SUMMARY
The proteolytic arm of the protein homeostasis network is maintained by both the ubiquitin-proteasome system (UPS) and autophagy. A well-balanced crosstalk between the two catabolic pathways ensures energy efficient maintenance of cellular function. Our current understanding of the crosstalk between the UPS and autophagy is centered around substrate ubiquitination. Herein we report an additional method of crosstalk involving ubiquitin-independent 20S proteasome regulation of autophagosome-lysosome fusion. We found that enhancement of 20S proteasome activity increased the degradation of the disordered soluble N-ethylmaleimide-sensitive factor activating protein receptor (SNARE) proteins, synaptosomal-associated protein 29 (SNAP29) and syntaxin 17 (STX17), but not vesicle-associated membrane protein 8 (VAMP8). This resulted in a reduction of autophagosome-lysosome fusion, which was ameliorated upon overexpression of both SNAP29 and STX17. In all, we herein present a mechanism of crosstalk between the proteasome and autophagy pathway that is regulated by ubiquitin-independent 20S proteasome-mediated degradation of SNAP29 and STX17.
Keywords: autophagy, proteasome, proteasome activation, autophagosome-lysosome fusion, SNARE proteins, SNAP29, Syntaxin17, UPS, ubiquitin-independent, 20S proteasome
INTRODUCTION
The ubiquitin-proteasome system (UPS) and the autophagy pathway are two complementary proteolytic systems that have evolved to degrade redundant, damaged and misfolded proteins, and worn-out organelles (Cohen-Kaplan et al., 2016). The UPS targets the degradation of mostly soluble, short-lived nuclear and cytosolic proteins, whereas the autophagy pathway enables cells to degrade protein complexes, protein aggregates and cellular organelles in a lysosome-dependent mechanism (Dikic, 2017).
In the UPS, the 26S proteasome is the main protease responsible for the degradation of proteins. The 26S proteasome is capped on each end with a 19S regulatory cap, which recognizes, unfolds and transports substrates into the cylindrical 20S core particle for degradation (Glickman and Ciechanover, 2002, Finley et al., 2016). While the 26S proteasome primarily degrades ubiquitinated substrates, the uncapped 20S core particle is limited to the degradation of unfolded proteins (i.e. intrinsically disordered and oxidatively damaged proteins) in a ubiquitin- and ATP- independent manner (Ben-Nissan and Sharon, 2014). The assembly and disassembly of the 26S and 20S proteasome complexes therefore exist as a dynamic equilibrium to address specific proteolytic needs (Bhattacharyya et al., 2014).
Autophagy is an intracellular lysosome-dependent degradation pathway characterized by the formation of an autophagosome (Kaur and Debnath, 2015, Glick et al., 2010). In the initial step, an isolated membrane forms through specific autophagy effectors such as the microtubules-associated protein light chain 3 (LC3) that engulfs its targeted protein aggregates or damaged organelles. Elongation and closure of the crescent-shaped structure (phagophore) forms the autophagosome. Fusion of the autophagosome with the lysosome is regulated by various SNARE proteins (Itakura et al., 2012), to form the autolysosome. Upon fusion, several hydrolases act as endopeptidases under the highly acidic condition (pH 4.5–5.0) of the autolysosome and degrade the cargo (Mindell, 2012).
At the protein level, autophagy appears to be induced to compensate for cellular stresses not addressed by proteasomal clearance (Cohen-Kaplan et al., 2016). A well-balanced crosstalk between the two catabolic pathways ensures energy efficient maintenance of cellular function and the cell’s amino acid reserve(Kwon and Ciechanover, 2017, Ji and Kwon, 2017). Dysregulation of these complementary proteolytic systems has been implicated in many human diseases, including cancer and neurodegenerative disorders (Limanaqi et al., 2018, Chung et al., 2018, Mooneyham and Bazzaro, 2017, Boland et al., 2018, Schmidt and Finley, 2014). A more complete understanding of the crosstalk between these two pathways may reveal new targets that can be explored for the treatment of proteostasis diseases (Onorati et al., 2018).
Our current understanding of the crosstalk between the UPS and autophagy is centered around substrate ubiquitination (Ji and Kwon, 2017, Peng et al., 2017). Shuttle proteins recognize ubiquitinated substrates and direct them towards either or both proteolytic pathways. For example, lysine-48 (K-48)-linked polyubiquitinated substrates are preferentially directed to the 26S proteasome by UB-associated (UBA)-Ub-Like (UBL) shuttling proteins, while p62/SQSTM-1 and neighbor of BRCA1 (NBR1) shuttle K-63-linked polyubiquitinated substrates to autophagic vacuoles(Cohen-Kaplan et al., 2016). Crosstalk is evident when improper processing of ubiquitin substrates by the 26S proteasome triggers an autophagic response. For example, accumulation of ubiquitinated proteins, following proteasome inhibition, results in ER stress, unfolded protein response (UPR), p53 and HDAC6 signaling, with ensuing activation of downstream pathways and upregulation of autophagy (ATG) genes (Zhu et al., 2010, Lee et al., 2003, Vousden and Ryan, 2009, Pandey et al., 2007). Induced clearance of ubiquitinated substrates, by inhibition of the de-ubiquitinase USP14, triggers conflicting outcomes in autophagic flux (Kim et al., 2018, Xu et al., 2016, Chakraborty et al., 2018).
The role of the 20S proteasome in autophagy regulation is largely unexplored, yet support for its relevance exists (Gao et al., 2010). In the present study, we report a previously unrecognized mechanism of cross-talk that implicates the 20S proteasome as a regulator of the autophagic SNARE proteins, SNAP29 and STX17. To elucidate the role of the 20S in autophagy regulation, we used the small molecule probe, TCH-165. TCH-165 is a 20S activator that enhances 20S-mediated proteolysis (Njomen et al., 2018). TCH-165 was also shown to regulate the equilibrium between 26S and 20S proteasome sub-complexes, in favor of the 20S, in a manner that does not involve induction of oxidative stress (Njomen et al., 2018). This regulation only significantly affects ubiquitin-dependent proteolysis at high concentration (≥30 μM) where the 26S proteasome is completely depleted. At ≤10 μM, residual 26S proteasome is maintained and the enhanced activity of the 20S proteasome could be monitored without significant perturbation of the ubiquitin-dependent pathway.
The enhanced 20S proteolytic activity allowed us to discover 20S-mediated regulatory events in cells and provide insight in the role of the 20S proteasome on proteasome-autophagy crosstalk. Herein, we report for the that the SNARE proteins SNAP29 and STX17 are degraded through a ubiquitin-independent pathway that is primarily mediated by the 20S proteasome. Enhancement of 20S proteasome activity, by TCH-165, decreased SNAP29 and STX17 protein levels and reduced autophagosome-lysosome fusion. Overexpression of both SNAP29 and STX17, but not the individual proteins ameliorated the effect of TCH-165-enhanced 20S proteolysis on autophagy. Collectively, our data suggest that the 20S proteasome regulation of SNAP29 and STX17 is a previously unrecognized mechanism of proteasome - autophagy crosstalk.
RESULTS
20S proteasome enhancer induces the accumulation of autophagic vacuoles
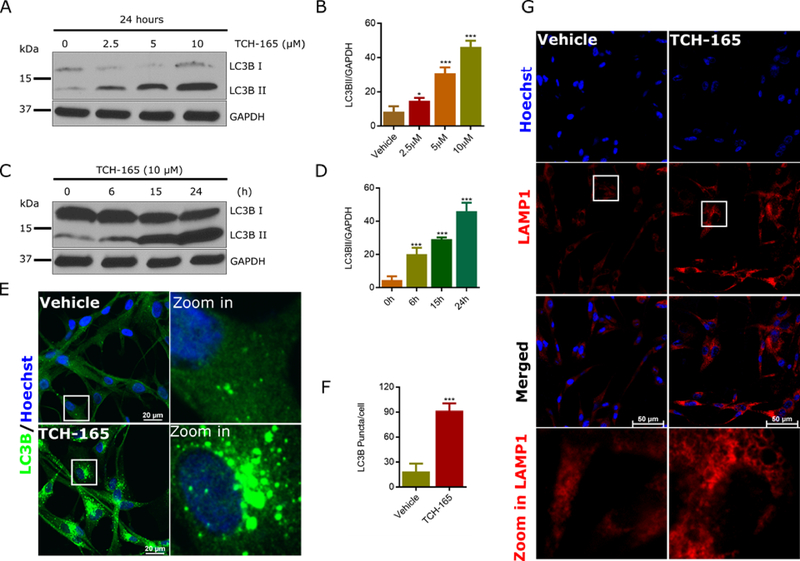
During autophagosome formation, microtubule-associated protein 1 light chain 3 (LC3B-I) is converted to LC3B-II via the conjugation of phosphatidylethanolamine (PE) to the C-terminal glycine of LC3B-I. LC3-II is then recruited to autophagosomal membranes and degraded upon autolysosome formation. Thus, the turnover of the autophagosomal substrate LC3B-II reflects autophagic activity (Kaur and Debnath, 2015, Glick et al., 2010). To elucidate the effect of proteasome activation on autophagy flux, glioblastoma cells (U-87 MG)(Jiang et al., 2009) were treated with the 20S proteasome enhancer, TCH-165, and the level of LC3B-II quantified by immunoblot and confocal microscopy. TCH-165 significantly increases the levels of LC3B-II in a concentration (Fig. 1A and 1B) and time (Fig. 1C and 1D) dependent manner. Furthermore, LC3B-I displays a diffuse staining pattern within the cytoplasm, while LC3B-II appears as small punctae in autophagic vacuoles (AVs)(Klionsky et al., 2016). Confocal immunofluorescence with anti-LC3B further confirmed the accumulation of LC3B positive vacuoles in cells treated with TCH-165 (10 μM) for 16h (Fig. 1E and 1F). Furthermore, staining the cells with antibody against lysosome associated membrane protein 1 (LAMP 1) showed increase in lysosome dilation (Fig. 1G).
