Abstract
Prostate cancer (PCa) is the most commonly diagnosed malignancy and the second leading cause of cancer-related death in American men. Androgen deprivation therapy (ADT) has become a standard treatment strategy for advanced PCa. Although a majority of patients initially respond to ADT well, most of them will eventually develop castration-resistant PCa (CRPC). Previous studies suggest that ADT-induced changes in the immune microenvironment (mE) in PCa might be responsible for the failures of various therapies. However, the role of the immune system in CRPC development remains unclear. To systematically understand the immunity leading to CRPC progression and predict the optimal treatment strategy in silico, we developed a 3D Hybrid Multi-scale Model (HMSM), consisting of an ODE system and an agent-based model (ABM), to manipulate the tumor growth in a defined immune system. Based on our analysis, we revealed that the key factors (e.g. WNT5A, TRAIL, CSF1, etc.) mediated the activation of PC-Treg and PC-TAM interaction pathways, which induced the immunosuppression during CRPC progression. Our HMSM model also provided an optimal therapeutic strategy for improving the outcomes of PCa treatment.
Author summary
Prostate cancer (PCa) is the second leading cause of cancer-related deaths in American men. Androgen deprivation therapy (ADT) is the first-line therapy for advanced PCa, yet a significant number of primary PCa patients treated with ADT eventually develop incurable castration-resistant prostate cancer (CRPC). Recent observations suggest that the immunosuppressive microenvironment of PCa might be responsible for the failures of various therapies. However, the role of immune system in CRPC progression is still unclear. To deeply understand the immunity leading to CRPC progression, we developed a unique systems biology approach (HMSM). Based on our analysis, we identified the key molecules (e.g. WNT5A, TRAIL, CSF1, etc.) mediating the communication of PCa and immune cells. Our HMSM system also revealed the optimal therapeutic strategy for PCa treatment. Collectively, our study provides a new insight to study tumor-related immune mechanisms and pave the way for the development of more effective treatments.
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related death in American men [1, 2]. Androgen deprivation therapy (ADT) has become standard therapy for the treatment of PCa. Although the majority of patients initially respond well to ADT, most patients will eventually become unresponsive, and the PCas recur within 1–3 years after ADT as castration-resistant prostate cancers (CRPC) [3].
Previous studies have demonstrated that androgen receptor (AR)-mediated signaling pathway plays a central role in CRPC cell survival and growth, constituting an attractive target for therapy [4, 5]. MDV3100 (enzalutamide), an FDA-approved drug, is a well-known AR antagonist that can effectively block androgen binding to AR, thereby preventing AR nuclear translocation and coactivator recruitment [6, 7]. However, prostate cancer treatment with AR antagonists can also acquire resistance through AR mutations [8–10], such as AR splice variants [11] and gene amplification. Therefore, there is an urgent need for the development of new therapeutic strategies. To date, immunotherapy represents an appealing option in prostate cancer treatment [12]. The FDA-approved vaccine (sipeucel-T [13]) and PD-1 inhibitor (e.g., pembrolizumab [14]) have been used to treat advanced PCa in clinical trials. However, recent phase III trials showed multiple failures of immunotherapy in PCa [15–18].
Recent observations suggest that the microenvironment (mE) of PCa is immunosuppressive, which appears to be responsible for the failures of various agents targeting the immune system in PCa [15, 19]. Escamila et al. found that tumor-associated macrophages (TAM) exert a negative impact on the treatment response of PCa after ADT [20]. ADT induced an increased expression of colony-stimulating factor 1 (CSF1) in prostate cancer cells (PCs), leading to a significant enhancement of TAM infiltration. The increased levels of IL10, VEGF, and EGF in TAMs, in turn, promote treatment resistance by enhancing immune suppression and tumor proliferation. Moreover, in the prostate-specific Pten-/- mouse model, Akins and colleagues found that Treg (Regulatory T cell) expansion occurred following ADT and the frequency and function of CD8+ T cells (CTLs) increased at the early stage but reduced after a late time point [21, 22]. In this study, we found that WNT5A activated AKT/AR signaling pathway and stimulated the PC proliferation, which might be a new mechanism of PC resistance to ADT. However, the precise cellular targets and the exact molecular mechanism of the immunity leading to CRPC remain unclear. Therefore, systematic understanding of the impact of androgen deprivation on the tumor-associated immune system will help to characterize novel cytokine networks and signal transduction pathways and develop more effective combined therapies for patients with advanced PCa. Taking above studies together, we hypothesize that 1) following ADT, the reactivation of AR signaling in PC cells and the altered immune mE contribute to the development of CRPC; 2) the communication between immune cells and PCs results in immune suppression and PCa progression; 3) targeting the immune-PC pathways mediated by cytokines after ADT may prevent CRPC development.
In recent years, some mathematical approaches have been developed to model the tumor growth, angiogenesis, and drug resistance, providing a new perspective way in exploring the molecular mechanisms of cancer treatment resistance [23–26]. Peng et al. developed an ODE-based model to characterize the effect of castration on the immune system and to predict the efficacy of combined therapy with ADT and vaccines on PCs [27]. However, the role of the immune system in CRPC progression was rarely studied. Based on the hypothesis described above, we developed a predictive 3D Hybrid Multi-scale Model (HMSM) with various types of data for systematically understanding the immunity leading to CRPC progression.
The HMSM model consists of a 3D agent-based model (ABM) and an ordinary differential equations (ODEs) model. The ABM is used for modeling tumor growth, angiogenesis, immune response in the prostate and lymph node compartments, and the ODEs model for dynamics of intracellular signal transduction. The HMSM model integrates key biological events spatially and temporally. Spatially, the simulated mE contains two components: prostate tumor space and lymph node. PCs and TAMs reside in the tumor space for tumor growth and angiogenesis, and CTLs and Tregs home in the lymph node and infiltrate to tumor bed once the initial immune response is activated. Temporally, we modeled the intracellular signaling dynamics (minutes to hours); cell division, apoptosis, migration, and immune infiltration (hours to days); drug response (days to weeks), and CRPC progression and tumor growth (weeks to months). After parameter tuning, the outcomes of our HMSM model in different conditions are fit with the experimental observations. Finally, we use this model to predict the effect of individual and combined treatments with WNT5A neutralization, CSF1R inhibition [20], IL-2 neutralization [22], and EGFR inhibition [28] on the development of CRPC. Our simulation indicates that suppression of Treg expansion with IL-2 antibody and blockade of PC-Treg and PC-TAM interactions appear to re-activate anti-tumor immune responses and to prevent CRPC occurrence. In summary, this study revealed the key cytokines/pathways-induced immunosuppression during CRPC progression and also provided an optimal therapeutic strategy for improving the outcomes of CRPC treatments.
Results
Inference of PC-Treg interactions
To model CRPC progression, we first identified the cell-cell interactions between PC and Treg based on the transcriptomic data. We calculated 1) the significantly overexpressed ligands- and receptors-encoded genes from the GEO (Gene Expression Omnibus) dataset GSE38043 [29] and GSE46218 [30]; 2) the directionality of cell-cell communication of ligand-receptor pairs based on the prior information in public databases, such as iRefWeb [31]. The interactions between PC and Treg were mainly inferred from above two GEO datasets using the approach reported previously [31]. The dataset GSE38043 was generated from isolated Treg cells of CRPC patients (3 patients VS. 3 control). Student T-test was used to filter the significantly overexpressed genes with a p-value < 0.05. In total, we filtered 18 ligand genes (e.g., WNT5A) and 26 receptor genes (e.g., DCR2, EGFR, etc.). The dataset GSE46218 was generated from prostate orthotopic xenograft models. We compared the gene expression profiles of castration-resistant prostate cancer and androgen-dependent prostate cancer, and obtained 23 overexpressed ligand-encoded genes and 39 overexpressed receptor-encoded genes from the castration-resistant PCa, such as FZD5, BMP6, TNFSF10 (TRAIL), etc. The calculation procedure was shown in S1 Fig. We did further filtration analysis for the identified ligand- and receptor-encoded gene pairs and found potential pairwise interactions between PC and Treg: Treg→WNT5A→PC, and PC→TRAIL→Treg (S2 Fig). All of the significantly overexpressed ligand- and receptor-encoding genes were listed in S1 and S2 Tables.
To determine the cell-cell interaction inferred above, we treated castration-resistant prostate cancer cells 22RV1 with WNT5A and generated RNA-seq data. Our analysis shows that WNT5A treatment up-regulates a group of genes in 22RV1 cells, e.g., AR, FZD5, SKP2, PKC, ERK, STAT3, and TNFSF10 (TRAIL), etc. (S3 Fig). Further functional analysis of the significantly expressed genes shows that some important pathways are enriched, including PI3K/AKT/AR pathway, Ras pathway, MAPK pathway, JAK/STAT pathway, prostate cancer pathway, and WNT pathway, etc (see the details in S3 Table). Thus, WNT5A appears to be a key factor in the activation of the survival and proliferation pathways in the castration-resistant PC cells. To further validate the results obtained from RNA-seq analysis and inferred WNT5A/TRAIL pathway loop, we treated 22RV1 cells with WNT5A and the gene expression and protein levels were measured using qRT-PCR and/or Western blot, including FZD5, TNFSF10 (TRAIL), BMP6, AR, BMP6, Skp2, Foxo1, and ERK. WNT5A receptor, FZD5 was significantly up-regulated at 1 hour after treatment (Fig 1A). In addition, WNT5A stimulation induced a sharp increase of TNFSF10 (TRAIL) (Fig 1B), which may further promote Treg expansion [32]. In addition, WNT5A led to a significant increase in the BMP6 level at 0.5, 1 and 3 hours following treatment (Fig 1C). This finding is consistent with the previous studies showing that WNT5A stimulates BMP-6 expression in metastatic prostate cancer (CaP) in the context of bone niche; and BMP-6 in turn stimulated the proliferation of CaP cells [33]. Most important, treating cells with WNT5A resulted in a dramatical and persistent increase in the transcript level of AR (Fig 1D). The protein levels of Skp2 and FOXO1 were increased at 3, 7 and 24 hours post-treatment (Fig 1E and 1F). These findings are consistent with reports that Skp2 and FOXO1 activation are associated with AR transactivation and tumorigenesis [34, 35]. Finally, increased ERK phosphorylation was observed at 0.5 and 7 hours (Fig 1G), also consistent with the early studies that MEK/ERK axis may promote CRPC development, leading to early relapse [36, 37]. Taken together, our experimental results demonstrated that WNT5A induced AR signaling activation and secretion of TRAIL, which potentially promotes CRPC development.
Fig 1. qRT-PCR and Western blot analysis to determine the effect of WNT5A on AR-signaling associated genes and proteins in androgen-resistant 22RV1 prostate cancer cells.
(A-D) The gene expression levels of FZD5, TNFSF10, BMP6 and AR at 0.5, 1, 3, 7 and 24h after treatment with WNT5A, respectively. (E-G) The effect of WNT5A on protein levels of skp2, Foxo1 and pERK, respectively.
Inference of PC-TAM interaction
We also identified the PC-TAM interactions based on the previous findings. Escamilla and coworkers found that CSF1 was significantly induced in the prostate cancer cells by ADT, leading to a significant increase in TAM [20]. TAM expresses elevated levels of VEGF, MMP-9, IL10, and EGF, thereby to promote the protumorigenic phenotype (such as angiogenesis and immune suppression) of macrophages [20, 38]. Tang et al. reported that Treg expansion in Pten-/- mice after castration was mediated by IL-2 [22].
