Abstract
Proliferation is tightly regulated during T cell development and is limited to immature CD4−CD8− thymocytes. The major proliferative event is initiated at the ‘β-selection’ stage following successful rearrangement of Tcrβ and is triggered by and dependent on concurrent signaling by Notch and the pre-TCR; however, it is unclear how these signals cooperate to promote cell proliferation. Here we found that β-selection-associated proliferation required the combined activity of two SCF ubiquitin ligase complexes that included as substrate recognition subunits the F-box proteins Fbxl1 or Fbxl12. Both SCF complexes targeted the cyclin-dependent kinase inhibitor Cdkn1b for ubiquitinylaton and degradation. We found that Notch signals induced the transcription of Fbxl1 whereas pre-TCR signals induced the transcription of Fbxl12. Thus, concurrent Notch and pre-TCR signaling induced the expression of two genes, Fbxl1 and Fbxl12, whose products functioned identically but additively to promote degradation of Cdkn1b, cell cycle progression, and proliferation of β-selected thymocytes.
A major aspect of the thymocyte maturation process is the precise regulation of cell proliferation. Rather than being a shared property of all or most developing thymocytes, proliferation is strictly limited to two stages of early CD4−CD8− (double negative; DN) thymocyte development. The initial proliferative phase, which occurs during the CD44+CD25− (DN1), CD44+CD25+ (DN2), and CD44−CD25+ (DN3) stages prior to initiation of V-[D]-J recombination at the TCRβ locus, is driven by thymus-expressed cytokines, specifically Kit ligand (stem cell factor) and IL-7, as well as signaling by Notch 1, 2, 3. The second proliferative phase coincides with ‘β-selection’, so is initiated in DN3 cells that have productively rearranged TCRβ and express the pre-TCR. The proliferative burst that accompanies β-selection takes place in CD44−CD25− (DN4) thymocytes, CD4−CD8+ intermediate single positive (ISP) thymocytes, and early CD4+CD8+ (double positive; DP) ‘blasts’, prior to rearrangement of TCRα and is estimated to result in a 100-200 fold expansion2, 4. The clonal expansion at this proliferative phase facilitates the diversification of the pre-selection TCR repertoire and is also required for the differentiation of DN thymocytes to the DP stage5.
Coordinated Notch-mediated and pre-TCR-mediated signaling is essential for β-selection-associated proliferation6, 7, 8, but precisely how the pre-TCR and Notch cooperate to regulate cell cycle entry and thymocyte proliferation has remained unclear9, 10. Prior to pre-TCR expression, the majority of DN3 thymocytes are retained in either the quiescent G0 phase or the ‘primed’ G1 phase of the cell cycle7. Transition of cells from G1 to the actively cycling S/G2/M phases is controlled by cyclins complexed with a cyclin-dependent kinase (CDK)11. The activity of cyclin-CDK complexes, and consequently cell cycle progression, is inhibited by members of the Cip/Kip family of CDK inhibitors, which include Cdkn1a (p21cip1), Cdkn1b (p27kip1) and Cdkn1c (p57kip2)11, with only Cdkn1b having a major role at the β-selection checkpoint12, 13, 14, 15, 16. Cdkn1b inhibits both cyclin A-CDK and cyclin E- CDK complexes11 and mice lacking Cdkn1b have an enlarged thymus14, 15, 16, whereas overexpression of Cdkn1b results in a block at the DN3 stage and a markedly reduced thymus size and cellularity17. Cdkn1b is highly expressed in quiescent ‘pre-β-selection’ DN3 thymocytes, but is down-regulated at the initiation of β-selection7.
Down-regulation of Cdkn1b occurs primarily through its poly-ubiquitinylation by a Skp-cullin-F-box (SCF) E3 ligase complex that includes the F-box substrate recognition protein Fbxl1, resulting in the proteasomal degradation of Cdkn1b18, 19, 20. Fbxl1−/− mice have a reduced thymus size, which is restored in Fbxl1−/− Cdkn1b−/− mice19. Notch signaling induces the expression of Fbxl1 in several cell lines, including T cell acute lymphoblastic leukemia cells, suggesting that Fbxl1 could be regulated by Notch in DN thymocytes21, 22, 23, 24. Pre-TCR signaling also induces the degradation of Cdkn1b7, 10; however, a direct regulatory role for the pre-TCR in the destabilization of Cdkn1b has not been established.
In this study, we investigated the regulation of Cdkn1b stability and its effect on cell cycle progression and proliferation at the β-selection checkpoint. Our findings identified a key role for the F-box protein Fbxl1 and an equally critical role for the related F-box protein, Fbxl12 in the destabilization of Cdkn1b. SCF complexes that contained Fbxl1 and SCF complexes that contained Fbxl12 cooperated in an additive fashion to target Cdkn1b for ubiquitinylation and proteasomal degradation and each was required for normal proliferation after β-selection. Notably, Fbxl1 and Fbxl12 were induced transcriptionally at the β-selection checkpoint by Notch signals and by pre-TCR signals, respectively. Together, these findings provide a regulatory mechanism for the β-selection proliferative burst that explains the requirement for and the cooperativity of Notch and pre-TCR signaling for this response.
Results
Cdkn1b and Fbxl1 control β–selection proliferation
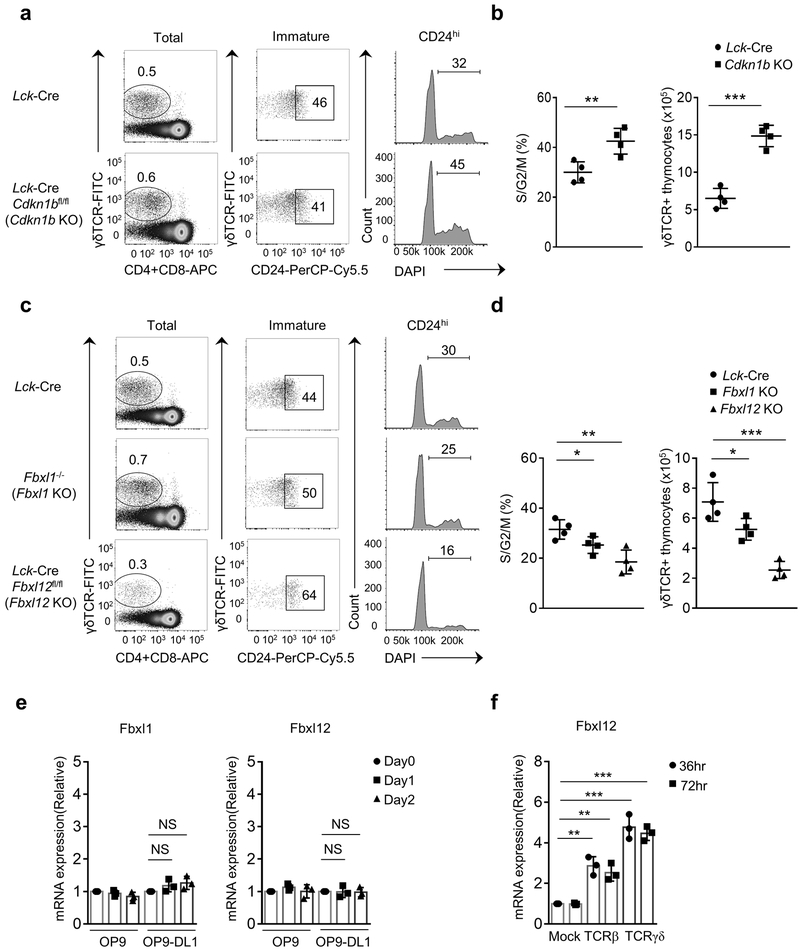
We generated Lck-Cre Cdkn1bfl/fl mice to induce deletion of Cdkn1b selectively in immature DN thymocytes. Similar to germline Cdkn1b−/− mice14, 15, 16, thymus size and cellularity a well as numbers of CD4+CD8− (CD4 single positive; CD4 SP) and CD4−CD8+ (CD8 single positive; CD8 SP) spleen T cells were increased by approximately 2-fold in Lck-Cre Cdkn1bfl/fl mice compared to Lck-Cre Cdkn1b+/+ mice (Supplementary Fig. 1a-d and data not shown). The percentage of cycling DN4 thymocytes, ISP thymocytes and DP blasts was significantly increased (approximately 1.3 fold) in Lck-Cre Cdkn1bfl/fl mice compared to Lck-Cre Cdkn1b+/+ mice (Supplementary Fig. 2a). Most Lck-Cre Cdkn1bfl/fl DP thymocytes (non-blasting) were quiescent (G0 or G1 phase), similar to Lck-Cre Cdkn1b+/+ DP thymocytes (Supplementary Fig.2a), indicating that they had successfully exited the cell cycle after the β-selection proliferative burst. Thus, deletion of Cdkn1b increased or extended post-β-selection proliferation, but did not induce proliferation in normally quiescent cell populations.
