Abstract
The mechanisms underlying interindividual variability in analgesic efficacy of nonsteroidal anti‐inflammatory drugs (NSAIDs) are not well understood. Therefore, we performed pain phenotyping, functional neuroimaging, pharmacokinetic/pharmacodynamic assessments, inflammation biomarkers, and gene expression profiling in healthy subjects who underwent surgical extraction of bony impacted third molars and were treated with ibuprofen (400 mg; N = 19) or placebo (N = 10). Analgesic efficacy was not associated with demographic or clinical characteristics, ibuprofen pharmacokinetics, or the degree of cyclooxygenase inhibition by ibuprofen. Compared with partial responders to ibuprofen (N = 9, required rescue medication within the dosing interval), complete responders (N = 10, no rescue medication) exhibited greater induction of urinary prostaglandin metabolites and serum tumor necrosis factor‐α and interleukin 8. Differentially expressed genes in peripheral blood mononuclear cells were enriched for inflammation‐related pathways. These findings suggest that a less pronounced activation of the inflammatory prostanoid system is associated with insufficient pain relief on ibuprofen alone and the need for additional therapeutic intervention.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ There is substantial interindividual variability in the analgesic efficacy of nonsteroidal anti‐inflammatory drugs (NSAIDs), but the mechanisms underlying this variability are not well understood.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This deep phenotyping study was conducted to identify factors associated with heterogeneity in analgesic response to ibuprofen after third molar extraction.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Our results demonstrate that there is substantial interindividual heterogeneity in the activation of the inflammatory prostanoid system in response to surgical trauma. A less pronounced activation is associated with insufficient pain relief on ibuprofen alone and the need for additional therapeutic intervention, such as an opioid analgesic.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Elucidation of the mechanisms underlying variability in analgesic efficacy may allow the identification of biomarkers that are predictive of the analgesic response to ibuprofen and other NSAIDs.
Although acute pain resulting from injury or other tissue damage can serve an adaptive function by promoting behaviors that limit the chance of further trauma, inadequate pain management in the postoperative setting can delay healing and adversely affect mental well‐being. Opioid analgesics are an important component of postoperative pain management in many patients. However, over‐prescription of opioids for surgical pain, typically 2–5 times more than patients actually use, has contributed to the opioid epidemic in the United States.1, 2 Thus, there is a need to consider alternative therapeutic options for those patients whose pain can be appropriately managed with nonaddictive analgesics, including nonsteroidal anti‐inflammatory drugs (NSAIDs).
NSAIDs exert their pharmacologic effects via inhibition of one or both cyclooxygenase (COX) enzymes (COX‐1 and COX‐2), which catalyze the first committed step in the synthesis of prostanoids (prostaglandin (PG)E2, PGD2, PGF2α, prostacyclin (PGI2), and thromboxane (Tx) A2) from arachidonic acid.3, 4 In the setting of inflammatory pain, these lipid mediators, particularly PGE2 and PGI2, act on their respective G protein coupled receptors to promote peripheral5, 6, 7, 8, 9 and central10, 11 sensitization. Because NSAIDs inhibit the formation of COX products that render the nociceptive system more excitable, rather than directly blocking pain signaling, they are effective in treating pain in which activation of the COX pathway is a key mechanism.
One such situation is surgical extraction of third molars, a procedure undergone by approximately 5 million patients per year in the United States.12, 13 The soft tissue and bony trauma associated with impacted third molar extraction surgery liberates key inflammatory mediators, including prostanoids, activating and sensitizing free nerve endings at the surgical site.6, 14 At the population level, NSAIDs are on average at least as effective as immediate release opioids following third molar extraction surgery.15, 16, 17, 18 However, there is considerable variability in the analgesic response to NSAIDs at the individual level, with 20–30% of patients requiring opioid rescue medication within 6 hours of the initial NSAID dose.19, 20 In order to avoid undertreating patients who will not respond adequately to NSAIDs, oral surgeons routinely prescribe opioids to be taken if needed, resulting in unnecessary prescriptions for a majority of patients who do not require them or only require minimal dosing.19, 21
The application of precision medicine approaches to pain management after third molar extraction may facilitate the optimization of NSAID therapy and limit opioid prescriptions to those patients who fail to attain adequate pain relief with NSAIDs. However, the development of such approaches requires a much better understanding of the molecular mechanisms that contribute to variation in analgesic efficacy of NSAIDs. Therefore, we performed a deep‐phenotyping study incorporating functional neuroimaging, pharmacokinetic/pharmacodynamic assessments, biochemical assays, and gene expression analysis to characterize the factors that are associated with interindividual variability in analgesic efficacy of ibuprofen following third molar extraction surgery.
Results
Study cohort
The study procedures are summarized in Figure 1. The study cohort included 29 healthy adults (16 men and 13 women) with a mean age of 24.9 ± 3.59 years. Ten subjects received placebo, and 19 subjects received an immediate release formulation of ibuprofen.22 The median maximum pain score before study drug administration (0–10 scale) was 7 (interquartile range (IQR): 5−8), and the study drug was administered 1.85 ± 0.55 hour (mean ± SD) after surgery. The first postsurgery and second postsurgery sample collections occurred at 1.51 ± 0.47 and 3.17 ± 0.42 hours after study drug administration, respectively.
Figure 1.
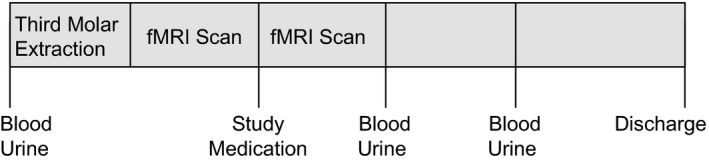
Study design. fMRI, functional magnetic resonance imaging.
Activity of ibuprofen
Ibuprofen was significantly more effective than placebo in relieving pain following third molar extraction. Thus, the median pain intensity difference (maximum pain score before study drug minus minimum pain score before rescue medication treatment) was 3 (IQR: 2−5) in the ibuprofen group, compared to −0.5 (IQR: −2 to 1.25) in the placebo group (P < 0.001). The patients’ global assessment of pain relief also favored ibuprofen with 16 of 19 subjects who received ibuprofen rating their pain as “much better” or “very much better” after study drug treatment, compared with 0 of 10 subjects who received placebo (P < 0.0001; Fisher exact test).
