Summary
Medulloblastoma is a malignant childhood cerebellar tumour comprised of distinct molecular subgroups. Whereas genomic characteristics of these subgroups are well defined, the extent to which cellular diversity underlies their divergent biology and clinical behaviour remains largely unexplored. We used single-cell transcriptomics to investigate intra- and inter-tumoural heterogeneity in twenty-five medulloblastomas spanning all molecular subgroups. WNT, SHH, and Group 3 tumours comprised subgroup-specific undifferentiated and differentiated neuronal-like malignant populations, whereas Group 4 tumours were exclusively comprised of differentiated neuronal-like neoplastic cells. SHH tumours closely resembled granule neurons of varying differentiation states that correlated with patient age. Group 3 and Group 4 tumours exhibited a developmental trajectory from primitive progenitor-like to more mature neuronal-like cells, whose relative proportions distinguished these subgroups. Cross-species transcriptomics defined distinct glutamatergic populations as putative cells-of-origin for SHH and Group 4 subtypes. Collectively, these data provide novel insights into the cellular and developmental states underlying subtype-specific medulloblastoma biology.
Introduction
Medulloblastoma (MB) comprises a series of molecularly and clinically diverse malignant childhood cerebellar tumours1. While advances in treatment have improved survival, many patients suffer from neurological sequelae or still succumb to their disease. Genomic studies of bulk patient cohorts have defined four consensus molecular subgroups (WNT, SHH, Group 3, and Group 4)2, each characterized by discrete genomic landscapes, patient demographics, and clinical phenotypes3–7. The association between genotypes, transcriptional profiles, and patient age at diagnosis suggests that distinct MB subgroups arise from the transformation of different cell types in precise spatio-temporal patterns. Such genotype to cell type associations have been partially investigated for WNT- and SHH-MBs, which are thought to originate from cells in the extracerebellar lower rhombic lip8 and from cerebellar granule neuron progenitors (GNPs)9,10, respectively. In contrast, cellular origins of Group 3- and Group 4-MB remain unconfirmed. Overlapping transcriptional and epigenetic signatures observed in bulk profiling studies have consistently hampered definitive classification of Group 3 and Group 4 tumours and suggest they may share common developmental origins3,11. Thus, a better understanding of MB cellular composition and substructure according to subgroup is a critical goal, especially for poorly characterized Group 3 and Group 4.
Single-cell RNA-seq (scRNA-seq) has emerged as a powerful method to comprehensively characterize cellular states within healthy and diseased tissues12. While in central nervous system malignancies scRNA-seq has been applied to decipher adult and pediatric gliomas13–16, such approaches have yet to be deployed across MB subgroups. Here, we applied full-length scRNA-seq across consensus MB subgroups to infer cellular trajectories, deconvolute bulk MB expression cohorts, and nominate developmental origins. We find that WNT, SHH, and Group 3 tumours exhibit subgroup-specific cellular trajectories that consist of malignant undifferentiated and differentiated neuronal-like populations, whereas Group 4 tumours recapitulate more differentiated populations of known lineage. Collectively, these data provide new insights into the molecular and cellular architecture of MB across all subgroups, with the potential to inform future studies aimed at improving patient outcomes.
Results
MB and cerebellar transcriptomes
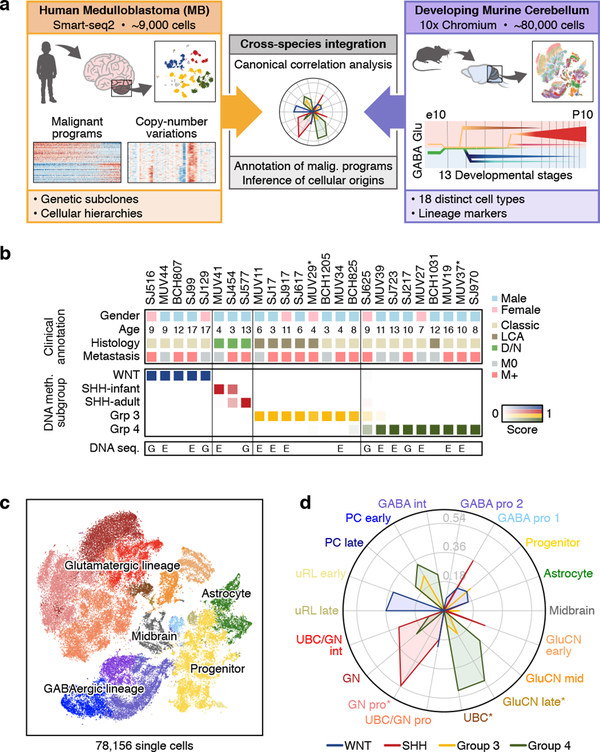
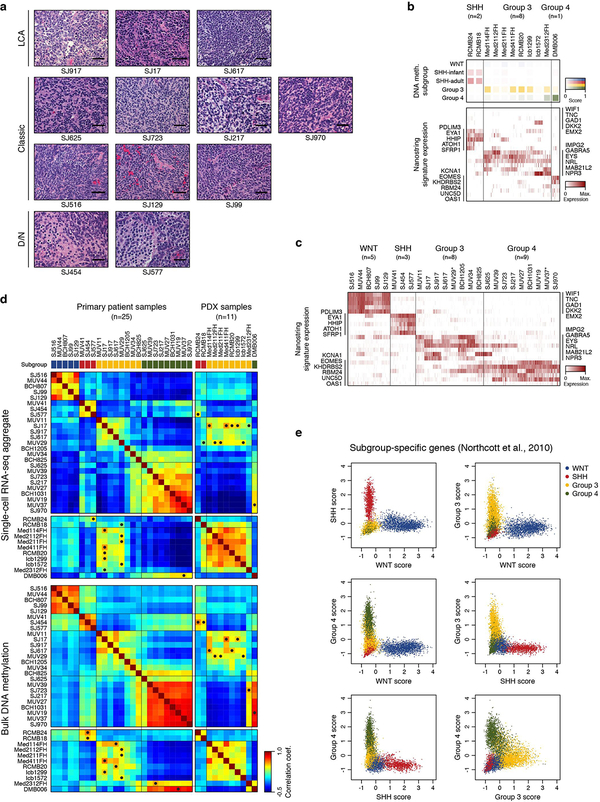
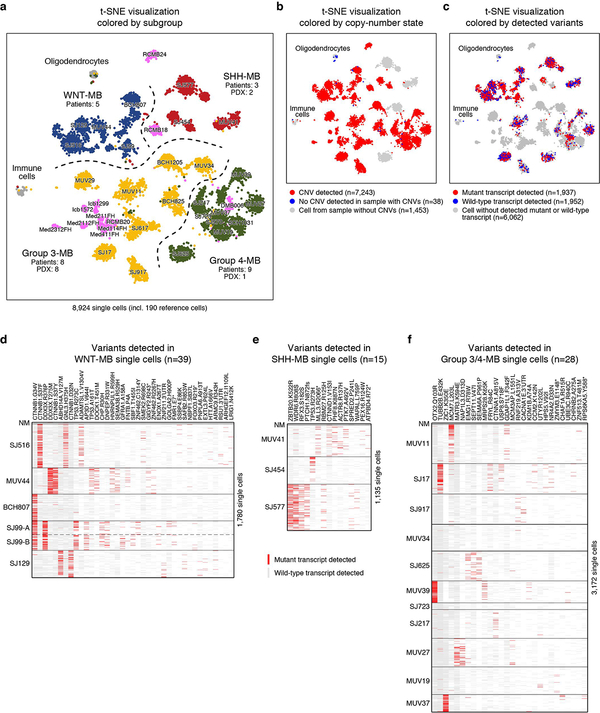
We prospectively obtained fresh surgical resections from 25 MB patients (23 diagnostic samples and 2 recurrences) and 11 patient derived xenograft (PDX) models (Figure 1a,b; ED Figure 1a,b; Supplementary Table 1a). Each tumour sample was molecularly classified using DNA methylation arrays17 (Figure 1b; ED Figure 1b). The majority of tumours were also characterized by whole-genome (n=5) or whole-exome (n=12) sequencing (Figure 1b; Supplementary Table 1b). To perform full-length scRNA-seq, cells were dissociated, sorted for viability and profiled using the Smart-seq2 protocol18 (see Methods). Analysis of known subgroup-specific signature genes19 demonstrated expected expression patterns (ED Figure 1b,c). Pairwise correlation of aggregated scRNA-seq and DNA methylation array data further substantiated subgroup classifications and PDX model fidelity (ED Figure 1d). Scoring single cells using published transcriptional signatures revealed that WNT and SHH tumours are exclusively comprised of cells scoring highly for their respective signatures. Conversely, cells derived from Group 3 and Group 4 tumours exhibited some degree of transcriptional overlap (ED Figure 1e). In total, 8,734 single cells passed quality controls, with a median of 4,561 genes detected per cell (Supplementary Table 1a).
Figure 1 |. Integrated analysis of MB and cerebellar single-cell transcriptomes.
a, Summary of human MB and developing murine cerebellar scRNA-seq datasets. b, Clinical and molecular details of the MB single-cell cohort. Asterisks indicate recurrent MB samples (n=2/25). Samples subjected to whole-genome (G) or whole-exome (E) sequencing are indicated. c, t-SNE representation of the cerebellar scRNA-seq dataset. Colors represent assigned cerebellar cell types. d, Radar plot showing the CCA coefficients between each murine cerebellar cell type (n=18) and bulk human MB expression data3. Asterisks indicate significant correlations (FDR corrected permutation test, P<0.05).
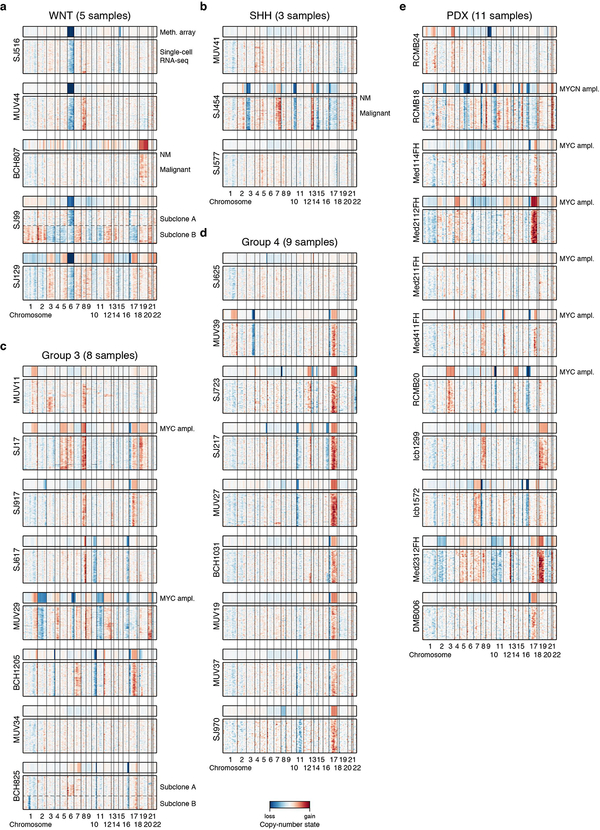
To classify single cells into malignant and non-malignant subsets, we used two complementary strategies. First, we inferred genome-wide CNVs from the scRNA-seq data as previously described13 (see Methods). This analysis identified large-scale genomic gains and losses in most patient samples (21/25), including hallmark alterations such as monosomy 6 (WNT) and isochromosome 17q (Groups 3 and 4; ED Figure 2a–e). Few cells (n=36) from these patients lacked discernable CNVs (see Methods). Second, we clustered single cells across all samples according to their transcriptional profiles. A minority of single cells in our cohort clustered with reference immune cells (n=6) or oligodendrocytes (n=22). (ED Figure 3a,b). All cells that lacked CNVs and/or clustered with normal reference populations were deemed as non-malignant and excluded from further analysis (n=43). Across individual tumours, 96–100% of cells were classified as malignant, consistent with prior estimates of high MB tumour cell fractions based on genome sequencing20. We further validated these assignments by quantifying genetic mutations identified by bulk tumour DNA sequencing in our scRNA-seq data (1,937 mutant and 1,952 wild-type transcripts detected; see Methods; ED Figure 3c–f).
