Summary
Aberrant activation of caspase-6 (C6) in the absence of other hallmarks of apoptosis has been demonstrated in cells and tissues from Huntington disease (HD) patients and animal models. C6 activity correlates with disease progression in HD patients and the cleavage of mutant huntingtin (mHTT) protein is thought to strongly contribute to disease pathogenesis. Here we show that the mHTT1–586 fragment generated by C6 cleavage interacts with the zymogen form of the enzyme, stabilizing a conformation that contains an active site and is prone to full activation. This shift towards enhanced activity can be prevented by a small molecule inhibitor that blocks the interaction between C6 and mHTT1–586. Molecular docking studies suggest that the inhibitor binds an allosteric site in the C6 zymogen. The interaction of mHTT1–586 with C6 may therefore promote a self-reinforcing, feed-forward cycle of C6 zymogen activation and mHTT cleavage driving HD pathogenesis.
Keywords: Huntington disease, caspase-6, allosteric, inhibitor, non-apoptotic
Graphical Abstract

eTOC blurb:
Cleaved mutant huntingtin stabilizes an active conformation in the proform of caspase-6, suggesting a feed-forward cycle as part of the molecular pathogenesis of Huntington’s disease. Small molecules targeting an allosteric site in pro-caspase-6 interrupt this activation and highlight the therapeutic potential of this mechanism.
Introduction
Increased activity of the executioner caspase-6 (C6) has been reported in a number of neurodegenerative conditions, including Alzheimer’s disease (AD) and Huntington’s disease (HD) (Graham, et al., 2011; LeBlanc, 2013). Interestingly, this activity is not directly associated with apoptotic cell death, since neurons with active C6 do not show apoptotic morphologies and other executioner caspases such as caspase-3 (C3) are not activated at the same time (Graham, et al., 2010; Graham, et al., 2011; LeBlanc, 2013). Non-apoptotic roles for caspases have been defined recently, which include neuron-specific processes such as the pruning of synapses, spines, and axons (Unsain and Barker, 2015).
Since caspase activity in these contexts does not lead to the demise of the whole cell, it must be tightly regulated (Unsain and Barker, 2015). Indeed, the slowly progressive neuronal dysfunction followed by eventual cell death observed in chronic neurodegenerative diseases, such as HD, can only occur if caspase activation is either incomplete, partially inhibited or locally confined (Unsain and Barker, 2015). Known mechanisms to maintain sublethal caspase activity include the interaction with endogenous inhibitor of apoptosis proteins (IAPs) or post-translational modifications (PTMs) of the enzyme (Unsain and Barker, 2015). While C6 cannot be inactivated by IAPs, its transcriptional regulation as well as differential phosphorylation or palmitoylation has been associated with increased activation in neurodegenerative disease (Cao, et al., 2012; Ehrnhoefer, et al., 2014; Skotte, et al., 2017; Velazquez-Delgado and Hardy, 2012).
HD is caused by an elongation in the polyglutamine tract of the huntingtin (HTT) protein above the toxic threshold of 36 (The Huntington’s disease Collaborative Research Group, 1993), and the length of the expansion determines the age of disease onset to a large degree (Duyao, et al., 1993; Stine, et al., 1993). Intriguingly, active C6 levels in human HD brain correlate with polyglutamine length and inversely correlate with age of onset (Graham, et al., 2010). Furthermore, elevated levels of active C6 are already observed in pre-symptomatic HD mutation carriers, suggesting a role in disease initiation (Graham, et al., 2010).
For full activation, the C6 zymogen requires cleavage through auto-processing or cleavage by other caspases, removing the pro-domain as well as the inter-subunit linker, an irreversible activation process observed in cells undergoing apoptosis (Fuentes-Prior and Salvesen, 2004). The substrate binding region around amino acid (aa) 130 can fluctuate between a non-canonical helical conformation and the canonical beta-strand conformation typical for caspases, which is only stabilized upon substrate binding (Dagbay, et al., 2017; Dagbay and Hardy, 2017). This suggests that the enzyme possesses higher structural flexibility than other, closely related caspase enzymes (Dagbay, et al., 2017). However, C6 can also be partially activated without proteolytic processing through a conformational change in the zymogen, which renders the active site accessible to a covalently binding chemical probe (Edgington, et al., 2012).
C6 cleaves HTT at aa 586, which is considered a crucial event in HD pathogenesis, since mice expressing C6 cleavage-resistant mutant HTT (C6R mice) do not develop any HD-like phenotypes (Ehrnhoefer, et al., 2018; Graham, et al., 2006; Metzler, et al., 2010; Milnerwood, et al., 2010; Pouladi, et al., 2009). Furthermore, C6R mice do not exhibit increased C6 activation compared to animals expressing cleavable mutant HTT (mHTT) (Graham, et al., 2010), suggesting that mHTT fragments generated by C6 cleavage may be required to initiate the toxic cycle that leads to neuronal dysfunction (Graham, et al., 2011).
Here, we further probe the interaction between C6 and HTT, and find that the mHTT fragment generated by C6 cleavage leads to increased activation of the C6 zymogen, providing a possible mechanism for the feedback loop of C6 activation and mHTT proteolysis in HD. Furthermore, we present a set of compounds capable of preventing the interaction of C6 with mHTT, which reduce C6 activation and mHTT cleavage and may represent a promising therapeutic strategy to disrupt the vicious cycle in HD.
Results
Caspase-6 interacts with huntingtin independent of proteolysis
While the cleavage site of a caspase substrate is mainly determined by a 4aa recognition sequence, we have previously shown that the protein context influences catalytic efficiency as well as specificity for the C6 substrate lamin A (Ehrnhoefer, et al., 2011). Such effects are likely mediated by enzyme-substrate interactions outside of the cleavage site (exosites). To determine whether similar interactions might be present between C6 and HTT, we designed a 4aa peptide substrate based on the cleavage site in HTT (IVLD), which releases aminoluciferin after proteolytic cleavage. This peptide was processed by C3 with approximately 35% efficiency compared to C6, as monitored by luminescence generated in the reaction, demonstrating low specificity for C6 (Fig. 1A). We then applied a TR-FRET-based (time-resolved fluorescence resonance energy transfer) method that uses an N-terminal antibody against HTT in combination with a neo-epitope antibody against the new C-terminus (D586) generated by caspase cleavage (Aharony, et al., 2015). In this assay, a HTT protein fragment comprising aa1–1212 was used as a substrate (HTT1–1212). While we observed a strong TR-FRET signal indicative of HTT proteolysis at D586 in the presence of C6, no HTT cleavage was observed using an equal amount of active C3 (Fig. 1B). This suggests that the context of the HTT protein increases the specificity for C6 cleavage at the IVLD recognition site, similar to our previous findings for lamin A (Ehrnhoefer, et al., 2011).
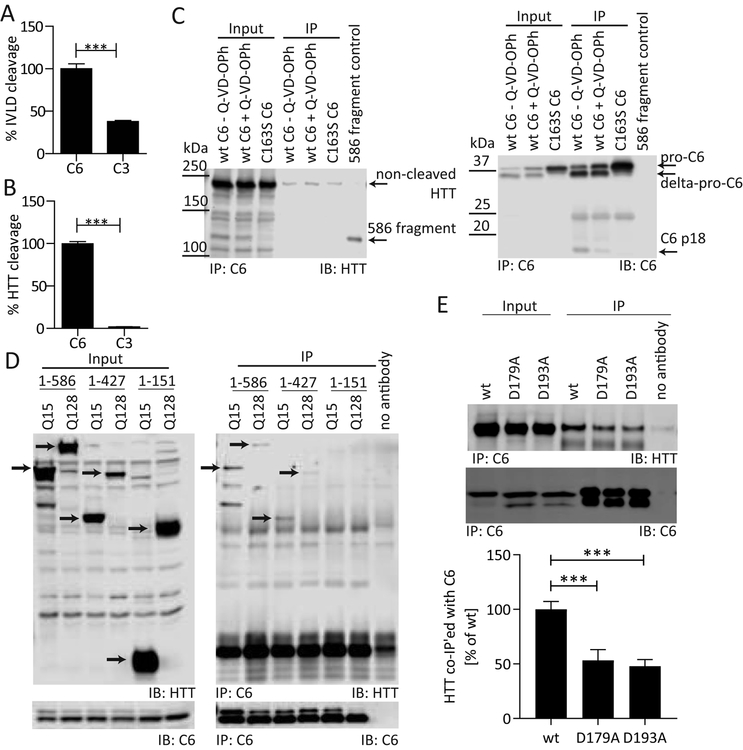
Figure 1: C6 interacts with HTT outside of the cleavage site at D586.
C3 processes an IVLD peptide substrate with approximately 35% efficiency compared to C6 (A), but does not cleave the IVLD sequence presented in the context of the HTT protein (B). (C) DDK-tagged wt or C163S mutant C6 were co-transfected into COS-7 cells with mHTT1–1212. Cells were treated with the pan-caspase inhibitor Q-VD-OPh (3 μM) as indicated. Lysates were immunoprecipitated with DDK antibody, and Western blots were probed for C6 with HD91 antibody and with antibody MAB2166 for co-immunoprecipitation of mHTT1–1212. Uncleaved mHTT1–1212 interacts with C6 irrespective of the C163S mutation or presence of an inhibitor, while the C6 mutation as well as the inhibitor Q-VD-OPh reduce the amount of the mHTT1–586 fragment as well as the amount of the active C6 fragment (p18). (D) DDK-tagged C6wt was co-transfected into COS-7 cells with wt or mHTT constructs of different lengths. Lysates were immunoprecipitated with DDK antibody, and Western blots were probed for C6 with HD91 antibody and with antibody MAB2166 for co-immunoprecipitation of HTT. Both wt and mHTT1–427 or longer interact with C6 (red arrows), while no interaction was observed for wt or mHTT1–151 despite similar levels of expression (black arrows). (E) DDK-tagged C6wt or mutants were co-transfected into COS-7 cells with mHTT1–586 and subjected to immunoprecipitation as in (D). The interaction with mHTT1–586 is reduced in the presence of the C6D179A and C6D193A linker mutations.
Graphed data are pooled results of three independent experiments graphed with S.E.M, Western blots are representative images of at least three independent experiments. Statistical significance was determined by Student’s t-test in (A) and (B), and by one-way ANOVA (p < 0.0001) with Tukey’s post-hoc correction in (E). ***: p <0.001.
To further probe the interaction between C6 and HTT, we next performed co-immunoprecipitation experiments from COS-7 cells co-transfected with human C6 and HTT1–1212 (Warby, et al., 2008). Non-cleaved HTT readily co-immunoprecipitates with wt C6, and this interaction is not prevented by incubation with the irreversible caspase inhibitor Q-VD-OPh (Fig.1C). Q-VD-OPh not only lowers HTT1–586 fragment levels, but also reduces the intracellular generation of active C6 subunits, suggesting that autoproteolytic processing of the zymogen is inhibited (Fig. 1C). The C163S active-site mutant version of C6, incapable of generating the mHTT-586 fragment or autoactivation, also co-immunoprecipitates HTT (Fig. 1C), confirming that HTT interacts with the zymogen form of the enzyme independent of active site binding.
To narrow down the protein region interacting with C6, we used truncation mutants for both wt (Q15) and mHTT (Q128). Co-immunoprecipitation experiments demonstrated that both wt and mHTT fragments interact with C6 up to a length of 427aa, while the 1–151aa fragments failed to co-immunoprecipitate (Fig. 1D). This suggests that the C6 interaction region on the HTT protein lies between aa 151 and 427, which is approximately 160 aa upstream of the cleavage site at D586 (Fig. 1D). For C6, the interaction with HTT was reduced when either D179 or D193 were mutated (Fig. 1E), suggesting that the linker region connecting the C6 subunits before full activation of the enzyme is important for the interaction.