The onset of pain relief was detectable between 15 and 30 minutes following drug administration, as indicated by a difference in the slope of pain scores between these time points ( Figure S1 a,b; P < 0.001 placebo vs. ibuprofen). Functional neuroimaging analysis was restricted to these early time points (predrug = 0, 15, and 30 minutes) because few patients in the placebo group were able to remain in the scanner without rescue medication longer than 30 minutes following placebo administration, whereas some patients in the ibuprofen group tolerated 60–75 minutes of postdrug scan time. Additionally, poor image quality, primarily due to motion artifacts, precluded analysis in 4 of 10 placebo group patients but only 1 of 19 ibuprofen group patients. Despite these limitations a significant change in cerebral blood flow (CBF) indicative of the analgesic effect of ibuprofen was detectable between 15 and 30 minutes in the summary analysis of the brain's pain processing regions ( Figure S1 c). This was primarily driven by perfusion changes in the insula, the anterior cingulate cortex, and the secondary somatosensory cortex.
Ibuprofen inhibited COX activity, as indicated by ex vivo whole blood assays and quantification of urinary PG metabolites ( Figure S2 ). COX‐1 activity ex vivo was inhibited by > 90% at both postsurgery time points in ibuprofen‐treated subjects (P < 0.001). COX‐2 activity ex vivo was also inhibited in ibuprofen‐treated subjects compared with placebo at the first postsurgery time point (ibuprofen: 25.3 ± 21.0% of baseline; N = 17 vs. placebo: 40.0 ± 19.3% of baseline; N = 10; P = 0.0404). Some subjects in both groups had received rescue medication prior to the first postsurgery sample collection, and acetaminophen, which was a component of the rescue medication (hydrocodone 5 mg/acetaminophen 325 mg), may also inhibit COX‐2 activity.23 When the comparison was restricted to subjects with plasma acetaminophen concentrations below the limit of quantitation, the inhibition of COX‐2 activity ex vivo at the first postsurgery time point remained apparent (ibuprofen: 24.7 ± 12.4% of baseline; N = 13 vs. placebo: 51.3 ± 12.6% of baseline; N = 5; P = 0.003). Ibuprofen‐treated subjects also exhibited significantly lower levels of in vivo markers of drug action, the urinary metabolites of PGE2, PGI2, PGD2, and TxA2 relative to baseline at the second postsurgery time point, compared to the placebo group (P < 0.01).
Variability in the ibuprofen response
The time to rescue medication treatment is shown in Figure 2 a. All 10 subjects in the placebo group requested opioid rescue medication, with a median time to rescue of 53 (IQR: 48−55) minutes after study drug administration. Nine subjects in the ibuprofen group requested opioid rescue medication and were categorized as partial responders. The time to rescue medication treatment was significantly longer in partial responders compared with the placebo group (P < 0.001; log‐rank test), with a median time to rescue of 105 (82−133) minutes after study drug administration. The remaining 10 subjects in the ibuprofen group did not require additional analgesic medication 4 hours after dosing and were categorized as complete responders. Pain intensity scores after study medication administration also differed among the response groups (Figure 2 b). The median pain intensity difference was 5 (IQR: 3−6.25) in complete responders, compared with 2 (IQR: 1−3.5) in partial responders and −0.5 (IQR: −2 to 1.25) in placebo‐treated subjects (P = 0.0003, Kruskal–Wallis test). The separation into the three groups was detectable as early as 30 minutes following study drug administration based on reported pain scores (Figure 2 c). Functional neuroimaging did not allow distinction between complete and partial responders within 30 minutes of drug administration, and the number of patients who did not tolerate prolonged scan time precluded sufficiently powered analysis at later time points (Figure 2 d).
Figure 2.
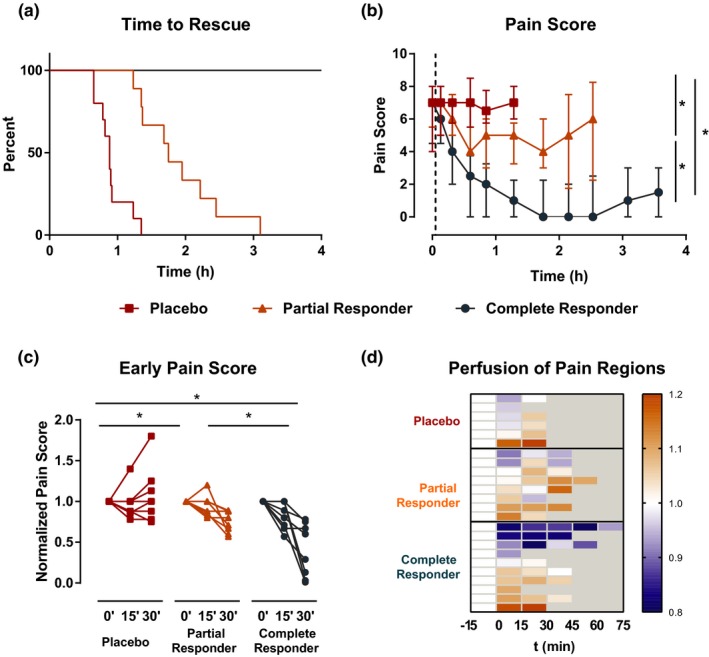
Interindividual variability in analgesic response to ibuprofen. (a) Kaplan–Meier plots depicting time to rescue medication administration by response group (placebo: n = 10; partial responders: n = 9; complete responders: n = 10; P < 0.001 for all comparisons in the log‐rank test). (b) Pain scores at each pain assessment prior to rescue medication administration by response group. Error bars indicate interquartile range (*P < 0.05; Kruskal–Wallis test). Vertical dashed line indicates time of study drug administration. (c) Change of pain scores relative to predrug (0 minutes) scores up to 30 minutes (*P < 0.05; Kruskal–Wallis test). (d) Heatmap depicting change from predrug of the integrated cerebral blood flow measurements in pain regions (NeuroSynth pain map) by individual patients.
Variability in the inflammatory response to surgery
No significant differences in demographic or clinical characteristics (e.g., number of extracted teeth, trauma score, etc.) were observed between complete and partial responders (Table 1), and baseline COX activity ex vivo and levels of PG metabolites did not differ among the response groups ( Table S1 ). Similar ibuprofen plasma concentrations and degree of inhibition of COX activity ex vivo were observed in complete and partial responders at both postsurgery time points ( Figure S3 ), suggesting that differences in the ibuprofen response cannot be explained by differences in pharmacokinetics. There was no significant difference in the frequency of CYP2C9*2 or CYP2C9*3 variant alleles, which can affect ibuprofen metabolism, between the response groups (data not shown).
Table 1.
