Abstract
Follicular regulatory T (Tfr) cells have specialized roles in modulating Tfh help to B cells. However, the precise role of Tfr cells in controlling antibody responses to foreign and auto-antigens in vivo is still unclear due to a lack of specific tools. We developed a Tfr-deleter mouse that selectively deletes Tfr cells, facilitating temporal studies. We found Tfr cells regulate early, but not late, germinal center (GC) responses to control antigen-specific antibody and B cell memory. Deletion of Tfr cells also resulted in increased self-reactive IgG and IgE. The increased IgE levels led us to interrogate the role of Tfr cells in house dust mite (HDM) models. We found Tfr cells control Tfh13 cell-induced IgE. In vivo, loss of Tfr cells increased HDM-specific IgE and lung inflammation. Thus, Tfr cells control IgG and IgE responses to vaccines, allergens and autoantigens and exert critical immunoregulatory functions prior to GC formation.
Introduction
Follicular helper T (Tfh) cells migrate to B cell follicles to stimulate antibody production by B cells in the germinal center (GC) reaction1. The GC reaction results in somatic hypermutation, affinity maturation and class switch recombination, although these processes may also occur outside GCs 2. Tfh cells provide essential costimulation (through ICOS and CD40L) and cytokines (such as IL-21 and IL-4) to help promote B cell responses3, 4. Tfh cells possess a degree of phenotypic plasticity that can be altered by the inflammatory milieu, causing Tfh cells to produce cytokines typically made by TH1, TH2 and TH17 cells5, 6, 7. Tfh cells are thought to be distinct from TH2 cells because TH2 cells can produce both IL-4 and IL-13 and express the transcription factor Gata3, but Tfh cells can only produce IL-4 and do not express IL-13 nor Gata38. Although TH2 cells can mediate IgE responses, Tfh cells might also play a role. Studies have suggested that the Tfh cell cytokine IL-21 is essential for IgE responses to house dust mite (HDM) antigen, and that Tfh cells may convert to TH2-like cells in the lung9, 10. IgE responses are not completely dependent on Gata3 expression, suggesting cells other than TH2 cells may promote IgE8. T regulatory (Treg) cells can inhibit allergic inflammation, possibly through suppressing TH2 cells11, 12.
Follicular regulatory T (Tfr) cells inhibit Tfh-mediated B cell responses13, 14. In vitro assays have shown Tfr cells can inhibit antibody secretion, class switch recombination and somatic hypermutation through metabolic reprogramming and epigenetic remodeling of B cells15, 16, 17. In addition, Tfr cells can suppress Tfh cell production of effector cytokines such as IL-4 and IL-21 in vitro, while maintaining the Tfh transcriptional program17. The role of Tfr cells in controlling Tfh-mediated B cell responses in vivo is less clear. Adoptive transfer studies into lymphopenic mice have shown that Tfr cells inhibit antigen-specific IgG levels16, 18, 19. However, studies using bone marrow chimera and/or genetic models in which the transcription factor Bcl6 was deleted in FoxP3+ cells have suggested that Tfr cells regulate non-antigen specific B cell responses but do not substantially affect GC B cells nor antigen-specific IgG levels; however results have been inconsistent20, 21, 22. Moreover, IL-10 produced by Tfr cells can promote, rather than inhibit, plasma cell formation23. One explanation for the variability between studies may be due to the models used since Bcl6 can be expressed on Treg subsets other than Tfr cells, Bcl6 might not be completely necessary for development of all Tfr cells, and compensatory effects may rescue Tfr deletion in non-inducible systems.
To determine the precise role of Tfr cells in controlling B cell responses we developed a Tfr-deleter mouse model to inducibly delete Tfr cells in intact hosts at specific time points during immune responses. We demonstrate that Tfr cells potently regulate antigen-specific and memory IgG levels early during responses before GC formation. Using a TH2-like HDM challenge model, we found that Tfr cells can regulate IL-13 production by Tfh cells and control IgE responses. Deletion of Tfr cells in vivo during HDM sensitization resulted in increased HDM-specific IgE and lung inflammation. Taken together, these data demonstrate that Tfr cells are key regulators of humoral and allergic immunity by controlling early GC responses.
Results
Development of a specific and inducible Tfr-deleter mouse model
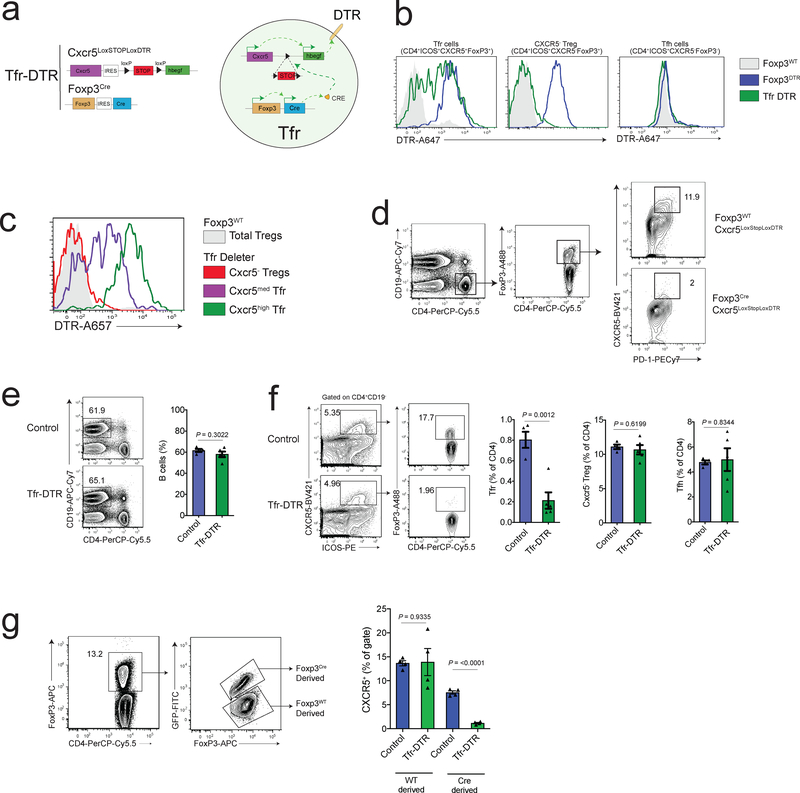
To study the role of Tfr cells during immune responses in vivo we created a mouse model to perturb Tfr cells in an inducible manner. To achieve this, we generated a mouse containing a Cxcr5IRES-LoxP-STOP-LoxP-DTR allele knocked into the Cxcr5 locus which was crossed to a FoxP3IRES-CreYFP allele-containing mouse to generate a Cxcr5IRES-LoxP-STOP-LoxP-DTR FoxP3IRES-CreYFP strain, referred to as the Tfr-DTR strain (Fig. 1a). In Tfr-DTR mice, FoxP3 expressing cells produce Cre recombinase which excises the stop cassette in the Cxcr5IRES-LoxP-STOP-LoxP-DTR allele resulting in an active Cxcr5IRES-DTR allele, and hence, DTR expression under the control of Cxcr5. Therefore, only cells expressing both FoxP3 and CXCR5, such as Tfr cells, express DTR on the cell surface, making them susceptible to deletion with diphtheria toxin (DT). We evaluated DTR expression on Tfr cells and CXCR5– Treg cells from wild type FoxP3, FoxP3-DTR24, or Tfr-DTR mice using an anti-DTR antibody. We found that Tfr cells (gated as CD4+ICOS+CXCR5+FoxP3+ cells) had substantial DTR expression in Tfr-DTR mice, albeit slightly lower compared to FoxP3-DTR mice (Figs. 1b, Supplementary Fig. 1a–b). Importantly, CXCR5– Treg cells expressed DTR only in FoxP3-DTR mice, and not in Tfr-DTR mice. Tfh cells (gated as CD4+ICOS+CXCR5+FoxP3– cells) did not express DTR, except for a very small population which are likely ex-Tfr cells (Figs. 1b, Supplementary Fig. 1a–b) 25. DTR expression was highest on Tfr cells expressing high levels of CXCR5, further demonstrating CXCR5-driven DTR expression (Fig. 1c).
Figure 1. Development of a Tfr-specific Deleter Model.
a) Schematic diagram of the Tfr-DTR strain. Allele details (left) and schematic of events leading to Tfr-specific DTR expression (right) are shown.
b) DTR expression on Tfr cells (left), CXCR5- Treg cells (middle) or Tfh cells (right) from control (Foxp3WT), Foxp3DTR, or Tfr-DTR mice.
c) DTR expression on CXCR5 negative, CXCR5-medium, or CXCR5-high Tfr cells from Tfr-DTR mice.
d) Quantification of Tfr cells from Tfr-DTR (Foxp3Cre Cxcr5LoxStopLoxDTR/WT) or control (Foxp3WT Cxcr5LoxStopLoxDTR/WT) mice which were immunized 7 days previously and received diphtheria toxin 2,4 and 6 days after immunization.
e) Quantification of B cells from Tfr-DTR or control mice (Foxp3Cre Cxcr5WT/WT) which were immunized 7 days previously and received diphtheria toxin 2, 4 and 6 days after immunization.
f) Quantification of Tfr, CXCR5- Treg and Tfh cells from mice as in (e).
g) Quantification of Tfr cells (by assessing CXCR5+ Treg cells) from Foxp3Cre/WT Cxcr5LoxStopLoxDTR/WT or control (Foxp3Cre/WT Cxcr5WT/WT) mice, gated on Cre-derived or WT-derived FoxP3 alleles.
