Abstract
CD8+ T cell exhaustion is a state of dysfunction acquired in chronic viral infection and cancer, characterized by the formation of Slamf6+ progenitor exhausted and Tim-3+ terminally exhausted subpopulations through unknown mechanisms. Here we establish the phosphatase PTPN2 as a novel regulator of the differentiation of the terminally exhausted subpopulation that functions by attenuating type 1 interferon signaling. Deletion of Ptpn2 in CD8+ T cells increased the generation, proliferative capacity, and cytotoxicity of Tim-3+ cells without altering Slamf6+ numbers during LCMV Clone 13 infection. Likewise, Ptpn2-deletion in CD8+ T cells enhanced Tim-3+ anti-tumor responses and improved tumor control. Deletion of Ptpn2 throughout the immune system resulted in MC38 tumor clearance and improved PD-1 checkpoint blockade responses to B16 tumors. Our results indicate that increasing the number of cytotoxic Tim-3+ CD8+ T cells can promote effective anti-tumor immunity and implicate PTPN2 in immune cells as an attractive cancer immunotherapy target.
Introduction
T cell exhaustion is a state of dysfunction observed in CD8+ T cells during chronic viral infection and cancer1, 2, 3, 4. During chronic viral infections, exhausted CD8+ T cells progressively lose functional capabilities such as cytokine production, cytotoxicity, and proliferative capacity5, 6. During the progression to exhaustion, T cells have reduced functional capacity compared to effector CD8+ T cells7, yet still have the potential to form memory cells if removed from chronic antigen stimulation8. The program of exhaustion9, 10, 11 is initiated during chronic antigen stimulation12, 13 and likely evolved as a mechanism to prevent excessive immunopathology during chronic antigenic insults14.
There are two subpopulations of exhausted CD8+ T cells, each with distinct functional properties. The progenitor population of exhausted cells, defined as PD-1int 15, CXCR5+ 16, 17, or Slamf6+ 4, possesses enhanced proliferative capacity, polyfunctional cytokine production, and serves as a reservoir of cells for the terminally exhausted population. The terminally exhausted population is defined as PD-1hi 15 or Tim-3+ 16, 17 and is cytotoxic, albeit having reduced proliferative capacity, longevity, and polyfunctional cytokine production. Eomes, Id2, Runx3, T cell receptor (TCR) stimulation, IL-2, IL-21, IL-12, and type 1 interferon (IFN-I) promote the formation of the terminally exhausted subpopulation15, 16, 18, 19, 20, 21, while Tbet, Tcf7, and Bcl6 enhance the formation of the progenitor exhausted subpopulation15, 17, 18. During responses to PD-1 immune checkpoint blockade (ICB), the progenitor population expands and gives rise to the terminally exhausted subset17. These subsets are found in murine and human tumors4, 18, 22, 23, 24, 25, 26, 27, and an increased ratio of progenitor to terminally exhausted cells is correlated with responsiveness to ICB in melanoma patients24. Given their distinct functions and responses to anti-PD-1, there is an urgent need to identify therapeutic targets that regulate the balance and functionality of exhausted subpopulations in cancer.
Here we identify PTPN2 as a regulator of the generation of the Tim-3+ subpopulation. Ptpn2 deletion is associated with enhanced IFN-α cytokine signaling and increased the number of Tim-3+ cytotoxic CD8+ T cells during LCMV Clone 13 infection without altering Slamf6+ cell numbers. Furthermore, Ptpn2 deletion promotes formation of the Tim-3+ subset and increases Tim-3+ CD8+ T cell responses in MC38 and B16 cancer models. This leads to complete clearance of MC38 tumors and improved PD-1 ICB responses to B16 tumors. Our data demonstrate that increasing the number of Tim-3+ cytotoxic CD8+ T cells can promote effective tumor immunity and provides rationale for PTPN2 as a cancer immunotherapy target.
Results
Loss of Ptpn2 promotes the early proliferation of CD8+ T cells during LCMV Clone 13 infection
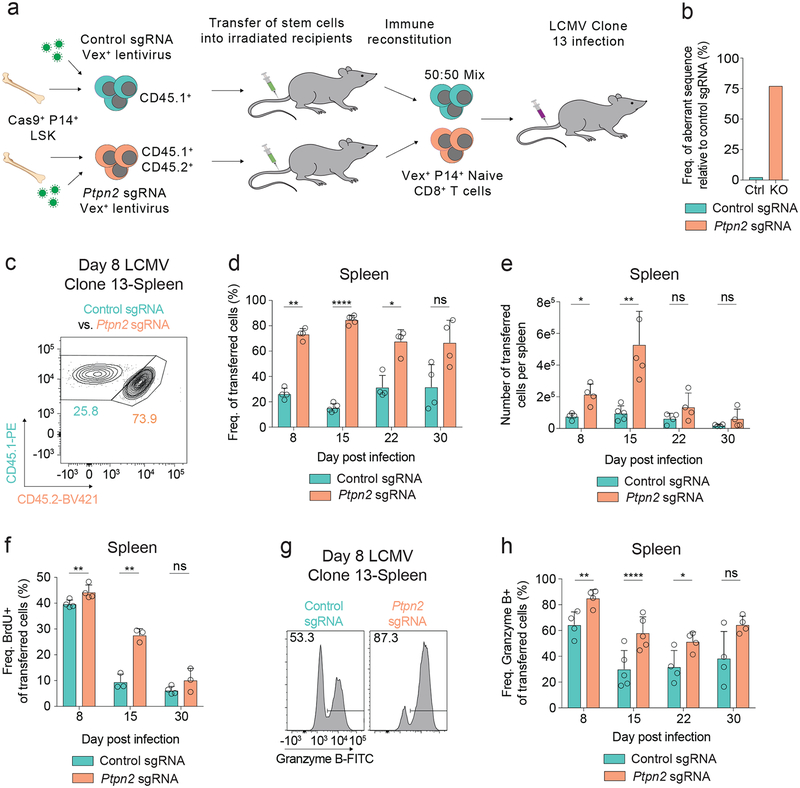
We recently conducted a pooled in vivo loss-of-function screen, and identified PTPN2 as a candidate regulator of CD8+ T cell responses28. To examine the role of PTPN2 in LCMV Clone 13 infection, we created bone marrow chimeras using CHIME (CHimeric IMmune Editing) – a chimera-based CRISPR-Cas9 delivery method (Figure 1a)28 to delete Ptpn2 in hematopoietic cells from P14 TCR transgenic mice (specific for the LCMV CD8 epitope GP33–41). We confirmed efficient deletion (~80%) of Ptpn2 in naive P14 CD8+ T cells using the TIDE assay29 (Figure 1b). To evaluate cell-intrinsic functions of PTPN2 in CD8+ T cells, we co-transferred congenically-marked naive P14 TCR transgenic Ptpn2 sgRNA-containing (Ptpn2-deleted) and control sgRNA-containing (control) CD8+ T cells to wild-type recipient mice and subsequently infected them with LCMV Clone 13. Given that Ptpn2 deficiency leads to alterations in thymocyte maturation and aberrant T cell activation at homeostasis30, we transferred only naive CD44– CD62L+ cells prior to LCMV infection (Supplementary Figure 1a). These cells did not express effector-related molecules, such as Granzyme B (Supplementary Figure 1b), or markers of cell proliferation, such as Ki-67, or BrdU incorporation (Supplementary Figures 1c, 1d). Following LCMV infection, there was a significant increase in the Ptpn2-deleted cells compared with control cells at days 8 and 15 post infection, but not day 30 post infection (Figures 1c–1e and Supplementary Figure 1e). The increased cell numbers at these time points were due, in part, to increased proliferation of Ptpn2-deleted cells (Figure 1f). Deletion of Ptpn2 also increased the percentage of Granzyme B+ cells at early time points (Figures 1g, 1h) but did not affect IFN-γ+ TNF+ production (Supplementary Figure 1f). Thus, Ptpn2 deletion provides CD8+ T cells with a transient advantage early during LCMV Clone 13 infection but does not prevent contraction at later time points.
Figure 1: Loss of Ptpn2 promotes the early proliferation of CD8+ T cells during LCMV Clone 13 infection.
(a) Schematic of co-transfer experiment during LCMV Clone 13 infections. Chimeric mice were generated using the CHIME system. (b) TIDE assay on naïve CD8+ T cells for a control and Ptpn2-targeting sgRNA. Representative of four independent experiments, n = 1 mouse. (c) Representative flow cytometry plot of co-transferred control or Ptpn2-deleted P14 T cells in the spleen 8 days post LCMV Clone 13 infection. Representative of eight independent experiments, n ≥ 4 mice. (d-e) Frequency of CD45.1+ transferred cells (d) and number (e) of control or Ptpn2-deleted P14 T cells in the spleen 8, 15, 22, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. (f) Quantification of BrdU incorporation for co-transferred control and Ptpn2-deleted P14 T cells 8, 15, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 3 mice. (g) Representative flow cytometry plots of Granzyme B expression from splenic control or Ptpn2-deleted P14 CD8+ T cells co-transferred at day 8 post LCMV Clone 13 infection as in (c). Representative of two independent experiments, n ≥ 4 mice. (h) Quantification of (g) days 8, 15, 22, and 30 post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. Bar graphs represent mean and error bars represent standard deviation. Statistical significance was assessed by two-sided Student’s paired t-test (d, e, f, h) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 1.
