Abstract
In B lymphopoiesis, activation of the pre-B cell antigen receptor (pre-BCR) is associated with both cell cycle exit and Igk recombination. Yet, how the pre-BCR mediates these functions remains unclear. Herein, we demonstrate that the pre-BCR initiated a feed-forward amplification loop mediated by the transcription factor IRF4 and the chemokine receptor CXCR4. CXCR4 ligation by CXCL12 activated the mitogen-activated protein kinase (MAPK) ERK which then directed the development of small pre- and immature B cells including orchestrating cell cycle exit, pre-BCR repression, Igk recombination and BCR expression. In contrast, pre-BCR expression and escape from interleukin 7 (IL-7) had only modest effects on B cell developmental transcriptional and epigenetic programs. These data demonstrate a direct and central role for CXCR4 in orchestrating late B cell lymphopoiesis. Furthermore, in the context of previous findings, our data provide a three-receptor system sufficient to recapitulate the essential features of B lymphopoiesis in vitro.
INTRODUCTION
B lymphopoiesis consists of alternating and mutually exclusive states of either stochastic immunoglobulin gene (Ig) recombination or cell proliferation with selection1. Pro-B cells initiate Ig heavy chain (Igh) recombination. Cells that successfully recombine Igh express Igμ and assemble a pre-B cell antigen receptor (pre-BCR) containing surrogate light chain (λ5 and VpreB). These cells first proliferate as large pre-B cells and then transit to become small pre-B cells where they exit cell cycle and recombine the Ig light chain loci (Igk then Igl)2, 3.
Proliferation in pro and large pre-B cells is driven by IL-7 receptor (IL-7R)-dependent activation of signal-transducer-and-activator-of-transcription 5 (STAT5) and phosphatidylinositol-3-OH kinase (PI(3)K)1. Active STAT5 induces expression of Ccnd3 (encoding Cyclin D3) while PI(3)K activation induces Myc and aerobic glycolysis4, 5, 6, 7, 8 . These signaling pathways also directly repress Igk recombination1, 9.10. In parallel, PI(3)K activation represses expression of the transcription factors FOXO1 and FOXO3, which induce Rag1 and Rag211, 12.
In contrast, canonically the pre-BCR transmits signals that initiate cell cycle exit and Igk recombination. Downstream activation of the protein kinase ERK induce expression of the transcription factors AIOLOS and IKAROS which repress Ccnd3 and Myc4, 13. ERK also induces the transcription factor E2A which binds and activates the Igk intronic enhancer (Eκi)9, 14, 15. Furthermore, escape from IL-7R signaling allows upregulation of FOXO1 and FOXO3. Interestingly, these differentiation mechanisms occur in small pre-B cells where there is concurrent repression of pre-BCR expression1, 16. Therefore, it is unclear if initial transient pre-BCR signaling is sufficient to execute the entire developmental program in small pre-B cells or if other signals are required.
The pre-BCR also upregulates CXCR4 that senses CXCL12 gradients and has been proposed to mediate movement of pre-B cells out of IL-7 rich bone marrow (BM) niches17, 18, 19. By controlling exposure to IL-7, CXCR4 is thought to control the balance between IL-7R and pre-BCR signaling1. However, CXCR4 transmits signals, including activating Ras-ERK, and in cancer has been implicated in multiple processes including invasion, epithelial-mesenchymal transition and proliferation20, 21, 22, 23, 24. Furthermore, in T cell development, CXCR4 synergizes with the pre-TCR to augment proliferation25. These data suggest that CXCR4 can mediate more than chemotaxis.
RESULTS
Small Pre-B cells contact CXCL12+ stroma
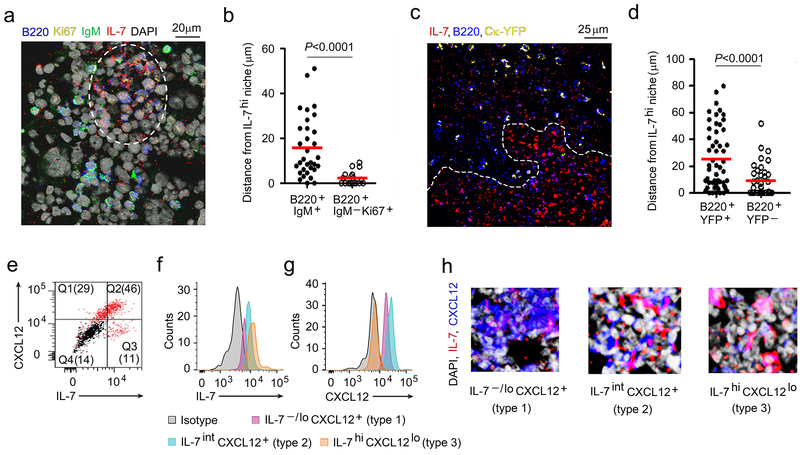
To understand the spatial relationships between proliferating and differentiating B cell progenitors and IL-7, we isolated intact BM cores from WT C57BL/6 mice and stained them with antibodies specific for IL-7, B220, Ki67 and IgM (Fig. 1a). Visualization of stained BM by multicolor confocal microscopy revealed that cycling pro and large pre-B cells (B220+IgM−Ki67+) localized in IL-7hi BM niches while B220+IgM+ B cell progenitors, that mostly include immature B cells, resided in IL-7−/lo BM regions (Fig. 1a,b, Supplementary Fig. 1a).
Figure 1. Location of proliferating and differentiating pre-B cells in the BM.
a, Confocal microscopy of WT BM section (8μm thick femur) stained with antibodies to IL-7 (red), B220 (blue), Ki67 (yellow), IgM (green) and DAPI (gray) to visualize the location of proliferating and IgM+ B cell progenitors. (Single color panels are presented in Supplementary Fig. 1a. The image is representative of 4 independent images from 3 WT mice) b, Distance of IgM+ and IgM−Ki67+ B cell progenitors from IL-7hif niches (white dashed line). Data were pooled from three independent experiments. Distance of each cell counted were shown with mean values (horizontal bars). P values were calculated by unpaired t-test. c, Visualization of B cell progenitors in BM sections (8μm thick femur) of Ck-YFP mice with antibodies to IL-7 (red) and CXCL12 (blue) by confocal microscopy. The image is representative of 3 independent images from 3 WT mice. d, Distances of YFP+ B cell progenitors from IL-7hi niches (white dashed line) in BM of Ck-YFP mice. Data were pooled from two independent experiments. Distance of each cell counted were shown with mean values (horizontal bars). P values were calculated by unpaired t-test. e, Flow cytometric analysis of IL-7 and CXCL12 expression by cultured BM stromal cells (n=3). f, g. Identification of stromal cells expressing IL-7 (i) and CXCL12 (j) by flow cytometry (n=3). k, Different areas of BM showing varying degrees of IL-7 and CXCL12 expressing stroma by Type 1, Type 2 and Type 3 stromal cells. Images are representative of 5 independent areas from 2 WT BM.
To identify the relative position of small pre-B cells in the BM, we used an Igk-YFP (yellow fluorescent protein) reporter mouse in which cells expressing Igk germline (Ck) and rearranged transcripts are marked with YFP26. YFP expression was low in pro-B cells with strong upregulation in small pre-B cells (Supplementary Fig. 1b). There is intermediate expression of YFP in large pre-B cells. However, IgM−YFP+Ki67+ cells were rare in the BM (data not shown) indicating YFP expression in large pre-B cells is insufficient to mark these cells in confocal micrographs. Examination of the position of B220+YFP+ B cell progenitors relative to IL-7 revealed they were excluded from IL-7hi niches (Fig. 1c,d). These studies suggest that small pre-B cells migrate away from IL-7 rich BM regions.
Flow cytometry of Ter119−CD45−BM stromal cells revealed that IL-7 and CXCL12 were usually co-expressed with IL-7intCXCR12+ more common than single-positive populations (Fig. 1e)18. Linear visualization of IL-7 and CXCL12 expression suggested the presence of three populations consisting of IL-7loCXCL12+ (Type 1 cells), IL-7intCXCL12+ (Type 2) or IL-7hiCXCL12lo (Type 3) (Fig. 1f, g). Examination of whole BM single planes revealed wide-spread distributions of each cell type (Supplementary Fig. 1c). However, individual high-power fields revealed distinct areas that were relatively enriched for Type 1, Type 2 or Type 3 cells (Fig. 1h and Supplementary Fig. 1d).
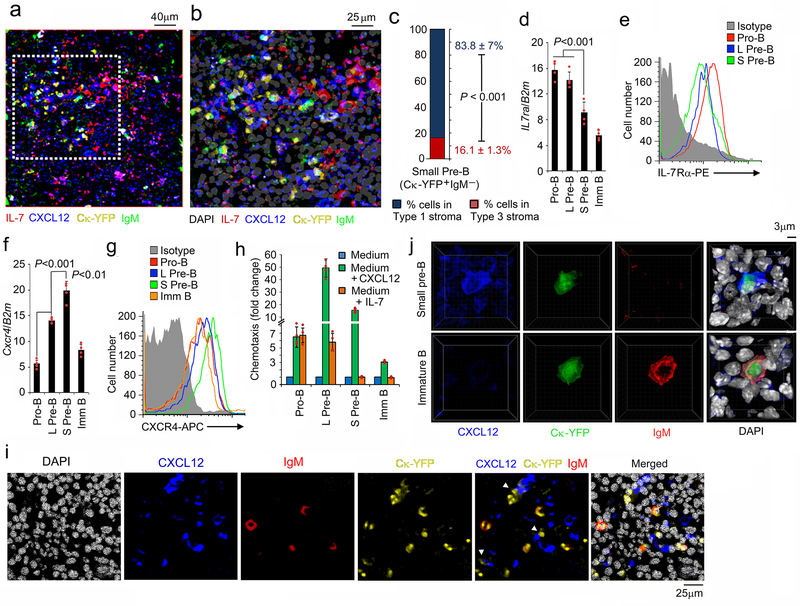
BM from Ck-YFP mice was then stained with antibodies specific for IgM, IL-7 and CXCL12 (Fig. 2a, b). Small pre-B (YFP+IgM−) and immature B (YFP+IgM+) cells resided in niches enriched for Type 1 (IL-7−/loCXCL12+) stromal cells (Fig. 2a-c).
