Abstract
There is an increasing trend in studies utilizing cell-specific deletion of genes through conditional gene deletion by CRE recombination. Despite numerous advantages, this strategy also has limitations such as ectopic CRE-expression and germline recombination. Two commonly used gonadotropin-releasing hormone (Gnrh)-driven CRE-expressing mice both target GnRH neurons. However, a direct comparison of the cells targeted and their phenotypic outcome have not yet been presented. To compare where recombination takes place, we crossed the Gnrh-cre and Lhrh-cre lines with the Rosa26-LacZ reporter mouse. Lhrh-cre allowed recombination of the Rosa26-LacZ gene in ~700 cells, which is comparable to the GnRH neuronal population. Surprisingly, there were >20 times more LacZ expressing cells in the adult Gnrh-cre:Rosa26-LacZ than the Lhrh-cre:Rosa26-LacZ brain. The greatest differences in targeting of the Gnrh-cre and Lhrh-cre lines were found in the septum, the suprachiasmatic nucleus, and the septohypothalamic area. This difference in cells targeted was present from embryonic day 12. A prior study using the Gnrh-cre to delete the transcription factor Otx2 found fewer GnRH neurons, leading to male and female sub-fertility. To recapitulate this study, we performed a fertility assay in Otx2:Lhrh-cre mice. We confirmed the requirement for Otx2 in GnRH neuron development, fertility and correct gonadotropin hormone release in Otx2:Lhrh-cre males, but the subfertility was more modest than in Otx2:Gnrh-cre and absent in female Otx2:Lhrh-cre. This suggests that ectopic expression of Gnrh-cre contributes to the reproductive phenotype observed. Finally, the Cre alleles caused germline recombination of the flox allele when transmitted from either parent, generating embryonic lethal knock-out offspring, producing smaller live litters.
Keywords: Lhrh, Gonadotropin-releasing hormone, Otx2, Fertility, Cre-LoxP
Introduction
Conditional gene deletion in mice by the use of the cre-loxP system is a powerful tool to address the specific role of genes in cellular function and disease [1]. To obtain a conditional knock-out mouse, 2 strains are crossed; the “flox” strain carries two 34 bp loxP sequence elements flanking the gene region to be excised from the DNA, and the “Cre” strain expresses the bacteriophage-derived CRE recombinase under the control of a specific regulatory region. As with most techniques, the cre-lox system comes with certain pitfalls and limits, which, if not considered, can lead to erroneous conclusions [2–4]. These limits include the possible cytotoxicity of Cre expression [5], germline recombination [6], inefficient gene deletion [1], as well as low or non-specific Cre expression [7]. To determine the specific cells or tissues expressing Cre, successful recombination can be evaluated by PCR, southern blot, or by identifying the expression of a reporter gene such as a ROSA26-driven reporter [8–12]. Reporter mice permit determination of the approximate time at which recombination takes place. However, the sensitivity of the ROSA26 allele, containing the loxP-flanked STOP cassette, might differ from the sensitivity of the loxP site of the experimental gene. In addition, both the site of Cre transgene insertion into the DNA, as well as the DNA region used to drive transgene expression impact the fidelity of Cre expression [13–15].
The role of well-defined transcriptional regulatory regions, which allow specific expression of a peptide to a limited cell population, are well studied for gonadotropin-releasing hormone (GnRH or LHRH) [13–18], a neuropeptide required for pubertal onset and fertility [11, 19–22]. Despite the careful characterization of the regulatory region of Gnrh1, it has proven difficult to create a Cre mouse that recapitulates the onset of GnRH expression and specifically targets this small neuronal population compromised of a little under 1,000 neurons in the adult mouse. These challenges are likely associated with the unique origin of GnRH neurons in the vomeronasal organ [23]. After GnRH neurons arise around embryonic day 10 (e10), they initiate their migration into the ventral forebrain, and localize throughout the anterior hypothalamic area where most GnRH neurons are found at ~e18–e19. Once at their final location, GnRH neurons project to the median eminence (ME) and release GnRH in a pulsatile pattern promoting luteinizing hormone (LH) and follicle-stimulating hormone (FSH) release from the anterior pituitary. The scattered location of GnRH neurons in the anterior hypothalamic area makes it challenging to specifically manipulate gene expression in these neurons by viral injections [24, 25], and has led to the generation of at least 4 different Gnrh-driven CRE expressing mice: the Tg(Gnrh1-cre)35Awo [26], the Tg(Gnrh1-cre)1Dlc [27], the Tg(Gnrh1-cre)1Gsc [28] and the Tg(Gnrh1-cre)1Rsp [29]. The Gnrh-cre (Tg[Gnrh1-cre]35Awo) mouse was generated by inserting a transgene that utilizes the mouse 3.4 Kb promoter to drive Cre recombinase expression in all GnRH neurons, in addition to some ectopic expression [26]. The Lhrh-cre (Tg[Gnrh1-cre]1Dlc) mouse, in which a mouse bacterial artificial chromosome containing the Gnrh1 gene driving Cre expression is inserted as a transgene into the genome, leads to Cre expression in ~96% of the GnRH neurons, with very limited off-target expression [16, 27].
Due to the known widespread ectopic expression of the Gnrh-cre (Tg[Gnrh1-cre]35Awo), we checked the extent to which this would be physiologically relevant. We hypothesized that the deletion of the developmental homeodomain transcription factor Otx2 using the Lhrh-cre (Tg[Gnrh1-cre]1Dlc, Otx2:Lhrh-cre), might lead to a less severe reproductive phenotype than Otx2 deletion using the Gnrh-cre (Otx2:Gnrh-cre) [30].
Materials and Methods
Mouse Breeding
Mouse experiments were approved by the Institutional Animal Care and Use Committee at the University of California, San Diego. Otx2-flox [31], Six6-flox (Mellon laboratory, unpublished), Lhrh-cre (Tg[Gnrh1-cre]1Dlc) [27], Gnrh-cre (Tg[Gnrh1-cre]35Awo) [26], Rosa26-LacZ (JAX #003309), and Rosa26-TdTo- mato (JAX #007909) mice were maintained on a C57BL/6J genetic background, and housed under a 12 h light-dark cycle, with food and water ad libitum. Mice were sacrificed by isoflurane or CO2 overdose, followed by cervical dislocation. All mice were systematically genotyped for germline recombination of the Floxed allele, and only mice without germline recombination were included in our study.
Fertility Assessments, Pubertal Onset, and Embryo Collection
Procedures for pubertal onset, embryo collection and fertility assessment were performed as described in detail previously [19, 32, 33]. For fertility assays, 8-week-old Otx2:Lhrh-cre mice were housed with Otx2-flox/flox mice. The number of litters born and the number of pups per litter were recorded over 180 days. Inspection of pubertal onset was initiated after weaning (20 days of age). Mice were inspected daily for vaginal opening in the females and preputial separation in the males. The day of preputial separation and vaginal opening was recorded. To generate embryos (e) of the desired age, timed-mated females were sacrificed at gestation day 11.5 (e11.5), 12.5 (e12.5), 13.5 (e13.5), and 17.5 (e17.5). Embryos were fixed in a solution composed of 10% acetic acid, 30% formaldehyde, 60% ethanol, overnight at 4 °C, and dehydrated in 70% EtOH before embedding in paraffin. Sagittal sections (10 μm) were floated onto SuperFrost Plus slides (Thermo Fisher Scientific) and processed for immunohistochemistry.
Hormone Measurements
Blood was collected by cardiac puncture. Serum was separated by centrifugation (RT, 2,300 g for 15 min), and serum stored at −20 ° C till RIA analysis of LH, FSH, and testosterone at the Ligand Assay and Analysis Core in the Center for Research in Reproduction at University of Virginia.