A mHTT1–586 fragment promotes C6 activation
C6 possesses an allosteric small molecule site that differs in zymogen and active forms and has thus been explored as a means to modulate C6 activation (Murray, et al., 2014). Since the binding of HTT may alter C6 activity through an allosteric mechanism, we decided to study C6 activity in cells co-transfected with C6 and wt and mHTT fragments of different lengths. Measuring lamin A cleavage by ELISA (Ehrnhoefer, et al., 2011), we were surprised to find that C6 activity was increased only in the presence of a mHTT1–586 fragment (Fig. 2A). No changes in C6-mediated lamin cleavage were observed when full-length HTT or shorter HTT fragments were overexpressed. Moreover, the HTT1–586 fragment without the pathogenic polyglutamine expansion (15Q) did not increase C6 activity in this system (Fig. 2A).
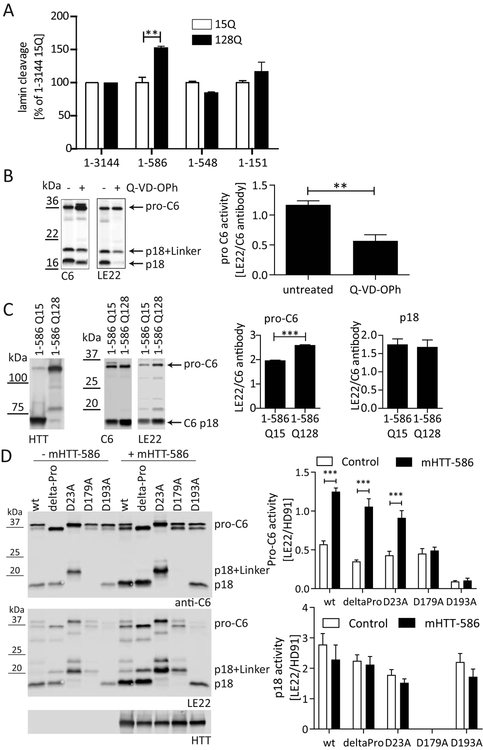
Figure 2: mHTT1–586 promotes an active site conformation in the C6 zymogen.
(A) C6wt was co-transfected into COS-7 cells with wt or mHTT constructs of different lengths. Co-transfection with mHTT1–586 specifically increased lamin A cleavage as measured by ELISA from cell lysates (Ehrnhoefer, et al., 2011). (B) COS-7 cells transfected with C6wt were treated with or without the caspase inhibitor Q-VD-OPh in the media, and lysates were incubated with the active-site binding probe LE22 (Edgington, et al., 2012). Western blotting demonstrates binding of LE22 to the C6 zymogen (detected with C6 antibody HD91) that is sensitive to inhibition by Q-VD-OPh. (C) C6wt was co-transfected into COS-7 cells with wt or mHTT1–586. Lysates were labelled with LE22 before analysis by Western blot using C6 antibody HD91. HTT was detected with antibody MAB2166. While LE22 labels both pro-C6 and the p18 subunit, the presence of mHTT1–586 increases LE22 labeling of pro-C6, but not p18. (D) C6wt and different C6 mutants were co-transfected into COS-7 cells with mHTT1–586 or the empty vector as indicated. Lysates were labelled with LE22 before analysis by Western blot using C6 antibody HD91. HTT was detected with antibody MAB2166. The increase in pro-C6 labeling in the presence of mHTT1–586 is observed for C6wt, C6deltaPro and C6D23A, but not for the C6D179A and C6D193A mutants. No increase in LE22 labeling was observed for the active C6 fragment (p18).
All graphed data are pooled results of three independent experiments graphed with S.E.M., Western blots are representative images. Statistical significance was determined by 2-way ANOVA with Tukey’s post-hoc correction in (A) polyglutamine length p = 0.0207, HTT construct length p = 0.0034 and (D) Pro-C6: HTT p <0.0001, C6 mutation p <0.0001, p18: HTT p = 0.1120, C6 mutation p = 0.0456, and by Student’s t-test in (B) and (C). **: p <0.01, ***: p <0.001.
We next used the chemical probe LE22 that binds to the active site of C6 and thus detects C6 activity directly (Edgington, et al., 2012). In agreement with published data (Edgington, et al., 2012), we observed binding of the probe LE22 not only to the p18 subunit, which harbours the active site, but also to partially and non-processed C6, suggesting that at least a subset of C6 zymogen already possesses an active site conformation capable of probe binding (Fig. 2B). When C6-transfected cells were treated with the irreversible pan-caspase inhibitor Q-VD-OPh prior to lysis, processing into the active subunit p18 was reduced and pro-C6 accumulated (Fig. 2B). Furthermore, the proportion of pro-C6 as well as p18-C6 labelled with LE22 decreased, consistent with blockage of the active site by the irreversible inhibitor (Fig. 2B). Using this approach, we found that co-transfections with mHTT1–586 (Q128), but not wt HTT1–586 (Q15) fragments specifically increased the fraction of pro-C6 labelled with LE22 (Fig. 2C). In contrast, no changes in LE22-labeling of the active p18 subunit were observed (Fig. 2C), indicating that the binding of mHTT1–586 specifically alters the conformation of pro C6 and either stabilizes the active site conformation or renders it more accessible for probe binding. This is in agreement with the notion that the p18 fragment preferentially or even exclusively exists in an active conformation, while only a fraction of the C6 zymogen occupies such a state (Edgington, et al., 2012) and the remainder is thus available for further activation by mHTT1–586.
mHTT1–586 promotes a conformation of pro-C6 capable of substrate binding
Activation of C6 is a multi-step process, where the pro-domain is removed through cleavage at D23 and cleavage at D179 and D193 excises the linker sequence separating the two subunits p18 and p10 (Dagbay and Hardy, 2017; Klaiman, et al., 2009). To determine, which part of the activation process of C6 is increased by mHTT1–586, we used C6 constructs with point mutations at these processing sites. Consistent with published data (Klaiman, et al., 2009), we find that the intracellular activation of C6 is prevented in a C6D179A cleavage resistant mutant, since the p18 fragment is not detected and there is also very little labelling of pro-C6 with LE22. This suggests that the zymogen form of the C6D179A mutant preferentially exists in a conformation incapable of substrate binding (Fig. 2D). On the other hand, removal of aa1–23 in the C6deltaPro mutant as well as the C6D193A mutation still allow for the generation of the p18 fragment (Fig. 2D), in agreement with previously proposed mechanisms of C6 activation (Klaiman, et al., 2009). The C6D23A mutation leads to the generation of a slightly larger p18 subunit, since the pro-domain remains attached, but also for this mutation the active site remains capable of binding the LE22 probe in both the unprocessed and the processed form of the enzyme (Fig. 2D). C6D193A on the other hand shows virtually no labelling with LE22 in the zymogen form, while the p18 subunit, once generated, maintains a normal conformation capable of probe binding (Fig. 2D).
Upon co-transfection of mHTT1–586, we observed a consistent shift towards more LE22 binding in the zymogen forms of C6wt, C6deltaPro, and C6D23A, whereas no change was observed for the C6D179A and C6D193A mutants (Fig. 2D). Similar to the data obtained with C6wt (Fig. 2C), no change in LE22 binding was found for the p18 subunits of the C6 mutant constructs, suggesting that the modulation of C6 activity by mHTT1–586 occurs mainly on the level of the zymogen form. The finding that mHTT1–586 does not increase the zymogen activity of C6D179A and C6D193A on the other hand may reflect the decreased binding of mHTT1–586 to these mutant enzymes (Fig. 1E).
A high-throughput screen with HTT as a substrate identifies irreversible allosteric C6 inhibitors
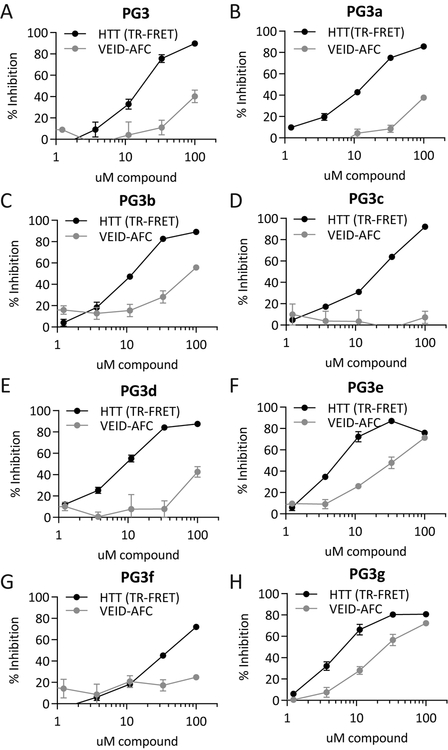
Since the context of the HTT sequence conferred high selectivity to cleavage by C6, we reasoned that a high-throughput screen using HTT as a substrate may also yield inhibitors with high selectivity. We therefore decided to perform a high-throughput screen using the TR-FRET-based C6 activity assay (Aharony, et al., 2015). 22644 compounds stemming mostly from a diverse library were screened in low volume 384-well microtiter plate format at 20 μM where possible. The screen made use of 69 microtiter plates yielding a median Z’ 0.62 (Zhang et al., 1999) (Suppl. Fig. S1), and we identified 291 hits that were associated with >30% inhibition. The primary screening hits were confirmed in triplicate followed by dose-response studies to determine the potencies of the compounds. From these initial hits, 75 were re-confirmed and 15 compounds showed dose-dependent inhibition (data not shown). Screening of available chemical analogues to hit compounds led to the prioritization of an active inhibitor series consisting of the parent compound PG3 and 7 analogues named PG3a-g (Suppl. Fig.S2), which (with the exception of PG3f) all showed similar inhibitory activities in the TR-FRET assay with IC50 values in the low micromolar range (Fig. 3, Suppl. Table S1). However, when we tested the inhibitory activity of PG3 family compounds in a C6 activity assay using the VEID-AFC peptide as a substrate, IC50 values were much higher, with some compounds demonstrating no inhibition at all at concentrations of up to 100 μM (Fig. 3, Suppl. Table S1). In particular, compounds PG3a-d and PG3f showed large shifts in IC50 (Suppl. Table S1).
Figure 3: PG3 family compounds inhibit the cleavage of HTT by C6.
(A) - (H) PG3 family compounds inhibit the C6-mediated cleavage of HTT1–1212 detected by TR-FRET, but are weak inhibitors of the C6-mediated cleavage of a VEID-AFC peptide substrate.
All graphed data are pooled results of three independent experiments graphed with S.E.M., IC50 values are listed in Suppl. Table S1. See also Suppl. Fig. S1 – S4.
To exclude the possibility that the inhibition observed by TR-FRET is due to an artefact of the screening system, we next treated HEK293 cells with different amounts of inhibitor in the presence of camptothecin in the medium to induce intracellular C6 activity (Ehrnhoefer, et al., 2011). The cleavage of lamin, a bona fide C6 substrate, was then quantified by ELISA (Ehrnhoefer, et al., 2011). We observed a dose-dependent inhibition of lamin cleavage upon treatment of cells with the PG3 compound series, yielding IC50 values in the low micromolar range, similar to the TR-FRET assay (Fig. 4A, Suppl. Table S1). Measurement of the intracellular ATP content in compound-treated HEK cells showed that the inhibitors were not toxic at concentrations up to 10 μM (Suppl. Fig. S2). Higher concentrations led to loss of viability for some compounds, thus preventing us from generating complete dose-response curves. Co-transfection experiments of C6 and HTT furthermore confirmed that PG3d, one of the compounds with the lowest IC50 value for intracellular lamin cleavage, also inhibited intracellular C6-mediated cleavage of HTT1–1212 (Fig. 4B). Taken together, we thus concluded that PG3 compounds, and in particular PG3d, are true C6 inhibitors that preferentially inhibit the cleavage of protein over peptide substrates both with purified proteins and in live cells. They may thus interrupt the binding at exosites such as the region between aa151–427 in HTT (Fig. 1D).