Figure 2. Small pre-B cells are in intimate contact with CXCL12+ stroma.
a,b, Distribution of small pre-B (YFP+IgM−) and immature (YFP+IgM+) B cell progenitors with respect to IL-7hi and CXCL12+ stromal cells in Ck-YFP BM sections (8μm thick femur) stained with antibodies to IL-7 (red), CXCL12 (blue), IgM (green) and DAPI (gray). The area indicated by white dashed line in ‘a’ was magnified in ‘b’. The image is representative of 4 independent images. c, Frequency of small pre-B (YFP+IgM−) cell progenitors in contact with type 1 (IL-7−/loCXCL12+) and type 3 (IL-7hiCXCL12lo) stromal cells. d-g, Quantitative real time PCR analyses of expression of Il7rα (d) and Cxcr4 (f) and flow cytometric analysis of corresponding cell surface expression of the IL-7Rα (e) and CXCR4 (g) on indicated B cell progenitor populations. (n=4); Data are presented as mean±SD. P values were determined by unpaired t-test. h, Chemotaxis of different B cell progenitors to IL-7 (10ng/ml) and CXCL12 (100ng/ml) by transwell migration assay. Data are normalized to migration with medium alone (n=4). Data are presented as mean±SD. i, Distribution of small pre-B (YFP+IgM−) and immature B (YFP+IgM+) cells in the CXCL12+ niches in BM (8μm thick femur) of Ck-YFP mice stained with antibodies to CXCL12 (blue), IgM (red) and DAPI (gray). The image is representative of 3 independent images. j, Three-dimensional reconstructions of small pre-B (YFP+IgM−) and immature B (YFP+IgM+) cells in the BM of Ck-YFP mice stained with antibodies to CXCL12 (blue), IgM (red) and DAPI (gray) using Imaris software (representative of n=9).
Examination of B cell progenitors for IL-7R and CXCR4 expression, by both mRNA and surface staining (Fig. 2d-g), revealed that IL-7R and CXCR4 were reciprocally expressed with small pre-B cells expressing relatively high surface densities of CXCR4 and low densities of IL-7R. Both IL-7R and CXCR4 were downregulated in immature B cells. Although pro-B and large pre-B cells can migrate along an IL-7 gradient, direct comparison of in vitro chemotaxis revealed that both large pre-B and small pre-B cells responded strongly to CXCR4 (Fig. 2h). Interestingly, large pre-B cells showed the strongest chemotaxis even though CXCR4 surface densities were higher on small pre-B cells. However, most small pre-B cells (YFP+IgM−) were in intimate contact with CXCL12+ stroma while IgM+ immature B cells resided in the same area but were not contacting CXCL12+ stroma (Fig. 2i). Additional HPFs with 3D reconstruction demonstrated that small pre-B cells were in tight contact with both CXCL12+ cells and high local accumulations of extracellular CXCL12 (Fig. 2j and Supplementary Fig, 1e). Furthermore, these small pre-B cells clearly had CXCL12 in their cytoplasm suggesting recent internalization of this ligand. This tight association suggests that CXCR4 might be doing more than positioning small pre-B cells away from IL-7.
CXCR4 directly regulates pre-B cell differentiation
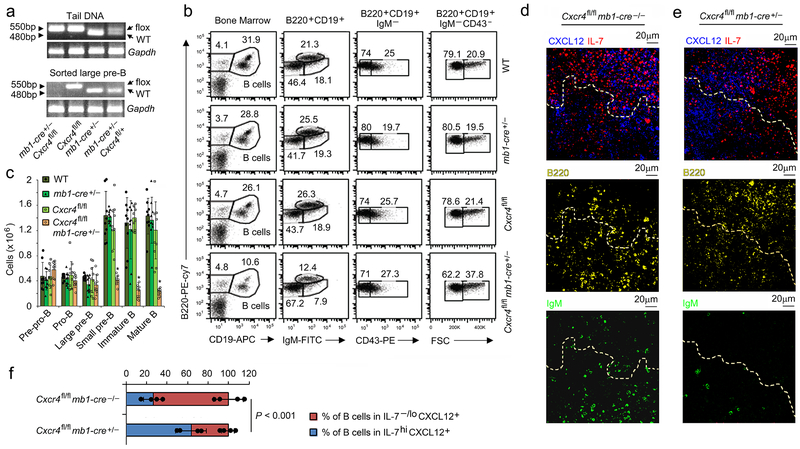
We next crossed Cxcr4fl/fl mice to mice expressing the Cre recombinase under the mb-1 promoter (mb1-Cre)27, 28. In these mice, deletion of Cxcr4 in large pre-B cells appeared complete with no detectable undeleted alleles in Cxcr4fl/fl mb1-Cre+/− (hereafter referred to as Cxcr4−/−) mice (Fig. 3a). BM was harvested from WT, mb1-Cre+/−, Cxcr4fl/fl and Cxcr4−/− mice and analyzed by flow cytometry. While the pre-pro to large pre-B cell compartments were normal in Cxcr4−/− mice (Fig. 3b-c), the number of small pre-B cells was decreased approximately three-fold while the number of immature and mature B cells was decreased five-fold. Multicolor confocal microscopy of BM from Cxcr4fl/fl and Cxcr4−/− mice revealed that almost all Cxcr4−/− IgM− B cell progenitors were near IL-7hi BM stroma with few IgM+ cells (Fig. 3d-f, Supplementary Fig. 2a,b). These data indicate that CXCR4 is required for normal development of small pre-B cells and their positioning away from IL-7hi BM niches
Figure 3. Cxcr4 is required for development of small pre-B cells.
a, Genomic PCR of WT and floxed alleles for Cxcr4 deletion in tail DNA and flow sorted large pre-B cells from indicated mice with Gapdh as control (n=3). b, Flow cytometric analysis of different developmental stages of B lymphopoiesis in the BM of wild-type (WT), mb1-cre+/−, Cxcr4fl/fl and Cxcr4fl/fl- mb1-cre+/− littermate control mice (n=9). B cell progenitors are defined as B220+CD19+, pro-B cells as B220+CD19+CD43+IgM−, large and small pre-B as B220+CD19+CD43−IgM−FSChi and B220+CD19+CD43−IgM−FSClo respectively and immature B cells as B220+CD19+CD43−IgM+. FSC, forward scatter. c, Absolute number of cells per mouse at different stages of B cell development in BM of WT, mb1-cre+/−, Cxcr4fl/fl and Cxcr4fl/fl-mb1-cre+/− mice (n=9)(Imm B, Immature B cell; Mat B, mature B cells. *P<0.001 compared to all the controls (WT, mb1-cre+/−, Cxcr4fl/fl). Data presented as average ± SD. d,e, Distribution of B cell progenitors in BM of Cxcr4fl/fl (CXCR4 sufficient; d) and Cxcr4fl/fl-mb1-cre+/− (CXCR4 deficient; e) mice by confocal microscopy of corresponding BM sections (8μm thick femur) stained with antibodies to IL-7 (red), CXCL12 (blue), B220 (yellow), IgM (green) and DAPI (gray) (n=4). f, Percentage of B cell progenitors (B220+) in proximity to IL-7−/loCXCL12+ and IL-7hiCXCL12+/−stroma (n=4 independent image). Data are presented as mean±SD. Each dot represents average distance obtained from each image of indicated genotype. P values were calculated by unpaired t-test.
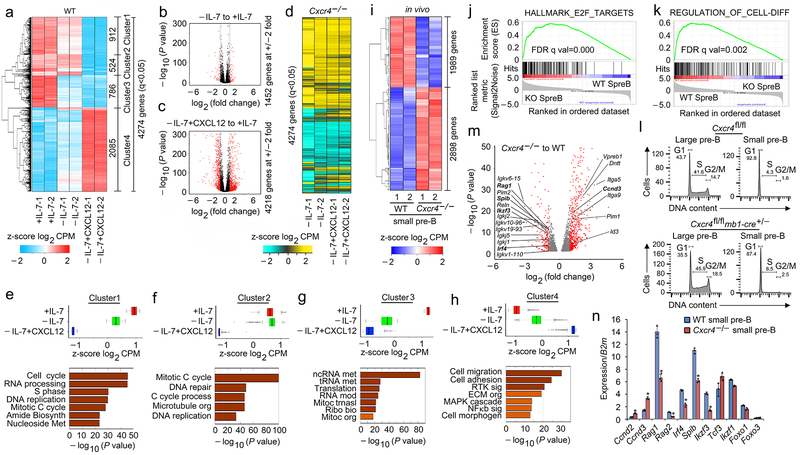
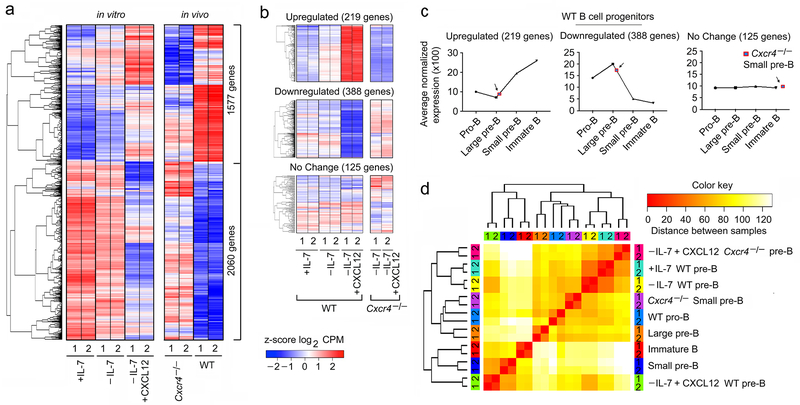
Previously, it has been reported that in vitro, withdrawal of IL-7 is sufficient to induce pre-BCR activation, cell cycle exit and Igk recombination. However, these experiments were done using the stroma feeder cell line OP9 which express CXCL124, 9, 17, 29, 30. Furthermore, there was no other positive control. Therefore, in a stroma cell free system, B220+IgM− progenitors from WT BM were grown in the presence of high IL-7 (16 ng/ml) for five days followed by two days of culture with IL-7 (16 ng/ml, +IL-7) or low IL-7 (0.2 ng/ml, referred hereafter as −IL-7) without or with CXCL12 (100 ng/ml, +CXCL12). Cells were harvested and subjected to RNA-Seq. Overall, the combination of withdrawing IL-7 and adding CXCR4 regulated 4,274 genes (Fig. 4a). Plotting fold-change versus significance (volcano plots) revealed that CXCL12 regulated many more genes by two-fold or more than IL-7 withdrawal alone (Fig. 4b,c). To confirm the direct role of CXCR4 in signaling, we cultured B220+IgM− progenitors from Cxcr4−/− BM and assayed for gene expression in cells withdrawn from IL-7 without or with CXCL12. As expected, addition of CXCL12 did not substantially regulate genes in Cxcr4−/− B cell progenitors (Fig. 4d).