Protein and Transcript Detection
Single immunohistochemistry and quantitative RT-PCR were performed as previously described [11, 19]. For double immuno-histochemistry, the same protocol as for single immunohisto-chemistry was used, with the modification that after developing and imagine the anti-CRE (1/1,000) staining using Impact DAB (brown), the slides were washed in PBS-Tween 0.02%, followed by 5 min in denaturing solution (Biocare #DNS001), after which the sections were blocked in 5% goat serum and Avidin-Biotin solutions (Vector Labs), and the anti-LHRH (1/1,000) antibody was applied O/N following the regular immunohistochemistry protocol, using Impact VIP (purple) to visualize GnRH expressing cells. All antibodies were validated using negative controls such as cre− tissue for the anti-CRE antibody, and Cre−:ROSA26-LacZ+ tissue for the anti-LacZ antibody. We have previously validated the GnRH and LacZ antibodies [11, 19, 32]. The primary antibodies used for single immunohistochemistry are rabbit anti-GnRH (Thermo Scientific #PA1–121, dilution 1/1,000, RRID:AB_325077), chicken anti-beta galactosidase (Abcam #AB9361, dilution 1/1,000, RRID:AB_307210), rabbit anti-CRE recombinase (Biolegend #908001, dilution 1/500, RRID: AB_2565079), and rabbit anti-RFP (Abcam #AB62341, dilution 1/500, RRID:AB_945213). For double immunohistochemistry, we used rabbit anti-CRE recombinase (Biolegend #908001, dilution 1/1,000, RRID: AB_2565079), and rabbit anti-LHRH antibody (Immunostar #20075, dilution 1/1,000, RRID: AB_572248). Primary antibodies were detected by secondary biotinylated antibodies (Vector Laboratories #BA-1000 and #BA-9010) and visualized using the Vectastain ABC elite kit with VIP, Impact VIP, or Impact DAB peroxidase (Vector Labs). To compare protein expression patterns between mouse lines, staining was done in parallel with identical conditions. Hypothalamic Gnrh1 content was determined as described previously [19, 34]. The enzymatic LacZ staining was performed by incubating the tissue overnight at 37 ° C in X-gal staining buffer (1 mg/mL 4-chloro-5-bromo-3-indolyl-β-galactosidase, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 2 mM MgCl2 in 100 mM PBS, pH 7.4).
Statistical Analysis
Data were analysed by Student t test or two-way ANOVA using GraphPad Prism 7 (Graph Pad Software, La Jolla, CA, USA) as noted in the figure legends. Significant differences were designated as p < 0.05.
Results
Substantial Difference in the Number of Cells Targeted by the Gnrh-cre versus the Lhrh-cre Allele
To determine whether the Lhrh-cre and Gnrh-cre alleles allow recombination in the same number of cells in the same locations, we performed LacZ staining in adult Lhrh-cre:Rosa26-LacZ and Gnrh-cre:Rosa26-LacZ mice and counted the number of LacZ expressing cells. We found >20 times more LacZ expressing cells in the Gnrh- cre:Rosa26-LacZ as compared to the Lhrh-cre:Rosa26-LacZ mice (Fig. 1a). The additional LacZ-expressing cells in the Gnrh-cre:Rosa26-LacZ were restricted to the ventral forebrain and were principally present in the septum (Fig. 1b, c). To determine if the additional LacZ positive cells expressed LacZ due to early recombination, but now did not actively transcribe the Cre allele, we performed staining for the CRE protein, which is only present in cells that are actively transcribing Cre. We found the CRE expression pattern to be similar to that of LacZ in both mouse lines (Fig. 1d), although with a slight reduction in the number of cells expressing CRE as compared to LacZ in adult Gnrh-cre:Rosa26-LacZ mice (Fig. 1b, c). To revalidate the targeting of GnRH neurons of the 2 studied Cre-alleles [26, 27], we performed double immunohisto-chemistry in adult brains of Lhrh-cre and Gnrh-cre mice. First, we validated the specificity of our antibodies performing double immunohistochemistry in a wild-type mouse, where CRE is not expressed (C57BL/6J), in a CRE-expressing mouse (Gnrh-cre+ mouse) and in a mouse model lacking GpRH expressing neurons [11]. We detected GnRH neurons in control mice (Fig. 1e, wild-type and Gnrh-cre+ mouse). This signal was specific as evidenced by the absence of GnRH staining in the negative control tissue [11] (Fig. 1e, GnRH lacking mouse). The anti-CRE antibody specifically detected CRE expressing cells (Fig. 1e, compare wild-type to Gnrh-cre+ mouse). The anti-CRE and anti-LHRH antibodies did not cross-react as evidenced by the distinction of cells expressing GnRH (purple) and CRE (brown) in the Gnrh-cre mouse (Fig. 1f). In agreement with previous studies [26, 27], we observed that Lhrh-cre targeted >95% of the GnRH expressing neurons and the Gnrh-cre targeted all the detected GnRH neurons in adulthood (Fig. 1f). Supporting our LacZ and CRE staining in Figure 1b and d, the Gnrh-cre, but not the Lhrh-cre allele, drove CRE expression in a high number of non-GnRH expressing cells (Fig. 1f, septum and POA). This high ectopic expression of Gnrh-cre was further supported by LacZ staining in Lhrh-cre:Rosa26-LacZ and Gnrh-cre:Rosa26-LacZ mice (Fig. 2), where LacZ staining reflects all living cells that have had the flox allele recombined prior to the time of imagine, and does not depend on active expression of the Cre-allele. We carefully compared the LacZ expression pattern in the adult brain of Lhrh-cre:Rosa26-LacZ and Gnrh-cre:Rosa26-LacZ mice and found major differences in the number of LacZ expressing cells, specifically we noted a significant number of LacZ expressing cells in the lateral septum (Fig. 2), preoptic area, the suprachiasmatic nucleus (SCN), the septohypothalamic nucleus, the olfactory tubercule, and the medial hypothalamus.
Fig. 1.