Column graphs represent the mean with error bars indicating standard error. P value indicates two-tailed student’s T test. Data are from individual experiments and are representative of 2 (b-d,g) or 4 (e-f) independent experiments with similar results.
To determine the efficiency of Tfr deletion, we immunized mice with NP-OVA and administered DT. We found Tfr cells were deleted from Tfr-DTR but not in either Cxcr5IRES-LoxP-STOP-LoxP-DTR FoxP3wt nor FoxP3Cre Cxcr5wt/wt control mice (Fig. 1d–f). Deletion of Tfr cells was selective in Tfr-DTR mice because neither B cells, Tfh cells, total CXCR5-Treg cells, nor activated Ki67+ Treg cells were deleted in Tfr-DTR mice (Fig. 1e–f, Supplementary Fig. 1c). Moreover, deletion of Tfr cells in Tfr-DTR mice resulted in the loss of FoxP3+ cells within individual GCs (Supplementary Fig. 1d). To determine whether deletion of Tfr cells was cell-intrinsic, we immunized Cxcr5IRES-LoxP-STOP-LoxP-DTR FoxP3Cre/wt mice (in which only ~50% of the Tfr cells will express DTR on the surface) and administered DT. DTR-expressing Tfr cells were deleted, but not non-DTR expressing Tfr cells from the same mouse, demonstrating cell-intrinsic deletion in the Tfr-DTR model (Fig. 1g).
Tfr cells potently regulate early germinal center formation
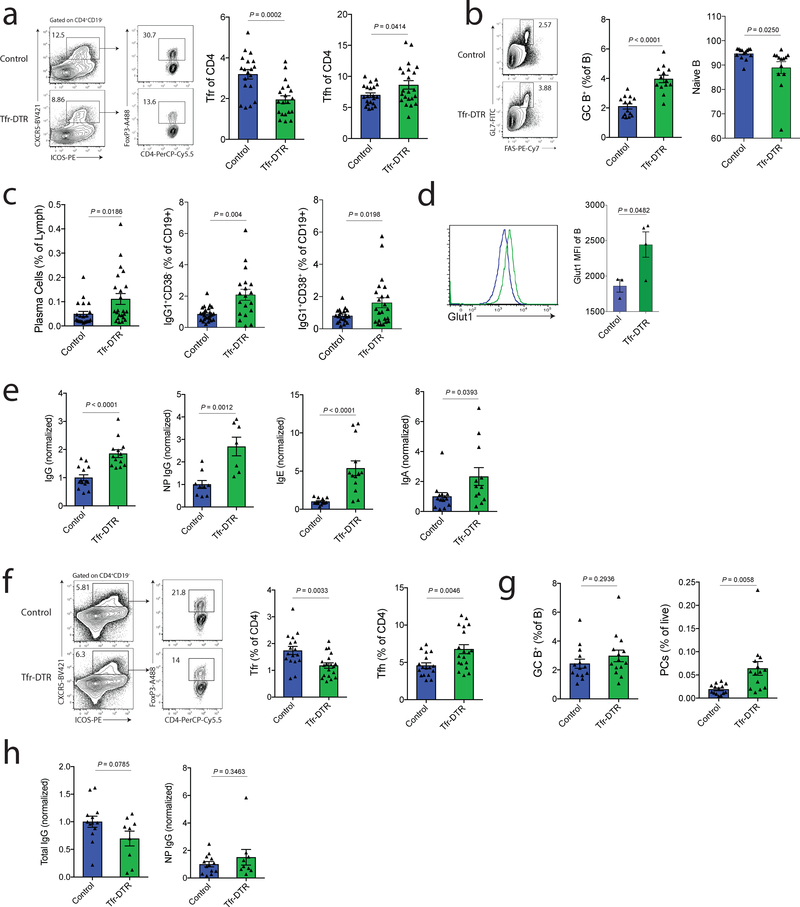
Our previous data suggest that Tfr cells can be limited by cytokines produced in GCs15. Moreover, Tfr cells seem to be less frequent in large, developed GCs (data not shown)26. These findings suggest that Tfr cells might regulate B cell responses most potently before mature GCs form. To assess the role of Tfr cells before GC initiation, we immunized Tfr-DTR or control (Foxp3creCxcr5wt/wt) mice with NP-OVA and deleted Tfr cells on days 5–9 with administration of DT, and assessed B cell responses at day 21. Tfr cells were largely attenuated, even 12 days after the last DT injection (Fig. 2a). There were minor, but significant, increases in the frequency of Tfh cells compared to control mice. The CD19+GL7+FAS+ GC B cell frequency was ~2-fold higher in Tfr-DTR mice compared to control mice, demonstrating that Tfr cells potently regulate initial GC formation (Fig. 2b). In addition, naive B cells, gated as CD38+IgG1- B cells, were slightly attenuated in Tfr-DTR mice. CD138+ plasma cells, IgG1+ class switched B cells, and IgG1+CD38+ memory-like B cells were also increased in Tfr-DTR mice suggesting Tfr cells regulate many arms of B cell effector responses (Fig. 2b–c). Tfr cells have previously been shown to cause metabolic reprogramming, including inhibition of glycolysis, in B cells15. We found Glut1 expression was higher in B cells from Tfr-deleted compared to control mice (Fig 2d). These results are consistent with a mechanism in which Tfr cells inhibit metabolism in B cells in vivo.
Figure 2. Tfr cells Potently Regulate Early Geminal Center Formation.
a) Quantification of Tfr (gated as CD4+ICOS+CXCR5+FoxP3+CD19−) and Tfh (gated as CD4+ICOS+CXCR5+FoxP3−CD19−) cells from dLNs of Tfr-DTR (Foxp3Cre Cxcr5LoxStopLoxDTR/WT) or Control (Foxp3Cre Cxcr5WT/WT) mice 21 days after immunization. Diphtheria toxin (DT) was administered on days 5,7 and 9 to delete Tfr cells before GC initiation.
b) Quantification of GC B cells (gated as CD19+GL7+FAS+) and naive B cells (gated as CD38+IgG1−) from dLNs at d21 after immunization as in (a).
c) Quantification of plasma cells (gated as CD138+), class switched B cells (gated as CD19+IgG1+CD38-), and memory-like B cells (gated as CD19+IgG1+CD38+) at d21 after immunization as in (a).
d) Glut1 expression on B cells from mice as in (a). Representative histogram (left) and quantification (right).
e) Quantification of total IgG (far left), NP-specific IgG (middle left), total IgE (middle right) and total IgA (far right) analyzed from serum of mice as in (a).
f) Quantification of Tfr (gated as CD4+ICOS+CXCR5+FoxP3+CD19−) and Tfh (gated as CD4+ICOS+CXCR5+FoxP3−CD19−) cells from dLNs of Tfr-DTR (Foxp3Cre Cxcr5LoxStopLoxDTR/WT) or Control (Foxp3Cre Cxcr5WT/WT) mice at d21 after immunization. Diphtheria toxin (DT) was administered on days 10, 12 and 14 to delete Tfr cells after GC formation.
g) Quantification of GC B cells (gated as CD19+GL7+FAS+) and plasma cells (CD138+) from dLNs at d21 after immunization as in (e).
h) Quantification of total IgG (left) and NP-specific IgG (left).
Column graphs represent the mean with error bars indicating standard error. P value indicates two-tailed student’s T test. Data are combined results from 4 (a-c,e) or 3 (f-h) independent experiments, or are from an individual experiment which is representative of two independent experiments (d).
To determine if Tfr cells regulate antigen-specific antibody production by regulating early GC responses, we assessed total and NP-specific antibody levels in Tfr-DTR mice. There was a 2-fold increase in total IgG and a ~2.5-fold increase in NP-specific IgG, demonstrating Tfr cells regulate antigen-specific antibody responses (Fig. 2e and Supplementary Fig. 2a). We also found moderate increases in total IgA and substantial increases in total IgE in Tfr-deleted mice (Fig. 2e). The latter result was unexpected since Tfr-DTR mice are on a C57bl/6 background. These results were not due to preferential deletion of a Tfr subset because the small number of Tfr cells remaining in Tfr-DTR mice phenotypically resemble Tfr cells from control mice (Supplementary Fig. 2b). We did find increased Ki67 expression in Tfr cells remaining in Tfr-DTR mice, likely due to a compensatory mechanism to overcome Tfr deficiency (Supplementary Fig. 2b).