Figure 1: TCH-165 induces the accumulation of LC3B II and autophagic vacuoles in U-87 MG cells:
(A) U-87 MG cells were treated with different concentrations of TCH-165 and immunoblotted with LC3B-specific antibody. (B) Image J quantification of A. (C) U-87 MG cells were treated with TCH-165 (10 μM) for 0, 6, 15, and 24h. Cell lysates were immunoblotted with LC3B-specific antibody and anti-GAPDH as a loading control. (D) Image J quantification of (C). (E) Confocal immunofluorescent analysis of U-87 MG cells, vehicle-treated (16h, top) or TCH-165-treated (10 μM, 16h; bottom) using rabbit anti-LC3B /Alexa Fluor 488 secondary (green). Blue = Hoechst (fluorescent DNA dye). (F) Mean LC3B punctae/cell for U-87 MG cells treated with vehicle or TCH-165 (E) was quantified by image J. (G) Confocal immunofluorescent analysis of U-87 MG cells, vehicle-treated (24h, left) or TCH-165-treated (10 μM, 24h; right) using rabbit anti-LAMP1 antibody/Alexa Fluor 488 secondary (pseudo red). Blue = Hoechst. All statistical analyses were performed on densitometry data (image J) of individual experiments. Data are graphed as mean ± SD (n=4 for B and D, n= 3 for F) and were analyzed by One-Way ANOVA with Bonferroni’s multiple comparison test (B and D or by unpaired student t-test (F) (ns=not significant, *p<0.05, **p<0.01, ***p<0.001).
20S proteasome enhancer inhibits autophagic flux
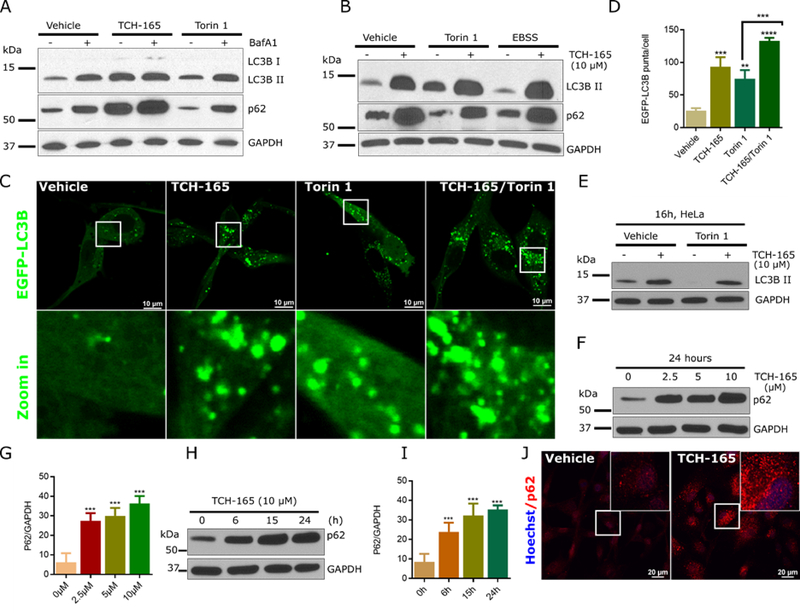
Increased levels of LC3B-II and accumulation of AVs could result from increased upstream autophagosome formation (activation) or impaired downstream degradation of basal level autophagosome (inhibition)(Mizushima et al., 2010). To distinguish between these two scenarios, we measured LC3B-II accumulation in the presence of the late stage autophagy inhibitor, bafilomycin A1 (BafA1, a V-ATPase inhibitor) (Mauvezin et al., 2015). Treatment with BafA1 or TCH-165 led to a significant increase in LC3B-II (Fig. 2A). As anticipated, the combination of BafA1 and autophagy activator torin1 (Zhao et al., 2015) increases LC3B-II levels (Fig. 2A, last two lanes vs. lane 1). However, treatment with TCH-165 for 16h followed by BafA1 for 4h did not increase LC3B-II levels above that induced by TCH-165 alone (Fig. 2A and S1), suggesting that TCH-165 inhibits autophagic flux.
Figure 2: TCH-165 inhibits autophagic flux in different cell lines:
(A) U-87 MG cells were treated with either vehicle, torin1 (500 nM) or TCH-165 (10 μM) for 16h, followed by treatment with bafilomycin A1 (100 nM) for 4h. Cell lysates were immunoblotted for LC3B, p62 and GAPDH. (B) U-87 MG cells were pre-treated with either vehicle or torin1 in complete medium or with vehicle in Earle’s Balanced Salt Solution, EBSS (starved), for 4h. TCH-165 (10 μM) was then added (+) and cells cultured for an additional 16h. Cell lysates were immunoblotted for LC3B, p62 and GAPDH. (C) Confocal immunofluorescence of EGFP-LC3B in U-87 MG cells; U-87 MG cells were treated with either vehicle, torin1 (200 nM), TCH-165 (10 μM) or a combination of torin 1/ TCH-165 for 16h and live-imaged with Nikon A1 confocal microscope using GFP settings. (D) Quantification of C by image J. (E) HeLa cells were treated with either vehicle, torin1 (200 nM), TCH-165 (10 μM) or combinations for 16h. Cell lysates were immunoblotted for LC3B and GAPDH. (F) U-87 MG cells were treated with different concentrations of TCH-165 and immunoblotted with p62-specific antibody. GAPDH was immunoblotted as a loading control. (G) Image J quantification of F. (H) U-87 MG cells were treated with TCH-165 (10 μM) for 0, 6, 15, and 24h. Cell lysates were immunoblotted with p62-specific antibody and anti-GAPDH as a loading control. (I) Image J quantification of H. (J) Confocal immunofluorescent analysis of U-87 MG cells, vehicle-treated (24h, left) or TCH-165-treated (10 μM, 24h; right) using mouse anti-p62 antibody/Alexa Fluor 594 secondary (red) and nuclei (blue) with Hoechst DNA dye. All statistical analyses were performed on densitometry data (image J) of three to five individual experiments. Data are graphed as mean ± SD (n=3 for D and 4 for G and I) and were analyzed by One-Way ANOVA with Bonferroni’s multiple comparison test (ns=not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). White boxes indicate zoom in area (Fig. 2C) or inserts (Fig 2J). See also figure S1 and S2.
20S proteasome enhancer inhibits late-stage autophagy
To determine whether the 20S proteasome enhancer, TCH-165, affects late stage autophagy, we evaluated the compound’s ability to inhibit torin1 or starvation (using Earle’s balanced salt solution, EBSS) induced autophagy. The combination of a late stage inhibitor and autophagy activators should significantly increase LC3B-II levels (Bjorkoy et al., 2009). As seen in Fig. 2B, TCH-165 treatment of U-87 MG cells increases the accumulation of LC3B-II in both torin1 and starvation induced autophagy. Furthermore, the combination of a late stage inhibitor and an autophagy activator should significantly increase the formation of AVs. For this study, U-87 MG cells expressing EGFP-LC3B were treated with torin1 and TCH-165 for 16h. Treatment with torin1 induced modest accumulation of AVs, while the combination of TCH-165 and torin1 significantly increased the number of AVs (Fig. 2C and 2D). This suggests that TCH-165 blocks the degradation of autophagosome induced by torin1. Similar activities were observed in HeLa (Fig. 2E), RPMI-8226 and HEK293T cells (Fig. S2A).
We evaluated the effects of TCH-165 on the autophagy substrate, p62. During autophagy, p62/SQSTM1 is incorporated into autophagosome via interaction with LC3B, where it serves as a receptor for the delivery of polyubiquitinated proteins to autophagosomes.(Pankiv et al., 2007, Johansen and Lamark, 2011). As such, its clearance also serves as a measure of autophagic flux. Treatment of U-87 MG cells with TCH-165 resulted in an increase in p62 in a concentration (Fig. 2F and 2G) and time (Fig. 2H and 2I) dependent manner. These data were further confirmed by confocal immunofluorescent imaging showing increased in p62 punctae following TCH-165 treatment (Fig. 2J). The accumulated p62 were found to be co-localized with LC3B in AVs (Fig. S2B). This further confirmed inhibition of autophagic flux in response to the 20S proteasome enhancer, TCH-165.
20S proteasome enhancer inhibits autophagosome-lysosome fusion
Increase in LC3B-II levels and accumulation of p62 in autophagic vacuoles implies that the 20S proteasome activator TCH-165 may inhibit late stage autophagy by targeting either the lysosome and/or autophagosome-lysosome fusion. To differentiate between the two plausible target sites, we used a tandem RFP-GFP-LC3B autophagy sensor to determine whether TCH-165- inhibits autophagy by interfering with autolysosome formation.