Germline deletion of the gene encoding Fbxl1 (Fbxl1), the substrate recognition subunit of the SCF E3 ligase complex that targets Cdkn1b for ubiquitylation and proteasomal degradation18, 25, resulted in a partial DN3-DN4 developmental block and a 2-fold reduction in the total number of thymocytes and splenic CD4 SP and CD8 SP T cells compared with Fbxl1+/+ mice (Supplementary Fig. 1a,c,d), confirming previous reports19. The percentage of cycling DN4 thymocytes, ISP thymocytes and DP blasts in Fbxl1−/− mice was reduced by 2-fold compared to Fbxl1+/+ mice (Supplementary Fig. 2a). Apoptosis was not increased (Supplementary Fig.2b), suggesting that the reduction in Fbxl1−/− thymocyte numbers was caused by reduced proliferation in response to β-selection signals. Fbxl1 interacted with cullin1 (Cul1) and therefore functioned as a subunit of an SCF complex (SCF-Fbxl1 hereafter) (Supplementary Fig. 2c). Fbxl1 bound to and destabilized Cdkn1b (Supplementary Fig. 2d), and this activity required the F-box domain of Fbxl1 (Supplementary Fig. 2e)18, 19, 20. Consistent with these findings, expression of Cdkn1b was increased in DN thymocytes from Fbxl1−/− mice compared to Fbxl1+/+ mice (Supplementary Fig.2f). The partial DN3-DN4 block, reduction in thymocyte cellularity, and cell cycle defects were completely reversed in Lck-Cre Cdkn1bfl/fl Fbxl1−/− mice (Supplementary Fig. 1a,c,d and Supplementary Fig. 2a)19 indicating that the developmental defects in Fbxl1−/− mice were caused by failure to down-regulate Cdkn1b. Together, these findings demonstrate that β-selection associated proliferation is regulated by SCF-Fbxl1-mediated degradation of Cdkn1b.
SCF-Fbxl12 regulates Cdkn1b and β-selection proliferation
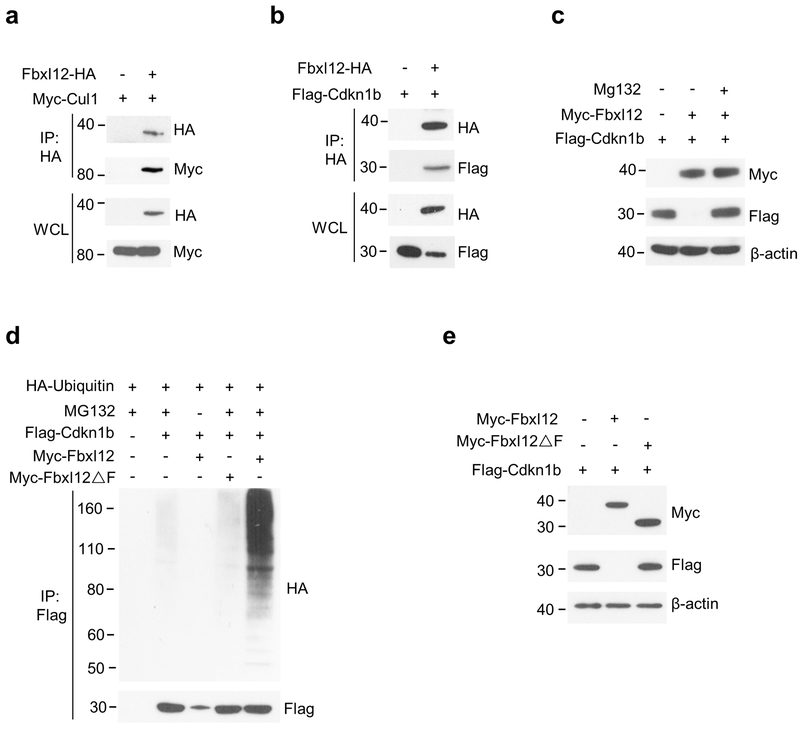
Compared to the moderate developmental defects observed in Fbxl1−/− mice, transgenic overexpression of Cdkn1b results in an almost complete DN3-DN4 block and a 10-fold reduction in thymocyte numbers17, suggesting that additional F-box protein(s) may regulate the turnover of Cdkn1b in immature thymocytes. Phylogenetic characterization and sequence and motif comparison of mammalian F-box proteins indicated that Fbxl1 is closely related to Fbxl12 suggesting that these proteins may target the same substrate26. Fbxl12 was highly and selectively expressed in thymocytes in both mouse and humans (Supplementary Fig. 3a,b), and, similar to Fbxl1, its expression was mostly limited to DN and DP thymocytes (Supplementary Fig. 3c). Co-transfection experiments in HEK-293T cells showed that, similar to Fbxl1, Fbxl12 bound to Cul1 (Fig. 1a) indicating that Fbxl12 functions as a subunit of an SCF complex (SCF-Fbxl12 hereafter). Fbxl12 also bound to Cdkn1b (Fig. 1b), and proteasome blockade with MG132 revealed that, similar to SCF-Fbxl1 complexes18, 19, 20, SCF-Fbxl12 complexes targeted Cdkn1b for polyubiquitylation and proteasomal degradation (Fig. 1c,d), and that this activity required the F-box motif (Fig. 1d,e).
Figure 1.
SCF complexes containing Fbxl12 (SCF-Fbxl12) target Cdkn1b for ubiquitination and proteasomal degradation. (a) Immunoprecipitation (IP) and immunoblot analysis showing the interaction of Fbxl12 and Cul1 in HEK-293T cells transfected for 48 h with plasmids encoding Myc-Cul1 and Fbxl12-HA. (b) IP and immunoblot analysis showing the interaction of Fbxl12 and Cdkn1b in HEK-293T cells transfected for 48 h with plasmids encoding Flag-Cdkn1b and Fbxl12-HA. (c) Immunoblot analysis showing the degradation of Cdkn1b in HEK-293T cells transfected with plasmids encoding Myc-Fbxl12 and Flag-Cdkn1b for 48 hr then treated or not with MG132 for 8 h. (d) IP and immunoblot analysis showing the ubiquitinylation of Cdkn1b by SCF-Fbxl12 complexes and its dependency on the Fbxl12 F-box domain in HEK-293T cells transfected with plasmids encoding Myc-Fbxl12 or Myc-Fbxl12ΔF (Myc epitope tagged Fbxl12 lacking the F-box domain), Flag-Cdkn1b and HA-ubiquitin (HA-Ub) for 48 hr then treated or not with MG132 for 8 h. (e) Immunoblot analysis showing degradation of Cdkn1b by SCF complexes containing Fbxl1 but not by SCF complexes containing Myc-Fbxl12ΔF in HEK-293T cells transfected with plasmids encoding Myc-Fbxl12 or Myc-Fbxl12ΔF and Flag-Cdkn1b then treated or not with MG132 for 8 h. All results are representative of three independent experiments.
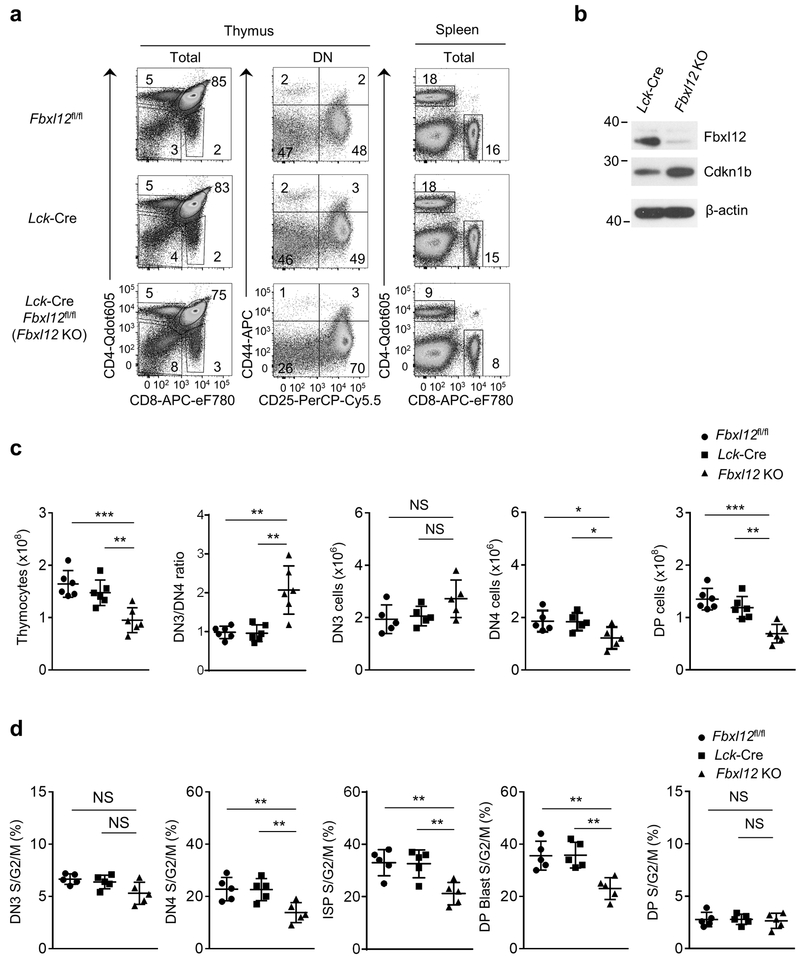
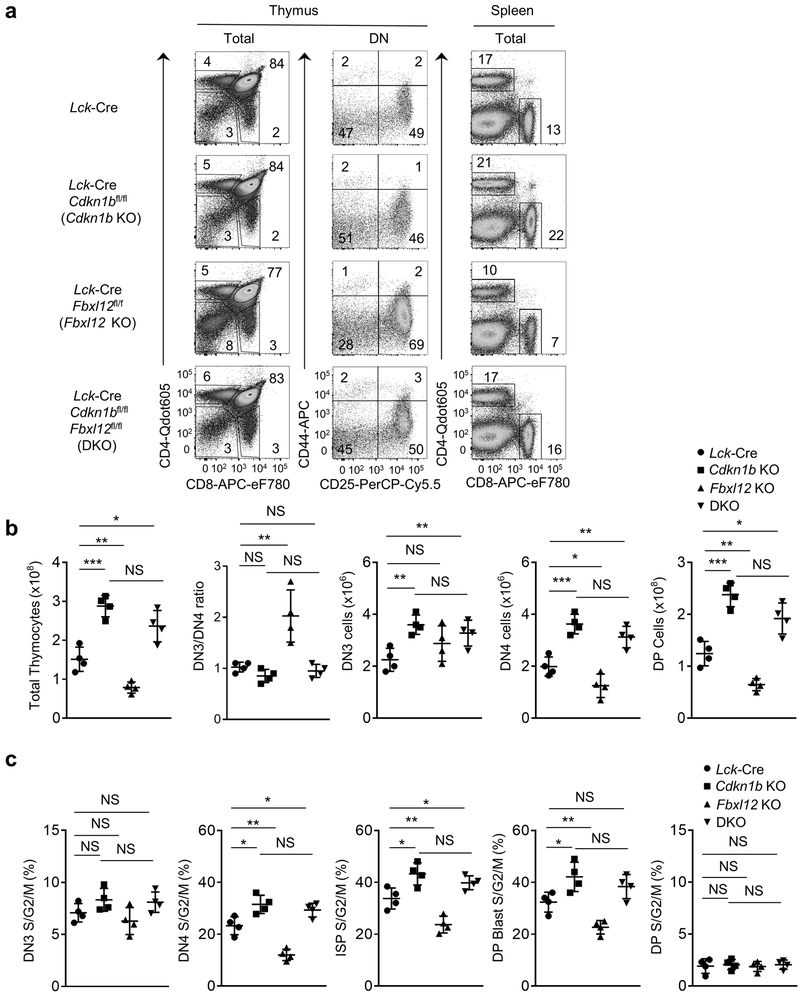