Next, we determined if Tfr cells can regulate B cell responses after GCs have already formed. We immunized Tfr-DTR or control (Foxp3creCxcr5wt/wt) mice with NP-OVA and administered DT on days 10–14 to delete Tfr cells. We found a reduction in Tfr cells in the Tfr-DTR mice, and slight increases in the frequency of Tfh cells, polarizing the follicular T cell subset towards Tfh cells (Fig. 2f). However, when we assessed GC B cells, we did not find any differences between Tfr-DTR and control mice (Fig. 2g). Likewise, we did not find any increases in total or NP-specific IgG, although plasma cells were slightly elevated as were levels of IgA and IgE (Fig. 2g–h and Supplementary Fig. 2c). The few Tfr cells remaining in Tfr-DTR mice had a similar phenotype to control mice except for elevated Ki67 (Supplementary Fig. 2d). The lack of increased antigen specific antibodies when Tfr cells were deleted post-GC formation was due to the stage of GC, and not total duration of Tfr deletion, because pre-GC deletion strategies have a phenotype as early as day 5 after Tfr deletion, and deletion of Tfr cells after GC formation does not result in a phenotype, even at day 26 (Supplementary Fig. 2e–f). These data demonstrate Tfr cells can regulate GC B cell development and antigen specific antibody responses early before GC formation and have less regulatory control after GCs have been initiated.
Tfr cells regulate autoreactive IgG and IgE antibodies
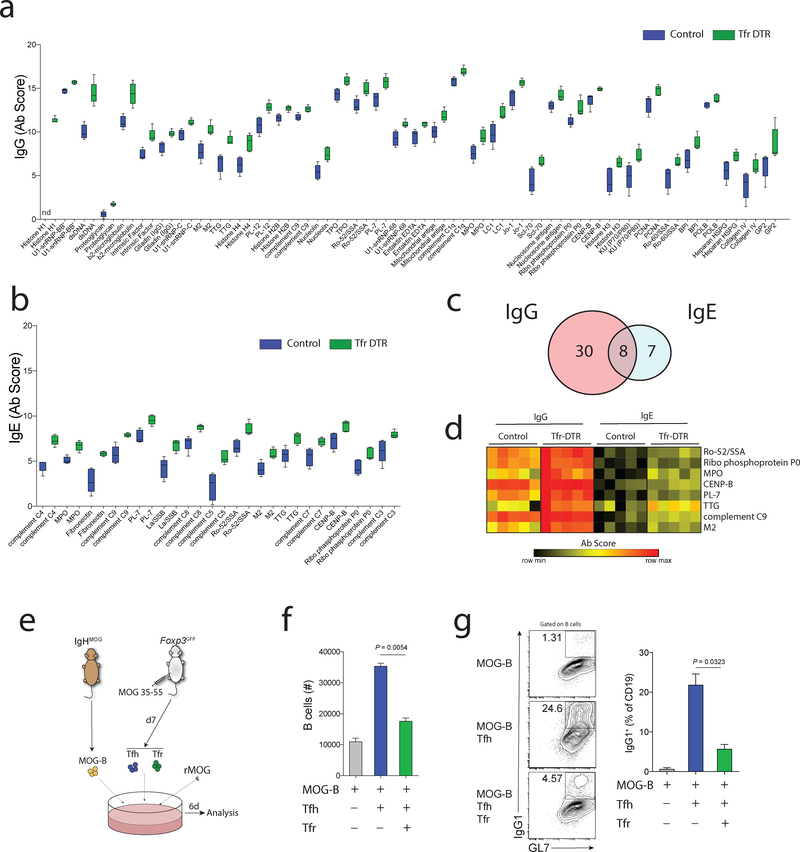
Next we assessed whether Tfr cells can regulate autoreactive antibodies. We deleted Tfr cells in Tfr-DTR mice before GC formation (at days 5–9), and analyzed sera at day 21 with autoantigen protein arrays. Autoreactive IgG was increased for one-third of the autoantigens in Tfr-DTR mice compared to control mice using a stringent cutoff (Mann-Whitney p<0.01) (Fig. 3a). In most cases, control mice had antibodies that recognize autoantigens, but levels were higher in the Tfr-DTR mice. However, in one example, there was substantial signal for anti-Histone H1 autoantibodies in Tfr-DTR mice, but no detectable signal in control mice (Fig. 3a). These data demonstrate that Tfr cells can regulate levels and formation of autoreactive IgG antibodies.
Figure 3. Tfr cells control autoreactive IgG and IgE during foreign antibody responses.
a) Quantification of IgG scores for indicated autoantigens from Tfr-DTR (Foxp3Cre Cxcr5LoxStopLoxDTR/WT) or Control (Foxp3Cre Cxcr5WT/WT) mice which were immunized with NP-OVA and given DT to delete Tfr cells before GC initiation as in Fig. 2a. Serum was collected at d21 after immunization. Autoantigens which showed a significant different difference between control and Tfr-DTR mice (38 out of 123, p<0.01, Mann-Whitney U test) are shown.
b) Quantification of IgE scores for indicated autoantigens as in (a). Autoantigens which showed a significant difference between control and Tfr-DTR mice (15 out of 123, p<0.01, Mann-Whitney U test) are shown.
c) Venn diagram showing overlap of differentially expressed autoantibodies for IgG and IgE isotypes in Tfr-DTR mice.
d) Heatmap showing IgG and IgE autoantibody scores for the 8 overlapping autoantigens Tfr-DTR compared to control mice.
e) Schematic of in vitro autoreactive B cell suppression assay. Tfh and Tfr cells from MOG35–55 immunized mice were cultured with IgHMOG B cells in the presence of rMOG for 6 days.
f) Relative count of B cells from suppression assays as in (e).
g) Quantification of class switching to IgG1 in suppression assays as in (e). Representative plots (left) and quantification (right) are shown.
Graphs are box and whisker plots with horizontal line indicating the mean and bars indicating range of values (a-b). Column graphs represent the mean with error bars indicating standard error (f-g). Data are from an individual experiment with 5 mice per group (a-b), or are replicate suppression assays from an individual experiment and is representative of 3 independent experiments (e-g). P value indicates two-tailed student’s T test.
Next, we determined if any of the substantial amounts of IgE in the Tfr-deleted mice were specific for autoantigens. Such autoreactive antibodies could be pathogenic, since patients with systemic lupus erythematosus (SLE) can generate autoreactive IgE responses, and autoreactive IgE can exacerbate disease in mouse models of lupus27, 28. 15 autoantigens were recognized to a higher degree by IgE in Tfr-DTR compared to control mice (Mann-Whitney p<0.01) (Fig. 3b). These autoantigens include La/SSB and Ro-52/SSA, which are targets of autoreactive IgE responses in SLE patients; anti-SSB and SSA IgE may be an indicator of immune complex mediated disease28. In addition, complement had higher IgE autoantibody scores in Tfr-DTR compared to control mice. When we used a less stringent cutoff of p<0.05, anti-β2-microglobulin and anti-GP2 IgE were present in Tfr-DTR mice, but not in control mice (data not shown). Using the stringent p<0.01 cutoff, there was evidence of increases in IgG and IgE targeting the same 8 autoantigens, including Ro-52/SSA, MPO, CENP-B, PL-7 TTG and M2 (Fig. 3c–d). Some of these IgG and IgE autoantibodies, such as anti-Ro-52/SSA are increased in SLE patients.
We next determined whether Tfr cells can regulate initial activation and class switch recombination of autoreactive B cells. We developed a Tfh-mediated antigen-specific autoreactive B cell class switch recombination assay. Myelin oligodendrocyte glycoprotein (MOG) was chosen as the model autoantigen because MOG immunization generates functional Tfh and Tfr cells18, 29, MOG-specific B cells cause a Devic-like disease in EAE models30, 31, and B cell depletion has large therapeutic benefit in multiple sclerosis32. For this assay, Tfh and Tfr cells were sorted from MOG35–55 immunized Foxp3GFP mice and cultured with B cells isolated from naive IgHMOG mice in the presence of rMOG (Fig. 3e). We found Tfh cells were able to stimulate IgHMOG B cells to expand and undergo class switch recombination (Fig. 3f–g). Importantly, Tfr cells were able to potently suppress IgHMOG B cell expansion and class switch recombination. These data demonstrate that Tfr cells can regulate initial autoreactive B cell activation and class switch recombination.