Baseline characteristics by response group
| Placebo | Partial responder | Complete responder | |
|---|---|---|---|
| N | 10 | 9 | 10 |
| Men/women (N) | 5/5 | 4/5 | 7/3 |
| Age, years (mean ± SD) | 26.1 ± 3.9 | 25.7 ± 2.9 | 23.1 ± 3.4 |
| Race (N) | |||
| White | 1 | 3 | 5 |
| Asian | 5 | 5 | 2 |
| African American | 4 | 0 | 0 |
| Other | 0 | 1 | 3 |
| Length of surgery (minutes (mean ± SD)) | 40.0 ± 15.0 | 36.6 ± 21.4 | 38.6 ± 32.2 |
| Number of teeth (N, median, (IQR)) | 4 (3,4) | 4 (4,4) | 3.5 (2.25,4) |
| Trauma score (N, median, (IQR)) | 8 (6.25,8) | 7 (6,8) | 7 (5.25,7.75) |
| Max pain score (N, median, (IQR)) | 7 (5,8) | 7 (6,8) | 7 (5,7.75) |
Interestingly, partial responders exhibited greater suppression of urinary 7‐hydroxy‐5,11‐diketotetranorprostane‐1,16‐dioic acid (PGEM; partial responders: 45.3 ± 14.9% of baseline vs. complete responders: 75.7 ± 23.7% of baseline; P = 0.0021), 2,3‐dinor 6‐keto‐PGF1α (PGIM; partial responders: 70.6 ± 43.1% of baseline vs. complete responders: 126.5 ± 43.7% of baseline; P = 0.0133), and tended that way for 11,15‐dioxo‐9α‐hydroxy‐2,3,4,5‐tetranorprostane‐1,20‐dioic acid (PGDM; partial responders: 66.5 ± 20.7% of baseline vs. complete responders: 83.1 ± 12.6% of baseline; P = 0.0947) at the first postsurgery time point, compared with complete responders (Figure 3). At the second postsurgery time point, the difference between partial and complete responders persisted for PGEM (partial responders: 21.2 ± 7.8% of baseline vs. complete responders: 38.4 ± 11.9% of baseline; P = 0.0082). Serum tumor necrosis factor‐α (TNF‐α; partial responders: 73.1 ± 26.8% of baseline vs. complete responders: 186.2 ± 143.3% of baseline; P = 0.0127) and interleukin (IL)‐8 (partial responders: 73.5 ± 40.6% of baseline vs. complete responders: 199.4 ± 88.1% of baseline; P = 0.0027) were induced to a greater extent in complete responders than in partial responders at the second postsurgery time point, whereas no significant differences between the groups were observed for IL‐6, IL‐10, and monocyte chemoattractant protein (MCP)‐1 (Figure 4). No significant differences in serum levels of these inflammatory mediators were observed at baseline ( Table S2 ).
Figure 3.
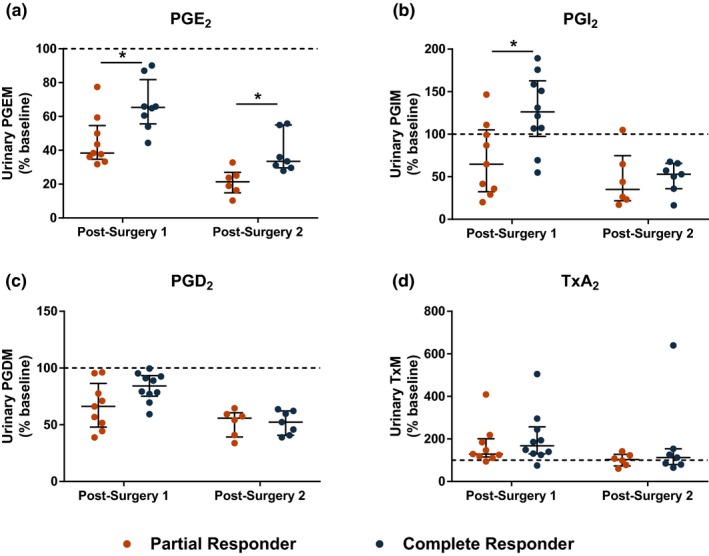
Comparison of urinary prostaglandin (PG) metabolite levels after surgery in partial and complete responders. Urinary metabolites of (a) prostaglandin E (PGE)2, (b) prostacyclin (PGI)2, (c) PGD2, and (d) thromboxane (Tx)A2 are expressed as percent change from baseline. Crossbars indicate median and interquartile range. *P < 0.05 by Wilcoxon rank‐sum test. PGDM, 11,15‐dioxo‐9α‐hydroxy‐2,3,4,5‐tetranorprostane‐1,20‐dioic acid; PGEM, 7‐hydroxy‐5,11‐diketotetranorprostane‐1,16‐dioic acid; PGIM, 2,3‐dinor 6‐keto‐PGF1α. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 4.
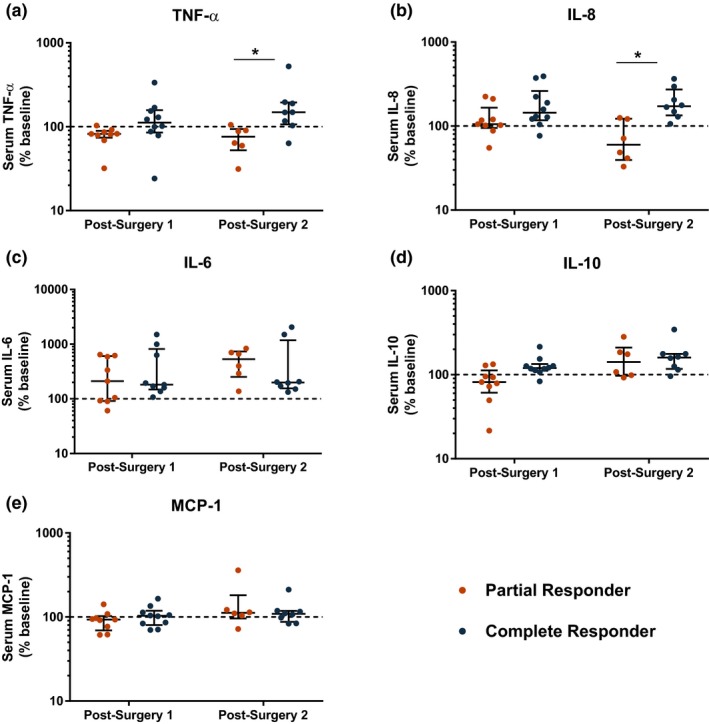
Comparison of serum inflammatory mediators after surgery in partial and complete responders. Serum levels of (a) tumor necrosis factor‐α (TNF‐α), (b) interleukin (IL)‐8, (c) IL‐6, (d) IL‐10, and (e) monocyte chemoattractant protein (MCP)‐1 are expressed as percent change from baseline. Crossbars indicate median and interquartile range. *P < 0.05 by Wilcoxon rank‐sum test. [Colour figure can be viewed at wileyonlinelibrary.com]
The placebo group showed the most changes in gene expression in peripheral blood mononuclear cells (PBMCs), with 431 genes at the first postsurgery time point and 2,653 genes at the second postsurgery time point differentially expressed compared to baseline (q < 0.05). Partial responders exhibited fewer changes in gene expression, with 99 differentially expressed genes (DEGs) at the first postsurgery time point and 1,092 DEGs at the second postsurgery time point (q < 0.05). Very few genes were differentially expressed compared to baseline in complete responders, with 7 DEGs at the first postsurgery time point and 47 DEGs at the second postsurgery time point (q < 0.05). Subsequent analyses focused on the second postsurgery time point because the majority of DEGs observed at the first postsurgery time point within each group were also differentially expressed at the second postsurgery time point (data not shown). There was marked overlap in the DEGs among the three groups (Figure 5 a). Pathway analysis of the DEGs indicated that these were enriched for pathways related to inflammation, but specific pathways differed among the ibuprofen response groups (Figures [Link] , [Link] , [Link] ). At the second postsurgery time point, 1,345 genes were differentially expressed between partial and complete responders (q < 0.2; Figure 5 b), with enrichment for inflammatory pathways (Table 2). Expression plots for select genes from these pathways are shown in Figure S7 . For the majority of these inflammatory genes, partial responders had significantly higher expression compared with complete responders.