By combining an acid insensitive RFP with an acid sensitive GFP, the conversion of an autophagosome (neutral pH, indicated by yellow fluorescence) to an autolysosome (acidic pH, indicated by red fluorescence) can be visualized by monitoring the specific loss of the GFP fluorescence (increase in the ratio of RFP to GFP positive punctae) upon acidification of the autophagosome following lysosomal fusion (Barth et al., 2010). U-87 MG (Fig. 3A and 3B) or HEK293T (Fig. S3) cells transiently expressing RFP-GFP-LC3B retained similar levels of GFP and RFP positive vacuoles upon TCH-165 treatment. This suggests that TCH-165 inhibits autophagy flux by interfering with autolysosome formation, a mechanism distinct from inhibition of lysosomal hydrolases (leupeptin as positive control, Fig. S3). Consistent with torin 1 promoting autophagy flux, torin 1 treated samples had significant increase in RFP to GFP punctae, which was reduced in the presence of TCH-165 (Fig. 3B). However, GFP and RFP positive punctae could also represent de-acidified autolysosome (Homewood et al., 1972), as seen with chloroquine treatment (Fig. S3). Thus, to distinguish inhibition of autolysosome formation from lysosome deacidification, we looked for changes in lysosomal pH using the acidotropic dye, lysotracker red, in cells expressing GFP-LAMP1. Unlike BafA1, which neutralizes the lysosome and reduces lysotracker red fluorescence, TCH-165 or torin 1 did not reduce lysotracker red signal. Consistent with dilation of the lysosome seen in Fig 1G, TCH-165 treatment results in a slight increase in red fluorescence (Fig. 3C and 3D). Cathepsin D maturation is sensitive to changes in lysosomal pH that might not be detectable by lysotracker red. Accordingly, we confirmed the lack of perturbation of lysosomal pH by measuring cathepsin D and E activity in cells treated with TCH-165 for 24h. Unlike chloroquine which alters lysosomal pH, or pepstatin A which directly inhibits cathepsin activity, TCH-165 did not affect cathepsin D activity (Fig. 3E). Consistently, immunofluorescence of LC3B and LAMP1 showed lack of co-localization between the autophagosome (LC3B) and the lysosome (LAMP1) in TCH-165 samples (Fig. 3F). Collectively, our data suggest that the 20S proteasome regulates autophagy flux at the autolysosome stage, through a mechanism that does not involve neutralization of lysosomal pH.
Figure 3: TCH-165 interferes with autolysosome formation:
(A) U-87 MG cells were transduced with 30 particles per cell of tandem-RFP-GFP-LC3B and cultured for 24h. Cells were then incubated with either vehicle, TCH-165 (10 μM), torin 1 (200 nM) or a combination of TCH-165 and torin1 for an additional 24h and imaged. (B) Quantification of RFP to GFP positive punctae of A (mean ± SD, n=3). (C) U-87 MG cells were transduced with 30 particles per cell of CellLight® Lysosomes-GFP (GFP-LAMP1) and cultured for 24h. Cells were treated with either vehicle, TCH-165 (10 μM), torin 1(100 nM) or Bafilomycin A1 (50 nM) for 16h. Cells were stained with 50 nM lysotracker red and imaged. (D) U-87 MG cells were treated with either vehicle, TCH-165 (10 μM), torin 1 (100 nM) or Bafilomycin A1 (50 nM) for 16h. Cells were stained with 50 nM lysotracker red and imaged. Data are presented as mean fluorescence ± SD (n=2) quantified by image J. (E) U-87 MG or HeLa Cells were treated with either vehicle, TCH-165 (10μM), torin 1 (100 nM), chloroquine (100μM) or pepstatin A (5 μM) for 24h. Cell lysates were assayed for cathepsin D and E activity using fluorogenic peptide substrate (mean ± SD, n=3). (F) U-87 MG cells were treated with either vehicle or TCH-165 (10 μM) for 24h and immunostained with rabbit anti-LC3B specific/Alexa Fluor 488 (green) antibody, mouse anti-LAMP1 specific/Alexa Fluor 594 (red) antibody and Hoechst DNA dye (blue). White arrows indicate colocalization of AVs with the lysosome(autolysosome), only present in vehicle control. All images were taken with an upright Nikon A1 confocal microscope using a 60x Plan Apo oil objective with standard filter sets for GFP and RFP. (ns=not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). See also figure S3
The 20S agonist regulates autophagosome-lysosome fusion through enhanced degradation of SNAP29 and STX17
To elucidate the mechanism by which TCH-165 inhibits autophagic flux, we evaluated some of the SNAREs and HOP proteins involve in autophagosome-lysosome fusion to identify those that are potential 20S substrates. For this, predictor of natural disorder region (PONDR) was used to identify the degree of disorder in these proteins (Xue et al., 2010).
These predictive calculations indicated that at least three of these proteins have a minimum of 40% overall disorder, with SNAP29 (87.6%) being almost completely disordered (Fig.4A) and thus was anticipated to be a plausible 20S proteasome substrate. Therefore, the cellular degradations of these proteins were evaluated. HeLa cells were treated with vehicle or torin1, with or without TCH-165. Interestingly, TCH-165 significantly reduced the protein levels of SNAP29 and STX17, but not VAMP8. However, VAMP8 was significantly reduced in torin1 treatment, but accumulated when torin1 was combined with TCH-165 (Fig. 4B). These observations suggest that unlike SNAP29 and STX17, VAMP8 is likely an autophagy substrate. These data are consistent with previous studies demonstrating that rapamycin, an autophagy inducer, promotes VAMP8 degradation (Fu et al., 2018). Similar results were obtained in HEK293T and RPMI-8226 cells (Fig.4C), suggesting that the observed effect is not cell type dependent.
Figure 4: TCH-165 stimulates proteasomal degradation of SNAP29 and STX17:
(A) Percent disorder in autophagy fusion proteins were predicted using predictor of natural disorder region (PONDR) software(Xue et al., 2010). (B) HeLa cells were treated with either vehicle, torin1 (200 nM), TCH-165 (10 μM) or combinations for 16h. Cell lysates were immunoblotted for STX17, SNAP29, VAMP8 and GAPDH. (C) HEK293T or RPMI-8226 cells were treated with either vehicle or TCH-165 (10 μM) for 16h and whole cell lysates immunoblotted for STX17, SNAP29, VAMP8 and GAPDH. (D) U-87 MG cells were treated with cycloheximide (100 μg/mL) plus vehicle, TCH-165 (10 μM), bortezomib (BTZ; 5 μΜ) or combinations for 8h. Whole cell lysates were immunoblotted for STX17, SNAP29, p62, LC3B and GAPDH. The degradation of SNAP29 (E) and STX17 (F) were quantified with image J software. Statistical analyses were performed on four independent experiments (n=4) using unpaired t-test on GraphPad Prism7. (G) U-87 MG cells were treated with vehicle or TCH-165 (10 μM) for 8h or 24h and mRNA quantified by RT-qPCR. Data are presented as mean ± SD of two independent experiments (n=2), each ran in triplicate. See also supplemental table S1
We investigated whether the changes in STX17 and SNAP29 were occurring post-translationally and at the level of the proteasome. Protein synthesis was pulsed with cycloheximide in the presence of vehicle, TCH-165, bortezomib (BTZ; proteasome inhibitor) or a combination of TCH-165 and BTZ for 8h. Immunoblot analyses indicated a significant increase in the degradation of SNAP29 and STX17 in TCH-165 treated sample, which was blocked by BTZ (Fig. 4D, 4E and 4F). These observations suggested that the change in the protein levels of SNAP29 and STX17 were due to enhanced degradation, most likely at the level of the proteasome. To be more certain, U-87 MG cells were treated with vehicle or TCH-165 for 8h or 24h and assayed for changes in total mRNA by RT-qPCR. At 8h, no significant changes were observed in the mRNA levels of STX17, SNAP29 and VAMP8. However, at 24h, the mRNA for SNAP29 and STX17 increased by about 3-fold for TCH-165 treated cells (Fig. 4G). These increases in mRNA levels were likely a feedback response to the enhanced degradation of these proteins. The mRNA level of VAMP8 was also upregulated by 2-fold, at 24h, probably in response to accumulation of AVs. As controls, the mRNA levels of LC3B and p62 were also quantified. Both genes showed significant upregulation in mRNA at both time points (Fig. 4G). Remarkably, changes in the mRNA of LC3B was more significant at 8h than at 24h. Given this significant upregulation in p62 and LC3B mRNAs at 8h, we immunoblotted for changes in protein levels of p62 and LC3B-II, following inhibition of protein synthesis with cycloheximide (CHX). As shown in Fig. 4D, both p62 and LC3B still accumulate when protein synthesis is blocked, suggesting that the upregulation in mRNA does not significantly contribute to the changes in protein levels of p62 and LC3B-II seen with TCH-165 treatment. However, the accumulation of p62 and LC3B-II are not as robust as in the absence of CHX. This could be due to limited level of basal autophagy as a result of limited protein synthesis (Watanabe-Asano et al., 2014). The upregulation of p62 and LC3B genes could therefore be a response to the accumulation of AVs or a general response to an exogenous stressor, given that these genes are components of a stress response pathway. Collectively, we have demonstrated that TCH-165 enhances proteasomal degradation of SNAP29 and STX17.
Co-overexpression of the intrinsically disordered fusion proteins, SNAP29 and STX17 restores autophagic flux
To verify that proteasomal degradation of SNAP29 and STX17 plays a role in regulating autophagy, SNAP29, STX17 or both were overexpressed in HeLa cells and autophagic flux monitored in the presence and absence of TCH-165. Here, we observed that overexpression of the individual proteins increased LC3B-II levels, probably due to increase flux, as p62 levels went down (Fig. 5A and B). When cells overexpressing either SNAP29 or STX17 were treated with TCH-165, LC3B-II accumulated further (Fig. 5A and B). However, when both proteins were overexpressed, TCH-165 failed to significantly increase LC3B-II nor p62 levels (Figs. 5A, 5B and S4) over the vehicle control. This suggests that their production outpaced their enhanced proteasomal degradation and autophagic flux was restored. Together, our data suggest that the 20S proteasome regulates autophagosome-lysosome fusion via proteolytic control of the critical fusion proteins, SNAP29 and STX17.
Figure 5: TCH-165 inhibits autophagy flux by enhancing ubiquitin-independent proteasomal degradation of SNAP29 and STX17.