Tfr cells regulate antigen-specific antibody during memory responses
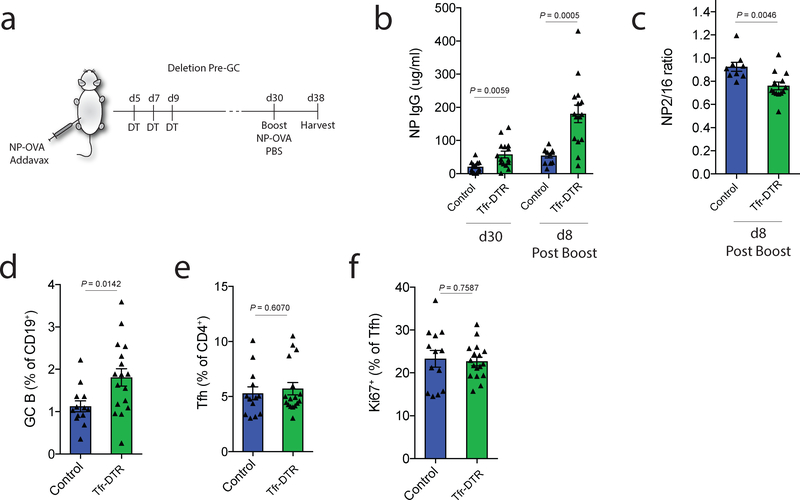
Next we determined if Tfr cells regulate memory B cell responses. To do this, we immunized Tfr-DTR or control (Foxp3Cre Cxcr5wt/wt) mice with NP-OVA containing a mild adjuvant, MF59-like Addavax™, and administered DT from days 5–9. At day 30, mice were boosted i.p. with unadjuvanted NP-OVA (Fig. 4a). We found that NP-specific antibody responses were ~2.8 times higher before, and ~3.3-fold higher after, the boost in Tfr-DTR compared to control mice (Fig. 4b). These data suggest antigen-specific memory responses are substantially increased when Tfr cells are deleted before GC initiation, suggesting Tfr cells control B cell memory to limit antibody responses. Next, we determined if the increased antigen-specific antibody had altered affinity since we hypothesized that Tfr cells can set thresholds on B cell responses. We performed ELISAs with low and high ratios of NP and calculated the NP2/16 ratio to approximate the affinity of NP-specific antibody. Antibody after boosting had a lower NP2/16 ratio in Tfr-DTR compared to control mice, suggesting lower affinity (Fig. 4c and Supplementary Fig. 3a). We did not find significant changes in the NP2/16 ratio in non-boosted experiments (Supplementary Fig. 3b–c).
Figure 4. Tfr cells regulate antibody memory responses.
a) Schematic of Tfr-deletion to assess memory responses. Tfr-DTR or control mice were immunized with NP-OVA in MF59 and DT was administered on days 5,7 and 9 to delete Tfr cells prior to GC formation. Mice received a boost of NP-OVA without adjuvant at d30. Mice were harvested on d38.
b) Analysis of NP-specific IgG levels before and after NP-OVA boost as in (a).
c) Quantification of the NP2/NP16 ratio in experiments as in (a).
d) Quantification of GC B cells (gated as CD19+GL7+FAS+) from dLN of mice at d38 as in (a).
e) Quantification of Tfh (gated as CD4+ICOS+CXCR5+FoxP3-CD19-) cells at d38 as in (a).
f) Quantification of Ki67 expression in Tfh cells gated as in (e).
Column graphs represent the mean with error bars indicating standard error. P value indicates two-tailed student’s T test. Data represent combined data from 3 independent experiments.
Memory B cell responses likely require secondary GCs to produce high affinity antibody33. Therefore, we next analyzed GC responses in Tfr-DTR mice in which Tfr cells were deleted and boosted with NP-OVA. We found increases in GC B cells in Tfr-DTR compared to control mice, suggesting augmented secondary GCs after rechallenge (Fig. 4d). However, it is important to note that this assay cannot distinguish GC B cells which form from memory B cells or naive B cells. In contrast to GC B cells, there were no significant increases in Tfh cell frequency or expression of Ki67 in Tfr-DTR mice after rechallenge (Fig. 4e–f). Taken together, these data indicate that Tfr cells restrain the quantity, but promote the affinity, of antigen-specific antibody during memory responses by regulating early GCs.
HDM antigen generates distinct populations of Tfh and Tfr cells
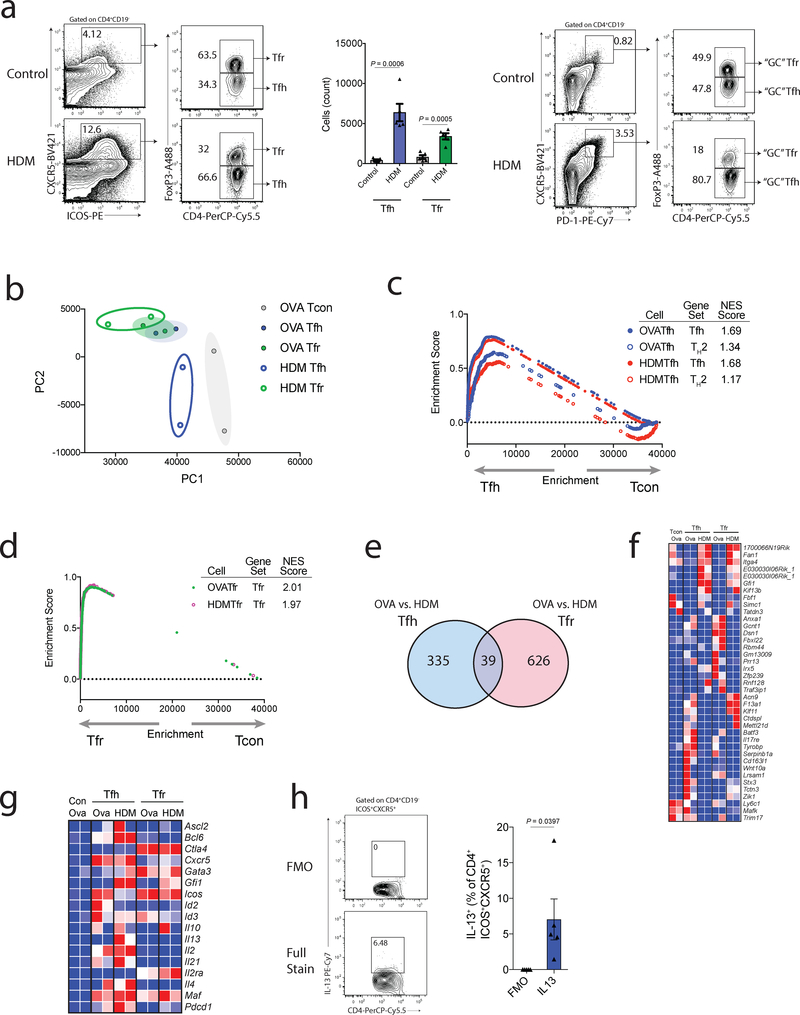
Our finding that Tfr-deletion results in increased IgE levels in mice immunized with NP-OVA suggested that Tfr cells may be able to regulate TH2-like responses. Therefore, we next determined if HDM exposure, a TH2-like response, generated Tfh and Tfr cells. We challenged C57bl/6 mice with HDM intranasally every 2 days and assessed mediastinal lymph nodes on day 7. Unimmunized mice had a very small ICOS+CXCR5+CD4+ population made up of ~60% Tfr cells, a ratio that is typical in basal states13 (Fig. 5a). In comparison, HDM exposed mice developed a substantial population of ICOS+CXCR5+CD4+ cells in which Tfr cells were only ~30%. Moreover, we found a subpopulation of Tfh and Tfr cells which assumed a GC-like phenotype, suggesting proper Tfh and Tfr effector differentiation.
Figure 5. House dust mite antigen generates distinct populations of Tfh and Tfr cells.
a) Quantification of Tfh and Tfr cells in response to HDM challenge. WT mice were challenged or not (control) with HDM intranasally on days 0, 2, 4 and 6. dLNs were harvested on d7. Gating strategy to identify Tfh and Tfr cells (left), total numbers of Tfh and Tfr cells (middle) and gating strategy for “GC” Tfh and Tfr cells (right) are shown.
b) Principal component analysis (PCA) showing relationship between transcriptional profiles of Tfh (CD4+ICOS+CXCR5+FoxP3-CD19-) and Tfr (CD4+ICOS+CXCR5+FoxP3+CD19-) cells generated in response to NP-OVA (subcutaneous) or HDM (intransal) challenge in Foxp3GFP mice.
c) Gene set enrichment analysis (GSEA) comparing Tfh cells generated in response to NP-OVA or HDM for Tfh or TH2 signatures (GSEA14308).
d) GSEA comparing Tfr cells generated in response to NP-OVA or HDM for Tfr signatures.
e) Venn diagram demonstrating the overlap of differentially expressed genes (p<0.05) between NP-OVA and HDM components for Tfh and Tfr cells.
f) Heatmap showing the 39 commonly differentially expressed genes in Tfh and Tfr cells in NP-OVA versus HDM challenge as in (e).
g) Heatmap of common follicular T cell and TH2 genes in Tfh and Tfr cells generated in response to NP-OVA or HDM challenge.
h) IL-13 production by HDM Tfh cells. Intracellular staining was performed on HDM treated mice as in (a). FMO= stain without anti-IL-13 antibody.
