SUMMARY
In complex genetic loci, individual enhancers interact most often with specific basal promoters. Here we investigate the activation of the Bicoid target gene hunchback (hb), which contains two basal promoters (P1 and P2). Early in embryogenesis, P1 is silent, while P2 is strongly activated. In vivo deletion of P2 did not cause activation of P1, suggesting that P2 contains intrinsic sequence motifs required for activation. We show that a two-motif code (a Zelda binding site plus TATA) is required and sufficient for P2 activation. Zelda sites are present in the promoters of many embryonically expressed genes, but the combination of Zelda plus TATA does not seem to be a general code for early activation or Bicoid-specific activation per se. Because Zelda sites are also found in Bicoid-dependent enhancers, we propose that simultaneous binding to both enhancers and promoters independently synchronizes chromatin accessibility and facilitates correct enhancer-promoter interactions.
In Brief
Ling et al. identify a two-motif sequence code that is required and sufficient for activation of a specific promoter by the transcription factor Bicoid. They suggest that binding a single factor to both enhancers and promoters increases the probability that they will interact.
Graphical Abstract
INTRODUCTION
Regulatory elements (enhancers) direct spatial and temporal expression patterns by activating the basal promoters of their associated transcription units (van Arensbergen et al., 2014; Ong and Corces, 2011; Vernimmen and Bickmore, 2015). In the genomes of higher eukaryotes, enhancers significantly outnumber genes. For example, in Drosophila, there are 50,000 to 100,000 developmentally regulated enhancers, or roughly four enhancers per expressed gene (Kvon et al., 2014). Each enhancer is positioned between two basal promoters along the genomic sequence, but more than 90% are dedicated to choosing one promoter and regulating one gene (Kvon et al., 2014). Interfering with enhancer-promoter interactions can lead to developmental defects and disease states, including cancer (Lupiáñez et al., 2015; Zheng, 2013). Thus, promoter activation is a highly regulated process, and studies have begun to elucidate the mechanisms involved.
A hierarchy of mechanisms controls which basal promoters are regulated by individual enhancers. Chromatin conformation capture strategies have identified topologically associating domains (TADs) that divide the genome into neighborhoods containing relatively large numbers of genes and their associated enhancers (Dixon et al., 2012; Hou et al., 2012; Nora et al., 2012; Sexton et al., 2012). Protein complexes at the TAD boundaries are thought to prevent inter-TAD interactions between enhancers and promoters (Dixon et al., 2016). Within TADs, protein-DNA complexes called insulators have been shown to prevent enhancers associated with one gene from interacting with the basal promoters of nearby genes (Burgess-Beusse et al., 2002; Hnisz et al., 2016; Phillips-Cremins and Corces, 2013).
An alternative hypothesis to insulation is promoter tethering, which involves attractive interactions between enhancers and basal promoters (Calhoun and Levine, 2003; Kwon et al., 2009; Swanson et al., 2010). Support for this hypothesis comes from genome-wide promoter analyses, which showed that the promoters of developmentally regulated genes contain different sequence signatures compared with the promoters of ubiquitously expressed housekeeping genes (Zabidi et al., 2015). Within developmentally regulated genes, there is some evidence that enhancers bound by specific proteins prefer basal promoters that contain specific sequence motifs (Butler and Kadonaga, 2001; Juven-Gershon et al., 2008; Ohtsuki and Levine, 1998; Zehavi et al., 2014). For example, the IAB5 enhancer of the bithorax complex prefers to activate promoters containing the sequence motif TATA at a specific position upstream of the transcription start site (TSS) (Ohtsuki and Levine, 1998). Another example is an enhancer bound by the Caudal transcription factor, which was shown to prefer a basal promoter containing a downstream promoter element (DPE) over a TATA-containing promoter with no DPE (Juven-Gershon et al., 2008). Correct promoter usage is also critical for the normal regulation of single genes containing multiple basal promoters. A study estimates that more than 30% of all genes in Drosophila contain at least two basal promoters (Rach et al., 2009), but the mechanisms controlling intra-gene promoter usage are not well understood.
Here we study intra-gene promoter preference at the hunchback (hb) locus. hb contains two basal promoters (P1 and P2) (Schröder et al., 1988; Tautz et al., 1987; Figure 1A). In early zygotes (nuclear division cycle [NC] 10–13), the P2 promoter is in an open configuration (Blythe and Wieschaus, 2016; Figure 1B) and drives strong expression throughout the anterior half of the embryo (Figure 1F). P2 expression is controlled largely by the homeodomain (HD)-containing protein Bicoid (Bcd) (Driever and Nüsslein-Volhard, 1989; Struhl et al., 1989), which is distributed in a long-range gradient with the highest levels near the anterior pole (Driever and Nüsslein-Volhard, 1988). There are two Bcd-dependent enhancers in the hb locus (Figure 1A): a proximal (Prox) enhancer adjacent to the P2 promoter (Driever and Nüsslein-Volhard, 1989; Struhl et al., 1989) and a distal (Dist) enhancer located 4 kb upstream (Perry et al., 2011; Chen et al., 2012). In contrast, the P1 promoter is in a closed chromatin configuration during these early stages (Figure 1B), and P1-specific transcripts are barely detectable (Figure 1E; Margolis et al., 1995; Wu et al., 2001). Slightly later (NC14), this promoter is activated in a two-striped expression pattern in response to an enhancer that is not activated by Bcd (Margolis et al., 1995). The P1 promoter lies only 3 kb upstream of the P2 promoter (Figure 1A) in a position where it could block interactions between the Dist enhancer and the P2 promoter. However, a previous study showed that the activities of both the Dist and the Prox enhancers are required for robust expression from P2 (Perry et al., 2011). Altogether, these results suggest that the Dist enhancer bypasses the P1 promoter to help activate P2, probably because P1 is in a closed chromatin configuration.
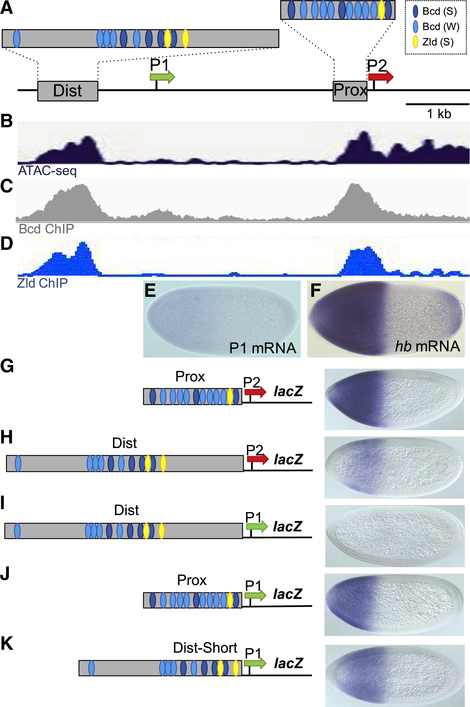
Figure 1. Testing Intrinsic Enhancer-Promoter Compatibilities at hb.
(A) Schematic of a 5.6 kb fragment from the hb locus showing two Bcd-dependent enhancers (gray boxes labeled Dist and Prox), approximate positions of Bcd and Zld binding sites in the two enhancers, and the P1 and P2 basal promoters (shown as green and red arrows, respectively). These conventions are used throughout this paper. See Data S1 for a detailed annotation of the hb locus.
(B) Read profiles of ATAC sequencing (ATAC-seq) in embryos at 18 min into NC13 (Blythe and Wieschaus, 2016).
(C) Binding profile of Bcd in NC9–NC14 embryos at the hb locus (Xu et al., 2014).
(D) Binding profile of Zld in NC13–NC14 embryos (Nien et al., 2011; Sun et al., 2015).
(E and F) Expression patterns of P1 transcripts (E) and hb transcripts (F) early in NC14.
(G–K) lacZ expression patterns driven by five enhancer-promoter combinations.
All embryos in this paper are oriented with anterior to the left and dorsal up.