Figure 4. Pre-B cell differentiation is directly regulated by CXCR4 signaling.
a, Hierarchical clustering of differentially regulated genes (RNA-Seq) identified at q<0.05 in WT pre-B cells (replicates shown), cultured for 48 hrs with 16ng/ml of IL-7 (+IL-7) or 0.2ng/ml of IL-7 (−IL-7) or 0.2ng/ml of IL-7 with 100ng/ml of CXCL12 (−IL-7+CXCL12) without stromal cells. b, c, Volcano plots of differential expression of genes in WT pre-B cells cultured with or without IL-7 (b) or −IL-7+CXCL12 versus −IL-7 (c) (n=2 RNA-seq; FDR<0.01). Red dots are genes with significantly increased (right side) or decreased (left side) expression. −log 10 of the P values from Kal’s statistical test. d. Heat map of same genes in same order shown in (a) from RNA-Seq of Cxcr4−/− pre-B cells (replicates shown), cultured as indicated. e-h, Average expression of all differentially expressed genes in Cluster 1 (e), Cluster 2 (f), Cluster 3 (g) and Cluster 4 (h) of WT pre-B cells in +IL-7 or −IL-7 or −IL-7+CXCL12 culture conditions and corresponding gene ontology analysis. Only terms with FDR <5%, P<0.05 and over log2 2 fold enrichment were reported. Gene expression for individual RNA-seq replicates were averaged and the z-scores are plotted. Box represents interquartile range (IQR; Q1 -Q3 percentile) with black horizontal line representing the median. Maximum and Minimum are defined as Q3+1.5*IQR and Q1-1.5*IQR respectively. Outliers are indicated as black dots along the whiskers. −log 10 of the P values are from Kal’s statistical test. i, Heat map of RNA-Seq with clustering of upregulated and downregulated genes in flow-purified Cxcr4fl/fl-mb1-cre+/− (Cxcr4−/−) and WT small pre-B cells (q<0.05, replicates shown). j, k, Gene set enrichment analysis (GSEA) of ‘HALLMARK_E2F_TARGETS’ (j) and ‘REGULATION_OF_CELL_DIFFERENTIATION’. GSEA reports q-values based on the median of the p-value distribution. (k) pathways enriched in Cxcr4−/− (KO) small pre-B cells compared to WT small pre-B cells. l, Cell cycle analysis of flow purified large and small pre-B cells from Cxcr4fl/fl and Cxcr4fl/fl-mb1-cre+/− (Cxcr4−/−) mice (n=2). m, Volcano plot of differentially expressed genes in Cxcr4−/− versus WT small pre-B cells. Red dots are genes with significantly increased (right side) or decreased (left side) expression in Cxcr4−/− small pre-B cells compared to WT (FDR<0.01). −log 10 of the P values are from Kal’s statistical test. n, Quantitative real time PCR analyses of the expression of Ccnd2, Ccnd3, Rag1, Rag2, Irf4, Spib, Ikzf3 (encodes AIOLOS), Tcf3 (encodes E2A), Ikzf1 (encodes IKAROS), Foxo1 and Foxo3 in flow purified WT and Cxcr4−/− small pre-B cells (n=3). Data presented as average ± SD. P values were determined by unpaired t-test. (*P<0.001 compared to expression in WT small pre-B cells). (Primers and probes are presented in Supplementary Table 1)
There were four distinct gene clusters regulated by CXCL12 and IL-7 withdrawal (Fig. 4a). In Cluster 1 (Fig 4e), downregulation of genes was primarily dependent on CXCL12. Gene ontology analysis revealed that these genes were enriched for cell cycle, metabolism and RNA processing programs. Among the cell cycle genes, Ccnd2, Ccnd3 (Supplementary Fig. 3a), Myc (Supplementary Fig. 3b) and MYC-dependent genes (Supplementary Fig. 3c, gene set enrichment analysis, GSEA) were strongly repressed by CXCL12. In Cluster 2 (Fig. 4f), gene repression was exclusively dependent on CXCL12 and these genes were enriched for mitosis and DNA replication and repair pathways. In Cluster 3 (Fig. 4g), gene repression was mostly dependent upon IL-7 withdrawal with an enrichment for RNA metabolism. CXCL12 primarily induced genes in Cluster 4 (Fig. 4h) implicated in cell migration, cell adhesion and signaling including the NF-κB (Supplementary Fig. 3d,e) and MAPK pathways. Transcription factors critical for late B lymphopoiesis were also induced including Irf4, Irf8, SpiB and Ikzf3 (encoding AIOLOS) (Supplementary Fig. 3f). CXCL12 did not induce Tcf3 (encodes E2A), Pax5, Foxo1, Foxo3 or Ikzf1 (IKAROS) (Supplementary Fig. 3g). Also, within genes induced in Cluster 4 were Rag1, Rag2 and Brwd110, 31 (Supplementary Fig. 3h, i). Overall, withdrawal of IL-7, and addition of CXCL12, were associated with repression of cell cycle programs, including E2F (Supplementary Fig. 3j) and induction of cell differentiation programs (Supplementary Fig. 3k).
We next isolated WT and Cxcr4−/− small pre-B cells and subjected them to RNA-Seq (Fig. 4i). There were 4,887 differentially expressed genes (q<0.05) in Cxcr4−/− small pre-B cells with increased expression being more common than decreased expression. Gene ontology analysis revealed that CXCR4 was required for the repression of cell cycle, metabolic pathways and DNA replication and repair pathways (Supplementary Fig. 3l). In contrast, CXCR4 was required to upregulate signaling pathways including Ras, NF-κB, cell motility and cell adhesion (Supplementary Fig. 3m). GSEA confirmed that CXCR4 was necessary for repressing cell cycle genes, including E2F targets, and inducing differentiation programs (Fig. 4j,k). More Cxcr4−/− large and small pre-B cells were progressing through cell cycle than WT cells (Fig. 4l). Volcano plots confirmed that CXCR4 was required for broadly repressing cell cycle genes and inducing differentiation genes (Fig. 4m). Many of these genes were confirmed by qPCR (Fig. 4n). Expression of some B cell lineage and maintenance TFs including, Tcf3, Ikzf1, Foxo1 and Foxo3 were not different in Cxcr4−/− small pre-B cells10, 29, 32, 33.
Repression of surrogate light chain is critical for subsequent antigen selection and tolerance in immature B cells16. The presence or absence of IL-7 had no significant effect on Vpreb1, Vpreb2 or Igll1 (λ5) transcription (Supplementary Fig. 4a-c). In contrast, addition of CXCL12 repressed Vpreb1, Vpreb2 and essentially silenced Igll1 transcription. CXCL12-mediated repression was dependent on CXCR4. In vivo, Vpreb1, Vpreb2 and Igll1 expression were highest in pro-B and large pre-B cells and low in small pre-B cells (Supplementary Fig. 4d-f). Furthermore, neither Vpreb1, Vpreb2 nor Igll1 were repressed in Cxcr4−/− small pre-B cells. Consistent with these data, in in vitro cultured pre-B cells CXCR4, but not IL-7 withdrawal, strongly repressed pre-BCR expression (Supplementary Fig. 4g). In toto, these data indicate that CXCR4 directly regulates specific developmental programs of late B lymphopoiesis.
CXCR4 signaling determines small pre-B cell identity
Next, we compared the transcriptional programs of WT in vitro cultured B cell progenitors under different conditions to those genes differentially expressed by Cxcr4−/− and WT small pre-B cells. Approximately, 85% of genes differentially regulated in vitro (3637 of 4274 genes) were also differentially expressed in vivo. Clustering of these genes revealed that the direction of regulation by the CXCL12/CXCR4 axis, was similar in vitro and in vivo (Fig. 5a).
