Abstract
The cornea provides a functional barrier separating the outside environment from the intraocular environment, thereby protecting posterior segments of the eye from infection and permanent damage. Pathological changes that compromise the structure or integrity of the cornea may occur as a result of injury or disease and can lead to debilitating effects on visual acuity. Over 10 million people worldwide are visually impaired or blind due to corneal opacity. Thus, physiologically relevant in vitro approaches to predict corneal toxicity of chemicals or effective treatments for disease prior to ocular exposure, as well as to study the corneal effects of systemic, chronic conditions, such as diabetes, are needed to reduce the use of animal testing and accelerate therapeutic development. We have previously bioengineered an innervated corneal tissue model using silk protein scaffolds to re-capitulate the structural and mechanical elements of the anterior cornea, as well as model the functional aspects of corneal sensation with the inclusion of epithelial, stromal, and neural components. The purpose of this unit is to provide a step-by-step guide for the preparation, assembly, and application of this 3D cornea tissue system to enable the study of corneal tissue biology.
Keywords: tissue engineering, cornea, sensory nerves, toxicology, 3D in vitro model, silk biomaterials
3D Corneal Tissue Model
Introduction
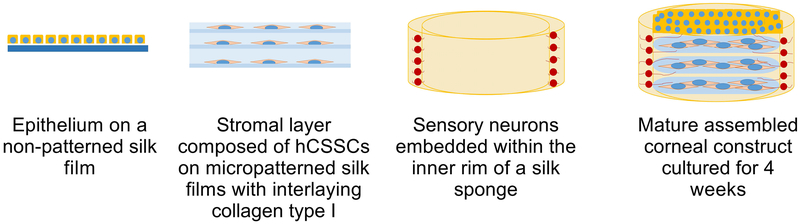
The highly innervated nature of the cornea and the presence of multiple cell types have made the development and application of relevant in vitro tissue models limited in scope and complexity. To address this need in the field, we have bioengineered an advanced, innervated human corneal tissue model containing a stratified corneal structure of significant strength and rigidity, transparency, and sustainable culture conditions (Wang et al., 2017; Siran et al., 2018). This tissue model takes advantage of the highly versatile nature of the silk protein as a biocompatible scaffold to construct a multi-layered, pre-assembled tissue structure. The cellular components of the model consist of human corneal epithelial cells (hCECs) seeded onto a silk film mimicking Bowman’s layer, followed by an underlying stromal layer composed of human corneal stromal stem cells (hCSSCs) seeded on 3 silk films, and differentiated sensory nerves innervating the central cornea from the periphery (Figure 1). Addition of a collagen type I coating between the layers and on the surface serves to mediate cohesiveness of the construct. In our previous work, we have applied different sensory sources, including dorsal root ganglion (DRGs) (Wang et al., 2015; Wang et al., 2017) and human-induced neural stem cells (hiNSCs) (Deardorff et al., 2018; Siran et al., 2018) to promote corneal sensory innervation in co-cultures of hCECs and hCSSCs. Utilization of hiNSCs has many advantages, including well-defined differentiation protocols that generate a gradual transition to differentiated nerves with elongated neuronal fibers (Cairns et al., 2016; Deardorff et al., 2018). Herein, we describe the application of hiNSCs in this system and detail culture conditions for isolation, subculture, and differentiation protocols.
Figure 1.
Schematic depicting the assembly of the different layers that compose the corneal tissue model. Each scaffold component, e.g. the non-patterned silk film, micropatterned silk films, and silk sponge are prepared from isolated silk fibroin and functionalized via a collagen coating, RGD-peptide, or poly-D-lysine coating, respectively, prior to cell seeding. Maturation of the construct characterized by stromal ECM deposition and innervation of the sensory nerves into the stromal layer occurs over an ideal 4-week timeframe.
The scaffold implemented in this corneal tissue model is protein-based using fibroin (silk), a naturally-occurring biomaterial produced by the silkworm, Bombyx mori. Silk is biocompatible, non-immunogenic, and robust for sustainable long-term culture conditions, therefore serving as a useful scaffold to control cell alignment, provide a tangible surface for stacking into 3D structures, and remains transparent as thin films (Abbott et al., 2016). Functionalization of silk scaffolds with Arginine-Glycine-Aspartate (RGD)-peptide, a binding motif for extracellular matrix (ECM) domains, promotes cell attachment via a number of integrin classes, including integrins α2β1, α3β1, and αvβ3 (Ruoslahti, 1996), which among many others are naturally expressed on the naïve keratocyte (Stepp, 2006). In our corneal tissue model, we utilize RGD-functionalized micropatterned films to establish a direct anchor to mediate keratocyte organization similar to the keratocyte network present in the adult cornea, thereby allowing for native ECM deposition between biomimetic silk lamellae (Ghezzi et al., 2017). Micropatterned silk films guide keratocyte and fibroblast alignment along microgrooves in conventional 2D cultures (Lawrence et al., 2009; Gil, Mandal, et al., 2010; Gil, Park, et al., 2010; Wu, Rnjak-Kovacina, et al., 2014) and in 3D microenvironments (Chen et al., 2017; Ghezzi et al., 2017), thus serving as a useful means to model keratocyte-ECM interaction s in vitro.
Tissue-engineered models aim to reconstruct the in vivo condition to study biological mechanisms in a defined, yet physiological microenvironment. This approach allows one to dissect the contribution of specific cell types to physiological and disease processes, which may be difficult to identify in a complex animal model. Using the described corneal tissue model, we have previously reported characterization of the activation of pain-related signaling pathways in response to chemical irritation (Siran et al., 2018) and nerve loss following hyperglycemia (Deardorff et al., 2018), two biological events that show parallel endpoints to the in vivo condition. A key advantage of this corneal tissue model system is the compartmentalization of individual cell populations into separable scaffolds, e.g. the epithelium seeded on the anterior silk film, the stroma seeded on posterior silk films, and the sensory nerves seeded in the peripheral silk sponge. While the construct reaches maturation over 4 weeks of culturing with the outgrowth of nerve fibers into the stroma and ECM production by resident keratocytes, the cell bodies remain distinct from one another, similar to the human cornea in vivo with the separation of the epithelium and stroma interfaced by a basement membrane and localization of neuronal somas outside of the cornea. This design provides a technical advantage in allowing for protein analysis of each cell type following stimulation to determine factors regulated by chemical or mechanical stimuli. Characterization of the pain response can apply to studying phenotypic changes in the corneal epithelium (e.g. pro-inflammatory factor expression, matrix metalloproteinase (MMP) expression), keratocytes (e.g. fibrotic markers, collagen and proteoglycan secretion, and ECM thickness), and nerve populations (e.g. fiber density and tortuosity) (McKay et al., 2018).
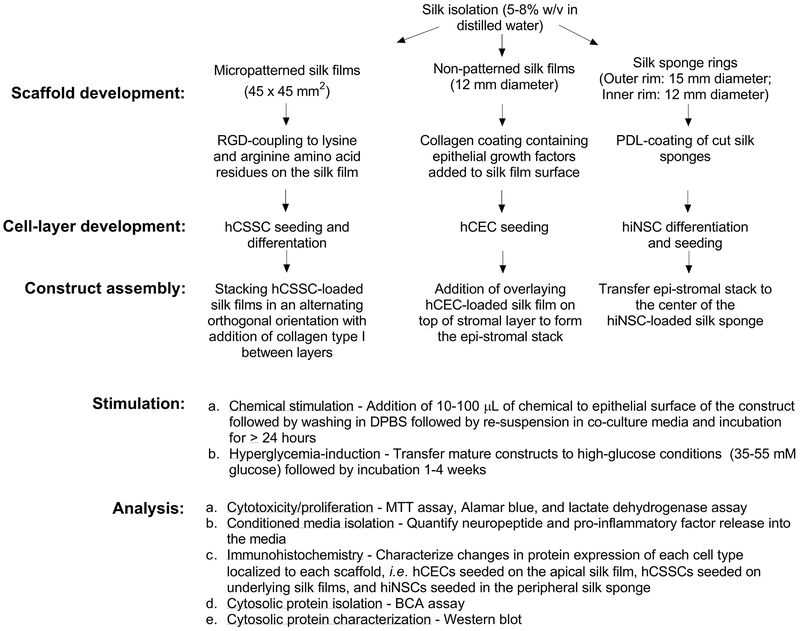
The purpose of this unit is to describe the methodology in detail regarding the preparation, assembly, and maintenance of the 3D corneal tissue model (Figure 2). The methods are divided into three major sections:
-
1)
Assembly - Basic Protocol 1 describes the assembly of the corneal construct using functionalized silk scaffolds and primary human-sourced cells. Support Protocols 1–3 detail the preparation and functionalization of silk films and sponges for cell seeding. We also provide information regarding sub-culture conditions and differentiation approaches for each cell type used in the model (Support Protocols 4–6).
-
2)
Application - Basic Protocol 2 describes potential applications of the model with methods focusing on chemical stimulation of the corneal construct.
-
3)
Characterization - Basic Protocol 3 describes isolation and characterization of the corneal construct in terms of viability as a measure of applied chemical toxicity. Support Protocols 7–11 elaborate on methods for further characterizing the biological response to stimuli, including isolation of conditioned media, fixation and immunohistochemistry, and protein characterization via Western blotting.
Figure 2.
Flowchart of the experimental set-up involving scaffold development, cell culture approaches, assembly, stimulation, and analysis of the corneal construct.