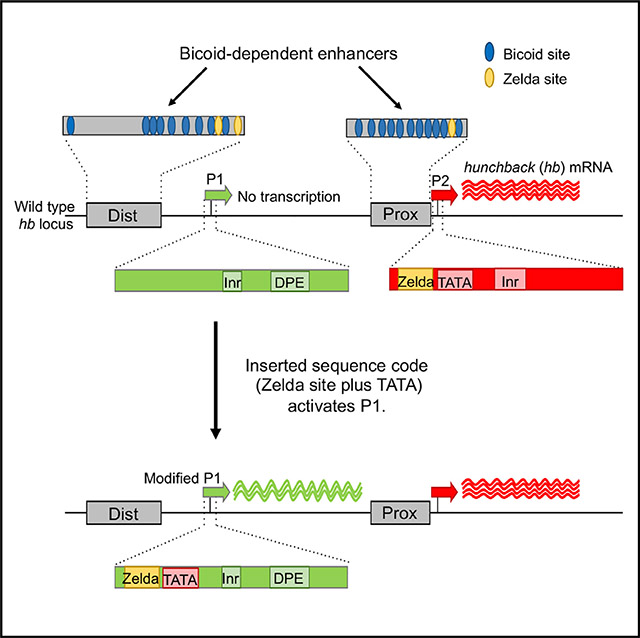
We used a combination of reporter genes and CRISPR-generated deletion mutants to unravel the mechanisms that activate P2 and prevent activation of P1. We showed that P2 contains a combination of sequence motifs (TATA and a closely linked Zelda binding site) that are required for activation and sufficient to partially convert P1 into a Bcd-dependent promoter. We also showed that inserting P1 into an open chromatin region results in activation. In all our experiments, promoter activation depended on strong binding motifs for the ubiquitous factor Zelda (Zld), which has been previously proposed as a pioneer factor that opens chromatin (Foo et al., 2014; Li et al., 2014; Schulz et al., 2015; Sun et al., 2015). We propose that the Zld site in P2 helps generate an open promoter configuration that is independent of and synchronized with enhancer activation.
RESULTS
Testing Intrinsic Enhancer-Promoter Compatibilities at hb
Two hypotheses can explain the differential responses of P1 and P2 to Bcd-dependent activation: First, the P1 basal promoter may lack specific sequence motifs that allow P2 to attract enhancer-bound Bcd activators. Second, P1 might be inactivated because of sequences outside the basal promoter that prevent activation. To test these hypotheses, we tested a series of reporter genes, each of which contained a different enhancer-promoter combination. As expected from previous studies (Driever and Nüsslein-Volhard, 1989; Struhl et al., 1989), a construct containing only the 270 bp Prox enhancer and the P2 promoter (Prox-P2) directed strong reporter gene expression throughout the anterior half of the embryo (Figure 1G). A reporter containing the 850 bp Dist enhancer and the P2 promoter (Dist-P2) also activated expression in this region (Figure 1H), but both the levels and the spatial extent of the pattern were decreased compared with the Prox-P2 construct. These results suggest that each enhancer can independently activate the P2 promoter. To determine whether the Dist or Prox enhancer can activate the P1 promoter in isolation, we constructed Dist enhancer and P1 promoter (Dist-P1) and Prox enhancer and P1 promoter (Prox-P1) transgenes and tested them in vivo. The Dist-P1 reporter failed to activate expression (Figure 1I), but surprisingly, the Prox-P1 reporter gene directed activation in anterior regions of the embryo (Figure 1J).
Both the inactive Dist-P1 and the active Prox-P1 transgenes contain strong clusters of Bcd binding sites (Figure 1A). However, the cluster in Dist-P1 (Figure 1I) is positioned more than 300 bp upstream of P1, while the cluster in Prox-P1 (Figure 1J) is adjacent to the P1 promoter. Perhaps the intervening sequences in Dist-P1 prevent activation. To test this, we removed them, which resulted in activation of the P1 promoter by the shortened Dist enhancer (Dist-Short-P1) (Figure 1K). Altogether, these results show that P1 can be activated when placed immediately downstream of either Bcd-dependent enhancer. They further suggest that sequences between the Dist enhancer and P1 help insulate it from activation. However, these intervening sequences do not prevent activation of the P2 promoter in the Dist-P2 construct (Figure 1H). This suggests that intrinsic sequence differences between the two promoters also contribute to their differential usage in the early embryo.
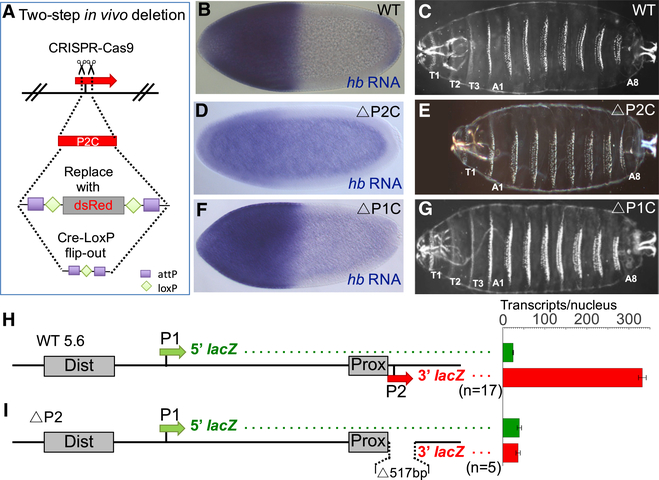
CRISPR/Cas9 Deletion of P2 Does Not Lead to the Activation of P1 In Vivo
Because both P1 and P2 can be activated by Bcd-dependent enhancers when tested in isolation, perhaps the two promoters compete for interactions with Bcd-bound enhancers in the endogenous locus. If this model is correct, deleting P2 might result in the activation of P1. To test this, we deleted 114 bp of the P2 core promoter sequence (−40 to +74; TSS = +1) from the endogenous gene (ΔP2C) (Figure 2A; Table S1; STAR Methods). When the ΔP2C mutant was homozygosed or tested in trans to an amorphic hb allele (hb14F), Bcd-dependent hb expression was nearly abolished (Figure 2D; Figure S1), and an embryonic lethal phenotype was produced. Cuticle preparations showed that the unhatched larvae failed to form the meso- and metathoracic segments (T2 and T3) (Figure 2E), an anterior phenotype previously observed for zygotic null hb mutants (Hülskamp et al., 1994; Lehmann and Nüsslein-Volhard, 1987). This result indicates that the P2 promoter is required for hb’s zygotic functions in anterior patterning and that Bcd cannot activate P1 even when it is the only available promoter. As a control, we deleted 135 bp of sequence spanning the P1 TSS (ΔP1C, −60 to +75) (Table S1; STAR Methods). Homozygosing this allele or putting it in trans to the hb14F null allele did not affect the level of Bcd-dependent hb RNA expression (Figure 2F; Figure S1) or cause detectable disruption of the larval cuticle (Figure 2G). These results indicate that transcription from P1 is not required for hb’s functions in anterior embryonic patterning. However, ΔP1C mutants fail to survive to adulthood (data not shown), suggesting that P1-driven expression provides important functions later in development. Altogether, these results argue strongly against the promoter competition hypothesis and strengthen the idea that Bcd-dependent activation of P2 is a promoter-intrinsic property.
Figure 2. Testing Whether Deletion of P2 Causes the Activation of P1.
(A) Schematic of the two-step strategy for CRISPR/Cas9-based deletion of P2 core (P2C) sequences. (B–G) Analysis of mutant hb alleles carrying P2 and P1 deletions (for precise breakpoint sequences, see Table S1). Expression patterns of hb mRNA are shown for a wild-type embryo (B), a ΔP2C homozygote (D), and a ΔP1C homozygote (F). hb mRNA levels are also quantified in Figure S1. Cuticle preparations of first instar larvae (C, E, and G) are shown to the right. Each cuticle image is a merged composite of photographs of the anterior and posterior regions of a single larva. Thoracic segments (T1–T3) are labeled when present.
(H and I) Analysis of a wild-type dual reporter gene (H) and an identical reporter containing a 517 bp deletion of the P2 promoter (I). See STAR Methods, Figures S2 and S3, Table S2, and Data S2. Schematics of reporter genes are shown on the left and individually labeled.
All reporter genes in this paper contain the 5′ and 3′ halves of the lacZ transcription unit cloned immediately downstream of the P1 and P2 basal promoters, respectively. Green and red bars on the right represent levels of 5′ and 3′ lacZ transcripts, respectively, driven by each construct. The levels are shown as number of transcripts per nucleus (see STAR Methods). Error bars represent SEM. The number of embryos measured for each construct is shown below each reporter schematic.