(A) HeLa cells were transfected with N-terminal FLAG-SNAP29, N-terminal FLAG-STX17 or both. Transfected and wild type cells were treated with vehicle or TCH-165 (10 μM) for 16h and whole cell lysate immunoblotted with anti-FLAG, anti-LC3B, anti-p62 and anti-GAPDH. (B) Image J quantification of LC3BII and GAPDH in A. Statistical analyses were performed on three independent experiments (mean ±SD, n=3) using unpaired t-test on GraphPad Prism7 (ns=not significant, *p<0.05, **p<0.01, ***p<0.001). (C) HeLa cells expressing N-terminal FLAG-SNAP29 were pretreated with either vehicle or bortezomib (20 μM) for 2h, followed by treatment with vehicle or TCH-165 (10 μM) for 16h. Whole cell lysates were immunoprobed with anti-Flag, anti-GAPDH or anti-K-48 ubiquitin (left column). Samples were also immunoprecipitated (IP) with anti-Flag and immunoblotted (IB) with anti-SNAP29 or anti-K-48 ubiquitin (right column). (D) Same as in C, except with cells expressing N-terminal FLAG-STX17. (E) HeLa cells were transfected with either control siRNA or PSMD2 (Rpn1) siRNA for 72h, followed by treatment with vehicle or TCH-165 for 16h. Whole cell lysates were immunoblotted for the indicated proteins. Numbers are densitometry of signal as a percentage of vehicle/control siRNA treatment. LE: Long exposure, SE: Short exposure. See also figure S4
The degradation of SNAP29 and STX17 are ubiquitin-independent
Some proteins are susceptible to both ubiquitin-dependent and ubiquitin-independent proteasomal degradation (Yang et al., 2004, Tsvetkov et al., 2010). To verify whether STX17 and SNAP29 belong to this group of proteins, SNAP29 and STX17 were pulled-down and probed for K-48 ubiquitin, which is the form recognized by the 26S proteasome for degradation. For this, cells expressing either Flag-SNAP29 (Fig. 5C) or Flag-STX17 (Fig.5D) were treated with either vehicle, TCH-165, bortezomib (BTZ; proteasome inhibitor) or their combinations. Consistently, SNAP29 and STX17 were rapidly degraded in the presence of TCH-165 but accumulated in the presence of bortezomib, with or without TCH-165 (Fig. 5C, and 5D), further implicating the proteasome as its target protease. Moreover, pull-down with anti-Flag revealed free SNAP29 and STX17, but not their ubiquitin conjugates upon immunoblotting with anti-K-48.
Although ubiquitin-independent degradation is primarily mediated by the 20S, ubiquitin-independent degradation by the 26S proteasome cannot necessarily be excluded, yet. Therefore, to determine the possible involvement of the 26S proteasome in the degradation of SNAP29 and STX17, siRNA knockdown of the 19S cap of the 26S proteasome was used. Knockdown of the 19S PSMD2 (Rpn1) subunit has been validated by others in the determination of 19S-dependent degradation of proteins (Winkler et al., 2013). For this study, HeLa cells were transfected with either control siRNA or Rpn1 siRNA for 72h and then treated with either vehicle or TCH-165 for 16h. Immunoblot revealed significant stabilization of SNAP29 in vehicle control of Rpn1 knockdown sample. However, under the knockdown condition, the degradation of SNAP29 was still enhanced by TCH-165 (Fig. 5E). This suggests that although the 26S proteasome may contribute in the degradation of SNAP29, the 20S is primarily responsible for its proteolysis. Surprising, STX17 which is less disordered than SNAP29 was not stabilized in the Rpn1 knockdown/vehicle control. This suggests that STX17 is primarily degraded by the 20S proteasome. Furthermore, Rpn1 knockdown/TCH-165 treatment showed a more robust enhanced degradation of STX17 compared to control siRNA/TCH-165 sample (Fig. 5E). This is consistent with an increasing pool (with Rpn1 knockdown) of activated 20S proteasome (upon TCH-165 exposure). Consistent with the role of STX17 in autophagy flux, LC3B II accumulated more in Rpn1 knockdown/TCH-165 compared to control siRNA/TCH-165. Predictably, Rpn1 knockdown (26S inhibition) lead to p62 (also a 26S substrate) accumulation compared to the control siRNA.
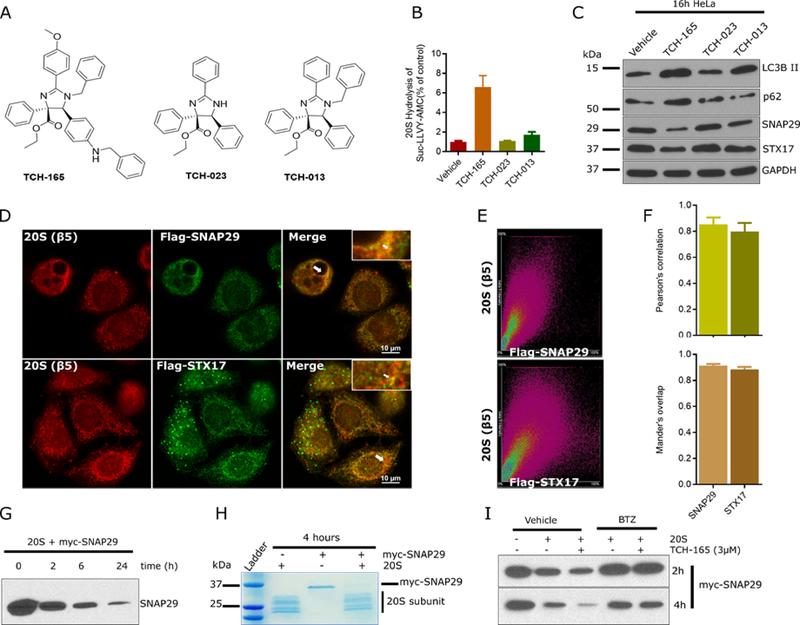
20S proteasome activity inversely parallels autophagic flux
We used three different imidazoline analogues to determine whether 20S proteasome enhancement parallels autophagy inhibition. For this, we used TCH-165 (potent 20S agonist), TCH-023 (inactive analogue) and TCH-013 (weak 20S agonist) (Fig. 6A) (Njomen et al., 2018, Lansdell et al., 2013). Indeed, the ability of these molecules to enhance 20S-mediated proteolysis of the peptide substrate, Suc-LLVY-AMC (Fig. 6B), directly correlates with the degradation of SNAP29, STX17 and the accumulation of autophagic substrates; LC3B-II and p62 (Fig. 6C). Predictably, pull-down experiment to verify the interaction of these proteins with the proteasome was unproductive, given that the interaction is likely weak and transient. However, immunofluorescence of cells expressing Flag-SNAP29 or STX17 revealed significant overlap with the proteasome (Fig. 6D–F). Considering that SNAP29 is readily available, its interaction with the 20S proteasome as well as its ubiquitin-independent proteolysis was confirmed by in an in vitro biochemical assay. SNAP29 containing a C-terminal myc- tag was degraded by purified 20S proteasome, as observed by immunoblot (Fig. 6G) and by Coomassie staining (Fig. 6H). Moreover, the degradation of SNAP29 in this assay was enhanced by TCH-165 and blocked by the proteasome inhibitor, bortezomib (Fig. 6I). Together, these data suggest that the SNARE proteins, SNAP29 and STX17, are 20S substrates and that ubiquitin-independent 20S proteasome-mediated proteolysis of these proteins regulates, at least in part, autophagic flux.
Figure 6: TCH-165 stimulates 20S proteasomal degradation of SNAP29 and STX17.
(A) Chemical structures of imidazolines TCH-165 (a potent 20S agonist), TCH-023 (inactive analogue) and TCH-013 (a moderate 20S agonist). (B) Degradation of the chymotrypsin-like fluorogenic peptide substrate, Suc-LLVY-AMC (10 μM) with 20S proteasome (1 nM) treated with vehicle (control) or 10 μM of TCH-165, TCH-023 or TCH-013. Data are presented as percentage of vehicle control. (C) HeLa cells were treated with vehicle or 10 μM of TCH-165, TCH-023 or TCH-013 and whole cell lysate immunoblotted for LC3B, p62, SNAP29, STX17 and GAPDH. (D) HeLa cells expressing Flag-SNAP29 or Flag-STX17 were immunostained with mouse anti-β5/Alexa fluor 546 (red) and rabbit anti-Flag/ Alexa Fluor 488 (green) and imaged with an upright Nikon A1 confocal microscope using a 60x Plan Apo oil objective. Arrows indicate areas of colocalization visible by naked eye. (E) Sample 2-D intensity histogram for D. (F) Pearson’s correlation and Mander’s overlap for SNAP29/ β5 and STX17/ β5 were quantified with Nikon software. (G) Purified c-myc/DDK-SNAP29 (200 nM) was degraded with 20S proteasome (20 nM) for 0h, 2h, 6h and 24h and analyzed by immunoblot with anti-SNAP29. (H) Purified c-myc/DDK-SNAP29 (200 nM) was degraded with 20S proteasome (20 nM) for 4h and analyzed by Coomassie instant blue staining following SDS-PAGE. (I) Purified c-myc/DDK-SNAP29 (200 nM) was degraded with 20S proteasome (20 nM) treated with vehicle, TCH-165 (3 μM), bortezomib (5 μM), or combinations, for 2h and 4 h and analyzed by immunoblot with anti-SNAP29.
Reversibility of cellular effects following TCH-165 treatment
We previously published that TCH-165 has low single digit micromolar potency against U-87 MG and other cancer cell lines (Njomen et al., 2018). To evaluate the reversibility of TCH-165 on cell viability, U-87 MG cells were treated with different concentrations of TCH-165. Cells were either unwashed (control) or washed at 8h or 24h and cultured for a total of 72h. Washing off TCH-165 after 8h incubation (but not 24h) reduces its 72-hour cytotoxicity by about 3-fold (Fig. S5A and S5B). Furthermore, washing the cells at 4h, 8h and 12h post treatment and allowing it to culture for a total of 24h (Fig. S5C) reduces LC3BII and p62 accumulation, compared to 24h treatment (Fig. S5D–F). At the level of LC3B, the cells appear to be recovering from TCH-165 treatment, as the unwashed controls at those same time points have higher LC3B accumulation (Fig. S5E). However, this result should be interpreted with caution, as this could also be the result of a reduced amount of TCH-165 present in the cells post-washing.