Column graphs represent the mean with error bars indicating standard error. P value indicates two-tailed student’s T test. Data are representative of 3 independent experiments (a,h), or are combined data from 2 independent experiments (b-g).
To determine if Tfh and Tfr cells after HDM exposure transcriptionally resemble Tfh and Tfr cells, we performed transcriptional analysis. We immunized Foxp3IRES-GFP mice with NP-OVA (emulsified in CFA) subcutaneously on d0 or HDM intranasally on days 0, 2, 4 and 6, and sorted Tfh (gated as CD4+ICOS+CXCR5+FoxP3−CD19−), Tfr (gated as CD4+ICOS+CXCR5+FoxP3+CD19–) or T conventional (‘Tcon’, gated as CD4+ICOS−CXCR5–FoxP3–CD19–) cells on day 7 and performed RNAseq transcriptional analysis. By principal component anlysis (PCA), most follicular T cell populations clustered separately from Tcon cells, however HDM Tfh cells clustered closer to Tcon cells than other cells (Fig. 5b). Next, we determined if Tfh cells from HDM challenged mice transcriptionally resemble Tfh cells or if they take on a TH2-like phenotype. OVA and HDM-specific Tfh cells had a similar enrichment for Tfh genes, and HDM Tfh cells did not have enrichment for TH2 genes (Fig. 5c). Similarly, Tfr cells from both OVA and HDM challenges had strong enrichment for Tfr genes. (Fig. 5d).
Although Tfh and Tfr cells from HDM challenged mice had intact transcriptional programs, we did find differentially expressed genes between these cells and their OVA challenge counterparts. There were 374 genes differentially expressed (p<0.05) between OVA and HDM Tfh cells, and 665 genes differentially expressed (p<0.05) between OVA and HDM Tfr cells with 39 genes being differentially expressed in both Tfh and Tfr cells (Fig. 5e). When we assessed these 39 genes in more detail, we found a subset of genes expressed in HDM Tfh and Tfr cells, but not OVA populations, such as Gfi1 (Tfh p=0.0130, Tfr p=0.0424) which has roles in stabilizing TH2 cells34 (Fig. 5f). We also evaluated genes commonly expressed in Tfh and Tfr cells. Some genes such as Icos and Id2, seemed to be expressed less in HDM Tfh cells compared to OVA Tfh cells, although only Id2 was statistically significant (p=0.0400)(Fig. 5g). We found a low, but positive, transcript for Il13 in HDM Tfh cells which was not present in OVA Tfh cells. In addition, HDM Tfh cells expressed more Gata3. To assess whether a subset of Tfh cells could produce IL-13, we performed intracellular cytokine staining and found a small proportion of Tfh cells that produced IL-13 (Fig. 5h). Taken together, these data demonstrate that Tfh and Tfr cells from HDM challenge have Tfh and Tfr transcriptional programs, but also some distinct transcriptional characteristics, such as an IL-13 transcript in HDM Tfh cells. Since HDM Tfh cells have an intact Tfh, but not TH2, program yet express IL-13, we refer to these cells as ‘Tfh13-like’ cells.
Tfr cells regulate Tfh13-mediated IgE responses to HDM in vitro
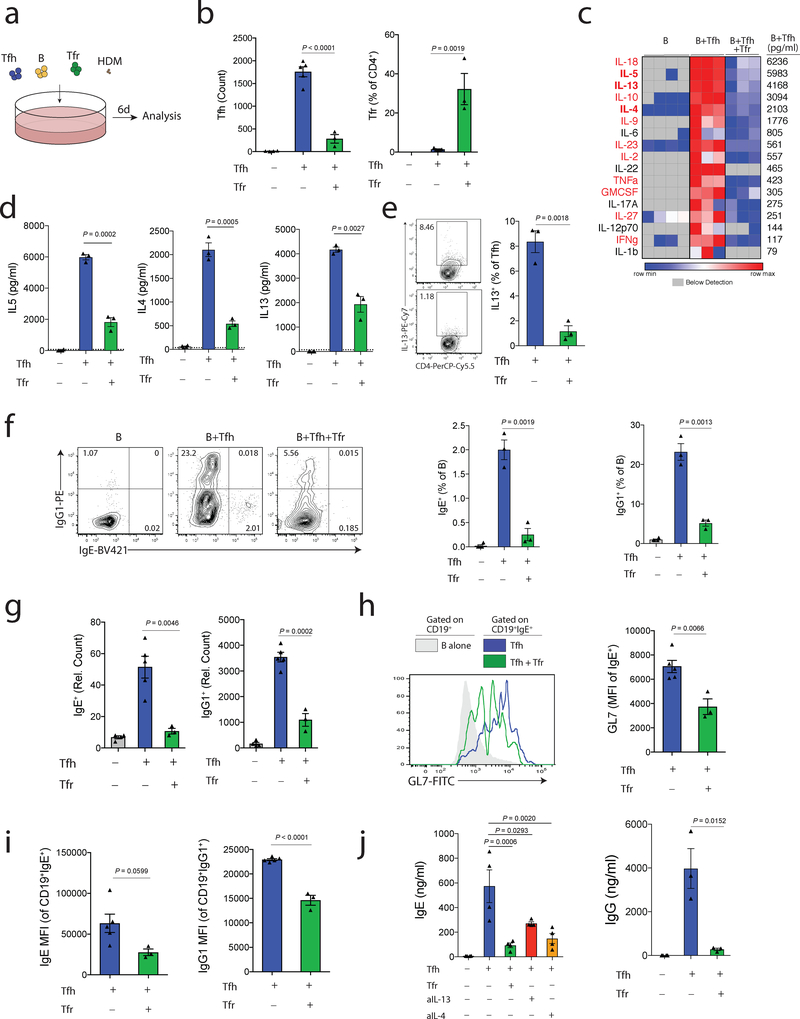
Since we found that Tfh13-like cells from HDM challenged mice expressed IL-13, and Tfr-deleted mice had elevated levels of autoreactive IgE, we hypothesized that Tfr cells may regulate IL-13 and IgE responses in the context of TH2-like allergic responses. To test this hypothesis, we developed an in vitro suppression assay in which Tfh13-like cells mediate class switching of B cells to IgE in response to HDM antigen. We administered HDM intranasally to Foxp3IRES-GFP mice every 2 days and on day 7 sorted total B, Tfh (gated as CD4+ICOS+CXCR5+FoxP3–CD19–) and Tfr (gated as CD4+ICOS+CXCR5+FoxP3+CD19–) cells from mediastinal lymph nodes. Cells were cultured together along with HDM for 6 days (Fig. 6a). Tfr cells inhibited Tfh cell proliferation and were still present at the end of the culture (Fig. 6b). We found that cultures containing Tfh13 and B cells contained large amounts of TH2-like cytokines including IL-5, IL13 and IL-4, all of which were suppressed by the addition of Tfr cells (Fig. 6c–d). In addition, Tfr cells suppressed the frequency of Tfh cells (gated as CD4+FoxP3− cells) expressing IL-13 protein (Fig. 6e). Tfh13 cells stimulated B cells to undergo class switching to IgG1, and to a lesser extent, IgE (Fig. 6f). Importantly, addition of Tfr cells resulted in near-complete reduction in IgE+ B cells and a substantial reduction in IgG1+ B cells (Fig. 6f–g). We did not find evidence of class switching to IgE when HDM was omitted from the wells, nor when we performed similar cultures using cells from NP-OVA immunization (Supplementary Fig. 4 and data not shown). Although HDM Tfr cells suppressed class switching of NP-OVA B cells to IgG1, NP-OVA Tfr cells may suppress Tfh13-mediated IgE class switching of B cells less potently than HDM Tfr cells (Supplementary Fig. 4).
Figure 6. Tfr cells regulate Tfh13-mediated IgE responses in vitro.
a) Schematic of experimental design for an in vitro HDM suppression assay. Total B, Tfh and Tfr cells were purified from the dLN of mice that received HDM on days 0,2,4 and 6 and added to culture wells along with HDM for 6 days.
b) Quantification of total Tfh cells (left) and the percentage of Tfr cells (FoxP3+ of CD4+IA-CD19- cells) (right) from cultures as in (a).
c) Quantification of cytokines in culture supernatants from cultures as in (a). Cytokines listed in red have levels statistically lower in cultures containing Tfr cells compared to cultures without Tfr cells.
d) Column graphs of IL5, IL4 and IL13 from data in (c).
e) Intracellular cytokine staining of IL-13 in Tfh cells from cultures as in (a).
f) Analysis of class switching to IgG1 and IgE in cultures as in (a). Representative gating (left, pregated on CD19+IA+CD4- cells), IgE+ B cell quantification (middle) and IgG1+ B cell quantification (right) are shown.
g) Counts of total IgE+ (left) and IgG1+ (right) B cells expressed as relative counts from experiments as in (a).
h) Expression of GL7 on IgE+ B cells from indicated cultures as in (a).
i) Expression of IgE on IgE+ B cells (left) and IgG1 on IgG1+ B cells are shown from cultures as in (a).
j) Levels of IgE (left) and IgG (right) in culture supernatants from cultures as in (a). Anti-IL13 or anti-IL4 were added to indicated wells.