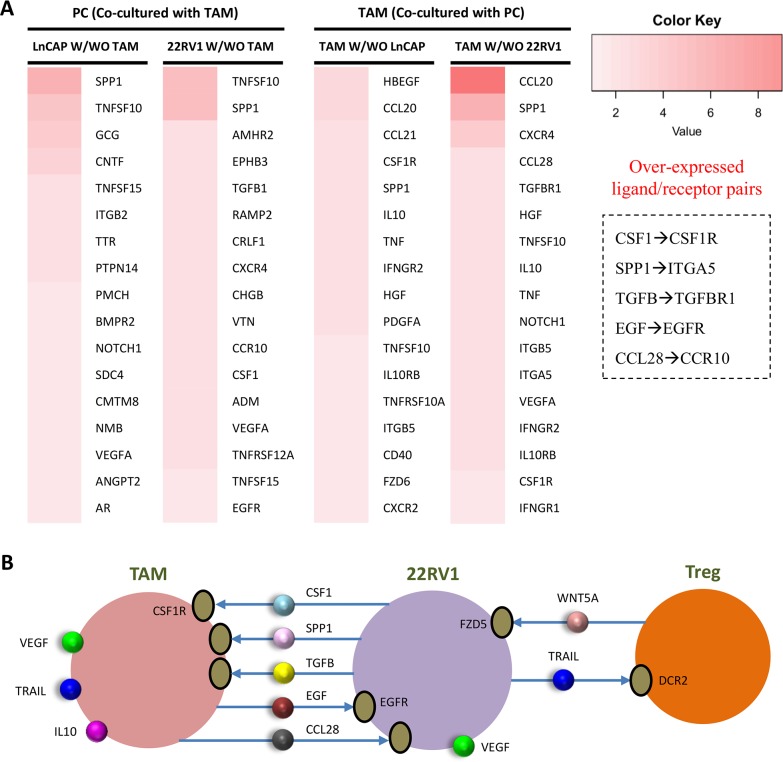
In order to validate the PC-TAM interactions inferred above, we performed coculture experiments (Materials and Methods). The in vitro experiments were designed to test the interactions of induced M2 macrophages with LNCaP cells (androgen-sensitive) or 22RV1. The RNA-seq data from the co-culture of M2 macrophages with LNCaP or 22Rv1 cells was used to validate the PC-TAM interactions. With a defined FC value > 1.3 (fold change of presence TAM to absence TAM), we totally obtained 11 over-expressed ligand genes (e.g., TNFSF10, VEGFA) and 6 receptor genes from the co-cultured LNCAP cells; and 13 ligand genes (TNFSF10, SPP1, etc.) and 12 receptor genes (e.g., EGFR) in the co-cultured 22RV1 cells. At the presence of TAMs, we found that 1) LNCaP positively expressed AR signaling axis; 2) 22RV1 secreted CSF1 and TNFSF10 (TRAIL), which potentially induced TAM recruitment and polarization, and Treg proliferation. Similarly, we obtained 27 overexpressed ligand genes (e.g., IL10) and 30 receptor genes (e.g., CSF1R) from M2 macrophages co-cultured with LNCAP cells, compared with the M2 cells without co-culture. Also, 31 ligand genes (IL10, TNFSF10, and VEGFA, etc.) and 46 receptor genes (CSF1R, TGFBR1, etc.) were over-expressed in M2 macrophage co-cultured with 22RV1 cells. Fig 2A shows the top-ranked overexpressed ligand and receptor genes in these three types of cells (S1 Data). As described in the above section, we determined the potential directional connections with high confidence scores (from iRefWeb) and obtained 5 ligand/receptor pairs between TAMs and 22RV1s (Fig 2A), including the positive loop PC→CSF1→TAM and TAM→EGF→PC demonstrated by other researchers [20]. Combing the above findings, Fig 2B revealed the cell-cell interaction network between TAM, Treg, and 22RV1. All the enriched genes corresponding to Fig 2A were presented in S4 Table.
Fig 2. Inference of TAM-PC interactions with RNA-Seq data.
(A) The left panel shows the RNA-seq data from the cocultured macrophage and PC LnCap and 22RV1 cells. Prostate cancer cells (LNCaP or 22RV1) were co-cultured with or without M2 macrophage (TAM) for 48 h and RNA samples were collected for RNA-seq analysis. All of the gene expression data (fold change value) were normalized with non-co-cultured counterpart cells. For example, LNCaP W/WO TAM shows the gene expression ratio of LNCaP cells co-cultured with TAM to LNCaP cells not co-cultured with TAM. The top-ranked overexpressed genes with FC>1.3 are presented. Five enriched ligand-receptor pairs were highlighted. (B) The inferred cell-cell interaction networks between TAM, Treg, 22RV1.
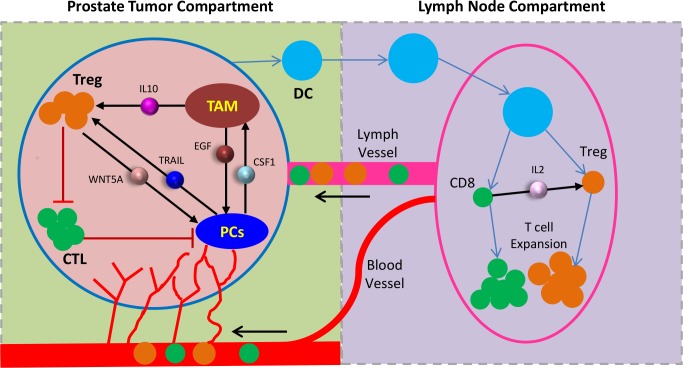
Taken together, our analyses show that two potential cell-cell interaction loops appear to involve in the development of CRPC. The first loop is the secreted WNT5A from Tregs and macrophages triggers the activation of signaling pathways of cell survival and proliferation (e.g., WNT5A signaling, PI3K/AKT/AR and MAPK pathways, etc.) in androgen-resistant PCa cells. TRAIL secreted from PCs promotes Treg proliferation [32]. The second loop is ADT-induced CSF1 expression in the tumor cells stimulates TAM infiltration. Increased TAM activation leads to increased secretion of EGF and VEGF, which in turn activate AR signaling and promote angiogenesis, respectively. Combining the above information of cell-cell communications, we highlighted an integral system in the immune mE of prostate cancer that may lead to CRPC development (Fig 3).
Fig 3. The system modeling diagram of CRPC development.
The HMSM model includes two components: prostate cancer compartment (left) and lymph node compartment (right). The arrows represent cell-cell communications, which were inferred from our data or other public datasets.
Model development
Hypothesis
Except the genomics data described above for the prediction of cell-cell interactions, the cell population data quantified from animal models was also needed for tumor growth modeling. Akins and Tang et al. developed a prostate-specific Pten-/- mouse model [21, 22] and observed the dynamical changes of CD8+ T cells and Tregs at 2.5wk and 5wk after castration. Their studies showed that 1) castration induced an accelerated Treg expansion in mice; 2) the frequency and function of CD8+ T cells (CTL) was significantly increased at the early stage after castration and dropped at 5 weeks post-castration; 3) blockage of IL-2 abolished the increased expansion of Treg in lymph node following ADT. Based on the above findings, we generated the following hypothesis for modeling the immune mE during CRPC progression, as shown in Fig 3. At the early stages after castration, androgen deprivation induces apoptotic death of PCs and antigen secretion from the dying prostate tumor cells. Dendritic cells (DCs) process these antigens and present them on the cell surface. Once activated, DCs migrate to the lymph nodes and interact with T cells to initiate immune response [39, 40]. As a result, a number of activated immune cells (CD8+ and Treg) are quickly cloned in the prostate draining lymph nodes and infiltrated through lymph vessels. CTLs are expanded at the early stage and induced the lysis of tumor cells. However, the number of CTLs is declined at a late time after ADT due to the accelerated amplification of Tregs [22, 41]. IL-2, produced by activated CD8+ T cells contributes to Treg proliferation [42]. Moreover, ADT induces the PC-Treg and PC-TAM positive signaling loops, which promote the resistance of PCa and elevates the expression of key cytokines in the PCa mE [43, 44].
Systematically modeling the immunity leading to CRPC progression
To systematically understand cell-cell interactions, we established a predictive 3D Hybrid Multi-scale Model (HMSM), which combines a 3D multi-scale agent-based model (ABM) for tumor growth and immune response, and an ODE system for dynamic signal transduction (Fig 4). Our HMSM model includes five types of cells (PC, TAM, CTL, Treg, and EC (Endothelial Cell)), which were represented by five types of cell agents of intracellular signaling events and interfaces.
Fig 4. Schematic representation of computational framework of HMSM model.
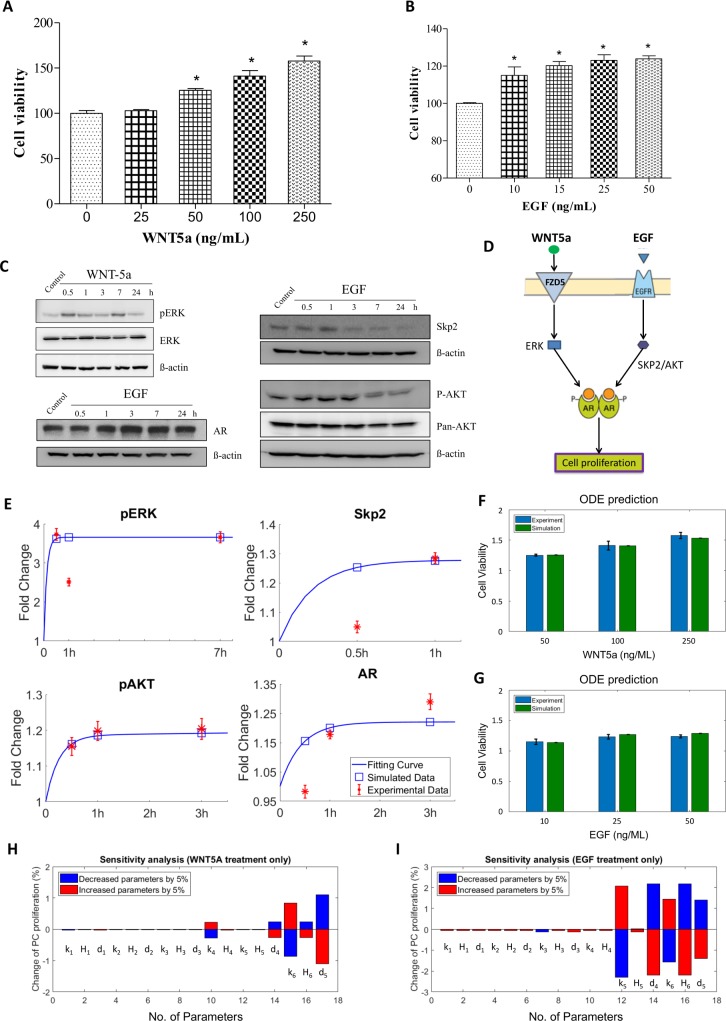
(1) PC agents represent prostate tumor cells with two possible states: androgen-dependent (sensitive), and androgen-independent (resistant). The development mechanism from androgen dependent to androgen independent PCs (CRPC) is associated with AR reactivation through AR mutations, AR variants, and gene amplifications after ADT [45, 46]. After castration, the concentration of androgen in blood and tumor tissues decreases to ~10% and 20% of the pre-treatment values, respectively [47, 48]. However, CRPC cells can synthesize dihydrotestosterone (DHT), thereby reactive AR [49]. In PC agents, we not only took into account of the androgen-independent pathway (triggered by EGF and WNT5A), but also DHT-mediated androgen-dependent signaling to determine the internal factors-associated tumor cell proliferation [46, 50]. The proliferation rates of PC cells were determined via Hill function for the androgen-dependent pathways and ODE system for the androgen-independent pathways (S1 Text). Therefore, we developed an ODE system to simulate WNT5A/EGF-triggered signal transduction of PC proliferation (Fig 5). Firstly, we observed the changes of PC proliferation treated by WNT5A or EGF with different doses (Fig 5A and 5B). Secondly, the key proteins associated with PC proliferation were measured using Western Blot (Fig 5C). We mainly used the signaling data from 0-3h for modeling in the ODE system because initial phosphorylation typically occurs around 2 hours after stimulated by extracellular ligands [51]. For Skp2, we only chose the observed values at 0.5 and 1h for model training as its expression didn’t change after 3h, compared with control. Finally, an ODE system was built on the signaling pathway network as shown in Fig 5D. The ODE system was described into details in “Materials and Methods”. All of the parameters involved in this system were estimated using GA algorithm [27, 52], and presented in the S5 Table. With the GA-guided optimal parameters, the ODE system fitted the experimental observations well and the ODE system stabilized after 1 hour (Fig 5E). In addition, Fig 5F and 5G indicate that the optimized ODE system recaptures the experimental observations shown in Fig 5A and 5B. Sensitivity analysis shows that the ODE system is quite stable when each parameter (S5 Table) was perturbed by a 5% increase or decrease (Fig 5H and 5I).