Considering the abundance and predominance of 20S-mediated proteolysis under conditions of oxidative stress, we next investigated whether the degradation of SNAP29 and STX17 are enhanced by hydrogen peroxide (H2O2). In this study, treatment of HeLa cells with 50 μΜ H2O2 significantly reduces the level of assembled 26S proteasome (Fig. S6A), with a corresponding increase in oxidized proteins (Fig. S6B). TCH-165 (at 10 μM) also reduced the level of 26S in HeLa cells (Fig. S6A) and in U-87 MG cells (Fig. S6C), while maintaining a significant amount of single capped 20S, as previously reported (Njomen et al., 2018). However, the degradation of SNAP29 and STX17, as well as changes in autophagy markers, LC3BII and p62, were not as robust following oxidative damage, compared to TCH-165 treatment (Fig. S6B). Consistent with 20S activation and the role of 20S in the degradation of oxidatively damaged proteins (Dal Vechio et al., 2014, Shringarpure et al., 2001), TCH-165 was able to enhance the degradation of oxidized proteins under basal conditions (Fig. S6B, lane 2). The combination of TCH-165 with H2O2 resulted in milder effects on the degradation of the SNARE proteins, and the accumulation of LC3BII and p62 (Fig. S6B, lane 4), compared to TCH-165 treatment only. This observation is most likely due to increase in proteasomal load (oxidized proteins) as a result of exposure to H2O2.
DISCUSSION
Our results described herein indicate that enhancement of 20S proteasome activity by TCH-165 reduces autophagic flux by preventing autophagosome-lysosome fusion. In the fusion step, the Qa soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Qa-SNARE), syntaxin 17 (STX17), is only recruited to completed autophagosome membrane (most likely from the cytosol) by immunity-related GTPase M (IRGM) (Kumar et al., 2018). STX17 then binds to its interacting partner SNAREs, snaptosomal-associated protein 29 (SNAP29, a Qbc-SNARE) and vesicle-associated membrane protein 8 (VAMP8, an R-SNARE) and promote autophagosome-lysosome fusion) (Itakura et al., 2012). Components of the homotypic fusion and protein sorting (HOPS)–tethering complex, vacuolar protein sorting 33A (VPS33A), VPS16, and VPS39 also assist in autophagosome-lysosome fusion, through interactions with STX17 (Jiang et al., 2014). We found that enhancement of 20S proteasome activity resulted in enhanced degradation of the disordered SNARE proteins SNAP29 and STX17, but not VAMP8. Overexpression of both SNAP29 and STX17, but not the individual proteins were needed to ameliorate the effect on autophagic flux. This amelioration is indicative of the reinstatement of the fusion process once production outpaces 20S degradation.
Although many of the proteins involved in the fusion process are membrane bound, SNAP29, is a cytosolic and highly disordered (87%) protein and therefore a likely candidate for unremitted 20S proteasome regulation (Tsvetkov and Shaul, 2012, Asher et al., 2006). STX17 does not appear as a typical 20S substrate as it is commonly thought of as a membrane associated protein. However, the relative susceptibility of disordered proteins to 20S-mediated proteolysis is dictated by their level of disorder, protein sequence, cellular compartmentalization and proteolytic accessibility (Biran et al., 2017, Tsvetkov and Shaul, 2012, Tsvetkov et al., 2009, Tsvetkov et al., 2008, Asher et al., 2006). Due to the poor hydrophobic nature of STX17’s membrane binding domain, a good fraction of STX17 are cytosolic and are only incorporated into the membrane of completed autophagosomes (Kumar et al., 2018, Jiang et al., 2014, Itakura et al., 2012, Minton, 2013). Furthermore, STX17 is recycled after autolysosome formation instead of being degraded like other autophagosome membrane proteins such as LC3B-II and p62 (Jiang et al., 2014). These observations in part suggest that STX17 is degraded through a non-lysosomal pathway. Moreover, the lack of their (SNAP29 and STX17) degradation following autophagy induction with mTOR inhibitors, as observed here and by others (Fu et al., 2018) further suggests a non-lysosomal mechanism of degradation. Indeed, we have shown here that STX17 and SNAP29 are 20S proteasome substrates. We further demonstrated that promoting the degradation of SNAP29 and STX17 through pharmacological stimulation of the 20S proteasome reduces autophagy flux by regulating autophagosome-lysosome fusion.
In terms of 20S proteasome target selectivity, the relative susceptibility of disordered proteins to 20S-mediated proteolysis is far more limited than perhaps anticipated, because it is dictated by the level of substrate disorder, protein sequence, cellular compartmentalization and proteolytic accessibility(Biran et al., 2017, Tsvetkov and Shaul, 2012, Tsvetkov et al., 2009, Tsvetkov et al., 2008, Asher et al., 2006). Moreover, small molecule 20S activation involves the induction of conformational changes that may be unique for each agonist, with the possibility of disparate pool of select target proteins for proteolytic degradation (Njomen and Tepe, 2019). Therefore, it is possible or perhaps likely, that different classes of 20S agonists may exert different substrate selectivity amongst proteolytically susceptible IDPs. Herein, we have used TCH-165 as a probe to implicate the 20S proteasome as an integral component of the autophagosome-lysosome fusion step. A schematic of the proposed signaling pathways consistent with our findings is depicted in in the graphical abstract.
In conclusion, we herein present an alternative mechanism of crosstalk between proteasome and autophagy pathway that is regulated by ubiquitin-independent, 20S proteasome-mediated degradation of SNAP29 and STX17.
SIGNIFICANCE
Proteins continually undergo degradation to maintain biological homeostasis. A well-balanced crosstalk between the ubiquitin-proteasome system (UPS) and the autophagy pathway ensures an energy efficient maintenance of cellular function via the degradation of reductant or damaged proteins and organelles. Pharmacological manipulation of this crosstalk has been recognized as a potential approach to treat many disorders including cancer and neurodegenerative disorders. A better understanding of the crosstalk between the two systems can identify alternative targets or maximize current therapeutic treatments of proteotoxic diseases. Our current understanding of the crosstalk between the UPS and autophagy is centered around substrate ubiquitination. We report herein an additional method of crosstalk involving ubiquitin-independent 20S proteasome regulation of autophagosome-lysosome fusion. In this work, we identified the disordered soluble N-ethylmaleimide-sensitive factor activating protein receptor (SNARE) proteins, synaptosomal-associated protein 29 (SNAP29) and syntaxin 17 (STX17), but not vesicle-associated membrane protein 8 (VAMP8) as 20S proteasome substrates. This provides a previously unrecognized mechanism of crosstalk between the two catabolic pathways.
Star Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to Tepe Jetze (tepe@chemistry.msu.edu).
Data and Code Availability
This study did not generate and/or analyze datasets or code. No data/code is deposited in repositories.
Experimental model and subject details
Cell culture
Human embryonic kidney cells (HEK293), HeLa or U-87 MG cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% Fetal Bovine Serum, and 100 U/mL Penicillin/Streptomycin, at 37°C with 5% CO 2. Human B lymphocyte (RPMI 8226) were maintained in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum, and 100 U/mL Penicillin/Streptomycin, at 37°C with 5% CO 2.
Method details
Transient transfection
BacMam plamids (Premo Autophagy Tandem Sensor RFP-GFP-LC3B and CellLight™ Lysosomes-GFP, BacMam 2.0) were directly added to cells in cover glass chamber 16h or 24h before treatment. For other transfections, the following protocol was followed. For plasmid transfection, plasmid to transfection reagent (sinofection reagent) was maintained at ratio of 1:6 (μg:μL). For siRNA transfection, a ratio of 2.5:1 (nM: μL) was used. Flag-SNAP29 and FLAGSTX17 were used at concentrations of 5 μg/million cells for overexpression experiments to ameliorate the effect of TCH-165 and at 2 μg/million cells for all other experiments. PSMD2 siRNA and control siRNA were used at concentrations of 250 nM. HeLa or U-87 MG cells were seeded at a density of 1million in 60 mm or 2.5 million in 100 mm dish overnight. Plasmids or oligonucleotides were mixed with 1 mL of serum free-DMEM medium. Sinofection transfection reagent was also mixed with 1 mL of serum free medium in a separate vial. The separate mixtures were combined and allowed to sit at RT for 15 minutes. For 60 mm dish, the mixture was directly added to the cells. For 100 mm dish, the mixture was further diluted with 2 mL medium before adding to cells, to cover the entire surface area. After incubating for 4h at 37°C, 5% CO 2, in a tissue culture incubator, fresh medium (4 mL for 60 mm and 10 mL for 100 mm dish) with 10%FBS was then added and allowed to culture. For Flag-SNAP29 and or STX-17 transfection, cells were cultured for a further 20h, then split into 60 mm dish each and cultured for 8h before treatment (at 32h post-transfection).For EGFP-LC3B, cells were cultured for a further 24h then split into cover glass chamber and cultured for 24h before treatment (at 48h post-transfection). For siRNA knockdown, cells were cultured for a further 48h then split into 60 mm dish each and cultured for 24h before treatment (at 72h post-transfection).