Figure 5.
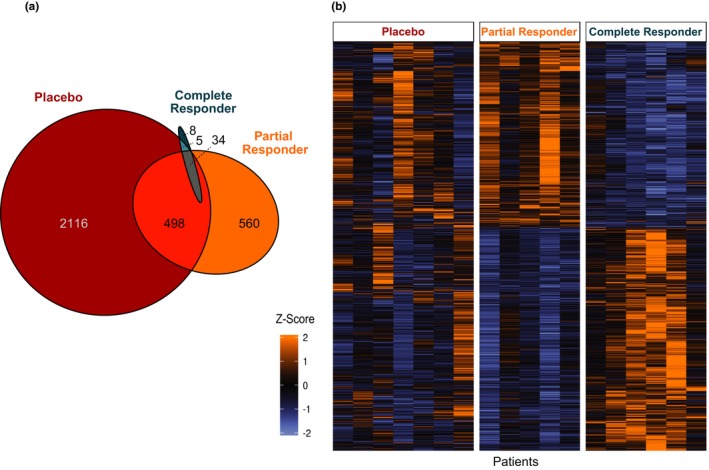
RNA sequencing analysis of peripheral blood mononuclear cells. (a) Venn diagram depicting overlap among the response groups in genes differentially expressed at the second postsurgery time point compared to baseline (q < 0.05). (b) Heatmap depicting differentially expressed genes between partial and complete responders at the second postsurgery time point (q < 0.2).
Table 2.
Pathway analysis of genes differentially expressed between partial and complete responders at the second postsurgery time point (q < 0.2)
| Pathway name | P value |
|---|---|
| Fc receptor‐mediated phagocytosis in macrophages and monocytes | 5.31 × 10−6 |
| TREM1 signaling | 5.98 × 10−6 |
| Neuroinflammation signaling pathway | 3.62 × 10−5 |
| CD28 signaling in T helper cells | 5.27 × 10−5 |
| Production of nitric oxide and reactive oxygen species in macrophages | 7.32 × 10−5 |
Discussion
Pain is a complex multidimensional experience that reflects the interaction between nociceptive, affective, and cognitive processes.24, 25 Given the diverse mechanisms that contribute to pain, it has long been recognized that there is substantial interindividual variability in the effectiveness of all analgesics, including NSAIDs.16, 19, 20, 26 For example, it has been estimated that only half of the patients with arthritis prescribed NSAIDs will have a moderate or better pain relief response.26 Studies in acute postsurgical pain following third molar extraction have demonstrated that, although NSAIDs are highly effective on average, 20–30% of patients required opioid rescue medication within 4–6 hours of the initial NSAID dose, indicating that these were individuals in whom NSAIDs failed to provide sufficient pain relief throughout the dosing interval.19, 20 Currently, pain therapy is configured on a trial‐and‐error approach, often involving several iterations of switching drugs and adjusting doses. However, the development of algorithms to personalize treatment based on genetic or nongenetic information is limited by our lack of understanding of the molecular mechanisms underlying interindividual variability in analgesic efficacy. Here, we demonstrate that variability in the response to ibuprofen following third molar extraction is detectable across multiple diagnostic domains—behavioral, brain imaging, and markers of systemic inflammation—indicating that partial and complete analgesic responses to ibuprofen reflect internally consistent phenotypes. In addition, we find that activation of the prostanoid biosynthetic pathway following surgical trauma differs between complete and partial responders, suggesting that the response phenotype relates to the mechanism of drug action.
Thus, a key strength of our study is the application of an array of complementary techniques to differentiate between partial and complete responders despite the small sample size. Pain and analgesic efficacy are challenging to quantify in a clinical setting due to the inherent subjectivity in the experience of pain and imprecision of pain rating scales.27 Functional neuroimaging has expanded the understanding of the neural basis of pain mechanisms and may provide an objective biomarker of efficacy of pain treatment. A prior study that used arterial spin labeling‐functional magnetic resonance imaging to quantify the effect of ibuprofen administration after third molar extraction demonstrated that the analgesic effect of ibuprofen was associated with decreases in CBF in brain regions known to be involved in the perception of postsurgical pain.28 We observed similar results in our ibuprofen‐treated subjects.
Notably, response to ibuprofen in our cohort could not be predicted based on clinical characteristics, ibuprofen pharmacokinetics, or pharmacodynamics. Rather, we observed that complete responders exhibited higher levels of urinary PG metabolites at the first postsurgery time point, compared with partial responders. Because the concentrations of PG metabolites in urine reflect COX activity in vivo over the entire collection interval, this measurement is influenced both by the degree of activation of the COX pathway in response to third molar extraction surgery, as well as inhibition of PG formation by ibuprofen. We observed a similar degree of COX inhibition ex vivo in both partial and complete responders at this time point; thus, the differences in urinary PG metabolite levels suggest that complete responders had greater activation of the COX pathway in vivo in response to surgery. These findings are consistent with prior studies in patients with rheumatoid arthritis and chronic pain demonstrating a greater response to NSAID treatment in patients with elevated PGE2 levels29, 30 and support the notion that NSAIDs are most effective in patients in whom activation of the COX pathway is a key mechanism contributing to their pain.