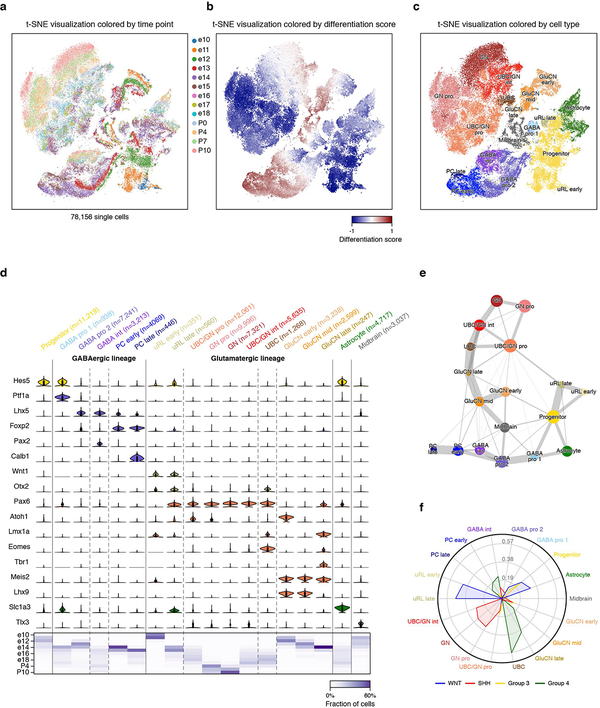
To relate MB single-cell profiles to normal developmental hierarchies, we leveraged recently generated scRNA-seq data for murine cerebellar development spanning 13 embryonic and early postnatal time points (total of 78,156 single cells; Figure 1a,c; ED Figure 4a–e)21. Canonical correlation analysis (CCA; see Methods) facilitated cross-species comparisons between our murine cerebellar single-cell, human MB single-cell, and bulk3 expression datasets. SHH-MB was highly correlated with GNP populations (cosine distance=0.54), consistent with literature9,10 supporting GNPs as the cell-of-origin for this subgroup (Figure 1d; ED Figure 4f). Notably, Group 4-MB was highly correlated with unipolar brush cells (UBC; cosine distance=0.50) and glutamatergic cerebellar nuclei (GluCN; cosine distance=0.49). In contrast, we did not detect high-confidence correlations between any cerebellar populations and either WNT or Group 3 subgroups.
Malignant trajectories within WNT-MB
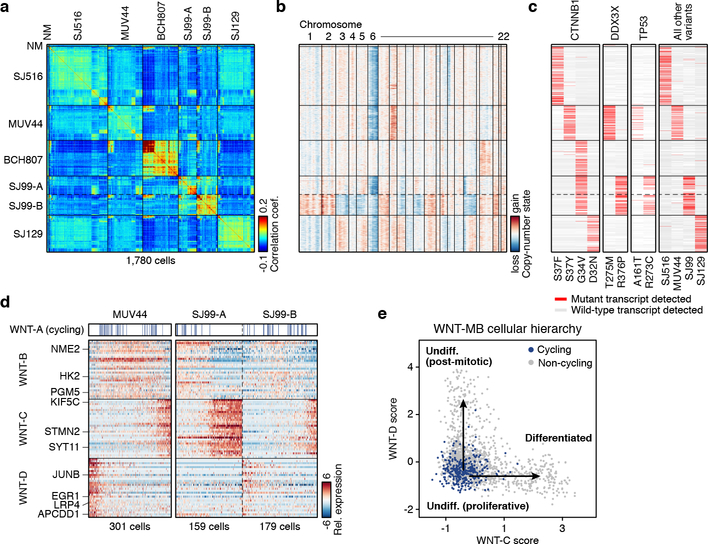
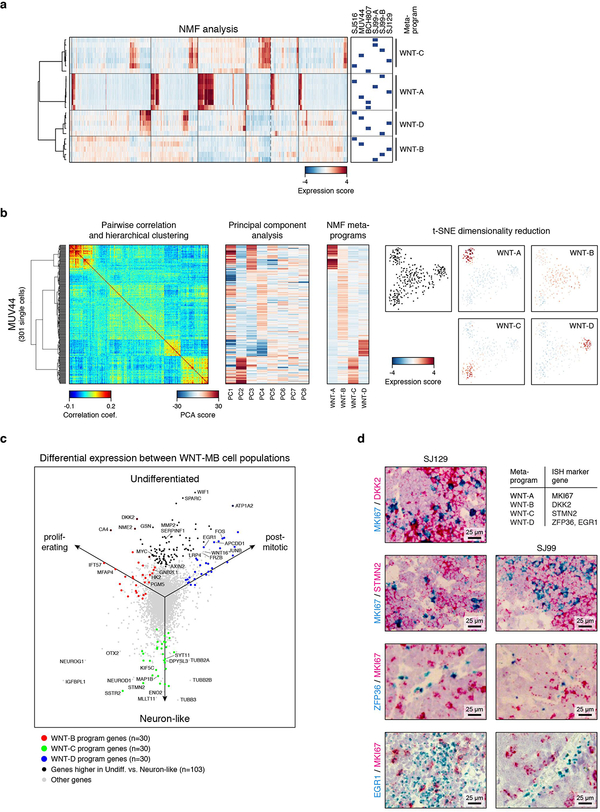
WNT subgroup patients account for ~10% of childhood MBs and have an excellent prognosis22. Somatic CTNNB1 mutations or germline APC mutations, both of which drive constitutive WNT signaling, are found in virtually all WNT-MBs3,23. Five WNT tumours were included in our dataset. Pairwise correlation analysis revealed multiple distinct transcriptional states that were consistently identified within these tumours (Figure 2a). Inferring CNVs from our scRNA-seq data identified four cases with monosomy 6 – a stereotypic genomic feature of this subgroup (Figure 2b; ED Figure 2a). The fifth case (BCH807) exhibited chromosome 19 gain and was negative for nuclear beta-catenin by immunohistochemistry (IHC; data not shown), both of which are atypical characteristics for this subgroup despite high-confidence molecular classification as WNT-MB (Figure 1b). SJ99 exhibited striking heterogeneity at both a transcriptional and genetic level, with evidence for two distinct subclones. Subclone SJ99-A exhibited monosomy 6 and chromosome 17p loss, whereas subclone SJ99-B exhibited broad gains and losses affecting nearly every chromosome. Investigation of genetically supported SNVs confirmed expression of mutant transcripts in 57.2% of cells (including key WNT-MB driver genes CTNNB1, DDX3X, and TP53; Figure 2c).
Figure 2 |. Intratumoural heterogeneity in WNT-MB.
a, Pairwise correlations between the expression profiles of all WNT-MB cells (n=1,780). Cells are ordered by hierarchical clustering within each sample or genetic subclone. NM, non-malignant. b, Single-cell derived CNVs by chromosome (columns). Cells are ordered as in panel (a). c, Mutant and wild-type transcripts detected in single cells. Cells are ordered as in panel (a). d, Relative expression of 90 genes representing WNT-MB meta-program (rows) across cells from MUV44 and SJ99. Cells positive for the cell cycle program (WNT-A) are indicated. e, Scatterplot of the WNT-C and WNT-D meta-program expression scores for all WNT-MB cells.
Non-negative matrix factorization (NMF) was applied to define underlying transcriptional programs specific to each tumour (ED Figure 5a,b; Supplementary Table 2a; see Methods). This analysis uncovered remarkably similar programs in all five WNT-MBs, which we grouped accordingly into four meta-programs (WNT-A/B/C/D). To interpret the characteristics of each meta-program, we evaluated their underlying gene signatures. WNT-A contained numerous markers of cell cycle activity (e.g. TOP2A, CDK1, and RRM2; P<0.001, Fisher’s exact test; Supplementary Table 2b). WNT-C was characterized by markers of neurogenesis/neuronal differentiation (STMN2, KIF5C, SYT11; P<0.001; Figure 2d). WNT-B consisted of ribosomal and metabolic genes (NME2, HK2, PGM5), while WNT-D contained select WNT pathway genes (LRP4, APCDD1) and immediate early response genes (JUNB, EGR1; Figure 2d). Cells scoring highest for both WNT-B and WNT-D expressed elevated levels of additional canonical WNT pathway genes (DKK2, AXIN2, WIF1) and MYC (ED Figure 5c; Supplementary Table 2c). We thus interpreted that these meta-programs reflect cell cycle activity (WNT-A), neuronal-like differentiation (WNT-C), and two WNT-driven states (WNT-B and WNT-D), with WNT-B characterized by elevated protein biosynthesis and metabolism (Figure 2d). RNA in situ hybridization (ISH) performed on the same tumours validated the expression of specific meta-program marker genes in subpopulations of cells (ED Figure 5d). Moreover, scoring each cell in our cohort for these four meta-programs defined a putative developmental trajectory for WNT-MB, with cell cycle activity restricted to cells that were both WNT-B high and WNT-C/D low (Fisher’s exact test, P<0.001), suggesting that this subpopulation fuels WNT-MB growth (Figure 2e). Importantly, each meta-program was identified in at least four samples (ED Figure 5a), suggesting that they reflect shared features of WNT-MB.