Immunoprecipitation
HeLa cells transiently transfected with Flag-SNAP29 (2 μg, plasmid) or Flag STX17 (2 μg, plasmid) in 60 mm dish (see transient transfection above) were pretreated with vehicle or bortezomib(20 μM) for 2h, followed by treatment with vehicle or TCH-165 (10 μM) for a further 16h. Cells were washed 2x with chilled DPBS buffer and scrapped with 200 μL of lysis buffer (50 mM TrisHCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, 1 mM Na3VO4, 1x sigmafast protease inhibitor cocktail). Cells were completely lyzed by sonication. Lysates were clarified by spinning at 14,000g for 20 minutes and supernatant collected into new tubes. Total protein was quantified by bicinchoninic acid (BCA) assay following standard protocol. Samples were normalized with lysis buffer to 0.5 mg/mL with lysis buffer. Lysates (200 μL or 100 μg) were mixed with 4 μL of Flag antibody (Cell Signaling Technology: 14793) and incubated at 4°C overnight with shaking. Pre-washed Pierce Protein A/G Magnetic Beads (25 μL or 0.25 mg) was then added and incubated at RT with shaking for 1h. Beads were collected with magnetic stand and washed 4x with 1 mL of lysis buffer. Flag-tagged proteins were eluted by shaking beads overnight at 4°C with 200 μL of 4 mg/mL 3x flag peptide in 50 mM Tris-HCl pH 7.5, 150 mM NaCl. Samples were collected with magnetic stand and boiled with 5x SDS buffer and blotted for SNAP29, STX17, and K-48 ubiquitin. Input lysates were also immunoblotted for Flag, K-48 ubiquitin, LC3B, P62, and GAPDH as above.
Immunoblot
Wild type or transfected cells at 70–80% confluency was treated with test compounds at the reported concentrations and time as indicated in figure legends. Samples meant for immunoblot only were washed 3x with warm DPBS buffer and scrapped with chilled RIPA buffer supplemented with protease inhibitor cocktail. Samples were briefly sonicated, spun and supernatant assayed for total protein with bicinchoninic acid (BCA) assay. Normalized and boiled samples were resolved on 4–20% Trisglycine SDS-PAGE and blotted onto a PVDF membrane. Membranes were blocked in 5% non-fat milk in TBST buffer for 60 minutes at RT. Membranes were then incubated with primary antibody in 2% non-fat milk TBST buffer, overnight at 4°C. Membranes were then washed 5x for 5 minutes each and incubated in secondary antibody at RT for 60 minutes. Membranes were washed again as above and developed with ECL clarity reagent. Images were captured with Hyblot films. All primary antibodies were used at a dilution of 1:1000, except for SNAP29 (1:2000) and STX17 (1:500). Goat anti-rabbit HRP and Goat anti-mouse HRP were also used at 1:1000.
OxyBlot (protein carbonyl quantification)
HeLa cells at 70% confluency in 100 mm dish were treated with vehicle, TCH-165(10 μM), H2O2 (50 μM), or a combination of TCH-165 (10 μM) and H2O2 (50 μM) for 16h.Cells were washed with warm DPBS and collected by trypsinization. Pellets were washed 3x with chilled DPBS buffer and split into two parts. One part was used for proteasome native gel as previously reported (Njomen et al., 2018) and the second half lyzed by sonication in RIPA buffer supplemented with sigmafast protease inhibitor cocktail and used for oxyblot. Lysates were clarified by spinning at 14,000g for 20 minutes and supernatant collected into new tubes. Total protein was quantified by bicinchoninic acid (BCA) assay following standard protocol. Samples were normalized with lysis buffer to 2 mg/mL with lysis buffer. One-part cell lysate (20 μL) was mixed with one part 12% SDS (20 μL) and incubated at RT for 10 minutes. Two parts (40 μL) 20 mM DNPH (2, 4-Dinirtophenylhyd solution in 10% TFA was then added, mixed and incubated at RT for 20 minutes. Reaction was then quenched with 1.5 part (35 μL) 2 M Tris/30% glycerol. Samples were resolved on 4–20% Tris/glycine gel at 4 °C, blotted onto PVDF membrane and probed with mouse anti DNP (SigmaAldrich, D8406 1:500) and anti-mouse HRP (1:1000). Underivatized lysates were boiled with SDS buffer and immunoblotted as above.
SiRNA knock down of PSMD2 (19S proteasome subunit)
HeLa cells were transfected with control and PSMD2 siRNA as described under transient transfection. Post-transfection (72h), cells were treated with either vehicle or TCH-165 (10 μM) for 16h. Samples were collected and immunoblotted as described under immunoblot.
qRT-PCR
Glioblastoma (U-87 MG) cells were treated with either vehicle or TCH-165 (10 μM) for 8h or 24h. Total mRNA was isolated by Trizol extraction and purified on a RNeasy column. Residual contaminating genomic DNA was digested with TURBO DNase and samples were cleaned with RNA clean-up columns. The cDNA was prepared from 200 ng of mRNA with random hexamers using iScript cDNA synthesis kit as per manufacturer instructions. cDNA (2 μL of PCR product) levels were then measured by quantitative real-time PCR (qRT–PCR) with 500 nM of forward and reverse primers each, using iTaq universal SYBR Green supermix on a Viia7 lightCycler (Applied Biosystems). Signals were normalized to that of GAPDH and quantified by the ΔΔCt method. Data are means ± SD of two independent experiments each ran in triplicates.
Proteasome activity assay
Activity assays were carried out in a 150 μL reaction volume. Imidazolines (10 μM) or DMSO (vehicle) were added to a black flat/clear bottom 96-well plate containing 1 nM of human 20S proteasome in 20 mM Tris-HCl, 100 mM NaCl, pH 7.4 and allowed to sit for 10 minutes at RT. The fluorogenic peptide substrate Suc-LLVY-AMC (10 μM) was then added and the enzymatic activity measured at 37°C on a SpectraMax M5e spectrometer by measuring increase in fluorescence unit per minute for 1h at 380/460 nm. The fluorescence units for the vehicle control was set to 100%, allowing for calculation of per cent activation as a ratio of drug-treated to vehicle control.
Cathepsin D and E Activity
U-87 MG or HeLa cells at ~70% confluency in 100 mm dish were treated with vehicle, TCH-165 (10 μM), Torin 1(100 nM), Pepstatin A (5 μM) or Chloroquine (100 μM) for 24h. Cells were washed 2x with chilled DPBS buffer and scrapped with 500 μL of lysis buffer (100 mM NaCl, 100 mM NaOAc, 1 mM EDTA, 1% Triton X-100, pH 5.0) into an Eppendorf tube. Cells were then lyzed by rocking the tubes on a shaker for 1h at 4οC. Lysates were clarified by spinning at 14,000g for 20 minutes and supernatant collected into new tubes. Total protein was quantified by bicinchoninic acid (BCA) assay following standard protocol. Samples were normalized with lysis buffer to 1 mg/mL, and then diluted to 0.1 mg/mL in assay buffer (100 mM NaCl, 100 mM NaOAc, pH 5.0). Each sample (100 μL or 10 μg) was transferred into quadruplicate well of a black-clear bottom 96 well plate. Cathepsin D and E substrate (BML-P145–0001, Mca-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys (Dnp)-D-Arg-NH2 [Mca= (7-methoxycoumarin-4-yl)acetyl; Dnp=dinitrophenyl]) was then diluted in assay buffer and 50 μL added to the wells to a final concentration of 20 μM. Plate was incubated at 37οC for 1h and fluorescence reading taken at 330/460 nm on a SpectraMax M5e. Data are presented as means ± SD of three independent experiments.
Cell viability assay (CellTiter-Glo)
U-87MG cells were seeded in a white solid bottom 96 well plate at a density of 5.0 × 103 cells/well and cultured overnight. Cells were treated with different concentrations of TCH-165 (or 1% DMSO). Cells were either not washed (control treatment) or washed with warm DPBS buffer at 8h or 24h and cultured in fresh medium alongside the control for a total of 72h. Cells were equilibrated to RT and CellTiter-Glo solution (100 μL) added and incubated with shaking for 10 minutes at RT. Assay plate was then allowed to equilibrate for 5 minutes at RT and luminescent readings taken on a SpectraMax M5e. Data are presented as a percentage of the vehicle control for each experimental condition.
In vitro degradation of SNAP29-c-Myc/DDK
Digestion of SNAP29-c-Myc/DDK was carried out in a 50 μL reaction volume made of 20 mM HEPES pH 7.4, 2 mM EDTA, 1 mM EGTA, 200 nM SNAP29, 20 nM purified human 20S proteasome with or without the test compounds. Briefly, 20S proteasome was diluted to 23 nM in the reaction buffer. Test compounds or vehicle (1 μL of 50x stock) were added to 44 μL of 23 nM 20S and incubated at RT for 20 minutes. The substrate (5 μL of 2 μM SNAP29-c-Myc/DDK) was then added to the reaction mixture and incubated at 37°C. Aliquots of 10 uL were taken out and mixed with 2x SDS loading buffer, at the indicated time. Samples were boiled and immunoblotted.
Confocal immunofluorescent
All reagents used for sample staining, except for methanol were from the immunofluorescence application solutions kit (Cell Signaling Technology cat# 12727). U87-MG or HeLa cells were plated in complete medium at 2.5×104 cells/well of a four-chamber glass slide overnight. Fresh medium (500 μL) containing the test compound was added and allowed to incubate for the desire time under cell culture conditions. Cells were washed with chilled immunofluorescent (IF) wash buffer (1x PBS, pH 8.0) and fixed with methanol at −20°C for 10 minutes. Cells were then washed 2x with IF wash buffer and incubated in blocking buffer (1x PBS, 5% normal goat serum, 0.3% Triton X-100 pH 8.0) for 60 minutes at RT. Blocking buffer was replaced with primary antibody diluted in dilution buffer (1x PBS, 1%BSA, 0.3%Triton X-100 pH 8.0) overnight (1:200) at 4°C. Primary antibody was washed off 3x and incubated in anti-rabbit Alexa Fluor 488 (1:500) and/or anti-mouse Alexa Fluor 594 for 60 minutes in the dark, at RT. Secondary antibodies were then washed off (3x) and nuclei stained with Hoechst (4 μg/mL) at RT, for 20 minutes. DNA dye was rinsed off and slides mounted in Prolong Gold and allowed to cure overnight at RT, in the dark. Slides were sealed with nail polish, and LC3B, LAMP1 and/or p62 puncta or fluorescence detected with a 60x Plan Apo oil objective on a Nikon C2+ or Nikon A1 confocal laser scanning microscope. For Flag SNAP29 or Flag-STX17 co-staining with proteasome β5, untreated cells were fixed with 4% paraformaldehyde at RT for 10minutes, and immunostained as above with rabbit anti-Flag/ Alexa Fluor 488 and mouse anti-β5/ Alexa Fluor 546.