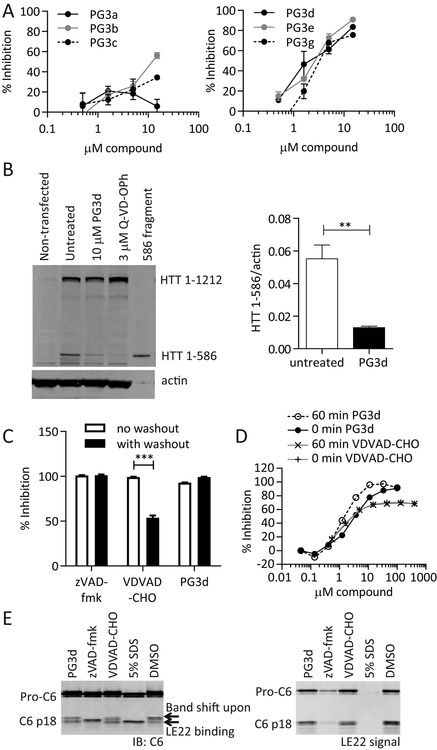
Figure 4: PG3 compounds inhibit intracellular C6 activity, PG3d is an irreversible, non-competitive C6 inhibitor.
(A) COS-7 cells were transfected with C6 and treated with different amounts of PG3 compounds in the media. The amount of intracellularly cleaved lamin A was analysed by ELISA in cell lysates (Ehrnhoefer, et al., 2011). (B) C6 was co-transfected with mHTT1–1212 into COS-7 cells, and cells were treated with PG3d or the cell-permeable pan-caspase inhibitor Q-VD-OPh in the media. Lysates were analysed by Western blotting with HTT antibody MAB2166, and the amount of cleaved HTT was quantified. (C) Recombinant C6 enzyme was incubated for 1h with 10 μM inhibitor, followed by two wash steps with ultrafiltration spin columns and a TR-FRET-based assay for HTT cleavage. While the reversible inhibitor VDVAD-CHO loses inhibition after washout, PG3d maintains its activity similar to the irreversible inhibitor zVAD-fmk. (D) 60 min pre-incubation of C6 with inhibitor leads to a shift in dose-response curves for PG3d, but not for the reversible inhibitor VDVAD-CHO in a TR-FRET based assay for HTT cleavage. (E) Recombinant C6 enzyme was incubated with 10 μM inhibitor or 5% SDS, followed by LE22 labeling and Western blotting. The irreversible active-site inhibitor zVAD-fmk and the denaturing SDS treatment prevent LE22 labeling, while the reversible inhibitor VDVAD-CHO is replaced by the irreversible binding of the LE22 probe. This leads to a signal in the LE22 channel as well as a band shift detected with the C6 antibody HD91. Although PG3d demonstrated irreversibility in (C), it does not prevent labelling of the active site with LE22, suggesting it is a non-competitive inhibitor.
IC50 values for (A) are listed in Suppl. Table S1. Graphed data are pooled results of three independent experiments graphed with S.E.M., Western blots are representative images. Statistical significance was determined by Student’s t-test in (B), and 2-way ANOVA (inhibitor p <0.0001, washout p <0.0001) with Tukey’s post-hoc correction in (C). **: p <0.01, ***: p <0.001.
We next decided to investigate the mode of inhibition using a washout experiment to distinguish between irreversible and reversible inhibition. C6 enzyme was pre-incubated with excess amounts of inhibitor expected to yield 100% inhibition, followed by extensive washing with buffer using spin filter columns. Inhibition of C6 activity was then analysed using the TR-FRET assay. Using the irreversible caspase inhibitor zVAD-fmk and the reversible inhibitor VDVAD-CHO as controls, we observed the expected reduction in inhibition after compound washout with VDVAD-CHO, but not zVAD-fmk (Fig. 4C). PG3d-mediated inhibition of C6 activity remained at 100% after the washout, suggesting that this inhibitor, similar to zVAD-fmk, is also irreversible (Fig. 4C). Irreversibly binders often show time-dependent inhibition, and we therefore checked whether different pre-incubation times of the compound with C6 would shift the dose-response curve for PG3d. As expected, the reversible peptide inhibitor VDVAD-CHO did not exhibit time-dependent behaviour, and the dose-response curves obtained with the TR-FRET assay did not differ for 0 min or 60 min pre-incubation time (Fig. 4D). For PG3d on the other hand we observed a shift towards a lower IC50 value when C6 was pre-incubated with the compound for 60 min before starting the assay, confirming the irreversible inhibitory mechanism (Fig. 4D).
Competitive irreversible inhibitors that target the C6 active site prevent the binding of the activity-based probe LE22 (Edgington, et al., 2012). To test for competitive binding, we thus pre-incubated C6 enzyme with PG3d, the irreversible peptide inhibitor zVAD-fmk or the reversible peptide inhibitor VDVAD-CHO, before exposing the enzyme to the LE22 label. C6 enzyme that was denatured by SDS was further included as a control, since the denaturing step should destroy the active site and also prevent LE22 binding. In samples treated with a reversible inhibitor, as well as in untreated controls, we observed a characteristic band shift of the p18 active fragment upon LE22 binding (Edgington, et al., 2012), as well as the fluorescent LE22 signal for both pro-C6 and p18 (Fig. 4E). While both denaturation by SDS and treatment with the irreversible inhibitor zVAD-fmk prevented labelling of C6 with LE22, PG3d treatment did not block accessibility of the active site, since the LE22 label was readily incorporated into both pro-C6 and the active p18 subunit in PG3d-treated samples (Fig. 4E). All data taken together therefore suggest that the compound PG3d is an irreversible allosteric inhibitor that binds outside the active site of C6. This is further supported by the minimal inhibitory effect of PG3d observed for the C6-mediated cleavage of small peptide substrates (VEID-AFC) in contrast to the strong inhibition of HTT and lamin A proteolysis (Suppl. Table S1).
Finally, we screened the PG compound series in a pre-clinical in-vitro pharmacology & ADME-Tox assay panel comprising the histone deacetylase (HDAC) 1, 3 and 8 enzymes, cytotoxicity (human A549 cell-line), cytochrome P450 inhibition (CYP2D6 and CYP3A4 isoforms) and cardiotoxicity (hERG). The PG compound series was found to inhibit each of these assays <25% at 10 μM (Suppl. Fig. S3), with the exception of PG3f.
PG3d prevents mHTT1–586-mediated activation of pro-C6
Since the PG3 series of inhibitors was identified with a screen using HTT as a substrate for C6, we next asked whether PG3d disrupts the interaction between the enzyme and its protein substrate. To this end, we co-transfected C6 and HTT and treated the cells with PG3d or the pan-caspase inhibitor Q-VD-OPh as a control. As seen before, HTT readily co-immunoprecipitates with C6 from untreated cells, and no significant difference in the amount of co-immunoprecipitated HTT was observed after Q-VD-OPh treatment (Fig. 5A). PG3d treatment, on the other hand, led to a 50% reduction in HTT co-immunoprecipitation (Fig. 5A), suggesting that although both compounds are C6 inhibitors, only PG3d prevents the binding of C6 to its protein substrate.
Figure 5: PG3d prevents the C6-HTT interaction and prevents mHTT1–586 mediated activation of C6.
(A) DDK-tagged C6wt was co-transfected into COS-7 cells with mHTT1–1212. Cells were treated with the pan-caspase inhibitor Q-VD-OPh (3 μM) or PG3d (10 μM) in the media as indicated. Lysates were immunoprecipitated with DDK antibody, and Western blots were probed for C6 with HD91 antibody and with antibody MAB2166 for co-immunoprecipitation of mHTT1–1212. Uncleaved mHTT1–1212 interacts with C6 irrespective of Q-VD-OPh treatment, while the incubation with PG3d significantly reduces the amount of co-immunoprecipitated mHTT. (B, C, D) C6wt, C6deltaPro and C6D23A were co-transfected into COS-7 cells with mHTT1–586 or the empty vector as control, and treated with PG3d (10 μM) in the media as indicated. Lysates were labelled with LE22 before analysis by Western blot using C6 antibody HD91. HTT was detected with antibody MAB2166. mHTT1–586 increases LE22 labelling of the C6 zymogen for C6wt, C6deltaPro and C6D23A, and this increase is prevented when cells are incubated in the presence of PG3d.
Graphed data are pooled results of three independent experiments graphed with S.E.M., Western blots are representative images. Statistical significance was determined by 1-way ANOVA (p = 0.0196), with Tukey’s post-hoc correction in (A), and by 2-way ANOVA with Tukey’s post-hoc correction in (B, C, D), wt C6: HTT p <0.0001, PG3d p <0.0001; deltaPro C6: HTT p = 0.0003, PG3d p = 0.0124; D23A C6: HTT p = 0.0003, PG3d p = 0.0104. *: p <0.05, **: p <0.01, ***: p <0.001.
As the interaction of C6 with mHTT1–586 specifically enhanced pro-C6 activity, we next asked whether this effect could be mitigated by PG3d. We therefore co-transfected cells with C6 and mHTT1–586 and exposed them to PG3d in the medium. As observed previously (Fig. 2), the presence of mHTT1–586 led to increased binding of the activity-based probe LE22 to pro-C6 (Fig. 5B). However, this effect was completely reversed by a treatment with PG3d in the culture medium (Fig. 5B). Furthermore, PG3d treatment also normalized the amount of LE22 labelling for deltaPro and D23A C6 (Fig. 5C and D), confirming that the compound prevented the mHTT1–586 mediated changes leading to the stabilization of the active conformation of the C6 zymogen.
PG3d fits into a previously described allosteric site in C6
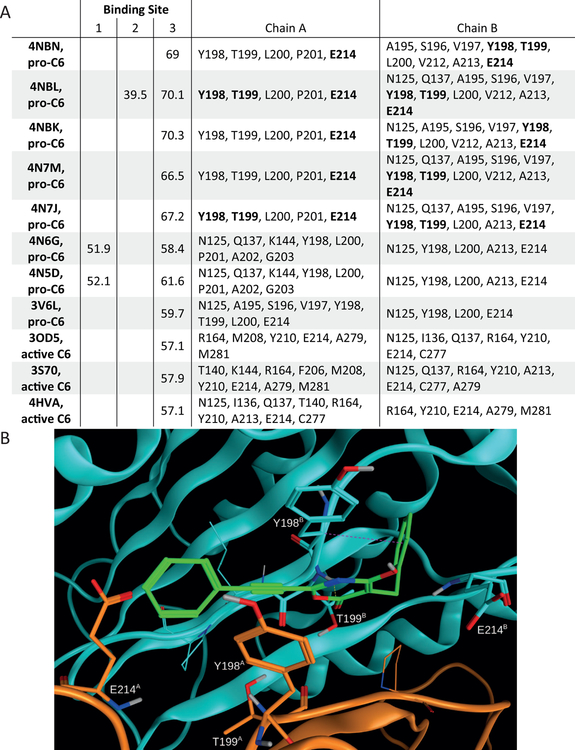
Having identified PG3d as an allosteric regulator of C6 activity, we next wanted to investigate potential binding sites and binding modes for the small molecule inhibitor. At first, we searched potential small molecule binding sites on the surface of 8 pro-C6 and 3 active C6 X-ray crystal structures using the Site Finder method. Subsequently, we docked PG3d into all identified pockets of relevant size using the molecular docking program GOLD in order to investigate the potential of PG3d to bind to these cavities. Based on this cross-docking approach, 3 sites emerged as potential PG3d binding pockets. Two of these sites showed poor docking scores for PG3d and were only present in 1 or 2 of the 8 pro-C6 structures (Fig. 6A). In contrast, the third site was predicted to bind PG3d in all 8 structures and the docking scores indicate a clear preference for PG3d for pro-C6 than for active C6 structures with the reorganized L2 loop. This observation is in agreement with our findings that PG3d mainly influences the formation of the active site in pro-C6.