Column graphs represent the mean with error bars indicating standard error. P value indicates two-tailed student’s T test (b-i, j; right) or One-way ANOVA with Tukey’s correction (j; left).
Data are from individual experiments and are representative of 3 independent experiments.
To determine if Tfr cells prevent class switching to IgE, suppress already class-switched IgE+ B cells, or both, we assessed levels of GL7, a GC B cell expressed molecule that is attenuated on B cells during Tfr suppression. We found that IgE+ B cells had lower expression of GL7 in the presence of Tfr cells, suggesting that IgE+ class switched B cells are less activated (Fig. 6h). We also analyzed protein levels of IgE and IgG1 within switched B cells. Both IgE and IgG1 expressing B cells had lower expression of IgE and IgG1, respectively, if Tfr cells were present, although this did not reach statistical significance for IgE (Fig. 6i). To determine if Tfh13 cytokines were essential for full IgE responses we added IL13 or IL4 blocking antibodies to cultures and assessed levels of IgE. Tfh cells stimulated large amounts of IgE secretion, which was strongly attenuated when IL-13 or IL-4 blocking antibodies were added, demonstrating cytokines produced by Tfh13 cells stimulate IgE (Fig. 6j). Importantly, IgE and IgG production was substantially suppressed by the presence of Tfr cells (Fig. 6j). Taken together, these data demonstrate that Tfr cells can suppress Tfh-mediated IL-13 and IgE responses in vitro.
Tfr cells regulate antigen specific IgE responses in vivo
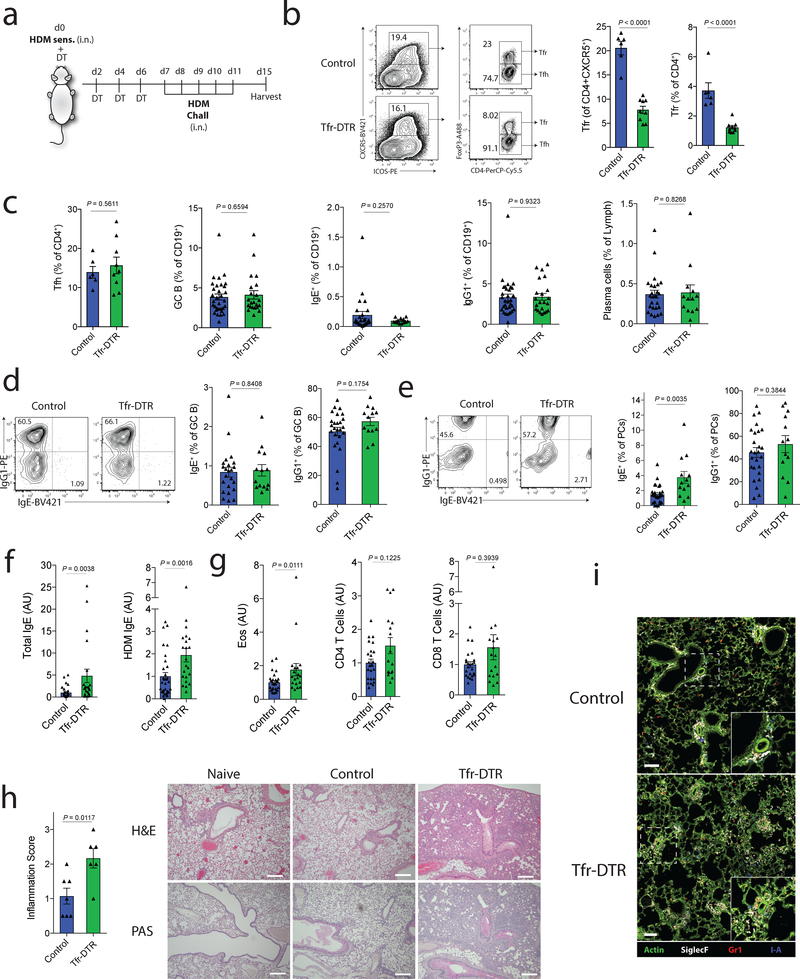
Next we assessed the role of Tfr cells in allergic immunity in vivo. For this, we used a HDM sensitization and challenge model that results in antigen-specific IgE responses and IL-13 dependent eosinophilic lung inflammation10, 35. We sensitized Tfr-DTR or control (Foxp3CreCxcr5wt/wt) mice with HDM and administered DT to delete Tfr cells. On day 7 we challenged mice with HDM (Fig. 7a). We found robust deletion of Tfr cells in Tfr-DTR mice both as a percentage of total CXCR5+CD4+ cells and of total CD4+ cells (Fig. 7b). We did not find any evidence of deletion of activated Treg cells in Tfr-DTR mice (Supplementary Fig. 5a). Deletion of Tfr cells did not result in altered frequencies of Tfh cells, GC B cells, IgG1+ B cells, IgE+ B cells, or plasma cells (Fig. 7c). In addition, deletion of Tfr cells did not alter the relative class switching to IgG or IgE in GC B cells (Fig. 7d). However, when we assessed plasma cells, we found that deletion of Tfr cells resulted in small increases in IgE plasma cells (Fig. 7e). Moreover, Tfr-DTR mice had substantially higher levels of total and HDM-specific IgE compared to control mice (Fig. 7f). These data demonstrate that Tfr cells can control HDM-specific IgE responses in vivo.
Figure 7. Tfr cells regulate HDM-specific IgE responses in vivo.
a) Schematic of HDM sensitization and challenge model to induce lung inflammation. Tfr-DTR (Foxp3Cre Cxcr5LoxStopLoxDTR/WT) or Control (Foxp3Cre Cxcr5WT/WT) mice received HDM sensitization at day 0 followed by DT administration at days 0,2,4 and 6. Mice were challenged with HDM on days 7–11 and harvested on day 15.
b) Analysis of Tfr cells from dLN of HDM challenged mice as in (a). Representative gating (left) and quantification (right) are shown.
c) Quantification of Tfh, GC B cells (CD19+GL7+FAS+), total IgE+ B cells (CD19+IgE+), total IgG1+ B cells (CD19+IgG1+) and total plasma cells (CD138+) from dLN of HDM challenged mice as in (a).
d) Quantification of IgG1 and IgE expression in GC B cells (CD19+GL7+FAS+).
e) Quantification of IgG1 and IgE expression in plasma cells (CD138+).
f) Quantification of total IgE or HDM-specific IgE from HDM challenged mice as in (a). (n=30, Control; n=22, Tfr-DTR)
g) Quantification of Eosinophils (left), CD4 T cells (middle) or CD8 T cells (right) from the BAL fluid of mice as in (a). (n=24, Control; n=17, Tfr-DTR)
h) Lung inflammation scores (left) and representative images of H&E or PAS staining (right) of lung samples. Scale bars = 500μM.
i) Immunofluorescence micrographs of lungs stained for Actin, SiglecF, Gr1 and I-A. Scale bars = 100μM.
Column graphs represent the mean with error bars indicating standard error. P value indicates two-tailed student’s T test (b-e) or Mann-Whitney (f-h).
Data are from individual experiments and are representative of 3 independent experiments (b, c left), are combined data from 4 independent experiments (c right, d-g), or are from one experiment (h-i).
To determine if deletion of Tfr cells results in altered lung inflammation, we analyzed bronchoalveolar lavage (BAL) fluid and found increases in eosinophils (Fig. 7g). Histological analysis of lungs showed increased inflammation consisting of cell infiltration to the airway/vessel walls and alveolar parenchyma in Tfr deleted mice compared to control mice (Fig. 7h). We found the immune cell infiltrate in the lungs of Tfr deleted mice were positive for Gr1 or SiglecF, suggesting the presence of granulocytes and eosinophils, respectively (Fig. 7i, Supplementary Fig. 5b). Taken together, these data demonstrate that Tfr cells regulate HDM-specific IgE responses in vivo and control immune cell infiltrate during HDM sensitization and challenge.
Discussion
The precise role of Tfr cells in modulating B cell responses has been elusive due to the lack of specific mouse models to study Tfr cells. In this study, we developed a novel Tfr-DTR mouse strain to study Tfr cells at distinct stages of immune responses. We found that Tfr cells potently regulate foreign and self-reactive IgG responses, especially prior to initial GC development. Surprisingly, we found a population of IL-13 expressing Tfh13-like cells during HDM challenge, and that Tfr cells potently regulated these cells to control IL-13 and HDM specific IgE responses. Taken together, these data indicate that Tfr cells have a dynamic role in controlling many types of B cell responses to foreign and self-reactive antigens, particularly before initial GC formation.