Mapping Sequences Required and Sufficient for Bcd-Dependent Activation
To identify individual promoter motifs that contribute to Bcd-dependent activation of P2, and to quantify their contributions, we constructed a dual reporter system using a 5.6 kb genomic fragment that contains both enhancers and both promoters (Figure 2H; Figure S2A; Data S1). Into this fragment, we inserted 5′ (1,402 bp) and 3′ (1,645 bp) sequences from the lacZ coding region immediately downstream of the P1 and P2 promoters, respectively (Figure 2H; Figure S2A; Data S2; STAR Methods). To quantify expression from each promoter, we used single-molecule fluorescence in situ hybridization (smFISH) with anti-sense oligonucleotide probe sets designed to differentially detect the 5′ and 3′ lacZ transcripts produced by P1 and P2 (STAR Methods; Figure S2).
In our first experiment, we quantified the number of 5′ and 3′ lacZ transcripts expressed by the wild-type reporter gene (see STAR Methods). The number of 5′ lacZ transcripts produced by P1 was 24.5 ± 1.2 per nucleus in anterior regions, while P2 produced more than 10 times as many 3′ transcripts (331.9 ± 9.3 per nucleus) (Figure 2H; Figure S2E). These results are generally consistent with previous studies of P1 and P2 expression from the endogenous locus (e.g., Figures 1E and 1F; Margolis et al., 1995; Wu et al., 2001). We also tested an identical reporter containing a deletion of P2 (STAR Methods; Table S2). This deletion nearly abolished expression of 3′ lacZ transcripts (Figure 2I), which is consistent with the results of deleting the P2 core promoter in the endogenous gene (Figures 2D and 2E; Figure S1). Altogether, these results suggest that the dual reporter system can be used for further quantitative analyses of hb promoter activities.
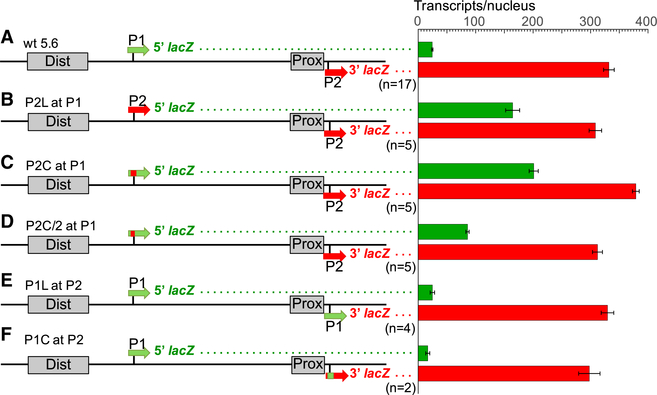
To test whether Bcd-dependent activation of P2 is controlled by intrinsic sequences, we manipulated the dual reporter gene by deleting sequences surrounding P1 (−3,284 to −2,724; P1 TSS = −3,218) and replacing them with sequences from P2 (P2 long fragment [P2L] at P1, −51 to +465). This manipulation caused a 6-fold increase in transcription of the 5′ lacZ mRNA (Figure 3B, significance was determined using the Student’s t test, p = 3.8795e−15). To narrow the regions critical for P2 activity, we precisely replaced only the P1 basal promoter sequence (−51 to +69) with P2 sequences (P2 core fragment [P2C] at P1), which caused a similar increase in activity (Figure 3C). We tested an even smaller P2 fragment (P2C/2 at P1, −51 to +8), which showed a significantly smaller increase (Figure 3D, compare with Figures 3B and 3C, p = 0.00024). These results indicate that the 120 bp P2 basal promoter can autonomously respond to Bcd-dependent activation.
Figure 3. Mapping Sequences Required and Sufficient for Bcd-Dependent Activation.
(A F) Analysis of a wild type dual reporter gene (A) and identical reporter genes containing replacements of P1 sequences with P2 sequences (B, C, and D), and vice versa (E and F). Reporter gene schematics are shown on the left and individually labeled (see also Table S2). P2L and P1L refer to long promoter fragments (~500 bp), while P2C and P1C refer to core (120 bp) promoter fragments. Green and red bars on the right represent levels of 5′ and 3′ lacZ transcripts (number per nucleus), respectively, driven by each construct. Error bars represent SEM. The number of embryos measured for each construct is shown below each reporter schematic.
In contrast, replacing the P2 promoter with P1 sequences (P1 long fragment [P1L] at P2 and P1 core fragment [P1C] at P2) caused a strong activation of that promoter at the P2 position (Figures 3E and 3F). This result is consistent with the activation of P1 when placed adjacent to the Dist and Prox enhancers in the Dist-Short-P1 and Prox-P1 minimal constructs (Figures 1J and 1K).
A Two-Motif Sequence Code Is Sufficient for Bcd-Dependent Promoter Activation
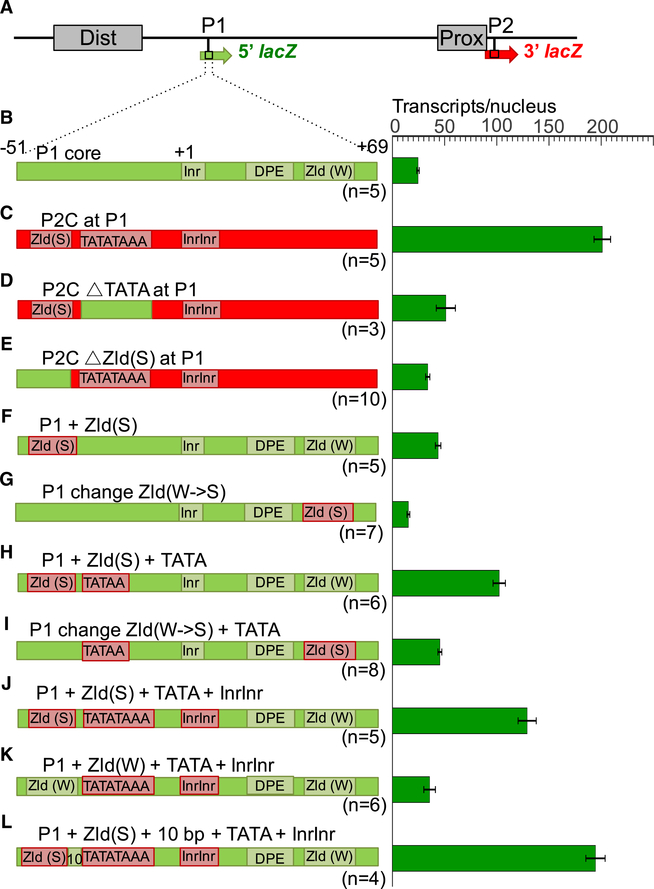
We examined the 120 bp promoter sequences (−51 to +69) from P1 and P2 for features that enable P2 to specifically respond to Bcd (Figures 4B and 4C; Table S3). P2 contains a strong TATA box that appears as two overlapping TATA sequences (TATATAAA), a double initiator (InrInr), and no DPE (Figure 4C), while P1 lacks TATA but contains a single initiator (Inr) and a DPE (Figure 4B). In addition to these differences in common basal promoter motifs, P2 has a strong binding site (CAGGTAG) for the ubiquitous factor Zld, located 5 bp upstream of TATATAAA in P2 (Figure 4C). The P1 promoter does not contain a strong Zld site (Figure 4B), but it has a variant motif (CAGGCAG) with a low affinity for Zld (Nien et al., 2011) located 10 bp downstream of its DPE. Previous chromatin immunoprecipitation (ChIP) experiments with anti-Zld antibodies showed a strong binding peak over the P2 region but no detectable binding to P1 (Harrison et al., 2011; Nien et al., 2011; Figure 1D), suggesting that the Zld site variant in P1 is not functional.
Figure 4. Testing Specific Promoter Motifs.
(A–L) Analysis of a wild type dual reporter gene (A and B) and identical reporter genes containing replacements of P1 core sequences (B) with P2 sequences (C–E), and conversions of P1 core sequences to insert specific promoter motifs from P2 (F–L). Reporter gene schematics are shown on the left and individually labeled. Complete sequences of each construct are shown in Table S3. Green bars on the right represent levels of 5′ lacZ transcripts (number per nucleus) driven by each construct. All of these constructs drive similar levels of 3′ lacZ transcripts, and we have omitted them from this figure for clarity. Error bars represent SEM. The number of embryos measured for each construct is shown below each reporter schematic.