Fig 5. ODE modeling of WNT5A-EGF/AR signaling and experimental validation.
(A) The effect of WNT5A on 22RV1 cell viability. (B) The effect of EGF on 22RV1 cell viability. (C) The effect of WNT5A and EGF on protein levels of Skp2, pERK, pAKT and AR. (D) The network topology of androgen-independent pathways in prostate cancer cells. WNT5A or EGF regulates the cellular proliferation by activating AR-related pathway. This network was represented as a series of ODE equations shown in Eq. (1–6) in the section “Materials and methods”. (E) The predicted values of four proteins fit the observation data well. (F, G) The ODE system-predicted PC proliferation at 72 hours perturbed by WNT5A (F) or EGF (G) with different doses. (H, I) Sensitivity analysis of the ODE system was performed under two conditions: WNT5A treatment only (H), and EGF treatment only (I). Each parameter was perturbed by increasing or decreasing 5%.
(2) TAM agents denote TAM cell population, which reside in the tumor space. TAMs are believed to promote tumor invasion and metastasis [20]. TAM infiltration is a significant unfavorable prognostic factor for prostate cancer patients [53, 54]. Our data analysis shows that the direct interactions between TAMs and PCs appear to be one of the main reasons for the resistance of PCs to androgen deprivation.
(3) CTL agents represent the CD8+ effector T cells, which mediate tumor cell lysis through cell-cell contact. Antigen-presenting DCs migrate to the draining lymph nodes and present the antigen to T cells, so that the CD4+ T helper and CD8+ effector cells are activated [55, 56]. After undergoing up to 8 generations of division, these T cells acquire effector functions, such as cytotoxicity [57]. Activated CTLs migrate to the tumor through lymph vessels or peripheral blood and contribute to tumor cell death either by releasing perforins that create holes in the target cell’s membrane or by triggering apoptosis in the target cells.
(4) Treg agents represent the immune-suppressive T cell population responsible for the maintenance of peripheral tolerance and have been implicated in the suppression of tumor immunity. Tregs have been shown to inhibit tumor-specific T cell functions, such as the cytotoxic effects of CTLs [58]. Tregs require ligand-specific activation and cell-cell communication to exert their suppressive activities [59]. The factor TRAIL, expressed in PC, can promote the growth of Tregs [32]. Also, the secreted IL-2 by CTLs promoted Treg expansion [42, 60]. In our HMSM model, Tregs in the tumor space was initially activated in lymph node and then infiltrated to the tumor to inhibit/prevent the cytotoxic effects of CTLs by suppressing CTL proliferation (Materials and Methods).
(5) EC agent is defined to simulate tumor angiogenesis in prostate cancer mE. After ADT, TAMs and PCs secret VEGF to induce new vessels to sprout from the pre-existing vasculature towards the center of a tumor, providing nutrients to the starving tumor cells and thereby stimulating tumor growth. VEGF spreads to the surrounding tumor tissues and is also consumed by endothelial cells [61]. The motion (branching or proliferation) of individual ECs located at the tip of a capillary sprout governs the movement of the whole sprout, and chemotaxis in response to VEGF gradients [62].
3D Multi-scale modeling
As shown in Fig 3, CRPC progression is simulated at intracellular, intercellular, and tissue levels in the HMSM model. Intracellular signal transduction was modeled by Hill functions or ODEs to represent the rates of proliferation and apoptosis (S1 Text).
The intercellular communication in HMSM reflects the relationship between cancer cells, immune cells and tumor mE evolution during CRPC progression through the following aspects. 1) ADT induces the prostate tumor cells to express CSF1, which promotes macrophage activation and infiltration [20]. 2) Enhanced infiltration of TAM results in the accumulation of VEGF, IL10, and EGF, promoting angiogenesis, tumor growth, and immune suppression [20, 38]. 3) The positive feedback loop between Tregs and PCs leads to PC proliferation stimulated by WNT5A and Treg growth induced by TRAIL [32]. 4) The production of IL-2 from CD8+ promotes Treg proliferation [63]. 5) CTLs recognize tumor cells in their local regions and migrate toward these target cells for clearance [64]. 6) Accumulation of Tregs potentially inhibits the proliferation of CTLs, resulting in the suppression of anti-tumor immune responses [58].
The tissue scale reflects 3D prostate tumor growth and cell responses to castration via various intercellular cell-cell interactions spatially and temporally as described above. At the tissue level, intracellular signaling pathways were triggered by the secreted cytokines, such as WNT5A, TRAIL, CSF1, EGF, IL-2, VEGF, and IL10, locally via the interfaces of cell agents, and result in the changes in the cells’ fate and behaviors, which in turn modulate the tumor mE for cell growth and response to treatment. In this scale, we defined the dynamic 3D distribution of the key cytokines secreted from four types of cells. VEGF can be secreted from both PCs and TAMs, leading to the initiation of tumor angiogenesis. In addition, the distribution of cell populations in tumor mE is defined by cell proliferation and migration in 3D ECM. In particularly, we defined the immune cell infiltration from lymph nodes to the tumor bed via lymph vessels.
Model implementation
At the beginning of simulation, 200, 100, 2, and 2 of PC, TAM, CD8+ T, and Treg cells were used, respectively, to mimic the initial stage for prostate tumor cells spreading to a new location. Based on all the observed data mentioned above, we manually tuned all the parameters associated with ABM part in HMSM for optimizing the data fitting (see the details in S1.9 Text). All the parameters of ABM model are presented in S6 Table. After optimization, we selected WNT5A antagonist, CSF1R inhibitor PLX3397 (PLX) [20], EGFR inhibitor, anti-IL2 mAb [22] as the representatives of agents, respectively, in HMSM to predict the therapeutic effects in silico. The number of each cell type in HMSM was recorded every 2 hours [25, 52, 65]. The drug effects were represented as the fold changes in the number of tumor and immune cells following treatments. Our HMSM model simulated a series of biological events up to 9 weeks, including prostate tumor growth, ADT-induced immune response, and the emergence of CRPC (S4 Fig).
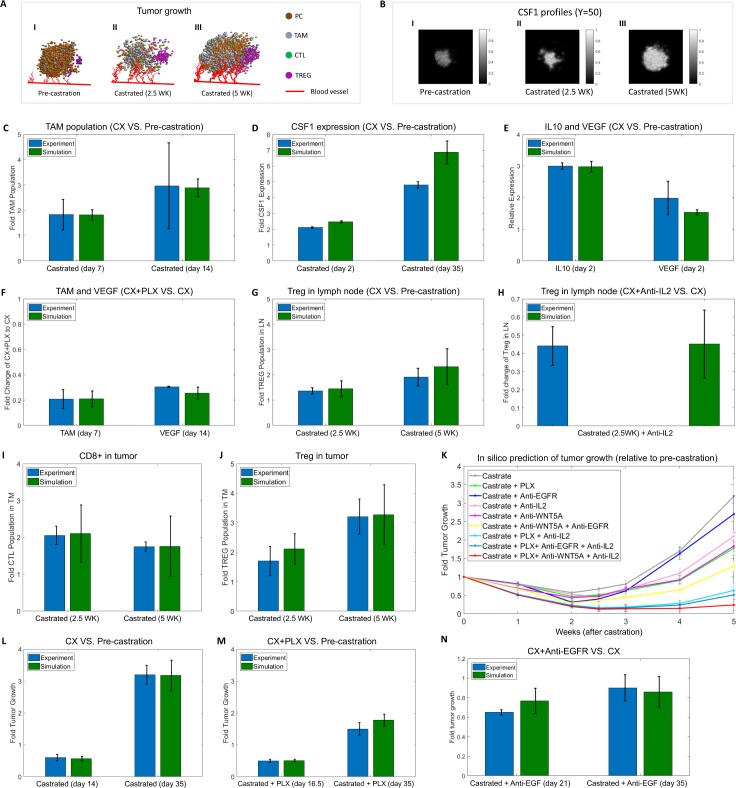
A simulation example of prostate tumor growth before and after castration in the immune suppressed mE is shown in Fig 6A. Panel I represents a simulated PC tumor status before castration. CD8+ T cells and Tregs gradually infiltrated to the tumor space from lymph nodes following the activation induced by prostate-specific antigen-presenting DCs. Castration leads to a rapid decrease of the androgen concentration and increase of immune infiltration, resulting in tumor regression. Panel II shows that most of the tumor cells are cleared at 2.5 wks after castration. After that, CRPC phase eventually emerges, manifested as the elevated TAM infiltration and increased Treg expansion, which appears to contribute to the tumor relapse and immune suppression in the prostate mE (Panel III). Fig 6B presents the dynamics of CSF1 profiles associated with CRPC development (S5–S7 Figs). CSF1 accumulation is transitorily decreased in the early stage after castration and rapidly rebounded as CRPC occurrence.
Fig 6. Experimental observation and in silico prediction of HMSM Model.
The bars with blue color are experimental-measured values, and the bars with green color are predicted values in the HMSM model. CX: Castration. Time 0 denotes pre-castration. (A) A simulation example of prostate tumor before/after castration. I) pre-castration; II) 2.5 weeks after castration; III) 5 weeks after castration. (B) A simulation example of cytokine profiles before/after castration. I) pre-castration; II) 2.5 weeks after castration; III) 5 weeks after castration. The slices are extracted when Y = 50. Y is the Y axis. (C) TAMs are elevated by ADT in prostate cancer (day 7 and day 14 castration). (D) CSF1 protein level was analyzed from castrated mice (day 2 and day 35 castration). (E) Relative gene expression of IL10 and VEGF at 48hours after ADT. (F) PLX lowered macrophage levels and VEGF expression after ADT. (G) The number of Treg cells is increased in lymph nodes at 2.5 weeks and 5 weeks post-castration. (H) Treg expansion in lymph nodes was reduced by IL-2 neutralization. (I) In silico prediction of CD8+ cells in the castrated tumor at 2.5 and 5 weeks. (J) In silico prediction of Treg cells in the castrated tumor at 2.5 and 5 weeks. (K) In silico prediction of single or combined treatment on PC growth after castration relative to pre-castration. (L-N) The predictions and experimental validations for prostate tumor growth with castration only (L) or plus CSF1R (M) or plus EGFR inhibition (N) after castration.