Germline deletion of Fbxl12 is embryonic lethal27. Therefore, we generated mice with a conditional (flox) deletion allele of Fbxl12 (Fbxl12fl/fl) and crossed these to Lck-Cre transgenic mice to delete Fbxl12 selectively in DN thymocytes (Supplementary Fig. 3d-f). The phenotype of Lck-Cre Fbxl12fl/fl mice was similar to that of Fbxl1−/− mice; specifically, there was a substantial, but incomplete block at the DN3-DN4 transition and an approximately 2-fold reduction in the number and percentage of cycling (S/G2/M phase) DN4, ISP and DP blasts compared to Lck-Cre Fbxl1+/+ mice (Fig. 2a-d and Supplementary Fig. 4a). Numbers of DP thymocytes and CD4 SP and CD8 SP thymocytes and spleen T cells were also reduced approximately 2-fold in Lck-Cre Fbxl12fl/fl mice compared to Lck-Cre Fbxl12+/+ mice (Fig. 2c and Supplementary Fig. 4b). Also similar to Fbxl1−/− mice, there was no increase in apoptosis of Lck-Cre Fbxl12fl/fl thymocytes compared to Lck-Cre Fbxl12+/+ thymocytes (Supplementary Fig. 4c). Expression of the orphan nuclear receptor RORγt, which regulates the expression of Cdkn1b and cell cycle entry in DP thymocytes28 was unaffected in Lck-Cre Fbxl12fl/fl thymocytes (Supplementary Fig. 5a). Moreover, expression of a TCRαβ transgene in Lck-Cre Fbxl12fl/fl thymocytes failed to reverse the partial DN3-DN4 block or restore normal thymocyte cellularity (Supplementary Fig. 5b,c), indicating that the developmental defect in Lck-Cre Fbxl12fl/fl thymocytes was not caused by a defect in TCRβ rearrangement. Expression of Cdkn1b was increased in both total (Supplementary Fig. 3f) and DN Lck-Cre Fbxl12fl/fl thymocytes (Fig. 2b) compared to Fbxl12+/+ thymocytes. As observed with Fbxl1−/− mice, the developmental defects in Lck-Cre Fbxl12fl/fl thymocytes were reversed in Lck-Cre Cdkn1bfl/fl Fbxl12fl/fl thymocytes (Fig. 3a-c). These results indicate that, similar to Fbxl1, the primary function of Fbxl12 in thymocytes was to regulate the turnover of Cdkn1b.
Figure 2.
Impaired β-selection-associated proliferation in Lck-Cre Fbxl12fl/fl mice. (a) Representative flow cytometry analysis showing the phenotype of thymocytes (left) or splenocytes (right) from mice of the indicated genotype. Thymus: left, CD4 vs CD8 staining of total thymocytes; center, CD44 vs CD25 staining of lineage-negative DN thymocytes. Spleen: CD4 vs CD8 staining of total splenocytes. (b) Immunoblot analysis showing absence of Fbxl12 and increased Cdkn1b expression in CD4−CD8− (DN) thymocytes from Lck-Cre Fbxl12fl/fl mice. (c) Cell numbers of the indicated thymocyte subsets and DN3/4 thymocyte ratio (n=6 mice per genotype). (d) Percentage of cycling S/G2/M stage cells in the indicated thymocyte subsets determined by staining for DAPI vs Ki-67 (n=5 mice per genotype). For all graphs, horizontal lines indicate the mean and vertical lines indicate standard deviation (±s.d.), P values were determined by unpaired two-tailed Student’s t-test. NS, not significant (P>0.05), * P<0.05, ** P<0.01, ***P<0.005. Data shown in (a) and (b) are representative of four or two independent experiments, respectively.
Figure 3.
Restoration of thymocyte development and β-selection associated proliferation in Lck-Cre Fbx12fl/fl mice by deletion of Cdkn1b. (a) Flow cytometry of cells from Thymus (left) or Spleen (right) from mice of the indicated genotype. Thymus: left, CD4 vs CD8 staining of total thymocytes; center, CD44 vs CD25 staining of lineage-negative DN thymocytes. Spleen: CD4 vs CD8 staining of total splenocytes. (b) Cell numbers of the indicated thymocyte subsets and DN3/DN4 ratio (n=4 mice per genotype). (c) Percentage of cycling S/G2/M stage cells in the indicated thymocyte subsets determined by staining for DAPI vs Ki-67 (n=4 mice per genotype). For all graphs, horizontal lines indicate the mean and vertical lines indicate the standard deviation (±s.d.), P values were determined by unpaired two-tailed Student’s t-test. NS, not significant (P>0.05), *P<0.05, **P<0.01, ***P<0.005, ****P<0.0001. Data shown in (a) are representative of four independent experiments.
SCF-Fbxl1 and SCF-Fbxl12 function additively
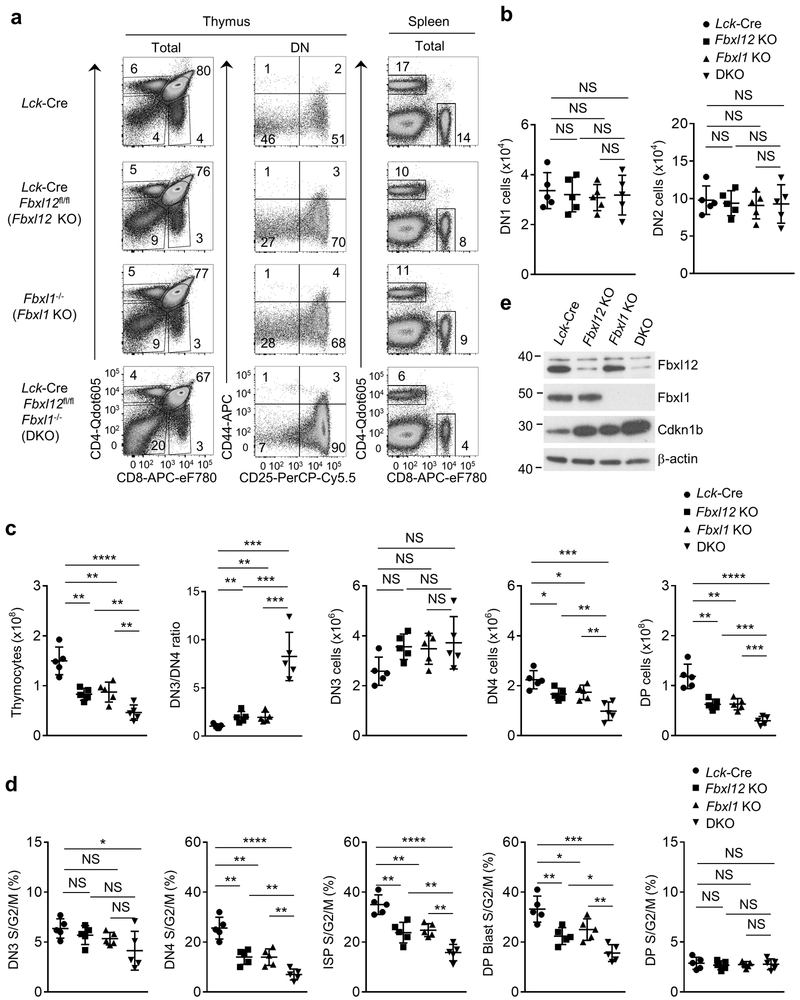
To determine if deletion of both Fbxl1 and Fbxl12 exacerbated the reduction in β-selection-associated proliferation compared to the individual gene deletions, we generated Lck-Cre Fbxl12fl/fl Fbxl1−/− mice. These mice had normal frequencies (Fig. 4a) and numbers (Fig. 4b) of early DN1 or DN2 thymocytes, but had a profound block at the DN3-DN4 transition (Fig.4a,c) and a much more severe reduction in the number and the proliferation of DN4, ISP and DP blasts compared to either Fbxl1−/− or Lck-Cre Fbxl12fl/fl mice (Fig.4c,d and Supplementary Fig. 6a), in addition to a significant further reduction in DP thymocytes and CD4 SP and CD8 SP thymocyte and spleen T cell counts (Fig. 4c and Supplementary Fig. 6b). Expression of Cdkn1b was increased further in Lck-Cre Fbxl12fl/fl Fbxl1−/− DN thymocytes compared to Fbxl1−/− or Lck-Cre Fbxl12fl/fl DN thymocytes (Fig. 4e), but did not result in an increase in thymocyte cell death (Supplementary Fig. 6c). To test whether the extent of β-selection-induced cell cycle progression and proliferation was sensitive to the amount of Cdkn1b protein, we generated Lck-Cre Fbxl12 f /+Fbxl1+/− mice in which Fbxl1 and Fbxl12 expression in DN thymocytes was reduced by approximately 50% compared to Fbxl12 +/+Fbxl1+/+ mice (Supplementary Fig. 7a,b). Lck-Cre Fbxl12 fl /+ Fbxl1+/− DN thymocytes had increased expression of Cdkn1b compared to Fbxl12 +/+ Fbxl1+/+ thymocytes (Supplementary Fig. 7b) and exhibited a phenotype that closely resembled that of Fbxl1−/− or Lck-Cre Fbxl12fl/fl mice (Supplementary Fig. 7a-d). These observations indicated that the proliferative response to β-selection was sensitive to cellular amounts of Fbxl1, Fbxl12 and Cdkn1b.