Although Tfr cells can be found inside and outside of GCs, there has been a debate in the field as to where and when Tfr cells regulate B cells. Tfr cells are less frequently found in GCs compared to the B cell follicle, and cytokines produced in high levels in GCs, like IL-21, can inhibit Tfr cells15, 26. Based on these observations, it has been suggested that Tfr cells do not substantially regulate B cell responses within GCs15. However, Tfr cells can regulate GC B cells in vitro17. Here we demonstrate that Tfr cells potently regulate antibody responses before, but not after, GC formation. Although our experiments cannot eliminate the possibility of Tfr cells having roles within very early GCs, the lack of a phenotype in mature GCs suggest that Tfr cells regulate GC development. However, it is important to note that Tfr cells may have more subtle roles in GCs such as facilitating GC resolution and promoting immune homeostasis.
We also found that Tfr cells regulate memory B cell responses. We propose that Tfr cells regulate memory B cell responses by preventing GC formation where some memory B cells originate. However, it is possible that non-GC memory B cells may also be regulated by Tfr cells in the B cell follicle. Since reactivation of memory B cells may require secondary GCs33, Tfr cells likely have two roles in modulating B cell memory: regulation of memory B cell formation and regulation of secondary GCs during memory B cell reactivation. We additionally found that Tfr cells promote antibody affinity during memory, suggesting that Tfr cells can modulate not only the quantity of antigen specific antibody but also the quality of the antibody. Previous studies found that deletion of Bcl6 in all Treg cell subsets from birth resulted in increased autoreactive antibodies, but these antibodies develop only after months22, 36. We found that deletion of Tfr cells resulted in increases in a variety of autoreactive IgG antibodies and surprisingly, autoreactive IgE antibodies. Autoreactive IgE has been found in autoimmune diseases such as systemic lupus erythematosus (SLE) and has been suggested to enhance autoimmune pathology27, 28, and also been found in models of epithelial damage37.
The role of Tfh and Tfr cells in TH2-like immunity has been unclear. Although Tfh cells are not thought to make IL-13, Tfh cells have been implicated in controlling IgE responses to HDM9, 10. Likewise, attenuated percentages of Tfr cells correlate with worse allergy in patients38. We found evidence of a small frequency of IL-13 producing Tfh cells in vivo after HDM administration. Using an in vitro HDM IgE assay, we showed that these Tfh cells could produce large amounts of IL-13, IL-5 and IL4, and potently stimulate B cell IgE responses. We have referred to these Tfh cells as Tfh13-like cells to distinguish them from previously described Tfh2 cells. Both IL-13 and IgE responses were potently suppressed by Tfr cells. Interestingly, deletion of Tfr cells in vivo during HDM sensitization/challenge resulted in higher levels of antigen specific IgE and increased lung inflammation. High affinity IgE responses occur preferentially through sequential switching of IgG1 to IgE39, 40. Since there was such a profound increase in IgE serum levels and alterations in IgE+, but not IgG1+, plasma cells, these data suggest that Tfr cells likely limit IgE plasma cell responses instead of sequential switching. However, more in depth studies are necessary to fully determine the role of Tfr cells in sequential switching. Taken together, these results demonstrate that Tfh cells can have roles in TH2-like responses to HDM and that these responses are controlled by Tfr cells. Therefore, Tfr cells likely have roles in regulating allergic inflammation and immunity to helminth infections, and modulating Tfr cells may help to control these responses.
Methods
Mice
Foxp3IRES-GFP mice on the C57Bl/6 background have been published previously41. Foxp3IRES-CreYFP and Foxp3DTR mice on the C57Bl/6 background were from Jackson Laboratories. IgHMOG mice were a kind gift from Hartmut Wekerle42. Cxcr5IRES-LoxP-STOP-LoxP-DTR knockin mice were generated by constructing a targeting vector in which an IRES-Frt-PGKNeo-Frt-LoxP-STOP-LoxP-hbegf sequence was placed directly downstream of the stop sequence of Cxcr5. The targeting vector was introduced into C57bl/6 ES cells by electroporation, and the resulting neomycin resistant ES cells were screened for homologous recombination. Positive clones were micro-injected into albino-B6 blastocysts and implanted into pseudopregnant female mice to generate chimeras. Germline transmission was achieved and mice were bred to FlpE carrying mice to remove the NEO cassette29. Resulting mice were then bred to Foxp3IRES-CreYFP mice to generate the Tfr-DTR colony. Mouse progeny were routinely screened for leakiness of the Foxp3IRES-CreYFP allele by flow cytometry. All mice were used according to Brigham and Women’s Hospital and Harvard Medical School Institutional Animal Care and Use Committee and National Institute of Health guidelines.
Immunization
Mice were immunized with 100μg NP-OVA (Biosearch Technologies) or 100μg MOG35–55 (UCLA Biopolymer Facility) emulsified in H37RA CFA s.c. in the mouse flanks as previously described 15, 43, unless otherwise noted. For memory studies, mice were immunized with 100μg NP-OVA mixed with Addavax-MF59-like adjuvant (Invivogen) s.c. in one flank. 30 days later mice received a boost of 100μg NP-OVA in PBS i.p. For allergy studies, mice were sensitized with 10μg HDM (Greer Labs) in PBS intranasally and then challenged with 10μg HDM in PBS intranasally on days 7,8,9,10 and 11, followed by harvesting at day 15. In some cases mice received 1μg of diphtheria toxin in PBS i.p. to delete Tfr cells at indicated timepoints.
Antibodies
The following antibodies were used for surface staining at 4oC for 30 minutes: anti-CD4 (Biolegend, 1:200, RM4–5), anti-ICOS (Biolegend,1:200, 15F9), anti-CD19 (Biolegend,1:200, 6D5), anti-PD-1 (1:200, RMP1–30), anti-CXCR5 biotin (BD Biosciences,1:100, 2G8), anti-GL7 (BD Biosciences,1:200, GL-7), anti-HB-EGF/DTR (RandD Systems, 1:200, AF-259-NA), anti-CD38 (Biolegend, 1:200, 90), anti-CD138 (Biolegend, 1:200, 281–2), anti-IA (Biolegend,1:200, M5/114.15.2), anti-SiglecF (BD Biosciences, 1:200, E50–2440), anti-CD8a (Biolegend, 1:200, 53–6.7), anti-CD11c (Biolegend, 1:200, N418), anti-CD11b (Biolegend, 1:200, M1/70). For CXCR5 detection, streptavidin-BV421 (Biolegend, 1:400, 405225) was used at 4oC. In some cases, anti-IgE (BD Biosciences, 1:200, R35–72) was included to block IgE bound to cell surfaces. For intracellular staining, samples were fixed with the Foxp3 Fix/Perm buffer set according to the manufacturer’s instructions (eBioscience). Samples were then intracellularly stained with anti-IgG1 (BD Biosciences, 1:200, A85–1), anti-IgE (BD Biosciences, 1:200, R35–72), anti-FoxP3 (eBiosciences, 1:200, FJK-16S), anti-Ki67 (BD Biosciences, 1:100, B56), or anti-Glut1 (Abcam, 1:200, EPR3915). In some cases, a donkey anti-goat A647 secondary was used (Invitrogen, 1:400, A-21447). See the “Life Sciences Reporting Summary” for more details.
Sorting
Draining lymph nodes were passed through 70 micron filters and resuspended in PBS supplemented with 1% FBS with 1mM EDTA. CD4+ cells were enriched by magnetic positive selection (Miltenyi Biotec). CD4+ enriched cells were then stained and sorted on an Aria II cell sorter (70μM nozzle) using optimal purity settings. Tfh cells were gated as CD4+ICOS+CXCR5+FoxP3–CD19– and Tfr cells were gated as CD4+ICOS+CXCR5+FoxP3+CD19–. B cells were isolated from flow-through from CD4+ selection, which was then positively selected using CD19 beads (Miltenyi Biotec).
Suppression assays
In vitro stimulation/suppression assays were performed by culturing 5×104 B cells, 3×104 Tfh and 1.5×104 Tfr cells (isolated from dLN of HDM, NP-OVA or MOG35–55 challenged mice) along with 20μg/ml HDM, NP-OVA or rMOG in 96-well culture plates for 6 days. In some cases 20μg/ml anti-IL-4 (Biolegend, 11B11) or anti-IL-13 (R&D Systems, MAB413) were added. Cells and supernatants were collected for flow cytometric and ELISA analysis.
ELISA
For total IgG and IgE levels in serum, Maxisorp (Nunc) plates were coated with anti-mouse Ig (SouthernBiotech) or anti-IgE (BD Biosciences, R35–72) respectively, and serum was incubated. Alkaline-phosphatase secondary reagents (SouthernBiotech) were added, followed by Phosphatase substrate (Sigma), and plates were read on a plate reader (Spectramax). For NP-specific antibody ELISAs, Maxisorp plates were coated with NP-BSA (Biosearch Technologies), followed by secondary reagents above. Anti-HDM IgE kits (Chondrex) were used for HDM specific antibody quantification.
