THE volume, variety, and velocity of medical imaging data is exploding, making it impractical for clinicians to properly utilize such available information resources in an efficient fashion. At the same time, interpretation of such large amount of medical imaging data by humans is significantly error prone reducing the possibility of extracting informative data. The ability to process such large amounts of data promises to decipher the un-decoded information within the medical images; develop predictive and prognosis models to design personalized diagnosis; allow comprehensive study of tumor phenotype, and; allow assessment of tissue heterogeneity for diagnosis of different type of cancers. Recently, there has been a great surge of interest on Radiomics [1], which refers to the process of extracting and analyzing several semi-quantitative and quantitative features from medical images with the ultimate goal of obtaining predictive or prognostic models. The Radiomics features can be extracted from different imaging modalities including Magnetic Resonance Imaging (MR); Positron Emission Tomography (PET), and; Computed Tomography (CT), among which the latter is considered as the most commonly used modality for lung cancer Radiomics due to its imaging sensitivity, high resolution, and isotropic acquisition in locating the lung lesions.
The 2018 Video and Image Processing (VIP) Cup is a student competition sponsored by IEEE Signal Processing Society (SPS). Each participating team is required to be composed of: (i) One faculty member (the Supervisor); (ii) At most one graduate student (the Mentor), and; (iii) At least three but no more than ten undergraduate students. Participation in the VIP-Cup is open to all teams from around the world satisfying the above mentioned eligibility criteria. Top three finalist teams were selected to present and compete at the final stage of the 2018 VIP-Cup, which was held at 2018 IEEE International Conference on Image Processing (ICIP) in Athens, Greece on October 7th, 2018. See “Winners of the 2018 VIP-Cup” for details. In the reminder of this article, we share an overview of the 2018 VIP-Cup experience including competition setup, technical approaches, statistics, and competition experience through organizers’ and finalist team members’ eyes.
Winners of the 2018 VIP Cup.
First Place: Team Markovian
Affiliation: Bangladesh University of Engineering and Technology.
Undergraduate students: Shahruk Hossain, Zaowad Rahabin Abdullah, A. K. M. Naziul Haque, Fahim Hafiz, Md. Farhan Shadiq, Md. Monayem Hassan, Md. Mushfiqur Rahman, Md. Tariqul Islam, Muhammad Suhail Najeeb, and Asif Shahriyar.
Supervisor: Mohammad Ariful Haque.
Technical approach: The Markovian team developed a pipeline involving a binary classifier frontend which determines the cancerous slices. The identified slices containing tumor are then passed to a segmentation model based on a 2D fully convolutional neural network (CNN) using dilated convolutions instead of pooling, to generate segmentation masks for the tumor. Finally, the predicted masks are passed through a post-processing block which cleans up the masks through morphological operations.
First Runner Up: Team Spectrum
Affiliation: Bangladesh University of Engineering and Technology.
Undergraduate students: Uday Kamal, Abdul Muntakim Raf, and Md. Rakibul Hoque.
Supervisor: Md. Kamrul Hasan.
Technical approach: The Spectrum team developed framework is based on the Recurrent 3D-DenseUNet, i.e., a new fusion of Convolutional and Recurrent neural networks. The developed approach is to train the network using image-volumes with tumor-only slices. A data-driven adaptive weighting method is also used to differentiate between tumorous and non-tumorous image-slices, which shows more promise than crude intensity thresholding. In other words, adaptive thresholding is used to generate binary images and then morphological dilation is applied as post-processing to overcome the random pixel missing and enlarge the boundary.

Team Markovian took the first place. (a) Presenting at the ICIP18 final competition on October 7th, 2018. (b) Pose with IEEE SPS Director Membership-Services and the organizer of 2018 VIP-Cup, Dr. Arash Mohammadi.
I. The 2018 VIP-Cup Challenge
Radiomics workflow [1], typically, consists of the following four main processing tasks: (i) Image acquisition; (ii) Image segmentation; (iii) Feature extraction and qualification, and; (iv) Statistical analysis and model construction. Segmentation (Item (ii)) is considered as a main critical step among different processing tasks within the Radiomics pipeline, and is the focus of the 2018 VIP-Cup competition. More specifically, the main task of the 2018 VIP-Cup is Segmentation of Tumor region in Lung CT Images. Segmentation (i.e., assigning a class label to each pixel) and Localization (i.e., providing the tumor bounding box) are considered as the critical step within the Radiomics workflow, as typically Radiomics features are extracted from the segmented sections. Although, manual delineation of the gross tumor is the conventional (standard) clinical approach, it is time consuming and extensively sensitive to inter-observer variability. Development of accurate and robust automatic segmentation methods, to minimize manual error and increase consistency of delineating regions, is of paramount importance and is the focus of this competition.