Figure 4.
Thymocyte development and β-selection-associated proliferation are strongly impaired in thymocytes lacking Fbxl1 and Fbxl12. (a) Flow cytometry analysis of cells from Thymus (left) or Spleen (right) from mice of the indicated genotype. Thymus: left, CD4 vs CD8 staining of total thymocytes; center, CD44 vs CD25 staining of lineage-negative DN thymocytes. Spleen: CD4 vs CD8 staining of total splenocytes. (b) Cell numbers of DN1 and DN2 thymocytes from mice of the indicated genoptype (n=5 mice per genotype). (c) Cell numbers of total thymocytes or the indicated thymocyte subsets and DN3/4 ratio (n=5 mice per genotype). (d) Percentage of cycling (S/G2/M) stage cells in the indicated thymocyte subsets determined by staining for DAPI vs Ki-67 (n=5 mice per genotype). (e) Immunoblot analysis showing Fbxl1, Fbxl12 and Cdkn1b expression in purified DN thymocytes from mice of indicated genotype. For all graphs, horizontal lines indicate the mean and vertical lines indicate standard deviation (±s.d.), P values were determined by unpaired two-tailed Student’s t-test. NS, not significant (P>0.05), *P<0.05, **P<0.01, ***P<0.005, ****P<0.0001. Data shown in (a) and (e) are representative of four or two independent experiments, respectively.
SCF-Fbxl1 and SCF-Fbxl12 target the same site on Cdkn1b
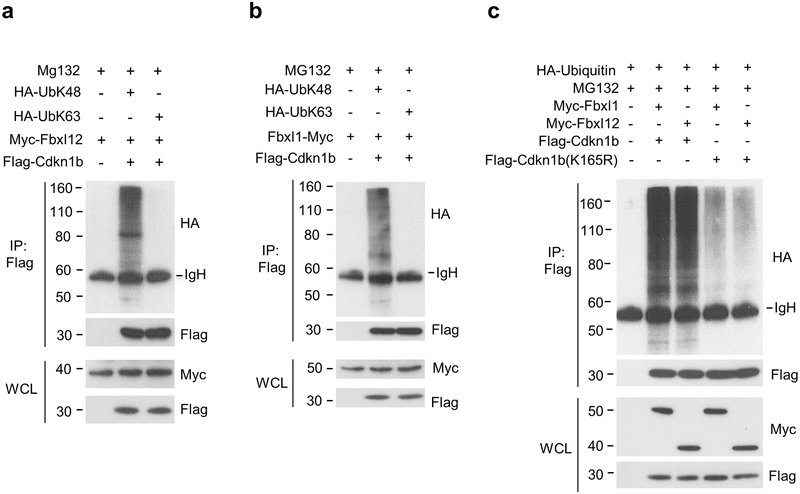
We next examined the type and specificity of Cdkn1b ubiquitinylation by SCF-Fbxl1 or SCF-Fbxl12 E3 ubiquitin ligase complexes. As expected, SCF-Fbxl1 and SCF-Fbxl12 each directed the K48 poly-ubiquitinylation of Cdkn1b (Fig. 5a,b), a modification that targets proteins for proteasomal degradation29. Consistent with the previous characterization of lysine (K) 165, which is conserved in mouse and human Cdkn1b, as the major, and possibly sole site of Cdkn1b K48-ubiquitinylation30, mutation of K165 to arginine (K165R) strongly reduced the poly-ubiquitinylation of Cdkn1b in HEK-293T cells co-transfected with Fbxl1 or Fbxl12 (Fig. 5c), indicating that both SCF-Fbxl1 and SCF-Fbxl12 ubiquitin ligase complexes directed the poly-ubiquitinylation of Cdkn1b at this site. These results, together with the phenotype of Lck-Cre Fbxl12fl/fl Fbxl1−/− mice compared to that of Fbxl1−/− or Lck-Cre Fbxl12fl/fl mice, suggested that SCF-Fbxl1 and SCF-Fbxl12 ubiquitin ligase complexes function identically but additively to target Cdkn1b for ubiquitinylation.
Figure 5.
Fbxl1 and Fbxl12 target the same site on Cdkn1b for K48 poly-ubiquitination. (a) Immunoprecipitation and immunoblot analysis showing K-48 but not K-63 ubiquitinylation of Cdkn1b by SCF-Fbxl12 complexes in HEK-293T cells transfected for 48 h with plasmids encoding Myc-Fbxl12, Flag-Cdkn1b and either HA-UbK48 or HA-UbK63 followed by treatment with MG132 for 8 h. (b) Immunoprecipitation and immunoblot analysis showing K-48 but not K-63 ubiquitinylation of Cdkn1b by SCF-Fbxl1 complexes in HEK-293T cells transfected for 48 h with plasmids encoding Myc-Fbxl1, Flag-Cdkn1b and either HA-UbK48 or HA-UbK63 followed by treatment with MG132 for 8 h. (c) Immunoprecipitation and immunoblot analysis showing ubiquitinylation of Cdkn1b at K165 by SCF-Fbxl1 and SCF-Fbxl12 complexes in HEK-293T cells transfected with plasmids encoding HA-Ub, Flag-Cdkn1b or Flag-Cdkn1b(K165R) and Myc-Fbxl1 or Myc-Fbxl12 for 48 h then treated with MG132 for 8 h. Results shown are representative of at least three independent experiments.
Notch and pre-TCR regulate Fbxl1 and Fbxl12 respectively
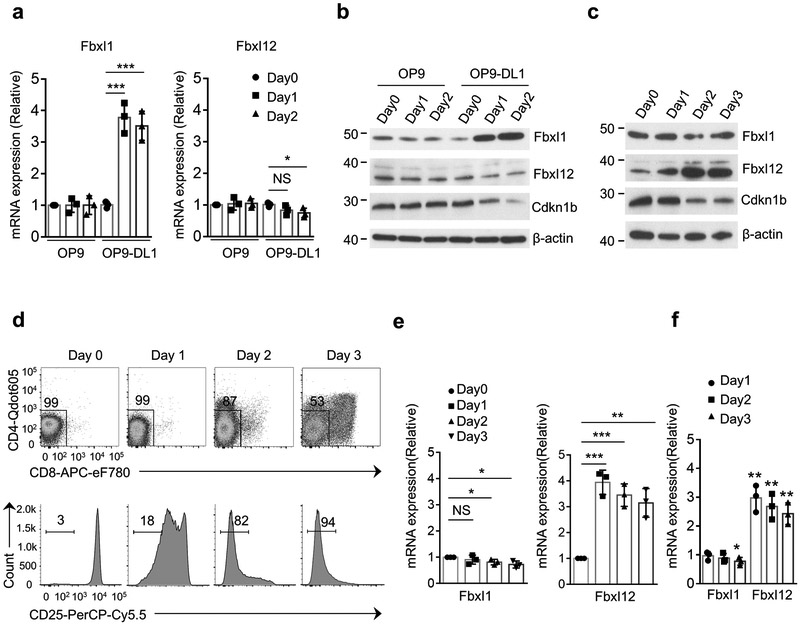
Because proliferation of DN thymocytes at the β-selection checkpoint is dependent upon coordinated signals transduced by Notch1 and the pre-TCR6, 31, 32, we next investigated if these inductive signals regulated the expression of Fbxl1 and/or Fbxl12. To evaluate the impact of Notch signaling on the expression of Fbxl1 and Fbxl12, we cultured Rag2−/− DN3 thymocytes, which are pre-TCR−, on OP9 stromal cells transduced with the Notch ligand Delta-like 1 (OP9-DL1)33. Rag2−/− DN3 cells cultured on OP9-DL1 cells, but not on OP9 cells, up-regulated Fbxl1 mRNA and protein after 1 or 2 days of culture relative to day 0, and this was associated with a reduction in the expression of Cdkn1b (Fig. 6a,b). Cell recovery was similar on OP9-DL1 and OP9 cells (data not shown), therefore the difference in Fbxl1 expression could not be attributed to differences in survival or proliferation. Induction of Fbxl1 mRNA in Rag2−/− thymocytes was observed 4 h after plating on OP9-DL1 cells (Supplementary Fig. 8a), indicating that this response did not require cell proliferation or transition to the DN4 stage. No increase in Fbxl12 mRNA or protein was detected in Rag2−/− DN3 cells cultured on either OP9 or OP9-DL1 cells at any time point between 4hrs and 2 days relative to day 0 (Fig. 6a,b and Supplementary Fig. 8a), indicating that Notch signaling induced the transcription of Fbxl1, but not Fbxl12.