Because previous studies showed positive correlations between developmentally regulated genes and TATA-containing promoters (Ohtsuki and Levine, 1998; Zabidi et al., 2015), we mutated the TATATAAA sequence in P2C at P1, which caused a 75% reduction of 5′ lacZ expression (Figure 4D, p = 1.0200e−5). We also tested the Zld site in P2C, based on the idea that Zld may play a pioneer-like role that increases chromatin accessibility (Foo et al., 2014; Li et al., 2014; Schulz et al., 2015; Sun et al., 2015). Mutating this site caused an 85% reduction in 5′ lacZ RNA (Figure 4E, p = 6.9918e−13). These results indicate that both the TATATAAA sequence and the Zld site are required for efficient activation of the P2 promoter when inserted into the position normally occupied by P1.
We next tested whether insertion of these motifs alone or in combination could convert the P1 promoter into a Bcd-responsive promoter. We focused on Zld site insertions because of the stronger effect caused by mutating that site in P2. First, we inserted a strong Zld motif into P1, keeping the distance between that site and the Inr the same as in P2, which caused a modest increase in 5′ lacZ transcripts (Figure 4F, p = 6.7511e−7). We also replaced the low-affinity Zld site downstream of the Inr with the consensus Zld site (CAGGTAG), which reduced expression below that observed with the unperturbed P1 (Figure 4G, compare with Figure 4B). These results suggest that one strong Zld site is not sufficient for generating an efficient promoter response to Bcd-dependent activation. We then added a single TATA box to the constructs containing inserted Zld sites upstream and downstream of the Inr. This extra insertion increased the expression of both constructs (Figures 4H and 4I) compared with those containing only one strong Zld site (Figures 4F and 4G, p = 1.4787e−5 and 8.5113e−9, respectively), indicating an additive effect between the two motifs.
The previous experiments tested the insertion of single TATA sequence into the P1 promoter, but the wild-type P2 promoter contains TATATAAA and an InrInr element. Both the double TATA and the double initiator are conserved among all sequenced Drosophila species (data not shown). We thus introduced the TATATAAA sequence and the InrInr sequence, together with a strong upstream Zld site into the P1 promoter. This triple insertion caused a modest but significant increase over adding the Zld site and single TATA alone (Figure 4J, compare with Figure 4H, p < 0.05). We then tested whether the strength of the Zld site is important by replacing the strong site upstream of the inserted TATATAAA with a weaker site (CAGGCAG), which caused a 75% reduction in activity (Figure 4K, compare with Figure 4J, p = 6.0733e−6). Finally, to directly test the distance relationship between the Zld site and the TATATAAA motif in P2 (normally 5 bp), we increased the distance between the two motifs to 10 bp. If there is a strict distance requirement between the two sites for promoter activity, this manipulation should disrupt or reduce promoter function. Instead, it increased expression (Figure 4L, compare with Figure 4J, p < 0.005), suggesting that some flexibility in the spacing between the two motifs can be tolerated.
Altogether, these results suggest that closely linked Zld and TATATAAA motifs contribute to P2 activation and that spacing between these motifs is a factor that helps determine promoter strength. However, the combination of Zld and TATATAAA, even with the InrInr (Figure 4J), does not lead to the level of activation observed when the whole P2C sequence is inserted into the P1 position (Figures 3B, 3C, and 4C), suggesting that other motifs inside the basal promoter are required for full activation.
The Zld Site in P2 Is Required for Activation by the Dist Enhancer
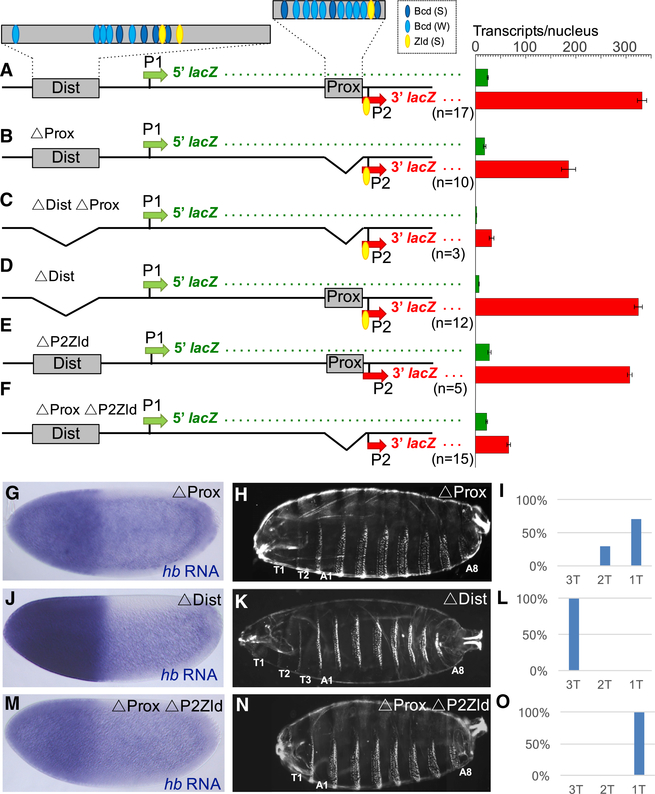
Previous studies and our experiments indicate that both Prox and Dist enhancers interact with P2 (Bothma et al., 2015; Perry et al., 2011). To quantify the contributions of each enhancer on transcription levels, we deleted each enhancer individually in the context of the dual reporter gene. Deletion of the Prox enhancer alone reduced levels of 3′ lacZ transcription from P2 by 44% (Figure 5B, p = 3.3597e−9). To test whether this expression is driven by the Dist enhancer, we deleted both enhancers, which abolished all lacZ expression from the reporter (Figure 5C). In contrast, deletion of the Dist enhancer alone did not significantly change the levels of 3′ lacZ driven by the dual reporter (Figure 5D).
Figure 5. Testing the Roles of the Prox and Dist Enhancers in Activating P2.
(A) Schematic of the 5.6 kb hb locus showing approximate positions of Bcd and Zld binding sites in the Dist and Prox enhancers (for precise positions, see Data S1).
(B–F) Analysis of dual reporter genes containing deletions of the Prox enhancer alone (B), both Prox and Dist enhancers (C), the Dist enhancer alone (D), the Zld site in P2 alone (E), and the Prox enhancer plus the Zld site in P2 (F). Sequences of deletion breakpoints are shown in Table S2. Green and red bars on the right represent levels of 5′ and 3′ lacZ transcripts (number per nucleus), respectively, driven by each construct. Error bars represent SEM. The number of embryos measured for each construct is shown below each reporter schematic. (G–O) Analysis of mutant hb alleles carrying deletions of the Prox enhancer alone (G–I), the Dist enhancer alone (J–L), and the Prox enhancer plus the Zld site in P2 (M–O) (see deletion breakpoints in Table S1). Expression patterns of hb mRNA (G, J, and M; for quantification, see Figure S4), cuticle preparations of first instar larvae (H, K, and N), and graphical representations of the percentage of larvae containing one, two, or three thoracic segments (I, L, and O) are shown for each construct. As in Figure 2, cuticle images (H, K, and N) are each merged composites of photographs of the anterior and posterior regions of single larvae.
We made similar deletions in the context of the endogenous hb gene to test whether the activities of each enhancer are required for hb function in vivo. When homozygosed or tested in trans with the hb14F null allele, deletion of the Prox enhancer (ΔProx) caused a significant reduction in hb expression (Figure 5G; Figure S4) and a variable loss of thoracic segments in the larval cuticle (Figures 5H and 5I). These results indicate that the Prox enhancer is required for Bcd-dependent hb activity in development. In contrast, CRISPR deletion of the Dist enhancer (ΔDist) did not cause a detectable change in hb expression levels (Figure 5J) or abnormality in cuticle preps (Figures 5K and 5L). ΔDist/hb14F flies are viable and fertile, suggesting that the Dist enhancer is dispensable for development under laboratory conditions.
The ΔProx described earlier leaves intact the Zld site plus TATATAAA combination at the P2 promoter. We hypothesized that the Zld site is critical for a productive long-range interaction between the Dist enhancer and P2 when the Prox enhancer is not present. To test this directly, we made two more deletions in the context of the dual reporter, one that removed only the Zld site from P2 (ΔP2Zld), and a second deletion that removed both the Prox enhancer and the P2 Zld site (ΔProx ΔP2Zld). Deleting the P2Zld site alone did not significantly change 3′ lacZ expression levels (Figure 5E). In contrast, deleting both the Prox enhancer and the P2 Zld site caused a 65% reduction of 3′ lacZ expression (Figure 5F) compared with ΔProx (Figure 5B, p = 1.7365e−9). We also used CRISPR editing to make ΔProx ΔP2Zld from the endogenous locus. This caused a stronger reduction of hb expression compared with ΔProx (Figure 5M; Figure S4), and an invariant cuticle phenotype lacking both T2 and T3 (Figures 5N and 5O). Altogether, these results suggest that the Dist enhancer cannot fully activate a Zld-less promoter from a distance.