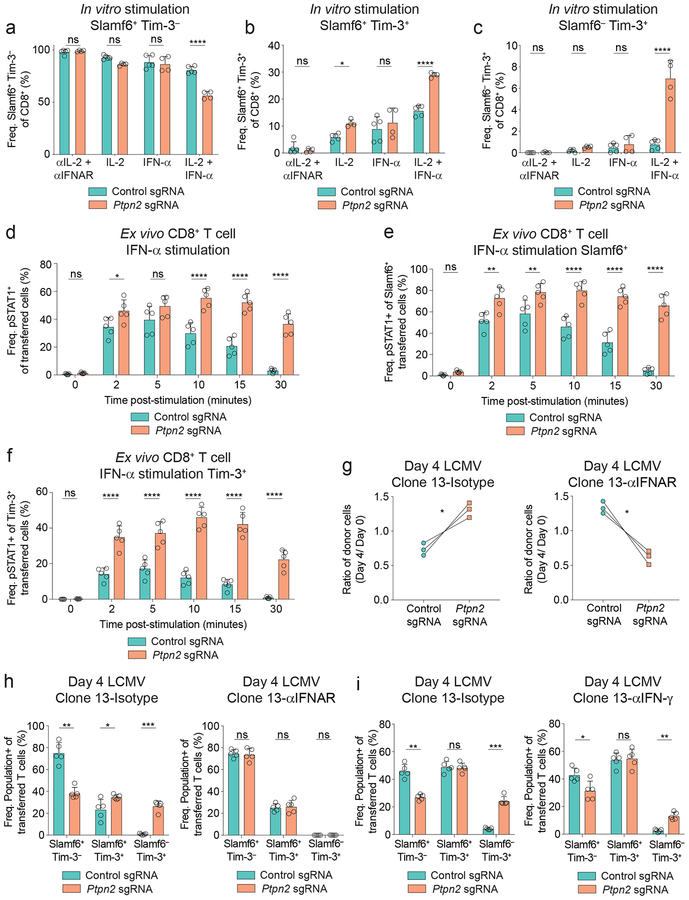
Deletion of Ptpn2 enhances formation of the Tim-3+ terminally exhausted subpopulation during LCMV Clone 13 infection
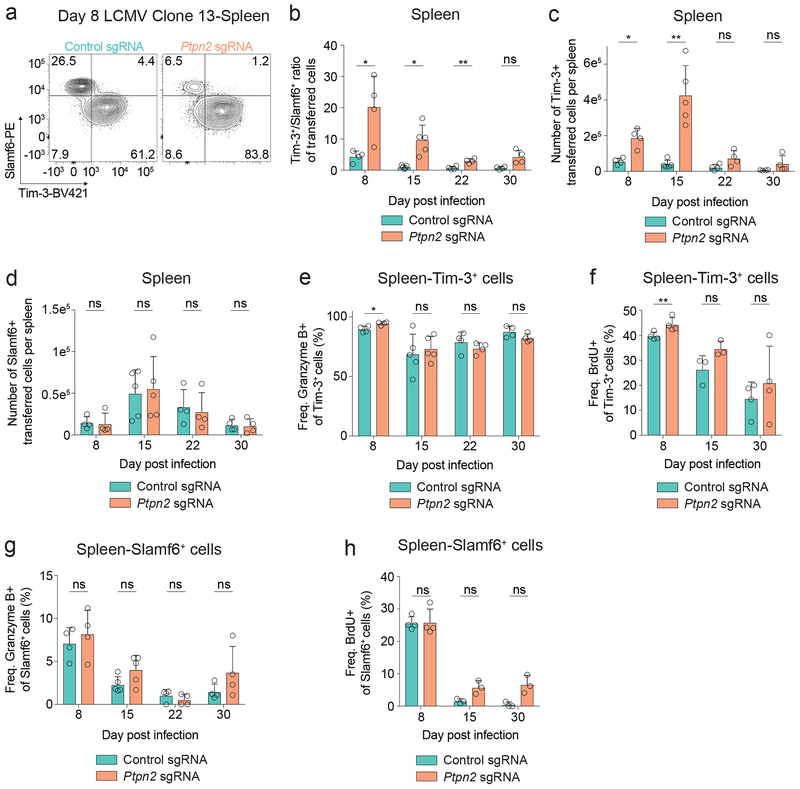
The changes in Granzyme B expression prompted us to examine the impact of Ptpn2 deletion on the generation of the Slamf6+ and Tim-3+ subpopulations. Ptpn2 deletion increased the ratio of Tim-3+ to Slamf6+ cells at days 8, 15, and 22 post infection (Figures 2a, 2b), despite transferring only Slamf6+ Tim-3– cells (Supplementary Figure 2a). Analysis of the populations using Tim-3 and a distinct progenitor marker (CXCR5)4, 16, 17 gave identical results (Supplementary Figures 2b, 2c). Moreover, following Ptpn2 deletion, we observed a significant reduction in expression of two additional markers of progenitor exhausted cells, CD127 and TCF1 (Supplementary Figure 2d). The increase in the Tim-3+ to Slamf6+ ratio was driven by a specific increase in the number of Tim-3+ cells following Ptpn2 deletion (Figure 2c), while there was no difference in the number of Slamf6+ cells (Figure 2d), nor CXCR5+ cells (Supplementary Figure 2e). Furthermore, in non-TCR transgenic mice, Ptpn2 deletion resulted in an increase in the percentage of Tim-3+ cells and a decrease in the percentage of Slamf6+ cells compared with control cells (Supplementary Figure 2f). Thus, deletion of Ptpn2 enhances formation of the Tim-3+ subset during LCMV Clone 13 infection.
Figure 2: Deletion of Ptpn2 enhances formation of the Tim-3+ terminally exhausted subpopulation during LCMV Clone 13 infection.
(a) Representative flow cytometry plots of Tim-3 and Slamf6 expression on control or Ptpn2-deleted P14 T cells in the spleen 8 days post LCMV Clone 13 infection. Representative of eight independent experiments, n ≥ 4 mice. (b) Ratio of Tim-3+/Slamf6+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, 22, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. (c) Number of Tim-3+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, 22, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. (d) Number of Slamf6+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, 22, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. (e) Quantification of Granzyme B expression for Tim-3+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, 22, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. (f) Quantification of BrdU incorporation for Tim-3+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 3 mice. (g) Quantification of Granzyme B expression for Slamf6+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, 22, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 4 mice. (h) Quantification of BrdU incorporation for Slamf6+ control or Ptpn2-deleted P14 T cells in the spleen 8, 15, and 30 days post LCMV Clone 13 infection. Representative of two independent experiments, n ≥ 3 mice. Bar graphs represent mean and error bars represent standard deviation. Statistical significance was assessed by two-sided Student’s paired t-test (b-h) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 2.
To determine if changes in Granzyme B expression and BrdU incorporation (Figures 1f–1h) were intrinsically changed in the Tim-3+ subpopulations, we compared co-transferred control and Ptpn2-deleted cells, and found only a minimal increase in Granzyme B expression in the Ptpn2-deleted Tim-3+ cells (Figure 2e). In addition, we found a nominal increase in BrdU incorporation between control and Ptpn2-deleted Tim-3+ cells (Figures 2f), indicating a similar increase in actively proliferating cells. We found no differences in Granzyme B expression nor BrdU incorporation in Slamf6+ control and Ptpn2-deleted cells (Figures 2g, 2h). These findings demonstrate that Ptpn2 deletion leads to a specific increase in the generation of the Tim-3+ subpopulation, while preserving the number of Slamf6+ cells, and that the altered ratio of Tim-3+ to Slamf6+ cells is primarily responsible for the observed increase in Granzyme B expression and BrdU incorporation.
Ptpn2 deletion promotes effector-skewed Slamf6+ and Tim-3+ subpopulations during LCMV infection
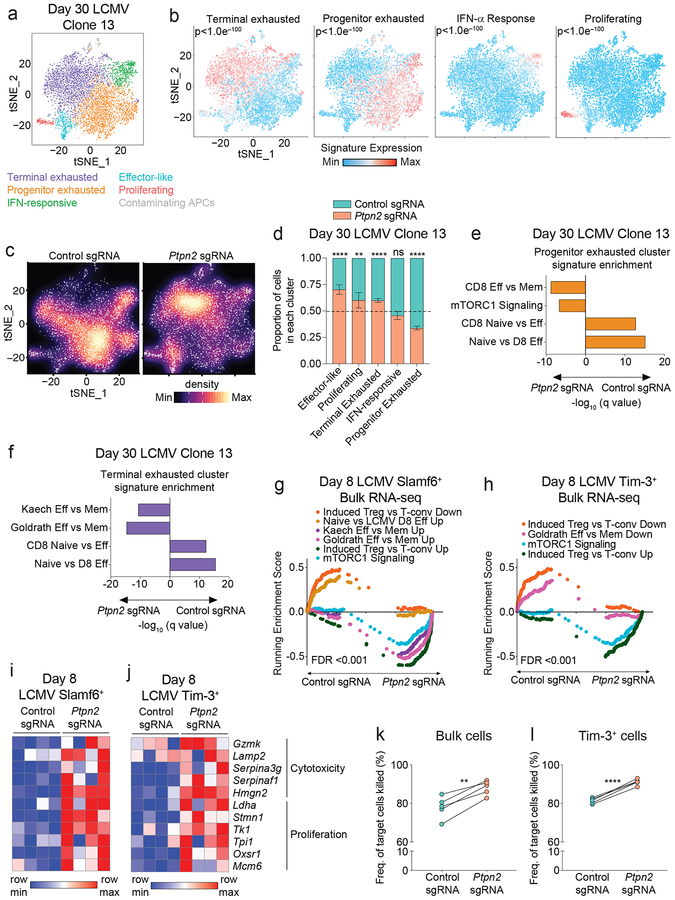
We next performed single-cell RNA-seq on control and Ptpn2-deleted cells day 30 post LCMV infection, as the canonical features of exhaustion are present at this time point5, 10. Unsupervised clustering of the cells revealed 6 subpopulations, which we identified by marker gene expression and previously-defined signature enrichment (Figures 3a, 3b and Supplementary Figures 3a, 3b). We recapitulated the previously described terminally exhausted, progenitor exhausted, proliferating, and effector-like populations4, and identified a distinct IFN-responsive cluster that contained both progenitor exhausted and terminally exhausted cells, suggesting it represented cells that were actively responding to IFN-α (Figure 3b and Supplementary Figure 3b). Further analysis of the distribution of the control or Ptpn2-deleted cells across the clusters revealed a significant skewing of the control cells into the progenitor exhausted cluster and the Ptpn2-deleted cells into the effector-like, proliferating, and terminally exhausted clusters (Figures 3c, 3d), consistent with our flow cytometry data (Supplementary Figure 3c). We also noted that within a subpopulation the Ptpn2-deleted cells or control CD8+ T cells tended to cluster together (Supplementary Figure 3d). Differential expression analysis between Ptpn2-deleted and control cells within the progenitor and terminally exhausted clusters revealed that the Ptpn2-deleted cells had increased expression of genes associated with terminally exhausted or effector cells such as Gzma, Cd160, Ccl4, and Ccl5 (adjusted p-values < 0.005), consistent with an enrichment of effector-related gene signatures (Figures 3e, 3f). Thus, at day 30 post LCMV infection, Ptpn2-deleted progenitor and terminally exhausted cells have increased transcription of effector-related genes.
Figure 3: Ptpn2 deletion promotes effector-skewed Slamf6+ and Tim-3+ subpopulations during LCMV infection.
(a) tSNE projection of single-cell RNA-seq profiles from 7,027 control or Ptpn2-deleted P14+ CD8+ T cells responding to day 30 LCMV Clone 13 infection. Clusters are distinct colors. Representative of one experiment, n = 4 pooled mice. (b) Enrichment of gene signatures in the clusters. Representative of one experiment, n = 4 pooled mice. (c) Plots depicting the inter-cluster density for control or Ptpn2-deleted cells. Representative of one experiment, n = 4 pooled mice. (d) Quantification of the proportion of control or Ptpn2-deleted cells in each cluster. Error bars represent the 95% confidence interval and the dotted line represents the proportion of Ptpn2-deleted cells among all cells projected. Representative of one experiment, n = 4 pooled mice. (e-f) Signature enrichments of co-transferred control and Ptpn2-deleted cells from the (e) progenitor or (f) terminally exhausted clusters. Representative of one experiment, n = 4 pooled mice. (g-h) GSEA curves for significantly enriched signatures in (g) Slamf6+ cells and (h) Tim-3+ cells for bulk RNA-seq of co-transferred control and Ptpn2-deleted cells day 8 post LCMV Clone 13 infection. Representative of one experiment, n = 2 mice with n = 2 technical replicates per mouse. (i-j) Row-normalized heat map of effector-associated genes in (i) Slamf6+ cells and (j) Tim-3+ cells for bulk RNA-seq of co-transferred control and Ptpn2-deleted cells day 8 post LCMV Clone 13 infection. Representative of one experiment, n = 2 mice with n = 2 technical replicates per mouse. (k) Quantification of frequency of tumor cells killed when co-cultured with control or Ptpn2-deleted CD8+ T cells (2:1 effector: target ratio) isolated from day 8 LCMV Clone 13 infection. Representative of two independent experiments, n = 5 mice. (l) Quantification of frequency of tumor cells killed when co-cultured with Tim-3+ control or Tim-3+ Ptpn2-deleted CD8+ T cells (2:1 effector: target ratio) isolated from day 8 LCMV Clone 13 infection. Representative of two independent experiments, n = 5 mice. Statistical significance was assessed by the two-sided Wilcoxon rank sum test (b, e, f), two-sided binomial test (d), two-sided Kolmogorov-Smirnov test (g-h), and two-sided Student’s paired t-test (k-l) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 3.