Competition Dataset:
The 2018 VIP-Cup dataset consists of images from non-small cell lung cancer (NSCLC) subjects provided by Dr. Andre Dekker and Dr. Leonard Wee from Maastricht Radiation Oncology (MAASTRO) Clinic. The introduced dataset is an updated and modified version of the NSCLC-Radiomics dataset [2], [3] available at The Cancer Imaging Archive (TCIA) [4]. In particular, the new dataset consists of improved and extended annotations and also contains missing annotations for the 108 subjects in the original release, which were used for evaluation purposes as the unseen dataset.
The initial training data released on May 26th, 2018 consisted of pre-treatment CT scans from NSCLC-Radiomics dataset [2], [3] divided into two categories: (i) 100 subjects with all slices including non-tumor ones, and; (ii) A selected set of 60 subjects with tumor-only slices. Each dataset contains manual delineations of the gross tumor volume provided by a Radiologist. Fig 1 illustrates an example of CT images of lung cancer patients with tumor contours. The final datasets that are provided for training, validation, and test purposes are from the newly introduced data as briefly outlined below:
-
D1.
Training Dataset: The 260 subjects for training purposes. The labels (RTStruct files) for these patients are provided.
-
D2.
Validation Dataset: The 40 subjects for validation purposes. The labels (RTStruct files) for these patients are provided, therefore, each team can evaluate their algorithms and report the results. Each team provides evaluation results based on these 40 subjects.
-
D3.
Test Dataset: The 40 subjects that are to be used for test purposes. The labels (RTStruct files) for these patients are NOT provided. For the 40 test subjects, as the true labels are not available publicly, each team provides the segmentation results in a binary image format.
Fig. 1.
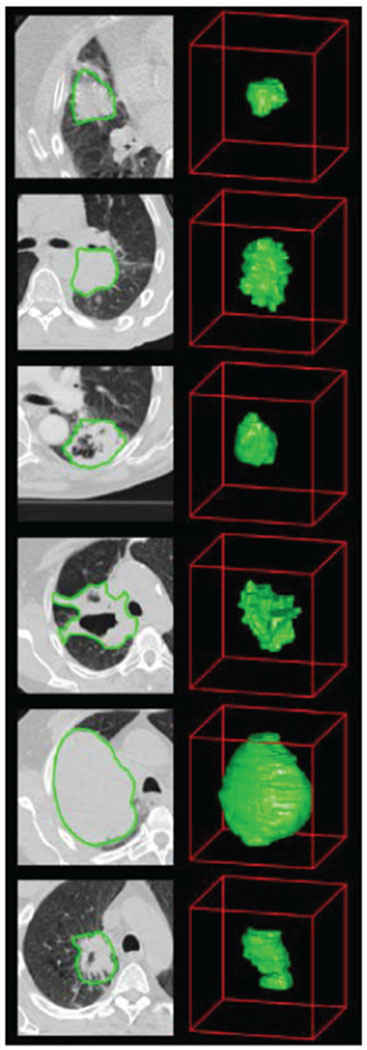
Example of the computed tomography (CT) images of lung cancer patients. CT images with tumor contours [3].
Submission Guidelines:
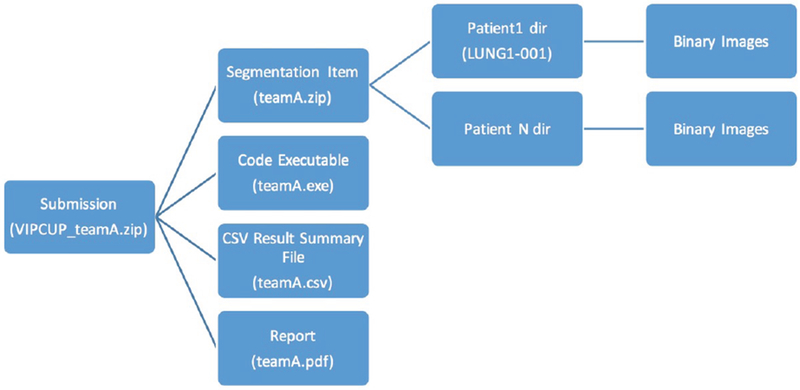
For the final competition stage, each participating team was required to submit a competition package consisting of four main items as specified in Fig. 2 and briefly outlined below:
-
S1.