Figure 6: GOLD docking studies suggest binding of PG3d to an allosteric pocket in the C6 zymogen.
(A) PG3d was docked into putative ligand binding sites (identified using Site Finder in the molecular modeling software suite Molecular Operating Environment) using GOLD. Docking scores for the putative small molecule binding site 3 are shown for all crystal structures analysed. For binding site 3 the residues interacting with docked PG3d are listed. (B) The top-ranked docking pose of protonated PG3d within the known allosteric pocket of CASP6 (PDB ID 4NBK, binding site 3) is predicted to strongly bind to C6 (score 70.3) and forms several non-covalent interactions with the protein (chain A in orange, chain B in cyan). Dashed lines indicate the presence of hydrogen bonds (yellow) and C-H-π interactions (magenta). Selected residues of C6 are shown in capped sticks representation. See also Suppl. Fig. S5.
The putative PG3d binding site 3 is located at the dimer interface between two pro-C6 molecules and contains the L2 loop region (Fig. 6B). Side chains of residues T199 (both chains) represent potential interaction partners for the irreversible binding of the compound. Based on the known reaction of oxazinone rings with hydroxyl group containing compounds (Lalaev, et al., 2010), a potential reaction mechanism for formation of a covalent bond between T199 and PG3d is shown in Suppl. Fig. S4. Docking of PG3D resulted in a compound binding mode with the reacting carbonyl group of the PG3d oxazinone ring positioned in all top-ranked docking poses in close proximity to the hydroxyl moiety of T199 (2.8–3.2 Å distance) on chain B.
Amino acids Y198, T199 and E214 of C6 were found to interact with PG3d in the 5 top-ranked docking poses for all C6 structures employed in the study (Fig. 6). These same residues were previously shown by X-ray crystallography to engage in the binding of small molecules designed for this site, and also shown to stabilize the pro-C6 protein (Murray, et al., 2014). We therefore decided to test whether binding of these allosteric compounds would counteract mHTT-mediated activation of pro-C6. Compound #12 was previously shown by co-crystallization to bind to pro-C6 at the dimer interface and contacts amino acids Y198, T199 and E214 (Murray, et al., 2014). We synthesized this compound and determined that it is not toxic in our COS7 cell model at concentrations up to 10 μM (Suppl. Fig. S5). When this dose was applied to COS7 cells co-transfected with C6 and mHTT1–586, the compound significantly reduced the activity of pro-C6 for both the wt, deltaPro and D23A enzymes (Fig. 7). In agreement with published data, compound #12 reduced the activity in single C6 transfections, but importantly also lowered excessive pro-C6 activity mediated by mHTT1–586 co-transfection (Fig. 7), confirming our hypothesis that allosteric modulation of the dimer interface prevents this interaction.
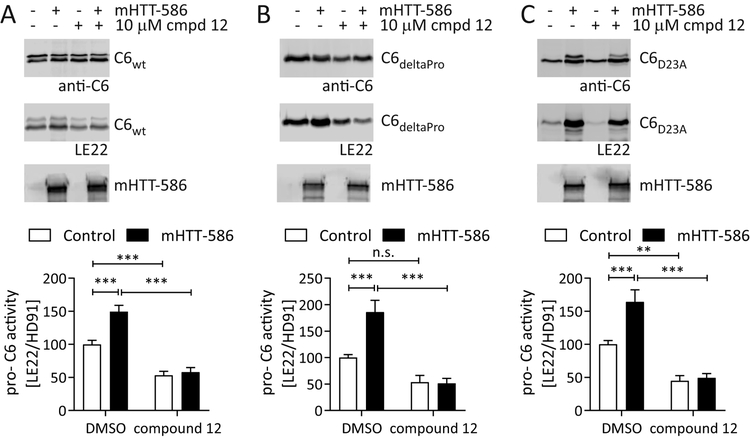
Figure 7: Inhibitor #12 prevents the activation of the C6 zymogen by mHTT1–586.
(A – C) C6wt, C6deltaPro and C6D23A were co-transfected into COS-7 cells with mHTT1–586 or the empty vector as control, and treated with inhibitor #12 (10 μM) in the media as indicated. Lysates were labelled with LE22 before analysis by Western blot using C6 antibody HD91. HTT was detected with antibody MAB2166. mHTT1–586 increases LE22 labelling of the C6 zymogen for C6wt, C6deltaPro and C6D23A, and this increase is prevented when cells are incubated in the presence of inhibitor #12. The compound furthermore reduces basal activity of the C6 zymogen in samples without mHTT1–586.
Graphed data are pooled results of three independent experiments graphed with S.E.M., Western blots are representative images. Statistical significance was determined by 2-way ANOVA, with Tukey’s post-hoc correction ), wt C6: HTT p = 0.0016, compound p <0.0001; deltaPro C6: HTT p = 0.0049, compound p <0.0001; D23A C6: HTT p = 0.0023, compound p <0.0001. *: p <0.05, **: p <0.01, ***: p <0.001, n.s.: not significant. See also Suppl. Fig. S6.
Discussion
HTT is a large protein that has multiple cellular functions, ranging from transcription and cell division to mitochondrial function as well as synaptic transmission, vesicle transport and autophagy among others (Saudou and Humbert, 2016). The polyglutamine expansion mutation in HD leads to disease phenotypes that are consistent with both a loss of normal HTT function as well a gain of toxic function (Cattaneo, et al., 2005). One example for a loss of function is the interaction of wt HTT with the palmitoyl transferase HIP14, which is required for HIP14 to be fully activated (Huang, et al., 2011). This interaction is disrupted in the presence of the HD mutation, leading to a loss of HIP14 activity in a mouse model of the disease (Huang, et al., 2011). For C6, our data are consistent with a gain-of-function model, since the C6 activation we observed was specific for a HTT1–586 fragment with an elongated polyglutamine stretch (Q128). In contrast, the same fragment with a polyglutamine length in the non-pathological range (Q15) did not activate C6, although it interacted with the enzyme as observed by co-immunoprecipitation.
We identified aa151–427 on HTT as the major interaction site with C6. This region contains multiple phosphorylation sites including S421, as well as a major site for palmitoylation (C214). HTT is subject to a large variety of post-translational modifications (PTMs), and these may regulate HTT’s subcellular localization and interaction partners, thus determining the function for each single HTT molecule within a cell (Ehrnhoefer, et al., 2011). Different PTMs may also interact and form a complex regulatory network. For HTT, it has been shown that phosphorylation events can fine-tune HTT ubiquitination, thus providing a reversible mechanism for the regulation of irreversible events such as proteasomal degradation (Kratter, et al., 2016; Thompson, et al., 2009). Similarly, phosphorylation at S421 and S434 can reduce HTT proteolysis at caspase cleavage sites D513, D552 and D586, again regulating an irreversible modification through a reversible PTM (Luo, et al., 2005; Warby, et al., 2009). HTT thus not only functions as a large scaffolding protein involved in multiple subcellular protein complexes (Saudou and Humbert, 2016; Shirasaki, et al., 2012), but also fine-tunes the activity of some of its enzymatic interaction partners such as HIP14 and C6, with reversible HTT PTMs adding a further layer of regulation. The HD mutation may disturb this function due to the reduced conformational flexibility observed in HTT fragments with polyglutamine stretches in the pathological range (Caron, et al., 2013) and alterations in the PTM regulatory network (Ehrnhoefer, et al., 2011). For C6, this could lead to an aberrant increase in the activity of the bound zymogen by promoting a conformation capable of substrate binding and autoactivation.
Here we show that the C6 zymogen can adopt a conformation in which the active site is accessible for binding to an activity-based probe. This is in agreement with published data demonstrating the presence of two conformational states for the C6 zymogen, which can be resolved by native gel electrophoresis (Edgington, et al., 2012). The fully active, cleaved C6 enzyme on the other hand migrates as a single peak on a native gel (Edgington, et al., 2012), consistent with our finding that no further activation of the C6 p18 subunit by mHTT1–586 is possible. X-ray crystallography data furthermore also demonstrate that C6 can exist in two conformations: a non-canonical and a canonical caspase conformation, which is stabilized upon substrate binding (Dagbay, et al., 2017; Vaidya, et al., 2011). Our data suggest that such a conformational change altering the accessibility of the C6 active site can be promoted by protein interactors, specifically the mHTT1–586 fragment. This model provides a simple explanation for the increase in C6 activity observed in HD, and corroborates the hypothesis of a feed-forward mechanism whereby C6 generates mHTT fragments that in turn lead to more C6 activation (Graham, et al., 2010; Graham, et al., 2011). Moreover, it is in agreement with the lack of C6 activation observed in the C6R mouse model, in which the generation of mHTT1–586 is prevented by a point mutation (Graham, et al., 2010).
Interestingly, we found that the activation of C6 by mHTT1–586 can be prevented using a small molecule inhibitor, PG3d. This molecule specifically inhibits the cleavage of protein, but not peptide substrates of C6, since we observed inhibition of lamin and HTT proteolysis but not of the processing of VEID-AFC (Suppl. Table S1). This phenomenon may be explained by an allosteric mechanism of action, as suggested by our molecular docking studies showing that PG3d fits into a previously reported allosteric site at the dimer interface of the C6 zymogen (Murray, et al., 2014). Allosteric sites have been reported for C3 and C7, as well, however these sites are present in the active enzymes (Hardy, et al., 2004; Scheer, et al., 2006). C6 is the only executioner caspase for which modulators targeting the zymogen have been described (Murray, et al., 2014; Stanger, et al., 2012), and our data demonstrate that one such compound prevents mHTT1–586-mediated activation of pro-C6 in a similar manner as PG3d. Nevertheless we cannot completely rule out the possibility that PG3d may bind pro-C6 at additional, yet to be discovered allosteric sites.
The active sites of different caspase family members are very similar and difficult to target specifically (Drag and Salvesen, 2010), which makes allosteric modulation a more tractable approach to achieving high specificity for C6 alone. We have identified a compound series with such a mode of action with low micromolar IC50 values for in vitro inhibition of C6, which can serve as tools to further probe the relationship between C6 and its interaction partners. Our data provides evidence that structurally distinct compounds that bind the allosteric site also inhibit activation of the C6 zymogen. This represents an important proof-of-concept that should spur further development and optimization of drug-like compounds that counteract the aberrant C6 activation caused by mHTT in HD. Most significant will be the identification of lead compounds with in vivo properties suitable to test this therapeutic hypothesis in animal models of Huntington disease.
Significance statement
The aberrant activation of caspase-6 (C6) is associated with a number of neurodegenerative diseases, including Huntington disease (HD). However, the cause for this activation is not well understood, since it occurs early in the disease process and is not associated with apoptotic cell death. Mutant huntingtin (mHTT), the protein causing HD, is subject to cleavage by C6 and proteolysis at the consensus site D586 has been linked to pathogenesis in a mouse model of the disease. Here we show that mHTT1–586 directly associates with C6 and leads to the stabilization of an active conformation in the pro-form of the enzyme. Such a mechanism could account for the slow disease progression and strengthens the hypothesis of a self-perpetuating cycle of C6 activation and mHTT proteolysis. We furthermore present a family of small molecule compounds capable of preventing the interaction between C6 and mHTT, and demonstrate that these inhibitors act through an allosteric mechanism that prevents the increase in pro-C6 activity, which would lead to a reduction in mHTT1–586 levels and disrupt the vicious cycle. Our work provides novel insights into the mechanism of C6 activation, demonstrating that auto-activation of C6 can be enhanced by interacting proteins through an allosteric mechanism. We furthermore suggest that disrupting the interaction between C6 and a mHTT fragment may be beneficial in HD.