We next asked whether Ptpn2 deletion impacted the cell states of Slamf6+ and Tim-3+ subpopulations at an earlier time point post LCMV infection. We performed RNA-seq on co-transferred Ptpn2-deleted or control CD8+ T cells 8 days post LCMV Clone 13 infection (Supplementary Table 1) and found that the Slamf6+ and Tim-3+ subpopulations clustered together regardless of Ptpn2 deletion (Supplementary Figure 3e). GSEA analysis revealed that both control and Ptpn2-deleted Slamf6+ cells were significantly enriched for the LCMV Slamf6 vs. Tim-3 Up signature4, whereas the control and Ptpn2-deleted Tim-3+ cells were significantly enriched for the LCMV Slamf6 vs. Tim-3 Down signature4 (Supplementary Figure 3f). Consistent with this, Ptpn2 deletion had almost no effect on the epigenetic state of either the Slamf6+ or Tim-3+ populations (<0.25% of open chromatin regions differentially expressed) 8 days post infection (Supplementary Figure 3g and Supplementary Table 2). Both control and Ptpn2-deleted CD8+ T cells still showed characteristic differences between the Slamf6+ and Tim-3+ populations (Supplementary Figure 3h). In addition, both control and Ptpn2-deleted cells showed chromatin accessible regions at the Tox locus (Supplementary Figure 3i), characteristic of T cell exhaustion10, indicating the cells were differentiating into the exhausted subpopulations.
Although the general states of the Slamf6+ and Tim-3+ cells were maintained at day 8, GSEA analysis of the Slamf6+ and Tim-3+ Ptpn2-deleted cells compared with control cells revealed an enrichment for effector-related gene signatures (Figures 3g, 3h). Within both the Slamf6+ and Tim-3+ subpopulations, Ptpn2-deleted cells had increased expression of effector-related genes pertaining to cytotoxicity (Gzmk, Lamp2, Serpina3g) and proliferation (Stmn1, Tk1, Tpi1), compared with control cells (Figures 3i, 3j). Thus, while Ptpn2 deletion does not fundamentally change the Slamf6+ and Tim-3+ subpopulations, it does lead to increases in Tim-3+ cell numbers and robustly increases effector-related genes both early and late post LCMV infection.
Consistent with the increase in effector-like profiles in the Ptpn2-deleted cells, Ptpn2-deleted CD8+ T cells had increased cytotoxic potential (Figure 3k). This increased killing was due to an intrinsic change within the Tim-3+ cells as Ptpn2-deleted Tim-3+ cells showed increased killing of target cells compared with control cells (Figure 3l). Overall, these findings validate the observed enrichment in cytotoxic genes seen by RNA-seq and demonstrate that Ptpn2 deletion improves the cytotoxic function of CD8+ T cells.
Ptpn2 deletion increases Tim-3+ cell differentiation and proliferation
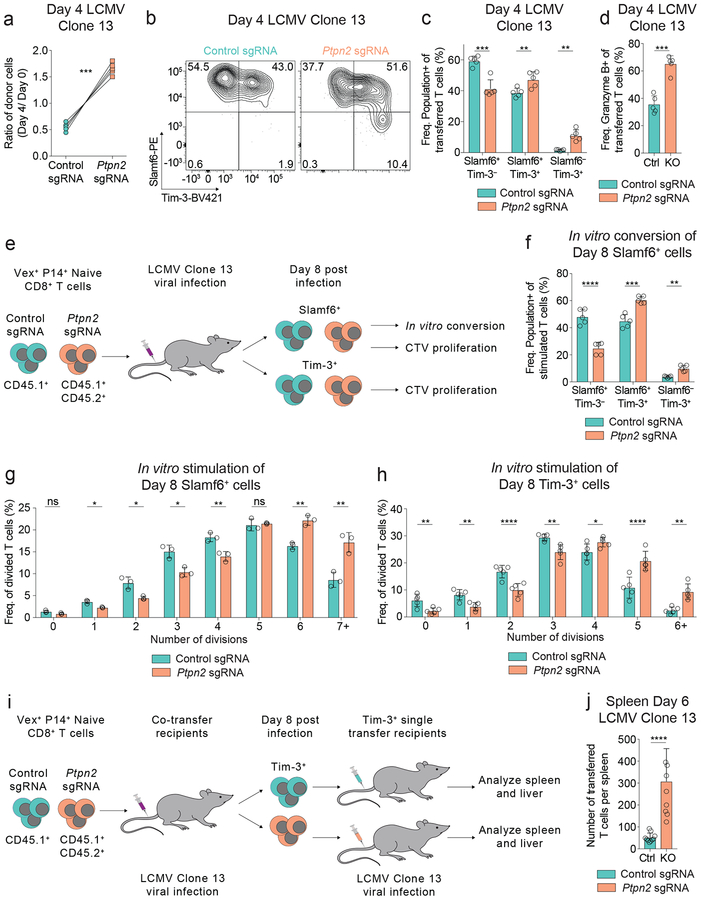
The increase in Tim-3+ cell numbers at day 8 post LCMV infection, coupled with the enhanced effector-related gene signatures, led us to ask whether PTPN2 was impacting the differentiation of these subpopulations. We found that Ptpn2-deleted CD8+ T cells had a significant competitive advantage over control cells at day 4 post infection (Figure 4a) and enhanced differentiation into Tim-3+ cells (Figure 4b). Ptpn2 deletion resulted in a decreased percentage of Slamf6+ Tim-3– cells and an increased percentage of Slamf6+ Tim-3+ cells and Slamf6– Tim-3+ cells (Figure 4c), as well as an increase in Granzyme B expression (Figure 4d). We next asked if the increase in the number and percentage of Tim-3+ cells was due to (1) increased conversion from Slamf6+ cells4, 16, 17, (2) increased proliferative capacity of either population, or (3) increased persistence (Figure 4e). First, to test conversion, we isolated antigen-experienced control or Ptpn2-deleted Slamf6+ cells (Figure 4e) and restimulated them in vitro with signals 1–3 (αCD3, αCD28, and IFN-α together)31, as well as IL-2, and found that Ptpn2-deleted Slamf6+ cells preferentially formed Slamf6+ Tim-3+ and Slamf6– Tim-3+ subsets compared to control cells (Figure 4f). Second, we tested proliferative capacity following restimulation, and found that antigen-experienced Ptpn2-deleted Slamf6+ cells underwent more rounds of division than control Slamf6+ cells (Figure 4g and Supplementary Figures 4a, 4b). When BrdU was a readout of proliferation there was comparable incorporation between control and Ptpn2-deleted Slamf6+ cells, indicating similar active proliferation (Figure 2h). Here, the increase in CTV dilution signifies enhanced proliferative capacity of the Ptpn2-deleted Slamf6+ cells compared to control cells. We also compared the proliferative capacity of antigen-experienced control and Ptpn2-deleted Tim-3+ cells following restimulation and found that Ptpn2-deleted Tim-3+ cells also underwent more rounds of division than control Tim-3+ cells (Figure 4h and Supplementary Figures 4c, 4d). Thus, while the Ptpn2-deleted Slamf6+ and Tim-3+ cells have a similar frequency of actively proliferating cells compared with control cells (Figures 2f, 2h), they had the greatest difference in proliferative capacity (Figures 4g, 4h).
Figure 4: Ptpn2 deletion increases Tim-3+ cell differentiation and proliferation.
(a) Quantification of frequencies of co-transferred control or Ptpn2-deleted CD8+ T cells day 4 post LCMV Clone 13 infection. Frequencies at day 4 were normalized to input frequencies at day 0. Representative of two independent experiments, n = 5 mice. (b) Representative flow cytometry plots of Slamf6 and Tim-3 expression on splenic CD8+ T cells day 4 post LCMV Clone 13 infection for co-transferred control and Ptpn2-deleted cells. Representative of two independent experiments, n = 5 mice. (c) Quantification of Slamf6+ Tim-3–, Slamf6+ Tim-3+, and Slamf6– Tim-3+ subsets in (b). Representative of two independent experiments, n = 5 mice. (d) Quantification of Granzyme B expression of cells as in (a). Representative of two independent experiments, n = 5 mice. (e) Schematic of in vitro conversion and CTV proliferation assays using co-transferred control or Ptpn2-deleted CD8+ T cells isolated at day 8 post LCMV Clone 13 infection. (f) Quantification of Slamf6+ Tim-3–, Slamf6+ Tim-3+, and Slamf6– Tim-3+ subsets following in vitro stimulation (αCD3/CD28 with IL-2 and IFN-α) of control or Ptpn2-deleted CD8+ T cells isolated at day 8 post LCMV Clone 13 infection. Representative of two independent experiments, n = 5 mice. (g-h) Quantification of frequency of divisions following in vitro stimulation (αCD3/CD28 with IL-2) of (g) CTV-labeled Slamf6+ or (h) CTV-labeled Tim-3+, control or Ptpn2-deleted CD8+ T cells isolated at day 8 post LCMV Clone 13 infection. Representative of two independent experiments, n = 3 mice (g), 5 mice (h). (i) Schematic of in vivo persistence assay of Tim-3+ cells. (j) Quantification of number of recovered Tim-3+ control or Ptpn2-deleted cells in the spleen day 6 post LCMV Clone 13 infection, following the transfer of these cells which were previously isolated at day 8 post LCMV Clone 13 infection. Representative of two pooled experiments, n = 8 mice. Bar graphs represent mean and error bars represent standard deviation. Statistical significance was assessed by two-sided Student’s paired t-test (a, c-d, f-h) or two-sided Student’s unpaired t-test (j) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 4.
To examine persistence of Tim-3+ cells in vivo, we transferred antigen-experienced control or Ptpn2-deleted Tim-3+ cells separately to new recipients infected with LCMV Clone 13 (Figure 4i). We recovered an increased number of Ptpn2-deleted Tim-3+ cells compared with control cells in the spleen and liver (Figure 4j and Supplementary Figure 4e). We also found a decrease in survival of the recovered Ptpn2-deleted Tim-3+ cells (Supplementary Figure 4f), suggesting that their increased cell number is likely due to a proliferative advantage (Figure 4h), consistent with the known role of PTPN2 in regulating the proliferative capacity of naive CD8+ T cells by dephosphorylation of kinases downstream of the TCR30. Thus, these findings demonstrate that Ptpn2 deletion affects the generation of Tim-3+ cells in multiple ways: by enhancing conversion of Slamf6+ cells into Tim-3+ cells and increasing the proliferative capacities of both Slamf6+ and Tim-3+ cells.
Ptpn2 deletion increases Tim-3+ cell differentiation through enhanced IFN-α signaling
We next determined the factors driving the increased differentiation of Slamf6+ cells into Tim-3+ cells following Ptpn2 deletion. We evaluated the necessity of IL-2 and IFN-α, since these cytokines have important roles in the differentiation of the Tim-3+ subpopulation during LCMV Clone 13 infection18, 20 and lead to phosphorylation of STAT5 and STAT1, respectively, both known targets of PTPN232, 33. We stimulated control or Ptpn2-deleted naive CD8+ T cells with αCD3/CD28 plus IL-2, IFN-α, both IL-2 and IFN-α, or blocking antibodies to abolish IL-2 and IFN-α signaling. Stimulation with IL-2, IFN-α, and IL-2 plus IFN-α led to increased CD25 expression on Ptpn2-deleted cells compared with control cells (Supplementary Figure 5a), indicating increased activation30. Consistent with antigen-experienced Slamf6+ cells, stimulation of naive Slamf6+ cells with IL-2 plus IFN-α resulted in a decreased percentage of Slamf6+ Tim-3– cells and an increased percentage of Slamf6+ Tim-3+ and Slamf6– Tim-3+ cells in the Ptpn2-deleted cells, compared with control cells (Figures 5a–5c and Supplementary Figure 5b). However, the addition of IL-2 or IFN-α separately was unable to recapitulate the marked increase of Slamf6– Tim-3+ cells observed following treatment with the combination (Figure 5c). Moreover, CD28 costimulation was also required for the differentiation of Slamf6– Tim-3+ cells (Supplementary Figure 5c). We also ruled out the contribution of soluble factors, as conditioned supernatant from Ptpn2-deleted cells did not increase the percentage of Slamf6– Tim-3+ cells compared with control cells (Supplementary Figure 5d). Thus, IL-2, IFN-α, and CD28 are all required for the enhanced generation of Tim-3+ cells in Ptpn2-deleted CD8+ T cells in our in vitro stimulation assay.