Code Executable: The executable file is prepared based on the following instructions: (i) Take as an input the path to the location of all patient folders to be analyzed; (ii) If the patient folders include associated labels (RTStruct files), then the output of the executable should be a file containing all results (as specified below), and; (iii) If the provided patient folders do not have any labels (RTStruct files), then the executable should return a folder containing all patient folders, where in each folder segmentation results are presented as Binary images.
-
S2.
Reporting Results: Results submitted as a “CSV” file providing the final evaluation results based on the three identified criteria (i.e., Eqs. (1), (3), and (5)) over The Validation Data Set (i.e., the 40 subjects for which the labels (RTStruct files) are provided). The CSV file included in the submission package is prepared based on the following specifications: (i) The CSV file needs to contain the results for all the 40 subjects provided based on the three identified criteria, and; (ii) For each subject report the average value of each criteria for all of the patients’ slices.
-
S3.
Segmentation Files: This item provides the segmentation results for The Test Data Set (i.e., the 40 subjects for which the labels (RTStruct files) are not provided). This segmentation file is prepared based on the following specifications: (i) The file must contain the Binary images in “.png” format and of the same resolution (size) as that of the input images, and; (ii) Each sub-folder within the segmentation folder needs to have binary images for all the slices provided for that specific patient.
-
S4.
Report: The report should be prepared in IEEE format providing: (i) Required background; (ii) Incorporated signal processing and pre-processing algorithms; (iii) Information regarding the algorithms used to perform the segmentation tasks; (iv) Explicitly provide details on other data sources, trained models, and/or similar items (if any) that is/are used for training purposes; (v) Potential novelties of the processing algorithms, and; (vi) Presents and discusses the evaluation results.
Fig. 2.
Different items to be included in the final submission package.
Evaluation Schemes:
The segmented contours computed by the participating teams are compared against the manual contours for all validation and test images based on the following thee evaluation criteria:
-
C1.Dice Coefficient, which is a measure of relative overlap (1 represents perfect agreement and 0 represents no overlap) computed as follows
Where ∩ denotes the intersection operator, X and Y are the ground truth and test regions. Please note that dice coefficient (D) has a restricted range of [0, 1]. The following two conventions are recommended for computation of Dice coefficient: (i) For True-Negative (i.e., there is no tumor and the processing algorithm correctly detected the absence of the tumor), the dice coefficient would be 1, and; (ii) For False-Positive (i.e., there is no tumor but the processing algorithm mistakenly segmented the tumor), the dice coefficient would be 0.(1) -
C2.Mean Surface Distance: The directed average Hausdorff measure is the average distance of a point in X to its closest point in Y, i.e.,
The (undirected) average Hausdorff measure is the average of the two directed average Hausdorff measures given by(2) (3) -
C3.Hausdorff Distance (95% Hausdorff distance): The directed percent Hausdorff measure, for a percentile r, is the rth percentile distance over all distances from points in X to their closest point in Y, e.g., the directed 95% Hausdorff distance is the point in X whose distance to its closest point in Y is greater or equal to exactly 95% of the other points in X. In mathematical terms, denoting the rth percentile as Kr, this is given by
The (undirected) percent Hausdorff measure is defined again with the mean:(4) (5)
This completes description of different components of the 2018 VIP-Cup challenge.
II. The 2018 VIP-Cup Statistics
The 2018 VIP-Cup started with a global engagement of more than 600 users from 42 countries (Fig. 3) to access competition data from all around the world, as shown in Fig. 4. At the starting line, the highest engagement was received from Bangladesh, Canada, India, and United States. At the registration stage, there were 129 members clustered into 28 teams from 10 countries including Australia, Bangladesh (11 teams), Canada, China, Hong Kong, India (3 teams), Iran (2 teams), Greece, Taiwan, and United States. Out of these 28 teams, 9 teams with a total of 56 members, from Australia, Bangladesh (three teams), India (two teams), Iran, Hong Kong, and Taiwan, made it to the final stage among which 6 teams had submitted all the required components and were evaluated to select the three finalist teams.
Fig. 3.