Figure 6.
Notch signaling and Pre-TCR signaling regulate Fbxl1 and Fbxl12 expression, respectively. (a) Real-time PCR quantitation of Fbxl1 (left) and Fbxl12 (right) mRNA expression in Rag2−/− DN3 thymocytes plated on OP9 or OP9-DL1 cells for the indicated times. mRNA expression is relative to Day 0. (b) Immunoblot analysis showing induction of Fbxl1 but not Fbxl12 by Notch signaling provided by OP9-DL1 stromal cells. (c) Immunoblot analysis showing induction of Fbxl12 but not Fbxl1 in total thymocytes from Rag2−/− mice that were injected (IP) with anti-CD3 antibody and harvested at the indicated timepoints. (d) Flow cytometry analysis of thymocytes from one experiment described in (c) showing CD4 vs CD8 staining and down-regulation of CD25 surface expression. (e) Real-time PCR quantitation of Fbxl1 (left) and Fbxl12 (right) mRNA expression in samples from the experiments described in (c,d). mRNA expression is relative to Day 0. (f) Real-time PCR analysis of Fbxl1 mRNA expression (left) and Fbxl12 mRNA (right) in Rag2−/− DN3 thymocytes transduced with retrovirus encoding GFP or TCRβ-IRES-GFP and plated OP9-DL4 cells for 1-3 days. Results are fold change of mRNA expression relative to GFP transduced cells at each time-point. For all graphs, horizontal lines indicate the mean, vertical lines indicate standard deviation (±s.d.), P values were determined by unpaired two-tailed Student’s t-test. NS, not significant (P>0.05), *P<0.05, **P<0.01, ***P<0.005. Data in (a,e,f) are combined results of three independent experiments. Data in (b-d) are representative of three independent experiments.
Injection of Rag2−/− mice, whose thymocytes have a complete block in development at the DN3 stage34, with mAb against CD3 mimics pre-TCR signaling by engagement of surface CD3 complexes that lack TCRβ and pre-TCRα, and induces a strong proliferative burst and transition of thymocytes to the DP stage35, 36, 37. Fbxl12 protein was modestly increased in Rag2−/− total thymocytes on day 1 after intraperitoneal injection of CD3 mAb and was strongly increased on day 2 and 3 compared to total thymocytes from un-injected Rag2−/− mice (Fig. 6c). Induction of Fbxl12 coincided with the down-regulation of Cdkn1b (Fig. 6c) and of CD25 (Fig. 6d) but preceded transition to the DP stage (Fig. 6c,d). Fbxl12 mRNA was induced in thymocytes 8 h after CD3 mAb injection and continued to be induced at 24 h and on day 2 and 3 (Fig. 6e and Supplementary Fig. 8b). However, Fbxl1 mRNA and protein were not up-regulated in Rag2−/− thymocytes in response to intraperitoneal injection of CD3 mAb (Fig. 6c,e and Supplementary Fig. 8b), indicating that pre-TCR signaling selectively induced the expression of Fbxl12. In addition, Rag2−/− DN3 thymocytes transduced with a TCRβ−IRES-GFP retrovirus to induce the expression of the pre-TCR and then cultured on OP9 cells expressing the Notch ligand DL4 (OP9-DL4 cells)6, 33 up-regulated Fbxl12 mRNA but not Fbxl1 mRNA compared to Rag2−/− DN3 thymocytes transduced with GFP retrovirus (Fig. 6f). These results demonstrated that Notch and pre-TCR signals selectively induced the expression of Fbxl1 and Fbxl12, respectively.
Fbxl1 and Fbxl12 can function interchangeably
To test whether either Fbxl1 or Fbxl12 alone would be sufficient to promote normal β-selection-associated proliferation if expressed at sufficiently high levels, we transduced Fbxl1−/− CD25+CD44−CD27hi (hereafter DN3b)38 pre-TCR+ post-β-selected thymocytes with GFP retrovirus (GFP) or with Fbx12-IRES-GFP (Fbx12-GFP) retrovirus to increase the amount of Fbxl12 protein in DN3b thymocytes lacking Fbxl1, as well as Lck-Cre Fbxl12fl/fl DN3b thymocytes with GFP retrovirus or with Fbxl1-IRES-GFP (Fbx12-GFP) retrovirus to increase the amount of Fbxl1 protein in DN3b thymocytes lacking Fbxl12, and then cultured the transduced cells for 3 days on OP9-DL1 stromal cells. GFP-transduced Fbxl1−/− or Lck-Cre Fbxl12fl/fl DN3b thymocytes proliferated less and generated fewer DP thymocytes compared to GFP-transduced Lck-Cre Fbxl1+/+ Fbxl12+/+ DN3b thymocytes, respectively (Fig. 7a,b and Supplementary Fig. 8c). However, the total cell numbers and the percent and number of DP thymocytes generated from Fbxl1−/− DN3b thymocytes transduced with Fbx12-GFP or from Lck-Cre Fbxl12fl/fl DN3b thymocytes transduced with Fbx1-GFP were significantly (2-fold or greater) increased relative to GFP-transduced Fbxl1−/− DN3b or Lck-Cre Fbxl12fl/fl DN3b thymocytes, respectively, and were similar to those generated by GFP-transduced Lck-Cre Fbxl1+/+ Fbxl12+/+ thymocytes (Fig. 7a,b). Notably, the percentage of proliferating (S/G2/M phase) DN and DP thymocytes was significantly increased (approximately 1.5 fold) in Fbxl12-GFP-transduced Fbxl1−/− DN3b cells and in Fbxl1-GFP-transduced Lck-Cre Fbxl12−/− DN3b cells compared to GFP-transduced Fbxl1−/− or Lck-Cre Fbxl12fl/fl DN3b thymocytes, respectively (Fig. 7b and Supplementary Fig. 8c). Thus, enhanced expression of Fbxl1 in the absence of Fbxl12 or enhanced expression of Fbxl12 in the absence of Fbxl1 was sufficient to promote normal or near-normal β-selection-associated proliferation and the generation of DP thymocytes in DN3 thymocytes that receive Notch and pre-TCR signals.
Figure 7.
Fbxl1 and Fbxl12 function interchangeably to promote proliferation but are not sufficient for β-selection. (a) Flow cytometry analysis showing generation of DP thymocytes by DN3b thymocytes from mice of indicated genotype transduced with retrovirus encoding GFP, Fbxl1-IRES-GFP or Fbxl12-IRES-GFP and plated on OP9-DL1 cells for 3 days. One representative of 4 experiments. (b) Enumeration of results from experiments shown in (a). Left to right: Number of total thymocytes, Percentage of DP thymocytes, Percentage of cycling (S/G2/M) DN cells, Percentage of cycling (S/G2/M) DP cells. (c) Flow cytometry analysis showing generation of DP thymocytes by DN3b thymocytes from B6 (WT) mice transduced with retrovirus encoding GFP (control) or Fbxl1-IRES-GFP and plated on OP9 or OP9-DL1 cells for 3days. (d) Number of total thymocytes (left) and percentage of DP stage cells (right) from the experiment in (c). (e) Flow cytometry analysis showing generation of DP thymocytes from DN3b thymocytes from Rag2+/+ (B6) mice transduced with retrovirus encoding GFP or DN3 thymocytes from Rag2−/− mice transduced with retrovirus encoding GFP or Fbxl12-IRES-GFP and plated on OP9-DL1 cells for 3 days. (f) Number of total thymocytes (left), and percentage of DP stage cells (right) in (e). For all graphs, horizontal lines indicate mean, vertical lines indicate standard deviation (±s.d.), P values were determined by unpaired two-tailed Student’s t-test. NS, not significant (P>0.05), *P<0.05, **P<0.01, ***P<0.005, ****P<0.0001. Data in (b,d,f)are combined results of three experiments. Data in (a,c,e) are representative of three independent experiments.