Figure 5. CXCR4 signaling determines small pre-B cell identity.
a, Hierarchical clustering of differentially regulated genes (RNA-Seq) identified at q<0.05 in WT pre-B cells, cultured as indicated compared to flow-purified Cxcr4fl/fl-mb1-cre+/− (Cxcr4−/−) and WT small pre-B cells (q<0.05). Common differentially expressed genes (both in in vitro and in vivo) were plotted in same order. b, Heat map of differentially regulated genes in vitro that changed by at least two-fold, were highly expressed (at least 1/10th of B2m expression), and in which differential regulation was statistically robust (P<10−5). Upregulated, downregulated and representative unchanged genes from indicated culture conditions were presented in the top, middle and bottom panels respectively. c, Average expression of the same upregulated (top panel), downregulated (middle panel) and unchanged genes (bottom panel) in different in vivo WT B cell progenitors indicated. d, Comparison of transcriptional programs regulated by CXCR4 both in in vivo and in vitro based on the expression levels of the 3637 genes that were differentially expressed under any condition. Hierarchical clustering was run on z-scored Log2 normalized expression levels, using a Euclidian correlation distance metric. The clustering dendrogram was plotted against the distance matrix for all indicated samples.
We then focused on those genes that were differentially regulated in vitro by at least two-fold, were highly expressed (>1/10th B2m expression), and in which differential regulation was statistically robust (P<10−5). With these parameters, withdrawal of IL-7 and addition of CXCL12 induced 219 genes and repressed 388 genes (Fig. 5b). As a control, 125 highly expressed genes were randomly chosen that demonstrated no significant change in transcription. Transcription of those genes induced by CXCL12 was dependent upon expression of CXCR4.
Plotting expression of each in vitro regulated gene group as a function of normal B cell development demonstrated that those genes strongly induced by CXCL12 in vitro were also strongly upregulated in vivo (Fig. 5c). The converse was true in that those genes repressed by CXCL12 decreased in expression upon transition to the small pre-B cell stage. Those genes that were not regulated in vitro, did not change in expression during B lymphopoiesis.
We next next captured all genes differentially regulated under any in vitro condition or in vivo in WT versus Cxcr4−/− small pre-B cells. These genes were then used to cluster the indicated different in vitro and in vivo populations (Fig. 5d). Interestingly, in vitro cultures in which IL-7 was present or absent, or cultures in which CXCL12 was added to Cxcr4−/− cells, largely clustered together. These in vitro conditions were most closely related to Cxcr4−/− small pre-B cells and then WT Pro-B cells. In marked contrast, those in vitro cultured cells in which IL-7 had been withdrawn, and CXCL12 added, strongly clustered with ex vivo small pre-B cells. These data indicate that withdrawal of IL-7 and addition of CXCL12 in vitro largely reconstitutes the in vivo developmental progression from proliferating pro-B cells to small pre-B cells.
CXCR4 dictates small pre-B cell chromatin accessibility
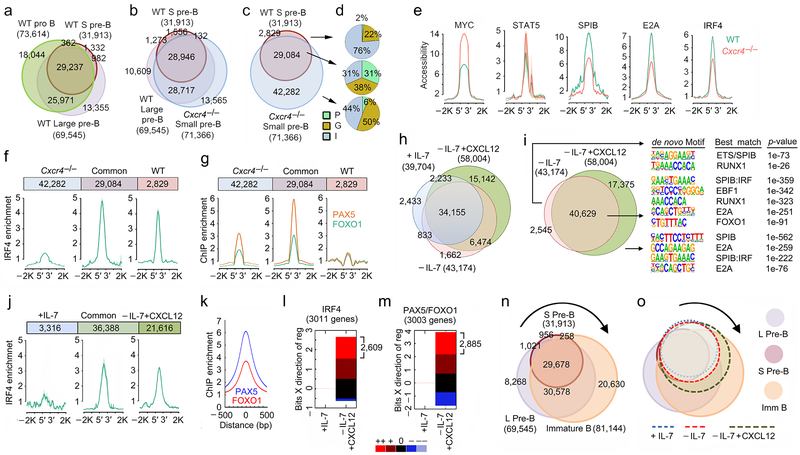
Examination of chromatin accessibility (ATAC-Seq) as a function of normal B cell development revealed a progressive loss of accessibility from pro-B cells through the small pre-B cell (31,913 peaks) stage (Fig. 6a)31. In contrast, in Cxcr4−/− small pre-B cells, there were 71,366 open peaks with 81% of these common with those of proliferative large pre-B cells (Fig. 6b).These data demonstrate that in the absence of CXCR4, small pre-B cells adopt a chromatin landscape with features of large pre-B cells.
Figure 6. CXCR4 signaling sets small pre-B cell epigenetic landscape.
a, Total and overlapping open chromatin regions (ATAC-Seq) in flow-purified WT pro-B, large pre-B and small pre-B cell progenitor populations. b, Total and overlapping open chromatin regions (ATAC-Seq) in flow-purified WT large pre-B, small pre-B and mb1-Cre+Cxcr4fl/fl (Cxcr4−/−) small pre-B cells. c, Overlap of accessible regions from flow-purified WT and Cxcr4−/− small pre-B cells. In a, b and c, total number of peaks for each population shown in parentheses with number in each Venn region indicated. d, Distribution of accessible regions across genome in promoters (P), gene bodies (G) and intergenic (I) regions. e, Average accessibility (ATAC-Seq) at MYC, STAT5, SPIB, E2A and IRF4 bound sites (determined from ChIP-Seq data) in WT and Cxcr4−/− small pre-B cells. f, Average enrichment at IRF4 binding (WT ChIP-Seq) in unique and common accessible regions of WT and Cxcr4−/− small pre-B cells. g, Average enrichment at PAX5 and FOXO1 binding (ChIP-Seq) in unique and common accessible regions of WT and Cxcr4−/− small pre-B cells. In ‘e’, ‘f’ and ‘g’ the plots represent average of two independent ChIP-seq and ATAC-seq data sets. h, Total and overlapping open chromatin regions (ATAC-Seq) in WT pre-B cells, cultured for 48 hrs with as indicated before (n=2). i, Total and overlapping accessible regions in −IL-7 and −IL-7+CXCL12 cultured WT pre-B cells (middle) and the de novo TF binding motifs associated with unique and common accessible regions. j, Average enrichment of IRF4 binding (ChIP-Seq) in unique and common accessible regions of +IL-7 and −IL-7+CXCL12 cultured WT pre-B cells. k, Average enrichment of PAX5 and FOXO1 binding (ChIP-Seq) in accessible regions of −IL-7+CXCL12 cultured WT pre-B cells. In ‘j’, and ‘k’ the plots represent average of 3 independent ChIP-seq data sets for IRF4 and 2 independent ChIP-seq data sets for FOXO1 and PAX5 respectively. l, m, EMBER analysis combining ChIP-Seq results for IRF4 (l) and PAX5/FOXO1 (m) with expression (RNA-Seq) assessing predominant expression patterns of genes within 100kb of IRF4 (l) and PAX5/FOXO1 (m) bound sites in WT pre-B cells cultured as indicated. Change in expression calculated relative to that in +IL-7 cultures WT pre-B cells. In some instances, more than one gene was within 100kb of a peak giving more genes than number of accessible peaks. Change in mean expression categorized as follows: --,>−3 s.d.; -, −1 s.d. to −3 s.d.; 0, −1 s.d. to +1 s.d.; +, 1 s.d. to 3 s.d.; ++, >3 s.d. (where s.d. is the sum of the s.d. values calculated for experimental replicates). n, Total and overlapping open chromatin regions (ATAC-Seq) in flow-purified WT large pre-B, small pre-B and immature B cell progenitors. Total number of peaks for each population shown in parentheses with number in each Venn region indicated. o, Total and overlapping open chromatin regions in WT pre-B cells cultured as indicated (+IL-7, −IL-7 or −IL-7+CXCL12) overlaid on accessibility observed in flow-sorted WT progenitors.
Comparison of accessible regions in Cxcr4−/− small pre-B cells to WT small pre-B cells revealed a preferential closing of intragenic and intergenic chromatin areas in WT small pre-B cells (Fig. 6c,d). Fifty-six percent of intergenic and 73% of intragenic accessibility peaks were in chromatin regions bearing epigenetic enhancer marks (H3K4me1+H3K27Ac+, data not shown). Of the 2,829 accessibility peaks that did not open in Cxcr4−/− small pre-B cells, only 16% were at enhancers (data not shown). These data suggest that CXCR4 preferentially silences large pre-B cell-specific enhancers.
Analysis of differentially regulated accessibility peaks for TF binding motifs (Supplementary Fig. 5a) revealed enrichment for those predicted to be recognized by IRF, SPIB, NF-κB and E2A in all accessible regions. However, CXCR4 appeared to specifically repress a subset of RUNX, STAT5, and MYC predicted binding sites, all of which are TFs important for earlier stages of B cell development9, 31, 34, 35. CXCR4 also repressed accessibility at sites known to bind STAT5 and Myc during B lymphopoiesis (Fig. 6e)9, 35. Conversely, CXCR4 was required to open binding motifs for the TFs E2A, SPIB, and IRF4 (Fig. 6e). CXCR4 also repressed accessibility at sites low or moderately enriched for IRF4, PAX5 and FOXO1 binding sites. It also opened sites poorly enriched for PAX5 and FOXO1 binding sites and highly enriched for IRF4 binding sites (Fig. 6f, g)29, 31. These findings suggest that in vivo, CXCR4 preferentially represses STAT5 and Myc binding while opening the genome at sites of IRF4 binding.