Basic Protocol 1:
ASSEMBLY AND MAINTENANCE OF THE CORNEAL TISSUE MODEL
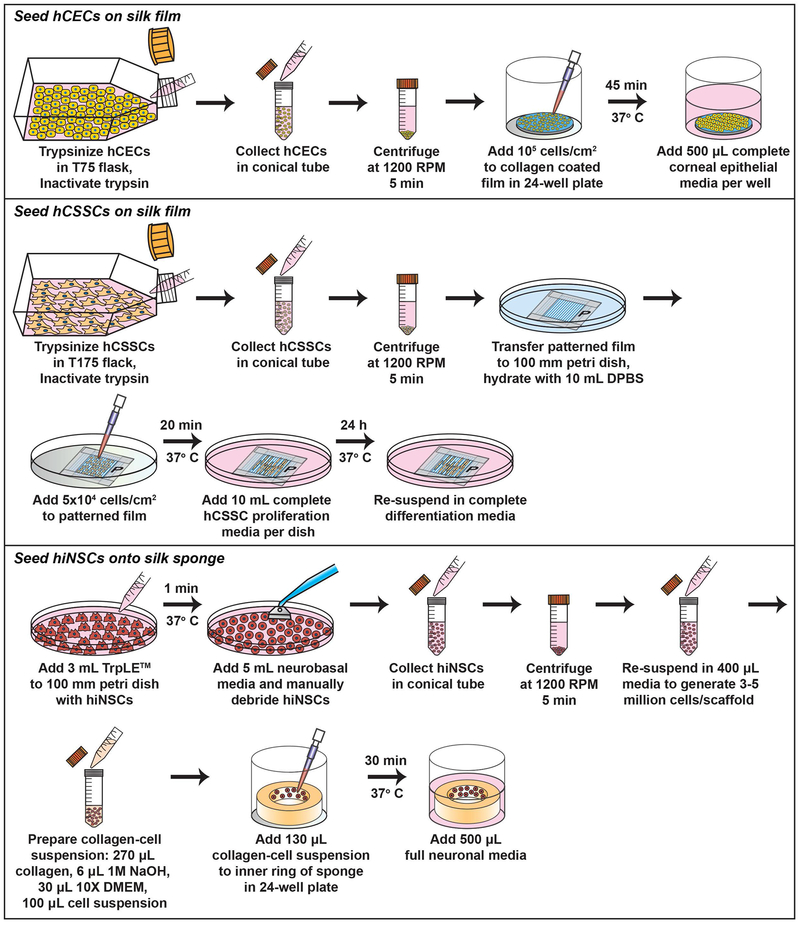
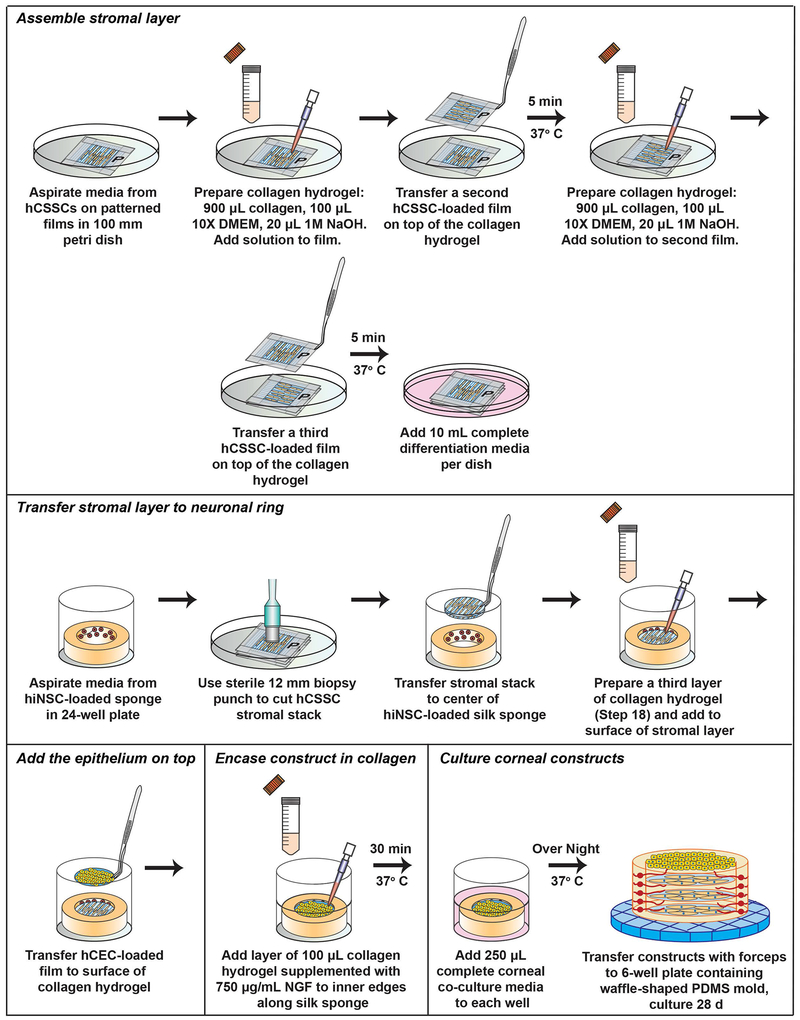
The assembly of the corneal construct requires prior isolation, preparation, and functionalization of silk scaffolds (described in Support Protocols 1–3), as well as culturing and seeding of primary cells onto the prepared silk biomaterials (Figure 3). Upon development of each cellular component, the tissue can be assembled and maintained stably for long-term conditions at 37°C/5% CO2 up to at least 4-8 weeks with media changes every other day. Following 24 hours post-assembly, the corneal construct is robust to manual transfer using forceps to different culture vessels or a bioreactor system (Siran et al., 2018) (Figure 4). Over time, the construct gains increased rigidity following ECM production within the interlaying stromal layers (Ghezzi et al., 2017; Wang et al., 2017), thus allowing for further manipulation without disrupting overall tissue structure.
Figure 3.
Schematic depicting the protocol for hCECs, hCSSCs, and hiNSCs seeding onto silk scaffolds.
Figure 4.
Schematic depicting the protocol for corneal construct assembly from the epithelial and stromal layers.
Materials
Laminar flow hood
Temperature- and CO2-controlled incubator
Primary human corneal epithelial cells (hCECs, ATCC, cat. no. ATCC® PCS-700-010™)
Complete corneal epithelial media (Refer to Reagents and Solutions Section)
T75 cell culture flask
Dulbecco’s phosphate buffered saline (DPBS) (Gibco, cat. no. 14190144)
Microscope
Pipette
0.05% w/v trypsin-EDTA (Gibco, cat. no. 25300054)
50 mL polypropylene centrifuge tube
Fetal bovine serum (FBS) (Gibco, cat. no. 16000)
Centrifuge
Collagen-coated silk films (Refer to Support Protocol 2)
24-well cell culture plates
Primary human corneal stromal stem cells (hCSSCs, primary cells isolated from limbal rim)
Complete hCCSC proliferation media (Refer to Reagents and Solutions Section)
T175 cell culture flask
Sterile micropatterned Arg-Gly-Asp (RGD)-coated silk films (Refer to Support Protocol 2)
100 mm petri dish
Forceps
Complete hCSSC differentiation media (Refer to Reagents and Solutions Section)
Micropipette
1.5 mL microcentrifuge tube
Rat-tail Collagen type I (Gibco, cat. no. A1048301)
10× DMEM (Sigma, cat. no. D2429)
1M NaOH (Sigma, cat. no. 795429)
Complete neurobasal media (Refer to Reagents and Solutions Section)
Human induced neural stem cells (hiNSCs)
TrpLE™ Express (Gibco, cat. no. 12604013)
Poly-D-lysine (PDL)-coated silk sponges (Refer to Support Protocol 3)
12 mm biopsy punch
Nerve growth factor (NGF)
Complete corneal co-culture media (Refer to Reagents and Solutions Section)
Waffle-patterned PDMS mold (Refer to Support Protocol 1, Step 5)
6-well cell culture plate
NOTE: All biological experiments should be performed in a Biological Safety Level 2 (BSL2)-approved laboratory environment under sterile conditions in a laminar flow hood. All reagents and biologics must be sterilized by autoclaving, filter-sterilization (0.2 μm filter), UV-light (>30 minutes), or ethanol-sterilization (70% v/v ethanol/water) prior to use in cell culture and maintained under aseptic conditions during the duration of the experiment.
Seed hCECs on silk film
-
1.
Sub-culture hCECs as described in Support Protocol 4. Upon ~90% confluence, trypsinize hCECs from T75 flask, inactivate trypsin with 2× volume of complete corneal epithelial media, and centrifuge at 1200 rpm for 5 min to pellet cells.
-
2.
Re-suspend hCECs in complete epithelial media and add 105 cells/cm2 to surface of collagen-coated silk film in a 24-well plate (Support Protocol 2, Step 16). Incubate at 37°C/5% CO2 for 45 minutes. Allow hCECs to attach to silk film as determined by visualization using a light microscope.
Increase incubation time up to 4 hours in minimal media to allow cell attachment, if needed. One hCEC-loaded film is used per construct.
-
3.Add complete corneal epithelial media to each well for 500 μL total and incubate at 37°C/5% CO2 overnight.
- hCECs will appear as flattened cells attached to the film at t=24 hours post-seeding.
Seed hCSSCs on silk film
-
4.
Sub-culture hCSSCs as described in Support Protocol 5. Upon 90% confluence, trypsinize hCSSCs from a T175 flask, inactivate trypsin with 10% FBS, and centrifuge at 1200 rpm for 5 minutes to pellet cells.
-
5.
Prepare RGD-functionalized silk films (Support Protocol 2, Step 13) for cell seeding by transferring to a 100 mm petri dish. Re-hydrate by adding 10 mL of DPBS to each dish and swirl gently to allow immersion. Aspirate DPBS.
-
6.
Re-suspend hCSSCs in complete hCSSC proliferation media and add 105 cells suspended in 500 μL to each silk film for a cell density of 5 × 104 cells/cm2. Incubate the silk film and cells in the 100 mm petri dish for 20 minutes at 37°C in a minimal volume of media to allow cell attachment to silk film.
-
7.
Confirm hCSSC attachment to silk film by visualizing under a light microscope. Add 10 mL of complete hCSSC proliferation media to each 100 mm dish and incubate overnight at 37°C/5% CO2.
-
8.
At t= 24 hours post-seeding, aspirate media and re-suspend hCSSC-loaded silk films in complete hCSSC differentiation media.
Confirm appropriate cell density on silk films. hCSSCs should appear as elongated spindles aligned along the microgrooves of the silk film. hCSSC-loaded silk films may be maintained in proliferation media if cell density or attachment is low to mediate cell proliferation on the silk film prior to construct assembly. Since hCSSC differentiation media is serum-free, cell proliferation is heavily reduced in this media.
Seed hiNSCs onto silk sponge
-
9.
Culture hiNSCs as described in Support Protocol 6. Upon 90% confluence, add 3 mL of TrpLE™ Express solution to a 100 mm petri dish to mediate detachment. Incubate for 1 minute at 37°C.
TrpLE™ is an animal-free replacement for trypsin that does not require re-suspension in FBS for inactivation.
-
10.
Add 5 mL of neurobasal media and manually remove cells by gently debriding the surface with a pipette tip or cell scraper.
-
11.
Pipette the cell solution gently and transfer to a 50 mL centrifuge tube. Rinse the remaining cells in the petri dish with 5 mL of DPBS to remove all cells and centrifuge for 5 minutes at 1200 rpm.
-
12.
Aspirate the media and re-suspend pellet in 400 μL of complete neurobasal media to generate 3-5 million cells per scaffold.
-
13.
Retrieve PDL-coated silk sponges (Support Protocol 3, Step 9). Aspirate PBS and transfer each one to a well of a 24-well plate with forceps.
-
14.Prepare a collagen-cell suspension using the following recipe added to a 1.5 mL microcentrifuge tube:
- 270 μL rat-tail collagen type I (3 mg/mL suspended in 20 mM acetic acid)
- 6 μL 1M NaOH
- 30 μL 10× DMEM
-
100 μL cell solutionAdd the cell solution to the mixture last following pH neutralization of the collagen by NaOH. The color of the solution should be light red rather than orange or yellow prior to addition of the cell solution.
-
15.
Gently mix collagen-cell suspension by pipetting up and down. Add 130 μL of collagen-cell suspension to each PDL-coated sponge using a cut pipette tip along the inner ring of the sponge.
-
16.
Incubate hiNSC-loaded sponges for 30 min at 37°C with minimal media to allow adhesion to the scaffold. Add 1 mL of complete neurobasal media and maintain at 37°C/5% CO2.
Assemble stromal layer
-
17.
Aspirate media from hCSSCs seeded on silk films (Step 8).
-
18.Prepare a collagen hydrogel using the following mixture in a 1.5 mL microcentrifuge tube at room temperature:
- 900 μL rat-tail collagen type I (3 mg/mL in 20 mM acetic acid)
- 100 μL 10× DMEM
- 20 μL 1M NaOH
-
19.Mix collagen mixture gently by pipetting up and down. Add carefully to the surface of hCSSC-loaded silk film by pipette.
- Take care to avoid introducing bubbles into the collagen mixture. Any bubbles introduced will be retained within the construct. The timing of this step is important to ensure that the hCSSC-loaded silk film adheres to the other silk films. Mix the collagen hydrogel and add to surface of the hCSSC-loaded silk film just prior to retrieval of the other hCSSC-loaded silk films.
-
20.Using sterile forceps, transfer a second hCSSC-loaded silk film on top of the collagen hydrogel with alignment of microgrooves oriented orthogonal to the bottom film. Incubate at 37°C/5% CO2 for 5 minutes.
- Use forceps to hold onto to edges of tape ensuring that the cell surface is undisturbed with transfer.
-
21.
Prepare a second collagen hydrogel mixture as before (Step 18). Add mixture slowly by pipette to the surface of the second hCSSC-loaded silk film. Incubate at 37°C/5% CO2 for 5 minutes.
-
22.
Transfer a third hCSSC-loaded silk film on top of the collagen surface with microgrooves oriented orthogonal to the second silk film.
Transfer stromal layer to neuronal ring
-
23.
Retrieve the hiNSC-loaded silk sponge rings seeded in a 24-well plate from the incubator and aspirate the media (Step 16).
-
24.Using a sterile 12 mm biopsy punch, cut the hCSSC stromal stack (Step 22) and transfer to the center of the hiNSC-loaded silk sponge.
- Take care to add the stromal layer (3 silk films loaded with hCSSCs) to the inner circle of the silk sponge. The stromal layer may fold up like a scroll during the transfer. Gently unfold and align the films using the fine forceps tip to create a seamless structure against the rim of the silk sponge. Each film of ~45 × 45 mm2 (minus the taped edges) should generate (4) 12 mm biopsies.
-
25.
Prepare a third layer of the collagen hydrogel (Step 18) and add to the surface of the stromal layer.