STAR Methods:
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dagmar Ehrnhoefer (ehrnhoefer@bio.mx).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
COS-7 cells were obtained from ATCC. They are originally derived from male Cercopithecus aethiops kidney tissue and immortalized by expression of the SV40 large T antigen (RRID:CVCL_0224). Cells were cultured at 37°C in a humidified incubator under a 5% CO2 atmosphere. Dulbecco’s modified Eagle medium was used and supplemented with 10% fetal calf serum and 2 mM L-Glutamine.
METHOD DETAILS
DNA constructs and reagents
HTT constructs were in the pcDNA backbone, and contained either 15 or 128 glutamine repeats, for wt and mHTT, respectively (Sanders, et al., 2014; Warby, et al., 2008). The length of the construct is indicated in amino acids for each figure.
C6 constructs are in the pCMV6 backbone (Origene RC201349), with a C-terminal Myc-DDK tag. C6 mutants were generated by site-directed mutagenesis using the following primers:
D23A For: 5’ - aca ttt ctc ttt tat aga agg cag ctg ttt ctg tca tgt ttt ctt cc - 3’
D23A Rev: 5’ - g gaa gaa aac atg aca gaa aca gct gcc ttc tat aaa aga gaa atg t - 3’
D179A For: 5’- cca act tct ctg tct gat tag cta cta cat cca aag gaa tg -3’
D179 Rev: 5’- cat tcc ttt gga tgt agt agc taa tca gac aga gaa gtt gg -3’
D193A For: 5’- tgt aaa cgg agg ctg cag cca cct cag tta tgt tg -3’
D193A Rev: 5’- caa cat aac tga ggt ggc tgc agc ctc cgt tta ca -3’
deltaPro For: 5’- gcc gcg atc gcc atg gcc ttc tat aaa aga gaa atg -3’
deltaPro Rev: 5’-cat ttc tct ttt ata gaa ggc cat ggc gat cgc ggc-3’
LE22 was a kind gift of Matthew Bogyo and Laura Edgington, and was synthesized as described (Edgington, et al., 2012) using solid-phase peptide synthesis, and the resulting peptide was coupled to the Cy5 fluorophore. Purity and identity was assessed by LC-MS.
Recombinant C6 and C3 enzymes were from Enzo Life Sciences (BML-SE170–5000 and BML-SE169–5000). The exact active site concentration of each caspase used was determined by titrating the enzymes against the pan-caspase inhibitor zVAD-fmk (Pop, et al., 2008). zVAD-FMK, VDVAD-CHO and Q-VD-OPh were from Enzo Life Sciences.
Compound library for inhibitor screen
The 22644 compounds screened included commercially available synthetic compounds originating from ChemBioNet (16671), Analyticon Discovery (2329) and ComGenex (2437), the ENZO SCREEN-WELL® FDA approved drug library (640), natural products from GEOMAR Helmholtz Centre for Ocean Research Kiel, Wischhofstr. 1–3, 24148 Kiel, Germany (80) and a focussed synthetic compound library from the University of Hamburg, Institut für Pharmazie, Bundesstraße 45, 20146 Hamburg, Germany (487). Stock solutions of each sample were prepared in 100% DMSO (v/v) and screened at 20 μM where possible.
IVLD-aminoluciferin, VEID-AFC and TR-FRET based caspase activity assays, lamin cleavage ELISA
The IVLD peptide substrate coupled to aminoluciferin was custom produced by Promega (Madison, WI) and stored as a 20 mM stock in DMSO. For the activity assay, IVLD-aminoluciferin was diluted to 20 μM in luciferase detection reagent (Promega, Madison, WI, #V859A) and incubated at room temperature for 30 min before recombinant caspases were added to a final concentration of 1 μM active site titrated enzyme. After further 30 min incubation at room temperature, luminescence was read on a Victor Plate Reader (Perkin Elmer, Waltham, MA).
VEID-AFC and TR-FRET based C6 activity assays as well as the lamin cleavage ELISA were performed as described previously (Aharony, et al., 2015; Ehrnhoefer, et al., 2011).
C6 activity measurements by TR-FRET, high-throughput screening format
The C6 TR-FRET assay was established in low volume 384 well microtiter plate format. Briefly, test compounds were added to assay plates using the Labcyte ECHO 550 (Labcyte, Sunnyvale, CA), after which 2.5 μl Caspase-6 (2 nM) using a MultiDrop Combi in 10 mM HEPES, pH 7.4; 100 mM NaCl, 0.05% gelatin, 0.1% Chaps, 2 mM DTT was added, followed by a pre-Incubation at 37 °C for 15 minutes. Subsequently, 3 μl of substrate solution (containing HTT1–1212 and the TR-FRET detection reagents). The assay plates were incubated for a further 90 minutes at 37 °C for 90 minutes and read on an EnVision Plate Reader (Perkin Elmer, Waltham, MA). The negative controls were located in column 24 and contain DMSO alone; the positive controls were located in column 23 and contain DMSO and 10 μM Ac-VEID-CHO. A total of 291 hits were identified yielding >30% inhibition. Hit confirmation (screening at the original compound concentration) and hit profiling (generating 11 point dose-response curves) experiments were performed in triplicate. All raw data were normalised relative to the negative/positive controls and the % inhibition calculated within ActivityBase XE (ID Business Solutions Ltd, UK). For dose-response experiments, the IC50 values were calculated by fitting the duplicate data to the four-parameter logistic equation in GraphPad Prism (version 5.02, GraphPad Software, Inc., USA) or ActivityBase XE. To test for optical interference, TR-FRET assays were run to completion and compounds added immediately before the TR-FRET read. Optical interference was defined as inhibition >20% in this assay.
Determination of reversible inhibition
2 μM inhibitor stock solution in DMSO were mixed with 20 μl recombinant C6 (0.125 U/μl) and incubated for 1 h at room temperature. Samples were then loaded onto a centrifugal filter unit (10kDa cut-off, Millipore UFC201024PL) and washed three times with 100 μl TR-FRET buffer each (10 mM HEPES pH 7.4, 100 mM NaCl, 0.05% gelatin, 0.1% CHAPS, 2mM DTT). The washed C6-inhibitor mix was recovered through centrifugation of the inverted tube and used for a TR-FRET C6 activity assay in a final volume of 50 μl as described previously (Aharony, et al., 2015).
Pre-clinical in-vitro pharmacology & ADME-Tox profiling
The PG compound series were screened at 10 μM in triplicate in the pre-clinical in-vitro pharmacology & ADME-Tox assay panel comprising histone deacetylase (HDAC) 1, 3 and 8 enzymes, cytotoxicity (human A549 cell-line), cytochrome P450 inhibition (2D6 and 3A4 isoforms) and cardiotoxicity (hERG binding assay) as described previously (Halley, et al., 2011; Linciano, et al., 2017). Microtiter plate bulk liquid handling was performed using a Cell Explorer HTS platform (PerkinElmer, Waltham, MA), Multidrop (Thermo, Waltham, MA) or Tecan Fluent (Tecan Group Ltd, Männedorf, Switzerland).
Histone deacetylase assay
Inhibition of histone deacetylase (HDAC) enzymes was determined using the homogeneous, single addition, bioluminogenic HDAC-Glo™ I/II assay (Promega Corp.) to monitor the activity of HDAC1, HDAC3 and HDAC8. The kit contains a pro-luminogenic substrate with an acetylated lysine peptide sequence derived from Histone 4 conjugated to aminoluciferin. HDAC mediated deacetylation of the lysine residue facilitates luminogenic substrate susceptibility to specific proteolytic cleavage by the enzyme in the Developer Reagent. The aminoluciferin product of that cleavage acts as a substrate for luciferase and the amount of light produced in this reaction is proportional to HDAC enzyme activity. Human recombinant HDAC enzymes were purchased from BPS Bioscience (San Diego, USA) and standard inhibitor trichostatin A (Sigma-Aldrich) was dissolved to a yield stock solution in 100% v/v DMSO and stored at –20 °C. To each well of a 384 well microtiter plate, test compounds (100 nl of 1 mM solution in 100% v/v DMSO), positive controls (trichostatin A with final concentration of 1 μM and 1% v/v DMSO) yielding 100% inhibition and negative controls (100 nl of 100% v/v DMSO) yielding 0% inhibition were added using the Echo 550® Liquid Handler. Assays were initiated by addition of 10 μl/well of the HDAC-Glo™ I/II assay Reagent (prepared by rehydration of lyophilised HDAC-Glo™ I/II substrate in 10 ml HDAC-Glo™ I/II assay buffer and 10 μl Developer Reagent) and mixed briefly by orbital shaking (500 – 700 rpm). The luminescence was measured at steady-state signal:background which was achieved after 20 min incubation at room temperature. The raw data were normalised relative to the positive and negative controls yielding the % inhibition for each compound.
Cytotoxicity assay
Compound mediated cytotoxicity of A549 cells were determined using the of CellTiter-Glo® (CTG) reagent (Promega Corp.). A549 cells were grown on surface modified T175 cell culture flasks in DMEM with 10% FCS, streptomycin (100 μg/ml) and 100 U/ml penicillin G. At about 80% confluency, cells were washed, trypsinised, resuspended and counted in RPMI-1640 medium before seeding into white 384 well microtiter plates (20 μl) at 500 cells/well and incubated at 37 °C in the presence of 5% CO2 and compounds for 24 h. Prior to addition of cells, to each well of a 384 well microtiter plate were added test compounds (200 nl of 1 mM solution in 100% v/v DMSO), positive controls (paclitaxel with final concentration of 10 μM and 1% v/v DMSO) yielding 100% inhibition and negative controls (200 nl of 100% v/v DMSO) yielding 0% inhibition using the Echo 550® Liquid Handler. This was followed by addition of 20 μl/well of CellTiter-Glo® (CTG) reagent (Promega Corp.) to each well and the luminescence signal detected using an EnVision® Multilabel 2103 Reader after 10 min incubation in the dark. The raw data were normalised relative to the positive and negative controls yielding the % inhibition for each compound.
Cytochrome P450 assay
Detection of compound mediated cytochrome P450 inhibition was made using the luminescence based P450-Glo™ (Promega Corp.) assay system. These assays made use of microsomal preparations of cytochrome P450 (2D6 and 3A4) from baculovirus infected insect cells (Corning Inc. NY), cytochrome c reductase (cytochrome b5 for cytochrome P450 3A4), luminogenic cytochrome P450 substrates (luciferin-derivatives of cytochrome P450 specific substrates which produce (4S)-4,5-dihydro-2-(6-hydroxybenzothiazolyl)-4-thiazolecarboxylic acid (D-luciferin) after cleavage by cytochrome P450 (2D6, Luciferin-ME EGE; 3A4, Luciferin-IPA), lyophilised Luciferin Detection Reagent and reconstitution buffer. To each well of a 384 well microtiter plate, test compounds (100 nl of 1 mM solution in 100% v/v DMSO), positive controls (2D6, quinidine; 3A4, ketoconazole) yielding 100% inhibition and negative controls (100 nl of 100% v/v DMSO) yielding 0% inhibition were added using the Echo 550® Liquid Handler. This was followed by addition of 5 μl/well of cytochrome P450/substrate mixture and incubation for 30 min at 37°C after which the reaction was initiated by addition of 5 μl/well NADPH regeneration system. The reactions were stopped by addition of 10 μl/well of Luciferin Detection Reagent, followed by an additional 30 min incubation at 37 °C with the luminescence signal detected using an Infinite® M1000 PRO plate reader. The raw data were normalised relative to the positive and negative controls yielding the % inhibition for each compound.
hERG cardiotoxicity assay
The Invitrogen Predictor™ hERG Fluorescence Polarisation Assay was used to determine compound mediated cardiotoxicity. To each well of a 384 well microtiter plate, test compounds (100 nl of 1 mM solution in 100% v/v DMSO), positive controls (E-4031, a blocker of hERG-type potassium channels) yielding 100% inhibition and negative controls (100 nl of 100% v/v DMSO) yielding 0% inhibition were added using the Echo 550® Liquid Handler. This was followed by addition of 5 μL homogenised membrane solution (undiluted) and 5 μl of the tracer (1 nM final concentration in assay). The plates were incubated for 2 h at 25 °C in a humidity controlled incubator and the Fluorescence Polarisation was measured using an EnVision® Multilabel 2103 Reader. The raw data were normalised relative to the positive and negative controls yielding the % inhibition for each compound.