We next asked how signaling through CXCR4 modulates chromatin accessibility in vitro. In contrast to the in vivo studies, addition of CXCL12 enhanced overall chromatin accessibility (Fig. 6h). The genomic sites specifically opened by CXCR4 signaling were enriched in SPIB, E2A and SPIB:IRF composite motifs (Fig. 6i and Supplementary Fig. 5b). This was similar to the TF motif sites enriched in WT small pre-B cells compared to Cxcr4−/− small pre-B cells. Increased accessibility at sites that can bind IRF4 was observed (Fig. 6j). In addition, CXCR4 signaling was associated with enrichment at sites that can bind PAX5 and FOXO1 (Fig. 6k). As an example, IRF4 binds strongly to the Jk proximal promoter and Eκi, which become accessible both in WT small pre-B cells and in in vitro pre-B cells cultured with CXCL12 (Supplementary Figure 5c). Brwd1 activation was associated with enhanced accessibility at sites that can bind IRF4 in the promoter and proximal intronic enhancer and enhanced accessibility at sites that can bind both PAX5 and FOXO1 in the proximal enhancer. FOXO1 alone binds the open 3’ intronic enhancer29. Overall, there was increased accessibility at important TF binding sites, including those for IRF, PAX5 and FOXO, in both promoters and enhancers (H3K4me1+H3K27Ac+) of WT small pre-B cells and of −IL-7+CXCL12 cultured pre-B cells (Supplementary Fig. 5d). These in vitro data suggest that CXCR4 signaling plays an additional role in opening PAX5 and FOXO sites in late B lymphopoiesis.
Analysis of gene expression within 100 kb of IRF4 bound peaks revealed 3011 genes. Of these, 2609 (86.7%) were strongly induced by IL-7 withdrawal and addition of CXCL12 (Fig. 6l). Similarly, there were 3,003 genes within 100 kb of a PAX5 and/or FOXO1 bound site. Of these, 2,885 (96.1%) were induced in −IL-7+CXCL12 treated small pre-B cells (Fig. 6m). These data suggest that withdrawal of IL-7 and addition of CXCL12 induce both IRF4 and FOXO1/PAX5 binding to regulate transcriptional programs of late B lymphopoiesis.
In vivo, CXCR4 appeared to repress overall chromatin accessibility while in vitro it enhanced accessibility. To understand this apparent paradox, we compared those accessibility peaks induced by CXCR4 in vitro to those of later stages of in vivo B cell development (Fig. 6n,o and Supplementary Fig. 5e-g). During normal B lymphopoiesis there was a radical increase in genomic accessibility from small pre-B cells to immature B cells. Most accessibility peaks opened by CXCR4 in vitro represent peaks found in immature B cells. These sites included both de novo accessibility peaks and peaks that were previously open in large pre-B cells. Our in vivo data reveals that CXCR4 signaling, and not IL-7 withdrawal, is largely responsible for repressing accessibility in large pre-B cells. Our in vitro data reveals an additional role for CXCR4, at the next stage of B cell development, in contributing to the open chromatin state of immature B cells.
CXCR4 signaling is essential for Igk recombination
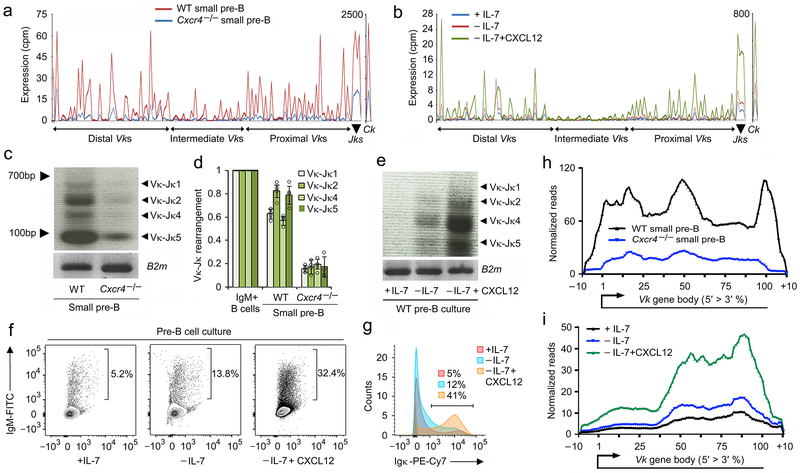
Initiation of Igk rearrangement is the hallmark of pre-B cell differentiation. Therefore, we next examined transcription at Igk in WT and Cxcr4−/− small pre-B cells. Transcription across the whole Igk locus was dependent upon CXCR4, both at the Vk and Jk gene segments (Fig. 7a). To determine the direct role of CXCR4, we examined the corresponding in vitro data (Fig. 7b). Withdrawal of IL-7 only induced modest transcription across the Igk locus. In contrast, addition of CXCL12 induced a pattern of transcription broadly similar to that observed in vivo in WT small pre-B cells.
Figure 7. CXCR4 signaling is necessary for Igk recombination.
a, Expression (RNA-Seq) of all Vks, Jks and Ck genes in WT and mb1-Cre+/−Cxcr4fl/fl (Cxcr4−/−) small pre-B cells (n=2). b, Expression (RNA-Seq) of all Vks, Jks and Ck genes in WT pre-B cells cultured as indicated (n=2). c, Semi-quantitative PCR analysis of Igk rearrangements in WT and Cxcr4−/− small pre-B cells. Data is representative of three experiments. d, Quantitative analysis of Vk-Jk rearrangements in WT and Cxcr4−/− small pre-B cells compared to WT immature B cells. (n=4). Data are presented as mean±SD. P values were determined by unpaired t-test. *P<0.001 versus WT small pre-B cells. e, Semi-quantitative PCR analysis of Igk rearrangements in cultured WT pre-B cells. Data is representative of three experiments. f, Surface IgM+ B cells after culture of WT pre-B cells as indicated. Data is representative of four experiments. g, Histogram of Igκ expression pre-B cells cultured as indicated (n=3). h,i, Quantified and integrated transcription across all Vk genes segments in in vivo B cell progenitors (h) and in vitro cultured pre-B cells (i) including immediately before and after Vk gene bodies.
We next examined Vκ-Jκ recombination. Semi-quantitative and quantitative PCR of Vκ-Jκ genomic recombination in Cxcr4−/− small pre-B cells revealed severely diminished recombination compared to either WT immature or small pre-B cells (Fig. 7c,d). Likewise, Vκ-Jκ recombination was dependent upon CXCL12 in vitro (Fig. 7e). Furthermore, staining in vitro differentiated B cell progenitor cultures for Igμ or Igκ demonstrated that addition of CXCL12 enhanced the frequency of cells expressing a BCR (Fig. 7f,g). These data demonstrate CXCR4 is required for efficient Vκ-Jκ recombination.
Diminished Vk transcription could be a consequence of reduced recombination and/or could reflect diminished accessibility prior to recombination. Therefore, we quantified and integrated transcription across all Vk gene segments including immediately before and after the Vk gene bodies (Fig. 7h,i). Transcription of the Vk gene bodies occurs both before and after recombination. In contrast, sequences immediately downstream of Vk are lost following recombination and therefore reflect pre-recombination transcription. As can be seen both in vitro and in vivo, Vk transcription is globally diminished including transcription in the immediate downstream flanking regions containing the nine nucleotide Vk recombination signal sequence (RSS) (beginning of 100-110% interval). The difference in transcription through the RSS is more pronounced in ex vivo cells. This difference in magnitude might reflect the higher rate of ongoing Igk recombination observed in vitro in the absence of CXCL12. Therefore, transcription pre-recombination, and therefore accessibility, is dependent upon CXCR4.
CXCR4-mediated ERK activation drives B cell development
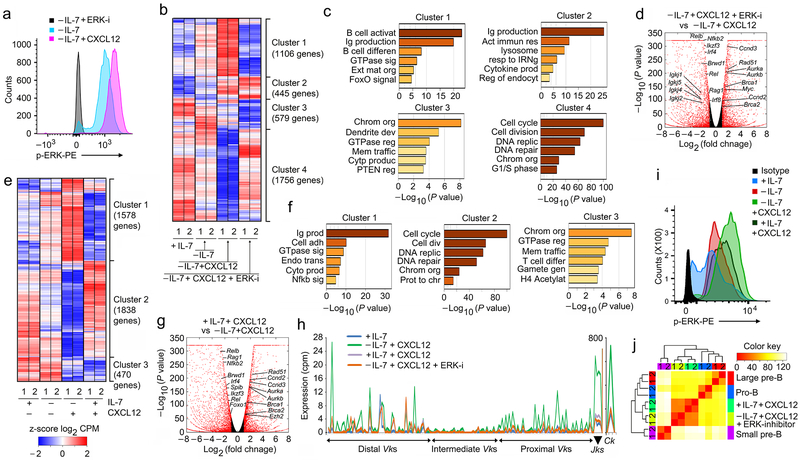
One of the principal signaling mediators of CXCR4 is ERK20, 22, 36. In vitro stimulation of differentiated B cell progenitors with CXCL12 in the absence of IL-7 increased intracellular phospho-ERK, which was blocked by ERK inhibitor (Fig. 8a). Therefore, B cell progenitors were then cultured in the absence of IL-7, with and without CXCL12, as well as with CXCL12 plus ERK inhibitor. Under these conditions, four transcriptional clusters were apparent (Fig. 8b,c). In the largest clusters, Clusters 1 and 4, treatment with ERK inhibitor largely reversed the transcriptional program induced by CXCL12. Cluster 2 transcription was mostly unaffected by ERK inhibitor while Cluster 3 was repressed by ERK inhibition. Gene ontology analysis of each cluster revealed that ERK both induced Ig production and B cell differentiation programs (Cluster 1) and repressed cell cycle programs (Cluster 4)(Fig. 8c). The magnitude of ERK-dependent transcription was readily apparent in a volcano plot of differentially regulated genes which confirmed that ERK was required to repress cell cycle genes and induce transcription of Igk and other differentiation genes (Fig. 8d). These data indicate that ERK mediates most of the transcriptional program downstream of CXCR4.