Model evaluation
To test the fit of our HMSM model to the training data, in silico simulations under several contexts were evaluated using the experimental data from our laboratory, as well as the data from previously reported studies (S2 Data). Firstly, we simulated the whole process of prostate tumor growth from the initial state to 6 wks after castration. The dynamic changes of TAM population, CSF1 expression, and TAM-secreted protumorigenic cytokines (e.g., IL10, and VEGF, etc.) in the simulated mE were predicted. Fig 6C shows that TAM population are increased by 1.826±0.2 folds at day 7 and continued to increase to 2.891±0.353 folds at the day 14 after castration. Fig 6D represents the simulated expression of CSF1 from prostate cancer cells. The expression of CSF1 in PCs is significantly increased after ADT, which is close to the measured results from the subcutaneous mouse model [20]. Moreover, the predicted expressions of IL10 and VEGF in TAMs have increased 2.98±0.171 and 1.54±0.078 folds at two days post-castration (Fig 6E). We also predicted the effect of the CSF1R inhibitor on the distribution of the cell population after castration. As shown in Fig 6F, inhibition of CSF1R with PLX after ADT results in a 5-fold reduction of the TAM population in the early stage, compared with the results in castration only. The CSF1R blockade appears to inhibit macrophage proliferation, and lower TAM-induced expression of VEGF, which potentially delays the emergence of CRPC. The simulated results shown in Fig 6C–6F are consistent with the experimental data reported in [20].
To examine the dynamical changes of immune responses, we calculated the changes of the Treg population after castration in the simulated system. As shown in Fig 6G, the number of Treg cells is significantly increased in lymph nodes at 2.5 weeks and 5 weeks post-castration compared to that from pre-castration. This simulation result is close to the previous findings reported by Tang et al. [22]. In addition, Fig 6H shows that Treg expansion is prevented by IL2 neutralization, suggesting the increased IL2 after castration and immunization promotes Treg expansion [22]. Taken together, we found that the HMSM model fits the observed data very well under different contexts.
Model validation
To further validate the reliability of our HMSM model optimized above, we compared the simulation results with additional experimental data (S2 Data) generated from Pten-/- prostate cancer mouse model [21, 27]. Fourteen weeks-old mice were castrated, and the relative changes of immune cells (Treg and CD8+ T cells) in tumor space were observed at the 2.5 weeks and 5 weeks after castration [21]. Fig 6I and 6J show that castration induces infiltration of Treg cells into the tumor area in the prostate tumor-bearing mice. However, the accumulation of functional CD8+ T cells in the prostate tumor is not long-lasting, evident at 2.5 weeks after castration but reduced at 5 weeks after castration. The measured CD8+ T cells at 2.5 weeks and 5 weeks after castration are around 2.05±0.25 and 1.75±0.125 folds of pre-castration, respectively [27]. Fig 6I indicates that the prediction of the CD8+ population (2.107±0.775, and 1.7606±0.8141 folds) in the HMSM model is consistent with the experimental observations. Moreover, the number of Tregs was significantly increased to 1.7±0.5 and 3.2±0.6 folds in the tumors at 2.5 and 5 weeks after castration, respectively [27]. HMSM model simulation shows that the predicted changes of the Treg population at 2.5 and 5 weeks after castration, and our predicted results are close to the experimental observations (Fig 6J). In summary, our testing experimental data further confirms that the outputs of HMSM model are reliable.
Prediction and validation of therapeutic outcomes
To identify the potential therapeutic targets of PCa in the immunosuppressive prostate cancer mE, we predicted the effects of single or combined treatments with castration on PCa growth using the established HMSM model (S2 Data). Our experimental data revealed WNT5A was a potential factor associated with CRPC development. Therefore, we simulated the effect of WNT5A neutralization on PC growth with our in silico model. Recent studies indicate that CSF1R inhibitor (PLX3397) [20, 66] and IL-2 neutralization [22] revealed the effects for immune re-activation after ADT. In addition, early studies have reported the efficacy of EGFR inhibitors (e.g. erlotinib, canertinib, and cetuximab, etc.) in castration-resistant prostate cancer in vitro and in vivo, and claimed that EGFR inhibition might improve the outcome of patients with CRPC [28, 67, 68]. Therefore, we mainly tested the anti-tumor effects of four representative agents in HMSM, including anti-WNT5A antibody, PLX3997, Anti-IL-2 antibody (IL-2 neutralization), and EGFR inhibitor.
Fig 6K shows the predicted outcomes from single or combined treatments relative to pre-castration. Prostate tumor cells were reduced sharply at the first 2 weeks after castration and then re-expanded continuously (silver curve). The combined treatment with castration and a single agent (Anti-WNT5A, PLX, Anti-IL-2, or EGFR inhibitor) yields a better treatment response than that from the castration only group. Comparing with Anti-IL-2 and EGFR inhibitor, a combination of castration with Anti-WNT5A or PLX yields better anti-tumor responses, indicating that blockade of the PC-Treg or PC-TAM interaction may effectively reduce tumor cell growth. In addition, the poor response was observed in the combined treatment group with castration plus EGFR inhibitor, compared with the other combined treatment groups. The optimal prediction outcome was achieved from the treatment group with a combination of PLX, Anti-WNT5A, and Anti-IL-2 (red curve) after castration, revealing that the activation of both Treg and TAM appears to contribute to CRPC development. Moreover, we compared the predicted results with the experimental observations reported previously. Fig 6L shows that the tumor growth rebounded approximately 3 times after castration, paralleling the emergence of CRPC observed in the clinical setting [20]. The addition of CSF1R inhibitor PLX3997 to castration resulted in a significant delay in the onset of CRPC (Fig 6M) [20]. Also, Anti-EGFR leads to 0.77±0.128 and 0.86±0.157 fold tumor growth at 3 and 5 weeks after castration relative to castration only (Fig 6N), which are close to the experimental observation of in vivo effects of EGFR inhibitors in 22RV1 xenografts mice model [28]. Above analyses indicate that the predictive capabilities of our HMSM model are high and the model-based predictions are reliable.
Discussion
The focus of this work is to explore CRPC progression in the immune mE and to develop optimal treatment strategies in silico to improve therapeutic responses of CRPC. To systematically understand the role of the immune system in CRPC development, we generated RNA-seq data and integrated it with the GEO datasets. Through the analysis of these data, we found the potential factors/cytokines (e.g., WNT5A, and TRAIL) associated with PC-immune interactions. Elevated levels of WNT5A have been reported in melanomas, lung cancer, breast cancer, and gastric cancer [69–73]. Lee, et al. investigated the Cap-bone stromal cells interaction, and reported that WNT5A secreted by bone stromal cells increases BMP-6 expression in Cap, thereby leading to Cap cell proliferation [33]. Our study demonstrated that WNT5A induced the activation of androgen-independent pathways and the elevated expression of TRAIL in CRPC cells after castration, indicating the enhancement of PC growth and immune suppression. As a type 2 membrane protein belonging to TNF superfamily, TRAIL is known to play a pivotal role in the immune regulation and antitumor immunity [74–76]. Early studies revealed that TRAIL has the potential to promote Treg proliferation in certain situations [77]. Ikeda and coworkers demonstrated that the proliferative effect of TRAIL on Tregs becomes apparent in autoimmunity [32]. The exploration of TRAIL function in prostate cancer may be of considerable significance for understanding CRPC mechanisms.
We are the first to systemically model the CRPC development in the immune mE using an integrated 3D system (S8 Fig). In our HMSM model, we simulated the PC growth before and after castration. The first stage covers a sequence of key biological events, including DC maturation, T cell activation, and division in lymph nodes triggered by DC, T cell migration and infiltration. The second stage denotes the initial castration therapy (5 weeks), in which the AR signaling reactivation appears around 2 weeks after castration (S4 Fig). Therefore, the proposed model provides a new way to present the dynamic changes in tumor growth, immune response, and drug treatment effect.
We also provide a novel computational platform to optimize the potential target therapy on the castrated PCs. ADT is a standard treatment for PC patients, including surgical castration, and AR disruption with pharmacologic interventions (such as MDV3100 (enzalutamide) [78]). However, clinical studies indicate that AR antagonist can induce AR T878A mutation and result in AR reactivation [79–81]. Our analysis of a representative GEO dataset (GSE67980) [82] also revealed that AR expression was increased when the patients with CRPC treated by enzalutamide (S9 Fig). In recent years, active immunotherapy, such as therapeutic vaccines, provide new strategies for overcoming tumor-mediated immune suppression [83]. Multifaceted approaches that combine vaccine with targeted therapies may have the potential to improve the current therapeutic outcomes by targeting the suppressive immune microenvironment and tumor survival. In the present study, we evaluated several new therapeutic strategies in silico with our optimized HMSM model. The simulated results showed that the optimal prediction outcome was achieved from the treatment group with a combination of PLX, Anti-WNT5A, and Anti-IL-2 after castration, revealing the important role of Treg and TAM activation.
Moreover, the proposed model includes a large number of parameters, and most of the parameters were tuned manually or determined based on the experimental results. In order to confirm the variability of the simulated results from the developed 3D hybrid multi-scale model, a parameter sensitivity analysis was performed by measuring the impact of a small perturbation (5% increase) of individual 34 key parameters on the prostate tumor cell populations (5wk after castration). We found that 1st and 2nd parameters (the basic proliferation rates caused by castration-dependent and castration-independent pathways in PCs) were more sensitive than others (S10 Fig). It indicates that ADT induced prostate cancer cells to progress and further express cytokines to promote CRPC occurence. The sensitivity analysis showed the changes in model outcomes were under 4%, indicating that the outcomes of the optimized model were stable. We also tested the effect of initial cell numbers and cut-off values in the ABM rules on the model variability. S11 Fig and S12 Fig show that simulated tumor growth is not sensitive to the perturbations on the initialization of cell number and the cut-off values in the ABM rules. We did additional analysis with the experimental time points overlaid as dots at the observed times, our results indicate that the optimized HMSM model is reliable (S13 Fig).
Although a number of mathematical approaches have been introduced to model the tumor growth and drug resistance in recent years, most of the well-defined 3D agent-based models not only neglect the stage-structured immune response during the tumor initialization and development, but also did not simulate the dynamics of intracellular pathways in the cell-cell communications [25, 65]. Solovyev et al. was the first to put forward the concept “hybrid model”, which combined ODE model and agent-based model to mimic signal transduction processes at the intracellular scale, stochastic cell behaviors at the intercellular scale, and the dynamic distribution of growth factors at the tissue scale [84]. However, their model was only designed for two-dimensional space so that it cannot be used in 3D tumor study. Our 3D Hybrid model (HMSM) overcomes the limitations of existing models described above, and creates a new paradigm for systematically understanding the immunity leading to CRPC.
There are several limitations of our HMSM model. We used some experimental data from in vitro 2D culture to model 3D microenvironment in this study. Ideally, experimental data obtained directly from 3D tissues can better reflect actual environmental status. However, such types of data are not easily available due to animal study settings. Moreover, using limited available animal data for ABM model training, validation, and prediction may not be enough for the validation of our large-scale-based ABM model. Incorporating much more observed data will increase the reliability of the model outcome. In the future, we will collect tumor tissue data from patients with PCa before and after castration to verify our 3D model. We will develop heterogeneity scoring approaches to evaluate cell-level heterogeneity (receptor expression) and tumor-level heterogeneity (cytokine levels, and geometry). We will extend our model to simulate the effect of new blood vessels on the tumor growth, e.g. modeling increased cancer cell migration and invasion. To better address clinically relevant issues, we will further improve our model in terms of varying-degree inhibition with inhibitors, enabling it to predict dose-related treatment outcomes.