Add the epithelium on top
-
26.Immediately retrieve the hCEC-loaded silk film (Step 3) using forceps. Aspirate media and carefully transfer the hCEC-loaded silk film to the surface of the collagen hydrogel using forceps (Step 25).
- The timing of this step is important to ensure that the hCEC-loaded silk film adheres to underlying stromal layers. Mix the collagen hydrogel and add to surface of the stromal layer just prior to hCEC-loaded silk film retrieval.
Encase the construct in collagen
-
27.Add a final layer of 100 μL of the collagen hydrogel (Step 18) supplemented with 750 μg/mL NGF to the inner edges of the topmost layer along the silk sponge to encase the construct. Incubate at 37°C/5% CO2 for 30 minutes.
- This collagen layer is important in holding the silk films to the silk sponge to enable robustness of the construct to transfer. Add NGF to the collagen mixture after neutralization with NaOH to prevent acid-mediated degradation of the growth factor.
Culture corneal constructs
-
28.
Add 250 μL of the complete corneal co-culture media to each well. Incubate at 37°C/5% CO2 overnight.
-
29.
At t=24 hours post-assembly, transfer constructs using forceps to a 6-well plate containing the custom waffle-shaped PDMS mold (Support Protocol 1, Step 7) to maintain an air-liquid interface. Add 2.5 mL of complete corneal co-culture media to each well.
-
30.Maintain corneal constructs at 37°C/5% CO2 for 4 weeks with media changes every other day.
- Aliquots of nerve growth factor should be added to fresh media immediately prior to use to minimize degradation.
SUPPORT PROTOCOL 1:
PREPARTION OF POLYDIMETHYLSILOXANE (PDMS) MOLDS
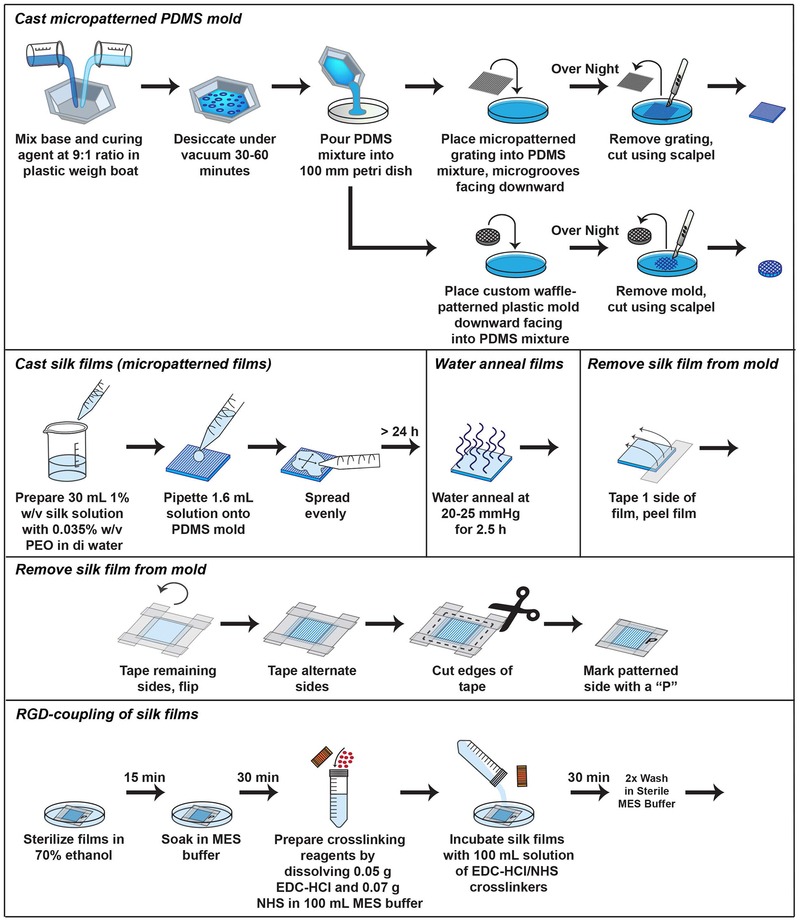
The PDMS molds provide re-useable substrates for casting micropatterned silk films (Figure 5). They can be cleaned with 70% v/v ethanol in water between uses and should be replaced if scratched or damaged. The waffle-patterned PDMS molds are used as stable, non-stick platforms for the corneal constructs during cultivation and fit in a 6-well culture plate. They may be autoclaved between uses for sterilization.
Figure 5.
Schematic depicting the protocol for casting PDMS molds and silk films.
Materials
45 × 45 mm2 micropatterned diffraction grating with microgrooves (500 nm depth and 3.5 μm width)
SYLGARD® 184 Silicone Encapsulant Clear (Components: 1) Base Part A and 2) Curing Agent Part B) (Krayden, cat. no. DC4019862)
Plastic weigh boat
Scale
Disposable pipette or glass stirrer
100 mm petri dish
Surgical scalpel
5 mm height × 5 cm Delrin® mold (McMaster-Carr)
Cast micropatterned PDMS mold
-
1.
Mix base and curing agent in a ratio of 9:1 in a plastic weigh boat and manually stir mixture with a disposable pipette or glass stirrer for 1 minute.
-
2.
Remove bubbles by desiccating mixture under vacuum for 30-60 minutes.
-
3.
Gently pour the PDMS mixture into a 100 mm petri dish. Place the micropatterned diffraction grating into the PDMS mixture with microgrooves facing downward. Incubate the mixture overnight at room temperature to allow solidification.
-
4.
Remove the glass grating from the surface of the mold. Cut the PDMS mold from the petri dish using a surgical scalpel. Denote the micropatterned surface by marking the side of the mold with an alcohol-resistant marker. Store PDMS mold at room temperature.
Cast waffle-patterned PDMS mold
-
5.
Use a custom-design, waffle-patterned plastic Delrin® mold with dimensions of 5 mm height × 5 cm diameter and 16 × 1 mm2 holes to generate replicate PDMS molds.
-
6.
As described above in step 3, add PDMS mixture to a 100 mm petri dish. Place custom plastic mold downward facing into PDMS mixture. Incubate the mixture overnight at room temperature to allow solidification.
-
7.
Remove the plastic mold from the surface and cut the PDMS mold along the edges of the imprint to generate a waffle-patterned PDMS mold. Autoclave PDMS mold prior to use in cell culture.
SUPPORT PROTOCOL 2:
PREPARATION OF SILK FILMS
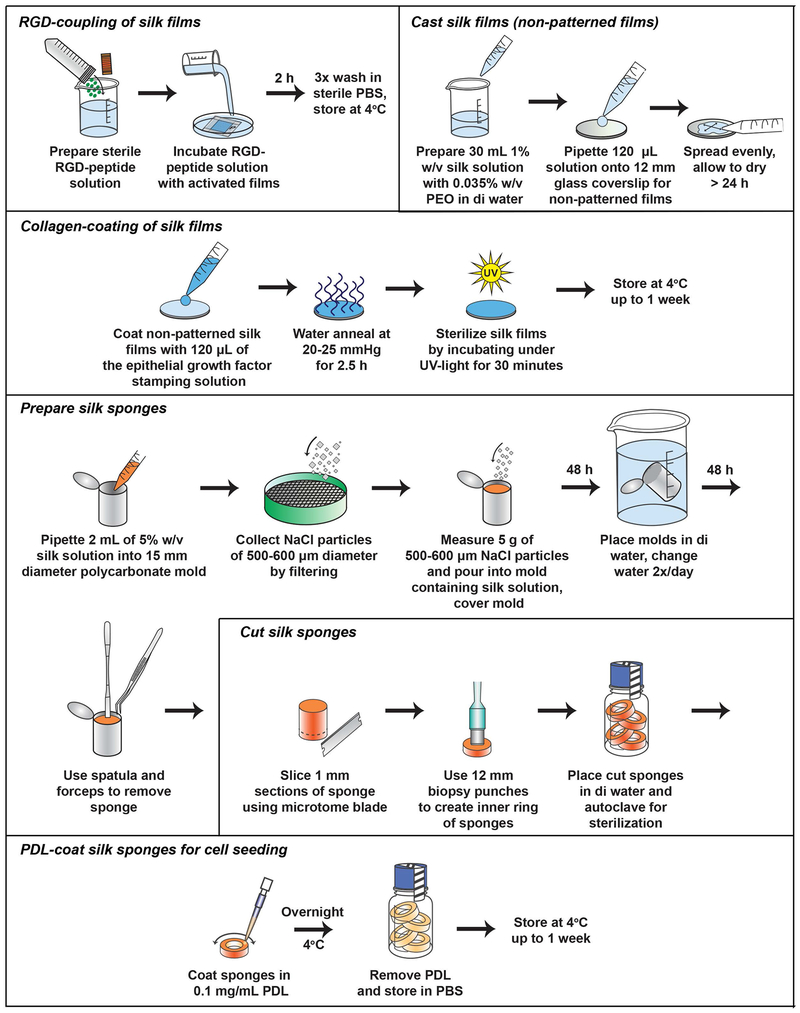
Detailed protocols for isolation of silk fibroin and preparation of silk films and sponges have previously been reported by our group (Rockwood et al., 2011). Thus, we refer the reader to the previously published protocol for details regarding general silk biomaterial preparation. We focus this section on detailed protocols required for functionalization of silk films specific for construction of the corneal tissue model (Figure 5). RGD-coupling to silk films via EDC-mediated coupling promotes a binding site for hCSSC attachment to the silk substrate (Figure 6). Alignment of hCSSCs along the microgrooves on the RGD-micropatterned surface can be visualized under the brightfield microscope. Collagen and proteoglycan deposition also align along the microgrooves similar to the corneal stroma in vivo (Wu, Rnjak-Kovacina, et al., 2014).
Figure 6.
Schematic depicting the protocol for conjugating an RGD-peptide to silk films and PDL-coating of silk sponges.
Materials
Silk solution (5-8% w/v in distilled water)
1% w/v poly-ethylene oxide (PEO) (Sigma, cat. no. 189464) in distilled water
Pipette
50 mL polypropylene centrifuge tubes
45 × 45 mm2 micropatterned PDMS molds (Refer to Support Protocol 1, Step 4)
12 mm glass coverslips (Optional)
Epithelial growth factor stamping solution (Refer to Reagents and Solutions Section)
Vacuum desiccator with temperature control
Tape
Pencil
Scissors
Phosphate buffered saline (PBS) (Sigma, cat. no. 11666789001)
2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.5) (ThermoScientific, cat. no. 28390)
1-ethyl-3-(dimethylaminopropyl) carbodiimide hydrochloride (EDC-HCl) (Sigma, cat. no. E6383)
N-hydroxysuccinimide (NHS) (Sigma, cat. no. 130672)
50 mL 0.2 μm filter-flask
Arginine-Glycine-Aspartic acid (RGD)-peptide
Light microscope
Cast silk films
-
1.Prepare 30 mL of a 1% w/v silk solution with 0.035% w/v PEO in distilled water for 18 PDMS molds with dimensions of 45 × 45 mm2. Mix solution gently by pipetting up and down.
- Add 1.05 mL 1% w/v PEO solution to 3.85 mL of 7.8% w/v stock in distilled water. Dilute with 25.1 mL distilled water for a final volume of 30 mL.
-
2.Pipette 1.6 mL solution onto the PDMS mold (Support Protocol 1, Step 4), spread evenly using a pipette tip or glass spreader, remove any bubbles gently by pipette, and incubate on a flat surface overnight at room temperature partially covered to allow water evaporation. Allow >24 hours to dry.
- Alternative: Silk films may be cast onto glass coverslips in a 24-well plate for preparation of non-patterned silk films. Addition of 120 μL of a 1% v/v silk solution and 0.035% w/v PEO onto a standard coverslip of 12 mm diameter may be used.
Water anneal films
-
3.
Place dried silk films on PDMS molds in a vacuum desiccator with temperature control. Add water to vessel at the bottom of the desiccator. Close the purge valve and open the vacuum valve to create an internal pressure of 20-25 mm Hg. Close both vacuum valves and incubate films for 2.5 hours at room temperature to water anneal.
Remove silk film from mold
-
4.