Figure 8. ERK signaling downstream of CXCR4 enables pre-B cell differentiation.
a, Flow-cytometry for intracellular phosphorylated ERK (p-ERK) in WT pre-B cells cultured as indicated. Negative control is -IL-7 cultured cells in presence of ERK inhibitor. Data is representative of three independent experiments. b, Hierarchical clustering of differentially regulated genes (RNA-Seq) identified both in vitro and in vivo at q<0.05 in WT pre-B cells (replicates shown), cultured as indicated. c, Gene ontology analysis of clusters identified in ‘b’. d, Volcano plot of differentially expressed genes in WT pre-B cells cultured as indicated. Red points mark genes with significantly increased (right side) or decreased (left side) expression in WT pre-B cells cultured −IL-7+CXCL12+ERK-I compared to −IL-7+CXCL12 (FDR<0.01). (n=2 RNA-seq; FDR<0.01). Red dots are genes with significantly increased (right side) or decreased (left side) expression. −log 10 of the P values from Kal’s statistical test. e, Hierarchical clustering of differentially regulated genes (RNA-Seq) identified in both in vitro and in vivo at q<0.05 in WT pre-B cells (replicates shown), cultured as indicated. f, Geneontology analysis of clusters identified in ‘e’. g, Volcano plot of differentially expressed genes in WT pre-B cells cultured in +IL-7+CXCL12 versus −IL-7+CXCL12 (FDR<0.01). (n=2 RNA-seq; FDR<0.01). Red dots are genes with significantly increased (right side) or decreased (left side) expression. −log 10 of the P values from Kal’s statistical test. h, Expression (RNA-Seq) of all Vks, Jks and Ck genes in WT pre-B cells cultured as indicated (n=2). i, Flow cytometry for intracellular p-ERK in WT pre-B cells cultured at +IL-7, −IL-7, +IL-7+CXCL12 and −IL-7+CXCL12+ERK-inhibitor conditions as indicated. Data is representative of three independent experiments. j, Comparison of differential transcriptional programs regulated by CXCR4 signals in WT pro-B, large pre-B and small pre-B cell progenitors and in vitro pre-B cells cultured in the presence or absence of IL-7 with CXCl12, and with or without ERK inhibitor. Hierarchical clustering of z-scored Log2 normalized expression levels, using a Euclidian correlation distance metric. The clustering dendrogram was plotted against the distance matrix for all indicated samples.
As sensing of CXCL12 is required for moving out of IL-7 rich niches, some B cell progenitors will be subjected to signals through both receptors. Furthermore, pro-B cells tightly associate with IL-7intCXCL12+ (Type 2) stroma18. To understand the consequences of this dual receptor signaling state, B cell progenitors cultured in IL-7 were stimulated with CXCL12 and subjected to RNA-Seq. Different combinations of IL-7 and CXCL12 stimulation yielded three gene clusters (Fig. 8e,f). In Clusters 1 and 2, stimulation with IL-7 and CXCL12 most closely resembled the gene transcription profile induced by IL-7 alone. Remarkably, in these clusters CXCL12 appeared to reinforce the IL-7-dependent transcriptional state. In contrast, in Cluster 3, IL-7 did not modulate normal CXCL12-mediated repression. Genes in Cluster 1, included transcribed Igk gene segments while Cluster 2 was enriched for cell cycle genes (Fig. 8f). Therefore, the main transcriptional programs regulated by CXCL12 were strongly repressed in the presence of IL-7. Only Cluster 3, which includes genes involved in chromatin organization and GTPase regulation, were regulated by CXCL12 in the presence of IL-7 (Fig. 8f). A volcano plot of differentially regulated genes confirms that IL-7 repressed most CXCL12-induced genes (Fig. 8g). These data indicate that for most gene targets, IL-7 is dominant over CXCR4.
We next examined how IL-7, CXCL12 and ERK signals integrate to regulate Igk transcription (Fig. 8h). The combination of IL-7 withdrawal and CXCL12 strongly induced broad transcription across the Igk locus. In contrast, either continual IL-7 signaling in presence and absence of CXCL12, or inhibition of ERK, failed to induce Igk locus transcription (Fig. 8h). These data indicate that only a precise combination of developmental cues, including escape from IL-7R signaling and ERK-dependent induction through CXCR4, will open the Igk locus to recombination.
We next examined how signaling through both the IL-7R and CXCR4 regulated ERK phosphorylation (Fig. 8i). Interestingly, withdrawal of IL-7 modestly increased p-ERK. In contrast, in the presence of IL-7 and CXCL12, p-ERK levels were intermediate between +IL-7 and −IL-7+CXCL12. Analysis of RNA-Seq data from cells treated with +IL-7+CXCL12 versus −IL-7+CXCL12+ERK-inhibitor revealed strong co-clustering together (Fig. 8j). These data suggest that one major function of IL-7, and the IL-7R, signaling is inhibition of ERK activation.
IRF4-mediated amplification of CXCR4 expression
Pre-BCR signaling induces IRF4 expression which has been postulated to drive CXCR4 upregulation17, 37. Indeed, Irf4 was induced in large pre-B cells expressing the pre-BCR and was further upregulated in small pre-B cells (Supplementary Fig. 6a). Increased expression was associated with enhanced accessibility at both Irf4 enhancers and the promoter (Supplementary Fig. 6b). During development from the pro-B to large pre-B to small pre-B stage there was progressive increase in accessibility at the Cxcr4 locus (Supplementary Fig. 6c) and high Cxcr4 expression was associated with binding of IRF4, FOXO1 and PAX5 at both the promoter and enhancers in small pre-B cells. These data is consistent with IRF4 driving Cxcr4 expression.
To determine if IRF4 could induce Cxcr4 expression, we infected B cell progenitors cultured with IL-7 with control retrovirus or retrovirus encoding IRF4. Infected cells were isolated by sorting for GFP and cultured. In the presence or absence of IL-7, cells expressing IRF4 upregulated Cxcr4 mRNA (Supplementary Fig. 6d) and CXCR4 surface expression in vivo was dependent upon IRF4 and IRF8 (Supplementary Fig. 6e). Furthermore, in vitro transwell migration assays revealed IRF4 transfected cells migrated more roubstly along a CXCL12 gradient than mock transfected control cells (Supplementary Fig. 6f). Finally, we isolated pro-B and large pre-B cells from Irf4−/−Irf8−/− mice and measured chemotaxis in response to CXCL12 (Supplementary Fig. 6g). As demonstrated, chemotaxis was significantly diminished in large pre-B cells in response to CXCL12. These data demonstrate that IRF4 induces Cxcr4 and enhances chemotaxis along CXCL12 gradients. As CXCR4 also induces IRF4, overall our data suggest a IRF4-CXCR4 feedforward loop that enhances migration into CXCL12-rich BM niches.
DISCUSSION
Previous models of late B cell development have evoked a two-receptor system in which the IL-7R drives proliferation in pro and large pre-B cells while expression of the pre-BCR directs cell cycle exit and Igk recombination1. Canonically, the pre-BCR does so both directly and by inducing CXCR4 expression which allows escape from IL-7 rich BM niches17. In this model, the balance between IL-7R and pre-BCR signaling determines if a cell proliferates or recombines Igk. By controlling cell positioning in the BM, it was thought that CXCR4 indirectly regulates the balance between these two receptor determiners of cell state. Here, we demonstrate that CXCR4 plays a direct and fundamental role in inducing the small pre-B cell state including upregulating the RAG proteins, repressing pre-BCR expression, and opening Igk to recombination. Indeed, many of the biological functions ascribed to the pre-BCR are directly mediated by CXCR4.
Until now, the essential functions of CXCR4 in B lymphopoiesis have not been appreciated. This is probably because previous in vitro systems used OP9 cells which secrete CXCL124, 9, 17, 29, 30. Without a defined in vitro system, one cannot dissect changes due to direct CXCR4 signaling versus indirect effects due to positioning relative to IL-7 rich niches. In such a defined system, IL-7 escape and CXCL12 are the essential, and sufficient, extrinsic cues needed to dictate the development of small pre-B cells.
In the BM, small pre-B cells occupied a unique niche closely associated with both CXCL12+ stroma cells and extracellular deposits of CXCL12. Furthermore, small pre-B cells in these niches were rich in intracellular CXCL12 suggesting recent endocytosis. This proximity is not simply a consequence of chemotaxis as large pre-B cells, which migrate more strongly towards CXCL12 in vitro, are rarely in direct contact with CXCL12+ stroma. Spatial proximity in the BM is predicted to ensure the strong delivery of CXCL12 specifically to small pre-B cells.
Late B lymphopoiesis requires coordinated induction of both TF networks and chromatin remodeling complexes that dictate which regulatory sites are open to TF binding1, 31. CXCR4 does both. It induces the expression of IRF4 and NF-κB which are both critical for late B lymphopoiesis17, 28. In addition, CXCR4 signaling enhances accessibility at sites bound by multiple mediators of late lymphopoiesis including IRF, E2A, SPIB, PAX5 and FOXO1. Conversely, sites bound by early mediators of B cell development, such as MYC and STAT5, are closed by CXCR4 signaling. It is likely that at least some CXCR4-dependent changes in chromatin accessibility are mediated by BRWD1 which both opens enhancers of late lymphopoiesis and represses those targeted by early TF developmental programs31.
Our data suggest a feed-forward mechanism that both amplifies CXCR4 signaling and enforces the small pre-B cell transcriptional state. Successful rearrangement of Igk, and expression of the pre-BCR, induces expression of IRF4. IRF4, in turn, induces expression of CXCR4 which both mitigates the effects of IL-7R signaling and induces a transcriptional program that includes IRF4. IRF4 then feeds forward to further increase CXCR4 expression and signaling. In addition, CXCR4 signaling strongly feeds back to silence pre-BCR expression. We propose that these feed-forward and feed-back loops function to separate proliferative and differentiative states, thereby ensuring genomic integrity1 and enforcing ordered developmental progression.