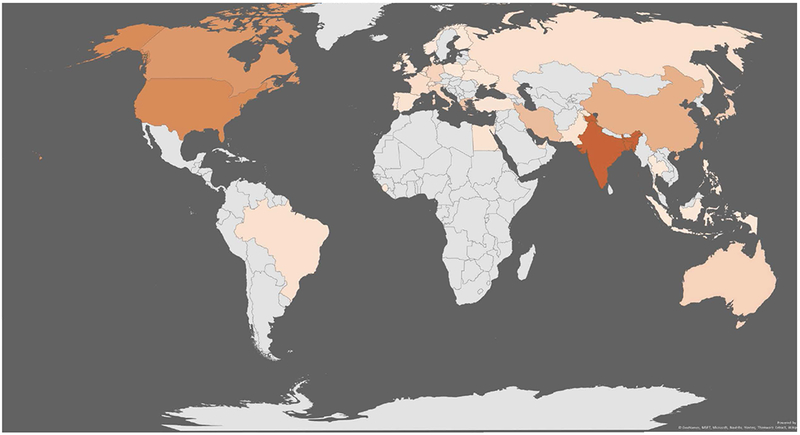
A global engagement map of the 2018 VIP-Cup.
Fig. 4.
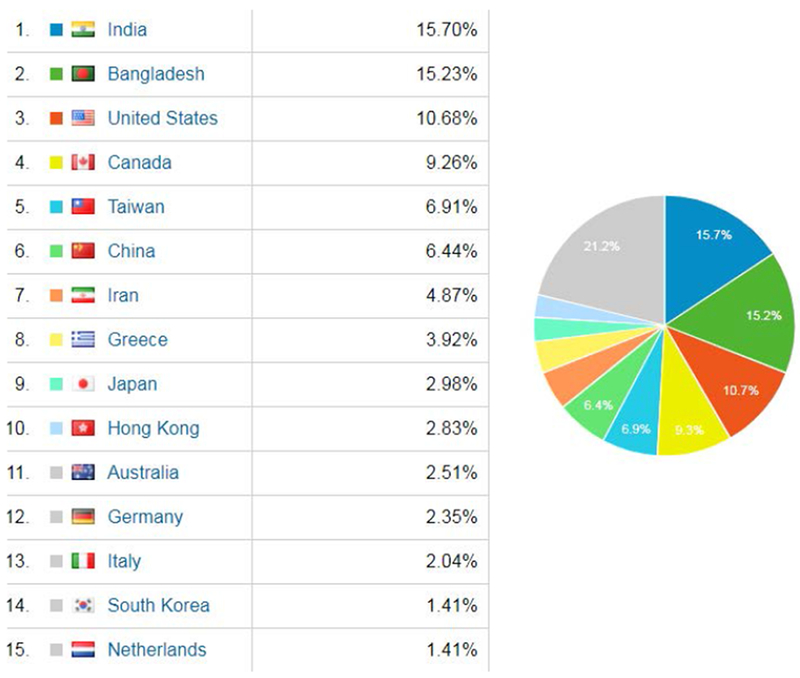
Country engagements in the 2018 VIP-Cup.
Final Stage of the Competition:
The 2018 VIP-Cup organizers evaluated the submissions and announced the three finalist teams: Markovian, NTU-MiRA, and Spectrum on September 10th, 2018. Evaluation was based on the results computed via the aforementioned three criteria (Eqs. (1)–(5)) over the validation and test datasets. Finalist teams were invited to the final competition stage at the 2018 ICIP, which was held on October 7th, 2018. Prior to the final competition and as the final challenge, the organizers have provided a list identifying the tumor-containing slices in the test data for the three finalists to apply their algorithms and provide the binary files for the tumor-only slices. The results are used as part of the ranking procedure of the top three teams. During the final competition on October 7th, the finalist teams have presented their work, where each team had 15 minutes for presentation and 5 minutes for Q & A. After the team presentations, the jury had an internal discussion to finalize the ranking. The jury included Dr. Farnoosh Naderkhani, Mrs. Parnian Afshar, Dr. Lucio Marcenaro, and Dr. Arash Mohammadi. In the ICIP’18 Student Career Luncheon on Monday, October 8th, 2018, Dr. Lucio Marcenaro, SPS’s Director of Student-Services, highlighted the second edition of the VIP-Cup and Dr. Arash Mohammadi, organizer of the 2018 VIP-Cup and SPS’s Director of Membership-Services, publicly announced the winners of the competition.
III. Highlights of Technical Approaches
All teams are composed of mainly undergraduate students supervised by a single faculty advisor and at most one graduate student mentor. Although different classical segmentation algorithms, such as Thresholding methods; Histogram-based methodologies; Morphological techniques, and Clustering approaches, were investigated by the participants, deep neural networks (DNNs) constitute the main building block of the processing pipeline developed by the finalists. Several different deep architectures were studied and implemented from encoder-decoder style models such as the U-Net [8], ResNet like architectures [10], DenseNet architectures [9] to finally LungNet architecture [7], which are proposed very recently in March 2018. A very interesting aspect of the processing techniques implemented by the finalists is that all three teams went one step ahead of what is currently introduced in the literature, and developed and designed hybrid and innovative architectures.