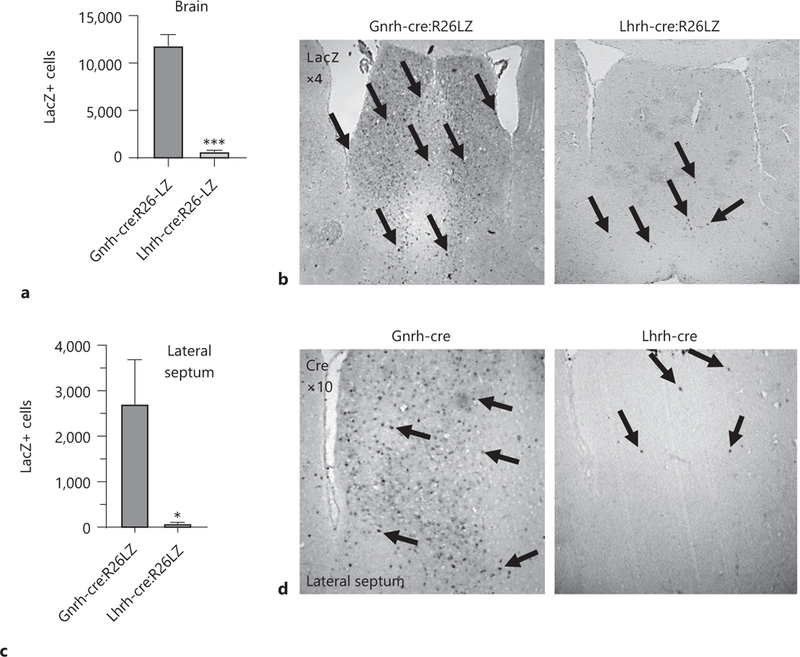
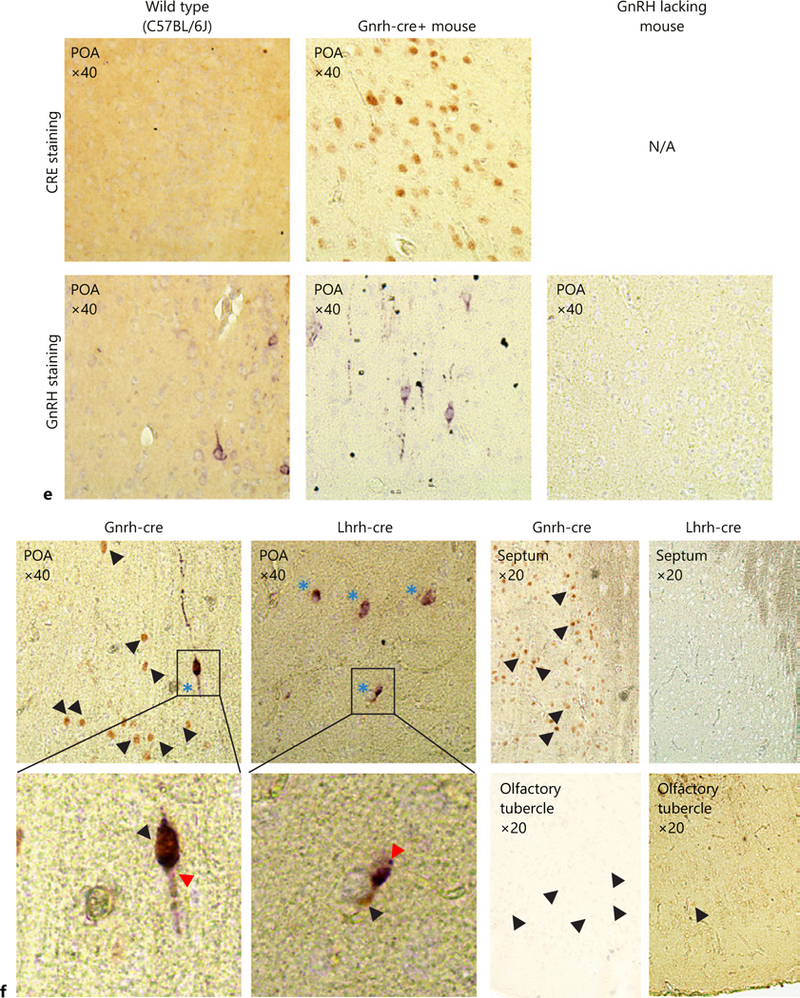
Quantification of LacZ positive (LacZ+) cells in the whole brain (a) and the lateral septum (c) from adult Gnrh-cre:Rosa26-LacZ and Lhrh-cre:Rosa26-LacZ mice, n = 3, Student t test; * p < 0.05. Immunohistochemical staining in the lateral septum for (b) LacZ in Gnrh-cre:Rosa26-LacZ and Lhrh-cre:Rosa26-LacZ (×4), and (d) CRE in Gnrh-cre and Lhrh-cre mice (×10). Arrows high-light LacZ or CRE expressing cells in coronal sections. e Negative controls for double immunohistochemistry, showing specificity of the anti-CRE and anti-LHRH antibodies. To assure that no cross-reaction of the antibodies occurred, the antibody validation was done following the entire double immunohistochemistry protocol where the anti-LHRH antibody was omitted for the CRE staining, and the anti-CRE antibody omitted for the GnRH staining. The GnRH-lacking mouse is the Gnrh-cre:Vax1-flox/flox mouse that we previously published as lacking GnRH expressing neurons in the hypothalamus as a negative control for the anti-LHRH antibody [11]. f Double immunohistochemistry for GnRH (purple, GnRH staining is indicated by a red arrow head) and CRE (brown, single staining is indicated by a black arrow head). Dual staining is indicated by blue stars. n = 3 adult brains.
Fig. 2.
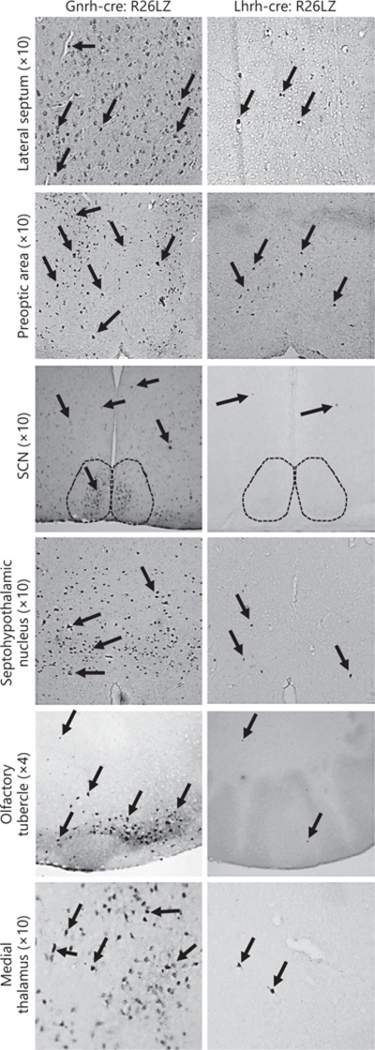
Illustrative immunohistochemical images of LacZ staining in coronal sections of adult Gnrh-cre:Rosa26-LacZ and Lhrh-cre:Rosa26-LacZ brains. Arrows highlight LacZ expressing cells. All images are ×10, except the olfactory tubercule, which is ×4. SCN, suprachiasmatic nucleus.
The Gnrh-cre Allele Targets More Cells than the Lhrh-cre Allele from e12.5 Onward
To determine when the difference in cells targeted by the 2 Cre alleles arose, we performed CRE staining in Gnrh-cre and Lhrh-cre embryos at embryonic day 11.5 (e11.5), e12.5, e13.5 and e17.5. We detected CRE expression as early as e11.5 in both mouse lines (Fig. 3, e11.5, nose). However, more cells expressed CRE at e12.5 in the Gnrh-cre than the Lhrh-cre embryos, a pattern we observed in all older age groups. Again, the major difference in expression was found in the developing septum, and hypothalamus, a difference that was prominent at e13.5 and e17.5 (Fig. 3).
Fig. 3.
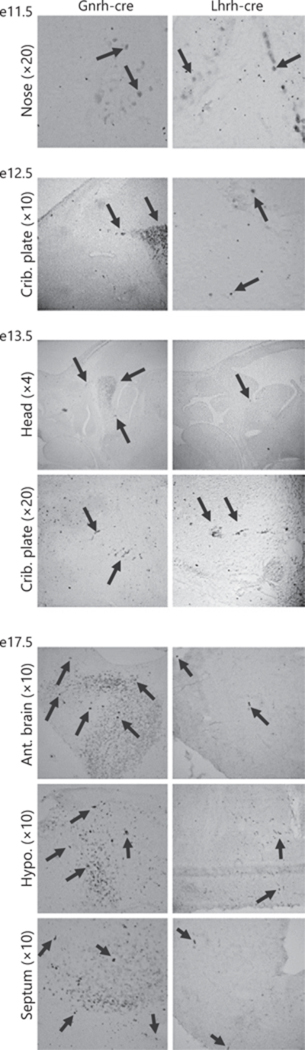
Early developmental differences in CRE expression in the Gnrh-cre and Lhrh-cre brains. Sagittal sections of Gnrh-cre and Lhrh-cre embryos were stained for CRE at embryonic day 11.5 (e11.5), e12.5, e13.5, and e17.5. Ant. brain, anterior brain; crib. plate, cribriform plate; hypo, hypothalamus. Images were taken at ×4, ×10, or ×20 as indicated in the figure.