Materials and methods
Experiment
1) Cell culture
LNCaP and THP1 cells were purchased from ATCC (Manassas, VA) and 22RV1 cells were a gift from Dr. Lin’s lab in Wake Forest Medical Center. LNCaP cells were maintained in RPMI-1640 medium supplemented with 10% bovine calf serum (FBS), 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 4500 mg/L glucose, 100 IU/mL penicillin, 100 μg/mL streptomycin and 1500 mg/L sodium bicarbonate, 22RV1 cells were maintained in DMEM medium containing 10% FBS, 2 mM L-glutamine, 100 IU/mL penicillin and 100 μg/mL streptomycin. THP1 cells were cultured with RPMI-1640 medium supplemented with 10% FBS, 2 mM L-glutamine 0.05 mM 2-mercaptoethanol, 100 IU/mL penicillin and 100 μg/mL streptomycin.
2) Cell viability analysis for the 22RV1 cells treated with EGF
2 X 104 22RV1 cells were plated in 24-well plates and incubated overnight. The cells were then treated w/wo EGF (1, 5, 10, 20, 50 ng/mL) for 72 hours. Cell viability was determined using a modified thiazolyl blue tetrazolium bromide (MTT) (Acros Organics, Thermo Fisher Scientific, New Jersey) method as described previously [85]. Briefly, 125 μL of 5 mg/mL MTT reagent in PBS was added to each well and incubated for 4 hours in a CO2 incubator. Cells were then lysed by adding 50 μL lysis buffer (20% SDS, 50% N, N, N-dimethyl formamide (DMF), PH4.7) at 37°C. The absorbance at 560 nm was measured using a spectrophotometer (Molecular Devices, Sunnyvale, CA).
3) Coculture of prostate cancer cells and macrophages
2 X 105 THP1 monocytes were seeded onto 6 well plates overnight and then treated with 10 ng/ml of 12-O-tetradecanoylphorbol-I3-acetate (PMA) for 24 h. The PMA containing medium was replaced with serum-free medium and culture for another 2 days. For the M2 cell differentiation, the cell was treated with 25 ng/ml of IL4 and 25 ng/ml of IL13 for 48 h. The differentiated cells were washed 3 times with PBS and 4 ml of RPMI medium containing 10% FBS, 2 mM L-glutamine, 100 IU/mL penicillin and 100 μg/mL streptomycin was added into each well. Two days prior to the co-culture experiment, 2 X 105 of LnCap and 22RV1 cells were seeded into 0.4 μM transwell inserts, respectively. For coculture, the culture medium in the inserts with prostate cells was removed and transferred onto the top of the 6-well plates with differentiated TPH1 cells. 2 mL of RPMI medium was added into each insert. Prostate cancer cells and M2 macrophage were co-cultured for an additional 48 hours, and cells were harvested for RNA analysis.
4) Treatment of 22RV1 cells with human recombinant WNT5A protein
The human recombinant WNT-5A protein was obtained from R&D System. 22RV1 cells were cultured in the 6-well plates. 80% confluence cells were incubated with serum-free medium for 24 h and then treated with 250ng/mL WNT5A for up to 24 h. The cells were then harvested for RNA and protein analysis using Western blot and quantitative real-time PCR.
5) Western blotting
The differentiated THP1 cells were lysed with 1 X RIPA buffer supplemented with protease and phosphatase inhibitor cocktail (Roche Applied Science, Indianapolis, IN) and stored in aliquots at -20°C until use. Twenty micrograms of cell lysates were denatured by boiling, and separated by SDS-PAGE. The separated proteins were then transferred to a nitrocellulose membrane (BioRad). The membranes were blocked using 5% non-fat dry milk for 1 h at room temperature and probed with antibodies overnight. After incubated with IgG horseradish peroxidase-conjugated secondary antibodies (Cell Signaling, Beverly, MA) for 2 h at room temperature, the immunoblots were developed using the enhanced chemiluminescence (ECL) reagent (Cell Signaling, Beverly, MA) and visualized using a FluroChemQ processor (Proteinsimple, Santa Clara, CA). The antibodies used in this study include CD206, ERK, pERK. AKT, pAKT, foxo1, Glut1, N-cad and skp2, which were obtained from Cell Signaling Technologies.
6) Quantitative Real-time PCR (qRT-PCR)
The qPCR amplification was performed in a 20uL reaction mixture containing 100 ng of cDNA, 10 uL 2 X All-in-OneTM qPCR mix (GeneCopoeia, Rockville, MD), 0.3 mM of upstream and downstream primers and nuclear-free water. The PCR reaction was conducted with 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, 40°C for 30 s and 60°C for 1 min, followed by dissociation curve analysis distinguishing PCR products. The expression level of a gene was normalized with the endogenous control gene β-actin. The relative changes of genes were calculated using the 2-ΔΔCT method and presented as mean ± SD (n = 3). The sequences of the paired sense and antisense primers for human Antigen receptor, TNF-10, FTZ1, and β-actin are listed in the S7 Table.
7) RNA isolation and sequencing
Total RNA was extracted from the cells using an RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The quality and quantity of total RNA were verified spectrophotometrically (NanoDrop 1000 spectrometer; Thermo Scientific, Wilmington, DE, USA) and electrophoretically (Bioanalyzer 2100; Agilent Technologies, Palo Alto, CA, USA). The mRNA libraries were prepared according to the TruSeq RNA Sample Prep Kit protocol (Illumina, San Diego, CA, USA) and sequenced using Illumina HiSeq2000 DNA sequence analyzer. RNA-seq reads were aligned with the reference genome using TopHat [86]. The resulting alignment files were input to Cufflinks to generate transcriptome assemblies, and the Fragments per Kilobase of Transcript per Million fragments mapped (FPKM) values of isoforms were calculated for individual genes [87].
ODE-based modeling of EGF-driven Androgen independent pathway
Androgen Receptor (AR) not only can be activated by extracellular growth factor through androgen-independent pathway network, but also by DHT from the tumor microenvironment through androgen dependent pathway. The activation or inhibition of these two ways depends on the stages of CRPC progression. In our study, we developed an ODE model to simulate the effect of WNT5A/EGF-triggered androgen-independent pathway on the proliferation of prostate cancer cells (Fig 5D). The extracellular concentration of WNT5A and EGF are the input variables for the ODEs. In the training of ODEs, the input parameters (WNT5A or EGF) were restricted to the range [0, 1], and the maximal value “1” represents the maximal dose of ligands we used in our experiment. The output is the fold change of proliferation rates relative to no stimulation. The ODE system has the following form:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
As mentioned above, our phosphor-proteomics data covered the key signaling proteins (pERK, pAKT, AR, and Skp2), which were involved in this androgen-independent signaling network (Fig 5C). The effect of WNT5A and EGF on 22RV1 cell proliferation were also presented in Fig 5A and 5B. All above parameters involved in this ODE system were estimated by optimizing formula (6) via the GA algorithm [27]:
| (6) |
Where and denote the measurement from the experiments and the theoretical results obtained from the ODE model of protein i at the time point t. The parameter vector θ = {k1,H1,d1,……,k6,H5,d5} in above formulas (1–5) can be obtained by formula (6). D1 and D2 represent the inhibitors of WNT5A and EGF pathways, respectively. The set I1 is the indexes of observed proteins in this signaling network, and time series set T1 = {0, 30min, 60min, 420min} covers all the time points related with experimental data (Fig 5C). S5 Table represents the estimated values of all parameters. The fitting accuracy of the predicted and measured values of key proteins is shown in Fig 5.
The agent-based model of CRPC progression
We defined five types of agents in the ABM model to represent PC, TAM, CTL, Treg, and EC, respectively (Fig 3). The ABM model simulates the effects of various cell-cell interactions on prostate tumor growth, angiogenesis, programmed immune response, and drug response in a simulated mE. We initialized the simulated microenvironment as a cuboid, which consists of two connected cubes. One is for the growth and proliferation of mixed PC, TAM, CTL, Treg, and EC compartments (tumor space) and the other is for the activation and division of T cells triggered by matured DC in lymph node and T cell infiltration from its lymph vessels. The proposed model simulated a series of key biological events involved in tumor growth, immune response, and CRPC development (S4 Fig). The details of time points related with these events were described in the S1 Text.
This multi-scale modeling includes intracellular, intercellular and tissue scales, which are illustrated in the S8 Fig, and described into details in the following sections. Detailed flowcharts of each agent were illustrated in the S1 Text. Individual cell behaviors were simulated by probability-based rule implementation [52, 65]. A cell senses the hints in its neighborhood such as local cytokines and drugs and adjusts itself with the embedded signaling pathways, and outputs the corresponding changes on its cell behaviors, including proliferation, survival, differentiation, migration, and cytokine secretion rate. Cell fate decision is then determined by rolling a dice and compared with the probability threshold of cell behavior (S14 Fig).
Intracellular level
In this study, the proliferation rate of PCs was determined by two pathways. Our hypothesis is that: the androgen concentration in blood and gland will be sharply decreased after castration, so that prostate tumor cell proliferation will be supported by WNT5A or EGF-mediated androgen-independent pathway until the occurrence of CRPC (AR-reactivation). The ODE system for cell proliferation of PCs has been described in the above section. Except the ODE system was applied to model the intracellular signaling network in PC cells, Hill functions were used to simulate the signal transduction of other cells to calculate apoptosis and proliferation rates, and further determine the cell behaviors.
Intercellular level
In response to the changes of WNT5A, EGF or DHT in its local mE, each prostate cancer cell will proliferate, migrate, become quiescent, or undergo death process. PCs secrete CSF1 to promote macrophage infiltration. Macrophage-derived EGF enhances tumor cell invasion [38]. TAMs also suppress the immune response of T cells by releasing the immunosuppressive factor, such as IL10 [88]. Similarly, the WNT5A-TRAIL positive loop between PCs and Tregs and the associated molecules are also considered as important modulating components for CRPC development and immune suppression. In addition, prostate tumor cells can be killed by CD8+ T cells. Treg cells can migrate towards CD8+ T cells locally and suppress the proliferation of these cells in a manner of cell cycle arrest or apoptosis [89].
Migration
A non-M-phase cell will migrate if it can find a free space nearby (S15 Fig). The ability of a cancer cell to undergo migration and invasion allows it to change position within the tissue. CD8+ T cells tend to move towards the places where tumor cells reside and try to eliminate residual cancer cells. Tumor cells can evade immune elimination through the loss of antigenicity and/or loss of immunogenicity and by coordinating an immunosuppressive microenvironment. Treg cells migrate to CD8+ T cells and suppress the proliferation of these effector cells. The migration was governed by space availability, migration speed, and stochastic effects using Hill functions and dice-casting simulation.
Programmed cytotoxic T cell response
A naïve T cell remains quiescent until it receives antigenic stimulation from dendritic cells through MHC-TCR interactions [55, 90]. After stimulation, naïve cells appear to be committed to a programmed response that causes them to divide and acquire effector functions [91]. For the first 19-24h, they do not replicate, but after this initial phase, they can rapidly undergo divisions with 7–10 generations [92]. After divisions, they acquire effector functions, such as cytotoxicity. In the ABM model, DC maturation started 12 hours later when they migrated into the lymph node. The initial immune response would be started 12 hours later after DC maturation. When T cells have received antigenic stimulation for 20 hours, they would rapidly divide (clonal expansion). For the clearance of cancer cells, each T cell might infiltrate to tumor space through the lymphatic vessel. After the stage of initial expansion, T cells continuously proliferate and to persist the population (T cell memory) [93]. We also simulated the activation and clonal expansion of naïve Tregs as CTLs in our model (S1 Text).