Once dry, place scotch tape on one edge of the film (~¼ inch), and gently pull the silk film off of the mold onto a clean surface.
-
5.
Gently tape the remaining edges of the film onto a clean surface. Remove the tape from the surface and turn the silk film over. Repeat addition of tape on the basolateral side of the film to cover up the tape glue. Write “P” on the corner of the tape with a pencil to denote the side that is patterned.
-
6.
Cut the edges of the tape and place films in a clean container for use.
-
7.
Check alignment of grooves using the 40× objective lens of a light microscope, and write arrows on tape with a pencil to denote orientation of grooves. Store dry at room temperature.
RGD-coupling of silk films
-
8.
Sterilize silk films prior to RGD-coupling by incubating films in 70% v/v ethanol in distilled water for 15 minutes at room temperature. Rinse films gently 5× in sterile DPBS to remove residual ethanol.
-
9.
Soak silk films in MES buffer for 30 minutes.
-
10.Prepare crosslinking reagents by dissolving 0.05 g EDC-HCl and 0.07 g NHS in 100 mL MES buffer. Filter-sterilize using a 0.2 μm filter.
- CAUTION: EDC and NHS are dangerous by inhalation. Weigh these chemicals in a chemical hood or transfer to a tared tube for measurements to reduce exposure. Prepare immediately prior to the reaction due to the labile nature of the reagents.
-
11.
Incubate silk films with 100 mL solution of EDC-HCl/NHS crosslinkers for 30 minutes at room temperature. Aspirate crosslinkers and wash silk films with MES buffer 2×.
-
12.
Prepare sterile RGD-peptide solution and incubate with activated films for 2 hours at room temperature or overnight at 4°C with gentle rocking.
-
13.
Aspirate the RGD-peptide solution and wash silk films in sterile PBS 3×. After coated, store silk films in PBS at 4°C.
Collagen-coating of silk films
-
14.
Prior to hCEC-seeding, coat non-patterned silk films (Step 2) with 120 μL of the epithelial growth factor stamping solution to enable cell adhesion.
-
15.
Water-anneal coated films at 20-25 mm Hg for 2.5 hours (Step 3).
-
16.
Sterilize silk films by incubating under UV-light for 30 minutes on each side. Store at 4°C for up to 1 week.
SUPPORT PROTOCOL 3:
PREPARATION OF SILK SPONGES
Materials
Silk solution (5-8% w/v in distilled water)
Pipette
15 mm diameter polycarbonate mold
Sodium chloride (NaCl) (Sigma, cat. no. S7653)
Scale
Stainless steel sieves with aperture size between 500 nm and 600 nm in diameter (Fisher Scientific, cat. no. 04-884-1AM (600 μm) and cat. no. 04-884-1AN (500 μm), respectively)
Distilled water
2 L plastic beaker
Spatula
Forceps
Microtome blade
12 and 15 mm biopsy punches
0.1 mg/mL poly-D-lysine (PDL) solution (Corning, cat. no. 354210)
Prepare silk sponges
-
1.
Pipette 2 mL of 5% w/v silk solution into each 15 mm diameter polycarbonate mold.
-
2.
Collect NaCl particles of 500-600 μm diameter by filtering via the 600 μm stainless steel sieve followed by the 500 μm sieve.
-
3.Measure 5 g of 500-600 μm NaCl particles and carefully pour into each mold containing the silk solution by gently tapping the bottom of the mold while adding the salt. Cover molds and store at room temperature for 2 days until solidification.
- Minimize the introduction of bubbles by adding the salt slowly while rotating the mold.
-
4.
After incubation, remove the cover and place molds into 2 L of distilled water in a plastic beaker to allow the salt to leach from the silk sponge. Change the water 2× per day for 2 days.
-
5.To remove the sponge from the mold, insert a clean spatula on the inside periphery of the mold and gently press the sponge inward to release from the mold. Use forceps to fully dislodge the sponge. Incubate the free sponge in distilled water for 1 day with 2 water changes.
- Sponges may be stored dry at room temperature indefinitely or at 4°C for several months.
Cut silk sponges
-
6.To cut the silk sponge into the proper dimensions, slice 1 mm sections using a microtome blade. Utilize commercial biopsy punches of 12 mm diameter and 15 mm diameter to create the inner and outer rims of the ring, respectively.
- Each whole sponge should generate at least 4 rings for neuronal seeding.
-
7.
Place cut sponges in distilled water and autoclave for sterilization.
PDL-coat silk sponges for cell seeding
-
8.
To PDL-coat the silk sponges prior to seeding, incubate sterile sponge in 0.1 mg/mL PDL solution in distilled water at 4°C overnight. Remove PDL prior to cell seeding.
-
9.
Re-suspend and store sponges in PBS at 4°C for up to 1 week.
SUPPORT PROTOCOL 4:
SUBCULTURING OF hCECs
Materials
Complete corneal epithelial media (Refer to Reagents and Solutions Section)
Pipette
T75 flask
Primary human corneal epithelial cells (hCECs, ATCC, cat. no. ATCC® PCS-700-010™)
FNC Coating Mix® (Athena Environmental Sciences, cat. no. 0407) (Optional)
500 mL cell culture flask
Seed and maintain hCECs
-
1.Add 15 mL of complete corneal epithelial media to a T75 flask.
- Optional: A fibronectin coating may be used to promote adhesion of hCECs to the flask by adding 3 mL of FNC Coating Mix® to flask followed by swirling to coat entire surface and then removal and addition of media.
-
2.
Thaw hCECs from a frozen stock culture by immersing the bottom of the vial in water at 37°C for 1-2 minutes.
-
3.
Immediately transfer hCECs to the T75 flask containing complete corneal epithelial media. Incubate at 37°C/5% CO2 for 24 hours to allow cell adhesion.
-
4.
Aspirate the media and supplement adhered hCECs with fresh epithelial media. Subculture hCECs upon 90% confluence up to passage 5.
SUPPORT PROTOCOL 5:
SUBCULTURING OF hCSSCs
Materials
Human corneal tissue
Surgical scalpel
0.5 mg/mL collagenase I solution (Sigma, cat. no. 10269638001)
Pipette
Centrifuge
Complete hCSSC proliferation media (Refer to Reagents and Solutions Section)
Complete hCSSC differentiation media (Refer to Reagents and Solutions Section)
T25, T75, and T175 flasks
Light microscope
NOTE: Institutional Review Board and federal guidelines must be followed regarding obtaining human corneal tissue.
Isolating and culturing hCSSCs
-
1.
Obtain corneal tissue according to institutional guidelines and isolate the limbal rim using a surgical scalpel based on previous protocols (Wu, Du, et al., 2014; Syed-Picard et al., 2018).
-
2.
Digest the corneal tissue using 0.5 mg/mL collagenase I solution overnight at 37°C followed by centrifugation at 1500 rpm for 5 minutes. Aspirate the supernatant and re-suspend the cell pellet in 2 mL of complete hCSSC proliferation media.
-
3.
Plate the cell solution in a T25 flask containing 5 mL of complete hCSSC proliferation media, gently mix, and incubate at 37°C/5% CO2 overnight.
-
4.
Check cell attachment and cell number using a brightfield microscope. Maintain hCSSCs in complete hCSSC proliferation media until 90% confluence and transfer to a T75 flask. Sub-culture hCSSCs up to passage 5 in a T175 flask.
-
5.To promote differentiation of hCSSCs to keratocytes, substitute complete hCSSC proliferation media with complete hCSSC differentiation media.
- Expression of keratocyte markers, keratocan and aldehyde dehydrogenase 3a, by hCSSCs can be verified by IHC, Western blot, or RT-PCR to confirm proper differentiation. Downregulation of stem cell markers, ABCG2 and Nestin, and an upregulation of keratocan and aldehyde dehydrogenase 3a by hCSSCs have been shown to occur by 1 week following culturing in complete differentiation media (Basu et al., 2014).
SUPPORT PROTOCOL 6:
SUBCULTURING OF hiNSCs
Materials
Mouse embryonic fibroblasts (MEFs) (ATCC, cat. no. ATCC® SCRC-1008™)
Pipette
DMEM containing 10% fetal bovine serum (FBS) and 1× antibiotic-antimycotic
0.1% w/v gelatin in distilled water (Sigma, cat. no. ES006B)
100 mm petri dishes
10 μg/mL of Mitomycin dissolved in DMEM containing 10% fetal bovine serum (FBS) and 1× antibiotic-antimycotic
Human induced neural stem cells (hiNSCs)
Complete neurobasal media (Refer to Reagents and Solutions Section)
Complete knockout media (Refer to Reagents and Solutions Section)
15 mL polypropylene centrifuge tube
Fibroblast growth factor-2 (FGF-2)
Sensory nerve media (Refer to Reagents and Solutions Section)
TrpLE™ Express (Gibco, cat. no. 12604013)
Dulbecco’s phosphate buffered saline (DPBS) (Gibco, cat. no. 14190144)
50 mL polypropylene centrifuge tubes
Centrifuge
Cell strainer
Subculture hiNSCs
-
1.
Culture MEFs on gelatin-coated 100 mm petri dishes in DMEM containing 10% FBS and 1× antibiotic-antimycotic.
-
2.Upon 100% confluence, inactivate MEFs by treating with 10 μg/mL of Mitomycin C for 2-3 hours at 37°C/5% CO2. Aspirate solution, wash 2× with DMEM media, and re-suspend in fresh DMEM containing 10% FBS and 1× antibiotic-antimycotic until hiNSC seeding.
- CAUTION: Mitomycin C is a carcinogen and must be handled with care. All waste should be disposed of in the appropriate biological and chemical waste streams.
-
3.
Retrieve hiNSCs from a frozen stock and add to 10 mL of complete neurobasal media in a 15 mL centrifuge tube. Centrifuge at 1200 rpm for 5 minutes. Aspirate the supernatant and re-suspend cells in complete knockout media.
-
4.
Aspirate the media from the inactivated MEF plate and carefully transfer the hiNSC cell solution to the MEF layer. Swirl the plate gently to distribute hiNSCs evenly over the feeder layer. Add 9 mL of additional complete knockout media containing 20 ng/mL FGF to the petri dish and maintain at 37°C/5% CO2 until multiple hiNSC colonies form on the surface of the MEF layer.
-
5.
Once confluent, aspirate the media and wash the cell layer gently with DPBS.
-
6.
Add 3 mL of TrpLE™ and incubate for 1 minute at 37°C.
-
7.
Transfer detached cells to a clean 50 mL centrifuge tube. To colonies remaining on plate that appear as white dome-shaped deposits, add a little DPBS gently, and use a pipette tip to mechanical remove cells. Transfer the cell suspension to a separate 50 mL tube. Centrifuge both tubes at 1500 rpm for 2-3 minutes.
-
8.
Add 8 mL of complete knockout media containing 20 ng/mL FGF to re-suspend the colony pellet.
-
9.
From a fresh inactivated MEF feeder plate, aspirate the media. Add 9 mL of additional knockout media containing 20 ng/mL FGF-2, and then transfer the colony cell pellet to the feeder plate and incubate at 37°C/5% CO2. Maintain hiNSCs on the MEF feeder layer until multiple colonies form.
-
10.
To single-cell suspension, add 12 mL of complete knockout media, vortex briefly, and strain using a plastic strainer to obtain a single-cell suspension.
-
11.
Add single-cell, filtered suspension to the gelatin-coated petri dish and incubate at 37°C/5% CO2 for 24 hours. Verify cell attachment and maintain in complete knockout media until differentiation.
Differentiate hiNSCs
-
12.
Differentiate hiNSCs to a sensory neuronal phenotype by incubating with complete sensory nerve media containing a number of growth factors and small molecule inhibitors.
-
13.
Aspirate complete knockout media and substitute with complete sensory nerve media.
-
14.Maintain hiNSCs in complete sensory nerve media on a gelatin-coated plate for 10 days at 37°C/5% CO2 with media changes every other day.
- Expression of nociceptor markers, e.g. transient-receptor potential cation channel (TRP) receptors TRPV1, TRPA1, and TRPM8, may be used to confirm successful sensory nerve differentiation.