Figure 5: Ptpn2 deletion increases Tim-3+ cell differentiation through enhanced IFN-α signaling.
(a-c) Quantification of (a) Slamf6+ Tim-3–, (b) Slamf6+ Tim-3+, and (c) Slamf6– Tim-3+ subsets following in vitro stimulation (αCD3/CD28) of control or Ptpn2-deleted naive CD8+ T cells in the presence of indicated cytokines or blocking antibodies. Representative of two pooled experiments, n ≥ 4 technical replicates. (d) Quantification of pSTAT1 expression of splenic CD8+ T cells day 6 post LCMV Clone 13 infection for co-transferred control and Ptpn2-deleted cells following ex vivo restimulation with IFN-α. Representative of two independent experiments, n = 5 biological replicates. (e-f) Quantification of pSTAT1 in (e) Slamf6+ or (f) Tim-3+ cells following ex vivo IFN-α restimulation of co-transferred control and Ptpn2-deleted cells as in (d). Representative of two independent experiments, n = 5 biological replicates. (g) Quantification of frequencies of co-transferred control and Ptpn2-deleted CD8+ T cells day 4 post LCMV Clone 13 infection following treatment with isotype (left graph) or IFNAR blocking antibody (right graph). Frequencies at day 4 were normalized to input frequencies at day 0. Representative of two independent experiments, n = 3 biological replicates. (h) Quantification of Slamf6+ Tim-3–, Slamf6+ Tim-3+, and Slamf6– Tim-3+ subsets day 4 post LCMV Clone 13 infection in mice that received co-transferred control and Ptpn2-deleted P14 CD8+ T cells and were treated with isotype (left) or IFNAR blocking antibody (right). Representative of two independent experiments, n = 5 biological replicates. (i) Quantification of Slamf6+ Tim-3–, Slamf6+ Tim-3+, and Slamf6– Tim-3+ subsets day 4 post LCMV Clone 13 infection in mice that received co-transferred control and Ptpn2-deleted P14 CD8+ T cells and were treated with isotype (left) or IFN-γ neutralizing antibody (right). Representative of two independent experiments, n = 5 biological replicates. Bar graphs represent mean and error bars represent standard deviation. Statistical significance was assessed by two-way ANOVA (a-c) or two-sided Student’s paired t-test (d-i) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 5.
Given the requirement for IFN-α in vitro and the known role for IFN-I signaling in the regulation of Tim-3+ and Slamf6+ subpopulations18, we next investigated the impact of Ptpn2 deletion on STAT1 phosphorylation. Ptpn2-deleted cells had an increased percentage and duration of pSTAT1 expression (Figure 5d), despite comparable levels of the type 1 interferon receptor (IFNAR1) (Supplementary Figure 5e). This increase in pSTAT1 was observed in both Slamf6+ and Tim-3+ subsets (Figures 5e, 5f). Consistent with this, IFN-I signaling was required for the early expansion of Ptpn2-deficient cells (Figure 5g), and the increased differentiation of Ptpn2-deleted cells into Tim-3+ cells (Figure 5h). Given Ptpn2 regulates common signaling molecules, such as STAT1 and JAK1, between the type 1 and 2 IFN pathways32, 33, we examined whether IFN-γ was required for the increased Tim-3+ subset differentiation of Ptpn2-deleted CD8+ T cells during LCMV Clone 13 infection. Following IFN-γ blockade, Ptpn2-deleted CD8+ T cells still had significantly increased expansion and elevated Slamf6+ to Tim-3+ subset differentiation compared with control cells (Supplementary Figure 5f and Figure 5i), but were decreased in percentage compared with isotype treated Ptpn2-deleted cells, indicating a non-essential role for IFN-γ in the generation of Tim-3+ cells (Supplementary Figure 5g). Overall, these findings indicate that type 1 IFN signaling plays a crucial role in the differentiation of Ptpn2-deleted cells into Tim-3+ cells.
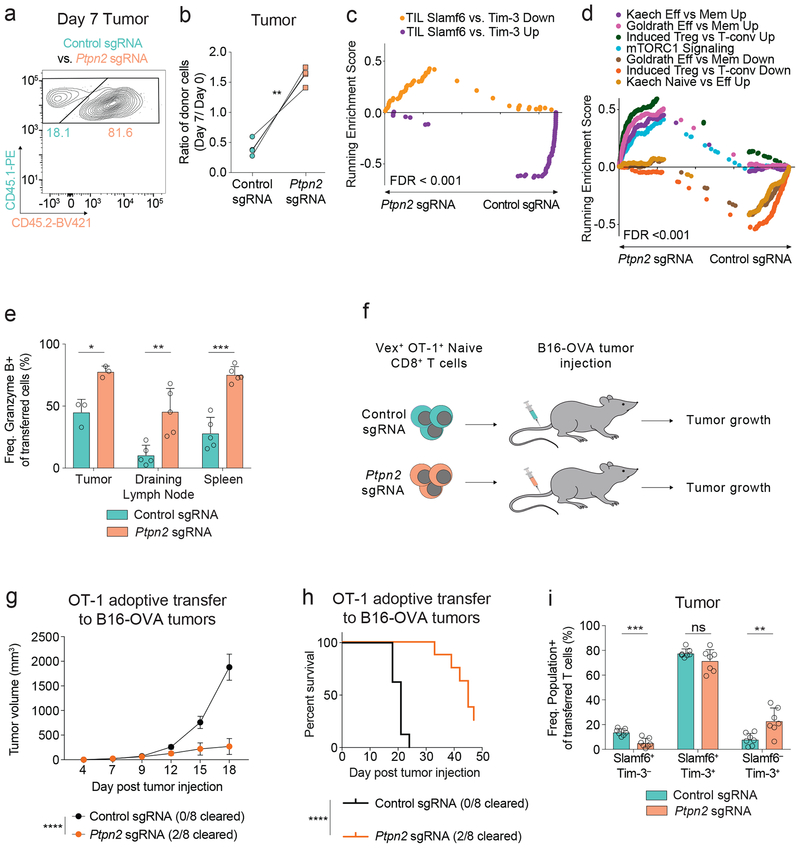
Loss of Ptpn2 enhances Tim-3+ CD8+ T cell differentiation in tumors
Given the importance of dysfunctional T cell subpopulations in tumors4, 18, 22, 23, 24, 25, 26, 27, we next asked if Ptpn2 also regulates the balance and functions of CD8+ T subpopulations in responses to tumors. We co-transferred congenically marked-naive OT-1 TCR transgenic (specific for the ovalbumin CD8 epitope OVA257–264) Ptpn2-deleted and control CD8+ T cells to wild-type recipients and subsequently challenged these mice with MC38-OVA tumors. Consistent with chronic LCMV infection, Ptpn2-deleted OT-1 CD8+ T cells significantly outcompeted control CD8+ T cells at day 7 in the tumor (Figures 6a, 6b and Supplementary Figure 6a). Ptpn2 deletion also led to an increase in CD25 and a decrease in CD127 expression in transferred CD8+ T cells in the tumor-draining lymph node (Supplementary Figures 6b, 6c), indicating increased activation of these cells. In addition, Ptpn2-deleted cells had increased generation of IFN-γ+ TNF– cells following peptide restimulation in vitro (Supplementary Figure 6d). Transcriptional profiling of control or Ptpn2-deleted CD8+ T cells revealed that Ptpn2-deleted CD8+ T cells were significantly enriched for the tumor infiltrating lymphocytes (TIL) Tim-3+ signature, whereas control cells were enriched for the TIL Slamf6+ signature (Figure 6c and Supplementary Table 3)4. In addition, Ptpn2-deleted cells were significantly enriched for mTORC1 signaling and several other effector-related signatures that were also enriched in the Ptpn2-deleted cells in the LCMV model (Figure 6d). Consistent with this, Ptpn2 deletion increased the percentages of Granzyme B-expressing OT-1 CD8+ T cells in the tumor, draining lymph node and spleen, compared with control OT-1 CD8+ T cells (Figure 6e). To determine the functional effect of these cells, we transferred control or Ptpn2-deleted OT-1 CD8+ T cells to recipient mice implanted with B16-OVA tumors (Figure 6f). Ptpn2-deleted CD8+ T cells had a significant effect on B16-OVA tumor growth and led to clearance of 25% of the tumors (Figures 6g, 6h). Moreover, when control and Ptpn2-deleted cells were co-transferred in the B16-OVA model, Ptpn2-deleted OT-1 T cells had an increased percentage of the Slamf6– Tim-3+ subset compared with control cells (Figure 6i). Thus, concordant with the LCMV Clone 13 model, Ptpn2-deleted CD8+ T cells outcompete control cells in the tumor, have elevated Tim-3 and Granzyme B expression, and possess increased effector function.
Figure 6: Loss of Ptpn2 enhances Tim-3+ CD8+ T cell differentiation in tumors.
(a) Representative flow cytometry plot of control or Ptpn2-deleted OT-1 T cells in the tumor 7 days post MC38-OVA injection. Representative of two independent experiments, n = 4 biological replicates. (b) Quantification of frequencies of co-transferred control or Ptpn2-deleted CD8+ T cells day 7 post MC38-OVA injection. Frequencies at day 7 were normalized to input frequencies at day 0. Representative of two independent experiments, n = 4 biological replicates. (c-d) (c) GSEA TIL Slamf6+ vs. Tim-3+ Up top 50 and TIL Slamf6+ vs. Tim-3+ Down top 50 signature enrichment and (d) GSEA effector signatures for co-transferred control or Ptpn2-deleted OT-1 T cells in MC38-OVA tumors 7 days post injection. Representative of one experiment, n = 3 pooled mice and 2 technical replicates. (e) Quantification of Granzyme B expression in co-transferred OT-1 CD8+ T cells day 7 post MC38-OVA implantation in the tumor, draining lymph node, and spleen for control and Ptpn2-deleted co-transferred mix as in (a). Representative of two independent experiments, n ≥ 3 mice. (f) Schematic of adoptive transfer of either control or Ptpn2-deleted naive OT-1 CD8+ T cells separately to mice challenged with B16-OVA one day post transfer of T cells. (g) Tumor growth curves for B16-OVA tumors following transfer of naive OT-1 control or Ptpn2-deleted CD8+ T cells separately into WT recipients that were implanted with B16-OVA cells. Representative of two independent experiments, n = 8 mice. (h) Survival curves of mice in (g). Representative of two independent experiments, n = 8 mice. (i) Quantification of Slamf6+ Tim-3–, Slamf6+ Tim-3+, and Slamf6– Tim-3+ subsets day 9 post B16-OVA implantation in the tumor for co-transferred control and Ptpn2-deleted cells as in (a). Representative of two independent experiments, n = 7 mice. Bar graphs represent mean and error bars represent standard deviation (except for g where error bars represent standard error). Statistical significance was assessed by two-sided Student’s paired t-test (b, e, i), two-sided Kolmogorov-Smirnov test (c, d), two-way ANOVA (g), or two-sided log-rank Mantel-Cox test (h) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 6.