Confocal Live imaging
For autophagosome-lysosome fusion, U-87MG or HEK293 cells cultured on cover glass slide were transduced with 30 particles per cell of tandem-RFP-GFP-LC3B and cultured for 24h. Cells were then incubated with either vehicle, TCH-165 (10 μM), chloroquine (100 μM) leupeptin A (200 μM), torin 1 (100 nM), or combination of torin 1 and TCH-165 for a further 24h. Cells were Imaged on an upright Nikon A1 confocal microscope using a 60x Plan Apo oil objective with standard filter sets for GFP and RFP. For LC3B puncta, U-87 MG cells were transiently transfected with EGFP-LC3B (2 μg/1million cells) using sinofection reagent as described above. Cells were then split at 24h post-transfection and seeded on cover glass slide for a further 24h. Cells were then treated with either vehicle, TCH-165 (10 μM), torin 1 (200 nM), or combination of torin 1 and TCH-165 for a further 16h and imaged as above with GFP filter. For lysosome pH, U-87 MG cells were seeded in a four-chamber cover glass overnight. Cells were treated with either vehicle, TCH-165 (10 μM) Bafilomycin A1 (50 nM) or Torin1 (100 nM) for 16h. Culture medium was replaced with fresh medium containing 50 nM Lysotracker red and allowed to incubate for 15 minutes under cell culture conditions. Cells were then imaged on an upright Nikon A1 confocal microscope using a 60x Plan Apo oil objective with an excitation and emission wavelengths of 560 nm and 570–620 nm, respectively. For co-staining with LAMP1, Cells were transduced with 30 particles per cell of CellLight™ Lysosomes-GFP, BacMam 2.0 and cultured for 24h, before treatment and lysotracker red staining.
Quantification and statistical analysis
Data are presented as mean ± standard deviation (SD). For each figure, the number of replicates is indicated in the figure legends. Statistical analysis was only performed on experiments with three or more n (biological replicates for cellular experiments or individual experiment for biochemical assays). Western blots and immunofluorescence (total fluorescence or puncta) quantification were performed with imageJ and colocalization (Mander’s overlab and pearson’s correlation) with Nikon software. For total fluorescence/colocalization, measurements were taken from an entire field of view. For puncta or AVs, mean puncta for an entire field of view per cell number in that field was used for each experiment. statistical analysis was performed with GraphPad Prism 7 software. Unpaired Student’s t-test was used for two samples while one-way analysis of variance with post hoc Bonferroni test was used for multiple comparisons of means. Effect was considered significant for *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Supplementary Material
Key Resource Table.
| REAGENT OR RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LC3B (D11) XP rabbit mAb | Cell Signaling Technology | Cat# 3868, RRID:AB_2137707 |
| SQSTM1/p62 (D5L7G) mouse mAb | Cell Signaling Technology | Cat# 88588, RRID:AB_2800125 |
| SQSTM1/P62 (D5E2) rabbit mAb | Cell Signaling Technology | Cat# 8025, RRID:AB_10859911 |
| LAMP1 (D2D11) XP rabbit mAb | Cell Signaling Technology | Cat# 9091, RRID:AB_2687579 |
| LAMP1 (D4O1S) mouse mAb | Cell Signaling Technology | Cat# 15665, RRID:AB_2798750 |
| SNAP29 (D-8) mouse mAb | Santa Cruz Biotechnology | Cat# sc-390801, RRID:AB_2800552 |
| STX17 rabbit pAb | Abcam | Cat# ab116113, RRID:AB_10903821 |
| VAMP8 rabbit pAb | Cell Signaling Technology | Cat# 13060, RRID:AB_2798103 |
| 20S proteasome α-3 (A-9) mouse mAb | Santa Cruz Biotechnology | Cat# sc-166205, RRID:AB_2253046 |
| 20S proteasome β-5 (A-10) mouse mAb | Santa Cruz Biotechnology | Cat# sc-393931, RRID:AB_2800553 |
| PSMD2 (A-11) mouse mAb | Santa Cruz Biotechnology | Cat# sc-271775, RRID:AB_10708257 |
| GAPDH (0411) HRP mouse mAb | Santa Cruz Biotechnology | Cat# sc-47724, RRID:AB_627678 |
| FLAG (D6W5B) rabbit mAb | Cell Signaling Technology | Cat# 14793, RRID:AB_2572291 |
| Goat anti-rabbit IgG HRP-linked | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Horse anti-mouse IgG HRP-linked | Cell Signaling Technology | Cat# 7076, RRID:AB_330924 |
| anti-rabbit IgG (H+L), F(ab’)2 Fragment (Alexa Fluor 488 Conjugate) | Cell Signaling Technology | Cat# 4412, RRID:AB_1904025 |
| anti-mouse IgG (H+L), F(ab’)2 Fragment (Alexa Fluor 594 conjugate) | Cell Signaling Technology | Cat# 8890, RRID:AB_2714182 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A10036, RRID:AB_2534012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TCH-165, TCH-023, TCH-013 | (Njomen et al., 2018) | N/A |
| Chloroquine | Cell Signaling Technology | Cat# 14774 |
| Torin 1 | Cell Signaling Technology | Cat# 14379 |
| Cycloheximide | Cell Signaling Technology | Cat# 2112 |
| Sigmafast inhibitor cocktail | Sigma Aldrich | Cat# S8820 |
| Bafilomycin A1 | Sigma Aldrich | Cat# B1793 |
| Sinofection transfection reagent | Sino Biological | Cat# STF02 |
| 3X FLAG Peptide | APExBIO | Cat# A6001 |
| N-Succinyl-Leu-Leu-Val-Tyr-7amido-4-methylcoumarin | BostonBiochem | Cat# S280 |
| Cathepsin D & E substrate (fluorogenic) | Enzo Life Sciences | Cat# BML-P145–0001 |
| Pepstatin A (synthetic) | Enzo Life Sciences | Cat# ALX-260–085-M005 |
| Human 20S proteasomes | BostonBiochem | Cat# E-360 |
| Bortezomib | Cayman Chemical | Cat# 10008822 |
| leupeptin | Sigma Aldrich | Cat# L2884 |
| Recombinant human SNAP29 with c-Myc tag | Origene | Cat# TP302179 |
| LysoTracker red | ThermoFisher Scientific | Cat# L7528 |
| TRIzol Reagent | ThermoFisher Scientific | Cat# 15596026 |
| Pierce™ Protein A/G Magnetic Beads | ThermoFisher Scientific | Cat# 88802 |
| TURBO DNase-free kit | ThermoFisher Scientific | Cat# AM1907 |
| iTaq universal SYBR green Supermix | Bio-Rad | Cat# 172–5120 |
| Fetal bovine serum | Sigma Aldrich | F2442 |
| RPMI-1640 Medium | ATCC | Cat# ATCC® 30–2001 |
| DMEM medium | ThermoFisher Scientific | Cat# 11965092 |
| Penicillin-Streptomycin | ThermoFisher Scientific | Cat# 15140122 |
| Critical Commercial Assays | ||
| BCA assay | ThermoFisher Scientific | Cat# 23225 |
| Immunofluorescence Application Solutions Kit | Cell Signaling Technology | Cat# 12727 |
| iScript cDNA synthesis kit | Bio-Rad | Cat# 1708891 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | Cat# G7570 |
| Experimental Models: Cell Lines | ||
| U-87 MG cells | ATCC | Cat# HTB-14, RRID:CVCL_0022 |
| RPMI 8226 | ATCC | Cat# CRM-CCL-155, RRID:CVCL_0014 |
| HeLa | ATCC | Cat# CCL-2, RRID:CVCL_0030 |
| HEK293 | ATCC | Cat# CRL-1573, RRID:CVCL_0045 |
| Oligonucleotides | ||
| PSMD2 siRNA (h) | Santa Cruz Biotechnology | Cat# sc-62900 |
| Signal Silence®Control siRNA (Unconjugated) | Cell Signaling Technology | Cat# 6568 |
| Premo Autophagy Tandem Sensor RFP-GFP-LC3B | ThermoFisher Scientific | Cat# P36239 |
| CellLight™ Lysosomes-GFP, Bac-Mam 2.0 | ThermoFisher Scientific | Cat# C10507 |
| N-FLAG-SNAP29 | Noboru Mizushima et al 2012 | Addgene plasmid # 45915 |
| N-FLAG-STX17 | Noboru Mizushima et al 2012 | Addgene plasmid # 45911 |
| EGFP-LC3B | Karla Kirkegaard et al 2005 | Addgene plasmid # 11546 |
| Primers for SNAP29, STX17, VAMP8, LC3B, p62 and GAPDH; see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| PONDR | Molecular Kinetics, Inc., 2003 | http://www.pondr.com/ |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 7 | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| ChemDraw Professional 17.1 | PerkinElmer | http://www.cambridgesoft.com/support/ |
The cells eliminate unwanted proteins through two major protein degradation systems called the proteasome and autophagy pathways. Njomen et al found that the ubiquitin-independent 20S proteasome system is a key regulator of the autophagy pathway. Using a small molecule activator of the 20S proteasome, the authors demonstrated that this new mechanism of crosstalk is achieved through 20S-mediated degradation of two key autophagy proteins; SNAP29 and STX17. These findings will likely open new area of drug discovery research for protein-associated disorders.
HIGHLIGHTS.