Basic Protocol 2:
CHEMICAL STIMULATION
The corneal construct may serve as an alternative pharmacological screen to animal models for evaluating chemical toxicity or biological responses to acute or chronic stimuli. Prior to application of this model for evaluating toxicological responses, we recommend assessing the cytotoxicity of the chemical of interest in a dose-dependent study using 2D monocultures. These experiments should provide a baseline for assessing the chemical of interest in the 3D corneal construct. The presence of the silk scaffold and ECM generated over the length of cultivation likely promote increased resistance to chemical-induced toxicity similar to the cornea in vivo in comparison to direct cell contact, as in 2D cultures. We have assessed the response of the corneal tissue model to different microenvironments and chemical stimulants with results summarized in Table 1.
Table 1.
Evaluating the effects of the microenvironment (elevated glucose or dynamic perfusion) and different chemical stimulants (capsaicin and sodium lauryl sulfate) on cell viability and gene and protein expression using the corneal tissue model. (Abbreviations: Interleukin-1 beta (IL-1β), Substance P (SP), Calcitonin gene-related peptide (CGRP); Gene names: CRCP -accessory protein for CGRP, SCN- sodium channel, KERA - keratocan (cornea-specific proteoglycan), LUM - lumican (proteoglycan abundant in the stroma)).
| Chemical | Condition Applied | Response | Reference |
|---|---|---|---|
| Elevated glucose | 35-55 mM Glucose | Concentration-dependent loss of hCECs and hCSSCs, nerve degeneration, ↑ IL-1β | (Deardorff et al., 2018) |
| Dynamic perfusion | Intraocular pressure of 15-20 mmHg and tear flow of 50 μL/min | ↑ CRCP, ↑ SCN, ↑ KERA, ↑ LUM | (Siran et al., 2018) |
| Capsaicin | 0.005% w/v in DMEM | ↑ SP, ↑ CGRP | (Siran et al., 2018) |
| Sodium lauryl sulfate | 10% v/v in PBS | ↑ SP, ↑ Tissue permeability | (Siran et al., 2018) |
Materials
Assembled corneal constructs (Refer to Basic Protocol 1)
Complete corneal co-culture media (Refer to Reagents and Solutions Section)
Scale
Chemical stimulant (e.g. capsaicin, sodium lauryl sulfate)
0.2 μm filter and syringe
15 mL polypropylene centrifuge tube
Pipette
Micropipette
Dulbecco’s phosphate buffered saline (DPBS) (Gibco, cat. no. 14190144)
Forceps
6-well plate
Chemical stimulation of construct
-
1.
Assemble and maintain the corneal construct in a 6-well plate in complete corneal co-culture media (Basic Protocol 1, Step 30).
-
2.
Prepare sterile chemical stimulants by measuring needed amount and filtering with a 0.2 μm filter via syringe into fresh 15 mL centrifuge tubes.
-
3.Apply 10 μL of chemical stimulant on the top epithelial surface of the construct and incubate for 10 minutes at 37°C/5% CO2.
- The volume of chemical stimulant added may be increased to cover the entire surface of the construct. Furthermore, the length of time of incubation may be decreased or increased accordingly. To evaluate the effects of the chemical on the neurons independent of the epithelial and stromal layers, the chemical stimulant may be injected into the silk sponge directly, thereby minimizing direct contact with the other cell types.
-
4.
Submerge the construct in 2 mL of DPBS, aspirate, and repeat wash 5×.
-
5.
Transfer construct to a clean 6-well plate and incubate in complete corneal co-culture media for at least 24 hours.
-
6.
Characterization of the biological response can be performed as described in Basic Protocol 3 and Support Protocols 7–11.
Basic Protocol 3:
ASSESSING VIABILITY BY MTT ASSAY
In order to determine the biological response to chemical stimuli, characterization of the corneal construct may be performed on the conditioned media, whole construct, or protein isolates from separate layers. Determining cell viability within the corneal construct using the MTT assay provides an optimal method to evaluate cell toxicity or proliferation in response to chemical stimulants. Limitations of this approach include the destructive nature of the assay and the requirement for use of the entire construct for analysis. A similar approach as described here may also be applied to evaluation of total double-stranded DNA via a Picogreen® assay. Further characterization of the protein content by enzyme-linked immunosorbent assay (ELISA), immunohistochemistry, and Western blot are described in Support Protocols 7–11.
Materials
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Invitrogen, cat. no. V13154)
Scale
Micropipette
Phosphate buffered saline (PBS)
Corneal construct (Refer to Basic Protocol 1, Step 30)
Dulbecco’s phosphate buffered saline (DPBS) (Gibco, cat. no. 14190144)
12-well cell culture plate
Forceps
Phenol red-free complete hCSSC differentiation media
1.5 mL microcentrifuge tubes
Dimethylsulfoxide (DMSO) (Sigma, cat. no. D8418)
10% w/v sodium dodecyl sulfate (SDS)-HCl solution (Optional)
96-well plate
Incubator
Foil
Vortex
Hand-held homogenizer (Optional)
Spectrophotometer for absorbance measurements at 540 nm
Incubate MTT reagent with construct
-
1.
Prepare MTT stock solution by dissolving 5 mg of MTT into 1 mL of PBS.
-
2.
To prepare constructs for incubation, transfer constructs to a 12-well plate using forceps. Aspirate phenol-red containing media, gently submerge construct in DPBS 2×, and add 500 μL of fresh phenol red-free complete corneal co-culture media.
-
3.
Add 10 μL of the MTT solution to each well containing the construct submerged in media. Protect from light by covering the plate with foil. Incubate constructs at 37°C/5% CO2 for 2 hours.
Isolate cell layers of the construct
-
4.
Using sterile forceps, transfer constructs to fresh wells containing 500 μL of PBS.
-
5.
Gently separate layers of the construct using forceps by removing the topmost silk film containing the epithelium. Transfer to a clean 1.5 mL microcentrifuge tube containing 100 μL of DMSO.
-
6.Repeat separation of the layers by removing the stromal layers (3 silk films with interlaying hCSSCs and collagen) from the silk sponge seeded with hiNSCs. Transfer layers to separate microcentrifuge tubes containing 100 μL of DMSO.
- ALTERNATIVE: SDS may be used to lyse cells by adding 100 μL of 10% w/v SDS-HCl solution (1 g SDS dissolved in 10 mL of 0.01 M HCl) and incubation up to 4 hours at 37°C.
- NOTE: The silk sponge retains media and liquid in its pores. Thus, the presence of excess solution within the scaffold itself may limit the accuracy of the quantification of cells within the sponge depending on the residual volume retained within the scaffold prior to cell lysis. This limitation must be considered when comparing cell viability between the epithelial, stromal, and neuronal layers.
-
7.Vortex tubes briefly to mix contents.
- OPTIONAL: Using a hand-held homogenizer, degrade the scaffolds by homogenizing for 30 seconds on ice 3×.
-
8.
Incubate cell layers and DMSO for 10 minutes at 37°C to solubilize the formazan salt.
-
9.
9. Transfer 100 μL of the DMSO-cell lyses solution to a 96-well plate. Measure absorbance at 540 nm using a spectrophotometer.
SUPPORT PROTOCOL 7:
PROTEIN ANALYSIS OF CONDITIONED MEDIA
Changes in secreted protein concentrations in response to stimuli may be evaluated by ELISA of common neuropeptides and pro-inflammatory factors. Depending on the protein of interest, stock media may contain basal levels of the protein, thus requiring special consideration in data analysis or experimental setup, such as utilization of complete media in standard curve preparation. The co-culture media formulation applied in these studies is serum-free, therefore, reducing many of the common serum-contaminants from the media. Common secreted markers for sensory nerve activation or tissue inflammation are included in Table 2.
Table 2.
Common secreted proteins related to nerve activation or tissue inflammation present in conditioned media isolated from corneal constructs. The relative concentration ranges of each protein detected in conditioned media in response to various chemical stimuli have been previously reported by our group (Deardorff et al., 2018; Siran et al., 2018). (Abbreviations: Substance P (SP), Calcitonin gene-related peptide (CGRP), Interleukin-1 beta (IL-1β), Tumor necrosis factor-alpha (TNF-α), Matrix metalloproteinase-9 (MMP-9)).
| Protein | Major Cell Source | Subclass | Concentration Detected in Conditioned Media | Reference |
|---|---|---|---|---|
| SP | Sensory Neuron (hiNSCs) | Neuropeptide | 100-4000 pg/mL | (Siran et al., 2018) |
| CGRP | Sensory Neuron (hiNSCs) | Neuropeptide | 300-800 pg/mL | (Siran et al., 2018) |
| IL-1β | Epithelium (hCECs) | Pro-inflammatory cytokine | 5-50 pg/mL | (Deardorff et al., 2018; Siran et al., 2018) |
| TNF-α | Epithelium (hCECs) | Pro-inflammatory cytokine | 20-350 pg/mL | (Deardorff et al., 2018; Siran et al., 2018) |
| MMP-9 | Epithelium (hCECs) | Matrix metalloproteinase | 2000-5000 pg/mL | (Deardorff et al., 2018) |
Materials
15 mL polypropylene centrifuge tubes
Pipette
Ice bucket
Dry ice or −80°C freezer (Optional)
Lyophilizer (Optional)
ELISA Kit (e.g. Substance P, CGRP, IL-1β, TNF-α, MMP-9)
Refrigerator
Rocker
Isolate conditioned media
-
1.
Collect 2 mL of the conditioned media of the corneal constructs in sterile 15 mL centrifuge tubes on ice. Re-suspend the constructs in fresh media and continue cultivation, as needed.
-
2.Depending on the protein of interest, protein analysis via ELISA may be performed on the media without further concentration or dilution.
- Optional: The media may be concentrated by placing sample in dry ice or incubating overnight at −80°C to freeze contents followed by lyophilization and re-suspension in 1 mL of PBS.
-
3.
Follow the manufacturer’s protocol for analysis.
SUPPORT PROTOCOL 8:
IMMUNOHISTOCHEMISTRY OF CORNEAL CONSTRUCTS
Assessing changes in cell density and morphology at the end timepoint requires fixation of the tissue and incubation with antibodies with stable fluorophores for confocal microscopy. Whole-mount analysis is ideal, thereby reducing the risk of tissue damage mediated via traditional paraffin-embedding and cutting.
Materials
Dulbecco’s phosphate-buffered saline (DPBS) (Gibco, cat. no. 14190144)
Micropipette
Phosphate-buffered saline (PBS) (Sigma, cat. no. 11666789001)
Paraformaldehyde (PFA) (4% w/v in PBS) (Santa Cruz Biotechnology, cat. no. sc-281692)
Permeabilization reagent: 0.1%−0.25% v/v Triton-X 100 (Sigma, cat. no. X100) in DPBS
Tris-buffered saline containing 0.1% v/v Tween-20 (TBST) (Sigma, cat. no. P1379)
Blocking buffer: 2-5% w/v bovine serum albumin (BSA) (Sigma, cat. no. 05470) in DPBS
Primary antibodies (BIII-tubulin)
Antibody diluent: 1% w/v BSA (Sigma, cat. no. 05470) in PBS
Secondary antibodies
6-well plate
Clean forceps
Surgical scalpel (Optional)
Mechanical rocker
Refrigeration
Foil
Confocal microscope
Isolate corneal constructs
-
1.Aspirate culture media and gently transfer construct to a clean 6-well plate. Re-suspend construct in 2 mL of sterile DPBS. Aspirate DPBS and repeat wash 2×.
- OPTIONAL: The construct may be cut at this point into smaller pieces using a sterile scalpel for other applications, including western blot or RT-PCR analysis.
Fix, permeabilize, and block
-
2.Add 1 mL of 4% PFA to each construct and incubate for 1 hour at room temperature or overnight at 4°C.
- CAUTION: This step must be completed in a chemical hood to minimize exposure to PFA fumes. The construct may be stored at 4°C in DPBS until further use. Long-term stability is dependent on the protein of interest.
-
3.
Aspirate PFA and wash construct with DPBS 3× to remove residual fixative.
-
4.Permeabilize the cells for antibody probing using 1 mL of 0.1%−0.25% v/v Triton-X-100 in DPBS by incubation for 15 minutes at room temperature (static).
- This step is dependent on the protein of interest. Permeabilization is required for cytosolic antigens requiring antibody perfusion into the cell. Probing for membrane proteins may not require permeabilization prior to antibody probing.