Autoantigen Arrays
Autoantibody reactivities against a panel of autoantigens were measured using an autoantigen microarray platform developed by University of Texas Southwestern Medical Center. Genepix Pro 6.0 software was used to analyze IgG or IgE fluorescence. Antibody score was generated through the following equation: log2(net fluorescence intensity)*signal to noise ratio+1). Mann-Whitney U test was used to determine statistical significance.
RNA-seq
RNA-seq was performed as described previously15. Briefly, RNA was isolated using MyOne Silane Dynabeads (Thermo Fisher Scientific). RNA was fragmented, and barcoded using 8bp barcodes in conjunction with standard Illumina adaptors. Primers were removed using Agencourt AMPure XP bead cleanup (Beckman Coulter/Agencourt) and samples were amplified with 14 PCR cycles. Libraries were gel purified and quantified using a Qubit high sensitivity DNA kit (Invitrogen) and library quality was confirmed using Tapestation high sensitivity DNA tapes (Agilent Technologies). RNA Sequencing reactions were sequenced on an Illumina NextSeq sequencer (Illumina) according to the manufacturer’s instructions, sequencing 50bp reads. Analysis was performed using the CLC Genomics Workbench version 8.0.1 RNA-seq analysis software package (Qiagen). Briefly, reads were aligned (mismatch cost=2, insertion cost=3, deletion cost=3, length fraction=0.8, similarity fraction=0.8) to the mouse genome and differential expression analysis was performed (total count filter cutoff=5.0). Results were normalized to reads per million. Gene-e (Broad Institute) was used to generate heatmaps. The datasets generated during the current study are available on Gene Expression Omnibus (GEO) (Accession number GSE134153) and are available from the corresponding author on reasonable request.
Gene Set Enrichment Analysis (GSEA)
RNA-seq data were compared to Tfh signatures25, Tfr signatures25, or TH2 signatures (GSE14308) using default settings in GSEA analysis software (Broad Institute).
Histology
Paraffin embedded lung sections were stained with hematoxylin and eosin (H&E) or periodic acid-Schiff (PAS). Images were taken on an Olympus BX41 microscope and Olympus DP26 camera, and images obtained using Olympus CellSens v1.1 software. Lung inflammation was scored as: no inflammation: 0, perivascular/peribronchial inflammation without infiltration of airway/vessel walls: 1, infiltration of airway/vessel walls without extension into alveolar parenchyma: 2, destruction of alveolar parenchyma: 3.
Statistics
Most statistical tests were performed using Prism 6.0 (GraphPad) utilizing Student’s two-tailed unpaired t test for normalized data, or Mann Whitney test for non-normal data as indicated. Statistics for RNA-seq was performed using CLC Genomics Workbench (Qiagen). Statistics for gene set enrichment was performed in GSEA (Broad Institute). All measurements were taken from distinct samples, except for memory experiments in which the same mice were bled before and after boost.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Transcriptomic data have been deposited in the Gene Expression Omnibus (GEO) with the accession code GSE134153.
Supplementary Material
Acknowledgements
We would like to thank T. Chatila, R. Anthony, D. Wesemann and M. Carroll for helpful discussions, the MICRON imaging core for help with microscopy, and H. Wekerle and S. Zamvil for reagents. This work was supported by US National Institute of Health through grants K22AI132937 (P.T.S.), P01AI56299 (A.H.S.), R37AI34495 (B.R.B.), R01HL11879 (B.R.B.) and the Evergrande Center for Immunologic Diseases.
Footnotes
Competing interests
The authors declare no competing interests
References
- 1.Crotty S Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29, 621–663 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Victora GD & Nussenzweig MC Germinal centers. Annu Rev Immunol 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Crotty S T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinuesa CG, Linterman MA, Yu D & MacLennan IC Follicular Helper T Cells. Annu Rev Immunol 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Cannons JL, Lu KT & Schwartzberg PL T follicular helper cell diversity and plasticity. Trends Immunol (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstein JS et al. TFH cells progressively differentiate to regulate the germinal center response. Nature immunology 17, 1197–1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luthje K et al. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nature immunology 13, 491–498 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Liang HE et al. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nature immunology 13, 58–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballesteros-Tato A et al. T Follicular Helper Cell Plasticity Shapes Pathogenic T Helper 2 Cell-Mediated Immunity to Inhaled House Dust Mite. Immunity 44, 259–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coquet JM et al. Interleukin-21-Producing CD4(+) T Cells Promote Type 2 Immunity to House Dust Mites. Immunity 43, 318–330 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Noval Rivas M & Chatila TA Regulatory T cells in allergic diseases. The Journal of allergy and clinical immunology 138, 639–652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curotto de Lafaille MA et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity 29, 114–126 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Sage PT & Sharpe AH T follicular regulatory cells. Immunol Rev 271, 246–259 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Maceiras AR, Fonseca VR, Agua-Doce A & Graca L T follicular regulatory cells in mice and men. Immunology 152, 25–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sage PT et al. Suppression by TFR cells leads to durable and selective inhibition of B cell effector function. Nature immunology 17, 1436–1446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sage PT, Paterson AM, Lovitch SB & Sharpe AH The coinhibitory receptor ctla-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity 41, 1026–1039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sage PT, Alvarez D, Godec J, von Andrian UH & Sharpe AH Circulating T follicular regulatory and helper cells have memory-like properties. J Clin Invest 124, 5191–5204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sage PT, Francisco LM, Carman CV & Sharpe AH The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nature immunology 14, 152–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollenberg I et al. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol 187, 4553–4560 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Wu H et al. Follicular regulatory T cells repress cytokine production by follicular helper T cells and optimize IgG responses in mice. Eur J Immunol 46, 1152–1161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linterman MA et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med 17, 975–982 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu W et al. Deficiency in T follicular regulatory cells promotes autoimmunity. J Exp Med 215, 815–825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laidlaw BJ et al. Interleukin-10 from CD4(+) follicular regulatory T cells promotes the germinal center response. Sci Immunol 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JM, Rasmussen JP & Rudensky AY Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature immunology 8, 191–197 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Hou S et al. FoxP3 and Ezh2 regulate Tfr cell suppressive function and transcriptional program. J Exp Med 216, 605–620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayin I et al. Spatial distribution and function of T follicular regulatory cells in human lymph nodes. J Exp Med 215, 1531–1542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dema B et al. Immunoglobulin E plays an immunoregulatory role in lupus. J Exp Med 211, 2159–2168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dema B et al. Autoreactive IgE is prevalent in systemic lupus erythematosus and is associated with increased disease activity and nephritis. PLoS One 9, e90424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sage PT et al. Dendritic Cell PD-L1 Limits Autoimmunity and Follicular T Cell Differentiation and Function. J Immunol 200, 2592–2602 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bettelli E, Baeten D, Jager A, Sobel RA & Kuchroo VK Myelin oligodendrocyte glycoprotein-specific T and B cells cooperate to induce a Devic-like disease in mice. J Clin Invest 116, 2393–2402 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnamoorthy G, Lassmann H, Wekerle H & Holz A Spontaneous opticospinal encephalomyelitis in a double-transgenic mouse model of autoimmune T cell/B cell cooperation. J Clin Invest 116, 2385–2392 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulero P, Midaglia L & Montalban X Ocrelizumab: a new milestone in multiple sclerosis therapy. Ther Adv Neurol Disord 11, 1756286418773025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McHeyzer-Williams LJ, Milpied PJ, Okitsu SL & McHeyzer-Williams MG Class-switched memory B cells remodel BCRs within secondary germinal centers. Nature immunology 16, 296–305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J, Jankovic D, Grinberg A, Guo L & Paul WE Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A 103, 18214–18219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomlinson KL, Davies GC, Sutton DJ & Palframan RT Neutralisation of interleukin-13 in mice prevents airway pathology caused by chronic exposure to house dust mite. PLoS One 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Botta D et al. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nature immunology 18, 1249–1260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crawford G et al. Epithelial damage and tissue gammadelta T cells promote a unique tumor-protective IgE response. Nature immunology 19, 859–870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y et al. Allergen immunotherapy improves defective follicular regulatory T cells in patients with allergic rhinitis. The Journal of allergy and clinical immunology (2019). [DOI] [PubMed] [Google Scholar]
- 39.He JS et al. IgG1 memory B cells keep the memory of IgE responses. Nature communications 8, 641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA & Lafaille JJ Sequential class switching is required for the generation of high affinity IgE antibodies. J Exp Med 209, 353–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only References
- 41.Bettelli E et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Litzenburger T et al. B lymphocytes producing demyelinating autoantibodies: development and function in gene-targeted transgenic mice. J Exp Med 188, 169–180 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sage PT & Sharpe AH In Vitro Assay to Sensitively Measure TFR Suppressive Capacity and TFH Stimulation of B Cell Responses. Methods Mol Biol 1291, 151–160 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Transcriptomic data have been deposited in the Gene Expression Omnibus (GEO) with the accession code GSE134153.