Next, we tested whether overexpression of Fbxl1 or Fbxl12 could substitute for Notch or pre-TCR signals, respectively, to promote β-selection-associated proliferation and DN-DP differentiation. DN3b thymocytes from wild-type B6 mice transduced with Fbxl1-GFP retrovirus did not differentiate into DP thymocytes when cultured on OP9 stromal cells for 3 days (Fig. 7c,d), but had significantly (1.5 fold) increased proliferation compared to GFP-transduced B6 DN3b thymocytes (Fig. 7d). Likewise, Rag2−/− (pre-TCR−) thymocytes transduced with Fbxl12-GFP did not differentiate into DP thymocytes when plated on OP9-DL1 stromal cells for 3 days (Fig. 7e,f) but had approximately 1.5 fold increased proliferation compared to GFP-transduced Rag2−/− thymocytes (Fig. 7f). These results indicated that forced expression of Fbxl1 or Fbxl12 could promote cell cycle progression and proliferation in the absence of Notch signals and pre-TCR signals, respectively, but were unable to substitute for Notch or the pre-TCR to promote the generation of DP thymocytes.
γδTCR+ thymocyte proliferation is controlled by Fbxl12
In contrast to αβ-lineage DN thymocytes which require both Notch and pre-TCR signals for maturation to the DP stage and for normal β-selection associated proliferation, immature CD24hi γδ-lineage thymocytes are less responsive to and dependent upon Notch signaling for maturation and proliferation32, 39. To examine the role of Fbxl1 and Fbxl12 in proliferation of γδ-lineage thymocytes, we first evaluated the effect of Cdkn1b deletion on cell cycle progression and proliferation of immature CD24hi γδTCR+ thymocytes39. The percentage of cycling CD24hi γδTCR+ thymocytes (Fig. 8a,b) and the total number of intra-thymic γδTCR+ thymocytes (Fig. 8b) were significantly (1.5-2 fold) increased. in Lck-Cre Cdkn1bfl/fl mice compared to Lck-Cre Cdkn1b+/+ mice. On the other hand, the percentage of cycling CD24hi γδTCR+ thymocytes as well as the total number of γδTCR+ thymocytes were decreased by approximately 1.5-2 fold in both Fbxl1−/− and Lck-Cre Fbxl12fl/fl mice compared to Lck-Cre Fbxl1+/+ Fbxl12+/+ mice (Fig. 8c,d). Notably, the reduction in percent cycling and the reduction in total number of γδTCR+ thymocytes was more severe in Lck-Cre Fbxl12fl/fl mice compared to Fbxl1−/− mice (Fig. 8c,d), suggesting that proliferation of immature γδ-lineage committed thymocytes was less dependent on Fbxl1 (Notch signaling) than on Fbxl12 (TCR signaling). Consistent with this observation, Fbxl1 mRNA was only slightly induced in γδTCR+ thymocytes cultured on OP9-DL1 cells compared to γδTCR+ thymocytes cultured on OP9 cells (Fig. 8e). We also detected little or no induction of Fbxl12 mRNA in γδTCR+ thymocytes cultured on either OP9 or OP9-DL cells (Fig. 8e), consistent with the fact that OP9 cells do not express ligands for most γδTCRs39. To compare the γδTCR- and pre-TCR-mediated regulation of Fbxl12, we retrovirally transduced Rag2−/− thymocytes with the KN6 γδTCR, which is engaged by ligands expressed by OP9 and OP9-DL stromal cells40 or with TCRβ to induce the expression of the autonomously signaling pre-TCR. Both TCRβ-transduced and KN6 γδTCR-transduced Rag2−/− thymocytes up-regulated Fbxl12 mRNA when plated on OP9-DL4 cells compared to mock transduced Rag2−/− thymocytes; however, induction of Fbxl12 was significantly greater in KN6 γδTCR-transduced than in TCRβ-transduced Rag2−/− thymocytes (Fig. 8f), suggesting that Fbxl12 expression is regulated quantitatively by TCR signal strength since γδTCR engagement is known to result in higher intensity signaling than autonomous pre-TCR signaling41 . Collectively, these results demonstrated that proliferation of γδ−lineage thymocytes is regulated mainly by TCR signaling-mediated induction of Fbxl12 and are less dependent upon Notch signaling-mediated induction of Fbxl1.
Figure 8.
Proliferation of immature γδTCR+ thymocytes is mediated primarily by TCR induced regulation of Fbxl12. (a) Flow cytometry analysis showing total γδTCR+ thymocytes (left) and percent immature CD24hi γδTCR+ thymocytes (center) from the indicated mice. Right panels show percent cycling (S/G2/M) CD24hi γδTCR+ thymocytes. (b) Quantitation of percentage of cycling (S/G2/M) γδTCR+ thymocytes (left) and number of total γδTCR+ thymocytes (right). (c) Flow cytometry analysis showing total γδTCR+ thymocytes from mice of the indicated genotype (left) and percent immature CD24hi γδTCR+ thymocytes (center) from the indicated mice. Right panels show percent cycling (S/G2/M) CD24hi γδTCR+ thymocytes. (d) Quantitation of percentage of cycling (S/G2/M) γδTCR+ thymocytes (left) and number of total γδTCR+ thymocytes (right). (e) Real-time PCR analysis showing quantitation of Fbxl1 (left) and Fbxl12 (right) mRNA in total γδTCR+ thymocytes form B6 (WT) mice plated on OP9 or OP9-DL1 cells for the indicated days . mRNA expression is relative to Day 0. (f) Real-time PCR analysis showing quantitation of Fbxl12 mRNA in Rag2−/− DN3 thymocytes transduced with retrovirus encoding GFP, TCRβ-GFP or KN6-TCRγδ-GFP then plated on OP9-DL4 cells for the indicated timepoints. mRNA expression is relative to mock infected 36 h sample. For all graphs, horizontal lines indicate mean, vertical lines indicate standard deviation (±s.d.), P values were determined by unpaired two-tailed Student’s t-test. NS, not significant (P>0.05), *P<0.05, **P<0.01, ***P<0.005, ****P<0.0001. Data in (a) and (c) are representative of three independent experiments. Data in (e.f) are combined results of three experiments.
Discussion
In this study, we demonstrated that two distinct SCF E3 complexes containing different F-box subunits (Fbxl1 or Fbxl12) were required for the normal proliferative response of αβ-lineage (pre-TCR+) thymocytes at the β-selection checkpoint. Transcription of Fbxl1 was regulated by Notch signaling whereas transcription of Fbxl12 was regulated by pre-TCR signaling and SCF-Fbxl1 and SCF-Fbxl12 complexes each directed the poly-ubiquitylation of the cyclin dependent kinase inhibitor Cdkn1b. The combined activity of SCF-Fbxl1 and SCF-Fbxl12 complexes was required to elicit the appropriate (normal) proliferative response to β-selection, as absence of either Fbxl1 or Fbxl12 significantly and similarly attenuated, and absence of both profoundly blocked proliferation in DN4, ISP and DP-blast thymocyte populations. The requirement for both SCF-Fbxl1 and SCF-Fbxl12 complexes was quantitative, because both were necessary for β-selection-associated proliferation and targeted the same amino acid residue in Cdkn1b (K165) for poly-ubiquitinylation. Moreover, if highly expressed, either Fbxl1 or Fbxl12 was able to direct the complete degradation of cellular Cdkn1b in cell lines, and enhanced expression of either Fbxl1 or Fbxl12 was sufficient to elicit a normal β-selection-associated proliferative response in DN3 thymocytes in the absence of the other F-box protein. These results support a model where Notch-induced Fbxl1 and pre-TCR-induced Fbxl12 function identically but also additively to degrade Cdkn1b to an extent necessary for optimal β-selection associated proliferation.
Cell cycling and proliferation in DN4, ISP and DP-blasts was significantly attenuated in Fbxl1+/−Fbxl12+/− mice, in which expression of Fbxl1 and Fbxl12 is reduced by approximately 50% and expression of Cdkn1b is increased by approximately 2-fold compared to Fbxl1+/+Fbxl12+/+ mice , indicating that the proliferative response was highly sensitive to cellular amounts of Fbxl1, Fbxl12 and Cdkn1b. These results are concordant with reports that a 2-fold reduction in Cdkn1b is sufficient to induce cell cycle progression in peripheral CD4 SP T cells.42 Both SCF-Fbxl1 complexes and SCF-Fbxl12 complexes have been reported to target several other proteins in addition to Cdkn1b27, 43, 44, 45, 46. Germline deficiency of Fbxl12 is embryonic or perinatal lethal and this has been attributed to trophoblast defects secondary to lack of SCF-Fbxl12 complex-mediated degradation of placental aldehyde dehydrogenase 3 (Aldh3)27. However, it is notable that T cell development was effectively restored in both Fbxl1−/− and Lck-Cre Fbxl12fl/fl mice by deletion of Cdkn1b, suggesting that Cdkn1b is the primary target of both SCF-Fbxl1 and SCF-Fbxl12 complexes relevant to β-selection-associated proliferation.