Developmental trajectories within SHH-MB
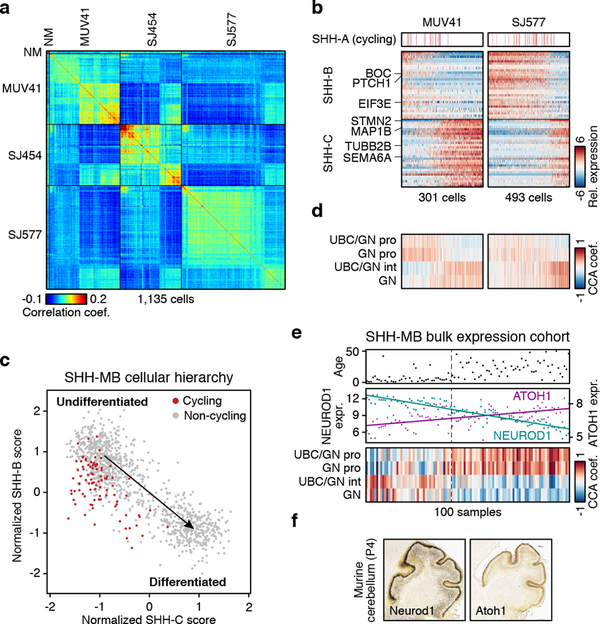
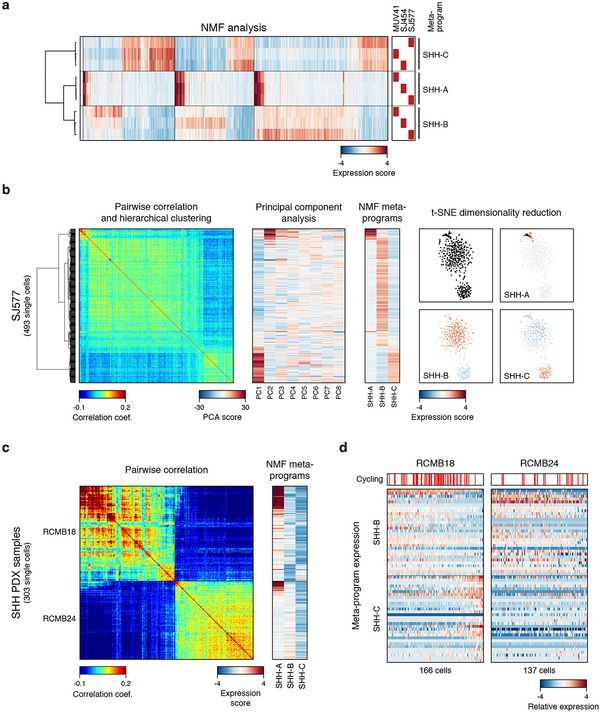
As the dominant subgroup in both infants (≤3yrs) and adults (≥18yrs)22, SHH-MB accounts for about one third of all MB patients. Outcomes are heterogeneous and associated with underlying genetics, demographics, and clinical features24. Our dataset included three SHH-MB patients, ranging from 3–13yrs (Figure 1b). Pairwise correlation and unsupervised NMF analysis uncovered three transcriptional programs (SHH-A/B/C) shared between these tumours (Figure 3a; ED Figure 6a,b; Supplementary Table 2a,b). SHH-A contained markers of cell cycle activity (e.g. TOP2A, CDK1, and RRM2; P<0.001, Fisher’s exact test). SHH-B was enriched for ribosomal genes and translational initiation/elongation factors (EIF3E, EEF1A1; P<0.001), and markers of canonical SHH signaling (PTCH1, BOC; P<0.001; Figure 3b). SHH-C was defined by markers of neuronal differentiation (STMN2, MAP1B, TUBB2B, SEMA6A; P<0.001; Figure 3b). We interpret that these programs reflect cell cycle activity (SHH-A), undifferentiated progenitors (SHH-B), and more differentiated neuronal-like programs (SHH-C). Scoring each SHH-MB cell for these programs defined a putative developmental trajectory, with proliferating cells restricted to undifferentiated progenitors (Figure 3c). These respective programs were partially recapitulated in SHH subgroup PDX models (ED Figure 6c,d).
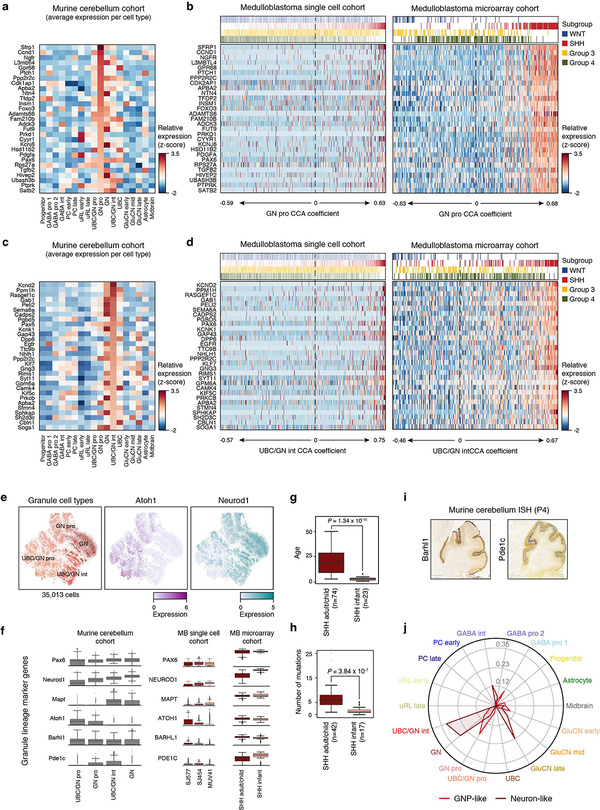
Figure 3 |. Age-associated developmental hierarchies in SHH-MB.
a, Pairwise correlations between the expression profiles of all SHH-MB cells (n=1,135). NM, non-malignant. b, Relative expression of 60 genes representing SHH-MB meta-program across cells from MUV41 and SJ577. Cells positive for the cell cycle program (SHH-A) are indicated. c, Scatterplot of the normalized SHH-B and SHH-C meta-program expression scores for all SHH-MB cells. d, CCA coefficients between SHH-MB single-cells and murine glutamatergic cell types. Cells are ordered as in panel (b). e, Patient age (upper panel), expression levels of ATOH1 and NEUROD1 (middle panel), and CCA coefficients between tumors and murine glutamatergic cell types (lower panel) for bulk SHH-MBs3 (n=100). f, Expression of Neurod1 (left panel) and Atoh1 (right panel) in the murine cerebellum (P4, postnatal day 4; data from the Allen Brain Atlas).
To investigate the developmental significance of these findings, we used CCA to compare SHH-MB meta-programs to murine cerebellar populations. SHH-B correlated with undifferentiated UBC/GNP and GNP populations, while SHH-C correlated with UBC/GN intermediate and differentiated granule neuron populations (Figure 3d; ED Figure 7a–d). To validate these observations in a larger cohort, we implemented a focused analysis of UBC, GNP, and granule neuron populations, assessing correlations between these cell types and bulk SHH-MB expression profiles (Figure 3e; ED Figure 7e). This analysis broadly split SHH-MBs into two age-associated categories: infant tumours correlated with intermediate and mature granule neurons (marked by high expression of NEUROD1), whereas adult tumours correlated with GNPs and mixed UBCs/GNPs (marked by high expression of ATOH1; Figure 3e,f; ED Figure 7f–j). Together, our data suggest that infant and adult SHH-MBs are enriched for temporally distinct GNP (or UBC) populations and/or have distinct differentiation capacities, further substantiating their divergent biology25–27.
Malignant programs within Group 3/4-MB
Group 3 and Group 4 tumours account for ~60% of MB diagnoses and remain the least understood with respect to disease biology and developmental origins7. Group 3 tumours are frequently metastatic at diagnosis and typified by genomic amplification/over-expression of MYC, which is associated with unfavorable outcomes11,28. Group 4 tumours are metastatic at diagnosis in approximately one third of patients and harbor recurrent chromatin modifier alterations28,29. Recent bulk profiling studies have demonstrated marked molecular and clinical heterogeneity in Group 3/4, with a subset of tumours exhibiting overlapping molecular signatures that confound robust classification3,30,31.
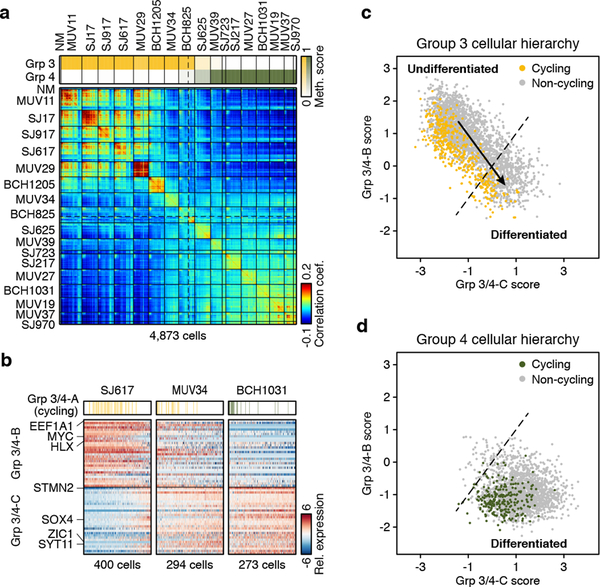
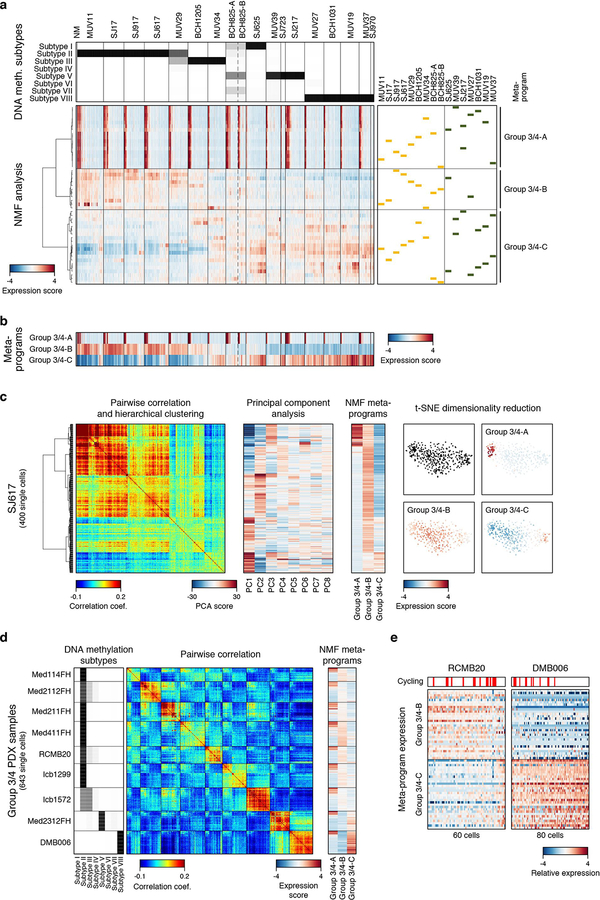
Given this prior knowledge, we performed a combined analysis of the scRNA-seq data for all 17 Group 3/4 tumours. Pairwise correlation analysis of single cells largely discriminated between subgroups, with a subset of ‘intermediate’ tumours exhibiting transcriptional ambiguity (MUV34, BCH825, SJ625; Figure 4a). NMF analysis performed on the combined series identified three distinct transcriptional programs (Group 3/4-A/B/C) (ED Figure 8a–c; Supplementary Table 2a,b). Group 3/4-A contained markers of cell cycle activity (e.g. TOP2A, CDK1, and RRM2; P<0.001, Fisher’s exact test). Group 3/4-B was primarily characterized by ribosomal and translational initiation/elongation genes (EIF3E, EEF1A1; P<0.001; Figure 4b) as well as by MYC and MYC target genes (e.g. HLX). Group 3/4-C contained well-recognized neuronal lineage markers (STMN2, SOX4, ZIC1, SYT11; P<0.01; Figure 4b). We interpret that these programs reflect cell cycle activity (Group 3/4-A), undifferentiated progenitor-like programs with high MYC activity (Group 3/4-B), and differentiated neuronal-like programs (Group 3/4-C; Figure 4b).