Third molar extraction promoted a systemic inflammatory response, as evidenced by induction of serum cytokines and chemokines and alterations in gene expression in PBMCs (composed primarily of monocytes and lymphocytes), which is consistent with studies in a variety of surgical models.31, 32 Although IL‐6, IL‐10, and MCP‐1 were induced to a similar degree regardless of response group, we observed greater increases in serum TNF‐α and IL‐8 levels in complete responders compared with partial responders at the second postsurgery time point. Prior studies have shown that PGE2 induces both TNF‐α33 and IL‐834, 35 in vitro. Thus, the induction of these inflammatory mediators in complete responders may be a consequence of greater PGE2 formation in response to surgery. However, it is also possible that these pathways are regulated in parallel, and additional studies are necessary to clarify whether there is a causal relationship between activation of the COX pathway and induction of TNF‐α and IL‐8 in humans. Because cytokines were measured in serum, they might have been released from activated cells rather than representing circulating levels. In contrast with some prior studies,36, 37 ibuprofen treatment did not decrease IL‐6 levels in our cohort. This may be due to differences in timing of sample collection, as our samples were collected ~ 3 and 5 hours after surgery, whereas effects of NSAIDs on cytokine levels have been observed at later time points (e.g., 12–24 hours after surgery). However, the results of our gene expression analysis support a marked early anti‐inflammatory effect of ibuprofen treatment. Complete responders exhibited much fewer differentially expressed genes after surgery, as well as lower inflammatory gene expression compared with partial responders. Taken together, these results suggest that differences in the regulation of the COX pathway and the inflammatory response to surgery contribute to interindividual variability in the analgesic efficacy of NSAIDs.
There are limitations to our study. Although demographic and clinical characteristics were not statistically different between partial and complete responders in our cohort, we cannot exclude the possibility that these factors contribute to variability in analgesic efficacy of ibuprofen in a larger population. Similarly, the small sample size precluded comprehensive investigation of genetic variants that might modulate ibuprofen response. Our cohort included only healthy young adults. Although this limits potential confounding due to effects of age and comorbidities, it precludes interrogation of the influence of these factors on ibuprofen response. In addition, we evaluated only a single dose of ibuprofen over a relatively short sampling period, so we cannot determine the efficacy of repeated dosing or evaluate the effects of ibuprofen on inflammatory mediators or gene expression beyond the acute postoperative period. Finally, our study evaluated ibuprofen response in the setting of acute inflammatory pain, and it is unknown whether similar mechanisms contribute to variation in analgesic efficacy in persistent or chronic pain.
Despite these limitations, our study serves as a necessary prerequisite for future studies to identify molecular mechanisms predictive of NSAID response. In light of the opioid crisis, there is an emphasis on the development of approaches to providing effective pain relief with nonaddictive analgesics, including NSAIDs.38 However, our results, as well as those of prior studies,16, 19, 20 underscore the heterogeneity in the analgesic response to NSAIDs. The ability to prospectively identify patients who would respond to NSAIDs would help limit unnecessary opioid prescriptions in those patients, as well as ensure that patients who would not achieve pain relief with NSAIDs alone have access to additional analgesics.
In conclusion, our results demonstrate that there is marked interindividual variability in the analgesic efficacy of ibuprofen following third molar extraction surgery that is not explained by overt differences in clinical characteristics, ibuprofen plasma concentrations, or degree of COX inhibition ex vivo. The differences in urinary PG metabolites, serum cytokines, and gene expression in PBMCs suggest that regulation of the inflammatory response to surgery differs between partial and complete responders. Elucidation of the molecular mechanisms underlying this variability may allow the identification of biomarkers that are predictive of the analgesic response to ibuprofen and other NSAIDs.
Methods
A detailed description of the methods is provided in the Supplementary Information .
Subjects
Healthy subjects (≥ 18 years of age) undergoing extraction of one or more partially or fully bony impacted third molars and who provided written informed consent were enrolled. Subjects were asked to abstain from analgesics, including products containing NSAIDs, aspirin, and acetaminophen, high‐dose vitamins, and nutritional supplements for 1 week prior to surgery.
Study procedures
The study protocol was approved by the University of Pennsylvania Institutional Review Board. Study procedures are summarized in Figure 1. Baseline blood and spot urine samples were collected, and subjects then underwent third molar extraction. After surgery, subjects reported pain intensity every 15 minutes using the 0–10 Numeric Rating Scale‐Pain Intensity, where 0 = no pain and 10 = worst pain imaginable. Approximately 45 minutes after surgery, subjects were placed in the magnetic resonance imaging (MRI) to begin functional imaging data collection. When the study subjects requested medication and reported a pain score ≥ 4 of 10 or indicated that their pain was no longer tolerable, they received a dose of ibuprofen sodium dihydrate (400 mg Advil Film Coated Tablets; Pfizer) or matching placebo by mouth. The blinded study medication and matching placebo were provided by Pfizer. After administering the study medication, MRI scanning continued for up to 60 minutes. Rescue medication (hydrocodone 5 mg/acetaminophen 325 mg) was allowed any time upon request. Immediately after the scanning session, postsurgery blood and spot urine samples were collected, and subjects were returned to the postoperative observation area for continued pain assessment. After the first seven subjects were enrolled, the study procedure was amended to include a second postsurgery blood and urine sample collection ~ 3 hours after study drug administration. This time point was added to evaluate the change in inflammatory response and ibuprofen concentrations over time during the acute postsurgical period. Once medically stable, the subjects were discharged home.
MRI acquisition and data processing
A 3 Tesla Siemens Trio MRI system with a 32‐channel phase arrayed head receiver was used. An anatomic scan was followed by sequential measurements of regional CBF using ASL‐MRI obtained with pseudo‐continuous labeling39, 40 and a background‐suppressed 3D stack‐of‐spirals readout.41 Perfusion MRI data were analyzed offline using established procedures by an investigator blinded to treatment assignment. The extracted CBF data from regions of interest were evaluated with standard statistical analyses to test for effects of ibuprofen vs. placebo on neural activity. Usable fMRI scans were obtained in 24 subjects (placebo: N = 6; ibuprofen: N = 18); others had machine or procedural issues or excess movement artifact sufficient to prevent appropriate analysis.