In particular, the team Markovian has developed a pipeline involving as the first step a binary classifier frontend which detects the tumorous slices. The identified slices containing the tumor are then passed to a segmentation model based on a 2D fully convolutional neural network (CNN) using dilated convolutions instead of pooling, to generate segmentation masks for the tumor. Finally, the predicted masks are passed through a post-processing block which cleans up the masks through morphological operations. The average dice coefficient of 0.627 over the validation dataset is reported. The average dice coefficient of 0.594 over the test set is computed. The team spectrum implemented framework is based on the Recurrent 3D-DenseUNet and a novel fusion of Convolutional and Recurrent neural networks that are introduced and incorporated for performing the competition tasks. The developed approach is to train the network using image-volumes with tumor only slices of size (256 × 256 × 8). A data-driven adaptive weighting method is then used to differentiate between tumorous and non-tumorous image-slices, which shows more promise than crude intensity thresholding. In other words, adaptive thresholding is used to generate binary images and then morphological dilation is applied as post-processing to overcome the random pixel missing and enlarge the boundary. The average dice coefficient of 0.74 over the validation dataset is reported. The average dice coefficient of 0.521 over the test set is computed. Team NTU-MiRA developed an end-to-end trainable network that performs lung cancer tumor segmentation in a supervised fashion. Inspired by the dense up-sampling layer proposed in [11], the dense down-sampling convolutional (DDC) layer is incorporated to reduce the computational burden. Dense lateral connections are introduced into the proposed model, which facilitates the network training and results in faster convergence. Finally, to handle the imbalance distribution between the foreground and background region, the dice loss function and the focal loss are used for optimization.
IV. Organizers’ Opinions
It was a challenging but, at the same time, an intriguing journey to organize the 2018 VIP-Cup. It was during the ICASSP’18, 15 to 20 April 2018, that we were set to organize this year’s VIP-Cup and given the limited time we had, the early stage was very challenging. In particular, the main initial issue was to find unseen data for test purposes due to common difficulties faced in acquiring and distributing labeled medical images. During the course of the competition and especially during the initial phase, organizing members had weekly conference calls to address the aforementioned issue and discuss different aspects of the competition. It was also very inspiring to meet the members of the finalists during the ICIP’18. Moreover, we are greatly impressed and encouraged during the course of the competition and at the final stage by the high technical level of implemented processing solutions developed and even proposed by the competitors and extensive dedication of the participating teams.
V. Participants’ Opinions
Throughout the 2018 VIP-Cup competition there was a great deal of interaction, not only through questions for the instructors posted to Piazza but also among the different students who often engaged in discussion over the provided responses. We, as organizers, were both encouraged and delighted to observe such a collaborative spirit among the participating students. Next, we provide an overview of some feedback and perspectives received from the winning teams.
Team Markovian
-
Thanks to IEEE Signal Processing Society (SPS) for organizing this wonderful competition regularly for the undergraduate students. We are learning a lot from these events.
— Dr. Mohammad Ariful Haque, Supervisor.
-
It was an amazing opportunity to participate in the IEEE VIP Cup for the second time; the first time was in 2017, where our team made it to 2nd Runner-Up. We learned from our previous experience and with the guidance of our supervisor, Dr. Mohammad Ariful Haque, we were able to capture the Champion title this time. Knowing that the task at hand had real world applications and could potentially help people around the world was a great motivating factor for me. Through the VIP Cup my understanding of neural networks, segmentation problems and biomedical image processing in general have grown quite a lot. I also learned a lot about teamwork, and what it takes to get things done under deadlines. Some of us will be graduating this year, but I am sure that Team Markovians will be rejuvenated with new members and comeback stronger in the next VIP Cup!
— Shahruk Hossain, Undergraduate.
-
Being able to take part in the VIP Cup for the 2nd time for Team Markovians has been a great experience. This challenge allowed us to dig deeper into the emerging world of Biomedical Image Processing. The new challenges, the short time frame and tight deadlines demanded immense patience and relentless work. Its great to see all the efforts pay off.
— Suhail Najeeb, Undergraduate.
-
This was my first VIP cup and it really helped me a lot understanding neural networks. The problem was challenging and I learned a great deal about using ML libraries in python. The team work I experienced was really something else. In the future I wish to work on biomedical image processing. The skills I improved participating in VIP cup 2018 certainly boosted my confidence in many ways.
