Abstract
The IFN‐stimulated gene ubiquitin‐specific proteinase 18 (USP18) encodes a protein that negatively regulates T1 IFN signaling via stearic inhibition of JAK1 recruitment to the IFN‐α receptor 2 subunit (IFNAR2). Here, we demonstrate that USP18 expression is induced by HIV‐1 in a T1 IFN‐dependent manner. Experimental depletion of USP18 by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated protein 9 (Cas9) gene editing results in a significant restriction of HIV‐1 replication in an induced pluripotent stem cell (iPSC)‐derived macrophage model. In the absence of USP18, macrophages have increased responsiveness to stimulation with T1 IFNs with prolonged phosphorylation of STAT1 and STAT2 and increased expression of IFN‐stimulated genes that are key for antiviral responses. Interestingly, HIV‐1 requires some signaling through the T1 IFN receptor to replicate efficiently because a neutralizing antibody that inhibits T1 IFN activity reduces HIV‐1 replication rate in monocyte‐derived macrophages. USP18 induction by HIV‐1 tunes the IFN response to optimal levels allowing for efficient transcription from the HIV‐1 LTR promoter while minimizing the T1 IFN‐induced antiviral response that would otherwise restrict viral replication and spread. Finally, iPSC and CRISPR/Cas9 gene targeting offer a powerful tool to study host factors that regulate innate immune responses.
Keywords: CRISPR, IFN, Jak/STAT
Short abstract
USP18 promotes HIV‐1 replication by negatively regulating expression of antiviral genes induced by T1 IFN signaling.
Abbreviations
- ART
antiretroviral therapy
- Cas9
CRISPR‐associated protein 9
- CRISPR
clustered regularly interspaced short palindromic repeats
- HIV‐1
Human immunodeficiency virus 1
- IFNAR
IFN‐α receptor
- iMacs
iPSC‐derived macrophages
- iPSC
induced pluripotent stem cells
- IRF
IFN regulatory factor
- ISG
IFN‐stimulated gene
- LCMV
lymphocytic choriomeningitis virus
- MDM
monocyte‐derived macrophage
- T1 IFN
type I IFN
- USP18
ubiquitin‐specific proteinase 18.
1. INTRODUCTION
It is well established that type I IFNs (T1 IFNs) can restrict acute HIV‐1 infection in vitro.1, 2, 3, 4, 5 In clinical trials treating human patients with recombinant IFN‐α2a, T1 IFNs can suppress viral replication in the absence of antiretroviral therapy (ART) in some patients.6, 7 However, this approach failed in long‐term treatment when study subjects became refractory to IFN treatment and viral loads returned to previous levels. T1 IFNs are important in establishing early control of HIV‐1 in an in vivo SIV rhesus macaque model,8 but chronic T1 IFN signaling correlates with chronic immune activation, immune exhaustion, elevated IFN‐stimulated gene (ISG) expression, and poor long‐term control of HIV‐1.9, 10
Two recently published studies have shown that in a humanized mouse model of HIV‐1 infection that experimental blockade of T1 IFN signaling resulted in restored immune function and rescued T cell function, including HIV‐1‐specific T cells.11, 12 In chronic lymphocytic choriomeningitis virus (LCMV) infection, blockade of IFN‐α receptor (IFNAR) also had significant benefit.13, 14 These studies highlight the importance of IFN regulation in the context of HIV‐1 infection specifically and viral infections in general. However, little is known about how HIV‐1 induces IFNs and why IFNs are unable to control infection in vivo.
Signaling through IFNAR results in the phosphorylation and activation of STAT proteins including STAT1 and STAT2. The consequence of STAT1/2 phosphorylation is induced expression of hundreds of ISGs.15, 16, 17 The protein products of ISGs then target host and viral machinery as a means to restrict viral replication. However, a small subset of the ISGs expressed are negative feedback mechanisms that turn off IFN signaling so that resolution of the immune response can occur. One of these important negative regulators is ubiquitin‐specific proteinase 18 (USP18).
USP18 is an IFN‐inducible deISGylating enzyme that specifically removes the ubiquitin‐like posttranslational modification, ISG15, from target proteins.18, 19, 20 During an IFN response, many newly synthesized proteins are ISGylated,21 which can have a variety of effects depending on the target. Some targets, such as IFN regulatory factor 3 (IRF3), are protected from ubiquitin‐mediated proteasomal degradation.22 It has also been shown that viral proteins, including HIV‐1 Gag, can be ISGylated.23 Durfee and colleagues21 propose that during an infection a small subset of viral structural proteins are ISGylated to disrupt the repeating structures found in viral capsids. Influenza B has evolved a mechanism to directly neutralize ISG15 with its NS1 protein24 and coronaviruses have a papain‐like protease that has deISGylase activity as a strategy to overcome ISG15,25, 26 indicating the importance of ISG15 in the antiviral response.