Although γδ-lineage (γδTCR+) thymocytes were previously thought to be relatively quiescent47, recent data based on cell cycle analysis have identified similar rates of proliferation in immature αβ-lineage (pre-TCR+) and ligand engaged γδTCR+ thymocytes, suggesting that the reduced number of γδ-lineage thymocytes relative to γδ-lineage thymocytes is rather explained by the lower frequency of in-frame γ+δ rearrangement compared to β-rearrangement and the requirement for ligand-mediated signaling by the γδTCR, but not the pre-TCR48. Whereas Notch signals are required for early (DN1-DN3) thymocyte development49 and for the DN-DP developmental transition regardless of the TCR complex expressed, most γδ-lineage committed thymocytes are relatively insensitive to Notch ligands and can complete their maturation in the absence of Notch signaling32, 39. Our observation that proliferation of γδTCR+ thymocytes was less impacted by deletion of Fbxl1 than by deletion of Fbxl12 ,and that, in contrast to pre-TCR+ thymocytes, γδTCR+ thymocytes did not up-regulate Fbxl1 on OP9-DL cells, are consistent with the idea that γδTCR+ thymocytes are relatively unresponsive to Notch ligands. However, we found that induction of Fbxl12 in response to ligand-mediated γδTCR signaling was superior to that elicited by pre-TCR signaling, explaining how ligand engaged γδ-lineage thymocytes can initiate a relatively robust proliferative response in the absence of Notch signaling.
In the absence of Cdkn1b, DP thymocytes successfully exit the cell cycle and become quiescent indicating that distinct molecular mechanisms are involved in cell cycle regulation and proliferation in DN and DP thymocytes. In the absence of the orphan nuclear receptor RORγt, β-selection appears to be unaffected, but DP thymocytes fail to exit the cell cycle and undergo apoptosis28. Deletion of the SCF F-box subunit Fbxw7 also does not affect DN thymocyte proliferation or β-selection, but instead results in increased DP thymocyte proliferation and failure to exit the cell cycle as result of elevated c-Myc50. Thus, whereas destabilization of Cdkn1b is the critical mediator of β-selection-induced proliferation in DN and ISP thymocytes and DP blasts, other regulatory proteins that include RORγt and Fbxw7 function as the key factors that enforce cell cycle exit and quiescence in DP thymocytes.
In summary, we identified a crucial role for destabilization of the cyclin dependent kinase inhibitor Cdkn1b for the proliferative burst that occurs in response to β-selection. Our results also demonstrated that cellular levels of Cdkn1b are controlled by the combined activity of two SCF ubiquitin ligase complexes, SCF-Fbxl1 and SCF-Fbxl12, that are independently regulated by Notch and pre-TCR signals, respectively, explaining the requirement for coordinated Notch and pre-TCR signaling for optimal thymocyte proliferation and differentiation at the β-selection checkpoint.
Online Methods
Mice.
Fbxl12 conditional knockout mice were generated with a targeting vector purchased from the Knockout Mouse Project (KOMP) repository (http:www.komp.org). The vector was linearized and transfected into B6 embryonic stem (ES) cells. Transfected ES cells were cultured with media containing neomycin and resistant clones were screened for homologous recombination by PCR. Blastocyst injections resulted in several chimeric mice, three of which gave germline transmission. Germline Fbxl12fl/+ mice were crossed to ROSA26::FLPe mice (Jackson labs; Stock no. 003946) to delete the Neo gene. Offspring were then crossed to generate Fbxl12fl/fl mice. Fbxl1−/− mice51 were provided by Dr.Liang Zhu (Albert Einstein College of Medicine). Cdkn1bfl/fl mice52 were purchased from Jackson Laboratory (Stock no. 027328). Lck-Cre transgenic mice, AND TCR-transgenic mice, Rag2−/− mice and CD45.1 C57BL/6 mice were obtained from Taconic Biosciences. Animal experiments were approved by the Animal Care and Use Committee of the National Institute of Child Health and Human Development.
Cell Lines.
HEK293T cells (ATCC) and Platinum-E Retroviral Packaging Cell Line (Cell Biolabs.Inc) were cultured in DMEM medium supplemented with 10% FBS, 2 mM L-glutamine, 50 IU/ml penicillin and 50 μg/ml streptomycin. OP9 cells expressing the Notch ligand Delta-like 1 (OP9-DL1) or Delta-like 4 (OP9-DL4), generated as previously described33, 53, were cultured in αMEM mediim, supplemented with 5% FBS (Gibco) and antibiotics (penicillin (100 ug/mL) + streptomycin (100 U/mL) (Invitrogen) (OP9 media). The GFP-TCRβ, -TCRγ, -TCRδ, -Fbxl1 and -Fbxl12 retrovirus producing GP+E cell lines were generated using pMIG-IRES-GFP as previously described6, 53.
Plasmids and constructs and retroviral transduction.
Fbxl12 was amplified from B6 thymocyte cDNA and cloned into pIRES-hrGFP-2a (Agilent) MSCV-IRES-GFP (Addgene) and pKMyc (Addgene) vectors. Fbxl1 was amplified from B6 thymocyte cDNA and cloned into MSCV-IRES-GFP (Addgene) vector. pcDNA3-myc-Fbxl1(Skp2), pGFP-E-Cdkn1b(p27), pcDNA3-DN-hCUL1-FLAG, and pEGFP-C1-FLAG-Ku80 were purchased from Addgene. pKMyc-Fbxl1 and pKMyc-Fbxl12 were used as a template to delete the F-box motif by PCR. Cdkn1b(K165R) was generated by site-directed mutagenesis with the Quick-Change Kit (Stratagene). Platinum-E Retroviral Packaging Cells were transfected with retroviral vectors. Retrovirus-containing medium was collected at 48 and 72h post-transfection. For transduction, 2.5×105 cells were incubated with 0.5ml retrovirus-containing medium for 16h and then replaced with fresh culture medium.
Flow cytometry and cell purification.
Single-cell suspensions were prepared in HBSS supplemented with 0.5% BSA and 0.5% NaN3. Cells were incubated with anti-FcR (2.4G2) for 10min followed by fluorochrome-conjugated antibody staining for 50min (4 °C ). For intracellular staining, after staining for surface antigens, cells were fixed in 2% paraformaldehyde (Polysciences) and permeabilizated with 0.1% Triton X-100 (Sigma-Aldrich), then stained with DAPI (Molecular Probes) and Ki-67 (BD Biosciences). Percent of apoptotic cells was determined by Annexin V (BD Biosciences) staining according to the manufacturer’s instructions. Samples were analyzed on an LSRII or Fortessa flow cytometer (BD Biosciences). DN thymocytes were purified by lineage marker negative selection using a magnetic bead/column system (MACS, Miltenyi Biotec). For DN3b cells, DN cells were further stained with CD27-PE, labelled with Anti-PE Microbeads and isolated by magnetic columns. For purification of γδTCR+ thymocytes, cells were first enriched by lineage marker (-TCRγδ) negative selection using a magnetic bead/column system (Miltenyi) followed by staining with TCRγδ-PE and positive selection with anti-PE mAb conjugated microbeads and isolation by magnetic columns. Antibodies used for flow cytometry: The lineage marker (Lin) mixture for DN cells included the following antibodies: CD4 (GK1.5), CD8α (53-6.7), TCRβ (H57-597), TCRγδ (GL3), CD19 (1D3), B220 (RA3-6B2), Gr1 (RB6-8C5), Ter119, CD49b (Dx5), NK1.1 (PK136), all purchased from BD Bioscience. Other antibodies used for staining included: CD4 (GK1.5 eBioscience), CD8 (53-6.7 eBioscience), CD24(M1/69 BD Bioscience), CD25(PC61 BD Bioscience), CD44 (S7 BD Bioscience), CD45.1 (A20 BD Bioscience), CD45.2 (A104 BD Bioscience), CD62L(MEL-14 BD Bioscience), CD69(H1.2F3 eBioscience).
Retroviral transductions of bone marrow-derived Rag2−/− progenitor T cells
Lineage- (CD3, CD11b, CD11c, CD19, CD45R, CD161, Ter119) PerCP-Cy5.5 negative CD117-APC positive progenitors were isolated from the bone marrow of Rag2−/− mice using flow cytometric cell sorting and co-cultured with OP9-DL4 cells in OP9 media, in the presence of IL-7 (5 ng/mL), SCF (50 ng/mL) and Flt3-L (1 ng/mL), for 7 days, to allow for T-cell differentiation to the CD44− CD25+ (DN3) stage. On day 7, differentiating T cells were cultured with GFP-, TCRβ-GFP or TCRγδ(KN6)-GFP retrovirus producing GP+E cell lines overnight (18 h) in OP9 media containing 4μg/ml Polybrene, SCF, Flt3-L and IL-7. Transduced DN3 (CD45-Alexafluor-700+ CD44-PerCP-Cy5.5- CD25-APC+ GFP+) cells were sorted by flow cytometry and cultured on OP9 or OP9-DL4 cells in the presence of the above-listed cytokines. Transduced CD45+GFP+ cells were harvested by flow cytometric cell sorting on days 1, 2, and 3 post-transduction.
Immunoprecipitation and immunoblot analysis.