Brain Areas with High CRE Expression in the Gnrh-cre Mouse also Express Otx2
Using in situ hybridization images obtained from www.brain-map.org (consulted April 2018), we mapped the Otx2 expression pattern in the developing mouse head. As expected at e13.5 Otx2 expression in the olfactory placode, cribriform plate, and anterior head colocalize with the expression pattern of Gnrh1 (Fig. 4, Gnrh1 e13.5, and Otx2 e13.5) and CRE expression in the Gnrh-cre mouse (Fig. 3, e12.5 and e13.5). At e15.5, Otx2 expression was high in the hypothalamus, a time when GnRH neurons start arriving in this brain area. At e18.5, Otx2 was highly expressed in the septum, an area with high CRE expression in the Gnrh-cre mouse at e17.5 (Fig. 3, e17.5). Otx2 expression was not observed in the hippo-campus or the SCN (Fig. 4, Otx2 e18.5).
Fig. 4.
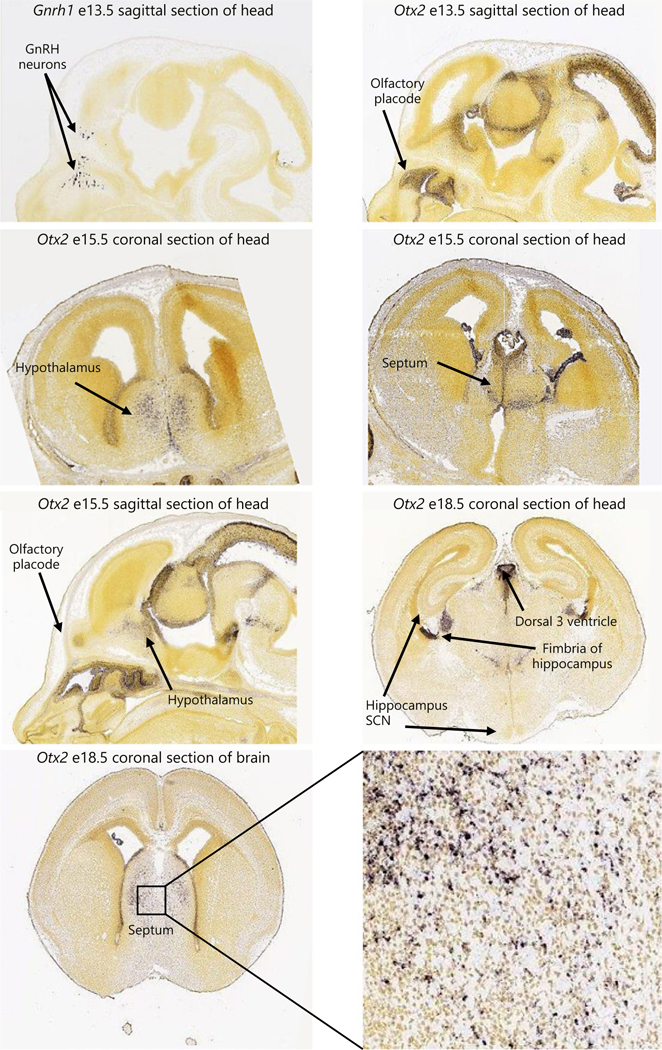
In situ hybridization images obtained from www.brain-map.org (consulted in April 2018) for Gnrh1 and Otx2 in brain sections from embryos (e) at the indicated ages. Positive staining is brown.
Otx2 Expression Is Required for GnRH Neuron Development
A prior study using the Gnrh-cre (Tg[Gnrh1-cre]35Awo) to delete the Otx2-flox allele (Otx2:Gnrh-cre) determined that the transcription factor Otx2 within GnRH neurons is required for proper GnRH neuron development and fertility [30]. Based on the different expression patterns of Gnrh-cre and Lhrh-cre (Fig. 1–3), and the overlap between CRE expression in the Gnrh-cre (Fig. 3) and Otx2 during development (Fig. 4), we decided to repeat this study, deleting the Otx2-flox allele using Lhrh-cre (Tg[Gnrh1-cre]1Dlc, Otx2:Lhrh-cre). We first asked how many GnRH neurons would be present in Otx2:Lhrh-cre mice at e13.5 and e17.5. As expected, in e13.5 controls, the full complement of GnRH neurons was located within the nasal and cribriform plate regions [23] (Fig. 5a, b). In contrast, at e13.5 in Otx2:Lhrh-cre, a reduction in the number of GnRH neurons was observed in all regions of migration (Fig. 5a, b), whereas an accumulation of GnRH neurons was found at the cribriform plate and a reduction in the hypothalamus and brain at e17.5 (Fig. 5c, d). The intensity of the GnRH staining in neuronal terminals at the ME was also substantially reduced in Otx2:Lhrh-cre embryos (Fig. 5c, ME). This reduction in GnRH was maintained into adulthood at the level of Gnrh1 transcription (Fig. 5e).
Fig. 5.
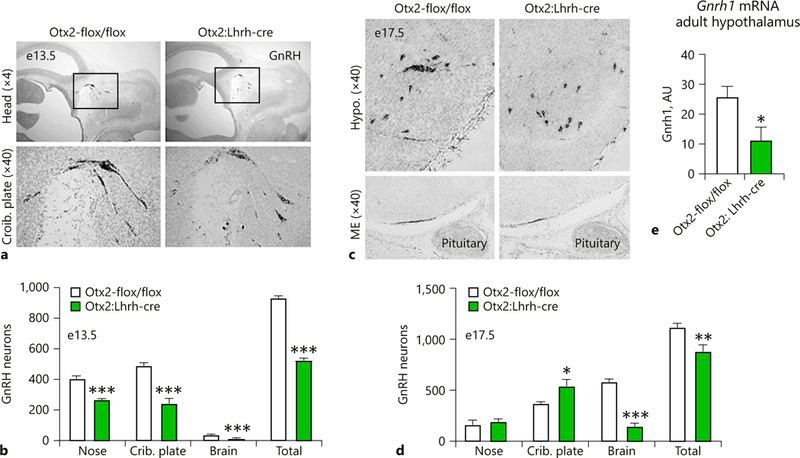
Otx2:Lhrh-cre embryos have abnormal GnRH neuron location in the head. Immunohistochemical staining and quantification of GnRH expressing cells on sagittal sections at (a, b) e13.5 and (c, d) e17.5. The black boxes in (a) indicate the area of the section that has been enlarged below. Crib. plate, cribriform plate; ME, median eminence. Two-way ANOVA, * p < 0.05, ** p < 0.01, and *** p < 0.001 as compared to control mouse, n = 3–4. (e) Quantitative RT-PCR analysis of Gnrh1 in adult male hypothalamus. Student t test, * p < 0.05, n = 4.