Figure 4 |. Malignant transcriptional programs within Group 3/4.
a, Pairwise correlations between the expression profiles of all Group 3/4-MB cells (n=4,873). NM, non-malignant. DNA Methylation-based subgroup prediction scores are indicated in the upper panel. b, Relative expression of 60 genes representing Group 3/4-MB meta-program from SJ617, MUV34, and BCH1031. Cells positive for the cell cycle program (Group 3/4-A) are indicated. c,d Scatterplots of the Group 3/4-B and Group 3/4-C meta-program expression scores for all Group 3 (c) and Group 4 (d) MB cells.
Scoring each Group 3/4-MB cell for these programs revealed striking patterns: prototypic Group 3 tumours were dominated (>88% of cells) by the undifferentiated progenitor-like program (Group 3/4-B), whereas the differentiated neuronal-like program (Group 3/4-C) was observed in virtually all cells (>95%) from prototypic Group 4 tumours, consistent with their neuronal differentiation phenotype11,28 (Figure 4c,d; Supplementary Table 2d). Group 3 tumours with MYC amplifications (SJ17, MUV29; ED Figure 2c) lacked neuronal differentiation altogether (<2% of cells), suggesting that oncogenic MYC expression may potentiate an undifferentiated progenitor-like state. Notably, Group 3/4 ‘intermediate’ tumours (MUV34, BCH825, SJ625) were comprised of a mixture of both malignant cell states, containing 12–20% of cells characterized by the undifferentiated program, with the remainder of cells characterized by the differentiated program. These transcriptional programs were also evident in 9 Group 3/4 PDX models (ED Figure 8d,e). Our results indicate that Group 3/4-MBs contain cells along a common continuum of neuronal differentiation.
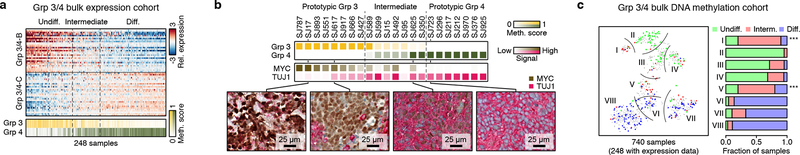
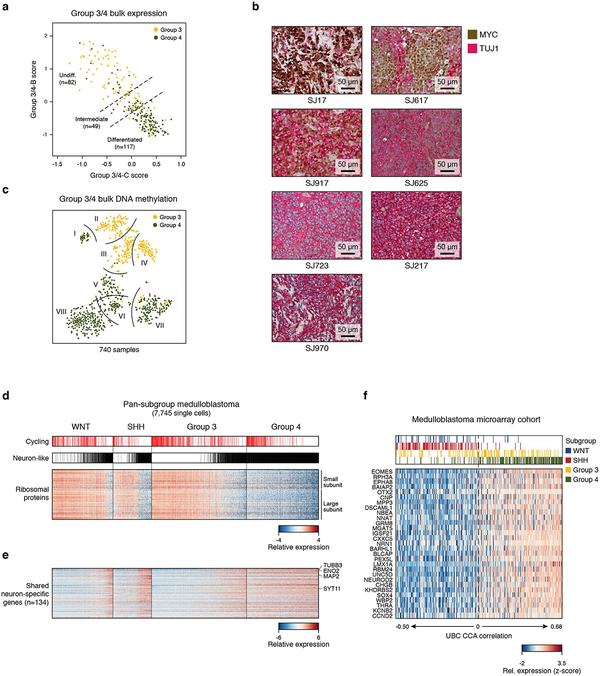
The observation that Group 3 and Group 4 MBs both contained cells scoring high for the neuronal-like differentiation program (Group 3/4-C) prompted us to examine whether varying proportions of cells with this shared program could underlie the molecular overlap seen in bulk tumour profiles. Quantifying the Group 3/4-B and C programs in bulk MB gene expression data3 (n=248 Group 3/4-MBs) recapitulated observations made in our single-cell cohort (Figure 5a). Sorting these profiles by their relative scores for these programs confirmed that prototypic Group 3-MBs were largely expressing characterized by the undifferentiated progenitor-like program (Group 3/4-B), while prototypic Group 4-MBs were dominated by the differentiated neuronal-like program (Group 3/4-C). A considerable fraction of tumours (18.8%) exhibited evidence of both programs (Figure 5a, ED Figure 9a). These intermediate tumours were characterized by elevated DNA methylation-based prediction scores (≥0.2) for both subgroups (OR=8.9, P<0.001, Fisher’s exact test). We validated these results by performing IHC on a series of twenty-two Group 3/4-MBs, using MYC and TUBB3 (TUJ1) as biomarkers of the Group 3/4-B and Group 3/4-C programs, respectively (Figure 5b; ED Figure 9b). Prototypical Group 3-MBs exhibited high expression of MYC and scant TUJ1 positivity, whereas prototypical Group 4-MBs were devoid of MYC-expressing cells and universally positive for TUJ1. Importantly, tumours classified as intermediate Group 3/4-MB by DNA methylation contained varying proportions of both MYC-expressing and TUJ1-expressing cells, consistent with our single-cell results.
Figure 5 |. Cellular composition of Group 3/4-MBs.
a, Relative expression of 60 genes representing Group 3/4-MB meta-programs across bulk Group 3/4-MBs. DNA Methylation-based subgroup prediction scores are indicated in the lower panel. b, DNA Methylation-based subgroup prediction scores (top panel) and relative IHC-based expression levels of MYC and TUJ1 (encoded by TUBB3; middle panel) in 22 Group 3/4-MBs. Four representative IHC images are shown (bottom panel). c, t-SNE representation of bulk Group 3/4-MBs classified according to published DNA methylation subtypes3,32 (n=740; left panel). Subtypes are delineated by curved lines. Samples are colored by differentiation state as defined in (a). Right panel shows quantification of undifferentiated, intermediate, or differentiated expression state per subtype. Asterisks indicate enrichment of intermediate samples (Fisher’s exact test, P<0.001).
We next investigated whether recently described3,32 DNA methylation-based subtypes of Group 3/4-MB were related to the meta-programs inferred from scRNA-seq. We found that DNA methylation subtypes I and V, both of which contain a mixture of Group 3- and Group 4-MBs, were significantly enriched for tumours with intermediate expression patterns (P<0.001, Fisher’s exact test; Figure 5c; ED Figure 9c). These results suggest that a continuum of cellular states accounts for the molecular substructure seen in Group 3/4 that complicates accurate consensus classification.
Lineage-specific correlates of Group 4-MB
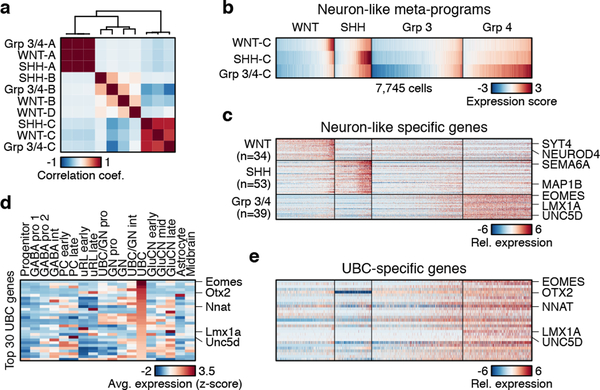
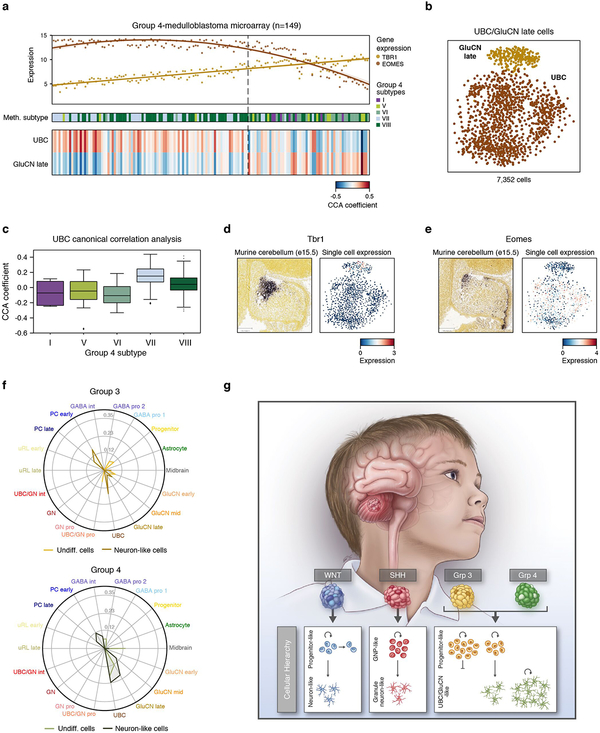
We next sought to compare and inter-relate the different subgroup-specific meta-programs. To this effect, we applied all observed meta-programs (n=10) to all 7,745 malignant cells in our dataset. Pairwise correlation of expression scores confirmed high similarity among cell cycle programs (WNT-A, SHH-A, Group 3/4-A; average r=0.99) (Figure 6a). The undifferentiated progenitor-like programs (WNT-B, SHH-B, Group 3/4-B) exhibited low correlations (average r=0.23), in agreement with their distinct underlying biology. In contrast, the neuronal-like differentiation programs (WNT-C, SHH-C, and Group 3/4-C) were highly correlated (average r=0.77; Figure 6a,b; ED Figure 9d), consistent with shared capacity for neuronal differentiation across subgroups. We reasoned that the neuronal-like differentiation programs defined in each subgroup consist of general neuronal differentiation markers, potentially masking markers of specific lineages. To elucidate markers that might inform developmental origins, we compared genes specific to neuronal-like cells in the different subgroups (n=260; relative to undifferentiated cell populations; see Methods). Half of these genes (52%) were shared between at least two subgroups and included general markers of neuronal differentiation (e.g. ENO2, SYT11, TUBB3, MAP2), while the remainder were exclusive to individual subgroups (13–20%; Figure 6c, ED Figure 9e, Supplementary Table 3). Glutamatergic lineage-specific transcription factors (TFs) EOMES and LMX1A ranked amongst the most differentially expressed genes specific to the Group 3/4-C program (Figure 6c; Supplementary Table 3). In mice, these TFs play essential roles in defining neuronal cell fates in the embryonic upper rhombic lip (uRL), including UBCs and glutamatergic cerebellar nuclei (GluCN), both of which are born out of the uRL during cerebellar morphogenesis33,34. Since our prior CCA analysis identified both UBCs and GluCN as being highly correlated with Group 4-MB expression datasets (Figure 1d, ED Figure 4f), we performed a deeper analysis into these correlations. Discriminatory UBC markers were specifically expressed in Group 4 single-cells and bulk tumour profiles, implicating a possible developmental link between UBCs and Group 4-MB (Figure 6d,e; ED Figure 9f). Similar results were observed for GluCN, although the highest correlations were limited to a subset of Group 4 tumours (ED Figure 10a–f). Collectively, these associations further implicate UBCs and GluCN of the embryonic cerebellum as candidate cells-of-origin for Group 4-MB.