Cell culture, transfections, Western blotting
COS-7 cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum and 2 mM L-Glutamine. Cells were transfected using the Xtreme gene reagent (Roche) according to manufacturer’s instructions. For viability measurements, intracellular ATP content was determined using the Cell TiterGlo kit (Promega) according to manufacturer’s instructions. For Western blotting and immunoprecipitation, cells were harvested 24 h after transfection scraping and centrifugation. For experiments without LE22 labeling, cell pellets were lysed in 50 mM HEPES pH 7.4, 100 mM NaCl, 1% Igepal, 1 mM EDTA and 10% glycerol with 4.2 mM Pefabloc and ‘Complete’ protease inhibitor cocktail (Roche) on ice, and protein concentrations were determined in the cleared lysates after centrifugation using the DC protein assay kit (Bio Rad, USA). Equal amounts were separated on 12% Bis-Tris gels (Invitrogen, USA), protein was transferred to PVDF Immobilon-FL membranes by electroblotting and membranes were developed with primary antibodies in 5% bovine serum albumin/phosphate buffered saline. The following antibodies were used for immunoblotting: anti-HTT 2166 (1:1000) from Millipore (MAB2166), anti-C6 (HD91, 1:100) generated in-house (Wong, et al., 2015), anti-actin (1:10,000) from Millipore (MAB1501R). Fluorescently labelled secondary antibodies conjugated to either 700 or 800 IR dye (1:5000; Rockland, USA) and the LiCor Odyssey Infrared Imaging system were used for detection.
LE22 labeling
Cell pellets were suspended in 20 mM HEPES pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA and 1 mM EGTA with 4.2 mM Pefabloc and ‘Complete’ protease inhibitor cocktail (Roche), incubated on ice for 30min and subjected to sonification at 30% amplitude for 10 sec. Protein concentrations were determined in the cleared lysates after centrifugation using the DC protein assay kit (Bio Rad, USA). Equal amounts of protein were mixed with DTT (1 mM final concentration) and Cy5-labeled LE22 probe (1 μM final concentration) (Edgington, et al., 2012), and incubated at 37 °C for 2 h. Samples were separated on 12% Bis-Tris gels (Invitrogen, USA), protein was transferred to PVDF Immobilon-FL membranes by electroblotting and membranes were developed with primary antibodies in 5% bovine serum albumin/phosphate buffered saline. Fluorescently labelled secondary antibodies conjugated to 800 IR dye (1:5000; Rockland, USA) were used, the Cy5 signal was imaged in the red (700 nm) detection channel on the LiCor Odyssey Infrared Imaging system.
Immunoprecipitation
Cells transfected with DDK-tagged human C6 were lysed in SDP buffer as described above. 200 μg total protein were subjected to immunoprecipitation over night at 4 °C using 2.5 μl anti-DDK antibody (Origene TA50011) coupled to 20 μl magnetic protein G beads (Dynal, Invitrogen). Immunoprecipitates and cell lysates were subjected to SDS-PAGE and Western blot as described above.
Molecular Docking
High-resolution X-ray crystal structures of pro-C6 (PDB IDs 3V6L, 4N5D, 4N6G, 4NBK, 4NBL, 4NBN, 4N7J, 4N7M) and active C6 (3OD5, 3S70, 4HVA) were obtained from the Protein Data Bank. Potential allosteric ligand binding sites within the CASP6 X-ray crystal structures were identified using the Site Finder algorithm implemented in the molecular modeling software suite Molecular Operating Environment version 2016.0802 (MOE, Chemical Computing Group Inc., Montreal, Canada). All protein structures were prepared with MOE. Three-dimensional coordinates of PG3d in its protonated and deprotonated form were generated using MOE.
Molecular docking was carried out using GOLD version 5.4.1 (Cambridge Crystallographic Data Centre, Cambridge, UK) and the GoldScore scoring function. 50 docking runs were conducted and the early termination option was switched off.
Synthetic protocols for inhibitors
General procedure for 4-Hydroxy-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG)
Variant A.
The corresponding malonyl chloride [(chlorocarbonyl)ethylketene] (3.1 mmol) was added to a stirred solution of 3-arylprop-2-ynamide (3 mmol) in anhydrous Et2O (30 mL) at r.t. The mixture was cooled in the refrigerator for 12–14 h. During this time a solid deposited that was filtered off, washed with Et2O, and dried to deliver pure PG3, PG3a-e. For an additional portion of product the filtrate was evaporated under reduced pressure at r.t. and the remaining residue was recrystallized (MeCN).
Compound PG3f was purified by chromatography over a silica gel column (elution with ethyl acetate:petrol ether 20:80).
Variant B.
The corresponding malonyl dichloride [(chlorocarbonyl)ethylketene] (3.3 mmol) was added to a stirred solution of 3-arylprop-2-ynamide (3 mmol) in anhydrous dichlorethane (20 mL) at r.t. The reaction mixture was heated under reflux for 1–1.5 hours. During this procedure a yellow solid deposited. The solid separated upon cooling was filtered off and washed with diethyl ether and dried to get pure PG3d, e, g.
Inhibitor #12.
The compound was synthesized as described previously (Murray, et al., 2014).
General methods for chemical analysis
Melting points of the compounds were determined using a SMP1 apparatus (Stuart Scientific). Mass spectra were obtained with a Finnigan MAT 311A instrument (EI, 70 eV). Elemental analyses were performed with a Heraeus CHN-O-Rapid instrument. IR spectra were recorded using a Varian 800 FT-IR spectrometer. 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were recorded utilizing a Bruker AV 400 spectrometer using tetramethylsilane as an internal standard and DMSO-d6, CDCl3 as solvent. Full and unambiguous assignment for all NMR signals was achieved by combined application of standard NMR techniques such as DEPT 13C NMR, H,H-COSY, HSQC (1J (13C, 1H) = 145 Hz), HMBC (n>1J (13C, 1H) = 10 Hz).
Characterization of chemical and biomolecular materials
5-Butyl-4-hydroxy-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG3). (Petina, et al., 2013)
Yield: 65% (0.53 g) (Variant A); yellow crystals; mp: 157–159 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 12.71 (br s, 1H, OH), 7.70 (m, 2H, o-CH-Ar), 7.61 (m, 1H, p-CH Ar), 7.52 (m, 2H, m-CH-Ar), 2.28 (m, 2H, CH2), 1.42 (m, 2H, CH2), 1.29 (m, 2H, CH2), 0.88 (m, 3H, CH3) ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 164.9 (C6), 161.0 (C4), 147.2 (C2), 132.7 (o-CH-Ar), 131.5 (p-CH-Ar), 129.2 (m-CH-Ar), 118.4 (i-C-Ar), 97.3 (C5), 91.2 (C2′), 80.7 (C1′), 29.3 (C2′′), 22.3 (C1′′), 21.9 (C3′′), 13.7 (C4′′) ppm; IR (KBr): υ 2222 (C≡C), 1743 (C=O), 1628 (C=N) cm−1. MS: m/z (%) 269 (10) [M]+, 226 (18) [M–C3H7]+, 165 (14), 130 (10), 129 (100) [PhC≡CCO]+, 128 (20), no other peaks >10%; analysis (calcd., found for C16H15NO3): C (71.36, 70.97), H (5.61, 5.46), N (5.20, 5.14).
4-Hydroxy-5-methyl-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG3a). (Petina, et al., 2013)
Yield: 74% (0.32 g) (Variant A); yellowish crystals; mp: 184–186 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 12.74 (br s, 1H, OH), 7.71 (m, 2H, o-CH-Ar), 7.61 (m, 1H, p-CH-Ar), 7.52 (m, 2H, m-CH-Ar), 1.80 (s, 3H, CH3) ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 164.8 (C6), 161.3 (C4), 146.9 (C2), 132.7 (o-CH-Ar), 131.5 (p-CH-Ar), 129.2 (m-CH-Ar), 118.4 (i-C-Ar), 92.8 (C5), 91.2 (C2′), 80.7 (C1′), 8.1 (CH3) ppm; IR (KBr): υ 2215 (C≡C), 1747 (C=O), 1635 (C=N) cm−1. MS: m/z (%) 227 (14) [M]+, 171 (20), 130 (11), 129 (100) [PhC≡CCO]+, 128 (12), 83 (14), 77 (13) [Ph]+, 75 (29), 74 (14), 70 (12), 63 (11), no other peaks >10%; analysis (calcd., found for C13H9NO3): C (68.72, 68.72), H (3.99, 4.01), N (6.16, 6.26).
5-Ethyl-4-hydroxy-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG3b). (Petina, et al., 2013)
Yield: 70% (0.51 g) (Variant A); yellowish crystals; mp: 177–178 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 12.77 (br s, 1H, OH), 7.71 (m, 2H, o-CH-Ar), 7.61 (m, 1H, p-CH-Ar), 7.53 (m, 2H, m-CH-Ar), 2.29 (q, 3JHH = 7.42 Hz, 2H, CH2), 1.01 (t, 3JHH = 7.42 Hz, 3H, CH3) ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 164.6 (C6), 160.8 (C4), 147.2 (C2), 132.7 (o-CH-Ar), 131.7 (p-CH-Ar), 129.1 (m-CH-Ar), 118.4 (i-C-Ar), 98.6 (C5), 91.1 (C2′), 80.6 (C1′), 16.1 (CH2), 12.0 (CH3) ppm; IR (KBr): υ 2221 (C≡C), 1744 (C=O), 1633 (C=N) cm−1. MS: m/z (%) 241 (28) [M]+, 129 (100) [PhC≡CCO]+, 128 (19), 75 (15), 51 (10), no other peaks >10%; analysis (calcd., found for C14H11NO3): C (69.70, 69.42), H (4.60, 4.53), N (5.81, 5.84).
4-Hydroxy-5-isopropyl-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG3c). (Petina, et al., 2013)
Yield: 62% (0.47 g) (Variant A); colourless crystals; mp: 202–203 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 12.76 (br s, 1H, OH), 7.70 (m, 2H, o-CH-Ar), 7.61 (m, 1H, p-CH-Ar), 7.52 (m, 2H, m-CH-Ar), 3.01 (sept, 3JHH = 7.00 Hz, 1H, CH), 1.17 (d, 3JHH = 7.00 Hz, 6H, CH) ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 164.4 (C6), 160.0 (C4), 147.4 (C2), 132.8 (o-CH-Ar), 131.6 (p-CH-Ar), 129.2 (m-CH-Ar), 118.5 (i-C-Ar), 101.9 (C5), 91.2 (C2′), 80.6 (C1′), 23.7 (>CH), 19.5 (CH3) ppm; IR (KBr): υ 2226 (C≡C), 1744 (C=O), 1627 (C=N) cm−1. MS: m/z (%) 255 (35) [M]+, 240 (54) [M–CH3]+, 172 (11), 130 (10), 129 (100) [PhC≡CCO]+, 128 (20), 75 (11), 69 (16), no other peaks >10%; analysis (calcd., found for C15H13NO3): C (70.58, 70.38), H (5.13, 5.32), N (5.49, 5.43).