The primacy of CXCR4 in directing late B lymphopoiesis begs a reconsideration of pre-BCR function. Clearly, a principal function of the pre-BCR is to induce the IRF4/CXCR4 feed-forward loop. It is also likely that the pre-BCR initiates molecular pathways that complement those activated by CXCR4. Most notably, the pre-BCR induces expression of the anti-apoptotic molecule MCL-135, 38. In contrast, CXCR4 does not appear to regulate any component of the apoptotic pathway except for mildly repressing Bcl2l11 (encodes BIM) and Bid (data not shown). The pre-BCR also induces IKAROS and AIOLOS which silence Ccnd34, 9, 13, 39. Ikaros is not regulated by CXCR4. Therefore, the pre-BCR provides both complementary and permissive signals that enable CXCR4-mediated differentiation.
From these studies, and previous work, a new model of late B cell development emerges in which coordinated signals through three receptors dictate differentiation from the pro-B to immature B cell stages1. Proliferation in pro-B and large pre-B cells is driven by the IL-7R which also represses Igk accessibility. Expression of the pre-BCR, and escape from IL-7R signaling, initiates a differentiation program. However, it is the induction of CXCR4 by IRF4, and the delivery of strong CXCL12-dependent signals, that completes differentiation into small pre-B cells robustly undergoing Igk recombination. Pre-BCR signaling, rather than providing transit across a discrete “checkpoint”, initiates a complex CXCR4-dependent program that guides B cell progenitors through a precise developmental program. Therefore, it is the constant interplay and integration of environmental cues, in the context of pre-BCR expression, which determines cell fate and orchestrates B lymphopoiesis.
Methods
Mice.
Wild-type (WT), Cκ-YFP (WT), RaDR-GFP (WT), mb1-Cre+/− Cxcr4fl/fl and mb1-Cre+Cxcr4fl/fl mice were housed in the animal facilities of the University of Chicago. Male and Female mice were used at 6-12 weeks of age, and experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of Chicago.
Isolation, culture and flow cytometry of bone marrow B cell progenitors.
Bone marrow (BM) was collected from WT, Cκ-YFP, mb1-Cre+/−, Cxcr4fl/fl or mb1-Cre+Cxcr4fl/fl mice and cells resuspended in staining buffer (3% (vol/vol) FBS in PBS). Erythrocytes were lysed and cells stained with anti-IL-7Rα (CD127, SB/199), anti-CXCR4 (2B11), anti-CD43 (S7), IgM (R6-60.2), IgD (11-36), anti-CD19 (1D3) and anti-B220 (RA3-6B2l); all from BD Biosciences) as described previously9,10. Pre-pro-B (Lin−CD19−B220+IgM−), pro-B cells (Lin−CD19+B220+CD43+IgM−), large pre-B cells (Lin−B220+CD43−IgM−FSChi), small pre-B cells (Lin−B220+CD43−IgM−FSClow) and immature B cells (Lin−B220+CD43−IgM+) were isolated by cell sorting with a FACSAria II (BD).
B cell progenitors (B220+IgM−) from WT, RaDR-GFP or mb1-Cre+Cxcr4fl/fl mice were isolated from bone marrow with a MACS separation column (Miltenyi Biotec) and cultured in complete Opti-MEM containing 10% (vol/vol) FBS and IL-7 (16 ng/ml) for five days. Further culture for 48 h with 16 ng/ml of IL-7 (+IL-7) or 0.2 ng/ml of IL-7 (−IL-7) or 0.2ng/ml of IL-7 with 100 ng of CXCL12 (−IL-7+CXCL12) without any stromal cells were performed before analyses.
For intracellular staining with antibodies to phosphorylated ERK (20A; Cat#612593, BD Biosciences Pharmingen), cells were fixed with BD Cytofix buffer and were made permeable with BD Phosflow Perm Buffer II (Cat#558030, BD Biosciences Pharmingen). The level of phosphorylation was determined by flow cytometry. 5 μM Erk inhibitor II (FR 180204, CAS 865362-74-9, Millipore) was used for last 4 h of the 48 h culture before the assay as negative control.
BM microscopy.
Both femurs of WT, Cκ-YFP, Cxcr4fl/fl or mb1-Cre+Cxcr4fl/fl mouse were isolated and put in DMEM+ 10% FBS medium. After removing ends, the body of the femur was connected to a syringe filled with media. Intact BM was flushed out gently and transferred into a mold containing OCT and frozen in 2-methylbutane in dry ice immediately. Frozen BM was sectioned (8-10 μm) serially. For tissue staining, the sections were fixed in 4% paraformaldehyde, blocked with 10% normal donkey serum and stained with primary antibodies to B220 (clone RA3-6B2, eBioscience and ab64100, Abcam), IgM (clone ll/41, eBioscience and FITC-conjugated Goat ant-mouse Jackson 115-097-020/115-096-075), Ki-67 (clone SP6, Ab16667, Lot#GR59808-1;, Abcam), IL-7 (M-19 polyclonal, sc1268, Lot#C2408,Santa Cruz Biotechnology), CXCL12 (FL-93 polyclonal, Santa Cruz Biotechnology and ab18919, Lot#GR116-13, Abcam) in various combinations and thereafter incubated with fluorochrome-conjugated secondary antibodies specific to the primary species and isotypes (Invitrogen). Finally, DAPI (Invitrogen) was applied for nucleus. Images were acquired at 12-bit depth, 1,024×1,024 pixel size, utilizing either the SP5 or SP8 laser scanning confocal microscope (Leica). Raw images were stored in manufacturer-specified .lif format and were converted to multichannel .tif images for further use. z-stack images were obtained using the Leica SP8 laser scanning confocal microscope in 0.5-μm increments and processed by Imaris software for 3D reconstruction.
Quantitative PCR, RNA-sequencing and analysis.
Total cellular RNA was isolated with an RNeasy kit (Qiagen) and RNA was reverse-transcribed with SuperScript III reverse transcriptase (Invitrogen). Then quantitative PCR in quadruplicate was performed using SYBR Green PCR Master Mix (Applied Biosystems) was analyzed. Gene expression was analyzed with an ABI PRISM 7300 Sequence Detector and ABI Prism Sequence Detection Software version 1.9.1 (Applied Biosystems). Results were normalized by division of the value for the unknown gene by that obtained for B2m. Sequences of primers were presented in Supplementary Table 1.
For RNA-Sequencing mRNA was isolated by oligo-dT beads and library was prepared using standard Illumina library protocol (Kit: RS-122-2101 TruSeq® Stranded mRNA LT-SetA). Libraries were sequenced on the Illumina Hiseq2500. QC-passed reads then aligned to the mm9 reference genome in a splice-aware manner using the STAR aligner. Gene expression was quantified using cuffquant, and normalized expression levels and differential expression levels were generated with cuffnorm and cuffdiff, respectively (version 2.2.1) as described previously31. Differential expression statistics (fold-change and p-value) were computed using DESeq2 and edgeR, on raw expression counts obtained from quantification. P-values were adjusted for multiple testing using the false discovery rate (FDR) correction of Benjamini and Hochberg. All heatmaps use hierarchical clustering based on the zscore of log 2 transformed normalized expression between various experimental conditions.
IL-7 and CXCL12 expression by Bone Marrow stromal cells (BMSCs).
BM was isolated from WT mice. Red blood cells were lysed and the remaining cells were plated in 6-well plates in complete medium to allow stromal cells to adhere to the bottom of the well. Nonadherent cells were aspirated, and flow cytometry was performed on the remaining adherent cells. Contaminating hematopoietic lineage cells and red blood cells were first excluded from our analysis using the CD45 and Ter119 markers, respectively. Then CXCL12 and IL-7 concentrations were assessed on the remaining bone marrow stromal cells by flow cytometry using the same antibodies used for microscopy (IL-7, M-19, sc1268, Lot#C2408, Santa Cruz goat anti-mouse and CXCL12, ab25117, Lot#GR116-13 both from Abcam).
Transwell Migration Assay.
Migration assays were performed as described previously19. A total of MACS column purified WT B220+ 0.25 to 0.5 × 106 cells/100 μl were placed in the upper compartment of a transwell chamber (5 μm pore size, Corning 3421) with 600 μl of medium containing 20 ng/ml recombinant IL-7 (cat#407-ML, R&D Systems) or 100 ng/ml recombinant CXCL12 (cat# 460-SD-010, R&D Systems). The number and proportion of cells before (as input in the upper chamber) and after 3 h that migrated into the lower chamber was measured by flow cytometry. The chemotaxis was expressed either as percentage or fold change relative to the number of input cells.
Cell-cycle analysis.
Cells were incubated in a solution containing propidium iodide and then were analyzed by flow cytometry as described before8. The proportion of cells in the G1, S and G2-M phases of the cell cycle was analyzed with FlowJo.
PCR analysis of Igk rearrangements.
Genomic DNA (or cDNA) was isolated from small pre-B cells of WT, Cxcr4fl/fl and mb1-Cre+Cxcr4fl/fl mice and from WT pre-B cells cultured for 48 h without stoma as described above8-10 (primers were shown in Supplementary Table 1). B2m expression was used to control for the amount of genomic DNA. The intensity of the band for each rearrangement product was normalized whenever necessary to values obtained from IgM+ B cells, given a value of 1.
Retroviral transduction.