DISCUSSION
Specific Promoter Activation at the hb Locus
The promoter deletion and insertion experiments in this paper show that Bcd-dependent promoter usage at the hb locus is controlled by intrinsic DNA sequences that lie in the interval between −51 and +69 with respect to the P2 TSS. Two sequence motifs (a strong Zld site plus TATATAAA) are critical for the efficient activation of the P2 promoter, and inserting them together into the inactive P1 promoter is sufficient to convert it to a partially active Bcd-dependent promoter. Because deletion of the P2 promoter does not result in the activation of the normally inactive P1 promoter, these motifs appear to function by actively and specifically promoting transcription, and there is little competition between P2 and P1 for Bcd-dependent activation.
Mechanism of P2 Activation
Understanding P2 regulation is complicated by the Zld site’s position immediately downstream of the Bcd-dependent Prox enhancer and by both the enhancer and the promoter being contained in a contiguous 390 bp fragment (Driever and Nüsslein-Volhard, 1989; Struhl et al., 1989). One specific issue is whether the Zld site upstream of the TATATAAA sequence should be considered part of the Prox enhancer or part of the P2 promoter. Three considerations suggest that it is an integral part of the promoter. First, the Zld site extends from position −41 to −35 bp with respect to the hb TSS and only 5 bp upstream of the TATA sequence at position −30. Second, our Prox enhancer deletion experiments suggest that the Zld site is required for strong activation by the Dist enhancer (Figure 5). Third, a study showed that at least 55 developmentally regulated promoters in Drosophila contain consensus Zld motifs that form a meta-peak ~50 bp upstream of the TSS (Chen et al., 2013). Altogether, we propose that Zld binding sites should be considered core promoter motifs for a subset of genes that are activated during the mid-blastula transition in the Drosophila.
Because Zld may function as a pioneer factor (Li et al., 2014; Sun et al., 2015), its binding to the P2 promoter might loosen chromatin by displacing nucleosomes. Such a mechanism has been proposed for Zld sites in enhancer elements (ten Bosch et al., 2006; De Renzis et al., 2007; Satija and Bradley, 2012; Harrison et al., 2011; Foo et al., 2014; Xu et al., 2014). In particular, the hb gene contains Zld sites in both its Bcd-dependent enhancers and in the P2 promoter. We propose that binding Zld generates an open chromatin configuration at both types of elements, which would synchronize the binding of Bcd to the enhancers and the binding of TFIID and other basal transcription factors to the P2 promoter. Because of the prevalence of Zld sites in the enhancers and promoters of embryonically expressed genes, this is likely to be a general mechanism that facilitates correct pairings between enhancers and promoters.
Mechanisms that Prevent P1 Activation
The P1 promoter does not contain either a strong Zld motif or a canonical TATA sequence, and it is in a closed chromatin configuration when Bcd-dependent activation of P2 occurs. This suggests that P1 is immune to Bcd-dependent activation but that when placed adjacent to either the Prox or the Dist-Short enhancer (Figures 1J and 1K) or inserted into the position of P2 in the dual reporter (Figures 3E and 3F), this promoter is efficiently activated. One explanation is that all three of these experiments position strong Zld sites in the enhancers within 100 bp of the P1 promoter. It is possible that these sites help organize a region of accessible chromatin that spreads into the adjacent P1 promoter, facilitating its activation, even in the absence of a canonical TATA box. To test this, we increased the distance between the nearest Zld site and the P1 promoter to more than 300 bp (Dist-P1) (Figure 1I), which resulted in the abolishment of expression. Perhaps this distance places the promoter beyond the range of spreading chromatin mediated by the Zld sites. In the endogenous hb gene, the P1 promoter is positioned more than 1 kb downstream of the nearest Zld site in the Dist enhancer and is inactive at this time.
Insertion of the 5′ half of the P2 core, which contains the Zld site, the TATATAAA sequence, and the InrInr, causes significant activation of P1 (Figure 3D), but this activation is only about half that seen when the 120 bp P2 core sequence is inserted intact into the P1 position (Figure 3C). This suggests that motifs downstream of the TSS are required for generating the transcription rates mediated by P2 in its normal position. Even the activation by the 120 bp P2 core sequence is less than two-thirds of that seen when the P2 is in its original position (Figure 3C). It is possible that the Zld site in the Prox enhancer, which is not included in either P2 insertion experiment, augments P2 expression or that sequences between the Dist enhancer and P1 contribute to a region of compacted chromatin that represses the ability to activate at this stage. Future experiments will be required to test these hypotheses.
Zld+TATA Is Not a General Code for Bcd-Specific Promoter Usage
Several published studies suggested that promoters containing specific sequence motifs might attract interactions with enhancers bound by specific proteins (Butler and Kadonaga, 2001; Juven-Gershon et al., 2008; Ohtsuki and Levine, 1998; Zehavi et al., 2014), and it is tempting to speculate that the two-motif code discovered here is a common feature of promoters activated by Bcd-bound enhancers. This does not seem to be the case. For example, of the 24 embryonic promoters that contain Zld sites and TATA boxes mentioned earlier, only one is activated by a Bcd-dependent enhancer. To test this idea more rigorously, we conducted a survey of 25 well-annotated Bcd-dependent target promoters (Table 1; STAR Methods). About half of these target genes (11, including hb) were previously classified as pre-mid-blastula transition (MBT) genes because they rank among the first zygotically activated genes (Chen et al., 2013). Of these, seven contain TATA in their promoter sequences, and two of these contain Zld sites within 100 bp upstream of the TSS. A third promoter contains a single Zld site at −90 but no TATA. The other 14 Bcd target genes are activated slightly later and were classified as mid-blastula transition zygotic (MBT-Zyg) or mid-blastula transition maternal (MBT-Mat) genes (Chen et al., 2013). Of these, only two have TATA-containing promoters, and only one of these also contains a canonical Zld site close to the TATA box. In summary, these results suggest a bias toward having TATA sequences in the promoters of the earliest expressed Bcd target genes, but they do not support the idea that Zld sites or TATA elements (or the combination of both) mediate Bcd-dependent activation per se.
Table 1.
Motif (Zld, TATA, and Inr) Searches of 25 Bcd-Dependent Promoters
| Gene | Expression stage | Promoter peak | Distance to the nearest enhancer (bp) | Zld site | TATA box | Inr |
|---|---|---|---|---|---|---|
| hbP2 | pre-MBT | narrow | 52 | GTACCTG—47bp—CTACCTG–5bp–TATATAAA—20bp———TCAGTCAGTC | ||
| gt | pre-MBT | narrow | 4978 | TATAAA—25bp———TCAGTT | ||
| slp1 | pre-MBT | narrow | 710 | TATAAA—25bp———TCAGCT | ||
| knirps | pre-MBT | N/A | 54 | TATATATA—24bp———TCAGTT | ||
| eve | pre-MBT | N/A | 1074 | TATAAA—23bp———GCACAC | ||
| ftz | pre-MBT | narrow | 3219 | TATATA—24bp———TCATTC | ||
| odd | pre-MBT | N/A | 2086 | TCAGTT | ||
| Bsq25D | pre-MBT | narrow | 32 | CTACCTG—18bp—CAGGTAG–17bp—TATAAA—20bp———GCAGTT | ||
| run | pre-MBT | narrow | 2448 | TCAGTT | ||
| h | pre-MBT | narrow | 3964 | TAGGTAG——————93bp——————TCAGTC | ||
| OS | pre-MBT | N/A | 4160 | TCAGTT | ||
| Kr | MBT-zyg | narrow | 856 | TATA—24bp———TCAGTC | ||
| btd | MBT-zvP | N/A | 2693 | TCAGTC | ||
| hbn | MBT-zvq | N/A | 872 | TCAGTC | ||
| prd | MBT-zyg | N/A | 2954 | TTACCTG—————84bp——————TCAGTC | ||
| otd | MBT-zyg | N/A | 4145 | TCATTT | ||
| ems | MBT-zvq | narrow | 6789 | TTATTC | ||
| CG3502 | MBT-zvq | N/A | 577 | * TATACCAGGTAA—13bp—TCAGTT | ||
| bowl | MBT-zvq | narrow | 2927 | TCAGTC | ||
| edl | MBT-zvq | narrow | 850 | TCAGTT | ||
| salm | MBT-zvq | narrow | 10265 | TCACTC | ||
| D | MBT-zvq | N/A | 3669 | TCAGTC | ||
| tsh | MBT-zvq | narrow | 48038 | TCAGTCAGTC | ||
| bancal | MBT-mat | narrow | 4747 | TTAGTT | ||
| cnc | MBT-mat | N/A | 1987 | TCAGTCAGTCAGTT | ||
25 Bcd-dependent promoters were scanned for motifs (Zld site, TATA box, and Inr) within the window of −220 to +20 bp with respect to the TSS (See STAR Methods). The columns from left to right are: gene symbol, class of expression stage, initiation pattern (narrow/focused or widespread promoter peak), the distance to the nearest Bcd-dependent enhancer, and motifs with spacing in between. The sequences were aligned according to the position of the Inr in each promoter, which exactly overlaps or is in close proximity to the annotated TSS. Abbreviations: MBT – midblastula transition; zyg – zygotic; mat – maternal; N/A – data not available.