Figure 6 |. Subgroup-specific transcriptional programs correlate with distinct neuronal lineages.
a, Pairwise correlation of expression scores of meta-programs defined separately in each subgroup and applied across cells from all subgroups. b, Expression scores for neuronal differentiation meta-programs across all cells (n=7,745). c, Relative expression levels of genes specific to neuron-like cells in different subgroups (n=126). Cells are ordered as in panel (b). d, Average expression levels of 30 UBC-associated genes (rows) in identified mouse cerebellar cell types (columns). Genes are ordered by their relative expression in UBCs. e, Relative expression of homologous genes in panel (d) in all cells from the MB single-cell cohort (columns). Cells are ordered as in panel (b).
Discussion
Despite extensive characterization of MB genomic landscapes, effective subgroup-specific therapies have yet to emerge, suggesting that a deeper understanding of the biological and cellular basis of MB is essential. This is particularly urgent for Group 3- and Group 4-MB, which often bear inferior outcomes. As a first challenge, these subgroups have proven difficult to accurately classify, confounded by transcriptional and epigenetic ambiguity. Our combined single-cell analysis of Group 3/4-MBs confirmed that prototypic Group 3-MBs are dominated by undifferentiated progenitor-like cells, whereas prototypic Group 4-MBs are comprised almost exclusively of more differentiated neuronal-like cells. Importantly, we identified a subset of ‘intermediate’ tumours characterized by varying proportions of both undifferentiated and more differentiated populations (ED Figure 10g). These findings offer a novel molecular and cellular explanation for the challenges associated with Group 3 and Group 4 sub-classification and provide a framework for future classifications that incorporate population heterogeneity.
Cellular origins for WNT- and SHH-MB have been mostly informed from genetically faithful mouse models8,9. Cross-species transcriptional analyses performed here confirmed significant correlations between SHH-MB and GNPs of variable differentiation states that were associated with patient age. Moreover, our analyses identified UBCs and GluCN as cellular correlates of Group 4-MB subtypes, building on prior studies that have implicated glutamatergic cellular origins for Group 44. For WNT-MB, we failed to identify significant correlation between malignant single-cell programs and cerebellar populations, compatible with an extracerebellar origin for this subgroup8. No significant correlations were detected between Group 3-MB and our cerebellar dataset. This observation may be attributed to transformation and cellular reprogramming driven by specific oncogenes (i.e. MYC) or may imply that Group 3-MBs have an extracerebellar origin. It is also plausible that our murine reference atlas was incomplete and lacked populations pertinent to either WNT- or Group 3-MB origins. Technical limitations of comparing single-cell datasets between species should not be underestimated, warranting future studies of the cellular correlates between human cerebellar and MB single cells.
In conclusion, our work provides the first cellular atlas of MB across all subgroups and a cross-species comparison to cerebellar development, shedding light on putative subgroup-specific origins. Our analyses also define the cellular states underlying each MB subgroup, disentangling determinants of intra- and inter-tumoural heterogeneity. These findings will enable future studies to assess translational opportunities and to evaluate the impact of therapeutic approaches on the spectrum of cellular states that drive MB.
Methods
Tissue sample collection and dissection
Human primary tumours
Patients and their parents at Boston Children’s Hospital, the Medical University of Vienna and St. Jude Children’s Research Hospital gave consent preoperatively according to Institutional Review Boards. Fresh tumours were collected at the time of surgery and processed directly. Tumour samples from Boston Children’s Hospital and the Medical University of Vienna were mechanically and enzymatically dissociated using a papain-based brain tumour dissociation kit (Miltenyi Biotec). Tumour samples from St. Jude Children’s Research Hospital were precut and dissociated for 30 minutes at 37°C in papain solution (10 units/ml, Worthington, LS003126) containing N-acetyl-L-cysteine (160 ug/ml, Sigma-Aldrich, A9165) and DNase I (12 ug/ml, Sigma-Aldrich, DN25), rinsed in Neurobasal medium and filtered using a 40 μm strainer.
Murine cerebellum
Murine cerebellar tissue from Crl:CD1 (ICR) mice at 13 distinct developmental time points was previously isolated21. Embryonic time points include each day between e10 to e18 and postnatal time points include P0, P4, P7, and P10. Two biological replicates were included at each time point and three at e14. Cerebella were dissociated as previously described.
Patient derived xenografts (PDXs)
PDXs were acquired from Robert Wechsler-Reya (Sanford Burnham Prebys Medical Discovery Institute), Xiao-Nan Li (Baylor College of Medicine) and the Brain Tumour Resource Laboratory (www.btrl.org). PDXs were injected into the cerebellum of NSG mice. Mice were observed daily and were euthanized at the apparition of signs of sickness including lethargy and neurological abnormalities. Low passage PDXs (<10) were dissected, precut and dissociated for 30 minutes at 37°C in papain solution (10 units/ml, Worthington, LS003126) containing N-acetyl-L-cysteine (160 ug/ml, Sigma-Aldrich, A9165) and DNase I (12 ug/ml, Sigma-Aldrich, DN25), rinsed in Neurobasal medium and filtered using a 40 μm strainer. The experiments were conducted in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals and according to the guidelines established by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee. Procedures in the protocol were approved by the Animal Care and Use Committee (ACUC) of SJCRH (Animal Assurance Number: A3077–01).
Fluorescence-activated cell sorting (FACS)
Dissociated tumour cells (from fresh primary tumours and PDXs) were resuspended in cold 1% bovine serum albumin in phosphate buffered saline (PBS-BSA1%). Cells were first stained first with CD45-Vioblue direct antibody conjugate (Miltenyi Biotec, 130–092–880) in PBS-BSA1% for 20 min at 4°C, washed and then costained with 1μM calcein AM (Life Technologies, C3100MP) and 0.33 μM TO-PRO-3 iodide (Life Technologies, T3605) in PBS-BSA1%. Sorting was performed with FACSAria Fusion (Becton Dickinson) using 488nm (calcein AM, 530/30 filter), 640nm (TO-PRO-3, 670/30 filter) and 405 nm (Vioblue, 450/50 filter) lasers. Non-stained controls were included with all tumours. CD45-positive cells were counterselected for the St. Jude samples only and viable medulloblastoma cells were identified by staining positive with calcein AM but negative for TO-PRO-3. Forward scatter area (FSC-A) versus side scatter width (SSC-W) criteria were used to discriminate doublets and select single cells. Single cells were sorted into 96-well plates containing cold TCL buffer (Qiagen, 1031576) containing 1% beta-mercaptoethanol, snap frozen on dry ice, and then stored at −80°C prior to whole transcriptome amplification, library preparation and sequencing.
Generation and processing of DNA methylation data
All single-cell patient and PDX samples were analysed using Illumina Infinium Methylation EPIC BeadChip arrays according to the manufacturer’s instructions. Data was generated from both freshly frozen and formalin-fixed paraffin-embedded (FFPE) tissue samples. Medulloblastoma subgroup predictions were obtained from a web-platform for DNA methylation-based classification of central nervous system tumours (www.molecularneuropathology.org, version 11b435). Resulting assignment of samples to WNT, SHH, Group 3, and Group 4 subgroups were used for all downstream analyses. A similar classification system was used for predicting medulloblastoma subtypes32. Copy-number variation (CNV) analysis from EPIC methylation array data was performed using the conumee Bioconductor package. Identified CNVs were compared to those predicted from the single-cell data (shown in EDF2).
Generation of whole-exome and whole-genome sequencing data
Human genomic whole-exome sequencing libraries were generated using the SureSelectXT kit specific for the Illumina HiSeq instrument (Agilent Technologies), followed by exome enrichment using the SureSelectXT Human All Exon V5 without UTRs bait set. The resulting exome enriched libraries of tumour and normal samples were then sequenced using paired end 100 cycle sequencing on a NovaSeq 6000 (Illumina) according to manufacturer’s instructions. Whole-genome sequencing libraries were constructed using the TruSeq DNA PCR-free sample preparation kit according to the manufacturer’s instructions. Tumour and normal samples were sequenced on an Illumina HiSeq 2500 instrument as previously described36. Somatic SNVs and INDELs were determined via the Mutect2 algorithm as implemented in GATK version 4.0. Coding and splice-related variants were subsequently annotated utilizing the Medal Ceremony annotation pipeline. Additionally, all reported somatic variants were manually curated in IGV.
Human single-cell RNA-seq data generation and processing
Whole transcriptome amplification, library construction and sequencing were performed as previously described following the Smart-seq2 modified protocol13. Expression levels were quantified as Ei,j=log2(TPMi,j/10+1), where TPMi,j refers to transcript-per-million for gene i in sample j, as calculated by RSEM37. TPM values were divided by 10 since we estimate the complexity of single cell libraries in the order of 100,000 transcripts and would like to avoid counting each transcript ~10 times, as would be the case with TPM, which may inflate the difference between the expression level of a gene in cells in which the gene is detected and those in which it is not detected.
To filter out poor quality cells, we first removed cells for which less than 2,500 genes were detected. For each processed 96-well plate, we then determined the average number of genes detected per cell minus two times its standard deviation. We then additionally filtered out the cells that were below that threshold. For the remaining cells, we calculated the aggregate expression of each gene as Ea(i)=log2(average(TPMi,1…n)+1), and excluded genes with Ea<4. In each subgroup (WNT, SHH, and Group 3/4), we defined relative expression by centering the expression levels, Eri,j=Ei,j-average[Ei,1…n] for the remaining cells and genes. On average, we detected ~4,500 genes per cell. Gene expression values were uploaded to the Gene Expression Omnibus (accession number GSE119926).
Pearson correlation coefficients between expression profiles of cells that passed quality filtering was calculated using centered gene expression levels (for each subgroup separately, shown in Figures 2a, 3a, 4a). Cells were ordered by hierarchical clustering using 1-correlation coefficient as the distance and Ward’s linkage, within each sample or genetic subclone (for samples SJ99 and BCH825).
Identification of copy number variations in single-cell data
Copy number variations were estimated as previously described13 by applying a moving average to the relative expression, with a sliding window of 100 genes within each chromosome after sorting genes by their chromosomal location (shown in EDF 2). Non-malignant tumour cells were determined by unsupervised clustering of all single-cell-derived copy-number profiles for each sample with 190 copy-number profiles derived from two non-malignant cell types (tumour-associated oligodendrocytes and immune cells14). For the majority of tumours (21/25), most of the cells did not cluster with the non-malignant cells but formed their own cluster(s) and showed clear evidence of CNVs. A small fraction of tumor cells clustered with the non-malignant cells (<4%). Given the high percentage of malignant cells in these tumors, we decided to classify all cells from the remaining four tumours (MUV41, SJ577, MUV34, and SJ625) as malignant. For two samples (SJ99 and BCH825) genetic subclones were identified based on their CNV profiles.
Identification of single-nucleotide variants in single-cell data
To detect mutant transcripts in our full-length scRNA-seq expression data (shown in Figure 2c and ED Figure 3c–f), sequencing reads were first aligned to the human genome build hg19 using STAR version 2.5.1b. RefSeq gene annotations were supplied to guide alignment. Variants were then quantified in each single cell at the genomic position in which they were detected in the WGS/WES data using samtools mpileup version 1.3. For some genes multiple variants were detected (e.g. four different variants were detected for CTNNB1 in WNT-MB) and quantified separately. To detect mutant and wild-type transcripts we required one or more supporting reads. We then filtered variants that were detected as mutant in less than three cells, or that were considered erroneously called as they were detected at elevated frequency in samples in which they were not detected in the genome sequencing data. A total of 82 variants remained after this filtering step. Mutations in highly expressed transcripts were detected in the majority of cells from the respective sample (e.g. OTX2 Q103R mutation in MUV39). Mutations in less highly expressed transcripts were detected less frequently. Only a small number of mutant transcripts were detected in samples in which they were not detected by genome sequencing (e.g. only 10 mutant transcripts were detected for the respective other CTNNB1 variants in the WNT-MB single cells; Figure 2c), illustrating the high specificity of our approach.