The cDNA encoding mouse IRF4 was subcloned into retroviral vector MIGR1-GFP8. Retroviruses containing constructs were produced by transient transfection of PLAT-E packaging cell lines. Infection of B cell progenitors from Irf4−/−Irf8−/−done as described8,9. After 48 h, GFP+ cells were isolated by cell sorting and B cell progenitors were cultured in complete medium with IL-7 at a high (16 ng/ml) or low concentration (0.2 ng/ml) and expression Cxcr4 and chemotaxis to CXCL12 were measured by quantitative real-time PCR and transwell migration assay respectively as described above.
Gene set enrichment analysis (GSEA).
Normalized counts per million (CPM) log2 value from RNA-Seq data and the HALLMARK database which had 50 pathways were used as done previously31. Gene set enrichment analysis (GSEA) was performed using program offered at this website http://software.broadinstitute.org/gsea/index.jsp using default options. Enrichment map was used for visualization of the GSEA results.
Assay for Transposase-Accessible Chromatin using Sequencing (ATAC-Seq).
ATAC-Seq was performed as described10,31. All raw sequence data was quality trimmed to a minimum phred score of 20 using trimmomatic40. Alignment to reference genome mm9 was done with BWA; for ATAC-Seq data, read pairs where one pair passed quality trimming but the other did not were aligned separately and merged with the paired-end alignments. PCR duplicates were removed using Picard Mark Duplicates and alignments with an edit distance greater than 2 to the reference, or that were mapped multiple times to the reference, were removed.
ATAC-Seq analysis followed the procedure described previously10,31. We used the peak calling results as a guide to identify regulatory elements, then quantified enrichment in these regulatory elements and ran differential analysis to compare samples. Reproducibility was factored into the differential statistics that we calculated using estimates of dispersion.
ATAC-Seq and ChIP-Seq peak calling and motif analysis.
Peaks for ChIP-Seq samples were called using MACS2 as done previously10,31. HOMER software (hypergeometric optimization of motif enrichment) for de novo motif discovery and next-generation sequencing analysis was used for new prediction of motifs in the peaks.
Comparison of ChIP-Seq data and mRNA expression.
The EMBER program1 was used for the identification of genes targeted by IRF4, PAX5 and FOXO1 binding and how those genes acted over the in vitro culture in presence (+IL-7) and absence (−IL-7) of −IL-7 and with presence of CXCL12 at attenuated IL-7 condition (−IL-7+CXCL12). EMBER integrates transcription factor–binding data with RNA-Seq expression data and uses an unsupervised learning algorithm to identify genes targeted by the transcription factor. This is done by defining a set of pair-wise comparisons, making the changes in expression mathematically discrete and searching for over-represented patterns among these data for the genes within 100 kb of the transcription factor–binding sites. Only genes that match an expression pattern were selected; therefore, not all transcription factor–binding sites were assigned to a target gene.
Statistical analysis.
Data were analyzed with the unpaired t-test and analysis of variance, followed by the test of least-significant difference for comparisons within and between groups. All categories in each analyzed experimental panel were compared P values below 0.05 were considered significant. All P values below 0.001 were rounded to facilitate comparisons of results.
GEO accession codes for our publicly available data sets.
ATAC-Seq and RNA-Seq for WT pro-B, WT large pre-B, WT small pre-B and WT Immature B (GSE103057). IRF4 ChIP-seq for WT Small pre-B (GSE103057). RAG1, RAG2 and H3K4me3 ChIP-Seq for small pre-B cells (GSE69478). MYC ChIP-seq (GSE40173) and STAT5 ChIP-Seq (Ref 13). H3K4me1 ChIP-seq in CH12 cells (ENCSR000DHQ). H3K27ac ChIP-Seq in CH12 cells (GSE31039), Bone marrow H3K4me1, H3K4me3 and H3K27ac ChIP-seq (GSE31039).
Data availability
The sequences, ATAC-Seqs for WT small pre-B, mb1-Cre+Cxcr4fl/fl (Cxcr4-KO) small pre-B, +IL-7, −IL-7, −IL-7+CXCL12 cultured WT pre-B; −IL-7 and −IL-7+CXCL12 cultured Cxcr4−/− small pre-B and all the corresponded RNA-Seqs including +IL-7+CXCL12 and −IL-7+CXCL12+ERK-inhibitor cultured pre-B cells were deposited in the GenBank database (accession number GSE129311)
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Olson and D. Leclerc for cell-sorting services; and the ImmGen Consortium for data assembly. We thank H, Singh (University of Pittsburgh) for providing the PAX5, FOXO1 ChIP-Seq data and M. Schlissel (University of Michigan) for providing the Cκ-YFP reporter mice. This work is supported by the US National Institutes of Health Grants AI120715, AI128785, and AI143778 (to M.R.C), AI120715-02, AI128785-01A1 (to M.M.), F32AI143120 (to D.E.K.), T32GM007281 (K.C.M), UL1TR002003 (M.M-C.) T32HD007009 (M.K.O.). Part of the bioinformatics analysis described was performed by the UIC Research Informatics Core, supported in part by NCATS through Grant UL1TR002003.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Clark MR, Mandal M, Ochiai K & Singh H Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol 14, 69–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herzog S, Reth M & Jumaa H Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol 9, 195–205 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Clark MR, Cooper AB, Wang L & Aifantis I The pre-B cell receptor in B cell development: recent advances, persistent questions and conserved mechanisms. Curr Top Microbiol Immunol 290, 87–104 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Mandal M et al. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol 10, 1110–1117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper AB et al. A unique function for cyclin D3 in early B cell development. Nat Immunol 7, 489–497 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Lazorchak AS et al. Sin1-mTORC2 suppresses rag and il7r gene expression through Akt2 in B cells. Mol Cell 39, 433–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rathmell JC et al. Akt-directed glucose matabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol 23, 7315–7328 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottlob K et al. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev 15, 1406–1418 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandal M et al. Epigenetic repression of the Ig-kappa locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol 12, 1212–1220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandal M et al. Histone reader BRWD1 targets and restricts recombination to the Ig-kappa locus. Nat Immunol 16, 1094–1103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herzog S et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol 9, 623–631 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Amin RH & Schlissel MS Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol 9, 613–622 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heizmann B, Kastner P & Chan S Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J Exp Med 210, 2823–2832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YC et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11, 635–643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakamoto S et al. E2A and CBP/p300 act in synergy to promote chromatin accessibility of the immunoglobulin kappa locus. J Immunol 188, 5547–5560 (2012). [DOI] [PubMed] [Google Scholar]
- 16.van Loo PF, Dingjan GM, Maas A & Hendriks RW Surrogate-light-chain silencing is not critical for the limitation of pre-B cell expansion but is for the termination of constitutive signaling. Immunity 27, 1–13 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Johnson K et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity 28, 335–345 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Fistonich C et al. Cell circuits between B cell progenitors and the IL-7+ mesenchymal progenitor cells control B cell development. J Exp Med 215, 2596–2599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tokoyoda K, Egawa T, Sugiyama T, Choi BI & Nagasawa T Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity 20, 335–344 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Tian Y et al. CXCL12 induces migration of oligodendrocyte precursor cells through the CXCR4activated MEK/ERK and PI3K/AKT pathways. Mol Med Rep 18, 4374–4380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song ZY, Wang F, Cui SX, Gao ZH & Qu XJ CXCR7/CXCR4 heterodimer-induced histone demethylation: a new mechanism of colorectal tumorigenesis. Oncogene 38, 1560–1575 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Pozzobon T, Goldoni G, Viola A & Molon B CXCR4 signaling in health and disease. Immunol Lett 177, 6–15 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Zheng N et al. CXCR7 is not obligatory for CXCL12-CXCR4-induced epithelial-mesenchymal transition in human ovarian cancer. Molecular carcinogenesis 58, 144–155 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee S, Behnam Azad B & Nimmagadda S The intricate role of CXCR4 in cancer. Adv Cancer Res 124, 31–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trampont PC et al. CXCR4 acts as a costimulator during thymic beta-selection. Nat Immunol 11, 162–170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amin RH et al. Biallelic, ubiquitous transcription from the distal germline Ig-kappa locus promoter during B cell development. Proc Nat Acad Sci, USA 106, 522–527 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobeika E et al. Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci U S A 103, 13789–13794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derudder E et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat Immunol 10, 647–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochiai K et al. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol 13, 300–307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagergren A et al. The Cxcl12, periostin, and Ccl9 genes are direct targets for early B-cell factor in OP-9 stroma cells. J Biol Chem 282, 14454–14462 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Mandal M et al. BRWD1 orchestrates epigenetic landscape of late B lymphopoiesis. Nat Commun 9, 3888–3902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essafi A et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24, 2317–2329 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Nodland SE et al. IL-7R expression and IL-7 signaling confer a distinct phenotype on developing human B-lineage cells. Blood 118, 2116–2127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malin S et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol 11, 171–179 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bossen C et al. The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol 16, 775–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montresor A et al. CXCR4- and BCR-triggered integrin activation in B-cell chronic lymphocytic leukemia cells depends on JAK2-activated Bruton’s tyrosine kinase. Oncotarget 9, 35123–35140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson EC et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26, 335–344 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Vikstrom IB et al. MCL-1 is required throughout B-cell development and its loss sensitizes specific B-cell subsets to inhibition of BCL-2 or BCL-XL. Cell Death Dis 7, e2345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma S et al. Ikaros and Aiolos inhibit pre-B cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol 30, 4149–4158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences, ATAC-Seqs for WT small pre-B, mb1-Cre+Cxcr4fl/fl (Cxcr4-KO) small pre-B, +IL-7, −IL-7, −IL-7+CXCL12 cultured WT pre-B; −IL-7 and −IL-7+CXCL12 cultured Cxcr4−/− small pre-B and all the corresponded RNA-Seqs including +IL-7+CXCL12 and −IL-7+CXCL12+ERK-inhibitor cultured pre-B cells were deposited in the GenBank database (accession number GSE129311)