Deletion of Ptpn2 enhances CD8+ T cell responses to tumors and checkpoint blockade efficacy
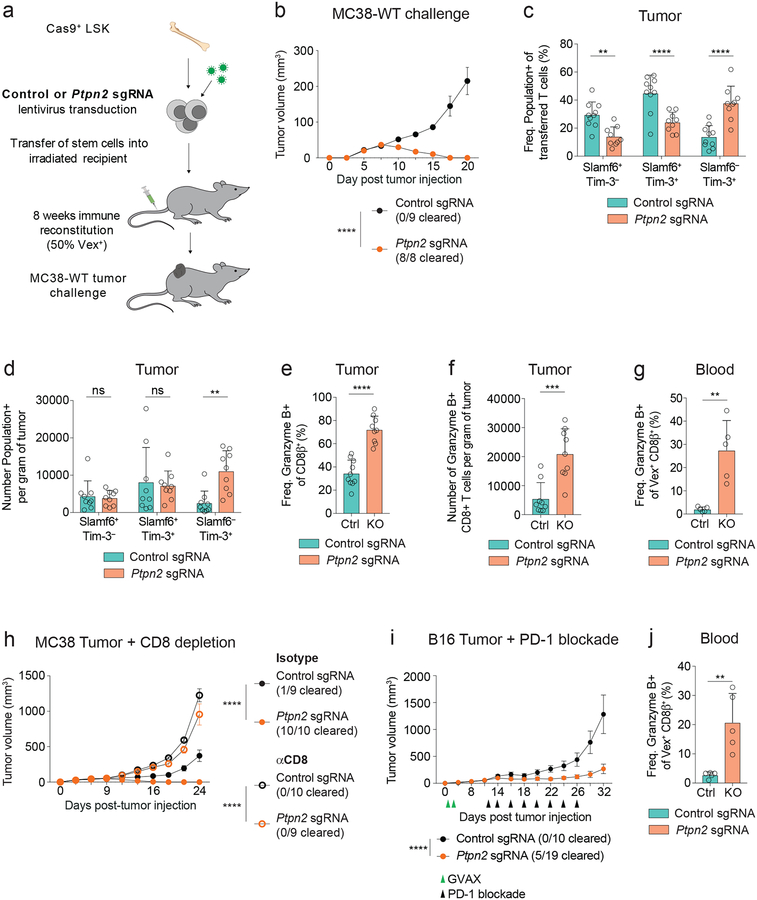
PTPN2 is ubiquitously expressed in the hematopoietic compartment and has roles in myeloid, T, and B cell development and function34. Thus, therapeutic targeting of PTPN2 could potentially affect multiple immune subtypes. To model this, we investigated whether Ptpn2 deletion in all hematopoietic cells would attenuate tumor growth by subcutaneously implanting MC38 tumors directly into the chimeras (Figure 7a and Supplementary Figure 7a). Given PTPN2 has a role in myeloid and T-regulatory cells35, 36, we examined these cell types in the blood of the chimeras at steady state and found similar frequencies of myeloid and regulatory T cell subsets in the blood, but elevated MHC-II+ monocytes in the Ptpn2-deleted chimeras, compared with controls (Supplementary Figure 7b). In addition, the Ptpn2-deleted chimeras did not have elevated inflammatory cytokine levels compared with controls (Supplementary Figure 7c).
Figure 7: Deletion of Ptpn2 enhances CD8+ T cell responses to tumors and checkpoint blockade efficacy.
(a) Schematic for MC38-WT tumor challenge in chimeric mice where approximately 50% of immune cells express a control sgRNA or a Ptpn2-targeting sgRNA. (b) Tumor growth curves for control or Ptpn2-deleted chimeric mice following 1 × 106 cell MC38-WT challenge. Representative of two independent experiments, n ≥ 8 mice. (c-d) Quantification of (c) frequency and (d) number of Slamf6+ Tim-3–, Slamf6+ Tim-3+, and Slamf6– Tim-3+ subsets in CD8+ T cells infiltrating day 9 MC38 tumors in control or Ptpn2-deleted bone marrow chimeras. Representative of two independent experiments, n ≥ 9 mice. (e-f) Quantification of (e) frequency and (f) number of Granzyme B-expressing CD8+ T cells infiltrating day 9 MC38 tumors implanted in control or Ptpn2-deleted bone marrow chimeras. Representative of two independent experiments, n ≥ 9 mice. (g) Quantification of frequency of Granzyme B+ CD8+ T cells from the blood of control or Ptpn2-deleted bone marrow chimeras day 14 post MC38 tumor implantation, pregated on CD8β+ Vex+ cells. Representative of two independent experiments, n = 5 mice. (h) Tumor growth curves for mice as in (a) challenged with 1 × 106 MC38-WT tumor cells following treatment with CD8-depleting antibody or isotype control. Representative of two independent experiments, n ≥ 9 mice. (i) Tumor growth curves for control or Ptpn2-deleted bone marrow chimeras challenged with 1 × 106 B16 tumor cells treated with GVAX (green triangles) on days 1, 4 and αPD-1 (black triangles) on days 12, 14, 16, 18, 20, 22, 24, and 26. Representative of two independent experiments, n ≥ 9 mice. (j) Quantification of frequency of Granzyme B+ CD8+ T cells from the blood of chimeras in (i) day 14 post B16 tumor implantation, pregated on CD8β+ Vex+ cells. . Representative of two independent experiments, n = 5 mice. Bar graphs represent mean and error bars represent standard deviation (except for b, h, i where error bars represent standard error). Statistical significance was assessed by two-way ANOVA (b-d, h-i), or two-sided Student’s unpaired t-test (e-g, j) (ns p>.05, * p≤.05, ** p≤.01, *** p≤.001, **** p≤.0001). See also Supplementary Figure 7.
We next evaluated tumor growth in these chimeras and found that deletion of Ptpn2 led to complete MC38 tumor clearance in all Ptpn2-deleted chimeras, whereas there was progressive tumor growth in control chimeras (Figure 7b and Supplementary Figure 7d). There were no differences in the absolute numbers of CD4+ T cells, CD8+ T cells, and myeloid cells in the MC38 tumors prior to clearance (Supplementary Figure 7e). However, the frequency and number of Slamf6– Tim-3+ CD8+ T cells was increased in tumors of Ptpn2-deficient chimeras compared with control chimeras (Figures 7c, 7d), whereas the number of Slamf6+ Tim-3– CD8+ T cells was unchanged. Furthermore, the frequency and number of Granzyme B+ CD8+ T cells was also increased in tumors of Ptpn2-deficient chimeras compared with control chimeras (Figures 7e, 7f). In addition, peripheral blood CD8+ T cells of Ptpn2-deleted chimeras had significantly more Granzyme B+ cells, fewer CD127+ cells, more Slamf6– Tim-3+ cells, and more CD44+ CD62L– cells (Figure 7g and Supplementary Figures 7f–7i). CD8+ T depletion studies revealed that CD8+ T cells are required for clearance of MC38 tumors in Ptpn2-deleted mice (Figure 7h and Supplementary Figure 7j). Furthermore, Ptpn2-deleted chimeras that completely eliminated primary tumors could clear a larger secondary challenge of MC38 tumor cells in a CD8+ T cell dependent manner (Supplementary Figures 7k, 7l).
We next asked whether Ptpn2 deficiency in the immune system could improve PD-1 ICB responses to a more immune-refractory model, B16 melanoma. Treatment of B16-challenged Ptpn2-deficient chimeras with PD-1 ICB and therapeutic vaccination (GVAX)37 resulted in attenuated tumor growth and increased survival compared with control chimeras (Figure 7i and Supplementary Figure 7m). In addition, 26% of the anti-PD-1 treated Ptpn2-deleted chimeras completely cleared their tumors, in contrast to progressive tumor growth in all control chimeras. This enhanced response to B16 melanoma was accompanied by an increase in Granzyme B+ CD8+ T cells in peripheral blood (Figure 7j). These findings demonstrate that Ptpn2 deficiency in the immune system increases the cytotoxic CD8+ T cell response in the tumor and ultimately leads to a CD8+ T cell dependent clearance of MC38 tumors and improved PD-1 ICB responses to B16 tumors.
Discussion
The mechanisms that govern the generation and balance of the terminally exhausted and progenitor exhausted subpopulations in chronic infection and cancer remain unknown. Here we demonstrate that deletion of Ptpn2 in CD8+ T cells enhances anti-tumor immunity by increasing the formation of the Tim-3+ subset. We find at early and late time points that Ptpn2 deletion promotes effector-related gene expression in both the progenitor (Slamf6+) and terminally exhausted (Tim-3+) subsets, indicating Ptpn2 deletion alters both the balance and functionality of the exhausted subpopulations. Deletion of Ptpn2 also promotes IFN-α signaling, which accelerates Tim-3+ cell differentiation at an early time point. Furthermore, deletion of Ptpn2 in the immune system leads to complete clearance of MC38 tumors and improves PD-1 ICB responses to B16 tumors. Overall, these findings improve our understanding of the differentiation and role of the Tim-3+ subpopulation during anti-tumor immune responses.
Our work implicates PTPN2 as a new regulator of the balance between the Tim-3+ and Slamf6+ subpopulations. PTPN2 has a crucial effect on this balance primarily at early time points during LCMV Clone 13 infection, where the Tim-3+ and Slamf6+ subpopulations are differentiating from early effector cells into exhausted cells. PTPN2 has a multitude of targets to dephosphorylate within the TCR, IL-2, IL-7, and IFN signaling cascades30, 38. Here we show that Ptpn2 deletion leads to enhanced IFN-I signaling, which is required for the early competitive advantage seen in Ptpn2-deleted CD8+ T cells, as well as the enhanced early differentiation of Slamf6+ Tim-3– cells into Slamf6+ Tim-3+ and Slamf6– Tim-3+ cells. These findings are consistent with IFN-I signaling attenuating the TCF1-Bcl6 axis during LCMV infection, resulting in an increase in the percentage of Tim-3+ cells18, and highlight a crucial role for IFN-I signaling early in the differentiation of terminally exhausted cells39, 40. We also find a non-essential role for IFN-γ signaling in promoting the differentiation of Tim-3+ cells in Ptpn2-deleted CD8+ T cells, consistent with its shared signaling pathway members as IFN-I. Additional studies will be needed to determine if IFN-γ has a direct role on CD8+ T cells or an indirect role on MHC-I expression in the host. Moreover, we find an essential role for IL-2 in vitro for enhancing Tim-3+ subset formation in Ptpn2-deleted cells, where it likely works in conjunction with IFN-α to support proliferation41. This is consistent with a requirement for proliferation of CXCR5+ progenitor cells to differentiate into Tim-3+ terminally exhausted cells during LCMV Clone 13 infection17. Overall, these findings help to elucidate the molecular mechanisms controlling CD8+ T cell fate decisions into progenitor or terminally exhausted subpopulations in response to LCMV infection42.
Currently, it is believed that an increase in the progenitor exhausted subpopulation promotes the efficacy of PD-1 ICB in chronic infection and cancer4, 16, 17, 24. However, the Tim-3+ subpopulation is the primary cytotoxic population4, 15, and thus also plays an important role in immune responses. It is likely that both progenitor exhausted and terminally exhausted cells are required for an effective immune response that balances cytotoxic potential and longevity. Our work represents a novel scenario where Ptpn2 deletion causes an early increase in the cytotoxic Tim-3+ subpopulation without altering the number of progenitor Slamf6+ CD8+ T cells. This occurs because: (1) Ptpn2-deleted Slamf6+ cells have increased conversion into Tim-3+ cells, (2) Ptpn2-deleted Slamf6+ cells have increased proliferative capacity and thus can replenish the Slamf6+ cell pool, and (3) Ptpn2-deleted Tim-3+ cells have increased proliferative capacity, which further expands their numbers. This early increase in the number of cytotoxic cells and their inherent cytotoxic potential results in clearance of MC38 tumors and improved responses of B16 tumors to PD-1 blockade. These findings can be reconciled with data showing that an increase in the progenitor subpopulation improves checkpoint blockade responses by considering that following PD-1 blockade, the progenitor exhausted subpopulation expands and converts into terminally exhausted cytotoxic Tim-3+ cells4, 16, 17. Our findings suggest that both the progenitor and terminally exhausted subpopulations can promote anti-tumor immunity and that Ptpn2 deletion leads to improved tumor immunity through an increase in Tim-3+ cytotoxic cells without depleting the progenitor subpopulation.