Identification of intra-tumour NMF programs and cellular hierarchies
Transcriptional programs were determined as previously described14 by applying Non-negative Matrix Factorization (NMF) to the centered expression data38. Negative values were converted to zero. Analysis was performed for each sample and subclone individually (excluding samples for which less than 100 cells were profiled), using only the malignant cells and setting the number of factors to 4 for WNT and 3 for SHH and Group 3/4 tumours. For each of the resulting factors we considered the 30 genes with the highest NMF scores to be characteristic of that factor (provided in Supplementary Table 2a). All single cells within the WNT, SHH, or Group 3/4 subgroups where then scored for these NMF programs (as described below, shown in ED Figures 5a, 6a, and 8a). Hierarchical clustering, with one minus Pearson correlation as the distance metric and Ward’s linkage, of the scores for each program revealed four (WNT subgroup) or three (SHH and Group 3/4 subgroups) main correlated sets of programs. The 30 genes with the highest average NMF score within each correlated program set (excluding ribosomal protein genes) were then used to define a total of 10 subgroup-specific meta-programs (provided in Supplementary Table 2b).
To interpret the characteristics of each meta-program, we manually inspected their underlying gene signatures. Additionally, we tested for enrichment of described gene sets (GO biological processes cell cycle and neuron differentiation, KEGG hedgehog signaling pathway, and manually curated ribosomal proteins and translational initiation/elongation factors) in each meta-program using Fisher’s exact test.
Generation of single-cell program expression scores
Single-cell expression scores were generated in a similar way as described previously13. Given a set of genes (Gj) reflecting a NMF program or meta-program, we calculate for each cell i, a score, SCj(i), quantifying the relative expression of Gj in cell i, as the average relative expression (Er) of the genes in Gj, compared to the average relative expression of a control gene-set (Gjcont): SCj(i)=average[Er(Gj,i)]-average[Er(Gjcont,i)]. The control gene-set contains 100 genes with the most similar aggregate expression level. In this way, the control gene-set has a comparable distribution of expression levels to that of the considered gene-set, and the control gene set is 100-fold larger, such that its average expression is analogous to averaging over 100 randomly-selected gene-sets of the same size as the considered gene-set.
Single cells were assigned to different cell populations based on the maximum expression score for their respective subgroup-specific meta-programs, excluding the cycling programs. The fraction of cells per tumour sample assigned to each cell population is provided in Supplementary Table 2d. Scores for the cycling programs were binarized into cycling and non-cycling (larger and smaller than 1, respectively). For illustration of the cellular hierarchies in SHH-MB, scores were normalized by minimizing the average minimum difference of all cells per sample to −1 or 1 (shown in Figure 3c). For the pan-subgroup analysis of all malignant medulloblastoma cells we re-centered expression values across the dataset and calculated expression scores for each of the 10 meta-programs. The pairwise correlation of expression scores is shown in Figure 6a.
Determination of cell population-specific genes
For comparison of cell populations in WNT-tumours, we calculated the average expression level of all cells per population (log2-transformed, un-centered expression levels were used). For this analysis we excluded cells from BCH807, as it was very different from the other four WNT-MBs and represents an atypical case (without cells scoring highest for the WNT-B meta-program, highly proliferative, negative staining for nuclear beta-catenin and lacking monosomy of chromosome 6). We then determined all genes with a difference smaller or larger than 1 between the average log2-transformed expression levels when comparing the undifferentiated proliferative population (highest for meta-program WNT-B) against the neuron-like population (WNT-C) or undifferentiated post-mitotic population (WNT-D). A total of 511 genes were identified in this way (provided in Supplementary Table 2c; ED Figure 5c).
For comparison of neuron-like cell populations between medulloblastoma subgroups, we first determined genes that were specific to any of the neuron-like populations. For every subgroup (WNT, SHH, and Group3/4), the average expression level of all neuron-like cells was compared to the average expression level of all undifferentiated cells from each subgroup, determining genes with a difference larger than 1 between the average log2-tranformed expression levels. This way we determined a total of 260 genes that were specific to the neuron-like cell population of at least one subgroup (provided in Supplementary Table 3). Genes that were specific to two or three subgroups were grouped as shared genes (shown in Figure 6c and ED Figure 9e).
RNA in situ hybridization
Paraffin-embedded tissue sections from two WNT-MB tumours of the single-cell cohort (SJ99 and SJ129) were obtained from St. Jude Children’s Research Hospital. Sections were mounted on glass slides and stored at −80°C. Slides were stained using the RNAscope 2.5 HD Duplex Detection Kit (Advanced Cell Diagnostics (ACD), 322430). Slides were baked for 1 hour at 60°C, deparaffinized and dehydrated with xylene and ethanol. The tissue was pretreated with RNAscope Hydrogen Peroxide (ACD, 322335) for 10 minutes at room temperature and RNAscope Target Retrieval Reagent (ACD, 322000) for 15 minutes at 98°C. RNAscope Protease Plus (ACD, 322331) was then applied to the tissue for 30 minutes at 40°C. Hybridization probes were prepared by diluting the C2 probe (red) 1:50 into the C1 probe (green). Advanced Cell Technologies RNAscope Target Probes used included Hs-MKI67 (ACD, 591771 and 591771-C2), Hs-DKK2 (ACD, 531131-C2), Hs-STMN2 (ACD, 525211-C2), Hs-ZFP36 (ACD, 427351) and Hs-EGR1 (ACD, 457671). Probes were added to the tissue and hybridized for 2 hours at 40°C. A series of 10 amplification steps were performed using instructions and reagents provided in the RNAscope 2.5 HD Duplex Detection Kit. Tissue was counterstained with Gill’s hematoxylin for 25 seconds at room temperature followed by mounting with VectaMount mounting media (Vector Laboratories).
Immunohistochemistry
Double labeling immunohistochemistry was performed using a 1:8000 dilution of anti-tubulin beta3 (clone TUJ1, Biolegend) and 1:25 dilution of anti-MYC (clone Y69, Abcam) diluted in Ventana antibody diluent (Roche Tissue Diagnostic, 251–018) and detected using the UltraView Red (Roche Tissue Diagnostics, 760–501) and UltraView DAB (Roche Tissue Diagnostics, 760–500) detection kits, respectively. Each target was evaluated using a semi-quantitative system to construct an H-score, obtained by multiplying the intensity of the stain (0: no staining; 1: weak staining; 2: moderate staining, and 3: strong staining) by the percentage (0 to 100) of cells showing that staining intensity (H-score range, 0 to 300).
Murine single-cell RNA-seq data generation and processing
Single cells from developing murine cerebellar tissue were processed using the microfluidics-based 10x Chromium protocol, as previously described21. In brief, single cells were prepared using the Chromium v1 Single Cell 3’ Library and Gel Bead Kit according to the manufacturers specifications. Quantification and quality checks for the library were performed using an Agilent Technologies DNA 1000 chip. Libraries were sequenced on an Illumina HiSeq 2500 machine. Raw sequencing data has been uploaded to the European Nucleotide Archive (accession PRJEB23051).
Murine developing cerebellum cells were filtered and normalized using the scanpy python package39. Genes expressed in less than 50 cells and cells expressing less than 200 genes were removed. Additionally, cells with less than 524 and greater than 3,206 total counts (±3 MADs) were removed. Furthermore, those cells with greater than 5% of their total counts mapping to mitochondrial genes were removed. Gene expression values were then divided by the total number of transcripts and multiplied by 10,000. Normalized values were calculated by natural-log transforming these values. We calculated scaled expression (z-scores for each gene) for downstream analysis.
Identification of cell types in developing murine cerebellum
The scanpy package implemented in python was applied to identify cell types among 82,228 cells expressing a total of 16,475 genes. After two rounds of clustering (using the Louvain method), populations predicted to be of non-cerebellar origins were excluded. Removed populations were enriched for haemoglobin, oligodendrocycte, and/or immune associated genes. The remaining 78,156 cells were visualized by t-SNE (using the first 100 principle components as input) and clustered a third time. We then merged clusters if mantel spearman correlation between gene distance matrices (using Manhattan distance) was greater than 0.9 (shown in Figure 1c and ED Figure 4c). Resulting clusters, in conjunction with marker genes, were used to identify major cell types in the developing cerebellum.
Integrated analysis of murine and human data sets
Gene expression matrices for human and mouse data sets were restricted to the 16,919 high-confidence homologous genes with gene order conservation (GOC) and whole genome alignment scores (WGA) greater than 75 percent, as defined by Ensembl. We removed genes without expression in at least 200 cells and filtered out those with gene dispersion across cells/samples less than equal to zero in each dataset. We also regressed out individual-specific effects in the single-cell data.
For canonical correlation analysis (CCA) the first 30 canonical correlation vectors were calculated to project each expression matrix into the maximally correlated subspace, as similarly described previously40. Briefly, CCA is implemented as singular value decomposition, by implicitly restarted Lanczos bidiagonalization algorithm, of a distance matrix between two gene expression matrices.
We adopted a correlation of differential expression approach to measure similarity between biological groups in two different studies. Such a procedure has previously been shown to be effective in implicating cellular origins for WNT and SHH medulloblastoma subgroups41. Gene expression for each cell, or centroids for each cluster when at the cluster level, is subtracted by mean gene expression of all other cluster centroids to determine differential expression. Cosine distance is then used to calculate correlations between differential expression vectors between studies as a metric for similarity (shown in Figures 1d, 3d, 3e, and ED Figures 4f, 7j, 10a, 10f). Significance is assessed by 10,000 permutations, followed by FDR correction, for cluster labels of interest. Genes driving CCA differential correlations between human and murine datasets were investigated by identifying genes both differentially expressed in the cell type of interest (Mann-Whitney U test) and correlated with the CCA correlation (Pearson correlation). Significant genes were those predicted to drive CCA differential correlations.
Non-negative Matrix Factorization applied to the centered murine expression data, with negative values assigned to zero and rank set to two, determined an undifferentiated and differentiated program. Both programs were projected onto a centered dataset of interest, scaled to a range of zero and one, then differentiated programs were subtracted from undifferentiated programs to calculate differentiation scores (shown in ED Figure 4b42).