5-Benzyl-4-hydroxy-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG3d). (Petina, et al., 2013)
Yield: 61% (0.55 g) (Variant A); Yield: 65% (0.59 g) (Variant B); yellow crystals; mp: 179–181 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 13.03 (br s, 1H, OH), 7.71 (m, 2H, o-CH-Ar), 7.60 (m, 1H, p-CH-Ar), 7.52 (m, 2H, m-CH-Ar), 7.25 (m, 4H, H2′′, H3′′), 7.17 (m, 1H, H4′′), 3.61 (s, 2H, CH2) ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 165.4 (C6), 161.0 (C4), 147.7 (C2), 139.3 (C1′′), 132.7 (o-CH-Ar), 131.6 (p-CH-Ar), 129.2 (m-CH-Ar), 128.21 and 128.18 (C2′′ and C3′′), 126.0 (C4′′), 118.4 (i-C-Ar), 96.5 (C5), 91.5 (C2′), 80.7 (C1′), 28.3 (CH2) ppm; IR (KBr): υ 2221 (C≡C), 1746 (C=O), 1627 (C=N) cm−1. MS: m/z (%) 302 (55) [M]+, 176 (28), 158 (74), 131 (15), 130 (65), 129 (100) [PhC≡CCO]+, 128 (20), 103 (12), 102 (15), 91 (27) [PhCH2]+, 77 (22) [Ph]+, 75 (13), 51 (12) [C4H3]+, no other peaks >10%; analysis (calcd., found for C19H13NO3): C (75.24, 75.27), H (4.32, 4.32), N (4.62, 4.65).
4-Hydroxy-5-phenyl-2-(phenylethynyl)-6H-1,3-oxazin-6-one (PG3e). (Komarov, et al., 2005; Petina, et al., 2013)
Yield: 87% (0.76 g) (Variant A); yield: 89% (0.77 g) (Variant B); yellow crystals; mp: 187–189 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 13.18 (br s, 1H, OH), 7.74 (m, 2H, o-CH-Ar), 7.63 (m, 1H, p-CH-Ar), 7.54 (m, 2H, m-CH-Ar), 7.52 (m, 2H, H2′′), 7.38 (m, 2H, H3′′), 7.29 (m, 1H, H4′′). ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 164.8 (C6), 160.0 (C4), 147.9 (C2), 132.8 (o-CH-Ar), 131.7 (p-CHAr), 130.6 (C1′′), 130.0 (C2′′), 129.2 (m-CH-Ar), 127.6 (C3′′), 127.2 (C4′′), 118.3 (i-C-Ar), 97.3 (C5), 91.8 (C2′), 80.8 (C1′) ppm; IR (KBr): υ 2220 (C≡C), 1747 (C=O), 1620 (C=N) cm−1. MS: m/z (%) 289 (35) [M]+, 218 (14), 190 (12), 172 (12), 130 (10), 129 (100) [PhC≡CCO]+, 118 (18), 89 (16), 77 (11) [Ph]+, 75 (13), 51 (10) [C4H3]+, no other peaks >10%; analysis (calcd., found for C19H13NO3): C (74.73, 74.79), H (3.83, 4.01), N (4.84, 4.75).
2-((2-Chlorophenyl)ethynyl)-4-hydroxy-5-methyl-6H-1,3-oxazin-6-one (PG3f).
Yield: 41% (0.32 g) (Variant A); yellow crystals; mp: 175–178 °C; 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 11.72 (br s, 1H, OH), 7.83 (m, 1H, CH-6”-Ar), 7.81 (m, 1H, CH-3”-Ar), 7.67 (m, 1H, CH-4”-Ar), 7.61 (m, 1H, CH-5”-Ar), 1.79 (s, 3H, CH3). 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 165.1 (C6), 161.3 (C4), 146.7 (C2), 135.9 (CCl-Ar), 135.0 (CH-6”-Ar), 133.1 (CH-4”-Ar), 129.9 (CH-3”-Ar), 127.9 (CH-5”-Ar), 118.7 (i-C-Ar), 92.8 (C5), 87.0 (C2′), 85.0 (C1′), 8.3 (CH3) ppm; IR (KBr): υ 2221 (C≡C), 1687 (C=O), 1628 (C=N) cm−1. MS: m/z (%) 263/261 (19/55) [M]+, 226 (13), 206 (13), 165/163 (33/100) [ClC6H4C≡CCO]+, 99 (24), 83 (20), 70 (15) no other peaks >10%; analysis (calcd., found for C13H8ClNO3): C (59.67, failed), H (3.08, failed), N (5.35, failed).
2-((4-Chlorophenyl)ethynyl)-4-hydroxy-5-phenyl-6H-1,3-oxazin-6-one (PG3g).
Yield: 85% (0.83 g) (Variant B); yellow crystals; mp: 185–187 °C (dec); 1H NMR (400 MHz, DMSO-d6, 25 °C): δ 13.19 (br. s, 1H, OH), 7.77 (m, 2H, o-CH-Ar), 7.62 (m, 1H, m-CH-Ar), 7.52 (m, 2H, H-2”), 7.37 (m, 2H, H-3”), 7.29 (m, 1H, H-4”) ppm. 13C NMR (100 MHz, DMSO-d6, 25 °C): δ 164.7 (C-6), 160.0 (C-4), 147.6 (C-2), 136.6 (p-CCl-Ar), 134.5 (o-CH-Ar), 130.5 (C-1”), 130.0 (C-2”), 129.4 (m-CH-Ar), 127.5 (C-3”), 127.2 (C-4”), 117.2 (i-C-Ar), 97.4 (C-5), 90.3 (C-2’), 81.5 (C-1’) ppm; IR (KBr): υ 2220 (C≡C), 1768 (C=O), 1621 (C=N) cm−1. MS: m/z (%) 325/324/323 (15/9/47) [M]+, 252 (22), 225 (10), 224 (14), 206 (16), 165/163 (32/100) [ClC6H4C≡CCO]+, 118 (28), 99 (18), 90 (17), 89 (21), 77 (9) [Ph]+, 70 (12), 63 (10) [C4H3]+, no other peaks >10%; analysis (calcd., found for C18H10ClNO3): C (66.78, 66.89), H (3.11, 3.10), N (4.33, 4.39).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was assessed using Student’s t-test for comparison of two groups, one-way ANOVA with post-hoc Tukey’s correction for the comparison of one variable between more than two groups, and two-way ANOVA with post-hoc Bonferroni correction for the comparison of two variables between groups. Variances between groups were similar. All analyses were performed using the GraphPad Prism 5.01 software package. Details on number of independent replicates (n), tests used and p-values can be found in the figure legends, all figures are graphed as mean ± SEM.
DATA AND CODE AVAILABILITY
The published article includes all data generated or analyzed during this study.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HTT-2166 | Millipore | Cat. no. MAB2166, RRID:AB_2123255 |
| Caspase-6 (HD91) | Wong, et al, 2015 | N/A |
| actin | Millipore | Cat. no. MAB1501R, RRID:AB_2223041 |
| DDK tag | Origene | Cat. no. TA50011, RRID:AB_2622345 |
| HTT D586 neo-epitope | Warby, et al, 2008 | N/A |
| N-terminal HTT antibody BKP1 | Warby, et al, 2008 | N/A |
| cleaved lamin A | Cell Signaling Technology | Cat. no. 2036, RRID:AB_2136275 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| LE22, Cy5-labeled | Edgington, et al, 2012 | N/A |
| recombinant caspase-6 enzyme | Enzo Life Sciences | Cat. no. BML-SE170 |
| recombinant caspase-3 enzyme | Enzo Life Sciences | Cat. no. BML-SE169 |
| VDVAD-CHO | Enzo Life Sciences | Cat. no. ALX-260–058 |
| Q-VD-OPh | Enzo Life Sciences | Cat. no. ALX-260–159 |
| zVAD-fmk | Enzo Life Sciences | Cat. no. ALX-260–138 |
| PG, a-g | this manuscript | N/A |
| caspase-6 inhibitor #12 | Murray, et al., 2014 | N/A |
| Critical Commercial Assays | ||
| Cell TiterGlo viability assay | Promega | Cat. no. G7570 |
| Deposited Data | ||
| pro-C6 crystal structure | www.rcsb.org, Murray, et al., 2014, Cao, et al., 2012 | accession nos. 4NBN, 4NBL, 4NBK, 4N7M, 4N7J, 4N6G, 4N5D, 3V6L |
| active C6 crystal structure | www.rcsb.org, Wang, et al., 2008, Liu, et al., 2011, Heise, et al., 2012 | accession nos. 3OD5, 3S70, 4HVA |
| Experimental Models: Cell Lines | ||
| COS-7 cells | ATTC | RRID:CVCL_0224 |
| Oligonucleotides | ||
| D23A For: 5’ - aca ttt ctc ttt tat aga agg cag ctg ttt ctg tca tgt ttt ctt cc - 3’ | this manuscript | N/A |
| D23A Rev: 5’ - g gaa gaa aac atg aca gaa aca gct gcc ttc tat aaa aga gaa atg t - 3’ | this manuscript | N/A |
| D179A For: 5’- cca act tct ctg tct gat tag cta cta cat cca aag gaa tg -3’ | this manuscript | N/A |
| D179 Rev: 5’- cat tcc ttt gga tgt agt agc taa tca gac aga gaa gtt gg -3’ | this manuscript | N/A |
| D193A For: 5’- tgt aaa cgg agg ctg cag cca cct cag tta tgt tg -3’ | this manuscript | N/A |
| D193A Rev: 5’- caa cat aac tga ggt ggc tgc agc ctc cgt tta ca -3’ | this manuscript | N/A |
| deltaPro For: 5’- gcc gcg atc gcc atg gcc ttc tat aaa aga gaa atg -3’ | this manuscript | N/A |
| deltaPro Rev: 5’-cat ttc tct ttt ata gaa ggc cat ggc gat cgc ggc-3’ | this manuscript | N/A |
| Recombinant DNA | ||
| pCIneo - HTT Q15 - 1-3144 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q128 - 1-3144 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q15 - 1-586 | Warby, et al., 2008 | N/A |
| pCIneo - HTT Q128 - 1 -586 | Warby, et al., 2008 | N/A |
| pCIneo - HTT Q15 - 1 -548 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q128 - 1 -548 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q15 - 1-427 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q128 - 1 -427 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q15 - 1-151 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q128 - 1-151 | Sanders, et al., 2014 | N/A |
| pCIneo - HTT Q15 - 1-1212 - 4c | Warby, et al., 2008 | N/A |
| pCMV6 - wt C6 | Origene | Cat. no. RC201349 |
| pCMV6 - C6 C163S | this study | N/A |
| pCMV6 - C6 D179A | this study | N/A |
| pCMV6 - C6 D193A | this study | N/A |
| pCMV6 - C6 D23A | this study | N/A |
| pCMV6 - C6 deltaPro | this study | N/A |
| Software and Algorithms | ||
| Site Finder algorithm in Molecular Operating Environment, version 2016.0802 | MOE, Chemical Computing Group Inc. | N/A |
| GOLD version 5.4.1, GoldScore | Cambridge Crystallographic Data Centre | N/A |
| GraphPad Prism 5.01 | GraphPad Software, San Diego, CA | N/A |
Highlights:
Caspase-6 binds its substrate huntingtin at an exosite, increasing specificity
Cleaved mutant huntingtin stabilizes an active conformation of pro-caspase-6
Increased activity of pro-caspase-6 is prevented by allosteric inhibitors
Acknowledgements:
M.R.H. was supported by the Canadian Institutes of Health Research (CIHR 20R90174), the Baden-Württemberg-Stiftung through the program “Internationale Spitzenforschung” and a sponsored research agreement with Teva Pharmaceuticals. A.R.R. acknowledges funding from the NIH (AI135489), D.E.E. and N.H.S held postdoctoral fellowships from the CIHR. D.E.E. was also supported by a Schrödinger fellowship from the Austrian Science funds (FWF). S.L. was supported by a doctoral scholarship from CIHR. N.S.C. was supported by postdoctoral fellowships from CIHR and the James Family. The authors thank Matthew Bogyo and Laura Edgington for their gift of LE22, and Andrea LeBlanc for the gift of C6 mutants. We are furthermore indebted to Robert Young, Richard Dean and David Percival for providing their help and expertise in triaging hit compounds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
M.R.H., D.E.E., O.P., M.K. and D.G. are inventors on patent WO2016020732, “Modulators of caspase-6”, which encompasses PG3 family compounds. M.R.H. was an employee of Teva Pharmaceuticals, Inc during the study; Teva did not play a role in the design or interpretation of this study. All other authors declare no competing financial interests.