-
5.Block non-specific binding sites with 2 mL of 5% w/v BSA in PBS for 1 hour at room temperature with gentle rocking.
- This step may be increased to overnight at 4°C, if necessary. The concentration of BSA may be decreased to 2% w/v BSA in PBS.
Incubate with antibodies
-
6.Prepare the primary antibody solution according to the manufacturer’s recommendation. Incubate 1 mL of primary antibody solution with the construct overnight at 4°C with gentle rocking.
- Commonly used concentrations include 1:50-1:200 (primary antibody: 1% w/v BSA/PBS). The total volume of antibody solution added to each construct is dependent on the proportion of construct used for IHC analysis. The construct may be transferred to a 12- or 24-well plate to reduce the volume of antibody solution required.
-
7.
Wash construct gently with 1× TBST up to 3×. Aspirate wash buffer.
-
8.Prepare the secondary antibody solution according to the manufacturer’s recommendation. Incubate 2 hours at room temperature with rocking or overnight at 4°C with gentle rocking.
- Commonly used concentrations include 1:1000-1:5000 (secondary antibody:PBS). Secondary antibodies conjugated to a fluorophore are light-sensitive, thus, this step should be performed in the dark, and samples should be covered with foil during incubation to reduce photobleaching.
Confocal microscopy
-
9.Imaging of whole-mounts should be performed on a confocal microscope.
- The average thickness of the corneal constructs may vary depending on ECM-production by hCSSCs, length of incubation, and collagen hydrogel thickness. Our studies have measured thicknesses of roughly 500-1000 μm based on a 4-6 week cultivation period.
Measuring nerve fiber length
-
10.To quantify nerve fiber length, utilize the ImageJ plugins, known as NeuroJ (Meijering et al., 2004), for 2D images or Simple Neurite Tracer (Longair et al., 2011) for 3D stacks of the region of interests.
- Follow the available manuals for converting the tiff images to 8-bit black/white images and automated analysis. Average n>3 region of interests per construct for technical replicates with n>3 biological replicates from different constructs.
- NOTE: The average length can vary depending on the region of the scaffold imaged, i.e. focusing on the sponge region versus the stromal region. Thus, the comparison between treatments and controls should be consistently quantified based on similar regions of interest.
SUPPORT PROTOCOL 9:
ISOLATE TOTAL PROTEIN CONTENT FROM EACH CELL LAYER
The amount and content of protein isolated from each layer of the corneal construct can be determined by separating the silk scaffolds containing the appropriate cell population, e.g. hCECs seeded on the apical silk film, hCSSCs and the underlying silk films, and hiNSCs seeded on the peripheral silk sponge. This separation is advantageous in allowing for biochemical analysis of each cell type to determine variances in cell-specific responses to stimulation while maintaining a complex tissue microenvironment during the experiment. Protein isolation can be carried out in a similar method to isolation of tissue explants using forceps to separate layers.
Materials
Phosphate buffered saline (PBS) (Sigma, cat. no. 11666789001)
Forceps
2 mL microcentrifuge tube
Micropipette
Ice bucket
1× M-PER™ Mammalian Protein Extraction Reagent (Thermofisher, cat. no. 78501) with added 1× protease inhibitor cocktail (Sigma, cat. no. P8340)
1× RIPA lysis buffer (Sigma, cat. no. 20- 188) with added 1× protease inhibitor cocktail (Sigma, cat. no. P8340) (Optional)
Hand-held homogenizer
Vortex
Centrifuge with refrigeration
Isolate corneal construct
-
1.
Aspirate the media and gently wash construct in 2 mL of PBS 3×.
-
2.Prepare M-PER™ lysis buffer containing 1× protease inhibitor cocktail, vortex to mix, and add 200 μL of lysis buffer to each 2 mL microcentrifuge tube on ice.
- Alternative: Utilize 1× RIPA buffer containing 1× protease inhibitor cocktail instead of M-PER™ lysis buffer.
-
3.
Gently separate the topmost silk film and epithelium using forceps and transfer to a labelled microcentrifuge tube containing lysis buffer on ice.
-
4.
Gently separate the stromal layers (3 silk films and interlaying hCSSCs-ECM) from the silk sponge using forceps and transfer to a labelled microcentrifuge tube containing lysis buffer on ice.
-
5.
Transfer the remaining the silk sponge containing the differentiated hiNSCs into a labelled microcentrifuge tube containing lysis buffer on ice.
-
6.Using a hand-held homogenizer with a blunt attachment, homogenize each cellular fraction for 30 s each time for a total of 3× with 1 minute in between. Maintain tubes on ice during homogenization. Vortex briefly to mix and incubate samples on ice for 20 minutes to ensure complete cell lysis.
- Ensure that samples do not overheat by keeping tubes on ice at all times. Also, ensure that samples are submerged in lysis buffer and add additional lysis buffer if needed so that all cells are adequately lysed.
-
7.Centrifuge samples at 14,000 rpm at 4°C for 15 minutes to pellet insoluble debris. Transfer the supernatant to clean, labelled tubes and store at −20°C until further use.
- Aliquot protein samples (25 μL per fraction) to minimize freeze-thaw cycles and maintain protein integrity.
SUPPORT PROTOCOL 10:
MEASURE ISOLATED PROTEIN CONCENTRATION
Materials
Isolated protein solution in lysis buffer (Refer to Support Protocol 9, Step 7)
1.5 mL microcentrifuge tubes
Micropipette
Lysis buffer (M-PER™ Mammalian Protein Extraction Reagent (Thermofisher, cat. no. 78501) with added 1× protease inhibitor cocktail or 1× RIPA lysis buffer (Sigma, cat. no. 20- 188) with added 1× protease inhibitor cocktail (Sigma, cat. no. 20-188))
Bovine serum albumin (BSA) Standards (2000 μg/mL, 1000 μg/mL, 500 μg/mL, 250 μg/mL, 125 μg/mL, 62.5 μg/mL, 31.25 μg/mL, 15.63 μg/mL)
BCA Protein Assay (Reagent A- contains sodium carbonate, sodium bicarbonate, bicinchoninic acid, sodium tartrate, and 0.1M sodium hydroxide; Reagent B- contains 4% w/v cupric sulfate) (ThermoScientific, cat. no. 23250)
Spectrophotometer to measure absorbance at 562 nm
Prepare standard curve
-
1.
Isolate total protein from each cellular layer of the construct as described in Support Protocol 9.
-
2.Prepare the BSA standard curve by dissolving 0.002 g of BSA into 1 mL of lysis buffer. Prepare a 2-fold dilution to generate a total of 7 standards ranging from 2000 μg/mL to 15.63 μg/mL.
- Briefly vortex each sample in between dilutions to mix. Change pipette tips between samples.
-
3.Prepare the working reagent by mixing Reagent A and Reagent B in a ratio of 50:1 (A:B).
- Perform measurements with at least 2 technical replicates. Prepare a total of 5.6 mL of working reagent for 6 biological replicates, 7 standards, and 1 blank.
-
4.
Add 200 μL of the working reagent to each well of a 96-well plate. Add 10 μL of each standard, unknown sample, and blank to the appropriate wells. Pipette the solution up and down to mix.
-
5.
Incubate the samples at 37°C for 30 minutes. Measure the absorbance at 562 nm using a spectrophotometer.
-
6.Plot the standard concentration relative to absorbance based on a linear correlation to generate the standard curve. Extrapolate unknown concentrations based on the standard curve.
- Technical replicates should be consistent between standards and samples with little variability.
SUPPORT PROTOCOL 11:
WESTERN BLOT ANALYSIS OF PROTEIN ISOLATED FROM CORNEAL CONSTRUCTS
Protein analysis of the corneal constructs can provide useful information regarding signal transduction pathways affected by chemical stimulation. By isolating each cellular layer independently, one can determine the specific effects on the epithelium, stroma, and neuronal cell populations. Key phenotypic protein markers are listed in Table 3 for each cell type. Standard loading controls include glyceraldehyde dehydrogenase (GAPDH), β-actin, and β-tubulin.
Table 3.
Phenotypic markers of each cell type present in the corneal construct. (Abbreviations: Transient receptor potential cation channel subfamily V member 1 (TRPV1), Transient receptor potential cation channel subfamily A member 1 (TRPA1), Transient receptor potential cation channel subfamily M member 8 (TRPM8)).
| Cell Type | Protein Marker |
|---|---|
| Epithelial (hCECs) | Keratin 3, Keratin 12 |
| Keratocytes (hCSSCs) | Keratocan, Lumican, Aldehyde dehydrogenase 3a, Decorin, Collagen type I and V |
| Sensory Neurons (hiNSCs) | BIII-tubulin, TRPV1, TRPA1, TRPM8 |
Materials
Isolated protein solution in lysis buffer (Refer to Support Protocol 9)
Micropipette
4× Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (Refer to Reagents and Materials Section)
Vortex
Heat block
Centrifuge
Polyacrylamide gel (4-20% Tris-glycine gradient gel) (Invitrogen, cat. no. XP0420A)
1× Tris-glycine running buffer (Refer to Reagents and Materials Section)
1× Tris-glycine transfer buffer (Refer to Reagents and Materials Section)
Nitrocellulose membrane
SDS-PAGE running and transfer boxes
Ice bucket
Bovine serum albumin (BSA)
Tris-buffered saline containing 1% v/v Tween-20 (TBST) (Refer to Reagents and Materials Section)
Rocker
Primary and secondary antibodies
Refrigeration
Foil
Gel imager
Perform SDS-PAGE
-
1.
Prepare samples for SDS-PAGE by transferring 21 μL of protein isolate to 7 μL of 4× SDS-PAGE loading buffer with a final protein concentration between 20-50 μg/mL (Support Protocol 9–10).
-
2.
Vortex samples briefly and centrifuge at 14,000 rpm for 30 seconds at room temperature to sediment solution.
-
3.
Heat samples on a hot block at 60°C for 10 minutes followed by centrifugation at 14,000 rpm for 30 seconds.
-
4.
Load 20-50 μg/mL of total protein per well of a polyacrylamide gel as determined by a BCA assay (Support Protocol 10). Add 1× Tris-glycine running buffer to chamber.
-
5.
Run gel at 120 V for 1.5 hours or until bromophenol band reaches the bottom of the gel.
Transfer protein to nitrocellulose membrane
-
6.
Isolate gel and soak in transfer buffer briefly. Prepare the Western blot setup by generating the transfer sandwich from bottom to top: filter paper, gel, nitrocellulose membrane, and filter paper.
-
7.
Setup the transfer box in 1× Tris-glycine transfer buffer at 100 V for 1 hour on ice.
Block and incubate with antibodies
-
8.
Following transfer, carefully remove the nitrocellulose membrane using forceps and block in 5% w/v BSA in TBST for 1 hour at room temperature with rocking.
-
9.
Incubate with primary antibody solution (1:1000) overnight at 4°C with rocking. After incubation, wash membrane 3× with TBST and incubate with the secondary antibody (1:10,000) for 1-2 hours rocking at room temperature and covered with foil.
-
10.
Wash membrane 3× with TBST for 5 minutes each time with rocking.
-
11.
Image fluorescence (for fluorescent secondary antibodies) or light emissions (Horseradish peroxidase secondary antibody) from probed membrane using a gel imager.
Materials
REAGENTS AND SOLUTIONS
These reagents are available from standard commercial vendors. We have specified the manufacturers for critical materials within the text. Use distilled water or Milli-Q-purified water for all recipes. Filter-sterilize all media solutions upon mixing using a 0.2 μm filter flask. After preparation, store solutions at 4°C under sterile conditions until further use. Growth factors should be dissolved and aliquoted upon receipt to minimize freeze-thaw cycles. Avoid continual heating of media to avoid growth factor degradation.
Complete corneal epithelial media
Corneal epithelial media (ATCC, cat. no. ATCC® PCS-700-030™)
Corneal epithelial cell growth kit: apo-transferrin (5 µg/mL), epinephrine (1.0 µM), extract P (0.4%), hydrocortisone (100 ng/mL), L-glutamine (6 mM), rh insulin (5 µg/mL), and CE Growth Factor (proprietary formulation, ATCC) (ATCC® PCS-700-040™)
1× antibiotic-antimycotic
Add corneal epithelial media to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 month of preparation.