Extended Data
ED Figure 1 |. Characteristics of the MB single-cell cohort.
a, Hematoxylin and eosin stained sections from all St. Jude single-cell samples (n=12). Tumours demonstrated large cell/anaplastic morphology (LCA, top), classic morphology (middle), or desmoplastic/nodular morphology (D/N, bottom). The scale bars represent 50 μm. b, Detailed characterization of the patient-derived xenograft (PDX) single-cell dataset. Subgroup prediction scores35 derived by DNA methylation profiling are indicated in the top panel (light shade: low probability, dark shade: high probability). The heatmap shows expression levels of previously described subgroup-specific marker genes19 in 946 PDX-derived single-cells. c, Heatmap shows expression levels of previously described subgroup-specific marker genes19 in 7,788 tumour-derived single-cells. d, Heatmaps show pairwise correlation of aggregated single-cell RNA-seq data (top) and bulk DNA methylation data (bottom) of all patient (n=25) and PDX (n=11) samples. For each PDX sample, the patient sample with the highest correlation coefficient is indicated by a black circle. e, Scatterplots show expression scores for published subgroup-specific gene sets for all single-cells in the patient cohort (n=7,788). Cells from WNT and SHH subgroups score only for their respective gene set. Some overlap is observed between cells from Group 3 and 4 subgroups and their respective gene sets, warranting the combined analysis of these subgroups in this study.
ED Figure 2 |. Copy-number analysis distinguishes malignant from non-malignant single-cells.
a-e, Heatmaps show single-cell RNA-seq-derived copy-number profiles of every cell in each sample (y-axis) along the genome (x-axis) for WNT (a), SHH (b), Group 3 (c) and Group 4 (d) patient MB as well as PDX (e). Copy-number profiles derived from array-based DNA methylation profiling from the same sample are shown above. Copy-number variations (CNVs) are observed in 21/25 patient tumor samples (all except MUV34, MUV41, SH577, and SJ625). Generally, we observe a high concordance between single-cell and DNA methylation array-derived copy number profiles. Genetic subclones at the level of broad copy-number changes are detected in samples SJ99 and BCH825. Cells without detected CNVs from samples that showed CNVs in the majority of cells (non-malignant cells; NM) are indicated for samples in which at least four non-malignant cells were detected (BCH807 and SJ454). Amplifications of the MYC and MYCN oncogenes detected by DNA methylation array are indicated.
ED Figure 3 |. Unsupervised clustering and detection of expressed SNVs in MB single-cells.
a, t-SNE visualization of the entire single-cell data set (n=8,924 cells). WNT (blue), SHH (red), Group 3 (yellow) and Group 4 (green) patient samples are indicated. Patient-derived xenograft (PDX) models are shown in pink. Non-neoplastic oligodendrocytes and immune cells are included for comparison. Generally malignant cells are expected to cluster by patient sample, whereas non-malignant cells are expected to cluster by cell type. Only few cells from different samples cluster with oligodendrocytes (n=22) or immune cells (n=6) and were classified as non-malignant. No additional clusters of cells from different samples were identified, indicating the absence of additional non-malignant cell populations in our dataset. b, Identical t-SNE visualization as in panel a, colored by copy-number state. CNVs were detected in most single-cells, facilitating their classification as malignant. A small number of cells did not show CNVs, even though CNVs were detected in the majority of cells from the respective sample (n=38). These cells were classified as non-malignant. Most cells with without CNVs clustered with normal oligodendrocytes (n=21), supporting their initial classification as non-malignant. Remaining cells without CNVs did not form clusters and likely represent poor quality cells. c, Identical t-SNE visualization as in panel (a), colored by detected mutant and wild-type transcripts. Cells classified as non-malignant are depleted for mutant transcripts (P < 0.01, Binomial test), supporting their initial classification. d, Heatmap shows detected mutant and wild-type transcripts for 39 variants (columns) in each cell (n=1,780, rows) of the WNT-MB dataset. If both mutant and wild-type transcripts are detected in a single cell, only the mutant transcript is shown. Variants were initially detected by genome sequencing and subsequently quantified in the scRNA-seq data. Sample BCH807 was not subjected to genome sequencing, and the CTNNB1 variant was manually detected by examining scRNA-seq alignments. Mutations are detected almost exclusively in single cells from samples in which they were detected by genome sequencing, illustrating the high specificity of single-cell variant detection. e, Heatmap shows mutant and wild-type transcripts for 15 variants in each cell (n=1,135, rows) of the SHH-MB dataset. Sample SJ454 was not subjected to genome sequencing, and the TP53 mutation was manually identified by examining scRNA-seq alignments. f, Heatmap shows mutant and wild-type transcripts for 28 variants in each cell (n=3,172, rows) of the Group 3/4-MB samples that were subjected to genome sequencing.
ED Figure 4 |. Single-cell mapping of murine cerebellar development.
a, b, c, Two-dimensional representation of the cerebellar (CB) single-cell RNA-seq data set by t-SNE. Each dot represents one cell. In (a), colors represent 13 different embryonic and early postnatal time points. In (b), colors indicate the differentiation score across the entire data set. In c, colors indicate cell types identified by Louvain clustering using the top 3,000 overdispersed genes. The main CB lineages were assigned based on published lineage markers. d, Annotation of 18 CB cell types based on the expression of lineage specific marker genes shown as violin plot. Violin plots represent kernel density estimation showing the distribution shape of the data. e, Lineage tree reconstruction using PAGA. The abstracted graph shows all cell types (nodes) as identified in panels (c) and (d). The size of the nodes is related to the number of cells in the defined cell type. The width of edges connecting cell types reflects the probability of the path. f, Radar plot showing CCA coefficients between each murine CB cell type and human MB subgroup single-cell RNA-seq.
ED Figure 5 |. Characterization of WNT-MB single-cell programs.
a, Expression scores for individual programs identified by unsupervised NMF analysis in each sample. Cells are ordered as in Figure 2a (n=1,780). Meta-programs WNT-A, WNT-B, WNT-C, and WNT-D were identified by hierarchical clustering of individual programs. b, Heatmaps show pairwise correlation (left), principal component analysis (PCA, center), and expression scores for NMF-derived meta-programs (right) for 301 cells from WNT-MB sample MUV44. The ordering of cells (rows) is maintained between the heatmaps. A two-dimensional representation of the same cells using t-SNE is shown on the far right (colored by expression scores for each meta-program). This analysis shows that the same programs and cell populations that are identified by the NMF analysis are also supported by PCA and t-SNE clustering. Furthermore, no additional programs and cell populations are identified (starting from PC5 components are less informative). c, Scatter plot shows isometric projection of average gene expression levels for cells with highest expression score for WNT-B (undifferentiated, proliferating), WNT-C (neuron-like), or WNT-D (undifferentiated, post-mitotic). WNT-B meta-program genes are indicated in red, WNT-C meta-program genes are indicated in green, and WNT-D meta-program genes are indicated in blue. Genes that are higher in both undifferentiated cell populations compared to neuron-like cells are indicated in black. d, Images show RNA in-situ hybridization experiments of five marker genes representative for the four WNT-MB meta-programs in two samples of the single-cell cohort. Results confirm expression of these genes independently of the scRNA-seq experiments.
ED Figure 6 |. Characterization of SHH-MB single-cell programs.
a, Expression scores for individual programs identified by unsupervised NMF analysis in each sample. Cells are ordered as in Figure 3a (n=1,135). Meta-programs SHH-A, SHH-B, and SHH-C were identified by hierarchical clustering of individual programs. b, Heatmaps show pairwise correlation (left), principal component analysis (PCA, center), and expression scores for NMF-derived meta-programs (right) for 493 cells from SHH-MB sample SJ577. The ordering of cells (rows) is maintained between the heatmaps. A two-dimensional representation of the same cells using t-SNE is shown on the far right (colored by expression scores for each meta-program). This analysis shows that the same programs and cell populations that are identified by the NMF analysis are also supported by PCA and t-SNE clustering. Furthermore, no additional programs and cell populations are identified (starting from PC3 components are less informative). c, Pairwise correlations between the expression profiles of 303 single-cells (rows, columns) from two SHH PDX samples (RCMB18 and RCMB24) (left panel). Expression scores for each of the NMF-derived meta-programs SHH-A, SHH-B, and SHH-C (columns) (right panel). Cells are ordered as in the left panel (rows). d, Heatmaps show the relative expression of the 60 genes representing the meta-programs SHH-B and SHH-C (rows), across 303 cells for RCMB18 and RCMB24. Cells are sorted by the difference between the two scores. Cells positive for the cell cycle program (SHH-A) are indicated by red bars. Similar cell populations as in the primary samples (undifferentiated GNP-like and differentiated neuron-like cells) are identified in RCMB18. No differentiated cells are identified RCMB24.
ED Figure 7 |. Cross-species mapping of SHH-MB origins.
a, Heatmap shows average expression levels of 30 GNP associated genes (rows) in cell types identified in the mouse CB dataset (columns). Genes are ordered by their relative expression in GNPs. b, The left panel shows the relative expression of orthologous genes in panel (a) in all cells from the single-cell cohort (n=7,745; columns). Cells are ordered by increasing GNP CCA cosine correlation coefficients. Cells expressing high levels of GNP associated genes are predominantly from SHH tumours. The right panel shows the relative expression of the same genes in the bulk microarray cohort (n=392). c-d, Heatmaps as in panels (a) and (b), but showing genes associated with the UBC/Granule intermediate cell type. e, Two-dimensional representation of GNPs/granule neurons from the cerebellar atlas by t-SNE. Each dot represents one cell (n=35,013). Colors represent the assigned cerebellar cell types (left panel), as well as the expression of Atoh1 and Neurod1 (middle and right panel). f, Boxplots of select granule lineage marker genes in the murine CB cohort (left panel), MB single-cell cohort (middle panel), and MB bulk microarray cohort (right panel). g, Boxplot of patient age associated with infant and adult/child subtypes of SHH-MB. h, Boxplot of the number of coding mutations associated with SHH-MB subtypes. The median is shown as a thick line; box limits are 25th and 75th percentiles; whiskers denote 1.5 times the interquartile range. i, In situ expression of Barhl1 (left panel) and Pde1c (right panel) at P4 during CB development (images obtained from the Allen Brain Atlas). j, Radar plot showing the CCA cosine correlation coefficients between each murine CB cell type and the MB single-cell cohort from cells scoring highest for meta-programs SHH-B (GNP-like cells) and SHH-C (Granule neuron-like cells).
ED Figure 8 |. Characterization of Group 3/4-MB single-cell programs.
a, Top panel shows Group 3/4 subtype prediction scores derived by DNA methylation profiling32 (light shade: low probability, dark shade: high probability). Expression scores for individual programs identified by unsupervised NMF analysis in each sample are indicated in the lower panel. Cells are ordered as in Figure 4a (n=4,873). Meta-programs Group 3/4-A, Group 3/4-B, and Group 3/4-D were identified by hierarchical clustering of individual programs. b, Expression scores across 4,873 single cells (columns) for each of the NMF-derived meta-programs Group 3/4-A, Group 3/4-B, and Group 3/4-C (rows). Cells are ordered as in panel (a). c, Heatmaps show pairwise correlation (left), principal component analysis (PCA, center), and expression scores for NMF-derived meta-programs (right) for 400 cells from Group 3-MB sample SJ617. The ordering of cells (rows) is maintained between the heatmaps. A two-dimensional representation of the same cells using t-SNE is shown on the far right (colored by expression scores for each meta-program). This analysis shows that the same programs and cell populations that are identified by the NMF analysis are also supported by PCA and t-SNE clustering. Furthermore, no additional programs and cell populations are identified (starting from PC4 components are less informative). d, Pairwise correlations between the expression profiles of 643 single cells (rows, columns) from nine patient-derived xenograft models (Med114FH, Med2112FH, Med211FH, Med411FH, RCMB20, Icb1299, Icb1572, Med2312FH, DMB006). Left panel shows Group 3/4 subtype prediction scores derived by DNA methylation profiling. Expression score for the NMF-derived meta-programs Group 3/4-A, Group 3/4-B, and Group 3/4-C (columns) are indicated in the right panel. e, Heatmaps show the relative expression of the 60 genes representing the meta-programs Group 3/4-B and Group 3/4-C (rows) across 140 cells for RCMB20 and DMB006. Cells are sorted by the difference between the two scores. Cells positive for the cell cycle program (Group 3/4-A) are indicated by red bars. Group 3 PDX samples are predominantly undifferentiated, with the exception of Med2312FH which is predominantly differentiated (classified by DNA methylation array as intermediate Group 3/4 sample). This parallels the high frequency of MYC amplifications in our Group 3 PDX cohort (5/8). Group 4 PDX sample DMB006 is also predominantly differentiated. These results are supportive of the cellular compositions detected in primary Group 3/4 samples.