— Zaowad Rahabin Abdullah, Undergraduate.
-
My VIP cup experience was a very exciting one as I arrived at Greece only three hours before the presentation. I was not sure whether I was going to make it or not. Special thanks to my fellow team mate Suhail who prepared some comprehensive slides so that I could prepare for the presentation within one hour. Also thanks to Shahruk for joining me during the Q/A session. While presenting, I just kept in mind that I had to explain to the judges the pros and cons of our model and illustrate its effectiveness. But I was very uncertain about my own presentation due to not getting enough time for preparing. However, when the results were published, I couldn’t believe it. All our efforts were worth it, I felt. It seems that the simplest solution often performs the best.
— Asif Shahriyar Sushmit, Undergraduate.
Team Spectrum
-
This was my 2nd time participation in IEEE VIP cup. The problem itself was very challenging. To train a robust model from scratch using very limited amount of data was the toughest part of the competition. I gained some valuable experience and learnt a lot by working on the challenge. Above all, I would like to thank all specially the organizers, our respected supervisor, and my parents for their constant help and support.
— Uday Kamal, Undergraduate.
-
What I learnt from this years IEEE VIP Cup is that eventually it comes down to the robustness of the whole pipeline. We were provided separate train, validation and test data in the competition. We trained our model with the train data only. But while selecting threshold for the output probabilities of the model we used the validation data to choose the best threshold. This somehow overfit our results to some degree on the validation data and we couldn’t attain our expected dice scores in the test data. This was a great experience for me. I would like to thank the organizers for this thrilling problem and exceptional dataset.
— Abdul Muntakim Rafi, Undergraduate.
-
It was an awesome experience. I am glad that parts of my work had been presented before the world through this competition. This challenging competition has helped me to develop a research oriented mindset under our respected supervisor Prof. Md. Kamrul Hasan Sir. Also, the organizers this year were very friendly and helpful as well. Last of all, I would like to thank my friends and family who constantly supported me and my team members for their dedication and hard work.
— Rakibul Hoque, Undergraduate.
Team NTU-MiRA
-
This was my first time to participate in an international competition. I would like to thank the organizers to provide such a challenging stage for us, and I have learned many techniques about deep learning and medical images. I am also grateful for our supervisor and my teammates to overcome many difficulties together. I believe that this experience can make us grow. Wish the 2019 VIP-Cup delightful and successful.
— Jhih-Yuan Lin, Undergraduate.
-
Accomplishing this challenging tumor segmentation task is a milestone to me. I learned lots of deep learning technologies about 3D medical image processing from the 2018 VIP-Cup, and tried all my best to overcome each problem, like the position and scale variants, the data imbalance, and the limitation of the hardware resources, to build an accurate, robust, and efficient model. On top of that, the experience of sharing our efforts on an international competition is the most precious thing. I’m very glad that we won third place. At last, I want to thank our supervisor Prof. Winston H. Hsu, my teammates, and all the friends who have helped us.
— Min-Sheng Wu, Undergraduate.
-
It’s an unforgettable experience for me to present at the international conference. I learned a lot about medical image segmentation skills during the process of the competition. And I’m thankful that the organizers provided such a challenging and exciting competition.
— Yu-Cheng, Chang, Undergraduate.
-
As an undergrad student, participating in an application oriented challenge that involves competitors from all over the world is an exciting, intriguing, and challenging experience. While many of the existing methods have been proposed to address various medical tasks from different aspects, many of the methods are built upon the pre-trained models or transfer learning mechanisms, which are prohibited in this competition. In the interest of the limited duration, we conducted extensive pilot studies and found many methods applicable to the given task. Driven by these methods, we developed an accurate and efficient network to address the task at hand in a 3D learning fashion. In conclusion, we sincerely thank the organizers for holding this competition. In the future, we hope there will be more competitions being held so that people from different backgrounds can all be involved.
— Yun-Chun Chen, Undergraduate.
-
I am an undergraduate student in my last year of study. My research mainly focuses on the deep generative models, but I always hoped to have a chance to work on image segmentation problem especially in the 3D domain. This contest provides me with a perfect opportunity. This contest is very challenging. We need to take a lot of time to visualize each data, read a lot of paper and discuss with each other to come up with the solution. I really enjoy the contest since it makes me grow and have a chance to work with some good teammates. I am also very happy that we won third place in the contest.
— Chao-Te Chou, Undergraduate.