Specific Deletion of Otx2 from GnRH Neurons Leads to Male Subfertility
To determine if the 56% reduction in Gnrh1 in adult Otx2:Lhrh-cre mice (Fig. 5e) resulted in subfertility, we performed a fertility assay. We found that the number of litters generated by Otx2:Lhrh-cre males, but not Otx2:Lhrh-cre females, was reduced (Fig. 6a). Surprisingly, we noted a high rate of pup mortality when mating male Otx2-flox/flox mice to female Otx2-flox/WT:Lhrh-cre mice. The reduction of litter size was due to some pups being born with severe cranial abnormalities, a developmental defect of full body knock-out Otx2 mice [35–37]. This suggests that Lhrh-cre transgene expression was occurring within the ovary, allowing germline recombination and transmission of a recombined null allele. Genotyping of progeny, where the Cre transgene was obtained from the female, demonstrated recombination within tail DNA of the offspring (Fig. 6b). Interestingly, in many cases, progeny of Otx2:Lhrh-cre exhibited recombination in tail DNA, independent of whether they inherited the Cre allele, suggesting that recombination occurred frequently during female germ cell development (Fig. 6b). To determine the frequency of germline recombination, we designed primers allowing us to analyze the percent of mono- and bi-allelic (heterozygote and homozygote recombination respectively) germline recombination of the Six6-flox allele, a mouse model that is fully viable as both a heterozygote and homozygote germline knockout mouse [19]. Analyzing our data from Six6:Lhrh-cre and Six6:Gnrh-cre mice, we found that both Lhrh-cre and Gnrh-cre caused germline recombination (mono- or bi-allelic) of the Six6-flox allele when the Cre-allele was transmitted from either the mother or the father (Fig. 6c). The recombination occurred more frequently when the Cre-allele was transmitted from the mother than from the father, and germline recombination of the Six6-flox allele was more frequent in the Lhrh-cre mice than the Gnrh-cre mice (Fig. 6c, Two-way ANOVA, overall effect of sex p = 0.0053, overall effect of mouse strain p = 0.0022). The germline recombination was obvious in our R26Rfp reporter mice, where Gnrh-cre induced R26Rfp recombination generating mice with a red-pinkish color (Fig. 6d). To determine how frequently homozygous recombination of the Six6-flox allele occurred, we calculated the percentage homozygous recombination (Fig. 6e). When the Lhrh-cre allele was transmitted from the mother, almost 1/5 of the offspring had homozygous recombination of the Six6-flox allele (Fig. 6e). Homozygous recombination of the Six6-flox allele happened in < 5% of the pups generated from Gnrh-cre males, females, and Lhrh-cre males (Fig. 1e, Two-way ANOVA, overall effect of sex p = 0.0206). The high ratio of germline recombined pups from Lhrh-cre mothers was validated using staining of the gonads where the germline recombination would cause functional recombination of a tracer gene such as R26LacZ or R26Rfp. LacZ or RFP staining of gonads confirmed that recombination was occurring in some oocytes (Fig. 6f, arrows). We were unable to see recombination in the sperm probably due to the high compaction of the sperm head (Fig. 6f, testis). The frequent homozygous recombination in pups from female Lhrh-cre mice caused Otx2:Lhrh-cre females to produce smaller litters than controls, due to the lethality of Otx2 knock-outs [30]. In contrast, males fathered litters the same size as controls (Fig. 6g). As the only fertility defect detected in Otx2:Lhrh-cre females was a decrease in litter size, which we attributed to the lethality of the offspring to germline recombination, we next focused on the Otx2:Lhrh-cre males.
Fig. 6.
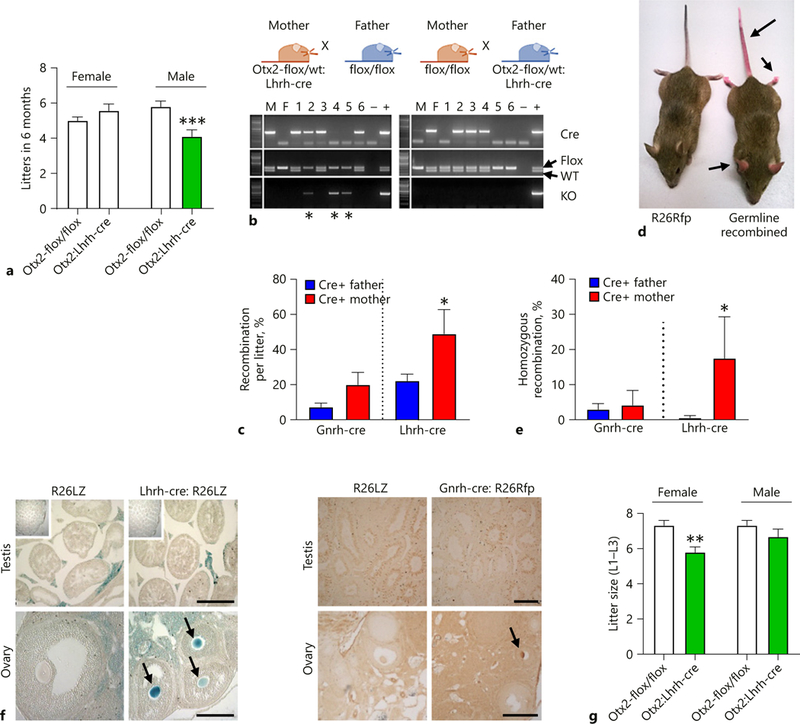
Otx2:Lhrh-cre males, but not females, are subfertile. a A fertility assay established the number of litters born in 6 months (n = 7–15). b Example of PCR showing germline recombination when Lhrh-cre is transmitted by the dam (*) in the Otx2:Lhrh-cre strain. Due to lethality of pups with homozygous recombination of the Otx2 allele, we were unable to calculate whether the recombination was mono- or bi-allelic in this mouse strain. To establish the re-combination frequency and whether it was mono- or bi-allelic, we analyzed recombination in Six6:Lhrh-cre and Six6:Gnrh-cre mice because Six6-flox mice with homozygous recombination is viable. Histograms showing (c) the average percentage of pups with germ-line recombination of the Six6-flox allele when the indicated Cre-alleles were transmitted from the father (blue) or mother (red), and (e) the percentage of pups with homozygous recombination. n = 3–14 litters. Two-way ANOVA followed by a Tukey’s multiple comparison test. * p < 0.05 as compared to opposite sex of the same mouse strain. d Picture showing germline recombination when the Gnrh-cre was transmitted by the dam in the Gnrh-cre:Rosa26-Td-Tomato mouse line (arrows). f X-gal staining in Lhrh-cre:Rosa26-LacZ (left images, blue staining) and immunohistochemistry to detect the TdTomato protein in Gnrh-cre:Rosa26-TdTomato (right images, brown staining) revealed recombination within the female germline (arrows, ovary). The square in the top left corner of the testis shows intact testis morphology (×4). Scale bar 50 μm. g Average litter sizes generated of Otx2-flox/flox (control) and Otx2:Lhrh-cre mice (n = 40–76). M, male; F, Female; WT, wild-type allele; KO, knock-out allele; Flox, floxed allele; –, negative control; +, positive control. a, g Student t test compared to control of either sex, ** p < 0.01; *** p < 0.001.
Otx2:Lhrh-cre Males Have Decreased LH Serum Levels and Reduced Sperm Production
GnRH stimulates the release of the gonadotropins FSH and LH, which are the 2 hormones required for fertility. Circulating levels of both LH and FSH were significantly reduced in Otx2:Lhrh-cre males (Fig. 7a, b). This led to a slight, non-significant, reduction in testosterone (Fig. 7c). Despite a reduction in hormone levels, testes weight (Control = 6.4 ± 0.4 mg, Otx2:Lhrh-cre 6.5 ± 0.5 mg, n = 5, Student t test, p > 0.05) and seminal vesicle weights (normalized to body weight) were comparable between control and Otx2:Lhrh-cre males (Control = 0.0064 ± 0.0006, Otx2:Lhrh-cre, 0.0006 ± 0.001, n = 5, Student t test, p > 0.05). In contrast, the total number of sperm was significantly reduced in Otx2:Lhrh-cre (Fig. 7d).
Fig. 7.