Tissue level
Modeling of tumor angiogenesis
Tumor-induced angiogenesis is the process by which new blood vessels develop from existing vasculature, through endothelial cell sprouting and proliferation (S16 Fig). This process is essential for tumor growth and spread [94]. Solid tumors are known to progress through two distinct phases of growth: the avascular phase and the vascular phase [95]. The transition from the avascular stage to the vascular stage, wherein the tumor possesses the ability to invade surrounding tissue and metastasis to distant parts of the body, depends on the ability of the tumor to induce new blood vessels from the surrounding tissue to sprout towards and then gradually surround and penetrate the tumor, thus providing it with an adequate blood supply and microcirculation. Tumor-induced angiogenesis is believed to start when a small avascular tumor exceeds a critical diameter (~2mm) [95]. At this stage, the tumor cells lacking nutrients and oxygen. In response, the tumor cells secrete a number of tumor angiogenic factors (TAF), such as VEGF [25]. Endothelial cells (EC) respond to the TAF concentration, forming sprouts, proliferating and migrating towards the tumor [62]. It takes approximately 10–21 days for the growing network to link the tumor to the parent vessel [96, 97].
In our ABM model, we also simulated tumor-induced angiogenesis. As shown in S4 Fig, the prostate tumor at the start point is in the avascular phase, and the initial tumor size is around ~1mm radius. With the distribution of VEGF in the tumor microenvironment [25], sprout branching and anastomosis were developed from an existing parent vessel (S16 Fig). The generation of new sprout (branching) occurs only from existing sprout tips. The newly formed sprouts are unlikely to branch immediately and that there must be a sufficient number of endothelial cells, near the sprout tip, for new sprouts to form (branching age is 18 hours [62]). We assume that the density of endothelial cells required for branching is inversely proportional to the concentration of TAF. In order to accomplish vascularization, the endothelial cells must proliferate and subsequently migrate the whole distance to the tumor [98]. After 4 weeks, the simulated prostate tumor will reach to ~4mm radius [99], and it will undergo castration. The details related to the modeling of tumor-induced angiogenesis were described in S1 Text.
T-cell activation and infiltration
Several studies have reported that tumor-associated antigens can induce T cell responses [100] and the production of antibodies, indicating that with the optimum balance of immune effectiveness over immune suppression [58, 59], and the immune system can fight cancer.
In this study, the proposed model simply simulated the cycle of generating an antitumor response. Dendritic cells engulf antigens from dying prostate tumor cells. As these dendritic cells migrate to the draining lymph node, they present the prostate-specific antigen (PSA) to naïve T cells (CD8+ effector T cells, regulatory T cells, etc.) for their clonal expansion and differentiation in the lymph node [101, 102]. Activated CD8+ effector T cells migrate to the tumor bed through the blood vessels or the lymphatic vessels [55]. When CD8+ T cells encounter tumor cells, they initiate programmed events leading to tumor cell death. Tregs similarly return to the tumor and suppress effector T cell killing efficiency to guard against overt inflammation and normal tissue damage. Within 2 days, these DCs lose their motility as they become integrated into the network of lymph-node DCs and die rapidly [56].
Fig 3 defines the entire simulated microenvironment consisting of tumor space and lymph node, which are connected with one lymphatic vessel. There is also a lymphatic vessel generated from lymph node, and it involves in the cycle of peripheral blood. In our model, activated CTLs and Tregs are migrated mainly through lymphatic vessel, moved to and accumulated in tumors. Each T cell agent is randomly selected and then infiltrated into the tumor tissue; its coordinates will be updated immediately. A schematic illustration of tumor growth and immune infiltration simulated by our HMSM model is displayed in S17 Fig.
Model implementation
The framework of the ABM model was designed using the conception of “Object-Oriented Programming” and achieved with C++. The ODE system of intracellular signaling pathways in PC was established with C and solved by the Fortran ODE Solver (DLSODE [103]), and called in the ABM model of HMSM (S1.10 Text). The ABM model was debugged and implemented under Linux environment on the cluster platform of Demon in Wake Forest Baptist Medical Center and Texas Advance Computing Center (TACC). All of the parameters in the ABM model were tuned by running the system 100 times for each candidate solution. The model with optimal parameters should fit the training data well. For addressing the stochastic results from the ABM, we evaluated the model outcomes after replicating simulations (repeat 100 times) on a fixed model. Both average and standard deviation were used to present the results.
Supporting information
(DOCX)
(TIF)
Twenty-three overexpressed ligand genes and 39 overexpressed receptor genes were identified from the dataset GSE46218, respectively. Eighteen overexpressed ligand genes and 26 receptor genes were identified from the dataset GSE38043. Based on the public ligand-receptor interaction database (iRefWeb) with a high confidence socore, a potential interaction pair was found: Treg→WNT5A→PC, and PC→TRAIL→Treg.
(TIF)
22RV1 cells were treated with 250ng WNT5A and RNA samples collected at 1, 3, 7, and 24 hours.
(TIF)
(TIF)
Two slices are presented: Y = 40, and Y = 50. Y is the Y axis (0≤Y≤100).
(TIF)
Two slices are presented: Y = 40, and Y = 50. Y is the Y axis (0≤Y≤100).
(TIF)
Two slices are presented: Y = 40, and Y = 50. Y is the Y axis (0≤Y≤100).
(TIF)
(TIF)
Primary tumor represents that the human tumor tissues were from PCa patients without treatment. CRPC (enzalutamide-naïve) indicates the human tumor tissues were from the PCa patients with CRPC occurrence who had not received enzalutamide. CRPC (progressed on enzalutamide represents the human tumor tissues were from the PCa patients who had received enzalutamide treatment after CRPC occurrence.
(TIF)
Sensitivity analysis was performed by measuring the impact of a small perturbation (5% increase) of individual 34 key parameters on the tumor cell (PC) population.
(TIFF)
“Control” denotes the simulation without perturbation. The results are comparable with those in Fig 6L. The variability of the average value is in the range of -1.1139% to 1.7024%.
(TIF)
The fold change of tumor growth on day 35 (5 weeks) after castration was examined by a 5% increase or decrease in the individual cut-off value in the migration rules. “Control” denotes the simulation without perturbation. The results are comparable with those in Fig 6L. The variability of the average value for increase or decrease is in the range [-1.07%, 0.12%] and [-1.94%, 1.26%], respectively.
(TIF)
(A) The fold change of TAM population after castration relative to pre-castration. (B) The fold change of CSF1 expression after castration relative to pre-castration. (C-D) the fold change of IL10 and VEGF expressions after castration relative to pre-castration. (E-F) The fold change of TAM population and VEGF expression after the treatment with castration plus PLX comparted to castraton only. (G) The fold change of Treg population in lymph nodes after castration relative to pre-castration. (H) The fold change of Treg population in lymph nodes after the treatment with castration plus IL-2 neutralization compared to castration only. (I-J) The fold change of CD8+ and Treg population in tumor space after castration relative to pre-castration. (K) The fold change of tumor growth after castration relative to pre-castration. (L) The fold change of tumor growth after treatment with castration plus PLX compared to pre-castration. (M) The fold change of tumor growth after treatment with castration plus EGFR inhibitor to castration only.
(TIF)
(TIF)
A cell at position Xi searches for the candidate locations within the distance R, and several empty positions (red dots) are identified. The probability (M) of a cell moving from Xi to Xj was determined by: 1) the moving offset (|di|); 2) the number of occupied cells (blue dots) around the new position; and 3) the type of the occupied cells. In our model, R equals 2 for migration, and 1 for proliferation.
(TIF)
If 0≤x≤14 or 86≤x≤100, ρ = 0; otherwise, ρ follows a normal distribution (14<x<86).
(TIFF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Acknowledgments
The authors acknowledge the Texas Advanced Computing Center (TACC) at the University of Texas at Austin (http://www.tacc.utexas.edu) and the DEMON high performance computing (HPC) cluster at Wake Forest University School of Medicine for providing HPC resources that have contributed to the research results reported within this paper.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by National Institutes of Health U01CA166886, U01AR069395-01A1, R01CA241930 and R01GM123037 (Zhou). Funding for open access charge: Dr Carl V. Vartian Chair Professorship Funds to Dr. Zhou from the University of Texas Health Science Center at Houston. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. 10.3322/caac.20073 . [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. 10.3322/caac.21166 . [DOI] [PubMed] [Google Scholar]
- 3.Fusi A, Procopio G, Della Torre S, Ricotta R, Bianchini G, Salvioni R, et al. Treatment options in hormone-refractory metastatic prostate carcinoma. Tumori. 2004;90(6):535–46. WOS:000226741000001. [DOI] [PubMed] [Google Scholar]
- 4.Devlin HL, Mudryj M. Progression of prostate cancer: multiple pathways to androgen independence. Cancer Lett. 2009;274(2):177–86. 10.1016/j.canlet.2008.06.007 . [DOI] [PubMed] [Google Scholar]
- 5.Lamont KR, Tindall DJ. Minireview: Alternative activation pathways for the androgen receptor in prostate cancer. Mol Endocrinol. 2011;25(6):897–907. 10.1210/me.2010-0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Amm J, Aragon-Ching JB. The changing landscape in the treatment of metastatic castration-resistant prostate cancer. Ther Adv Med Oncol. 2013;5(1):25–40. 10.1177/1758834012458137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene. 2015;34(14):1745–57. 10.1038/onc.2014.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taplin ME, Rajeshkumar B, Halabi S, Werner CP, Woda BA, Picus J, et al. Androgen receptor mutations in androgen-independent prostate cancer: Cancer and Leukemia Group B Study 9663. J Clin Oncol. 2003;21(14):2673–8. 10.1200/JCO.2003.11.102 . [DOI] [PubMed] [Google Scholar]
- 9.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. 10.1016/j.ccr.2010.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. 10.1038/ng.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo ZY, Yang X, Sun F, Jiang RC, Linn DE, Chen HG, et al. A Novel Androgen Receptor Splice Variant Is Up-regulated during Prostate Cancer Progression and Promotes Androgen Depletion-Resistant Growth. Cancer Research. 2009;69(6):2305–13. 10.1158/0008-5472.CAN-08-3795 WOS:000264541300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drake CG. Prostate cancer as a model for tumour immunotherapy. Nat Rev Immunol. 2010;10(8):580–93. Epub 2010/07/24. 10.1038/nri2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amato RJ, Stepankiw M. Clinical Efficacy of TroVax in the Treatment of Progressive Castration-resistant Prostate Cancer. Clin Med Insights Oncol. 2012;6:67–73. Epub 2012/01/19. 10.4137/CMO.S7654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med. 2015;372(26):2521–32. Epub 2015/04/22. 10.1056/NEJMoa1503093 . [DOI] [PubMed] [Google Scholar]
- 15.Maia MC, Hansen AR. A comprehensive review of immunotherapies in prostate cancer. Crit Rev Oncol Hematol. 2017;113:292–303. Epub 2017/04/22. 10.1016/j.critrevonc.2017.02.026 . [DOI] [PubMed] [Google Scholar]
- 16.Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, van den Eertwegh AJ, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2014;15(7):700–12. Epub 2014/05/17. 10.1016/S1470-2045(14)70189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh P, Pal SK, Alex A, Agarwal N. Development of PROSTVAC immunotherapy in prostate cancer. Future Oncol. 2015;11(15):2137–48. Epub 2015/08/04. 10.2217/fon.15.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redman JM, Gulley JL, Madan RA. Combining immunotherapies for the treatment of prostate cancer. Urol Oncol. 2017;35(12):694–700. Epub 2017/11/18. 10.1016/j.urolonc.2017.09.024 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasero C, Gravis G, Guerin M, Granjeaud S, Thomassin-Piana J, Rocchi P, et al. Inherent and Tumor-Driven Immune Tolerance in the Prostate Microenvironment Impairs Natural Killer Cell Antitumor Activity. Cancer Res. 2016;76(8):2153–65. Epub 2016/05/20. 10.1158/0008-5472.CAN-15-1965 . [DOI] [PubMed] [Google Scholar]
- 20.Escamilla J, Schokrpur S, Liu CN, Priceman SJ, Moughon D, Jiang ZY, et al. CSF1 Receptor Targeting in Prostate Cancer Reverses Macrophage-Mediated Resistance to Androgen Blockade Therapy. Cancer Research. 2015;75(6):950–62. 10.1158/0008-5472.CAN-14-0992 WOS:000351941400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akins EJ, Moore ML, Tang S, Willingham MC, Tooze JA, Dubey P. In situ vaccination combined with androgen ablation and regulatory T-cell depletion reduces castration-resistant tumor burden in prostate-specific pten knockout mice. Cancer Res. 2010;70(9):3473–82. 10.1158/0008-5472.CAN-09-2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang S, Moore ML, Grayson JM, Dubey P. Increased CD8(+) T-cell Function following Castration and Immunization Is Countered by Parallel Expansion of Regulatory T Cells. Cancer Research. 2012;72(8):1975–85. 10.1158/0008-5472.CAN-11-2499 WOS:000302905700009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tridane A, Kuang Y. Modeling the interaction of cytotoxic T lymphocytes and influenza virus infected epithelial cells. Math Biosci Eng. 2010;7(1):171–85. 10.3934/mbe.2010.7.171 . [DOI] [PubMed] [Google Scholar]
- 24.Eikenberry S, Thalhauser C, Kuang Y. Tumor-immune interaction, surgical treatment, and cancer recurrence in a mathematical model of melanoma. PLoS Comput Biol. 2009;5(4):e1000362 10.1371/journal.pcbi.1000362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Zhang L, Jing C, Ye G, Wu H, Miao H, et al. Multi-scale agent-based modeling on melanoma and its related angiogenesis analysis. Theor Biol Med Model. 2013;10:41 Epub 2013/06/27. 10.1186/1742-4682-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang L, van de Ven AL, Guo D, Andasari V, Cristini V, Li KC, et al. Computational modeling of 3D tumor growth and angiogenesis for chemotherapy evaluation. PLoS One. 2014;9(1):e83962 10.1371/journal.pone.0083962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng H, Zhao W, Tan H, Ji Z, Li J, Li K, et al. Prediction of treatment efficacy for prostate cancer using a mathematical model. Sci Rep. 2016;6:21599 10.1038/srep21599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gravina GL, Marampon F, Piccolella M, Biordi L, Ficorella C, Motta M, et al. Antitumor effects of carnertinib in castration resistant prostate cancer models: a comparative study with erlotinib. Prostate. 2011;71(14):1481–91. Epub 2011/03/30. 10.1002/pros.21363 . [DOI] [PubMed] [Google Scholar]
- 29.Huen NY, Pang AL, Tucker JA, Lee TL, Vergati M, Jochems C, et al. Up-regulation of proliferative and migratory genes in regulatory T cells from patients with metastatic castration-resistant prostate cancer. Int J Cancer. 2013;133(2):373–82. Epub 2013/01/16. 10.1002/ijc.28026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang N NY, Flores-Morales A. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46218. 2013.
