60×), and the detection pinhole diameter of the confocal optics was configurable in the software. For the AR-TIF2 HCS assay the IXU was set up to sequentially acquire two images per well using a 20×/0.45 NA ELWD objective in each of three fluorescent channels. In the Hoechst channel (Ch1) the 405 laser was set at 10% power and the PMT gain was 550. In the TIF2-GFP channel (Ch2) the 488 laser was set at 10% power, and the PMT gain was 625. In the ARRFP channel (Ch3) the 561 laser was set at 10% power, and the PMT gain was 625. On average, the IXU scanned a single 384-well plate, two images per channel, in 90 min using these settings. The ImageXpress Micro (IXM) is an automated field-based high content imaging platform integrated with the MetaXpress software. The IXM optical drive includes a 300 W Xenon lamp broad spectrum white light source and 2/3″ chip Cooled CCD Camera and optical train for standard field of view imaging and an IXM transmitted light option with phase contrast. The IXM is equipped with a 4× Plan Apo 0.20 NA objective, a 10× Plan Fluor 0.3 NA objective, a 20× Ph1 Plan Fluor ELWD DM objective, a 20 S Plan Fluor ELWD 0.45×, NA objective, a 40, S Plan Fluor ELWD 0.60 NA objective, and a single slide holder adaptor. The IXM is equipped with the following ZPS filter sets; DAPI, FITC/ALEXA 488, CY3, CY5, and Texas Red.
Analyze the images of the three fluorescent channels (Hoechst Ch1, TIF2-GFP Ch2, and AR-RFP Ch3) of the AR-TIF2 PPIB using the translocation enhanced image analysis module of the MetaXpress software (Molecular Devices LLC, Sunnyvale, CA), as described above (Fig. 2) (see Note 11). The Ch3 average inner fluorescent intensity of AR-RFP within the TIF2-GFP positive nucleoli masks generated from the Ch2 images is utilized to quantify the formation or disruption of AR-TIF2 PPIs (Fig. 2b).
4. Notes
Typically better responses are obtained when the AR and TIF2 adenovirus biosensors are used to co-infect U-2 OS cells harvested from tissue culture flasks that are ≤70% confluent.
The optimal volume of each lot of recombinant adenovirus biosensor per 106 U-2 OS cells is determined empirically in virus titration experiments [4]. Increasing amounts of virus are incubated with the same number of cells and then the levels of biosensor expression and % of cells that are co-infected are determined on the HCS platform after 24 h in culture. Performing infections in cells suspended in a low volume of media combined with periodic inversion (every 10 min) to maintain cells in suspension enhances both the rate of infection and expression levels. In addition to the U-2 OS osteosarcoma cell line, the adenovirus biosensors have been used to co-infect several prostate cancer cell lines; DU-145, PC-3, LNCaP, C4–2, and 22Rv1. Compared to U-2 OS cells, the infection of CaP cell lines required significantly more (>10-fold) adenovirus and the rates of co-infection with both viruses for DU-145, LNCaP, and 22RV1 cell lines were typically <20%, compared to >90% in U-2 OS cells. Since the AR-TIF2 PPIB assay requires co-expression of both biosensors, the much lower co-infection rates in these CaP cell lines are a serious limitation. In addition, LNCaP cells are poorly adherent and were not retained after cell-washing procedures. We successfully titrated the adenoviruses high enough to achieve >50% co-infection in the PC-3 cell line and demonstrated that the assay can be adapted to this CaP cell background.
Determining the optimal cell seeding density is a critical assay development parameter for all cell-based assays, including HCS assays. The goal is to minimize the cell culture burden while ensuring that sufficient cells are captured per image to give statistical significance to the image analysis parameters of interest. Typically, variability is inversely related to the number of cells captured and analyzed; in HCS assays variability generally increases as the number of cells analyzed decreases.
The optimal length of time in culture post viral infection should be determined empirically for each adenovirus biosensor. In general, we have found 24 h post infection to be optimal for most recombinant adenovirus biosensor constructs.
Agonist concentration response and time-course experiments should be conducted to determine the optimal DHT concentration and length of exposure required to induce AR-TIF2 PPIs, and to determine how stable AR-TIF2 PPIs are over time. These data are used to select a DHT concentration and treatment time for the maximum plate controls to provide a robust and reproducible assay signal window, relative to DMSO minimum plate controls, with acceptable signal-to-background ratios, typically ≥3-fold, and Z′-factor coefficients ≥0.5.