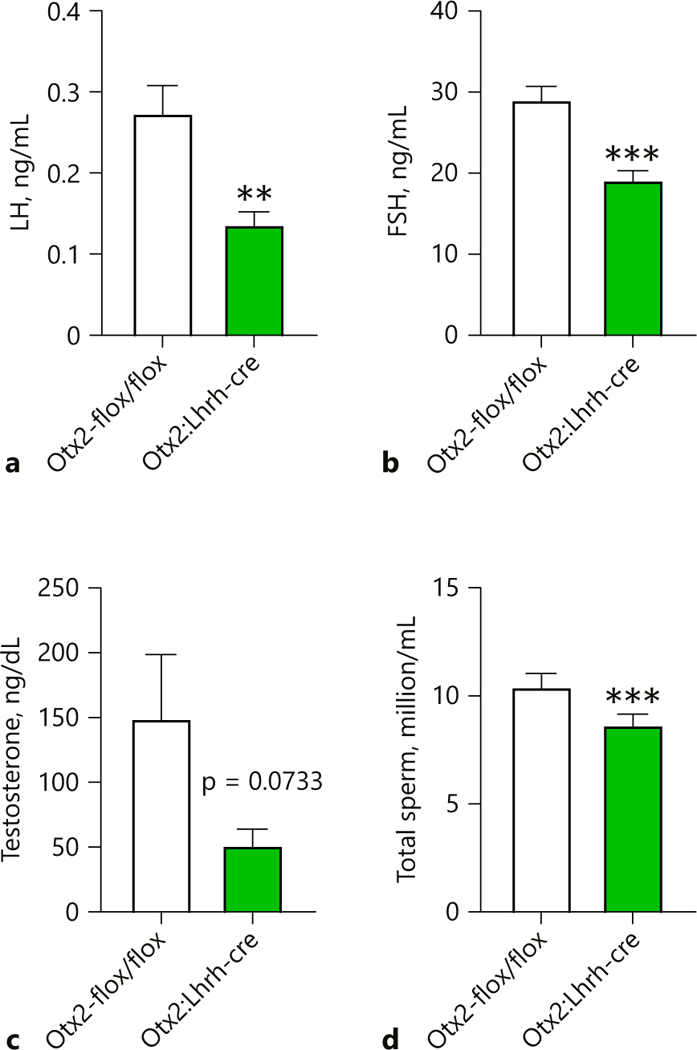
Loss of Otx2 expression results in decreased LH serum levels and fewer sperm. Average serum (a) LH and (b) FSH levels of 6–8-month-old male mice (n = 5–7). c Average testosterone level of 6–8-month-old males (n = 5). d Average number of sperm per epididymis of 6–8-month-old males (n = 5). Student t test, ** p < 0.01, *** p < 0.01.
Discussion
Lhrh-cre Recombination in the Ovary of Otx2:Lhrh-cre Results in Smaller Litters
The surprising difficulty in reproducing scientific findings is a major concern in research [38], and an increasing focus of the National Institutes of Health (NIH; http://grants.nih.gov/grants/RigorandReproducibilityChart508.pdf). Although very few techniques and experimental designs come without pitfalls, understanding and recognizing such limits is important to correctly interpret data. While the Lhrh-cre transgenic mouse line offers an excellent percentage of recombination in GnRH neurons, targeting over 96% of adult GnRH neurons [27], and efficiently deletes the floxed allele [10, 39], our study demonstrates that CRE activity is not just limited to the brain in these mice. We observed CRE recombinase activity in the female oocyte of both Lhrh-cre and Gnrh-cre, which resulted in germline recombination in progeny derived from females. Based on other reports, it is not unexpected that the GnRH promoter could drive transgene expression within the oocyte [40, 41]. Although we did not detect recombination in sperm using staining, based on our PCR results, up to 20% of the offspring generated from a Cre+ father experienced germline recombination. When the Lhrh-cre allele was transmitted from the mother, ~50% of pups had the flox-allele recombined in the entire body (germline recombination), where ~1/5 of these pups experienced homozygous recombination (bi-allelic recombination), generating a full body, germline knock-out mouse. The frequency of germline recombination was significantly lower when the Lhrh-cre allele was transmitted from the father and the frequency of homozygous recombination when the Lhrh-cre allele was transmitted from the father was a rare event. In both the Gnrh-cre and the Lhrh-cre mice, the frequency of germline re-combination was higher when the Cre-allele was transmitted from the female, as compared to the male. Overall, germline recombination, whether mono- or bi-allelic was more frequent in Lhrh-cre than Gnrh-cre mice. Because Otx2 knock-out is lethal [35–37], we attribute the reduced litter size of Otx2:Lhrh-cre females to the high frequency of homozygous germline recombination when the Lhrh-cre allele was transmitted from the mother, resulting in a reduction in the litter size. This study clearly illustrates that germline recombination is prominent in the 2 studied mouse lines, and if not considered, can cause wrongful conclusions.
Extensive Ectopic Expression of the Gnrh-cre Allele in the Brain
The cre-lox system is a wonderful tool to generate conditional knock-out and/or knock-in mice and allows for efficient cell-tracing. However, in some instances, lack of consideration for the limits of the cre-lox system can lead to overstated or misleading conclusions. Germline re-combination and ectopic expression of different Gnrh-driven cre mouse lines has previously been described in the literature. However, prior to this study, a careful side-by-side comparison of the expression pattern of the widely used Lhrh-cre (Tg[Gnrh1-cre]1Dlc) [10, 27] and Gnrh-cre (Tg[Gnrh1-cre]35Awo) [26, 42] alleles had not been performed. Our side-by-side comparison of the number of cells targeted by Gnrh-cre and Lhrh-cre confirmed previous reports of a greater number of cells targeted by the Gnrh-cre allele than the Lhrh-cre allele in the brain [26, 27]. The difference in CRE expression could be detected from e12.5 onward in the developing mouse brain. The early increase in CRE expression in non-GnRH neurons is important to consider when the studied flox-allele deletes genes important in septum and ventral forebrain development [15]. To determine the number of cells that had been targeted by both the Lhrh-cre and Gnrh-cre, we used lineage tracing, which allows the expression of a tracer gene (here LacZ) in all cells that have expressed CRE prior to the time of staining. We found >20-fold more cells expressing LacZ in the Gnrh-cre:Rosa26-LacZ as compared to the Lhrh-cre:Rosa26-LacZ brain. The additional LacZ expression in the Gnrh-cre:Rosa26-LacZ mouse was restricted to the ventral forebrain, and principally observed in the septum, a brain area known to be targeted by both the Lhrh-cre and Gnrh-cre promoters [13, 26, 27, 39, 43], as well as the hypothalamus, including the SCN. A part of the Gnrh1 promoter-driven CRE expression in the lateral septum most likely reflects a neuronal population, which transiently activates the Gnrh1 promoter [15]. To validate that no genetic derivation or mouse strain differences [44] had impacted the CRE or GnRH expression in our mice, we performed double immunohistochemistry for CRE and GnRH in adult Gnrh-cre and Lhrh-cre mice. As expected, both Lhrh-cre and Gnrh-cre targeted 95–100% of GnRH expressing neurons in adulthood. The dual immunohistochemistry confirmed that the Gnrh-cre allele drove CRE expression in a significant number of non-GnRH expressing cells, with the most profound expression pattern in the lateral septum and the broader hypothalamic area. In agreement with others, we found fewer CRE, than LacZ, expressing cells in the adult septum. This transient expression of CRE has previously been noted [10, 45], and suggests some GnRH neurons might silence the Gnrh-promoter prior to adulthood. However, it should be noted, that once the Cre-allele has been expressed in a cell, allowing the recombination of the “flox” sequences, this DNA re-combination is permanent. Therefore, using lineage tracing to understand the number of cells targeted at any given time point prior to staining more accurately reflects the cells where the gene of interest has been deleted. Thus, to determine where the Otx2-flox/flox allele could have been deleted, in addition to within GnRH neurons, we compared Otx2 in situ hybridization images from www.brain-map.org. Comparing the overlap between Gnrh-cre targeted areas and Otx2 expressing areas during development, showed that Gnrh-cre targets areas that later express Otx2 including the preoptic area of the hypothalamus, an area where GnRH neurons are located in adult-hood, and the lateral septum, a brain area known to be critical for sexual behavior [46, 47]. It will thus be of interest in future studies to determine if Otx2:Gnrh-cre mice have abnormal sexual behavior.
The Number of GnRH Neurons Required for Male and Female Fertility
Otx2 is expressed in GnRH neurons, where it regulates the expression of GnRH as well as the survival of these neurons [22, 30, 34, 36, 41, 48, 49]. Despite the increased number of cells targeted by the Gnrh-cre allele from as early as e12.5 (see paragraph 5.2), GnRH neuron distribution and the number of GnRH neurons was comparable between Otx2:Lhrh-cre (this study) and Otx2:Gnrh-cre [30]. The reduction of brain/hypothalamic GnRH neurons in Otx2:Lhrh-cre at e17.5, and the decreased targeting to the ME, was a defect also noted in Otx2:Gnrh-cre mice [30], supporting the validity of both mouse lines for the study of GnRH neuron development.