Quantification of COX activity and plasma drug concentrations
COX‐1 activity ex vivo was evaluated by quantifying serum TxB2 levels, as previously described.42 COX‐2 activity ex vivo was evaluated by quantifying plasma PGE2 levels following lipopolysaccharide stimulation in whole blood, as previously described.43 COX activity in vivo was determined by quantification of major urinary prostanoid metabolites: PGIM, PGEM, PGDM, and 2,3‐dinor TxB2, by liquid chromatography mass spectrometry, as previously described.44 Plasma concentrations of ibuprofen and acetaminophen were quantified by liquid chromatography mass spectrometry, as previously described.45
Serum cytokines
Concentrations of IL‐6, IL‐1β, IL‐8, IL‐10, TNF‐α, and MCP‐1 in serum were quantified by MILLIPLEX multiplex assay (Millipore) by the Radioimmunoassay and Biomarkers Core at the Diabetes Research Center at the University of Pennsylvania. The levels of IL‐1β were below the limit of detection in the majority of samples, so further statistical analysis was not performed for this analyte.
CYP2C9 genotyping
Subjects were genotyped for CYP2C9*2 and CYP2C9*3 using TaqMan SNP Genotyping assays (ThermoFisher).
Gene expression analysis
Total RNA was isolated from PBMCs using the RNeasy Miniprep Kit (QIAGEN) and converted to sequencing libraries using the SMARTer Stranded Total RNA‐Seq Kit version 2—Pico Input Mammalian (Clontech) using a unique dual‐barcode combination. Reads were aligned to GRCh38 build of the human reference genome using STAR version 2.6.0c46 and gene models from version 92 of the Ensembl annotation.47 Data were normalized and quantified using PORT version 0.8.5b‐beta (https://github.com/itmat/Normalization), run at the gene level in strand‐specific mode and provided with gene models from version 92 of the Ensembl annotation. Pairwise differential expression (DE) analyses were performed on the gene‐level read counts using voom‐limma software package version 3.34.0.48 The data are accessible through Gene Expression Omnibus Series accession number GSE120596. Only genes with > 0 reads across all samples in at least one of the two compared groups were used for pairwise DE analyses. Pathways enriched in each of the DE gene lists were identified using Ingenuity Pathway Analysis (Qiagen).49
Statistical analysis
The objective of the study was to investigate the factors associated with variability in the analgesic response to ibuprofen across multiple phenotypic domains, including pain ratings, consumption of rescue medicine, fMRI, pharmacokinetic and pharmacodynamics measures, inflammatory biomarkers, and gene expression profiles. The fMRI measurements were selected as the primary end point, and sample size was determined based on the number of usable scans needed to assess differences in cerebral blood flow within subjects between the pain state and the treated state. Because there were no studies to indicate what the variation might be in the important brain regions as pain relief on ibuprofen was achieved, this study was described as exploratory and a sample size consistent with the functional brain imaging literature chosen (n = 10 placebo, n = 19 ibuprofen). Retrospective sample size analysis for the outcome that provided the rationale to investigate inflammation as the distinguishing factor between partial and complete responders, the urinary PGEM, shows a group size n < 10 provided 80% power to detect a significant difference (P < 0.05, t‐test). Data are reported as mean ± SD or median (25th percentile, 75th percentile). Baseline characteristics and biochemical measurements were compared by t‐test or analysis of variance or their nonparametric equivalents, as appropriate. Time to rescue medication treatment was evaluated by log‐rank test. Postsurgery measurements of COX activity ex vivo, urinary PG metabolite levels, and serum inflammatory mediators were normalized to baseline values for each subject and compared at each time point by Wilcoxon rank‐sum test. P < 0.05 was considered statistically significant.
Funding
This work was partially supported by funds for investigator initiated research from Pfizer (J.T.F and E.V.H.), a Precision Medicine Accelerator Grant from the University of Pennsylvania Center for Precision Medicine (T.G.), a Translational Medicine and Therapeutics post doctoral fellowship from the Pharmaceutical Research and Manufacturers of America Foundation (K.N.T.), the National Institute of Biomedical Imaging and Bioengineering (P41 EB015893; J.A.D.) and the National Heart, Lung, and the Blood Institute funded Personalized NSAID Therapeutics Consortium (HL117798; T.G. and G.A.F.). The Radioimmunoassay and Biomarkers Core at the University of Pennsylvania Diabetes Research Center is supported by National Institutes of Health (NIH) DK19525.
Conflicts of Interest
J.T.F. has received research support from the US Food and Drug Administration, and the National Institutes of Health, Data Safety Monitoring Board service compensation from National Institutes of Health and Cara Therapeutics, and consulting fees from Analgesic Solutions, Aptinyx, Biogen, Campbell Alliance, Daiichi Sankyo, DepoMed, Evadera, Jansen, Lilly, Novartis, and Pfizer. T.G. has received consulting fees from Novartis, Bayer, and PLx Pharma. G.A.F. received research support from the National Institutes of Health, the American Heart Association, the Volkswagen Foundation, Calico Laboratories, and Amgen. He received consulting fees from Amgen, GSK, Tremeau Pharmaceuticals, and Heron Therapeutics. E.V.H. received research funding from Pfizer and Consumer Healthcare Products Association. X.L. became an employee of Eli Lilly and Merck following completion of his work on this study. All other authors declared no competing interests for this work.
Author Contributions
K.N.T., E.V.H., G.A.F., J.T.F., and T.G. wrote the manuscript. K.N.T., J.A.D., G.A.F., E.V.H., T.G., and J.T.F. designed the research. S.A.S., K.N.T., E.V.H., J.T.F., X.L., E.J.G., H.G., L.M.L., and T.G. performed the research. H.L., J.A.D., N.F.L., and G.R.G. analyzed the data.
Supporting information
Figure S1. Cerebral blood flow (CBF) measured by ASL‐fMRI.
Figure S2. Comparison of COX‐1 and COX‐2 activity ex vivo and PG urinary metabolite levels after surgery in placebo (red) and ibuprofen‐treated (blue) subjects.
Figure S3. Comparison of ibuprofen plasma concentrations and COX‐1 and COX‐2 activity ex vivo after surgery in partial (gold) and complete (teal) responders.
Figure S4. Heatmap and pathway analysis of genes (rows) differentially expressed at the second postsurgery time point compared to baseline in the placebo group.
Figure S5. Heatmap and pathway analysis of genes (rows) differentially expressed at the second postsurgery time point compared to baseline in the partial responders.
Figure S6. Heatmap and pathway analysis of genes (rows) differentially expressed at the second postsurgery time point compared to baseline in the complete responders.
Figure S7. Expression of select genes detected by RNA‐seq at the second postsurgery time point.
Supplementary Methods and Tables.
Acknowledgments
Dr FitzGerald is the McNeil Professor of Translational Medicine and Therapeutics.