CG3502 has a combined TATA-Zld motif.
Contributions from Two Enhancers and Zld+TATA Motifs Optimize P2 Expression Levels
Previous studies suggested that the Prox and Dist hb enhancers work together to maximize expression levels of hb (Bothma et al., 2015; Perry et al., 2011). The data presented in this paper substantially extend these studies. First, our data show that both enhancers make productive interactions with P2. Deleting the Prox enhancer alone in the context of the dual reporter caused a 44% reduction in P2 expression, and deleting both the Prox and the Dist enhancers virtually abolished expression, confirming that the Dist enhancer can contribute significantly in the absence of the Prox enhancer. In vivo, deletion of the Prox enhancer causes a strong reduction in hb expression, causing lethality and the loss of two thoracic segments from the larval cuticle. Thus, the amount of hb produced by the Dist enhancer alone is insufficient to provide in vivo hb function. In contrast to previous studies (Perry et al., 2011), we did not detect a significant effect on P2 expression levels when the Dist enhancer was deleted from the reporter gene. Furthermore, deleting the Dist enhancer did not lead to a mutant phenotype, suggesting that it is dispensable for development under normal laboratory conditions. Thus, the Prox enhancer is critical for hb function, and although the Dist enhancer can interact with P2 to some degree, the level produced by this enhancer alone cannot replace that normally provided by the Prox enhancer.
Finally, our results show that the Zld site and TATATAAA each contribute quantitatively to the level of transcription driven by the P2 promoter. Furthermore, our attempts to convert P1 into a Bcd-responsive promoter resulted in many output levels (Figure 4). Constructs carrying the Zld+TATA code (Figures 4H, 4J, and 4L) were expressed at higher levels than those containing a single Zld or TATA site (Figures 4F and 4K). In addition, a construct carrying the TATATAAA motif and the double initiator (Figure 4J) was expressed at higher levels than one carrying a simple TATA sequence and an Inr (Figure 4H). Finally, changing the spacing between the Zld site and the TATA motif strongly affected expression levels (Figure 4J, compare with Figure 4L). Altogether, these experiments suggest that basal promoter sequences have critical roles in precisely determining levels of transcription, in addition to mediating specific enhancer-promoter interactions.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stephen Small (sjs1@nyu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly strains
The following stocks were used in these experiments: y[1]w[1118] (wild-type), ΦC31 (y+); 38F1 (w+), y[1] M{vas-Cas9.RFP-}ZH-2A w [1118] (BDSC_55821), y[1]w[67c23] P{y[+mDint2] = Crey}1b; D[*]/TM3, Sb[1] (BDSC_851), and hb14F/Tm3B, Sb, hb-lacZ. ΦC31 (y+); 38F1 (w+) was used for all transgenesis. Transgenic flies used in this paper all carried two copies of the relevant transgene. y[1] M {vas-Cas9.RFP-}ZH-2A w[1118] was used for CRISPR/Cas9 genome editing. y[1]w[67c23] P{y[+mDint2] = Crey}1b; D[*]/TM3, Sb[1] was used for Cre/loxP recombination to remove DsRed from CRISPR-generated DsRed insertions. For in situ hybridization and smFISH, the CRISPR mutant alleles were made heterozygous with a TM3 Sb balancer containing a hb-lacZ reporter by crossing to the hb14F/Tm3B, Sb, hb-lacZ. All flies were kept on standard fly cornmeal-molasses-yeast media.
METHOD DETAILS
Reporter gene cloning and transgenesis
For minimal constructs, enhancer and promoter fragments were cloned sequentially into a piB-HC-lacZ vector (Chen et al., 2012) using standard cloning techniques. For dual reporters, the 5.6kb genomic fragment from the hb locus (−5157 to +465 bp; Data S1) was amplified by PCR in two pieces: one extending from the Dist enhancer to the P1 promoter, and the other from sequences downstream of P1 to the P2 promoter (Data S2). The two fragments were cloned into a modified piB-HC-lacZ vector containing a polylinker flanked by the attB-recombination sequence and the 3′ half (starts at 1403bp) of the lacZ coding sequence (AscI/XhoI). The two hb fragments were inserted using AscI/NcoI and SpeI/XhoI respectively. The 5′ half (stops at 1402bp) of lacZ and a 3′UTR from a-tubulin were joined together by PCR and ligated into the plasmid using NcoI and SpeI. The entire sequence of the dual reporter gene can be found in the Data S2. hb fragments carrying deletions, sequence insertions, and promoter sequence conversions were generated by PCR or synthesized by Integrated DNA Technologies and inserted using the same restriction sites as the wild-type fragments. Sequence details of all manipulations are included in Tables S2 and S3. All constructs were integrated into the 38F1 landing site on chromosome II using ΦC31 integrase-mediated cassette exchange (Bateman et al., 2006).
Generation of hb mutants by CRISPR
hb mutants were generated by CRISPR/Cas9 gene editing essentially as described (Gratz et al., 2013, 2014). gRNA sequences were inserted into the pU6-BbsI-chiRNA vector (Table S1). Two homology arms flanking the cleavage sites were inserted into the multiple cloning sites of a modified pHD-DsRed-attP vector (pHD-DsRed-2XattP), which was used as a donor template. The modified pHDDsRed-attP has a second attP sequence inserted after the loxP. A mixture of the donor vector (500ng/μl) and two gRNA vectors (100ng/μl each) was injected into embryos of y[1] M{vas-Cas9.RFP-}ZH-2A w[1118]. F1 flies were mated with yw, and F2s were screened for the dsRed+ transformants. For transformants, sequences at the junctions between deletion break point and the inserted dsRed marker were amplified by PCR and verified by sequencing. The transformants were balanced and then crossed to y[1]w [67c23] P{y[+mDint2] = Crey}1b; D[*]/TM3, Sb[1] to remove the dsRed marker in the mutant allele. The attP-loxP-attP sequence (~150 bp) remained at the deletion site after the excision of the dsRed cassette.
in situ hybridization and cuticle preparations
Embryos were collected for 1–3hr after egg laying at room temperature. Embryos were dechorionated in Clorox for 2 minutes and shaken in 4% formaldehyde (1 X PBS) for fixation for 25 minutes. A mixture of methanol and heptane was used to remove the vitelline membrane. In situ hybridization was performed as previously described (Small, 2000) using digoxigenin- or fluorescein-labeled antisense RNA probes and the standard alkaline phosphatase assay. Embryos were mounted with Aqua-Poly/Mount and imaged on a Zeiss Axioskop microscope. For CRISPR-generated mutant lines, each mutant allele was made heterozygous with a TM3 Sb balancer containing a hb-lacZ reporter. To identify mutant homozygotes as embryos, progeny were first stained for lacZ expression, and those embryos not stained by lacZ were separated from the rest, and subsequently stained for hb expression. For cuticle preps, embryos were aged 20–24hr, and fixed in a 1:4 mixture of glycerol and acetic acid at 65°C overnight, and mounted in a 1:1 mixture of Hoyer’s medium and lactic acid as previously described (Wu et al., 1998).
smFISH
smFISH was performed as previously described (Little et al., 2013). Embryos were collected for 2–2.5hr after egg laying to maximize the number of embryos at mitosis of NC13. Embryos were dechorionated in Clorox, fixed in 4% formaldehyde, and devitellinized in methanol. The hybridization step was performed overnight at 37°C. Atto-565 conjugated probe sets complementary to 5′ lacZ and Atto-633 conjugated probe sets complementary to 3′ lacZ were used for the 5.6kb hb dual reporters (Figure S2A). Atto-565 hb and Atto-633 lacZ were used for CRISPR mutants. Mutant homozygotes were identified as embryos that showed no detectable lacZ expression. DAPI staining was used to visualize nuclear cycles.