GnRH neuron migration is halted around birth; thus, any delay in GnRH neuron migration causes a reduction of hypothalamic GnRH neurons and this has serious consequences to GnRH neuron targeting to the ME and fertility [10, 19, 30, 32, 45]. We found an increase in the number of GnRH neurons at the cribriform plate in e17.5 old Otx2:Lhrh-cre embryos. This increase in GnRH neurons at the entrance to the brain (cribriform plate) was associated with a reduction of GnRH neurons in the brain, although the total number of GnRH neurons in these embryos was only decreased by 21%, a reduction not expected to impact fertility [19, 32, 36, 50, 51]. However, our study was not designed to address if the differential distribution of GnRH neurons in Otx2:Lhrh-cre mice was caused by GnRH neuron death, abnormal migration, or a change in proliferation. The reduction of GnRH neurons in the brain at e17.5 in Otx2:Lhrh-cre mice was in agreement with the overall reduction of hypothalamic Gnrh1 gene expression in adulthood, and caused reduced circulating LH and FSH in adult males. The reduction of LH and FSH in Otx2:Lhrh-cre males led to subfertility, supporting previous findings using Otx2:Gnrh-cre animals [30]. Otx2:Lhrh-cre male subfertility was characterized by a decrease in the number of litters sired and fewer sperm, a phenotype recapitulated in Otx2 heterozygote males [36]. This supports that correct dosage of Otx2 within GnRH neurons is critical for normal reproductive function. The decreased expression of Gnrh1 was associated with a reduction in LH and FSH, and a slight reduction in testosterone, probably causing the subfertility. Surprisingly, unlike Diaczok et al. [30], we did not observe any fertility phenotype in the Otx2:Lhrh-cre females, which have normal ovaries (not shown), suggesting correct release of LH and FSH. The maintained fertility of Otx2:Lhrh-cre females is supported by other studies showing that fewer than 34% of GnRH neurons are required for estrous cycling and fertility [32, 50]. The greater subfertility of the Otx2:Gnrh-cre females [30], as compared to the Otx2:Lhrh-cre (this study) might be the result of germline recombination in the female Otx2:Gnrh-cre oocytes, incomplete recombination of the Otx2-flox allele when using the Lhrh-cre allele, or ectopic expression of the Gnrh-cre allele.
Conclusion
We have confirmed the importance of Otx2 within GnRH neurons for establishing a correctly located GnRH neuronal population, and its control of male fertility. The comparison of CRE expression during development and adulthood between Gnrh-cre and Lhrh-cre mice identified a significant difference in the number of cells targeted as early as e12.5. Specifically, the Gnrh-cre has high ectopic expression in the preoptic area and the septum. Finally, both the Gnrh-cre and Lhrh-cre alleles can recombine in both male and female germline cells, thus allowing the generation of mosaic and full body-knock out off-spring.
Acknowledgments
We thank Susan Mayo, Sunamita Leming, Ikuo Kimura, Ichiko Saotome, Erica Pandolfi, Duong Nguyen, and Jason D. Meadows for technical assistance. We thank Qingbo Tang and Carolyn Kelley for assistance with the early stages of this research. We thank Dr. Siew-Lan Ang (MRC NIMR, London, UK) for the Otx2-flox mice, Dr. Catherine Dulac (Harvard University, Cambridge, MA, USA) for the Lhrh-cre mice, and Dr. Andrew Wolfe (Johns Hopkins University) for the Gnrh-cre mice.
Funding Sources
This work was supported by NIH Grants R01 HD072754 and R01 HD082567 (to P.L.M.). It was also supported by NICHD/NIH P50 HD012303 as part of the National Centers for Translational Research in Reproduction and Infertility (P.L.M.). P.L.M. was also partially supported by P30 DK063491, P30 CA023100, and P42 ES101337. H.M.H. was partially supported by K99/R00 HD084759 and the USDA National Institute of Food and Agriculture Hatch project 1018024. C.T. was partially supported by the Endocrine Society and R.H. was partially supported by the Howell Foundation and the Frontiers of Innovation Scholars Program, UC San Diego. D.D.C. was partially supported by T32 GM008666 and T32 DK007541 and was a student in the UCSD Biological Sciences Graduate Program. The University of Virginia, Center for Research in Reproduction, Ligand Assay and Analysis Core, is supported by the Eunice Kennedy Shriver NICHD/NIH Grant P50 HD028934.
Footnotes
Ethics Statement
Animal experiments conform to internationally accepted standards and have been approved by the appropriate institutional review body.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
- 1.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004. January;6(1):7–28. [DOI] [PubMed] [Google Scholar]
- 2.Harno E, Cottrell EC, White A. Metabolic pitfalls of CNS Cre-based technology. Cell Metab. 2013. July; 18(1):21–8. [DOI] [PubMed] [Google Scholar]
- 3.Heffner CS, Herbert Pratt C, Babiuk RP, Sharma Y, Rockwood SF, Donahue LR, et al. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun. 2012;3(1):1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magnuson MA, Osipovich AB. Pancreas-specific Cre driver lines and considerations for their prudent use. Cell Metab. 2013. July; 18(1): 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA. 2000. December;97(25):13702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rempe D, Vangeison G, Hamilton J, Li Y, Jepson M, Federoff HJ. Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis. 2006. January;44(1):44–9. [DOI] [PubMed] [Google Scholar]
- 7.Dubois NC, Hofmann D, Kaloulis K, Bishop JM, Trumpp A. Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis. 2006. August;44(8):355–60. [DOI] [PubMed] [Google Scholar]
- 8.Messina A, Langlet F, Chachlaki K, Roa J, Rasika S, Jouy N, et al. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci. 2016. June;19(6):835–44. [DOI] [PubMed] [Google Scholar]
- 9.Soriano P Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999. January;21(1):70–1. [DOI] [PubMed] [Google Scholar]
- 10.Babwah AV, Navarro VM, Ahow M, Pampillo M, Nash C, Fayazi M, et al. GnRH Neuron-Specific Ablation of Gαq/11 Results in Only Partial Inactivation of the Neuroendocrine-Reproductive Axis in Both Male and Female Mice: In Vivo Evidence for Kiss1r-Coupled Gαq/11-Independent GnRH Secretion. J Neurosci. 2015. September;35(37):12903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL. Deletion of Vax1 from Gonadotropin-Releasing Hormone (GnRH) Neurons Abolishes GnRH Expression and Leads to Hypogonadism and Infertility. J Neurosci. 2016;36:3506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann HM, Mellon PL. A small population of hypothalamic neurons govern fertility: the critical role of VAX1 in GnRH neuron development and fertility maintenance. Neurosci Commun (Houst). 2016;2:2. [PMC free article] [PubMed] [Google Scholar]
- 13.Lawson MA, MacConell LA, Kim J, Powl BT, Nelson SB, Mellon PL. Neuron-specific expression in vivo by defined transcription regulatory elements of the GnRH gene. Endocrinology. 2002. April;143(4):1404–12. [DOI] [PubMed] [Google Scholar]
- 14.Pape JR, Skynner MJ, Allen ND, Herbison AE. Transgenics identify distal 5′- and 3′-sequences specifying gonadotropin-releasing hormone expression in adult mice. Mol Endocrinol. 1999. December; 13(12):2203–11. [DOI] [PubMed] [Google Scholar]
- 15.Skynner MJ, Slater R, Sim JA, Allen ND, Herbison AE. Promoter transgenics reveal multiple gonadotropin-releasing hormone-I-expressing cell populations of different embryological origin in mouse brain. J Neurosci. 1999. July;19(14):5955–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iremonger K, Herbison A. Elucidating the Structure and Function of Gonadotropin-Releasing Hormone (GnRH) Neuron Dendrites In: Armstrong WE, Tasker JG, editors. Neurophysiology of Neuroendocrine Neurons. Chichester, UK: John Wiley & Sons, Ltd; 2014. Chapter 12. [Google Scholar]