Contributor Information
Tilo Grosser, Email: tilo@pennmedicine.upenn.edu.
John T. Farrar, Email: jfarrar@pennmedicine.upenn.edu.
References
- 1. Thiels, C.A. et al Wide variation and overprescription of opioids after elective surgery. Ann. Surg. 266, 564–573 (2017). [DOI] [PubMed] [Google Scholar]
- 2. Gauger, E.M. , Gauger, E.J. , Desai, M.J. & Lee, D.H. Opioid use after upper extremity surgery. J. Hand Surg. Am. 43, 470–479 (2018). [DOI] [PubMed] [Google Scholar]
- 3. Chen, L. , Yang, G. & Grosser, T. Prostanoids and inflammatory pain. Prostaglandins Other Lipid. Mediat. 104–105, 58–66 (2013). [DOI] [PubMed] [Google Scholar]
- 4. Grosser, T. , Theken, K.N. & FitzGerald, G.A. Cyclooxygenase inhibition: pain, inflammation, and the cardiovascular system. Clin. Pharmacol. Ther. 102, 611–622 (2017). [DOI] [PubMed] [Google Scholar]
- 5. Bickel, A. , Dorfs, S. , Schmelz, M. , Forster, C. , Uhl, W. & Handwerker, H.O. Effects of antihyperalgesic drugs on experimentally induced hyperalgesia in man. Pain 76, 317–325 (1998). [DOI] [PubMed] [Google Scholar]
- 6. Roszkowski, M.T. , Swift, J.Q. & Hargreaves, K.M. Effect of NSAID administration on tissue levels of immunoreactive prostaglandin E2, leukotriene B4, and (S)‐flurbiprofen following extraction of impacted third molars. Pain 73, 339–345 (1997). [DOI] [PubMed] [Google Scholar]
- 7. Petersen, K.L. , Brennum, J. & Dahl, J.B. Experimental evaluation of the analgesic effect of ibuprofen on primary and secondary hyperalgesia. Pain 70, 167–174 (1997). [DOI] [PubMed] [Google Scholar]
- 8. Kilo, S. , Forster, C. , Geisslinger, G. , Brune, K. & Handwerker, H.O. Inflammatory models of cutaneous hyperalgesia are sensitive to effects of ibuprofen in man. Pain 62, 187–193 (1995). [DOI] [PubMed] [Google Scholar]
- 9. Renner, B. , Walter, G. , Strauss, J. , Fromm, M.F. , Zacher, J. & Brune, K. Preoperative administration of etoricoxib in patients undergoing hip replacement causes inhibition of inflammatory mediators and pain relief. Eur. J. Pain 16, 838–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koppert, W. et al The cyclooxygenase isozyme inhibitors parecoxib and paracetamol reduce central hyperalgesia in humans. Pain 108, 148–153 (2004). [DOI] [PubMed] [Google Scholar]
- 11. Arendt‐Nielsen, L. , Egsgaard, L.L. & Petersen, K.K. Evidence for a central mode of action for etoricoxib (COX‐2 inhibitor) in patients with painful knee osteoarthritis. Pain 157, 1634–1644 (2016). [DOI] [PubMed] [Google Scholar]
- 12. Friedman, J.W. The prophylactic extraction of third molars: a public health hazard. Am. J. Public Health 97, 1554–1559 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levy, B. , Paulozzi, L. , Mack, K.A. & Jones, C.M. Trends in opioid analgesic‐prescribing rates by specialty, U.S., 2007–2012. Am. J. Prev. Med. 49, 409–413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swift, J.Q. , Garry, M.G. , Roszkowski, M.T. & Hargreaves, K.M. Effect of flurbiprofen on tissue levels of immunoreactive bradykinin and acute postoperative pain. J. Oral Maxillofac. Surg. 51, 112–116 (1993). [DOI] [PubMed] [Google Scholar]
- 15. Cooper, S.A. , Engel, J. , Ladov, M. , Precheur, H. , Rosenheck, A. & Rauch, D. Analgesic efficacy of an ibuprofen‐codeine combination. Pharmacotherapy 2, 162–167 (1982). [DOI] [PubMed] [Google Scholar]
- 16. Dionne, R.A. , Campbell, R.A. , Cooper, S.A. , Hall, D.L. & Buckingham, B. Suppression of postoperative pain by preoperative administration of ibuprofen in comparison to placebo, acetaminophen, and acetaminophen plus codeine. J. Clin. Pharmacol. 23, 37–43 (1983). [DOI] [PubMed] [Google Scholar]
- 17. Chang, D.J. , Fricke, J.R. , Bird, S.R. , Bohidar, N.R. , Dobbins, T.W. & Geba, G.P. Rofecoxib versus codeine/acetaminophen in postoperative dental pain: a double‐blind, randomized, placebo‐ and active comparator‐controlled clinical trial. Clin. Ther. 23, 1446–1455 (2001). [DOI] [PubMed] [Google Scholar]
- 18. Desjardins, P.J. et al A double‐blind randomized controlled trial of rofecoxib and multidose oxycodone/acetaminophen in dental impaction pain. J. Oral Maxillofac. Surg. 65, 1624–1632 (2007). [DOI] [PubMed] [Google Scholar]
- 19. Hersh, E.V. et al Dose‐ranging analgesic study of Prosorb diclofenac potassium in postsurgical dental pain. Clin. Ther. 26, 1215–1227 (2004). [DOI] [PubMed] [Google Scholar]
- 20. Hersh, E.V. et al Ibuprofen liquigel for oral surgery pain. Clin. Ther. 22, 1306–1318 (2000). [DOI] [PubMed] [Google Scholar]
- 21. Maughan, B.C. et al Unused opioid analgesics and drug disposal following outpatient dental surgery: a randomized controlled trial. Drug Alcohol Depend. 168, 328–334 (2016). [DOI] [PubMed] [Google Scholar]
- 22. Brain, P. , Leyva, R. , Doyle, G. & Kellstein, D. Onset of analgesia and efficacy of ibuprofen sodium in postsurgical dental pain: a randomized, placebo‐controlled study versus standard ibuprofen. Clin. J. Pain 31, 444–450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sciulli, M.G. et al Effects of acetaminophen on constitutive and inducible prostanoid biosynthesis in human blood cells. Br. J. Pharmacol. 138, 634–641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vardeh, D. , Mannion, R.J. & Woolf, C.J. Toward a mechanism‐based approach to pain diagnosis. J. Pain 17, T50–T69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Basbaum, A.I. , Bautista, D.M. , Scherrer, G. & Julius, D. Cellular and molecular mechanisms of pain. Cell 139, 267–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moore, R.A. , Moore, O.A. , Derry, S. , Peloso, P.M. , Gammaitoni, A.R. & Wang, H. Responder analysis for pain relief and numbers needed to treat in a meta‐analysis of etoricoxib osteoarthritis trials: bridging a gap between clinical trials and clinical practice. Ann. Rheum. Dis. 69, 374–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woodcock, J. , Witter, J. & Dionne, R.A. Stimulating the development of mechanism‐based, individualized pain therapies. Nat. Rev. Drug Discov. 6, 703–710 (2007). [DOI] [PubMed] [Google Scholar]