FISH-stained embryos were mounted with VECTASHIELD antifade mounting medium and imaged on a Leica SP8 confocal microscope under the photon-counting mode with a 63× HC PL APO CS2 1.3 NA glycerol immersion objective. Pixel size was 75 × 75um with a z-step size of 250nm. For dual reporter lines, a total volume of 38.44 × 38.44 × 20 μm3 was imaged at 25%–40% embryo length (EL, 0% = anterior tip) starting at the surface of the embryo (Figures S2B and S2C). For CRISPR mutants, a total volume of 153.76 × 38.44 × 20 μm3 was imaged in the anterior part of the embryo. Images were acquired exclusively from embryos in metaphase, anaphase, or telophase of NC13 (Figure S2B). There is no active transcription at these stages, which allows the direct quantification and comparison of the accumulated transcripts from 5′ and 3′ lacZ in the cytoplasm.
Promoter analysis
The Bcd-dependent promoters were either confirmed by searching the literature, or determined by comparing expression patterns of 66 Bcd-dependent enhancers (Chen et al., 2012) with the patterns of their nearest genes. For genes with alternative promoters, only promoters being transcribed during early embryonic stages (Chen et al., 2013) were used. The information of pre-MBT or MBT and narrow or wide promoter peak was from previous studies (Chen et al., 2013; Ni et al., 2010). The TSS position was determined using the FlyBase r5.47 annotation or the annotation made by Uwe Ohler’s lab (Ni et al., 2010). For the promoter motif search, sequences surrounding the TSS (−220 to +20) were scanned. For Inrs, we searched for sequences matching the consensus TCAGTY (Ohler et al., 2002) with at most one mismatch. For TATA, we searched for any sequence match TATA within the −100 to 0 window for each start site. For Zld sites, the six highest affinity TAGteam sites (Nien et al., 2011) were used in the sequence scan.
QUANTIFICATION AND STATISTICAL ANALYSIS
Processing of raw images and identification of candidate cytoplasmic spots was performed using the FISH Toolbox as described (Little et al., 2013). An intensity distribution histogram was generated for all candidates, where real cytoplasmic spots and noise appeared as two separate peaks (Figure S2D). The threshold separating real spots from noise was determined as the valley between the two peaks. The total number of identified cytoplasmic spots was counted as the number of transcripts produced from each promoter in the field of observation. The total number was then divided by the number of nuclei generating an average number of transcripts per nucleus (Figure S2E).
To compare detection efficiencies between the 5′ and 3′ lacZ probe sets, we compared their abilities to detect expression from the Prox-P2-lacZ transgene (Figure S3), which contains a full-length lacZ gene, and is expected to produce identical numbers of 5′ and 3′ lacZ mRNAs. Indeed, the two probe sets identified similar numbers of transcripts in this experiment (Figures S3B and S3C), which convinced us that these probe sets can be used to compare mRNA levels produced by P1 and P2 in the context of the 5.6 kb reporter gene.
For quantifying hb mRNA, the number of cytoplasmic spots were counted in 2.5% egg length bins along the AP axis from 25% to 50% (0% = anterior tip), and then averaged by the number of nuclei (Figures S1 and S4). In regions where cytoplasmic spots were too dense to be resolved well (> 350 per nucleus), the total fluorescence collected were converted to counts of spots according to their linear relationship in low-density regions. Error bars on all plots are standard error of the mean. The number of embryos measured can be found either in figures or figure legends. All p values in the text were calculated using the Student’s t test.
DATA AND CODE AVAILABILITY
Original data for smFISH in the paper is available at Zenodo Data (https://doi.org/10.5281/zenodo.3258890).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep anti-DIG-AP, Fab fragments | Roche | Cat# 11093274910, RRID:AB_2734716 |
| Sheep anti-Fluorescein-AP, Fab fragments | Roche | Cat# 11426338910 |
| Bacterial and Virus Strains | ||
| NEB 5-alpha competent E.coli | NEB | Cat# C2987I |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5’ lacZ Atto 565 smFISH probe | Shawn Little’s lab | Little et al., 2013 |
| 3’ lacZ Atto 633 smFISH probe | Shawn Little’s lab | Little et al., 2013 |
| hb Atto 565 smFISH probe | Shawn Little’s lab | Little et al., 2013 |
| lacZ ISH DIG RNA probe | This lab | N/A |
| hb ISH Fluorescein RNA probe | This lab | N/A |
| VECTASHIELD antifade mounting medium | Vector Laboratories | Cat# H-1000 |
| DAPI | Sigma-Aldrich | Cat# 9542 |
| NBT/BCIP Stock Solution | Roche | Cat# 11681451001 |
| Aqua-Poly/Mount | Polysciences | Cat# 18606–20 |
| Deposited Data | ||
| Raw data for smFISH | This paper | Zenodo Data https://doi.org/10.5281/zenodo.3258890 |
| Experimental Models: Drosophila Strains | ||
| y[1] w[1118] | Bloomington Drosophila Stock Center | Cat# 6598 |
| ΦC31 (y+); 38F1 (w+) | This lab | |
| y[1] M{vas-Cas9.RFP-}ZH-2A w[1118] | Bloomington Drosophila Stock Center | Cat# 55821 |
| y[1]w[67c23] P{y[+mDint2] = Crey}1b; D[*]/TM3, Sb[1] | Bloomington Drosophila Stock Center | Cat# 851 |
| hb14F/Tm3B, Sb, hb-lacZ | Christine Rushlow’s lab | N/A |
| Flies with transgene at 38F1 landing site on chromosome II | This paper | N/A |
| ΔP2C | This paper | N/A |
| ΔP1C | This paper | N/A |
| ΔProx | This paper | N/A |
| ΔDist | This paper | N/A |
| ΔProx, ΔP2 Zld | This paper | N/A |
| Oligonucleotides | ||
| ΔP2C gRNA 1: GTTATATATCGCTCAGGTAGA | This paper | N/A |
| ΔP2C gRNA 2: GATTTGTCGGGATTTTCGTA | This paper | N/A |
| ΔP1C gRNA 1: GTTGGCGAGTGCACTGTTGTG | This paper | N/A |
| ΔP1C gRNA 2: GCGTTTCGTCCTTTGGATGTT | This paper | N/A |
| ΔProx/ΔProx ΔP2Zld gRNA 1: GAGGAGTAGGCAGCTAGCGT | This paper | N/A |
| ΔProx gRNA 2: GGTAGACGGATGCACGCGTCA | This paper | N/A |
| ΔProx ΔP2Zld gRNA 2: GTTATATATCGCTCAGGTAGA | This paper | N/A |
| ΔDist gRNA 1: GTCTCAACAGAAGTTCCTTCG | This paper | N/A |
| ΔDist gRNA 2: GCCCAAATATTCCACATAAAC Recombinant DNA | This paper | N/A |
| Prox-P2-lacZ | This paper | N/A |
| Dist-P2-lacZ | This paper | N/A |
| Prox-P1-lacZ | This paper | N/A |
| Dist-P1-lacZ | This paper | N/A |
| Dist short-P1-lacZ | This paper | N/A |
| 5.6 kb hb dual reporter wt | This paper | N/A |
| 5.6 kb hb dual reporter ΔP2 | This paper | N/A |
| 5.6 kb hb dual reporter P2L at P1 | This paper | N/A |
| 5.6 kb hb dual reporter P2C at P1 | This paper | N/A |
| 5.6 kb hb dual reporter P2C/2 at P1 | This paper | N/A |
| 5.6 kb hb dual reporter P1L at P2 | This paper | N/A |
| 5.6 kb hb dual reporter P1C at P2 | This paper | N/A |
| 5.6 kb hb dual reporter P2C ΔTATA at P1 | This paper | N/A |
| 5.6 kb hb dual reporter P2C ΔZld(S) at P1 | This paper | N/A |
| 5.6 kb hb dual reporter P1 + Zld(S) | This paper | N/A |
| 5.6 kb hb dual reporter P1 +change Zld (W->S) | This paper | N/A |
| 5.6 kb hb dual reporter P1 +Zld(S) + TATA | This paper | N/A |
| 5.6 kb hb dual reporter P1 change Zld (W->S) + TATA | This paper | N/A |
| 5.6 kb hb dual reporter P1+ Zld(S) + TATA + InrInr | This paper | N/A |
| 5.6 kb hb dual reporter P1+ Zld(W) + TATA + InrInr | This paper | N/A |
| 5.6 kb hb dual reporter P1+ Zld(S) + 10bp + TATA +InrInr | This paper | N/A |
| 5.6 kb hb dual reporter ΔProx | N/A | |
| 5.6 kb hb dual reporter ΔDist ΔProx | This paper | N/A |
| 5.6 kb hb dual reporter ΔDist | This paper | N/A |
| 5.6 kb hb dual reporter ΔP2Zld | This paper | N/A |
| 5.6 kb hb dual reporter ΔProx ΔP2Zld | This paper | N/A |
| pHD-DsRed-2XattP Software and Algorithms | This paper | N/A |
| MATLAB | The Mathworks Inc. | https://www.mathworks.com |
| FISH Toolbox Other | Little et al., 2013 | N/A |
| Confocal microscope | Leica | SP8 |
| Microscope | Zeiss | Axioskop |
| Digital camera for microscopy | Zeiss | AxioCam MRc |
Highlights.