- 31.Qiao W, Wang W, Laurenti E, Turinsky AL, Wodak SJ, Bader GD, et al. Intercellular network structure and regulatory motifs in the human hematopoietic system. Molecular Systems Biology. 2014;10(7). 10.15252/msb.20145141 WOS:000340295800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda T, Hirata S, Fukushima S, Matsunaga Y, Ito T, Uchino M, et al. Dual effects of TRAIL in suppression of autoimmunity: the inhibition of Th1 cells and the promotion of regulatory T cells. J Immunol. 2010;185(9):5259–67. Epub 2010/10/06. 10.4049/jimmunol.0902797 . [DOI] [PubMed] [Google Scholar]
- 33.Lee GT, Kang DI, Ha YS, Jung YS, Chung J, Min K, et al. Prostate cancer bone metastases acquire resistance to androgen deprivation via WNT5A-mediated BMP-6 induction. Br J Cancer. 2014;110(6):1634–44. Epub 2014/02/13. 10.1038/bjc.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464(7287):374–9. Epub 2010/03/20. 10.1038/nature08815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Tindall DJ, Huang H. Modulation of androgen receptor by FOXA1 and FOXO1 factors in prostate cancer. Int J Biol Sci. 2014;10(6):614–9. Epub 2014/06/21. 10.7150/ijbs.8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee R, McGuinness DH, McCall P, Underwood MA, Seywright M, Orange C, et al. Upregulation of MAPK pathway is associated with survival in castrate-resistant prostate cancer. Br J Cancer. 2011;104(12):1920–8. Epub 2011/05/12. 10.1038/bjc.2011.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toren P, Kim S, Johnson F, Zoubeidi A. Combined AKT and MEK Pathway Blockade in Pre-Clinical Models of Enzalutamide-Resistant Prostate Cancer. PLoS One. 2016;11(4):e0152861 Epub 2016/04/06. 10.1371/journal.pone.0152861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel). 2014;6(3):1670–90. 10.3390/cancers6031670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez D, Vollmann EH, von Andrian UH. Mechanisms and consequences of dendritic cell migration. Immunity. 2008;29(3):325–42. Epub 2008/09/19. 10.1016/j.immuni.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seyfizadeh N, Muthuswamy R, Mitchell DA, Nierkens S, Seyfizadeh N. Migration of dendritic cells to the lymph nodes and its enhancement to drive anti-tumor responses. Crit Rev Oncol Hematol. 2016;107:100–10. Epub 2016/11/09. 10.1016/j.critrevonc.2016.09.002 . [DOI] [PubMed] [Google Scholar]
- 41.Shen CR, Yang WC, Chen HW. The fate of regulatory T cells: survival or apoptosis. Cell Mol Immunol. 2014;11(1):11–3. 10.1038/cmi.2013.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12(3):180–90. Epub 2012/02/22. 10.1038/nri3156 . [DOI] [PubMed] [Google Scholar]
- 43.Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: Potential targets of anti-cancer therapy. European Journal of Cancer. 2006;42(6):717–27. 10.1016/j.ejca.2006.01.003 WOS:000237377100005. [DOI] [PubMed] [Google Scholar]
- 44.Wyckoff J, Wang WG, Lin EY, Wang YR, Pixley F, Stanley ER, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Research. 2004;64(19):7022–9. 10.1158/0008-5472.CAN-04-1449 WOS:000224292400028. [DOI] [PubMed] [Google Scholar]
- 45.Karantanos T, Evans CP, Tombal B, Thompson TC, Montironi R, Isaacs WB. Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level. Eur Urol. 2015;67(3):470–9. 10.1016/j.eururo.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizokami A, Namiki M. Reconsideration of progression to CRPC during androgen deprivation therapy. J Steroid Biochem Mol Biol. 2015;145:164–71. 10.1016/j.jsbmb.2014.03.015 . [DOI] [PubMed] [Google Scholar]
- 47.Mizokami A, Koh E, Fujita H, Maeda Y, Egawa M, Koshida K, et al. The adrenal androgen Androstenediol is present in prostate cancer tissue after androgen deprivation therapy and activates mutated androgen receptor. Cancer Research. 2004;64(2):765–71. 10.1158/0008-5472.Can-03-0130 WOS:000188399300045. [DOI] [PubMed] [Google Scholar]
- 48.Labrie F, Dupont A, Belanger A. Complete androgen blockade for the treatment of prostate cancer. Important Adv Oncol. 1985:193–217. . [PubMed] [Google Scholar]
- 49.Labrie F. Intracrinology Mol Cell Endocrinol. 1991;78(3):C113–8. 10.1016/0303-7207(91)90116-a . [DOI] [PubMed] [Google Scholar]
- 50.Liao RS, Ma S, Miao L, Li R, Yin Y, Raj GV. Androgen receptor-mediated non-genomic regulation of prostate cancer cell proliferation. Transl Androl Urol. 2013;2(3):187–96. 10.3978/j.issn.2223-4683.2013.09.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu G, Vogel SM, Gao X, Javaid K, Hu G, Danilov SM, et al. Src phosphorylation of endothelial cell surface intercellular adhesion molecule-1 mediates neutrophil adhesion and contributes to the mechanism of lung inflammation. Arterioscler Thromb Vasc Biol. 2011;31(6):1342–50. Epub 2011/04/09. 10.1161/ATVBAHA.110.222208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji Z, Su J, Wu D, Peng H, Zhao W, Nlong Zhao B, et al. Predicting the impact of combined therapies on myeloma cell growth using a hybrid multi-scale agent-based model. Oncotarget. 2017;8(5):7647–65. 10.18632/oncotarget.13831 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Research. 2006;66(2):605–12. 10.1158/0008-5472.CAN-05-4005 WOS:000234915100003. [DOI] [PubMed] [Google Scholar]
- 54.Fang LY, Izumi K, Lai KP, Liang L, Li L, Miyamoto H, et al. Infiltrating Macrophages Promote Prostate Tumorigenesis via Modulating Androgen Receptor-Mediated CCL4-STAT3 Signaling. Cancer Research. 2013;73(18):5633–46. 10.1158/0008-5472.CAN-12-3228 WOS:000324806300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams JL, Smothers J, Srinivasan R, Hoos A. Big opportunities for small molecules in immuno-oncology. Nat Rev Drug Discov. 2015;14(9):603–22. 10.1038/nrd4596 . [DOI] [PubMed] [Google Scholar]
- 56.Bousso P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nat Rev Immunol. 2008;8(9):675–84. 10.1038/nri2379 . [DOI] [PubMed] [Google Scholar]
- 57.Chao DL, Davenport MP, Forrest S, Perelson AS. A stochastic model of cytotoxic T cell responses. Journal of Theoretical Biology. 2004;228(2):227–40. 10.1016/j.jtbi.2003.12.011 WOS:000221205400007. [DOI] [PubMed] [Google Scholar]
- 58.Bauer CA, Kim EY, Marangoni F, Carrizosa E, Claudio NM, Mempel TR. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J Clin Invest. 2014;124(6):2425–40. 10.1172/JCI66375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beyer M, Schultze JL. Regulatory T cells in cancer. Blood. 2006;108(3):804–11. 10.1182/blood-2006-02-002774 . [DOI] [PubMed] [Google Scholar]
- 60.Owen DL, Mahmud SA, Vang KB, Kelly RM, Blazar BR, Smith KA, et al. Identification of Cellular Sources of IL-2 Needed for Regulatory T Cell Development and Homeostasis. J Immunol. 2018;200(12):3926–33. Epub 2018/05/08. 10.4049/jimmunol.1800097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stokes CL, Lauffenburger DA. Analysis of the roles of microvessel endothelial cell random motility and chemotaxis in angiogenesis. J Theor Biol. 1991;152(3):377–403. 10.1016/s0022-5193(05)80201-2 . [DOI] [PubMed] [Google Scholar]
- 62.Anderson AR, Chaplain MA. Continuous and discrete mathematical models of tumor-induced angiogenesis. Bull Math Biol. 1998;60(5):857–99. 10.1006/bulm.1998.0042 . [DOI] [PubMed] [Google Scholar]
- 63.Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108(5):1571–9. 10.1182/blood-2006-02-004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–9. 10.1126/science.aae0491 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su J, Zhang L, Zhang W, Choi DS, Wen J, Jiang B, et al. Targeting the biophysical properties of the myeloma initiating cell niches: a pharmaceutical synergism analysis using multi-scale agent-based modeling. PLoS One. 2014;9(1):e85059 10.1371/journal.pone.0085059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. 10.1158/2159-8274.CD-10-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cathomas R, Rothermundt C, Klingbiel D, Bubendorf L, Jaggi R, Betticher DC, et al. Efficacy of cetuximab in metastatic castration-resistant prostate cancer might depend on EGFR and PTEN expression: results from a phase II trial (SAKK 08/07). Clin Cancer Res. 2012;18(21):6049–57. Epub 2012/09/15. 10.1158/1078-0432.CCR-12-2219 . [DOI] [PubMed] [Google Scholar]
- 68.Festuccia C, Gravina GL, Biordi L, D'Ascenzo S, Dolo V, Ficorella C, et al. Effects of EGFR tyrosine kinase inhibitor erlotinib in prostate cancer cells in vitro. Prostate. 2009;69(14):1529–37. Epub 2009/06/30. 10.1002/pros.20995 . [DOI] [PubMed] [Google Scholar]
- 69.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1(3):279–88. Epub 2002/06/28. . [DOI] [PubMed] [Google Scholar]
- 70.Huang CL, Liu D, Nakano J, Ishikawa S, Kontani K, Yokomise H, et al. Wnt5a expression is associated with the tumor proliferation and the stromal vascular endothelial growth factor—an expression in non-small-cell lung cancer. J Clin Oncol. 2005;23(34):8765–73. Epub 2005/11/30. 10.1200/JCO.2005.02.2871 . [DOI] [PubMed] [Google Scholar]
- 71.Pukrop R, Krischer M. Changing views about personality disorders: Comment about the prospective studies CIC, CLPS, and MSAD. J Pers Disord. 2005;19(5):563–72; discussion 94–6. Epub 2005/11/09. 10.1521/pedi.2005.19.5.563 . [DOI] [PubMed] [Google Scholar]
- 72.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66(21):10439–48. Epub 2006/11/03. 10.1158/0008-5472.CAN-06-2359 . [DOI] [PubMed] [Google Scholar]
- 73.Yamamoto H, Oue N, Sato A, Hasegawa Y, Yamamoto H, Matsubara A, et al. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29(14):2036–46. Epub 2010/01/27. 10.1038/onc.2009.496 . [DOI] [PubMed] [Google Scholar]