-
As a graduate student focusing on medical imaging, it was my first time to lead a team to compete with people from all over the world. From this challenge, I learned a lot about the domain of medical image segmentation techniques and found that there is still a long way to go in this domain. Noise, imbalance, and scarcity of data are the main issues make it challenging for us to build an accurate and robust segmentation model. I’m so excited to win the third place. There was more work than expected, but it was really a precious and rewarding experience. Many thanks to my hardworking team members and the organizers for holding this contest.
— Chun-Ting, Wu, Graduate.
Winners of the 2018 VIP Cup.
Second Runner Up: Team Markovians
Affiliation: National Cheng Kung University and National Taiwan University,
Undergraduate students: Chun-Ting Wu, Chao-Te Chou, Min-Sheng Wu, Jhih-Yuan Lin, Yun-Chun Chen, Yu-Cheng Chang,
Supervisor: Winston Hsu,
Technical approach: Team NTU-MiRA developed an end-to-end trainable data driven solution that learns to predict volumetric segmentation outputs. To handle the imbalance distribution between the foreground and background regions, the dice loss function and the focal loss are used for optimization.

First runner-up team Spectrum. (a) Present at the ICIP18 final competition on October 7th, 2018. (b) Pose with Dr. Arash Mohammadi. (c) A behind-the-scenes look.
Winners of the 2018 VIP Cup.

Second runner-up team NTU-MiTRA. (a) Present at the ICIP18 final competition on October 7th, 2018. (b) Pose with Dr. Arash Mohammadi. (c) Members of Team NTU-MiTRA at ICIP’18. (d) A group photo with the jury members, Dr. Farnoosh Naderkhani, Mrs. Parnian Afshar, Dr. Lucio Marcenaro, and Dr. Arash Mohammadi.
VI. Acknowledgment
The organizers of the 2018 VIP-Cup would like to express their utmost gratitude to all who made this adventure a reality including but not limited to the participating teams, the judging panel, the local organizers, and IEEE SPS Membership Board. This competition would have not been possible without the timely and prompt help and follow ups from members of The Cancer Imaging Archive (TCIA) and National Cancer Institute (NCI). In addition, great appreciation goes to Dr. Andre Dekker and Dr. Leonard Wee from Maastricht Radiation Oncology (MAASTRO) Clinic for providing the competition dataset. Special thanks goes in particular to Dr. Lucio Marce-naro, director student-services of IEEE SPS, for his support and great and exceptional follow ups and dedication that made it possible for members from all three finalist teams to attend ICIP’18 and present at the final stage. Finally, great appreciation goes to student organizers who were involved through out the coarse of the 2018 VIP Cup, specially, Parnian Afshar, Suzette Slim, and Wu Xin for their dedication and hard work in preparation of different competition datasets and performing the evaluations.
Biographies
VII. Authors
Arash Mohammadi (arash.mohammadi@concordia.ca) is an assistant professor with Concordia Institute for Information Systems Engineering (CIISE), Concordia University. He is the Director Membership-Services of IEEE SPS. He was the Lead Guest Editor for an Special Issue in IEEE Transactions on Signal and Information Processing Over Networks entitled ”Distributed Signal Processing for Security and Privacy in Networked Cyber-Physical Systems”; the Organizing Committee chair of “IEEE Signal Processing Society Winter School on Distributed Signal Processing for Secure Cyber-Physical Systems”, and; Co-Chair of “Symposium on Advanced BioSignal Processing for Rehabilitation & Assistive Systems”, at IEEE GlobalSIP’17. He is currently Co-Chair of “Symposium on Advanced Bio-Signal Processing and Machine Learning for Medical Cyber-Physical Systems”, at IEEE GlobalSIP’18.
Parnian Afshar (p_afs@encs.concordia.ca) is a Ph.D. candidate at Concordia Institute for Information System Engineering (CIISE). Her research interests include signal processing, biometrics, image and video processing, pattern recognition, and machine learning. She has extensive research/publication record in medical image processing related areas.
Amir Asif (amir.asif@concordia.ca) is a Professor with the ECE Department at Concordia University since 2014, where he is now serving as the Dean of the Faculty of Engineering and Computer Science. Previously, he was a professor with the EECS Department at York University, Canada. Dr. Asif has served on the editorial boards of numerous journals and international conferences, including Associate Editor for IEEE Transactions of Signal Processing (2014-18), IEEE Signal Processing letters (2002-2006, 2009-2013). He has organized four IEEE conferences on signal processing theory and applications.