- 28. Hodkinson, D.J. et al Cerebral analgesic response to nonsteroidal anti‐inflammatory drug ibuprofen. Pain 156, 1301–1310 (2015). [DOI] [PubMed] [Google Scholar]
- 29. Tokunaga, M. , Ohuchi, K. , Yoshizawa, S. , Tsurufuji, S. , Rikimaru, A. & Wakamatsu, E. Change of prostaglandin E level in joint fluids after treatment with flurbiprofen in patients with rheumatoid arthritis and osteoarthritis. Ann. Rheum. Dis. 40, 462–465 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eisenach, J.C. , Curry, R. , Rauck, R. , Pan, P. & Yaksh, T.L. Role of spinal cyclooxygenase in human postoperative and chronic pain. Anesthesiology 112, 1225–1233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cruickshank, A.M. , Fraser, W.D. , Burns, H.J. , Van Damme, J. & Shenkin, A. Response of serum interleukin‐6 in patients undergoing elective surgery of varying severity. Clin. Sci. (Lond.) 79, 161–165 (1990). [DOI] [PubMed] [Google Scholar]
- 32. Watt, D.G. , Horgan, P.G. & McMillan, D.C. Routine clinical markers of the magnitude of the systemic inflammatory response after elective operation: a systematic review. Surgery 157, 362–380 (2015). [DOI] [PubMed] [Google Scholar]
- 33. Renz, H. , Gong, J.H. , Schmidt, A. , Nain, M. & Gemsa, D. Release of tumor necrosis factor‐alpha from macrophages. Enhancement and suppression are dose‐dependently regulated by prostaglandin E2 and cyclic nucleotides. J. Immunol. 141, 2388–2393 (1988). [PubMed] [Google Scholar]
- 34. Neuschafer‐Rube, F. , Pathe‐Neuschafer‐Rube, A. , Hippenstiel, S. , Kracht, M. & Puschel, G.P. NF‐kappaB‐dependent IL‐8 induction by prostaglandin E2 receptors EP1 and EP4 . Br. J. Pharmacol. 168, 704–717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neuschafer‐Rube, F. , Pathe‐Neuschafer‐Rube, A. , Hippenstiel, S. & Puschel, G.P. Prostaglandin E2 enhanced TNFα‐mediated IL‐8 induction in monocytic cell lines and PBMC. Cytokine 113, 105–116 (2019). [DOI] [PubMed] [Google Scholar]
- 36. Mahdy, A.M. , Galley, H.F. , Abdel‐Wahed, M.A. , el‐Korny, K.F. , Sheta, S.A. & Webster, N.R. Differential modulation of interleukin‐6 and interleukin‐10 by diclofenac in patients undergoing major surgery. Br. J. Anaesth. 88, 797–802 (2002). [DOI] [PubMed] [Google Scholar]
- 37. Singh, P. et al A prospective study to assess the levels of interleukin‐6 following administration of diclofenac, ketorolac and tramadol after surgical removal of lower third molars. J. Oral Maxillofac. Surg. 14, 219–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collins, F.S. , Koroshetz, W.J. & Volkow, N.D. Helping to end addiction over the long‐term: the research plan for the NIH heal initiative. JAMA 320, 129–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dai, W. , Garcia, D. , de Bazelaire, C. & Alsop, D.C. Continuous flow‐driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn. Reson. Med. 60, 1488–1497 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu, W.C. , Fernandez‐Seara, M. , Detre, J.A. , Wehrli, F.W. & Wang, J. A theoretical and experimental investigation of the tagging efficiency of pseudocontinuous arterial spin labeling. Magn. Reson. Med. 58, 1020–1027 (2007). [DOI] [PubMed] [Google Scholar]
- 41. Vidorreta, M. , Wang, Z. , Chang, Y.V. , Wolk, D.A. , Fernandez‐Seara, M.A. & Detre, J.A. Whole‐brain background‐suppressed pCASL MRI with 1D‐accelerated 3D RARE Stack‐Of‐Spirals readout. PLoS One 12, e0183762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patrignani, P. , Filabozzi, P. & Patrono, C. Selective cumulative inhibition of platelet thromboxane production by low‐dose aspirin in healthy subjects. J. Clin. Invest. 69, 1366–1372 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Panara, M.R. et al Dose‐dependent inhibition of platelet cyclooxygenase‐1 and monocyte cyclooxygenase‐2 by meloxicam in healthy subjects. J. Pharmacol. Exp. Ther. 290, 276–280 (1999). [PubMed] [Google Scholar]
- 44. Yu, Y. et al Vascular COX‐2 modulates blood pressure and thrombosis in mice. Sci. Transl. Med. 4, 132ra54 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li, X. et al Differential impairment of aspirin‐dependent platelet cyclooxygenase acetylation by nonsteroidal antiinflammatory drugs. Proc. Natl. Acad. Sci. USA 111, 16830–16835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dobin, A. et al STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zerbino, D.R. et al Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ritchie, M.E. et al limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kramer, A. , Green, J. , Pollard, J. Jr & Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cerebral blood flow (CBF) measured by ASL‐fMRI.
Figure S2. Comparison of COX‐1 and COX‐2 activity ex vivo and PG urinary metabolite levels after surgery in placebo (red) and ibuprofen‐treated (blue) subjects.
Figure S3. Comparison of ibuprofen plasma concentrations and COX‐1 and COX‐2 activity ex vivo after surgery in partial (gold) and complete (teal) responders.
Figure S4. Heatmap and pathway analysis of genes (rows) differentially expressed at the second postsurgery time point compared to baseline in the placebo group.
Figure S5. Heatmap and pathway analysis of genes (rows) differentially expressed at the second postsurgery time point compared to baseline in the partial responders.
Figure S6. Heatmap and pathway analysis of genes (rows) differentially expressed at the second postsurgery time point compared to baseline in the complete responders.
Figure S7. Expression of select genes detected by RNA‐seq at the second postsurgery time point.
Supplementary Methods and Tables.