Epithelial growth factor stamping solution
50 μL of 4 mg/mL rat-tail collagen
200 ng/mL nerve growth factor (NGF)
200 ng/mL epithelial growth factor (EGF)
100 ng/ml hepatic growth factor (HGF)
100 ng/mL keratinocyte growth (KGF)
Add growth factors and collagen in a small 0.5 mL microcentrifuge tube. Mix gently by pipette. Store up to 1 week at 4°C.
Complete hCSSC Proliferation Media
DMEM low glucose
17.7 g/L MCDB-201 dissolved in distilled water (pH 6.67) (200 mL total volume)
2% v/v fetal bovine serum (FBS)
1% v/v AlbuMAX™ (lipid-rich bovine serum albumin)
0.1 mM ascorbic acid-2-phosphate
5 mg/mL transferrin
5 mg/mL insulin
5 ng/mL selenous acid
100 μg/ml penicillin
100 μg/mL streptomycin
5 μg/mL gentamicin
10 ng/mL epithelial growth factor (EGF)
10−8 M dexamethasone
100 ng/mL cholera toxin
Add DMEM low glucose to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 month of preparation.
Complete hCSSC Differentiation Media
Advanced DMEM
1 mM ascorbic acid-2-phosphate
10 ng/mL basic fibroblast growth factor-2 (FGF-2)
0.1 ng/mL transforming growth factor-β3 (TGF-β3)
100 μg/ml penicillin
100 μg/mL streptomycin
5 μg/mL gentamicin
Add Advanced DMEM to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 month of preparation.
Complete Corneal Co-Culture Media
Advanced DMEM
1 mM ascorbic acid-2-phosphate
10 ng/mL basic fibroblast growth factor-2 (FGF-2)
0.1 ng/mL transforming growth factor-β3 (TGF-β3)
100 μg/ml penicillin
100 μg/mL streptomycin
5 μg/mL gentamicin
50 ng/mL nerve growth factor (NGF)
Add Advanced DMEM to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 month of preparation.
Complete Neurobasal media
Neurobasal media
1% v/v GlutaMAX™
1× antibiotic-antimycotic
2% v/v B-27™ supplement
Add Neurobasal media to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 month of preparation.
Complete Knockout media
Knockout media
20% v/v knockout serum
0.1 mM β-mercaptoethanol
1% v/v GlutaMAX™
1× antibiotic-antimycotic
Add Knockout media to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 month of preparation.
Sensory nerve media
Neurobasal media
1× GlutaMAX™
1× antibiotic/antimycotic
25 ng/mL nerve growth factor (NGF)
25 ng/mL brain-derived neurotrophic factor (BDNF)
25 ng/mL glial cell-derived neurotrophic factor (GDNF)
10 μM DAPT (CAS # 208255-80-5)
10 μM SU-5402 (CAS # 215543-92-3)
3 μM CHIR99021 (CAS # 252917-06-9)
Add neurobasal media to combined contents for a total volume of 500 mL. Filter-sterilize using a 0.2 μm filter flask. Store media at 4°C and minimize heating to reduce growth factor degradation. Utilize media within 1 week of preparation.
1× RIPA lysis buffer
150 mM NaCl
1% v/v Nonidet P-40 (NP-40)
0.5% w/v sodium deoxycholate
0.1% w/v sodium dodecyl sulfate (SDS)
25 mM Tris-HCl (pH 7.4)
1× Protease inhibitor cocktail
Add distilled water for a final volume of 10 mL. Once mixed, store at 4°C and use within one week.
4× SDS-PAGE loading buffer
20 mM Tris-HCl (pH 6.8)
0.4 M dithiothreitol (DTT)
0.4% w/v bromophenol blue
40% v/v glycerol in distilled water
Add 10 mL distilled water for a total volume of 10 mL. Aliquot and store at −20°C for up to 6 months.
10× Tris-glycine running buffer
30 g Tris base
144 g Glycine
10 g Sodium dodecyl sulfate
Dilute contents in 1 L of distilled water while stirring. Maintain solution at room temperature or 4°C. Dilute to 1× concentration immediately prior to use.
20× Tris-glycine transfer buffer
24.2 g Tris base
150.1 g Glycine
Dilute contents in 1 L of distilled water while stirring. Maintain solution at room temperature or 4°C. Dilute to 1× concentration immediately prior to use.
1× TBS
137 mM NaCl
2.7 mM KCl
19 mM Tris-base
Dissolve in distilled water for a final volume of 1 L. Adjust final pH to 7.4. Add 1 mL of Tween-20 per 1 L of TBS to generate TBST. Store at room temperature for up to a month.
COMMENTARY
Background Information
Advantages of the application of this 3D co-culture approach over conventional 2D conventional methods include higher physiological relevance with the presence of a self-assembled ECM, co-culture conditions that collectively influence individual cell responses, the separation of functional units in distinct scaffolds for analysis of each cell type, and sustained cultivation for weeks and months to permit chronic assessments of treatments (versus only acute assessments). These factors may significantly impact the response to chemical toxins or microenvironmental changes, such as prolonged exposure to high glucose.
The corneal tissue model described contains relative dimensions consistent with the average diameter of the human cornea proper, e.g. 12 mm diameter for the inside rim of the sponge and the corneal diameter of the human eye (Rufer et al., 2005), respectively. In terms of corneal thickness, the average thickness of the human cornea may range from 0.5-0.6 mm with significant variability from person to person (Doughty et al., 2000). It is widely accepted that dissection and maintenance of cadaveric corneal tissue in culture media results in significant swelling that correlates to loss in transparency of the tissue (Meek et al., 2003) with an average thickness of over 1 mm ex vivo (Thuret et al., 2005). Likewise, the corneal tissue model is assembled with a thickness of 1 mm, however, relatively high tissue transparency is maintained in this system. De-swelling of the construct may be performed using standard dextran-containing media prior to in vivo transplantation, if needed (Borderie et al., 1997; Wolf et al., 2009).
While our model represents a recent advance in developing a functional corneal system, further studies are warranted to characterize the predictability of the construct as an in vitro platform to screen the safety and efficacy of pharmaceuticals and chemicals applied to the human cornea. Provided the diversity within the human population and the various animal models used in the study of corneal pain, identifying reliable pain biomarkers remains a common goal within the field (McKay et al., 2018). The application and further development of this human-focused in vitro model may aid in bridging this gap.
Critical Parameters and Troubleshooting
The preparation of silk scaffolds requires strict adherence to standard silk isolation and processing to ensure quality control. The isolated silk solution should be used within 1 month following dilution and stored at 4°C. Alternatively, lyophilized silk is much more stable for long periods of time and can be maintained at room temperature. Maintenance of the corneal constructs require proper culture conditions at 37°C/5% CO2 and aseptic approaches to minimize contamination. Since the model contains primary cells, only young passages of hCECs and hCSSCs (passage <4-5) should be utilized in order to allow sustainability for long culture times (>4 weeks). The chosen endpoint assessments will depend on the research question with multiple methods available to determine morphological, biochemical, and functional changes in the tissue structure following exposure to chemical irritants. Table 4 summarizes common troubleshooting issues that may arise during the assembly, maintenance, or characterization of the corneal constructs.
Table 4.
Troubleshooting guide for construct assembly and characterization.
| Problem | Possible Cause | Solution |
|---|---|---|
| hCECs do not attach to silk films and are floating or adhered to cell culture plate | • Lack of collagen coating on silk film • Older passage of hCECs • Addition of media to well prior to cell adhesion |
• Always prepare fresh stamping solution and growth factors prior to coating of film. • Use passages of <5 of hCECs. These cells are particularly sensitive to aging and thawing. • Visualize by microscope proper cell adherence to film prior to addition of media to well. |
| Lack of confluent hCEC layer | • Older passage of hCECs • Absence of epithelial growth factors in media • Poor cell proliferation |
• Utilize only passages of <5 of hCECs. These cells are particularly sensitive to aging and thawing. • Prepare fresh media containing standard epithelial growth factors, e.g. EGF, BPE, etc. • May substitute a corneal epithelial cell line, e.g. HCE-TJ, or limbal epithelial stem cell source to improve epithelial barrier integrity. |
| Poor hiNSC proliferation on the MEF feeder plate | • Older passage of MEFs • Lack of confluent feeder layer • Absence of fresh FGF in media |
• Utilize only passages of <5 of MEFs. • Seed MEFs at a high density and allow for 100% confluence prior to inactivation. • Prepare fresh media containing FGF immediately prior to use. |
| hCSSCs do not attach to RGD-coated films and are floating or adhered to cell culture plate | • Low pH of the film may cause cell death • Lack of RGD-functionalization to film • Addition of media to petri dish prior to cell adhesion |
• Ensure that the MES solution pH is neutral (pH 6.5-7). Wash films in PBS prior to use in cell culture. • Utilize freshly dissolved RGD peptide to crosslink to films. • Visualize by microscope proper cell adherence to film prior to addition of media to petri dish. |
| Frequent bacterial or fungal contamination | • Contamination of scaffolds prior to seeding • Lack of aseptic technique during cell culture |
• Autoclave silk sponges after cutting and immediately prior to use in cell culture. Sterilize silk films under UV-light (30 min each side) and immerse in 70% v/v EtOH in distilled water prior to RGD-coupling. Filter-sterilize (0.2 μm) crosslinking solutions and RGD-peptide prior to incubation with sterile films. Autoclave waffle-shaped PDMS mold prior to inclusion in system. • Adopt standard aseptic approaches during construct assembly and maintenance. |
| Low total protein isolated | • Inadequate cell lysis • Protein degradation • Cell loss during cultivation |
• Ensure that the isolated cell layer is submerged in the lysis buffer. • Include protease inhibitors in lysis buffer and maintain samples on ice during homogenization and incubation steps. Aliquot samples to reduce freeze-thaw cycles. • Monitor media color between media changes. A slight change should be noticeable indicating active metabolism. Ensure that cells properly attach to scaffolds prior to assembly of the construct. |
| Unusually high total protein isolated | • Inadequate removal of media prior to construct lysis • Bacterial or fungal contamination |
• Be sure to gently wash or immerse construct in PBS prior to isolation. • Monitor media color between media changes. A severe change in color to yellow or cloudiness of the media are common indicators of contamination. Visually inspect constructs for contamination. |
| Poor IHC images | • High silk autofluorescence in the green channel • Lack of antibody binding |
• Choose secondary antibodies with emissions wavelengths outside of the green channel range, e.g. λem=480-638 nm) • Incubate primary and secondary antibodies with construct overnight at 4°C with rocking to allow perfusion into the tissue. |
Anticipated Results
The assembly of the corneal tissue model is expected to yield intact corneal constructs that are stable for at least 4-8 weeks for applications in drug screening or biological studies. Cultures beyond 8 weeks may be possible but are limited by the passage number of the primary hCECs and hCSSCs. This timeframe is consistent with self-assembled stromal models that rely on de nova ECM deposition by hCFs and can be maintained from 4 to 11 weeks (Guo et al., 2007; Ren et al., 2008) followed by analysis of the biochemical responses to various chemical or environmental stimuli (McKay, Hjortdal, Priyadarsini, et al., 2017; McKay, Hjortdal, Sejersen, et al., 2017).
A total of 12 constructs are usually assembled per experiment allowing for sufficient technical replicates for each characterization. Assembly of 12 constructs requires at least 9 RGD-coated silk films seeded with hCSSCs based on isolation of 4 stacks per film. Confirmation of phenotypic marker expression for each cell type should be performed using IHC or western blot approaches to verify that successful differentiation has been achieved (Refer to Table 3). Furthermore, the cell morphology of each cell type can be validated by microscopy following seeding on silk materials and post-4 week incubation to verify stratification of the epithelial barrier and keratocyte quiescence. Primary epithelial cells have been found to proliferate slowly on silk films in the co-culture media, thus requiring high cell number density at seeding. Furthermore, multi-layer stratification of the epithelial layer may require utilization of limbal epithelial stem cells with highly proliferative capabilities, which may derived from pluripotent stem cells (Mikhailova et al., 2016; Zhang et al., 2017).
Time Considerations
Table 5 describes relative time considerations for construction and application of the corneal tissue model.
Table 5.