Finally, this work supports the development of PTPN2 inhibitors for cancer immunotherapy and the deletion of PTPN2 in CAR T cell-based therapies. PTPN2 has a cell-intrinsic role in CD8+ T cells in tumors, limiting their accumulation and expression of Granzyme B, consistent with Ptpn2 deficient CD8+ T cell responses in the LCMV model and the RIP-mOVA model of diabetes43. Moreover, adoptive transfer of Ptpn2-deleted CD8+ T cells potently attenuates the growth of B16-OVA tumors, demonstrating that the increase in Tim-3+ cells following Ptpn2 deletion has a therapeutic benefit on tumor immunity. Furthermore, deletion of Ptpn2 in the whole hematopoietic compartment leads to a CD8+ T cell-dependent complete clearance of MC38 tumors, accompanied by a significantly elevated systemic cytotoxic CD8+ T cell response, which could be beneficial for enhancing immunity to disseminated metastatic disease44, 45. Ptpn2 deletion in the immune system also improves PD-1 ICB responses to B16 tumors, indicating its potential use as a combination therapy with PD-1 blockade.
PTPN2 is a particularly attractive cancer immunotherapy target given its established tumor-intrinsic role in restraining anti-tumor immunity46. Inhibition of PTPN2 in a tumor-bearing host would enhance IFN-γ signaling within tumor cells, thereby increasing MHC-I expression and sensitivity to IFN-γ-mediated apoptosis46. Increased MHC-I expression also would promote TCR driven differentiation of exhausted T cells into the Tim-3+ population4. In addition, PTPN2 inhibition in CD8+ T cells would increase IFN-I signaling and further enhance the formation and effector function of the cytotoxic Tim-3+ population. Overall, these findings provide the rationale for combining PTPN2 inhibition and PD-1 ICB for cancer immunotherapy and for deletion of PTPN2 in CAR T cell therapies.
Online methods
Mouse breeding and production
7–10-wk-old female or male mice were used for all experiments and 7–14-wk-old female or male mice were used as donors for bone marrow chimera experiments. Wild-type C57BL/6J mice were purchased from The Jackson Laboratory. LoxP-STOP-loxP Cas9 mice (B6J.129(B6N)-Gt(ROSA) 26Sortm1(CAG-cas9*,-EGFP)Fezh/J)47 were a generous gift from Dr. Feng Zhang, Massachusetts Institute of Technology. These mice were bred to Zp3-Cre mice (C57BL/6-Tg(Zp3-cre)1Gwh/J) to delete the loxP-STOP-LoxP in the female germline. The resulting Cas9-expressing strain was then bred to OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J) or P14 (Taconic B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz backcrossed 10 generations to Jackson C57BL/6J) TCR transgenic mice on the CD45.1 (B6.SJL-Ptprca Pepcb/BoyJ) congenic background. All strains used were backcrossed at least 10 generations to Jackson C57BL/6J. Sample size was chosen to ensure the possibility of statistical analysis and minimize the use of animals. Data exclusion was not used. Age and sex-matched animals were used for each experiment. For chimerism experiments LSK donor, LSK recipients, and CD8+ T cell transfer recipients were sex matched. Animals were also co-housed when possible. All attempts to reproduce our findings were successful. The LCMV Clone 13 infection, MC38 tumor, and B16 tumor experiments (Figures 4i, 5g–5i, 6g–6h, 7b–7j and Supplementary Figures 2f, 4e–4f, 5f–5g, 7d–5m) were blinded during data collection. All experimental mice were housed in specific pathogen-free conditions and used in accordance with animal care guidelines from the Harvard Medical School Standing Committee on Animals and the National Institutes of Health.
Guide RNA design and cloning
The sgRNA oligonucleotides (Controls: GCGAGGTATTCGGCTCCGCG, GCTTTCACGGAG GTTCGACG and Ptpn2: GAATATGAGAAAGTATCGAA, CTCACTTCCATTATACCACC), were designed using the Broad CRISPR algorithm48. sgRNAs were cloned into our sgRNA vector using a BsmBI restriction digest. The plasmid and full sequence have been deposited on Addgene (Name: pXPR_053, Addgene ID: 113591).
Bone marrow isolation and chimera setup
Bone marrow cells were isolated and cultured as previously described28. In short, femurs, tibias, hips, and spines were isolated from donor mice, crushed, and lysed with Ammonium-Chloride-Potassium (ACK) lysing buffer (Gibco Cat# A1049201). LSK cells (lineage– Sca-1+ Kit+) were enriched with a CD117 MACS isolation kit (Miltenyi Cat# 130-091-224) and then sorted to purity. The LSK cells were spin transduced with lentiviral vectors on Retronectin-coated plates (Takara Bio Cat# T100B) at a multiplicity of infection (MOI) of 25. LSK cells were then transferred intravenously into irradiated CD45.2+ wild-type recipients.
Cell lines
MC38-OVA (gift from N. Collins, Dana Farber Cancer Institute), B16-OVA (gift from D. Sen, Dana Farber Cancer Institute), MC38-WT (gift from D. Vignali, University of Pittsburgh School of Medicine), B16.F10 and B16/GMCSF (both gifts from G. Dranoff, Novartis Institutes for Biomedical Research), 293× (gift from C. Kadoch, Dana Farber Cancer Institute, and MC38-GP33–41 cells49 were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 20 μg/ml gentamicin. MC38-OVA and B16-OVA cells were produced by transduction of parental MC38 and B16.F10 cells with the lentiviral vector TRC-pLX305 (Broad Institute) containing ovalbumin (OVA) protein. MC38-OVA and B16-OVA cells were selected for 2 days with 2 μg/mL puromycin prior to use to ensure expression of OVA (construct is OVA IRES Puromycin resistance). MC38-GP33–41 cells were monitored for expression of GFP to ensure expression of GP33–41 peptide (construct is GP33–41 IRES GFP). GMCSF secretion by B16/GMCSF cells was validated by ELISA. Parental MC38 and B16.F10 cell lines were validated by exome sequencing. BHK-21 cells (gift from E. John Wherry, University of Pennsylvania) were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 5% tryptose phosphate broth. Vero cells (gift from E. John Wherry, University of Pennsylvania) were cultured in EMEM supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were confirmed mycoplasma negative.
TIDE assay
The TIDE (Tracking of Indels by DEcomposition) assay was performed as previously described29. Briefly, DNA was extracted from cells (DNeasy Blood and Tissue kit, QIAgen Cat# 69506) and PCR was used to amplify the expected sgRNA target site, which was then purified (QIAquick PCR Purification kit, QIAgen Cat# 28106) and analyzed by Sanger sequencing. TIDE primers used for respective sgRNAs (sgRNA-1 Forward: GGGCACTGAGCAGCAAACTTTAT, sgRNA-1 Reverse: GTGACTAGCTTTCA TCTTTGCCTCTT, sgRNA-2 Forward: CTGGAAGGCTGGCTGTAGTGTT, sgRNA-2 Reverse: CTAACCTCCTCAGGCACCAGTC).
Antibodies used for flow cytometry and sorting
Flow cytometry analyses were performed on a BD LSR II or BD FACSymphony and cell sorting was performed on a BD Aria II. Antibodies and dyes were purchased from BD Biosciences (7-AAD Cat# 559925 (1:100 dilution), Slamf6 Clone 13G3 Cat# 561540 (1:100 dilution), BrdU Clone 3D4 Cat# 552598 (1:50 dilution), Ki67-PerCP-Cy5.5 Clone B56 Cat# 561284 (1:100 dilution)); Biolegend (B220 Clone RA3–6B2 Cat# 103208, 103226 (1:100 dilution), CD11b Clone M1/70 Cat# 101208, 101216 (1:100 dilution), CD127 Clone A7R34 Cat# 135014, 135024 (1:100 dilution), CD25 Clone PC61 Cat# 101904 (1:100 dilution), CD3ε Clone 17A2 Cat# 100220, 100308, 100336, 100328 (1:100 dilution), CD4 Clone RM4–5 Cat# 100516, 100531, 100543 (1:100 dilution), CD44 Clone IM7 Cat# 103008, 103028, 103030 (1:100 dilution), CD45.1 Clone A20 Cat# 110708, 110716, 110741 (1:100 dilution), CD45.2 Clone 104 Cat# 109824, 109832, 109830 (1:100 dilution), CD5 Clone 53–7.3 Cat# 100608 (1:100 dilution), CD62L Clone MEL-14 Cat# 104417 (1:100 dilution), CD8α Clone 53–6.7 Cat# 100737 (1:100 dilution), CD8β Clone YTS156.7.7 Cat# 126606, 126608, 126610, 126620, 126614 (1:100 dilution), c-Kit Clone ACK2 Cat# 135108 (1:100 dilution), CXCR5 Clone L138D7 Cat# 145509 (1:50 dilution), Gr-1 Clone RB6–8C5 Cat# 108408 (1:100 dilution), Granzyme B Clone GB11 Cat# 515403, 515406 (1:100 dilution), I-A/I-E Clone M5/114.15.2 Cat# 107614 (1:100 dilution), IFN-γ Clone XMG1.2 Cat# 505810 (1:100 dilution), IFNAR1 Clone MAR1–5A3 Cat# 127314 (1:100 dilution), Ly-6c Clone HK1.4 Cat# 128007 (1:100 dilution), PD-1 Clone 29F.1A12 Cat# 135206, 135209 (1:100 dilution), Sca-1 Clone D7 Cat# 108108, 108128 (1:100 dilution), TCR Vα2 Clone B20.1 Cat# 127814, 127806 (1:100 dilution), TCR Vβ5 Clone MR9–4 Cat# 139506 (1:100 dilution), TCR Vβ8 Clone KJ16–133.18 Cat# 118406 (1:100 dilution), TER-119 Clone TER-119 Cat# 116208 (1:100 dilution), TCF1 Clone 7F11A10 Cat# 655208 (1:100 dilution), Tim-3 Clone RMT3–23 Cat# 119703, 119723 (1:100 dilution), TNF Clone MP6-XT22 Cat# 506322 (1:100 dilution), TruStain fcX Clone 93 Cat# 101320 (1:50 dilution), Rat IgG2a κ Isotype Clone RTK2758 Cat# 400508 (1:100 dilution), Rat IgG2b κ Isotype Clone RTK4530 Cat# 400612 (1:100 dilution), Streptavidin Cat# 405225 (1:400)); Invitrogen anti-GFP (Polyclonal) Cat#A21311 (1:350 dilution); Thermo Fisher Scientific (Foxp3 Clone FJK-16s Cat# 48-5773-82 (1:50 dilution), Near-IR Fixable Live/Dead Cat# L34976 (1:500 dilution)); and Cell Signaling Technology pSTAT1 Clone 58D6 Cat# 9174 (1:100 dilution).
Adoptive T cell transfer
Spleens were isolated from chimeric mice (> 8 week post reconstitution) and naïve CD8+ T cells were purified using a naïve CD8+ MACS kit (Miltenyi Cat# 130-096-543, >95% purity). Cells were stained with lineage-specific antibodies (TER-119, B220, and Gr-1) and 7-Aminoactinomycin D (7-AAD) and then sorted (Lineage–, 7-AAD–, Vex+ cells). For LCMV co-transfer studies, cells were transferred (500:500 mix) to recipient mice on day −1, and mice were infected with LCMV Clone 13 (as below) on day 0. For tumor co-transfer studies, cells were transferred (1000:1000 mix) to recipient mice on day −1, and mice were injected with MC38-OVA or B16-OVA (as below) on day 0. For transfer studies to determine effects on tumor growth, either 5,000 control or Ptpn2-deleted CD8+ T cells were transferred alone into wild-type recipients on day −1, and mice were injected with B16-OVA (as below) on day 0.