Keyvan Farahani (farahank@mail.nih.gov) is a Program Director in the Image-Guided Interventions (IGI) Branch, Cancer Imaging Program of the National Cancer Institute (NCI). In this capacity he is responsible for the development of NCI initiatives that address diagnosis and treatment of cancer through integration of advanced imaging and minimally invasive therapies. Since 2002 Dr. Farahani has lead the NCI initiatives in Oncologic IGI with programs focused on industrial developments, early phase clinical trials, and image-guided drug delivery using nanotechnologies. He has led a series of NCI workshops that promote an open science model to develop, optimize and validate platforms for IGI. Prior to joining NCI in fall of 2001, Dr. Farahani was a faculty of the department of Radiological Sciences at the University of California, Los Angeles, where he obtained his MS (’89) and PhD (’93) degrees in Biomedical Physics.
Justin Kirby (kirbyju@mail.nih.gov) works at Frederick National Laboratory for Cancer Research which provides imaging informatics support to National Cancer Institute (NCI)’s Cancer Imaging Program. His current work focuses on creating open science resources to improve reproducibility and transparency in cancer imaging research. Most notably, his team manages The Cancer Imaging Archive (TCIA) which is a service aimed at helping researchers to share de-identified radiology and pathology images.
Anastasia Oikonomou
(anastasia.oikonomou@sunnybrook.ca) is the head of the Cardiothoracic Imaging Division at Sunnybrook Health Science Centre; Site Director of the Cardiothoracic Imaging Fellowship program at University of Toronto, and an Assistant Professor with the Department of Medical Imaging at the University of Toronto. Her research interests include imaging of pulmonary malignancies and interstitial lung diseases, Radiomics and machine learning methods in imaging of pulmonary disease.
Konstantinos N. Plataniotis (kostas@ece.utoronto.ca) is Bell Canada Chair in Multimedia, and a Professor with the ECE Department at the University of Toronto. He is a registered professional engineer in Ontario, Fellow of the IEEE and Fellow of the Engineering Institute of Canada. Dr. Plataniotis was the IEEE Signal Processing Society inaugural Vice President for Membership (2014-2016) and the General Co-Chair for the IEEE GlobalSIP 2017 (November 2017, Montreal, Q.C.). He co-chairs the 2018 IEEE International Conference on Image Processing (ICIP 2018), October 7-10, 2018, Athens Greece, and 2021 IEEE International Conference in Acoustics, Speech & Signal Processing (ICASSP 2021), Toronto, Canada.
References
- [1].Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A, Benali H “From Hand-Crafted to Deep Learning-based Cancer Radiomics: Challenges and Opportunities,” Submitted to IEEE Signal Processing Magazine, 2018. [Google Scholar]
- [2].Aerts, Hugo JWL, Velazquez Rios, Emmanuel, Leijenaar, Ralph TH, Parmar, Chintan, Grossmann, Patrick, Carvalho, Sara, … Lambin, Philippe, “Data From NSCLC-Radiomics,” The Cancer Imaging Archive, 2015. [Google Scholar]
- [3].Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, … Lambin P, “Decoding Tumour Phenotype by Noninvasive Imaging using a Quantitative Radiomics Approach,” Nature Communications, Nature Publishing Group, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F, “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository,” Journal of Digital Imaging, vol. 26, no. 6, pp. 1045–1057, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Oikonomou A, Khalvati F, et al. “Radiomics analysis at PET/ CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy,” Scientific reports, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F, “Radiomics-based Prognosis Analysis for Non-small Cell Lung Cancer,” Sci. Rep, vol. 7, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Anthimopoulos M, Christodoulidis S, Ebner L, Geiser T, Christe A, and Mougiakakou S, “Semantic Segmentation of Pathological Lung Tissue with Dilated Fully Convolutional Network,” https://arxiv.org/abs/1803.06167, 2018. [DOI] [PubMed] [Google Scholar]
- [8].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional Networks for Biomedical Image Segmentation,” https://arxiv.org/abs/1505.04597, 2015.
- [9].Huang G, Liu Z, Der Maaten LV, and Weinberger KQ, “Densely Connected Convolutional Networks,” CVPR, volume 1, page 3, 2017. [Google Scholar]
- [10].He K, Zhang X, Ren S, and Sun J, “Deep Residual Learning for Image Recognition,” IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778. [Google Scholar]
- [11].Wang P, Chen P, Yuan Y, Liu D, Huang Z, Hou X, and Cottrell G, “Understanding Convolution for Semantic Segmentation,” https://arxiv.org/abs/1702.08502, 2017.