ED Figure 9 |. Analysis of Group 3/4 intermediate samples and pan-subgroup comparison.
a, Scatterplot of the meta-program Group 3/4-C (x-axis) and Group 3/4-B (y-axis) expression scores for Group 3 and Group 4 bulk MBs3 (n=248; yellow and green dots, respectively). Samples that score similarly for both programs are classified as intermediate samples (n=49) b, Representative MYC and TUJ1 (encoded by TUBB3) IHC images of seven Group 3/4 samples. Four of these samples are shown at higher magnification in Figure 5b (SJ17, SJ617, SJ625, SJ723). c, Two-dimensional representation of 740 Group 3/4 MB samples analyzed by DNA methylation profiling using t-SNE3. Eight subtypes are delineated by curved lines. Samples are colored by their predicted subgroup35. d, Heatmap showing expression of transcripts coding for ribosomal proteins (n=75, rows). Cells positive for the cell cycle programs, and cells classified as neuron-like cells are indicated on top. Cells are ordered as in Figure 6b (n=7,745). e, Heatmap showing relative expression levels of genes that are specific to neuron-like cells and are shared between multiple subgroups (n=134, rows). Cells are ordered as in panel (e). f, Heatmap shows the relative expression of UBC-specific genes in Figure 6d (n=30; rows) in the bulk expression array cohort (n=392; columns). Samples are ordered by increasing CCA cosine correlation coefficient.
ED Figure 10 |. Cross-species mapping of Group 4-MB origins.
a, Top panel shows expression of TBR1 and EOMES in bulk Group 4-MB expression array data (n=149). Middle panel shows Group 3/4 DNA methylation-based subtype annotations for each sample. Bottom panel shows CCA scores from comparison of bulk MB expression data and UBCs and GluCN late populations from the cerebellar single-cell dataset. b, t-SNE visualization shows clustering of glutamatergic populations correlated with Group 4 MBs. c, Boxplot of CCA cosine correlation coefficients from comparison of bulk MB expression data and UBCs, according to Group 3/4 subtypes. The median is shown as a thick line; box limits are 25th and 75th percentiles; whiskers denote 1.5 times the interquartile range. d, e, Left panels show ISH data obtained from the Allen Brain Atlas for Tbr1 (d) and Eomes (e) in the developing murine cerebellum at the indicated time point. Right panels show the expression of Tbr1 (d) and Eomes (e) in the murine single-cell dataset according to the t-SNE structure shown in (b). f, Radar plot showing CCA cosine correlation coefficients between each murine CB cell type and Group 3-MB (upper panel) or Group 4-MB (lower panel) cells scoring highest for meta-programs Group 3/4-B or Group 3/4-C. g, Graphical summary of subgroup-specific cellular hierarchies identified in MB.
Supplementary Material
Supplementary Table 1: Cohort details
Supplementary Table 2: Transcriptional programs
Supplementary Table 3: Comparison of neuronal-like cells between MB subgroups
Acknowledgements
P.A.N. is a Pew-Stewart Scholar for Cancer Research (Margaret and Alexander Stewart Trust) and recipient of The Sontag Foundation Distinguished Scientist Award. P.A.N. was also supported by the National Cancer Institute (R01CA232143–01), American Association for Cancer Research (NextGen Grant for Transformative Cancer Research), The Brain Tumour Charity (Quest for Cures), the American Lebanese Syrian Associated Charities (ALSAC), and St. Jude. M.L.Su. was supported by grants from the Howard Goodman Fellowship at MGH, the Merkin Institute Fellowship at the Broad Institute of MIT and Harvard, the Wang Family Fund, the V Foundation for Cancer Research, the Swiss National Science Foundation Sinergia program, and the Alex Lemonade Stand. M.L.Su. is also recipient of The Sontag Foundation Distinguished Scientist Award. B.E.B. is the Bernard and Mildred Kayden Endowed MGH Research Institute Chair and an American Cancer Society Research Professor. This research was supported by a Pioneer Award from the NIH Common Fund and National Cancer Institute (DP1CA216873). V.H. is supported by a Human Frontier Science Program long-term fellowship (LT000596/2016-L). L.B. is supported by a Future Leaders Award from The Brain Tumour Charity (GN-000518). M.G.F. was supported by a Career Award for Medical Scientist from Burroughs Wellcome Fund, a K12 Paul Calabresi Career Award for Clinical Oncology (K12CA090354), a Harvard Brain Cancer SPORE - Career Enhancement Program Award, the National Institutes of Health (3P30 CA006516–53S6), The Cure Starts Now Foundation, Solving Kids’ Cancer, Inc/The Bibi Fund, The Andruzzi Foundation, and Alex’s Lemonade Stand Foundation. I.S., D.K., D.L. were supported by the Austrian National Bank (OeNB Jubiläumsfonds Project 15173). M.N.R. is supported by the ALSF, PBTF, AKBTC, and CBJOLF. We are indebted to the Flow Cytometry Core Lab (Department of Developmental Neurobiology, St. Jude) and the Core Flow Cytometry and Cell Sorting Shared Resource Facility (St. Jude). From St. Jude, we explicitly acknowledge the Hartwell Center, the Biorepository, members of the Clinical Genomics team, the Diagnostic Biomarkers Shared Resource in the Department of Pathology, and the Center for In Vivo Imaging and Therapeutics. We thank Dr. Stan Pounds (Department of Biostatistics, St. Jude) for valuable discussions. We thank Brandon Stelter for assistance with artwork.
B.E.B. discloses financial interests in Fulcrum Therapeutics, 1CellBio, HiFiBio, Arsenal Biosciences, Cell Signaling Technologies and Nohla Therapeutics.
Footnotes
Data availability
The single-cell RNA-seq and array-based DNA methylation data of 36 patient and PDX samples described in this study has been deposited in the Gene Expression Omnibus (GEO) with the accession code GSE119926. The single-cell RNA-seq data of the developing murine cerebellum has been deposited to the European Nucleotide Archive (ENA) with the accession code PRJEB23051.
References:
- 1.Gajjar AJ & Robinson GW Medulloblastoma-translating discoveries from the bench to the bedside. Nat Rev Clin Oncol 11, 714–722, doi: 10.1038/nrclinonc.2014.181 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Taylor MD et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 123, 465–472, doi: 10.1007/s00401-011-0922-z (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Northcott PA et al. The whole-genome landscape of medulloblastoma subtypes. Nature 547, 311–317, doi: 10.1038/nature22973 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CY et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62, doi: 10.1038/nature16546 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hovestadt V et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541, doi: 10.1038/nature13268 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Northcott PA et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56, doi: 10.1038/nature11327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Northcott PA et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer 12, 818–834, doi: 10.1038/nrc3410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson P et al. Subtypes of medulloblastoma have distinct developmental origins. Nature 468, 1095–1099, doi: 10.1038/nature09587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang ZJ et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 14, 135–145, doi: 10.1016/j.ccr.2008.07.003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver TG et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132, 2425–2439, doi: 10.1242/dev.01793 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Cho YJ et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 29, 1424–1430, doi: 10.1200/JCO.2010.28.5148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanay A & Regev A Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338, doi: 10.1038/nature21350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tirosh I et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313, doi: 10.1038/nature20123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Filbin MG et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360, 331–335, doi: 10.1126/science.aao4750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel AP et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401, doi: 10.1126/science.1254257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venteicher AS et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, doi: 10.1126/science.aai8478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hovestadt V et al. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol 125, 913–916, doi: 10.1007/s00401-013-1126-5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181, doi: 10.1038/nprot.2014.006 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Northcott PA et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol 123, 615–626, doi: 10.1007/s00401-011-0899-7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30, 413–421, doi: 10.1038/nbt.2203 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter RA et al. A Single-Cell Transcriptional Atlas of the Developing Murine Cerebellum. Curr Biol 28, 2910–2920 e2912, doi: 10.1016/j.cub.2018.07.062 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Kool M et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 123, 473–484, doi: 10.1007/s00401-012-0958-8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waszak SM et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: a retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol, doi: 10.1016/S1470-2045(18)30242-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih DJ et al. Cytogenetic prognostication within medulloblastoma subgroups. J Clin Oncol 32, 886–896, doi: 10.1200/JCO.2013.50.9539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Northcott PA et al. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta Neuropathol 122, 231–240, doi: 10.1007/s00401-011-0846-7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kool M et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25, 393–405, doi: 10.1016/j.ccr.2014.02.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merk DJ et al. Opposing Effects of CREBBP Mutations Govern the Phenotype of Rubinstein-Taybi Syndrome and Adult SHH Medulloblastoma. Dev Cell 44, 709–724 e706, doi: 10.1016/j.devcel.2018.02.012 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Northcott PA et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 29, 1408–1414, doi: 10.1200/JCO.2009.27.4324 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DT et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105, doi: 10.1038/nature11284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cavalli FMG et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 31, 737–754 e736, doi: 10.1016/j.ccell.2017.05.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwalbe EC et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol 18, 958–971, doi: 10.1016/S1470-2045(17)30243-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma T et al. Second-generation molecular subgrouping of medulloblastoma: an international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol, doi: 10.1007/s00401-019-02020-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chizhikov VV et al. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc Natl Acad Sci U S A 107, 10725–10730, doi: 10.1073/pnas.0910786107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Englund C et al. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci 26, 9184–9195, doi: 10.1523/JNEUROSCI.1610-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capper D et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474, doi: 10.1038/nature26000 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rusch M et al. Clinical cancer genomic profiling by three-platform sequencing of whole genome, whole exome and transcriptome. Nat Commun 9, 3962, doi: 10.1038/s41467-018-06485-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B & Dewey CN in BMC Bioinformatics Vol. 12 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaujoux R & Seoighe C A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11, 367, doi: 10.1186/1471-2105-11-367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15, doi: 10.1186/s13059-017-1382-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poschl J et al. Genomic and transcriptomic analyses match medulloblastoma mouse models to their human counterparts. Acta Neuropathol 128, 123–136, doi: 10.1007/s00401-014-1297-8 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Tamayo P et al. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci U S A 104, 5959–5964, doi: 10.1073/pnas.0701068104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Cohort details
Supplementary Table 2: Transcriptional programs
Supplementary Table 3: Comparison of neuronal-like cells between MB subgroups