- 74.Lunemann JD, Waiczies S, Ehrlich S, Wendling U, Seeger B, Kamradt T, et al. Death ligand TRAIL induces no apoptosis but inhibits activation of human (auto)antigen-specific T cells. J Immunol. 2002;168(10):4881–8. Epub 2002/05/08. 10.4049/jimmunol.168.10.4881 . [DOI] [PubMed] [Google Scholar]
- 75.Song K, Chen Y, Goke R, Wilmen A, Seidel C, Goke A, et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an inhibitor of autoimmune inflammation and cell cycle progression. J Exp Med. 2000;191(7):1095–104. Epub 2000/04/05. 10.1084/jem.191.7.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimberley FC, Screaton GR. Following a TRAIL: update on a ligand and its five receptors. Cell Res. 2004;14(5):359–72. Epub 2004/11/13. 10.1038/sj.cr.7290236 . [DOI] [PubMed] [Google Scholar]
- 77.Wang SH, Chen GH, Fan Y, Van Antwerp M, Baker JR, Jr. Tumor necrosis factor-related apoptosis-inducing ligand inhibits experimental autoimmune thyroiditis by the expansion of CD4+CD25+ regulatory T cells. Endocrinology. 2009;150(4):2000–7. Epub 2008/11/15. 10.1210/en.2008-1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Effect of MDV3100, an androgen receptor signaling inhibitor (ARSI), on overall survival in patients with prostate cancer postdocetaxel: Results from the phase III AFFIRM study. Journal of Clinical Oncology. 2012;30(5). WOS:000208892400002. [Google Scholar]
- 79.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. Epub 2015/05/23. 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rathkopf DE, Smith MR, Ryan CJ, Berry WR, Shore ND, Liu G, et al. Androgen receptor mutations in patients with castration-resistant prostate cancer treated with apalutamide. Ann Oncol. 2017;28(9):2264–71. Epub 2017/06/22. 10.1093/annonc/mdx283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prekovic S, van Royen ME, Voet AR, Geverts B, Houtman R, Melchers D, et al. The Effect of F877L and T878A Mutations on Androgen Receptor Response to Enzalutamide. Mol Cancer Ther. 2016;15(7):1702–12. Epub 2016/05/20. 10.1158/1535-7163.MCT-15-0892 . [DOI] [PubMed] [Google Scholar]
- 82.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–6. Epub 2015/09/19. 10.1126/science.aab0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tucker JA, Jochems C, Gulley JL, Schlom J, Tsang KY. Immunotherapy: shifting the balance of cell-mediated immunity and suppression in human prostate cancer. Cancers (Basel). 2012;4(4):1333–48. 10.3390/cancers4041333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Solovyev A, Mi Q, Tzen YT, Brienza D, Vodovotz Y. Hybrid equation/agent-based model of ischemia-induced hyperemia and pressure ulcer formation predicts greater propensity to ulcerate in subjects with spinal cord injury. PLoS Comput Biol. 2013;9(5):e1003070 10.1371/journal.pcbi.1003070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao W, Kridel S, Thorburn A, Kooshki M, Little J, Hebbar S, et al. Fatty acid synthase: a novel target for antiglioma therapy. Br J Cancer. 2006;95(7):869–78. Epub 2006/09/14. 10.1038/sj.bjc.6603350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kryczek I, Zou L, Rodriguez P, Zhu G, Wei S, Mottram P, et al. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J Exp Med. 2006;203(4):871–81. 10.1084/jem.20050930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30(5):636–45. Epub 2009/05/26. 10.1016/j.immuni.2009.04.010 . [DOI] [PubMed] [Google Scholar]
- 90.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2(5):415–22. 10.1038/87720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naive CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat Immunol. 2001;2(5):423–9. 10.1038/87730 . [DOI] [PubMed] [Google Scholar]
- 92.Wodarz D, Thomsen AR. Effect of the CTL proliferation program on virus dynamics. Int Immunol. 2005;17(9):1269–76. 10.1093/intimm/dxh303 . [DOI] [PubMed] [Google Scholar]
- 93.Murali-Krishna K, Lau LL, Sambhara S, Lemonnier F, Altman J, Ahmed R. Persistence of memory CD8 T cells in MHC class I-deficient mice. Science. 1999;286(5443):1377–81. 10.1126/science.286.5443.1377 . [DOI] [PubMed] [Google Scholar]
- 94.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. 10.1016/s0092-8674(00)81683-9 . [DOI] [PubMed] [Google Scholar]
- 95.Macklin P, McDougall S, Anderson ARA, Chaplain MAJ, Cristini V, Lowengrub J. Multiscale modelling and nonlinear simulation of vascular tumour growth. Journal of Mathematical Biology. 2009;58(4–5):765–98. 10.1007/s00285-008-0216-9 WOS:000262482600011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paweletz N, Knierim M. Tumor-related angiogenesis. Crit Rev Oncol Hematol. 1989;9(3):197–242. . [DOI] [PubMed] [Google Scholar]
- 97.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438(7070):932–6. 10.1038/nature04478 . [DOI] [PubMed] [Google Scholar]
- 98.Sholley MM, Ferguson GP, Seibel HR, Montour JL, Wilson JD. Mechanisms of neovascularization. Vascular sprouting can occur without proliferation of endothelial cells. Lab Invest. 1984;51(6):624–34. . [PubMed] [Google Scholar]
- 99.Geng Q, Liu J, Gong Z, Chen S, Chen S, Li X, et al. Phosphorylation by mTORC1 stablizes Skp2 and regulates its oncogenic function in gastric cancer. Mol Cancer. 2017;16(1):83 10.1186/s12943-017-0649-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 2002;296(5574):1876–80. 10.1126/science.1070945 . [DOI] [PubMed] [Google Scholar]
- 101.De Angelis G, Rittenhouse HG, Mikolajczyk SD, Blair Shamel L, Semjonow A. Twenty Years of PSA: From Prostate Antigen to Tumor Marker. Rev Urol. 2007;9(3):113–23. [PMC free article] [PubMed] [Google Scholar]
- 102.Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10(5):556–69. 10.1016/j.stem.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mathew S, Sundararaj S, Mamiya H, Banerjee I. Regulatory interactions maintaining self-renewal of human embryonic stem cells as revealed through a systems analysis of PI3K/AKT pathway. Bioinformatics. 2014;30(16):2334–42. Epub 2014/04/30. 10.1093/bioinformatics/btu209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
Twenty-three overexpressed ligand genes and 39 overexpressed receptor genes were identified from the dataset GSE46218, respectively. Eighteen overexpressed ligand genes and 26 receptor genes were identified from the dataset GSE38043. Based on the public ligand-receptor interaction database (iRefWeb) with a high confidence socore, a potential interaction pair was found: Treg→WNT5A→PC, and PC→TRAIL→Treg.
(TIF)
22RV1 cells were treated with 250ng WNT5A and RNA samples collected at 1, 3, 7, and 24 hours.
(TIF)
(TIF)
Two slices are presented: Y = 40, and Y = 50. Y is the Y axis (0≤Y≤100).
(TIF)
Two slices are presented: Y = 40, and Y = 50. Y is the Y axis (0≤Y≤100).
(TIF)
Two slices are presented: Y = 40, and Y = 50. Y is the Y axis (0≤Y≤100).
(TIF)
(TIF)
Primary tumor represents that the human tumor tissues were from PCa patients without treatment. CRPC (enzalutamide-naïve) indicates the human tumor tissues were from the PCa patients with CRPC occurrence who had not received enzalutamide. CRPC (progressed on enzalutamide represents the human tumor tissues were from the PCa patients who had received enzalutamide treatment after CRPC occurrence.
(TIF)
Sensitivity analysis was performed by measuring the impact of a small perturbation (5% increase) of individual 34 key parameters on the tumor cell (PC) population.
(TIFF)
“Control” denotes the simulation without perturbation. The results are comparable with those in Fig 6L. The variability of the average value is in the range of -1.1139% to 1.7024%.
(TIF)
The fold change of tumor growth on day 35 (5 weeks) after castration was examined by a 5% increase or decrease in the individual cut-off value in the migration rules. “Control” denotes the simulation without perturbation. The results are comparable with those in Fig 6L. The variability of the average value for increase or decrease is in the range [-1.07%, 0.12%] and [-1.94%, 1.26%], respectively.
(TIF)
(A) The fold change of TAM population after castration relative to pre-castration. (B) The fold change of CSF1 expression after castration relative to pre-castration. (C-D) the fold change of IL10 and VEGF expressions after castration relative to pre-castration. (E-F) The fold change of TAM population and VEGF expression after the treatment with castration plus PLX comparted to castraton only. (G) The fold change of Treg population in lymph nodes after castration relative to pre-castration. (H) The fold change of Treg population in lymph nodes after the treatment with castration plus IL-2 neutralization compared to castration only. (I-J) The fold change of CD8+ and Treg population in tumor space after castration relative to pre-castration. (K) The fold change of tumor growth after castration relative to pre-castration. (L) The fold change of tumor growth after treatment with castration plus PLX compared to pre-castration. (M) The fold change of tumor growth after treatment with castration plus EGFR inhibitor to castration only.
(TIF)
(TIF)
A cell at position Xi searches for the candidate locations within the distance R, and several empty positions (red dots) are identified. The probability (M) of a cell moving from Xi to Xj was determined by: 1) the moving offset (|di|); 2) the number of occupied cells (blue dots) around the new position; and 3) the type of the occupied cells. In our model, R equals 2 for migration, and 1 for proliferation.
(TIF)
If 0≤x≤14 or 86≤x≤100, ρ = 0; otherwise, ρ follows a normal distribution (14<x<86).
(TIFF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.