Determining the DMSO tolerance is a critical assay development step for all cell-based assays including HCS assays. DMSO has two notable effects on HCS assays [2–4, 13–18]. At DMSO concentrations ≥5% there is a significant cell loss due to cytotoxicity and/or reduced cell adherence. At DMSO concentrations >1% but <5%, cell morphologies can be significantly altered from a well spread and attached morphology to a more rounded loosely attached morphology that interferes with the ability of the image analysis algorithm to segment images into distinct cytoplasm and nuclear regions. The DMSO tolerance of the HCS assay and the compound library stock concentration are the major factors that influence the selection of the compound concentrations for primary screening, confirmation, and follow-up studies.
The optimal compound exposure period is a critical assay development parameter for all cell-based assays that should be empirically determined for each new assay. Longer compound exposure periods can result in elevated cytotoxicity levels that may significantly hinder the ability to make reliable measurements, or may obscure target-based activity.
Determining appropriate cell fixation and nuclear staining conditions is a critical assay development parameter for all end point HCS assays [2–4, 13–18]. Combining cell fixation with Hoechst nuclear staining in a single procedure saves time and reduces the number of steps in the protocol. Cell fixation is important because the scanning and acquisition of a full 96-well or 384-well assay plate on an automated imaging platform depends upon the number of fluorescent channels and images captured per well and the focusing options selected. Scanning times may range anywhere from 10–15 min to 2–3 h per plate depending upon the complexity of the image acquisition procedure and a time-dependent drift in the assay signal may occur in unfixed samples.
To control and reduce environmental exposure levels from formaldehyde and for long-term storage of fixed assay plates, we recommend using an automated dispensing and plate washing platform for dispensing formaldehyde and for the aspiration and washing steps.
Although we have described the image acquisition process on the ImageXpress Ultra and Micro platforms, most automated imaging platforms designed for HCS with similar light sources, objective lens magnification, and detectors should be capable of capturing images.
Although we have described the translocation enhanced image analysis module of the MetaXpress software provided with the ImageXpress Ultra and Micro platforms (Fig. 2), most automated imaging platforms designed for HCS provide similar image analysis algorithms that should be capable of analyzing these images. Alternatively, images could be analyzed using third-party image analysis software such as the open-source CellProfiler image analysis software.
Acknowledgments
The studies reported herein were funded by grant support from the National Institutes of Health (NIH); R21NS073889 Johnston (PI) from the NINDS, and R01CA160423 and RO1CA183882 Johnston (PI) from the NCI.
References
- 1.Colas P (2008) High-throughput screening assays to discover small-molecule inhibitors of protein interactions. Curr Drug Discov Technol 5:190–199 [DOI] [PubMed] [Google Scholar]
- 2.Dudgeon D, Shinde SN, Shun TY, Lazo JS, Strock CJ, Giuliano KA, Taylor DL, Johnston PA, Johnston PA (2010) Characterization and optimization of a novel protein-protein interaction biosensor HCS assay to identify disruptors of the interactions between p53 and hDM2. Assay Drug Dev Technol 8:437–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudgeon D, Shinde SN, Hua Y, Shun TY, Lazo JS, Strock CJ, Giuliano KA, Taylor DL, Johnston PA, Johnston PA (2010) Implementation of a 220,000 compound HCS campaign to identify disruptors of the interaction between p53 and hDM2, and characterization of the confirmed hits. J Biomol Screen 15:152–174 [DOI] [PubMed] [Google Scholar]
- 4.Hua Y, Shun TY, Strock CJ, Johnston PA (2014) High-content positional biosensor screening assay for compounds to prevent or disrupt androgen receptor and transcriptional intermediary factor 2 protein-protein interactions. Assay Drug Dev Technol 12:395–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalonde S, Ehrhardt DW, Loqué D, Chen J, Rhee SY, Frommer WB (2008) Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J 53:610–635 [DOI] [PubMed] [Google Scholar]