LCMV production and plaque assay
LCMV Clone 13 virus was produced by infecting BHK-21 cells with an LCMV Clone 13 stock at an MOI of 0.01 and harvesting viral supernatants 48 hours later. Viral titers were determined by plating diluted viral stocks on Vero cells with an agarose overlay. Four days later the Vero cells were stained with neutral red dye, and plaques quantified 14 hours later.
LCMV infection and analysis
Mice were infected with 4×106 PFU LCMV Clone 13 i.v., monitored for weight loss, and bled or sacrificed at day 8, 15, 22 or 30 post infection for flow cytometry analyses. To deplete CD4+ T cells, mice were injected i.p. with 200 μg αCD4 (GK1.5, BioXCell Cat# BE0003–1) on days −1 and 1 (relative to LCMV Clone 13 injection on day 0). To block IFNAR1, mice were injected i.p. with 1 mg αIFNAR (MAR1–5A3, BioXCell Cat# BE0241) or isotype (MOPC-21, BioXCell Cat# BE0083) on days −1 and 1 (relative to LCMV Clone 13 injection on day 0). To block IFN-γ, mice were injected i.p. daily with 250 μg αIFN-γ (XMG1.2 BioXCell Cat# BE0055) or isotype (HRPN BioXCell Cat# BE0088) on days 0–3 (relative to LCMV Clone 13 injection on day 0).
Isolation of liver-infiltrating lymphocytes
Livers were excised, mechanically minced, and filtered to single-cell suspension. Lymphocytes were enriched on a Percoll gradient (VWR Cat# 89428–524).
Tumor injection
Mice were anesthetized with 2.5% 2,2,2-Tribromoethanol (Avertin, Sigma Aldrich Cat# T48402–25G) and injected in the flank subcutaneously with 2×106 MC38-OVA tumor cells (co-transfer experiments), or 1×106 MC38-WT (chimera primary challenge). For secondary rechallenge experiments chimeras were allowed to rest for 60 days post primary tumor clearance and then were rechallenged with 5×106 MC38-WT tumor cells on the opposite flank subcutaneously (as above). For B16.F10 experiments, mice were challenged with 1×106 B16.F10 subcutaneously (day 0), followed by injections on the opposite flank of 1×106 irradiated B16/GM-CSF cells (days 1 and 4). Mice were then treated i.p. with 100 μg rat monoclonal αPD-1 (clone 29F.1A12) on days 12, 14, 16, 18, 20, 22, 24, and 26. For B16-OVA experiments mice were challenged with 2.5×105 B16-OVA tumor cells subcutaneously. Tumors were measured every 2–3 days once palpable using a caliper. Tumor volume was determined by the volume formula for an ellipsoid: 1/2 × D× d2 where D is the longer diameter, and d is the shorter diameter. Mice were sacrificed when tumors ulcerated, reached 2 cm3, or upon a body condition score >2. To deplete CD8+ T cells, mice were injected i.p. with 100 μg αCD8β (53–5.8 BioXCell Cat# BE0223) or isotype (HRPN BioXCell Cat# BE0088) on days −3, 0, 3, 6, 9 and 200 μg αCD8α (2.43 BioXCell Cat# BE0061) or isotype (LTF-2 BioXCell Cat# BE0090) on days 12, 15, 18, 21, 24 (relative to MC38 tumor injection on day 0). For depletion of CD8+ T cells in secondary rechallenge experiments, mice were injected i.p. with 100 μg αCD8α (53–6.7 BioXCell Cat# BE0004–1) or isotype (2A3 BioXCell Cat# BE0089) on days −3, 0, 3, 6, 9 (relative to MC38 rechallenge on day 0).
Tumor infiltrating lymphocyte isolation
Tumors were excised and mechanically minced. Tumors were then incubated in collagenase for 10 minutes at 37°C. Lymphocytes were enriched using an Optiprep gradient (Sigma-Aldrich Cat# D1556).
Monitoring of T cell responses in the blood
To monitor the stability of Vex transduction, mice were bled via the tail vein. Blood was then lysed twice using ACK lysis buffer, stained, and analyzed by flow cytometry. For monitoring of T cell responses to LCMV infection or tumor, mice were anesthetized with Isoflurane (Henry Schein Cat# 029404), retro-orbitally bled, and lymphocytes were isolated by centrifugation at 400g on a Histopaque-1083 gradient (Sigma-Aldrich Cat# 10831–6X100ML), and stained for flow cytometry.
In vitro T cell differentiation assay using naive CD8+ T cells
Naive CD8+ T cells were obtained from spleens of control and Ptpn2-deleted chimeric mice as described above. Naive CD8+ T cells were then activated on plate-bound αCD3 (OKT-3, BioXCell Cat# BE0001) (5 μg/ml) with or without αCD28 (37.51, BioXCell Cat# BE00015) (5 μg/ml), and supplemented with 200 U/mL IL-2 (R&D Systems Cat# 202-IL), 1000 U/mL IFN-α (PBL Assay Science Cat# 12105–1), 50 μg/mL αIL-2 (S4B6–1, JES61A12, BioXCell Cat# BE0043–1, BE0043), or 50 μg/mL αIFNAR blocking antibodies for 72 hours. For supernatant transfer experiments, supernatant was isolated from wells of control or Ptpn2-deleted stimulated cells (72 hours post stimulation as above). The supernatant was added to wild-type naive CD8+ T cells activated on plate-bound αCD3 (5 μg/ml) and αCD28 (5 μg/ml) and supplemented with 200 U/mL IL-2, 1000 U/mL IFN-α, 50 μg/mL αIL-2, or 50 μg/mL αIFNAR blocking antibodies for 72 hours.
In vitro cytotoxicity assay using antigen-experienced Tim-3+ CD8+ T cells
Naive P14 control and Ptpn2-deleted CD8+ T cells were co-transferred to wild-type recipient mice (500:500 mix) on day −1, and the mice were infected with LCMV Clone 13 on day 0 (as above). CD4+ T cells were depleted on days −1 and 1 (as above). Splenocytes were isolated on day 8 post infection and enriched for CD8+ T cells using the CD8α+ MACS kit (Miltenyi Cat# 130-117-044). Vex+ Tim-3+ CD8+ T cells (Tim-3+) or Vex+ CD8+ T cells (Slamf6+ Tim-3+) were sorted from both control and Ptpn2-deleted cells. MC38-GP33–41-GFP tumor cells were seeded with 20 ng/mL IFN-γ, one day prior to plating of the T cells. Tim-3+ or Slamf6+ Tim-3+ CD8+ T cells were then co-cultured with MC38-GP33–41-GFP tumor cells at a 2:1 effector to target ratio for 16 hours. After co-culture, cells were trypsinized, stained, and analyzed by flow cytometry to determine the number of remaining live tumor cells. Killing percentage was calculated by the formula:
100% × [1 – (# live tumor cells in well with T cells / # live tumor cells in well with no T cells)].
In vitro T cell conversion assay using antigen-experienced Slamf6+ CD8+ T cells
Naive P14 control and Ptpn2-deleted CD8+ T cells were co-transferred to wild-type recipient mice (500:500 mix) on day −1, and the mice were infected with LCMV Clone 13 on day 0 (as above). CD4+ T cells were depleted on days −1 and 1 (as above). Splenic control and Ptpn2-deleted CD8+ T cells were isolated from co-transfer recipient mice, and sorted for Slamf6+ Vex+ CD8+ T cells. 50,000 Slamf6+ Vex+ CD8+ T cells were stimulated on plate-bound αCD3 (5 μg/ml) and αCD28 (5 μg/ml), and were supplemented with 200 U/mL IL-2 and 1000 U/mL IFN-α for 96 hours. Cells were then stained and differentiation was assessed by flow cytometry.
In vitro CTV proliferation assay using antigen-experienced Slamf6+ and Tim-3+ CD8+ T cells
Naive P14 control and Ptpn2-deleted CD8+ T cells were co-transferred to wild-type recipient mice (500:500 mix) on day −1, and the mice were infected with LCMV Clone 13 on day 0 (as above). CD4+ T cells were depleted on days −1 and 1 (as above). Splenic control and Ptpn2-deleted CD8+ T cells were isolated from co-transfer recipient mice, and sorted for Slamf6+ Vex+ CD8+ T cells and Tim-3+ Vex+ CD8+ T cells. These cells were labeled with 1 μM Cell Trace Violet Proliferation dye (Thermo Fisher Scientific Cat# C34557) for 20 minutes at 37°C. 50,000 Tim-3+ Vex+ CD8+ T cells were stimulated on plate-bound αCD3 (0.5 μg/ml) and αCD28 (0.5 μg/ml) for 72 hours. Cells were then stained and proliferation was assessed by flow cytometry.
BrdU incorporation and detection
Mice were injected with 1 mg of BrdU i.p. 16 hours prior to sacrifice and analysis. Cells were processed and stained using the BrdU flow kit (BD Biosciences Cat# 552598).
Ex vivo restimulation and flow cytometry of phosphorylated proteins
Naive P14 control and Ptpn2-deleted CD8+ T cells were co-transferred to wild-type recipient mice (500:500 mix) on day −1, and the mice were infected with LCMV Clone 13 on day 0 (as above). CD4+ T cells were depleted on days −1 and 1 (as above). Splenocytes were isolated on day 6 post infection and enriched for CD8+ T cells using CD8α+ MACS kit. CD8+ T cells were stimulated on plate-bound αCD3 (10 μg/ml) and αCD28 (10 μg/ml), and with 1000 U/mL IFN-α for 0, 2, 5, 10, 15 and 30 minutes at 37°C. After stimulation, cells were pelleted at 800g, fixed in 2% methanol free-formaldehyde (Cell Signaling Technology Cat# 12606) and permeabilized with ice-cold 90% methanol for 20 minutes on ice. Cells were then washed with MACS buffer and stained with pSTAT1 antibody.
Quantification of serum cytokines
Serum was collected from control or Ptpn2-deleted chimeric mice at week 12 post reconstitution. Inflammatory cytokines (IL-6, IL-10, MCP-1, IFN-γ, TNF and IL-12p70) were measured using a BD Cytometric Bead Assay - Mouse Inflammation Kit (BD Biosciences Cat# 552364).
ATAC-seq library preparation and analysis
50,000 co-transferred control or Ptpn2-deleted Slamf6+ or Tim-3+ P14 T cells per replicate were sorted from spleens of day 8 LCMV Clone 13 infected mice into PBS with 10% FBS. Pelleted cells were incubated in 50μl of reaction mix (containing 2× Tagmentation DNA buffer, Tn5 enzyme, 2% digitonin in nuclease-free water) as previously described50. The transposase reaction was performed at 37°C for 30 minutes with agitation at 300 RPM. DNA was then purified using a MinElute Reaction Cleanup kit (QIAgen Cat# 28206). A post PCR cleanup was performed using Agencourt AMPure XP beads (Beckman Coulter Cat# A63880) and library quality was verified using Tapestation analysis. Samples were sequenced on an Illumina NextSeq500 sequencer using paired-end 37bp reads.
Quality trimming and primer removal within raw fastq files were done with Trimmomatic 0.33 using the following parameters: LEADING:15 TRAILING:15 SLIDINGWINDOW:4:15 MINLEN:36. Trimmed reads were aligned to mm9 with bowtie2.2.4 using a maximum insert size of 1000. Aligned bams were sorted, duplicates marked, and reads mapping to the blacklist region removed51. Peak-calling using MACS 2.1.1 was performed on merged bam files (samtools 1.3) from biological replicates using a q-value threshold of 0.001. Consensus peaks from all biological conditions were merged to create a single peak universe. Cut sites were extracted from each biological replicate and the number of cuts within each peak region were quantified to generate a raw count matrix. DESeq2 was used to normalize the counts matrix and perform differential accessibility analysis between all relevant comparisons. Tracks were visualized using Integrative Genomics Viewer 2.3.77 (Broad Institute).