A two-motif code (Zelda site + TATA) allows Bicoid to activate a specific promoter
Binding Zelda to enhancers and promoters synchronizes their ability to interact
The Zelda site + TATA code is not a general code for Bicoid-specific activation
ACKNOWLEDGMENTS
We thank Shawn Little for the 5′ and 3′ lacZ probe sets and advice on smFISH, Hongtao Chen for the smFISH protocol and codes for image analysis, Po-Ta Chen for the image analysis codes, and Jinshuai Cao for technical assistance. We thank Shelby Blythe, Christine Rushlow, Claude Desplan, Justin Blau, Richard Mann, Erika Bach, and Esteban Mazzoni for insightful discussions throughout the course of this work. Pinar Onal, Peter Whitney, Christine Rushlow, Esteban Mazzoni, and the anonymous reviewers provided invaluable feedback on the manuscript. This work was conducted in facilities constructed with support from Research Facilities Improvement Grant C06 RR-15518-01 from the NIH National Center for Research Resources and was supported by NIH grant RO1 GM51946 (to S.S.) and NYU Dean’s Undergraduate Research Fund Grants (to K.Y.U. and T.S.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2019.06.038.
REFERENCES
- Bateman JR, Lee AM, and Wu CT (2006). Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173, 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, and Wieschaus EF (2016). Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife 5, e20148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Ng S, Perry MW, Gregor T, and Levine M (2015). Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife 4, e07956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, and Felsenfeld G (2002). The insulation of genes from external enhancers and silencing chromatin. Proc. Natl. Acad. Sci. USA 99 (Suppl 4), 16433–16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JEF, and Kadonaga JT (2001). Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VC, and Levine M (2003). Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. USA 100, 9878–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu Z, Mei C, Yu D, and Small S (2012). A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell 149, 618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C, and Zeitlinger J (2013). A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2, e00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, and Wieschaus EF (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, and Ren B (2016). Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 62, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, and Nüsslein-Volhard C (1988). A gradient of bicoid protein in Drosophila embryos. Cell 54, 83–93. [DOI] [PubMed] [Google Scholar]
- Driever W, and Nüsslein-Volhard C (1989). The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337, 138–143. [DOI] [PubMed] [Google Scholar]
- Foo SM, Sun Y, Lim B, Ziukaite R, O’Brien K, Nien CY, Kirov N, Shvartsman SY, and Rushlow CA (2014). Zelda potentiates morphogen activity by increasing chromatin accessibility. Curr. Biol 24, 1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, and O’Connor-Giles KM (2013). Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194, 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, and Eisen MB (2011). Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Day DS, and Young RA (2016). Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell 167, 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, and Corces VG (2012). Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell 48, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Lukowitz W, Beermann A, Glaser G, and Tautz D (1994). Differential regulation of target genes by different alleles of the segmentation gene hunchback in Drosophila. Genetics 138, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, and Kadonaga JT (2008). Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 22, 2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kazmar T, Stampfel G, Yáñez-Cuna JO, Pagani M, Schernhuber K, Dickson BJ, and Stark A (2014). Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512, 91–95. [DOI] [PubMed] [Google Scholar]
- Kwon D, Mucci D, Langlais KK, Americo JL, DeVido SK, Cheng Y, and Kassis JA (2009). Enhancer-promoter communication at the Drosophila engrailed locus. Development 136, 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, and Nüsslein-Volhard C (1987). hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev. Biol. 119, 402–417. [DOI] [PubMed] [Google Scholar]
- Li XY, Harrison MM, Villalta JE, Kaplan T, and Eisen MB (2014). Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 3, e03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Tikhonov M, and Gregor T (2013). Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. (2015). Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JS, Borowsky ML, Steingrímsson E, Shim CW, Lengyel JA, and Posakony JW (1995). Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development 121, 3067–3077. [DOI] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, and Zhu J (2010). A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods 7, 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, and Rushlow C (2011). Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao G, Niemann H, and Rubin GM (2002). Computational analysis of core promoters in the Drosophila genome. Genome Biol. 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, and Levine M (1998). GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 12, 3325–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, and Corces VG (2011). Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet 12, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, and Levine M (2011). Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 108, 13570–13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, and Corces VG (2013). Chromatin insulators: linking genome organization to cellular function. Mol. Cell 50, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach EA, Yuan H-Y, Majoros WH, Tomancak P, and Ohler U (2009). Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 10, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, and Bradley RK (2012). The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 22, 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder C, Tautz D, Seifert E, and Jäckle H (1988). Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 7, 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, McKay DJ, and Harrison MM (2015). Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, and Cavalli G (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472. [DOI] [PubMed] [Google Scholar]
- Small S (2000). In vivo analysis of lacZ fusion genes in transgenic Drosophila melanogaster. Methods Enzymol. 326, 146–159. [DOI] [PubMed] [Google Scholar]
- Struhl G, Struhl K, and Macdonald PM (1989). The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell 57, 1259–1273. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nien CY, Chen K, Liu HY, Johnston J, Zeitlinger J, and Rushlow C (2015). Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Evans NC, and Barolo S (2010). Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Lehmann R, Schnurch H, Schuh R, Seifert E, Kienlin A, Jones K, and Jackle H (1987). Finger protein of novel structure encoded by hunchback, a second member of the gap class of Drosophila segmentation genes. Nature 327, 383–389. [Google Scholar]
- ten Bosch JR, Benavides JA, and Cline TW (2006). The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967–1977. [DOI] [PubMed] [Google Scholar]
- van Arensbergen J, van Steensel B, and Bussemaker HJ (2014). In search of the determinants of enhancer-promoter interaction specificity. Trends Cell Biol. 24, 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, and Bickmore WA (2015). The Hierarchy of Transcriptional Activation: From Enhancer to Promoter. Trends Genet. 31, 696–708. [DOI] [PubMed] [Google Scholar]
- Wu X, Vakani R, and Small S (1998). Two distinct mechanisms for differential positioning of gene expression borders involving the Drosophila gap protein giant. Development 125, 3765–3774. [DOI] [PubMed] [Google Scholar]
- Wu X, Vasisht V, Kosman D, Reinitz J, and Small S (2001). Thoracic patterning by the Drosophila gap gene hunchback. Dev. Biol 237, 79–92. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen H, Ling J, Yu D, Struffi P, and Small S (2014). Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 28, 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, and Stark A (2015). Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehavi Y, Kuznetsov O, Ovadia-Shochat A, and Juven-Gershon T (2014). Core promoter functions in the regulation of gene expression of Drosophila dorsal target genes. J. Biol. Chem 289, 11993–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J (2013). Oncogenic chromosomal translocations and human cancer (review). Oncol. Rep 30, 2011–2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data for smFISH in the paper is available at Zenodo Data (https://doi.org/10.5281/zenodo.3258890).