- 6.Pagliaro L, Felding J, Audouze K, Nielsen SJ, Terry RB, Krog-Jensen C, Butcher S (2004) Emerging classes of protein-protein interaction inhibitors and new tools for their development. Curr Opin Chem Biol 8:442–449 [DOI] [PubMed] [Google Scholar]
- 7.Reilly M, Cunningham KA, Natarajan A (2009) Protein-protein interactions as therapeutic targets in neuropsychopharmacology. Neuropsychopharmacology 34:247–248 [DOI] [PubMed] [Google Scholar]
- 8.Wells J, McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450:1001–1009 [DOI] [PubMed] [Google Scholar]
- 9.Koehler A (2010) A complex task? Direct modulation of transcription factors with small molecules. Curr Opin Chem Biol 14:331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S, Saradhi M, Chaturvedi NK, Tyagi RK (2006) Intracellular localization and nucleocytoplasmic trafficking of steroid receptors: an overview. Mol Cell Endocrinol 246:147–156 [DOI] [PubMed] [Google Scholar]
- 11.Stauber R, Afonina E, Gulnik S, Erickson J, Pavlakis GN (1998) Analysis of intracellular trafficking and interactions of cytoplasmic HIV-1 Rev mutants in living cells. Virology 251:38–48 [DOI] [PubMed] [Google Scholar]
- 12.Stauber R (2002) Analysis of nucleocytoplasmic transport using green fluorescent protein. Methods Mol Biol 183:181–198 [DOI] [PubMed] [Google Scholar]
- 13.Johnston P, Sen M, Hua Y, Camarco D, Shun TY, Lazo JS, Grandis JR (2014) High-content pSTAT3/1 imaging assays to screen for selective inhibitors of STAT3 pathway activation in head and neck cancer cell lines. Assay Drug Dev Technol 12:55–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston PA, Shinde SN, Hua Y, Shun TY, Lazo JS, Day BW (2012) Development and validation of a high-content screening assay to identify inhibitors of cytoplasmic Dynein-mediated transport of glucocorticoid receptor to the nucleus. Assay Drug Dev Technol 10:432–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickischer D, Laethem C, Trask OJ, Williams RG, Kandasamy R, Johnston PA, Johnston PA (2006) Development and implementation of three mitogen-activated protein kinase (MAPK) signaling pathway imaging assays to provide MAPK module selectivity profiling for kinase inhibitors: MK2-EGFP translocation, c-Jun, and ERK activation. Methods Enzymol 414:389–418 [DOI] [PubMed] [Google Scholar]
- 16.Trask O, Baker A, Williams RG, Nickischer D, Kandasamy R, Laethem C, Johnston PA, Johnston PA (2006) Assay development and case history of a 32K-biased library high-content MK2-EGFP translocation screen to identify p38 mitogen-activated protein kinase inhibitors on the ArrayScan 3.1 imaging platform. Methods Enzymol 414:419–439 [DOI] [PubMed] [Google Scholar]
- 17.Trask O, Nickischer D, Burton A, Williams RG, Kandasamy RA, Johnston PA, Johnston PA (2009) High-throughput automated confocal microscopy imaging screen of a kinase-focused library to identify p38 mitogen-activated protein kinase inhibitors using the GE InCell 3000 analyzer. Methods Mol Biol 565:159–186 [DOI] [PubMed] [Google Scholar]
- 18.Williams R, Kandasamy R, Nickischer D, Trask OJ, Laethem C, Johnston PA, Johnston PA (2006) Generation and characterization of a stable MK2-EGFP cell line and subsequent development of a high-content imaging assay on the Cellomics ArrayScan platform to screen for p38 mitogen-activated protein kinase inhibitors. Methods Enzymol 414:364–389 [DOI] [PubMed] [Google Scholar]
- 19.Knauer S, Moodt S, Berg T, Liebel U, Pepper-kok R, Stauber RH (2005) Translocation bio-sensors to study signal-specific nucleo-cytoplasmic transport, protease activity and protein-protein interactions. Traffic 6:594–606 [DOI] [PubMed] [Google Scholar]
- 20.Knauer S, Stauber RH (2005) Development of an autofluorescent translocation biosensor system to investigate protein-protein interactions in living cells. Anal Chem 77:4815–4820 [DOI] [PubMed] [Google Scholar]
- 21.Burd C, Morey LM, Knudsen KE (2006) Androgen receptor corepressors and prostate cancer. Endocr Relat Cancer 13:979–994 [DOI] [PubMed] [Google Scholar]
- 22.Chmelar R, Buchanan G, Need EM, Tilley W, Greenberg NM (2006) Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int J Cancer 120:719–733 [DOI] [PubMed] [Google Scholar]