Bulk RNA-seq analysis of T cells
Day 7 or 8 post tumor or virus injection respectively, co-transferred T cells were isolated from the tumor or spleen (LCMV) (as above) and replicates of 500 cells were sorted into 25μL of buffer RLT (QIAgen Cat# 79216) + 1% beta-mercaptoethanol v/v. After flash-freezing on dry ice and storage at −80°C, lysates were converted to cDNA following capture with Agencourt RNAClean beads (Beckman Coulter Cat# A63987) using the SmartSeq2 protocol as previously described52. The cDNA was amplified using 16 PCR enrichment cycles prior to quantification and dual-index barcoding with the Illumina Nextera XT kit. The libraries were enriched with 12 cycles of PCR, then combined in equal volumes prior to final bead clean-up and sequencing on a NextSeq500 by 37 bp paired-end reads. After demultiplexing, low quality base-reads were trimmed with Trimmomatic53 using the following parameters: LEADING:15, TRAILING:15, SLIDINGWINDOW:4:15, MINLEN:16. Trimmed reads were then aligned to the mm10 mouse genome using Bowtie2. HTSeq was used to map aligned reads to genes and to generate a gene count matrix. Normalized counts and differential expression analysis were performed using the DESeq2 R package. We performed gene set enrichment analysis as previously described54 using signatures from the Hallmark database, the MSigDB collection, and from our prior analysis of exhausted CD8+ T cells from the spleens of LCMV Clone 13 infected mice or from B16-OVA tumors4.
Single-cell RNA-seq library preparation and analysis
Co-transferred control or Ptpn2-deleted P14 CD8+ T cells were sorted from spleens of day 30 LCMV Clone 13 infected, CD4-depleted mice based on the markers CD8β, CD45.1, CD45.2, Vex, and fixable live/dead. Cells were counted and loaded onto the Chromium Controller (10× Genomics) for a target recovery of 5,000 single cells. Samples were processed per the manufacturer’s protocol and sequenced on an Illumina NextSeq500 sequencer using a 75bp kit with paired-end reads. The Cell Ranger analysis pipeline version 1.2 was used for sample demultiplexing, barcode processing, alignment, filtering, UMI counting, and aggregation of sequencing runs. The R Seurat package55 was used for downstream analyses.
For each cell, two quality control metrics were calculated: (1) the total number of genes detected and (2) the proportion of UMIs contributed by mitochondrially encoded transcripts. Cells were excluded from downstream analysis if fewer than 200 or greater than 2500 genes were detected and if mitochondrially encoded transcripts constituted greater than 5% of the total library, yielding an expression matrix of 7,027 cells by 13,133 genes. Each gene expression measurement was normalized by total expression within the corresponding cell and multiplied by a scaling factor of 10,000. Mean and dispersion values were calculated for each gene across all cells; 1,829 genes classified as highly variable. Highly variable genes were used for principal components analysis (PCA). Principal components were determined to be significant (P< 0.001) using the jackstraw method and tSNE was performed on these significant PCs (PCs 1–17) using default parameters for 1,000 iterations for visualization in two dimensions. Unsupervised clustering was performed using a shared nearest neighbor modularity optimization-based algorithm56. Single-cell signature scoring using FastProject57 was performed with the Hallmark database from MSigDB and using signatures of the subpopulations derived from our prior analysis of exhausted CD8+ splenocytes from LCMV Clone 13 infected mice4. Differential gene expression and signature enrichment analysis was performed using a two-sided Wilcoxon rank sum test. To determine the relative proportion of Ptpn2-deleted cells within each cluster, a two-sided binomial test was performed against the proportion of Ptpn2-deleted cells within the total dataset.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 software or R 3.3.1. Data were considered statistically significant with p values < 0.05 by two-sided paired Student’s t test for comparing two groups in co-transfer experiments, two-sided unpaired Student’s t test for comparing two groups, one-way ANOVA for single comparisons with groups greater than two, two-way ANOVA for repeated measures comparisons or for multiple comparisons within groups, and a two-sided log-rank Mantel-Cox test for survival analysis. For GSEA analysis of RNA-seq data a two-sided Kolmogorov-Smirnov test was used. For analysis of single-cell RNA-seq data a two-sided Wilcoxon rank sum test was used for signature enrichments and a two-sided binomial test was used to determine proportional differences of control or Ptpn2-deleted cells in the clusters. For ATAC-seq analysis a hypergeometric test was used. Please see the Life Sciences Reporting Summary for additional details.
Data availability statement
The data and materials that support the findings of this study are available from the corresponding author upon reasonable request. All sequencing data from this study has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession code GSE134413.
Supplementary Material
Acknowledgements
We thank N. Collins (Dana Farber Cancer Institute), C. Kadoch (Dana Farber Cancer Institute), D. Vignali (University of Pittsburgh), E. John Wherry (University of Pennsylvania), and G. Dranoff (Novartis Institutes for BioMedical Research) for sharing cancer cell lines; F. Zhang (Broad Institute of MIT and Harvard) for sharing loxP-STOP-loxP-Cas9 mice. This work was supported by funding from T32CA207021 to M.W.L., the 2016 AACR-Bristol-Myers Squibb Fellowship in Translational Immuno-oncology grant number 16-40-15-MILL, National Center for Advancing Translational Sciences/National Institutes of Health Award KL2 TR002542, and the Jane C. Wright, MD, Endowed Young Investigator Award from ASCO to B.C.M., P50CA101942 to G.J.F., U19AI133524 to A.H.S and W.N.H., and U54 CA224088, R01 CA229851, and P01 AI108545 to A.H.S.
Footnotes
Competing Financial Interests Statement
A.H.S. has patents on the PD-1 pathway licensed by Roche/Genentech and Novartis, consults for Novartis, is on the scientific advisory boards for Surface Oncology, Sqz Biotech, Elstar Therapeutics, Elpiscience, Selecta, and Monopteros, and has research funding from Merck, Novartis, Roche, Ipsen, UCB and Quark Ventures. W.N.H. has a patent application on T cell exhaustion-specific enhancers held by Dana-Farber Cancer Institute and now is employed by Merck. W.N.H. is also a founder of Arsenal Biosciences. A.H.S. and W.N.H. have a patent application on PTPN2 as a therapeutic target held/submitted by Dana-Farber Cancer Institute. G.J.F. has a consulting or advisory role for Novartis, Lilly, Roche/Genentech, Bristol-Myers Squibb, Bethyl Laboratories, Xios Therapeutics, Quiet Therapeutics, Seattle Genetics; patents, royalties, or other intellectual property from Novartis, Roche/Genentech, Bristol-Myers Squibb/Medarex, Amplimmune/Astrazeneca, Merck, EMD Serono, Boehringer Ingelheim and research funding from Bristol-Myers Squibb. The remaining authors declare no competing interests.
References
- 1.Zajac AJ et al. Viral Immune Evasion Due to Persistence of Activated T Cells Without Effector Function. The Journal of Experimental Medicine 188, 2205–2213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wherry EJ T cell exhaustion. Nature Immunology 12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Baitsch L et al. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest 121, 2350–2360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller BC et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R & Ahmed R Viral Persistence Alters CD8 T-Cell Immunodominance and Tissue Distribution and Results in Distinct Stages of Functional Impairment. Journal of Virology 77, 4911–4927 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day CL et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Wherry EJ et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Angelosanto JM, Blackburn SD, Crawford A & Wherry EJ Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J Virol 86, 8161–8170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doering TA et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 37, 1130–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen DR et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauken KE et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin H & Wherry EJ CD8 T cell dysfunction during chronic viral infection. Curr Opin Immunol 19, 408–415 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Utzschneider DT et al. High antigen levels induce an exhausted phenotype in a chronic infection without impairing T cell expansion and survival. J Exp Med 213, 1819–1834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornberg M et al. Clonal exhaustion as a mechanism to protect against severe immunopathology and death from an overwhelming CD8 T cell response. Front Immunol 4, 475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paley MA et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He R et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Im SJ et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu T et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Science Immunology 1, eaai8593–eaai8593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan Q et al. The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nat Immunol 18, 931–939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snell LM et al. CD8(+) T Cell Priming in Established Chronic Viral Infection Preferentially Directs Differentiation of Memory-like Cells for Sustained Immunity. Immunity 49, 678–694 e675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danilo M, Chennupati V, Silva JG, Siegert S & Held W Suppression of Tcf1 by Inflammatory Cytokines Facilitates Effector CD8 T Cell Differentiation. Cell Rep 22, 2107–2117 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Philip M et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brummelman J et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J Exp Med 215, 2520–2535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sade-Feldman M et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013.e1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thommen DS et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 24, 994–1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui I et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211 e110 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Kurtulus S et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(−)CD8(+) Tumor-Infiltrating T Cells. Immunity 50, 181–194 e186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaFleur MW et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat Commun 10, 1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinkman EK, Chen T, Amendola M & van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiede F et al. T cell protein tyrosine phosphatase attenuates T cell signaling to maintain tolerance in mice. J Clin Invest 121, 4758–4774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtsinger JM, Valenzuela JO, Agarwal P, Lins D & Mescher MF Cutting Edge: Type I IFNs Provide a Third Signal to CD8 T Cells to Stimulate Clonal Expansion and Differentiation. The Journal of Immunology 174, 4465–4469 (2005). [DOI] [PubMed] [Google Scholar]
- 32.ten Hoeve J et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol 22, 5662–5668 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, & McGlade CJ The T Cell Protein Tyrosine Phosphatase Is a Negative Regulator of Janus Family Kinases 1 and 3. Current Biology 12, 446–453 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Doody KM, Bourdeau A, & Tremblay ML T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling-lessons from the knockout mouse model and implications in human disease. Immunological Reviews 228, 325–341 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Spalinger MR et al. PTPN2 Regulates Inflammasome Activation and Controls Onset of Intestinal Inflammation and Colon Cancer. Cell Rep 22, 1835–1848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensson MN et al. Reduced expression of phosphatase PTPN2 promotes pathogenic conversion of Tregs in autoimmunity. J Clin Invest 129, 1193–1210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dranoff G et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A 90, 3539–3543 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleppe M et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet 42, 530–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson EB et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teijaro JR et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starbeck-Miller GR, Xue HH & Harty JT IL-12 and type I interferon prolong the division of activated CD8 T cells by maintaining high-affinity IL-2 signaling in vivo. J Exp Med 211, 105–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto M, Im SJ, Araki K & Ahmed R Cytokine-Mediated Regulation of CD8 T-Cell Responses During Acute and Chronic Viral Infection. Cold Spring Harb Perspect Biol 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiede F, Ziegler A, Zehn D & Tiganis T PTPN2 restrains CD8(+) T cell responses after antigen cross-presentation for the maintenance of peripheral tolerance in mice. J Autoimmun 53, 105–114 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Marabelle A et al. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J Clin Invest 123, 2447–2463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamarin D et al. Localized Oncolytic Virotherapy Overcomes Systemic Tumor Resistance to Immune Checkpoint Blockade Immunotherapy. Science Translational Medicine 6, 226ra232–226ra232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manguso RT et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 47.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juneja VR et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med 214, 895–904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trombetta JJ et al. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr Protoc Mol Biol 107, 4 22 21–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waltman L & van Eck NJ A smart local moving algorithm for large-scale modularity-based community detection. The European Physical Journal B 86 (2013). [Google Scholar]
- 57.DeTomaso D & Yosef N FastProject: a tool for low-dimensional analysis of single-cell RNA-Seq data. BMC Bioinformatics 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials that support the findings of this study are available from the corresponding author upon reasonable request. All sequencing data from this study has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series accession code GSE134413.