- 17.Wolfe A, Ng Y, Divall SA, Singh SP, Radovick S. Development of an immortalised, post-pubertal gonadotrophin-releasing hormone neuronal cell line. J Neuroendocrinol. 2008. September;20(9):1029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer AK, Miller NL, Yip K, Tran BH, Mellon PL. Enhancers of GnRH transcription embedded in an upstream gene use homeodomain proteins to specify hypothalamic expression. Mol Endocrinol. 2010. October;24(10): 1949–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 2011. January;31(2):426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason AJ, Hayflick JS, Zoeller RT, Young WS 3rd, Phillips HS, Nikolics K, et al. A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science. 1986. December; 234(4782):1366–71. [DOI] [PubMed] [Google Scholar]
- 21.Mason AJ, Pitts SL, Nikolics K, Szonyi E, Wilcox JN, Seeburg PH, et al. The hypogonadal mouse: reproductive functions restored by gene therapy. Science. 1986. December;234(4782): 1372–8. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann HM, Mellon PL. Regulation of GnRH Gene Expression In: Herbison AE, Plant TM, editors. The GnRH Neuron and its Control. Hoboken (N.J.): Wiley Blackwell; 2018. pp. 95–120. [Google Scholar]
- 23.Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989. March;338(6211):161–4. [DOI] [PubMed] [Google Scholar]
- 24.Campos P, Herbison AE. Optogenetic activation of GnRH neurons reveals minimal requirements for pulsatile luteinizing hormone secretion. Proc Natl Acad Sci USA. 2014. December; 111(51): 18387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore AM, Prescott M, Czieselsky K, Desroziers E, Yip SH, Campbell RE, et al. Synaptic Innervation of the GnRH Neuron Distal Dendron in Female Mice. Endocrinology. 2018. September; 159(9):3200–8. [DOI] [PubMed] [Google Scholar]
- 26.Wolfe A, Divall S, Singh SP, Nikrodhanond AA, Baria AT, Le WW, et al. Temporal and spatial regulation of CRE recombinase expression in gonadotrophin-releasing hormone neurones in the mouse. J Neuroendocrinol. 2008. July;20(7):909–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon H, Enquist LW, Dulac C. Olfactory inputs to hypothalamic neurons controlling re-production and fertility. Cell. 2005. November; 123(4):669–82. [DOI] [PubMed] [Google Scholar]
- 28.Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne HJ, Todman MG, et al. Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron. 2006. October;52(2):271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimshek DR, Kim J, Hübner MR, Spergel DJ, Buchholz F, Casanova E, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002. January;32(1):19–26. [DOI] [PubMed] [Google Scholar]
- 30.Diaczok D, DiVall S, Matsuo I, Wondisford FE, Wolfe AM, Radovick S. Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol. 2011. May;25(5):833–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J. A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development. 1996. January;122(1): 243–52. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann HM, Tamrazian A, Xie H, Pérez-Millán MI, Kauffman AS, Mellon PL. Heterozygous deletion of ventral anterior homeobox (Vax1) causes subfertility in mice. Endocrinology. 2014. October;155(10):4043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann HM. Determination of Reproductive Competence by Confirming Pubertal Onset and Performing a Fertility Assay in Mice and Rats. J Vis Exp. 2018. October;(140). 10.3791/58352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larder R, Mellon PL. Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors. J Biol Chem. 2009. June;284(25): 16966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, et al. Fore-brain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995. October;121(10):3279–90. [DOI] [PubMed] [Google Scholar]
- 36.Larder R, Kimura I, Meadows J, Clark DD, Mayo S, Mellon PL. Gene dosage of Otx2 is important for fertility in male mice. Mol Cell Endocrinol. 2013. September;377(1–2):16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995. November;9(21):2646–58. [DOI] [PubMed] [Google Scholar]
- 38.Freedman LP, Venugopalan G, Wisman R. Reproducibility2020: progress and priorities. F1000 Res. 2017. May;6:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirilov M, Clarkson J, Liu X, Roa J, Campos P, Porteous R, et al. Dependence of fertility on kisspeptin-Gpr54 signaling at the GnRH neuron. Nat Commun. 2013;4(1):2492. [DOI] [PubMed] [Google Scholar]
- 40.Ramakrishnappa N, Rajamahendran R, Lin YM, Leung PC. GnRH in non-hypothalamic reproductive tissues. Anim Reprod Sci. 2005. August;88(1–2):95–113. [DOI] [PubMed] [Google Scholar]
- 41.Kim HH, Wolfe A, Cohen RN, Eames SC, Johnson AL, Wieland CN, et al. In vivo identification of a 107-base pair promoter element mediating neuron-specific expression of mouse gonadotropin-releasing hormone. Mol Endocrinol. 2007. February;21(2):457–71. [DOI] [PubMed] [Google Scholar]
- 42.Tan CL, Sheard PW, Jasoni CL. Developing neurites from mouse basal forebrain gonadotropin-releasing hormone neurons use Sonic hedgehog to modulate their growth. Int J Dev Neurosci. 2018. May;68:89–97. [DOI] [PubMed] [Google Scholar]
- 43.Shimshek DR, Bus T, Grinevich V, Single FN, Mack V, Sprengel R, et al. Impaired reproductive behavior by lack of GluR-B containing AMPA receptors but not of NMDA receptors in hypothalamic and septal neurons. Mol Endocrinol. 2006. January;20(1):219–31. [DOI] [PubMed] [Google Scholar]
- 44.Rivera J, Tessarollo L. Genetic background and the dilemma of translating mouse studies to humans. Immunity. 2008. January;28(1):1–4. [DOI] [PubMed] [Google Scholar]
- 45.Pandolfi EC, Hoffmann HM, Schoeller EL, Gorman MR, Mellon PL. Haploinsufficiency of SIX3 abolishes male reproductive behavior through disrupted olfactory development, and impairs female fertility through disrupted GnRH neuron migration. Mol Neurobiol. 2018. November;55(11):8709–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baum MJ, Tobet SA, Starr MS, Bradshaw WG. Implantation of dihydrotestosterone propionate into the lateral septum or medial amygdala facilitates copulation in castrated male rats given estradiol systemically.. Horm Be-hav 1982. June;16(2):208–23. [DOI] [PubMed] [Google Scholar]
- 47.Menniti FS, Erskine MS, Tobet SA, Baum MJ. Dihydrotestosterone-induced inhibition of lordosis in estrogen-primed ovariectomized rats following 6-hydroxydopamine or electrolytic septal lesions. Pharmacol Biochem Behav. 1982. February;16(2):211–6. [DOI] [PubMed] [Google Scholar]
- 48.Kelley CG, Givens ML, Rave-Harel N, Nelson SB, Anderson S, Mellon PL. Neuron-restricted expression of the rat gonadotropin-releasing hormone gene is conferred by a cell-specific protein complex that binds repeated CAaTT elements. Mol Endocrinol. 2002. November;16(11):2413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bakke M, Zhao L, Parker KL. Approaches to define the role of SF-1 at different levels of the hypothalamic-pituitary-steroidogenic organ axis. Mol Cell Endocrinol. 2001. June;179(1–2): 33–7. [DOI] [PubMed] [Google Scholar]
- 50.Herbison AE, Porteous R, Pape JR, Mora JM, Hurst PR. Gonadotropin-releasing hormone neuron requirements for puberty, ovulation, and fertility. Endocrinology. 2008. February; 149(2): 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffmann HM, Pandolfi EC, Larder R, Mellon PL. Haploinsufficiency of homeodomain proteins Six3, Vax1, and Otx2, causes subfertility in mice via distinct mechanisms. Neuroendocrinology. doi: 10.1159/000494086. [DOI] [PMC free article] [PubMed] [Google Scholar]