- 23.Culig Z, Hobisch A, Bartsch G, Klocker H (2000) Androgen receptor–an update of mechanisms of action in prostate cancer. Urol Res 28:211–219 [DOI] [PubMed] [Google Scholar]
- 24.Culig Z, Klocker H, Bartsch G, Hobisch A (2002) Androgen receptors in prostate cancer. Endocr Relat Cancer 9:155–170 [DOI] [PubMed] [Google Scholar]
- 25.Gregory C, Johnson RT Jr, Mohler JL, French FS, Wilson EM (2001) Androgen receptor stabilization in recurrent prostate cancer is associated with hypersensitivity to low androgen. Cancer Res 61:2892–2898 [PubMed] [Google Scholar]
- 26.Evans RM (1988) The steroid and thyroid hormone receptor family. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna N, O’Malley BW (2002) Minire-view: nuclear receptor coactivators–an update. Endocrinology 143:2461–2465 [DOI] [PubMed] [Google Scholar]
- 28.McKenna N, O’Malley BW (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465–474 [DOI] [PubMed] [Google Scholar]
- 29.Culig Z, Santer FR (2012) Androgen receptor co-activators in the regulation of cellular events in prostate cancer. World J Urol 30:297–302 [DOI] [PubMed] [Google Scholar]
- 30.Culig Z, Santer FR (2013) Molecular aspects of androgenic signaling and possible targets for therapeutic intervention in prostate cancer. Steroids 78(9):851–859 [DOI] [PubMed] [Google Scholar]
- 31.Fujimoto N, Miyamoto H, Mizokami A, Harada S, Nomura M, Ueta Y, Sasaguri T, Matsumoto T (2007) Prostate cancer cells increase androgen sensitivity by increase in nuclear androgen receptor and androgen receptor coactivators; a possible mechanism of hormone-resistance of prostate cancer cells. Cancer Investig 25:32–37 [DOI] [PubMed] [Google Scholar]
- 32.Agoulnik I, Vaid A, Nakka M, Alvarado M, Bingman WE 3rd, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM, Weigel NL (2006) Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res 66:10594–10602 [DOI] [PubMed] [Google Scholar]
- 33.Feng S, Tang Q, Sun M, Chun JY, Evans CP, Gao AC (2009) Interleukin-6 increases prostate cancer cells resistance to bicalutamide via TIF2. Mol Cancer Ther 8:665–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakka M, Agoulnik IU, Weigel NL (2013) Targeted disruption of the p160 coactivator interface of androgen receptor (AR) selectively inhibits AR activity in both androgen-dependent and castration-resistant AR-expressing prostate cancer cells. Int J Biochem Cell Biol 45:763–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi X, Xue L, Zou JX, Gandour-Edwards R, Chen H, deVere White RW (2008) Prolonged androgen receptor loading onto chromatin and the efficient recruitment of p160 coactivators contribute to androgen-independent growth of prostate cancer cells. Prostate 68:1816–1826 [DOI] [PubMed] [Google Scholar]
- 36.Xu J, Li Q (2003) Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol 17:1681–1692 [DOI] [PubMed] [Google Scholar]
- 37.Bebermeier J, Brooks JD, DePrimo SE, Werner R, Deppe U, Demeter J, Hiort O, Holterhus PM (2006) Cell-line and tissue-specific signatures of androgen receptor-coregulator transcription. J Mol Med 84:919–931 [DOI] [PubMed] [Google Scholar]
- 38.Heemers H, Schmidt LJ, Kidd E, Raclaw KA, Regan KM, Tindall DJ (2010) Differential regulation of steroid nuclear receptor coregulator expression between normal and neoplastic prostate epithelial cells. Prostate 70:959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang P, Chandra V, Rastinejad F (2010) Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol 72:247–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonneveld E, Jansen HJ, Riteco JA, Brouwer A, van der Burg B (2005) Development of androgen- and estrogen-responsive bioassays, members of a panel of human cell line-based highly selective steroid-responsive bioassays. Toxicol Sci 83:136–148 [DOI] [PubMed] [Google Scholar]
- 41.Festuccia C, Gravina GL, Angelucci A, Millimaggi D, Muzi P, Vicentini C, Bologna M (2005) Additive antitumor effects of the epidermal growth factor receptor tyrosine kinase inhibitor, gefitinib (Iressa), and the nonsteroidal antiandrogen, bicalutamide (Casodex), in prostate cancer cells in vitro. Int J Cancer 115:630–640 [DOI] [PubMed] [Google Scholar]
- 42.Luo S, Martel C, LeBlanc G, Candas B, Singh SM, Labrie C, Simard J, Belanger A, Labrie F (1996) Relative potencies of flutamide and casodex: preclinical studies. Endocr Relat Cancer 3:229–241 [Google Scholar]