References
- Aharony I, Ehrnhoefer DE, Shruster A, Qiu X, Franciosi S, Hayden MR, and Offen D (2015). A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet 24, 2604–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Wang XJ, Liu CW, Liu DF, Li LF, Gao YQ, and Su XD (2012). Inhibitory mechanism of caspase-6 phosphorylation revealed by crystal structures, molecular dynamics simulations, and biochemical assays. J Biol Chem 287, 15371–15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron NS, Desmond CR, Xia J, and Truant R (2013). Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc Natl Acad Sci U S A 110, 14610–14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo E, Zuccato C, and Tartari M (2005). Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci 6, 919–930. [DOI] [PubMed] [Google Scholar]
- Dagbay KB, Bolik-Coulon N, Savinov SN, and Hardy JA (2017). Caspase-6 Undergoes a Distinct Helix-Strand Interconversion upon Substrate Binding. J Biol Chem 292, 4885–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagbay KB, and Hardy JA (2017). Multiple proteolytic events in caspase-6 self-activation impact conformations of discrete structural regions. Proc Natl Acad Sci U S A 114, E7977–E7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M, and Salvesen GS (2010). Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9, 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M, et al. (1993). Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nature genetics 4, 387–392. [DOI] [PubMed] [Google Scholar]
- Edgington LE, van Raam BJ, Verdoes M, Wierschem C, Salvesen GS, and Bogyo M (2012). An optimized activity-based probe for the study of caspase-6 activation. Chem Biol 19, 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Martin DDO, Schmidt ME, Qiu X, Ladha S, Caron NS, Skotte NH, Nguyen YTN, Vaid K, Southwell AL, et al. (2018). Preventing mutant huntingtin proteolysis and intermittent fasting promote autophagy in models of Huntington disease. Acta neuropathologica communications 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Skotte NH, Ladha S, Nguyen YT, Qiu X, Deng Y, Huynh KT, Engemann S, Nielsen SM, Becanovic K, et al. (2014). p53 increases caspase-6 expression and activation in muscle tissue expressing mutant huntingtin. Hum Mol Genet 23, 717–729. [DOI] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Skotte NH, Savill J, Nguyen YT, Ladha S, Cao LP, Dullaghan E, and Hayden MR (2011). A quantitative method for the specific assessment of caspase-6 activity in cell culture. PLoS One 6, e27680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrnhoefer DE, Sutton L, and Hayden MR (2011). Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry 17, 475–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, and Salvesen GS (2004). The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J 384, 201–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA, Metzler M, Bissada N, Wang L, Faull RL, et al. (2010). Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci 30, 15019–15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. (2006). Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125, 1179–1191. [DOI] [PubMed] [Google Scholar]
- Graham RK, Ehrnhoefer DE, and Hayden MR (2011). Caspase-6 and neurodegeneration. Trends in neurosciences 34, 646–656. [DOI] [PubMed] [Google Scholar]
- Halley F, Reinshagen J, Ellinger B, Wolf M, Niles AL, Evans NJ, Kirkland TA, Wagner JM, Jung M, Gribbon P, et al. (2011). A bioluminogenic HDAC activity assay: validation and screening. J Biomol Screen 16, 1227–1235. [DOI] [PubMed] [Google Scholar]
- Hardy JA, Lam J, Nguyen JT, O’Brien T, and Wells JA (2004). Discovery of an allosteric site in the caspases. Proc Natl Acad Sci U S A 101, 12461–12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise CE, Murray J, Augustyn KE, Bravo B, Chugha P, Cohen F, Giannetti AM, Gibbons P, Hannoush RN, Hearn BR, Jaishankar P, Ly CQ, Shah K, Stanger K, Steffek M, Tang Y, Zhao X, Lewcock JW, Renslo AR, Flygare J, Arkin MR (2012). Mechanistic and structural understanding of uncompetitive inhibitors of caspase-6. Plos One 7, e50864–e50864, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Sanders SS, Kang R, Carroll JB, Sutton L, Wan J, Singaraja R, Young FB, Liu L, El-Husseini A, et al. (2011). Wild-type HTT modulates the enzymatic activity of the neuronal palmitoyl transferase HIP14. Hum Mol Genet 20, 3356–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaiman G, Champagne N, and LeBlanc AC (2009). Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochimica et biophysica acta 1793, 592–601. [DOI] [PubMed] [Google Scholar]
- Komarov AV, Yakovlev IP, Zakhs AV, and Prep’yalov AV (2005). Reaction of Phenylmalonyl Dichloride with 3-Phenylpropynamide and Transformations of the Product by the Action of Some Nucleophiles. Russ J Gen Chem 75, 770. [Google Scholar]
- Kratter IH, Zahed H, Lau A, Tsvetkov AS, Daub AC, Weiberth KF, Gu X, Saudou F, Humbert S, Yang XW, et al. (2016). Serine 421 regulates mutant huntingtin toxicity and clearance in mice. The Journal of clinical investigation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaev BY, Yakovlev P, Kuz’mich NN, Ksenofontova GV, and Zakhs VE (2010). Reactions of 2-Alkylsulfanyl- and 2-Alkoxy-4-hydroxy-6H-1,3-oxazin-6-ones with O-Nucleophiles. Russian Journal of General Chemistry 80, 2043–2047. [Google Scholar]
- LeBlanc AC (2013). Caspase-6 as a novel early target in the treatment of Alzheimer’s disease. The European journal of neuroscience 37, 2005–2018. [DOI] [PubMed] [Google Scholar]
- Linciano P, Dawson A, Pohner I, Costa DM, Sa MS, Cordeiro-da-Silva A, Luciani R, Gul S, Witt G, Ellinger B, et al. (2017). Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery. ACS Omega 2, 5666–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang H, Wang X-J, Li L-F, Su X-D (2011). Get phases from arsenic anomalous scattering: de novo SAD phasing of two protein structures crystallized in cacodylate buffer. Plos One 6, e24227–e24227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Vacher C, Davies JE, and Rubinsztein DC (2005). Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J Cell Biol 169, 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M, Gan L, Mazarei G, Graham RK, Liu L, Bissada N, Lu G, Leavitt BR, and Hayden MR (2010). Phosphorylation of huntingtin at Ser421 in YAC128 neurons is associated with protection of YAC128 neurons from NMDA-mediated excitotoxicity and is modulated by PP1 and PP2A. J Neurosci 30, 14318–14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, Ko RW, Vasuta OC, Graham RK, Hayden MR, et al. (2010). Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron 65, 178–190. [DOI] [PubMed] [Google Scholar]
- Murray J, Giannetti AM, Steffek M, Gibbons P, Hearn BR, Cohen F, Tam C, Pozniak C, Bravo B, Lewcock J, et al. (2014). Tailoring small molecules for an allosteric site on procaspase-6. ChemMedChem 9, 73–77, 72. [DOI] [PubMed] [Google Scholar]
- Petina OA, Yakovlev IP, and Geffken D (2013). Preparation of arylpropynamides and their reaction with malonyl acid derivatives. Synthesis (Germany) 45, 803–809. [Google Scholar]
- Pop C, Salvesen GS, and Scott FL (2008). Caspase assays: identifying caspase activity and substrates in vitro and in vivo. Methods in enzymology 446, 351–367. [DOI] [PubMed] [Google Scholar]
- Pouladi MA, Graham RK, Karasinska JM, Xie Y, Santos RD, Petersen A, and Hayden MR (2009). Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain 132, 919–932. [DOI] [PubMed] [Google Scholar]
- Sanders SS, Mui KK, Sutton LM, and Hayden MR (2014). Identification of binding sites in Huntingtin for the Huntingtin Interacting Proteins HIP14 and HIP14L. PLoS One 9, e90669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, and Humbert S (2016). The Biology of Huntingtin. Neuron 89, 910–926. [DOI] [PubMed] [Google Scholar]
- Scheer JM, Romanowski MJ, and Wells JA (2006). A common allosteric site and mechanism in caspases. Proc Natl Acad Sci U S A 103, 7595–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki DI, Greiner ER, Al-Ramahi I, Gray M, Boontheung P, Geschwind DH, Botas J, Coppola G, Horvath S, Loo JA, et al. (2012). Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron 75, 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotte NH, Sanders SS, Singaraja RR, Ehrnhoefer DE, Vaid K, Qiu X, Kannan S, Verma C, and Hayden MR (2017). Palmitoylation of caspase-6 by HIP14 regulates its activation. Cell Death Differ 24, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger K, Steffek M, Zhou L, Pozniak CD, Quan C, Franke Y, Tom J, Tam C, Krylova I, Elliott JM, et al. (2012). Allosteric peptides bind a caspase zymogen and mediate caspase tetramerization. Nature chemical biology 8, 655–660. [DOI] [PubMed] [Google Scholar]
- Stine OC, Pleasant N, Franz ML, Abbott MH, Folstein SE, and Ross CA (1993). Correlation between the onset age of Huntington’s disease and length of the trinucleotide repeat in IT-15. Hum Mol Genet 2, 1547–1549. [DOI] [PubMed] [Google Scholar]
- The Huntington’s disease Collaborative Research Group. (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72, 971–983. [DOI] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O’Rourke JG, Khashwji H, et al. (2009). IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol 187, 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsain N, and Barker PA (2015). New Views on the Misconstrued: Executioner Caspases and Their Diverse Non-apoptotic Roles. Neuron 88, 461–474. [DOI] [PubMed] [Google Scholar]
- Vaidya S, Velazquez-Delgado EM, Abbruzzese G, and Hardy JA (2011). Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J Mol Biol 406, 75–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Delgado EM, and Hardy JA (2012). Phosphorylation regulates assembly of the caspase-6 substrate-binding groove. Structure 20, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Cao Q, Liu X, Wang K-T, Mi W, Zhang Y, Li L-F, Leblanc AC, Su X-D (2010). Crystal strucutre of active caspase-6 bound with Ac-VEID-CHO. EMBO Rep 11, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, Overall CM, and Hayden MR (2008). Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet 17, 2390–2404. [DOI] [PubMed] [Google Scholar]
- Warby SC, Doty CN, Graham RK, Shively J, Singaraja RR, and Hayden MR (2009). Phosphorylation of huntingtin reduces the accumulation of its nuclear fragments. Molecular and cellular neurosciences 40, 121–127. [DOI] [PubMed] [Google Scholar]
- Wong BK, Ehrnhoefer DE, Graham RK, Martin DD, Ladha S, Uribe V, Stanek LM, Franciosi S, Qiu X, Deng Y, et al. (2015). Partial rescue of some features of Huntington Disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol Dis 76, 24–36. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, and Oldenburg KR (1999). A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4, 67–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study.