Cells were lysed in RIPA lysis buffer (50mM Tris, 150mM NaCl,1% NP-40, 0.5% Sodium Deoxycholate,1%SDS) or NP40 lysis buffer (50mM Tris,137mM NaCl,0.5% NP-40, 1mM EDTA) with protease inhibitor cocktail (Roche). Cell lysates were pre-cleared with Gammabind G sepharose beads (GE healthcare) for 20 min then incubated with antibodies overnight, followed by a 2hr incubation with 30ul Gammabind G sepharose beads. Beads were washed three times with lysis buffer then boiled in LDS sample buffer (Invitrogen). For in vivo ubiquitination assays, 293T cells were transfected with the indicated plasmids including plasmid encoding Ub-HA. 48hr post transfection, cells were treated with 10μm MG132 for 8 h, then lysed in denaturing buffer (1% SDS, 50 mM Tris pH 7.5, 0.5 mM EDTA and 1 mM dithiothreitol). After incubation at 95 °C for 5 min, samples were processed for immunoprecipitation. For immunoblot analysis, proteins were fractionated in 4-12% bis-Tris gels (Invitrogen) then transferred to PVDF membranes (Merck Millipore). The membranes were blocked for 1 h in PBST containing 5% fat-free milk, then incubated with primary antibodies overnight followed by 3 washing steps and 1 h of incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies. Blots were developed with ECL (GE healthcare) and exposed to film (Kodak). Antibodies used: HA (12CA5), β-actin(AC-74), Flag(M2) were obtained from Sigma, Fbxl12(ab96831) was obtained from Abcam, ROR gamma (NBP2-24503) was obtained from Novus Bio. Skp2(H-435), Skp2(A-2), CUL-1 (H213), c-Myc (A-14), c-Myc (9E10), p21(C-19), p27(F-8), p27(C-19), Ku-86 (H-300), Fbxl12 (H-273), Goat anti-rabbit IgG-HRP (sc-2030), Goat anti-mouse IgG-HRP (sc-2031), mouse anti-rabbit IgG-HRP (sc-2357), Goat anti-mouse IgG-HRP (sc-2032), donkey anti-goat IgG-HRP (sc-2033) were purchased from Santa Cruz.
RNA isolation and Real-time PCR.
Total RNA was extracted from cells using Trizol (Invitrogen) and reverse transcribed with the SuperScript First-Strand Synthesis system (Invitrogen). Transcripts were quantified with a Roche LightCycler480. Gene-expression levels were calculated and presented as expression relative to control genes.
Statistical Analysis.
All data are presented as mean ± s.d. The unpaired, nonparametric Student’s t-test (Mann–Whitney test) was used for the statistic analysis. GraphPad Prism 7.0 was used for data analysis and presentation. P<0.05 was considered statistically significant.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgements
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver, NICHD (PEL: Project number 1ZIAHD001803-19), a grant from the Swedish Society for Medical Research (BZ), the Canadian Institutes of Health Research (JCZP: FND-154332) (JCZP), and the National Institutes of Health (JCZP: 1P01AI102853-01) (JCZP). JCZP is supported by a Canada Research Chair in Developmental Immunology and KY is supported by a Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada. The authors thank R. Bosselut for critical reading of the manuscript and R. Bosselut, A. Bhandoola, B.J. Fowlkes, N. Taylor and H. Petrie for helpful discussions on the project.
Footnotes
Competing Interests.
The authors declare no competing interests.
References
- 1.Rodewald HR, Ogawa M, Haller C, Waskow C & DiSanto JP Pro-thymocyte expansion by c-kit and the common cytokine receptor gamma chain is essential for repertoire formation. Immunity 6, 265–272 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Shortman K, Egerton M, Spangrude GJ & Scollay R The generation and fate of thymocytes. Semin Immunol 2, 3–12 (1990). [PubMed] [Google Scholar]
- 3.Wong GW, Knowles GC, Mak TW, Ferrando AA & Zuniga-Pflucker JC HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood 120, 1439–1448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penit C, Lucas B & Vasseur F Cell expansion and growth arrest phases during the transition from precursor (CD4-8-) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J Immunol 154, 5103–5113 (1995). [PubMed] [Google Scholar]
- 5.Kreslavsky T et al. beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity 37, 840–853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciofani M et al. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J Immunol 172, 5230–5239 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Hoffman ES et al. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev 10, 948–962 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Maillard I et al. The requirement for Notch signaling at the beta-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med 203, 2239–2245 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yashiro-Ohtani Y, Ohtani T & Pear WS Notch regulation of early thymocyte development. Semin Immunol 22, 261–269 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Aifantis I, Mandal M, Sawai K, Ferrando A & Vilimas T Regulation of T-cell progenitor survival and cell-cycle entry by the pre-T-cell receptor. Immunol Rev 209, 159–169 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Rowell EA & Wells AD The role of cyclin-dependent kinases in T-cell development, proliferation, and function. Crit Rev Immunol 26, 189–212 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Deng C, Zhang P, Harper JW, Elledge SJ & Leder P Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82, 675–684 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto A, Takeishi S & Nakayama KI p57 regulates T-cell development and prevents lymphomagenesis by balancing p53 activity and pre-TCR signaling. Blood 123, 3429–3439 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Nakayama K et al. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85, 707–720 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Kiyokawa H et al. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 85, 721–732 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Fero ML et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 85, 733–744 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Tsukiyama T et al. Down-regulation of p27Kip1 expression is required for development and function of T cells. J Immunol 166, 304–312 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Carrano AC, Eytan E, Hershko A & Pagano M SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1, 193–199 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Nakayama K et al. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell 6, 661–672 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Kossatz U et al. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev 18, 2602–2607 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarmento LM et al. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med 202, 157–168 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dohda T et al. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res 313, 3141–3152 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Del Debbio CB et al. Notch Signaling Activates Stem Cell Properties of Muller Glia through Transcriptional Regulation and Skp2-mediated Degradation of p27Kip1. PLoS One 11, e0152025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hristova NR, Tagscherer KE, Fassl A, Kopitz J & Roth W Notch1-dependent regulation of p27 determines cell fate in colorectal cancer. Int J Oncol 43, 1967–1975 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Tsvetkov LM, Yeh KH, Lee SJ, Sun H & Zhang H p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol 9, 661–664 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Jin J et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18, 2573–2580 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishiyama M, Nita A, Yumimoto K & Nakayama KI FBXL12-Mediated Degradation of ALDH3 is Essential for Trophoblast Differentiation During Placental Development. Stem Cells 33, 3327–3340 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Sun Z et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Clague MJ & Urbe S Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Oshikawa K, Matsumoto M, Oyamada K & Nakayama KI Proteome-wide identification of ubiquitylation sites by conjugation of engineered lysine-less ubiquitin. J Proteome Res 11, 796–807 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Tussiwand R et al. The preTCR-dependent DN3 to DP transition requires Notch signaling, is improved by CXCL12 signaling and is inhibited by IL-7 signaling. Eur J Immunol 41, 3371–3380 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC & von Boehmer H Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J Exp Med 203, 1579–1590 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt TM & Zuniga-Pflucker JC Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17, 749–756 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Shinkai Y et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992). [DOI] [PubMed] [Google Scholar]
- 35.Levelt CN, Mombaerts P, Iglesias A, Tonegawa S & Eichmann K Restoration of early thymocyte differentiation in T-cell receptor beta-chain-deficient mutant mice by transmembrane signaling through CD3 epsilon. Proc Natl Acad Sci U S A 90, 11401–11405 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinkai Y & Alt FW CD3 epsilon-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/− mice in the absence of TCR beta chain expression. Int Immunol 6, 995–1001 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Wiest DL, Kearse KP, Shores EW & Singer A Developmentally regulated expression of CD3 components independent of clonotypic T cell antigen receptor complexes on immature thymocytes. J Exp Med 180, 1375–1382 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taghon T, Yui MA, Pant R, Diamond RA & Rothenberg EV Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Ciofani M, Knowles GC, Wiest DL, von Boehmer H & Zuniga-Pflucker JC Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity 25, 105–116 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Haks MC et al. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity 22, 595–606 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Hayes SM & Love PE Strength of signal: a fundamental mechanism for cell fate specification. Immunol Rev 209, 170–175 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Rowell EA, Walsh MC & Wells AD Opposing roles for the cyclin-dependent kinase inhibitor p27kip1 in the control of CD4+ T cell proliferation and effector function. J Immunol 174, 3359–3368 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Jiang H et al. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell 18, 699–709 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Tedesco D, Lukas J & Reed SI The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev 16, 2946–2957 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SY, Herbst A, Tworkowski KA, Salghetti SE & Tansey WP Skp2 regulates Myc protein stability and activity. Mol Cell 11, 1177–1188 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Li X, Zhao Q, Liao R, Sun P & Wu X The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 278, 30854–30858 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Passoni L et al. Intrathymic delta selection events in gammadelta cell development. Immunity 7, 83–95 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Prinz I et al. Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nat Immunol 7, 995–1003 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Wilson A, MacDonald HR & Radtke F Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med 194, 1003–1012 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onoyama I et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med 204, 2875–2888 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakayama K et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 19, 2069–2081 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chien WM et al. Genetic mosaics reveal both cell-autonomous and cell-nonautonomous function of murine p27Kip1. Proc Natl Acad Sci U S A 103, 4122–4127 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohtashami M, Shah DK, Kianizad K, Awong G & Zuniga-Pflucker JC Induction of T-cell development by Delta-like 4-expressing fibroblasts. Int Immunol 25, 601–611 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.