Autophagosome-lysosome fusion is regulated, in part, by the 20S proteasome
SNAP29 and STX17 are degraded in a ubiquitin-independent manner
SNAP29 and STX17 are proteolytic substrates of the 20S proteasome
Crosstalk between the 20S proteasome and autophagy
ACKNOWLEDGMENTS
Financial support for this work was provided by the National Institute of Allergy and Infectious Diseases (1R21AI117018-01A1) and the National Institute of General Medical Sciences (T32GM092715) of the National Institutes of Health. The authors also gratefully acknowledge financial support from Michigan State University (SPG grant program) and the Clinical and Translational Science Institute (CTSI grant program). The authors will also like to thank the Center for Advanced Microscopy at MSU for letting us use the facility and for technical supports (Dr. Melinda Frame).
Footnotes
DECLARATION OF INTEREST
A provisional patent (US 62/539,049) related to compound TCH-165 has been filed by Michigan State University. The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
The supporting Information includes Supplemental Experimental Procedures,six figures, and a supplemental table referenced in the article and a key resource table in the star method.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Asher G, Reuven N & Shaul Y 2006. 20S proteasomes and protein degradation “by default”. Bioessays, 28, 844–9. [DOI] [PubMed] [Google Scholar]
- Barth S, Glick D & Macleod KF 2010. Autophagy: assays and artifacts. J. Pathol, 221, 117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nissan G & Sharon M 2014. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules, 4, 862–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Yu H, Mim C & Matouschek A 2014. Regulated protein turnover: snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol, 15, 122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran A, Myers N, Adler J, Broennimann K, Reuven N & Shaul Y 2017. A 20S Proteasome Recepetor for Degradation of Intrinsically Disordered Proteins. BioRxiv, 210898. [Google Scholar]
- Bjorkoy G, Lamark T, Pankiv S, Overvatn A, Brech A & Johansen T 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol., 452, 181–97. [DOI] [PubMed] [Google Scholar]
- Boland B, Yu WH, Corti O, Mollereau B, Henriques A, Bezard E, Pastores GM, Rubinsztein DC, Nixon RA, Duchen MR, Mallucci GR, Kroemer G, Levine B, Eskelinen EL, Mochel F, Spedding M, Louis C, Martin OR & Millan MJ 2018. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov, 17, 660–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty J, Von Stockum S, Marchesan E, Caicci F, Ferrari V, Rakovic A, Klein C, Antonini A, Bubacco L & Ziviani E 2018. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol. Med, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KM, Hernandez N, Sproul AA & Yu WH 2018. Alzheimer’s disease and the autophagic-lysosomal system. Neurosci. Lett [DOI] [PubMed] [Google Scholar]
- Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C & Ciechanover A 2016. The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int. J. Biochem. Cell Biol, 79, 403–418. [DOI] [PubMed] [Google Scholar]
- Dal Vechio FH, Cerqueira F, Augusto O, Lopes R & Demasi M 2014. Peptides that activate the 20S proteasome by gate opening increased oxidized protein removal and reduced protein aggregation. Free Radic. Biol. Med, 67, 304–13. [DOI] [PubMed] [Google Scholar]
- Dikic I 2017. Proteasomal and Autophagic Degradation Systems. Annu. Rev. Biochem, 86, 193–224. [DOI] [PubMed] [Google Scholar]
- Finley D, Chen X & Walters KJ 2016. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem. Sci, 41, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R, Deng Q, Zhang H, Hu X, Li Y, Liu Y, Hu J, Luo Q, Zhang Y, Jiang X, Li L, Yang C & Gao N 2018. A novel autophagy inhibitor berbamine blocks SNARE-mediated autophagosome-lysosome fusion through upregulation of BNIP3. Cell Death Dis, 9, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Gammoh N, Wong PM, Erdjument-Bromage H, Tempst P & Jiang X 2010. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy, 6, 126–37. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S & Macleod KF 2010. Autophagy: cellular and molecular mechanisms. J. Pathol, 221, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH & Ciechanover A 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev, 82, 373–428. [DOI] [PubMed] [Google Scholar]
- Homewood CA, Warhurst DC, Peters W & Baggaley VC 1972. Lysosomes, pH and the antimalarial action of chloroquine. Nature, 235, 50–2. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C & Mizushima N 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell, 151, 1256–69. [DOI] [PubMed] [Google Scholar]
- Ji CH & Kwon YT 2017. Crosstalk and Interplay between the Ubiquitin-Proteasome System and Autophagy. Mol. Cells, 40, 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, White EJ, Conrad C, Gomez-Manzano C & Fueyo J 2009. Autophagy pathways in glioblastoma. Methods Enzymol, 453, 273–86. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T & Mizushima N 2014. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell, 25, 1327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T & Lamark T 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy, 7, 279–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J & Debnath J 2015. Autophagy at the crossroads of catabolism and anabolism. Nat. Rev. Mol. Cell Biol, 16, 461–72. [DOI] [PubMed] [Google Scholar]
- Kim E, Park S, Lee JH, Mun JY, Choi WH, Yun Y, Lee J, Kim JH, Kang MJ & Lee MJ 2018. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep, 24, 732–743. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy, 12, 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Jain A, Farzam F, Jia J, Gu Y, Choi SW, Mudd MH, Claude-Taupin A, Wester MJ, Lidke KA, Rusten TE & Deretic V 2018. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol, 217, 997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT & Ciechanover A 2017. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci, 42, 873–886. [DOI] [PubMed] [Google Scholar]
- Lansdell TA, Hurchla MA, Xiang J, Hovde S, Weilbaecher KN, Henry RW & Tepe JJ 2013. Noncompetitive Modulation of the Proteasome by Imidazoline Scaffolds Overcomes Bortezomib Resistance and Delays MM Tumor Growth in Vivo. ACS Chem. Biol, 8, 578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC & Glimcher LH 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. U. S. A, 100, 9946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limanaqi F, Biagioni F, Gambardella S, Ryskalin L & Fornai F 2018. Interdependency Between Autophagy and Synaptic Vesicle Trafficking: Implications for Dopamine Release. Front. Mol. Neurosci, 11, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C, Nagy P, Juhasz G & Neufeld TP 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun, 6, 7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA 2012. Lysosomal acidification mechanisms. Annu. Rev. Physiol, 74, 69–86. [DOI] [PubMed] [Google Scholar]
- Minton K 2013. Membrane dynamics: How lysosomes SNARE autophagosomes. Nat. Rev. Mol. Cell Biol, 14, 65. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T & Levine B 2010. Methods in mammalian autophagy research. Cell, 140, 313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooneyham A & Bazzaro M 2017. Targeting Deubiquitinating Enzymes and Autophagy in Cancer. Methods Mol. Biol, 1513, 49–59. [DOI] [PubMed] [Google Scholar]
- Njomen E, Osmulski PA, Jones CL, Gaczynska M & Tepe JJ 2018. Small Molecule Modulation of Proteasome Assembly. Biochemistry, 57, 4214–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njomen E & Tepe JJ 2019. Proteasome Activation as a New Therapeutic Approach To Target Proteotoxic Disorders. J. Med. Chem, asap, DOI: 10.1021/acs.jmedchem.9b00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati AV, Dyczynski M, Ojha R & Amaravadi RK 2018. Targeting autophagy in cancer. Cancer, 124, 3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, Mccray BA, Ritson GP, Nedelsky NB, Schwartz SL, Diprospero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH & Taylor JP 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature, 447, 859–63. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G & Johansen T 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem, 282, 24131–45. [DOI] [PubMed] [Google Scholar]
- Peng H, Yang J, Li G, You Q, Han W, Li T, Gao D, Xie X, Lee BH, Du J, Hou J, Zhang T, Rao H, Huang Y, Li Q, Zeng R, Hui L, Wang H, Xia Q, Zhang X, He Y, Komatsu M, Dikic I, Finley D & Hu R 2017. Ubiquitylation of p62/sequestosome1 activates its autophagy receptor function and controls selective autophagy upon ubiquitin stress. Cell Res, 27, 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M & Finley D 2014. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta, 1843, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shringarpure R, Grune T & Davies KJ 2001. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell. Mol. Life Sci, 58, 1442–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P, Asher G, Paz A, Reuven N, Sussman JL, Silman I & Shaul Y 2008. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins, 70, 1357–66. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N & Shaul Y 2009. The nanny model for IDPs. Nat. Chem. Biol, 5, 778–81. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N & Shaul Y 2010. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ, 17, 103–8. [DOI] [PubMed] [Google Scholar]
- Tsvetkov P & Shaul Y 2012. Determination of IUP based on susceptibility for degradation by default. Methods Mol. Biol, 895, 3–18. [DOI] [PubMed] [Google Scholar]
- Vousden KH & Ryan KM 2009. p53 and metabolism. Nat. Rev. Cancer, 9, 691–700. [DOI] [PubMed] [Google Scholar]
- Watanabe-Asano T, Kuma A & Mizushima N 2014. Cycloheximide inhibits starvation-induced autophagy through mTORC1 activation. Biochem. Biophys. Res. Commun, 445, 334–9. [DOI] [PubMed] [Google Scholar]
- Winkler LL, Hwang J & Kalejta RF 2013. Ubiquitin-independent proteasomal degradation of tumor suppressors by human cytomegalovirus pp71 requires the 19S regulatory particle. J. Virol, 87, 4665–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, Li X, Liang W, Lu X, Qian L & Yuan J 2016. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev, 30, 1718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK & Uversky VN 2010. PONDR-FIT: a metapredictor of intrinsically disordered amino acids. Biochim. Biophys. Acta, 1804, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li CC & Weissman AM 2004. Regulating the p53 system through ubiquitination. Oncogene, 23, 2096–106. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhai B, Gygi SP & Goldberg AL 2015. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. U S A, 112, 15790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Dunner K Jr. & Mcconkey DJ 2010. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene, 29, 451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate and/or analyze datasets or code. No data/code is deposited in repositories.