Relative timeframe for completion of each step in the assembly, stimulation, and characterization of the corneal tissue model. The amount of time may vary significantly depending on the user and the total number of constructs generated. These timeframes are based on 12 constructs per experiment.
| Stage | Application | Active Time | Total Time |
|---|---|---|---|
| Scaffold Preparation | PDMS molds | 30 minutes | 1 day with overnight incubation |
| Silk isolation | 3-4 hours | 5-6 days | |
| Patterned, RGD-conjugated silk films | 3 hours | 2.5 days with 48 hour incubation | |
| Nonpatterned silk film with epithelial growth factor stamp | 2-3 hours | 2.5 days with 48 incubation | |
| Silk sponge | 2 hours | 5 days | |
| Cell Culture | Culture hCECs | 2 hours | 1 week from frozen stock + 2 days on silk film = ~9 days |
| Culture hCSSCs | 2 hours | 2-3 days from frozen stock + 2 days on silk film = ~5 days | |
| Culture hiNSCs | 5 hours | 1 week to culture MEF feeder plate + 0.5 day to inactivate + 1-2 weeks of hiNSC on MEFs + 10 day sensory differentiation = ~3-4 weeks | |
| Cultivation | Assemble construct | 2-3 hours | 0.5 day |
| Maintenance of construct | 12 hours | 4 weeks with media changes every other day | |
| Chemical Stimulation | Topical application of chemical to construct | 1 hour | 1 day with overnight incubation |
| Characterization | MTT toxicity assay | 1.5 hours | 4 hours |
| Conditioned media isolation and ELISA | 3 hours | 1.5 days with overnight incubation | |
| IHC | 2 hours + imaging | 2.5 days | |
| Cell lysis and protein isolation | 30 minutes | 2 hours | |
| Measuring protein concentration by BCA assay | 30 minutes | 1 hour | |
| Western blot | 1 hour | 1.5 days with overnight incubation |
Acknowledgements
We acknowledge funding support from the NIH (R01EY020856, P41EB002520) (DK).
Footnotes
Conflict of Interests
The authors declare no conflicts of interest.
Literature Cited
- Abbott RD, Kimmerling EP, Cairns DM, & Kaplan DL (2016). Silk as a Biomaterial to Support Long-Term Three-Dimensional Tissue Cultures. ACS Appl Mater Interfaces, 8(34), 21861–21868. doi: 10.1021/acsami.5b12114 [DOI] [PubMed] [Google Scholar]
- Basu S, Hertsenberg AJ, Funderburgh ML, Burrow MK, Mann MM, Du Y, … Funderburgh JL (2014). Human limbal biopsy-derived stromal stem cells prevent corneal scarring. Sci Transl Med, 6(266), 266ra172. doi: 10.1126/scitranslmed.3009644 [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference describes the isolation and characterization of hCSSCs from limbal rims.
- Borderie VM, Baudrimont M, Lopez M, Carvajal S, & Laroche L (1997). Evaluation of the deswelling period in dextran-containing medium after corneal organ culture. Cornea, 16(2), 215–223. [PubMed] [Google Scholar]
- Cairns DM, Chwalek K, Moore YE, Kelley MR, Abbott RD, Moss S, & Kaplan DL (2016). Expandable and Rapidly Differentiating Human Induced Neural Stem Cell Lines for Multiple Tissue Engineering Applications. Stem Cell Reports, 7(3), 557–570. doi: 10.1016/j.stemcr.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference details the development and characterization of stable hiNSC lines.
- Chambers SM, Qi Y, Mica Y, Lee G, Zhang X-J, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi S-H, & Studer L (2012). Combined small molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nature biotechnology, 30(7), 715–720. doi: 10.1038/nbt.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference describes the growth factor and inhibitor cocktail to promote sensory nerve differentiation from stem cells.
- Chen J, Zhang W, Kelk P, Backman LJ, & Danielson P (2017). Substance P and patterned silk biomaterial stimulate periodontal ligament stem cells to form corneal stroma in a bioengineered three-dimensional model. Stem Cell Res Ther, 8(1), 260. doi: 10.1186/s13287-017-0715-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff PM, McKay TB, Wang S, Ghezzi CE, Cairns DM, Abbott RD, … Kaplan DL (2018). Modeling Diabetic Corneal Neuropathy in a 3D In Vitro Cornea System. Sci Rep, 8(1), 17294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughty MJ, & Zaman ML (2000). Human corneal thickness and its impact on intraocular pressure measures: a review and meta-analysis approach. Surv Ophthalmol, 44(5), 367–408. [DOI] [PubMed] [Google Scholar]
- Ghezzi CE, Marelli B, Omenetto FG, Funderburgh JL, & Kaplan DL (2017). 3D Functional Corneal Stromal Tissue Equivalent Based on Corneal Stromal Stem Cells and Multi-Layered Silk Film Architecture. PLoS One, 12(1), e0169504. doi: 10.1371/journal.pone.0169504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil ES, Mandal BB, Park SH, Marchant JK, Omenetto FG, & Kaplan DL (2010). Helicoidal multi-lamellar features of RGD-functionalized silk biomaterials for corneal tissue engineering. Biomaterials, 31(34), 8953–8963. doi: 10.1016/j.biomaterials.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil ES, Park SH, Marchant J, Omenetto F, & Kaplan DL (2010). Response of human corneal fibroblasts on silk film surface patterns. Macromol Biosci, 10(6), 664–673. doi: 10.1002/mabi.200900452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Hutcheon AE, Melotti SA, Zieske JD, Trinkaus-Randall V, & Ruberti JW (2007). Morphologic characterization of organized extracellular matrix deposition by ascorbic acid-stimulated human corneal fibroblasts. Investigative ophthalmology & visual science, 48(9), 4050–4060. doi: 10.1167/iovs.06-1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence BD, Marchant JK, Pindrus MA, Omenetto FG, & Kaplan DL (2009). Silk film biomaterials for cornea tissue engineering. Biomaterials, 30(7), 1299–1308. doi: 10.1016/j.biomaterials.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, & Armstrong JD (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics, 27(17), 2453–2454. doi: 10.1093/bioinformatics/btr390 [DOI] [PubMed] [Google Scholar]
- McKay TB, Hjortdal J, Priyadarsini S, & Karamichos D (2017). Acute hypoxia influences collagen and matrix metalloproteinase expression by human keratoconus cells in vitro. PLoS One, 12(4), e0176017. doi: 10.1371/journal.pone.0176017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay TB, Hjortdal J, Sejersen H, & Karamichos D (2017). Differential Effects of Hormones on Cellular Metabolism in Keratoconus In Vitro. Sci Rep, 7, 42896. doi: 10.1038/srep42896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay TB, Seyed-Razavi Y, Ghezzi CE, Dieckmann G, Nieland TJF, Cairns DM, … Kaplan DL (2018). Corneal pain and experimental model development. Prog Retin Eye Res. doi: 10.1016/j.preteyeres.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference details the biological mechanisms and clinical features of corneal pain and the rationale and methodology for developing in vitro models to study this process.
- Meek KM, Leonard DW, Connon CJ, Dennis S, & Khan S (2003). Transparency, swelling and scarring in the corneal stroma. Eye (Lond), 17(8), 927–936. doi: 10.1038/sj.eye.6700574 [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, & Unser M (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A, 58(2), 167–176. doi: 10.1002/cyto.a.20022 [DOI] [PubMed] [Google Scholar]
- Mikhailova A, Ilmarinen T, Ratnayake A, Petrovski G, Uusitalo H, Skottman H, & Rafat M (2016). Human pluripotent stem cell-derived limbal epithelial stem cells on bioengineered matrices for corneal reconstruction. Exp Eye Res, 146, 26–34. doi: 10.1016/j.exer.2015.11.021 [DOI] [PubMed] [Google Scholar]
- Ren R, Hutcheon AE, Guo XQ, Saeidi N, Melotti SA, Ruberti JW, … Trinkaus-Randall V (2008). Human primary corneal fibroblasts synthesize and deposit proteoglycans in long-term 3-D cultures. Dev Dyn, 237(10), 2705–2715. doi: 10.1002/dvdy.21606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwood DN, Preda RC, Yucel T, Wang X, Lovett ML, & Kaplan DL (2011). Materials fabrication from Bombyx mori silk fibroin. Nat Protoc, 6(10), 1612–1631. doi: 10.1038/nprot.2011.379 [DOI] [PMC free article] [PubMed] [Google Scholar]; This reference provides a detailed guide to silk isolation and processing.
- Rufer F, Schroder A, & Erb C (2005). White-to-white corneal diameter: normal values in healthy humans obtained with the Orbscan II topography system. Cornea, 24(3), 259–261. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E (1996). RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol, 12, 697–715. doi: 10.1146/annurev.cellbio.12.1.697 [DOI] [PubMed] [Google Scholar]
- Siran W, Ghezzi CE, Cairns DM, Pollard RE, Chen Y, Gomes R, … Kaplan DL (2018). Human Corneal Tissue Model for Nociceptive Assessments. 7(19), e1800488. doi: 10.1002/adhm.201800488 [DOI] [PubMed] [Google Scholar]
- Stepp MA (2006). Corneal integrins and their functions. Exp Eye Res, 83(1), 3–15. doi: 10.1016/j.exer.2006.01.010 [DOI] [PubMed] [Google Scholar]
- Syed-Picard FN, Du Y, Hertsenberg AJ, Palchesko R, Funderburgh ML, Feinberg AW, & Funderburgh JL (2018). Scaffold-free tissue engineering of functional corneal stromal tissue. J Tissue Eng Regen Med, 12(1), 59–69. doi: 10.1002/term.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuret G, Manissolle C, Campos-Guyotat L, Guyotat D, & Gain P (2005). Animal compound-free medium and poloxamer for human corneal organ culture and deswelling. Invest Ophthalmol Vis Sci, 46(3), 816–822. doi: 10.1167/iovs.04-1078 [DOI] [PubMed] [Google Scholar]
- Wang S, Ghezzi CE, Gomes R, Pollard RE, Funderburgh JL, & Kaplan DL (2017). In vitro 3D corneal tissue model with epithelium, stroma, and innervation. Biomaterials, 112, 1–9. doi: 10.1016/j.biomaterials.2016.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ghezzi CE, Cairns DM, Pollard RE, Chen Y, Gomes R, McKay TB, Pouli D, Jamali A, Georgakoudi I, Funderburgh JL, Kenyon K, Hamrah P, & Kaplan DL (2018). Human Corneal Tissue Model for Nociceptive Assessments. 7(19), e1800488. doi: 10.1002/adhm.201800488 [DOI] [PubMed] [Google Scholar]; This reference details the development and application of the corneal tissue model.
- Wang S, Ghezzi CE, White JD, & Kaplan DL (2015). Coculture of dorsal root ganglion neurons and differentiated human corneal stromal stem cells on silk-based scaffolds. J Biomed Mater Res A, 103(10), 3339–3348. doi: 10.1002/jbm.a.35465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AH, Welge-Lussen UC, Priglinger S, Kook D, Grueterich M, Hartmann K, … Neubauer AS (2009). Optimizing the deswelling process of organ-cultured corneas. Cornea, 28(5), 524–529. doi: 10.1097/ICO.0b013e3181901dde [DOI] [PubMed] [Google Scholar]
- Wu J, Du Y, Mann MM, Funderburgh JL, & Wagner WR (2014). Corneal stromal stem cells versus corneal fibroblasts in generating structurally appropriate corneal stromal tissue. Exp Eye Res, 120, 71–81. doi: 10.1016/j.exer.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rnjak-Kovacina J, Du Y, Funderburgh ML, Kaplan DL, & Funderburgh JL (2014). Corneal stromal bioequivalents secreted on patterned silk substrates. Biomaterials, 35(12), 3744–3755. doi: 10.1016/j.biomaterials.2013.12.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Du L, Sun P, Shen L, Zhu J, Pang K, & Wu X (2017). Construction of tissue-engineered full-thickness cornea substitute using limbal epithelial cell-like and corneal endothelial cell-like cells derived from human embryonic stem cells. Biomaterials, 124, 180–194. doi: 10.1016/j.biomaterials.2017.02.003 [DOI] [PubMed] [Google Scholar]