In addition to its enzymatic activity, USP18 negatively regulates T1 IFN signaling.27 USP18 is recruited by STAT2 to the type I IFN receptor subunit, IFNAR2, where it binds to IFNAR2 and prevents phosphorylation of JAK1 by blocking the interaction of JAK1 and the IFNAR2 subunit.27, 28, 29 USP18 expression also plays a role in limiting TRAIL‐induced apoptosis and has also been shown to regulate the susceptibility of certain cancer cells to IFN‐α and drug‐induced apoptosis.30, 31
Macrophages play an important role in HIV‐1 as reservoirs and can contribute directly to HIV‐1 pathogenesis.32 HIV‐1 in the ART era can be seen as a chronic disease characterized by chronic immune activation and chronic inflammation with a higher risk of non‐AIDS‐related morbidities and mortalities. Macrophages play an important role in this process and can act as mediators of inflammation.33, 34 We recently reported that HIV‐1 replication in macrophages requires activity of STAT1, a protein usually associated with antiviral responses.35 The role of STAT1 was in postintegration expression of HIV‐1 mRNA from the long terminal repeat (LTR) promoter. This paradoxical mechanism where HIV‐1 usurps antiviral pathways as a means of driving its own replication suggests that a complex host–pathogen interplay ultimately determines if HIV‐1 can efficiently replicate in macrophages. In general, the role of macrophages in the HIV‐1 life cycle is important because eliminating persistently infected macrophages in addition to latently infected T cells will be necessary for a sterilizing cure to be achieved. Thus, it is critical that we understand the host–pathogen dynamics that regulate HIV‐1 replication in macrophages and understand how HIV‐1 subverts the innate immune response including the potent antiviral T1 IFN response.
Our data show that USP18 expression is induced by HIV‐1 in human macrophages. The goal of this study was to determine the role of USP18 in HIV‐1 infection/replication and determine if the IFNAR‐blocking effect of USP18 benefits HIV‐1. We hypothesized that USP18 suppresses antiviral pathways that would normally restrict HIV‐1 and that perturbing USP18 would restore and enhance these pathways leading to better control of the virus. To test this, we have utilized a novel clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR‐associated protein 9 (Cas9) knockout induced pluripotent stem cell (iPSC)‐derived macrophage model. Our data show that experimental depletion of USP18 in human macrophage models does restrict HIV‐1 replication due to an enhanced cellular antiviral response and increased sensitivity to the effects of T1 IFN. Thus, USP18 benefits HIV‐1 replication by striking a balance between weak IFN signals required for viral replication and strong IFN signals that would halt viral replication.
2. MATERIALS AND METHODS
2.1. Cell culture
2.1.1. Monocyte‐derived macrophages
PBMCs were prepared from leukopaks obtained from healthy donors from LifeSouth Community Blood Center (Gainesville, FL) under approval by the Institutional Review Board at the University of Florida. PBMCs were isolated by centrifugation on LymphoSep® Lymphocyte Separation Medium (MP Biomedicals, Santa Ana, CA) medium. Monocytes were isolated by positive selection on LS Columns (Miltenyi Biotec, Bergisch Gladbach, Germany) with human CD14 MicroBeads (Miltenyi Biotec). Freshly isolated monocytes were differentiated into macrophages in MDM medium (DMEM with 4.5 g/L glucose (Corning, Corning, NY), 10% heat‐inactivated human serum (Zen‐Bio, Research Triangle Park, NC), 2 mM l‐glutamine (Corning), and 100 IU penicillin–streptomycin (Corning)) supplemented with 10 ng/mL M‐CSF for 7–10 days. After differentiation, the medium was replenished without M‐CSF and the cells rested overnight before being used for experiments.
2.1.2. THP‐1 cells
THP‐1 cells (TIB‐202; ATCC, Manassas, VA) were cultured in THP‐1 medium containing RPMI 1640 with 2 mM l‐glutamine (Corning), 10% heat‐inactivated FBS (Sigma–Aldrich, St. Louis, MO), 100 IU penicillin–streptomycin (Corning), 10 mM HEPES (Corning), 4500 g/L d‐glucose (Sigma–Aldrich), 1 mM sodium pyruvate (Corning), and 0.05 mM 2‐mercaptoethanol (Gibco, Gaithersburg, MD). Cells were differentiated into adherent macrophages by adding 100 nM PMA (Sigma–Aldrich). After 2 days, the PMA was removed and the cells were washed in PBS and rested overnight in THP‐1 medium.
2.1.3. Other cell lines
Lenti‐X 293T cells (Clontech, Mountain View, CA) were cultured and maintained in 293T medium (DMEM (Corning), 10% heat‐inactivated FBS (Sigma–Aldrich), 100 IU penicillin–streptomycin (Corning), 2 mM l‐glutamine (Corning), and 0.05% sodium bicarbonate (Corning)). TZM‐bl cells, obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM‐bl from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc.,36, 37, 38, 39, 40 were cultured and maintained in 293T medium. ACH‐2 cells obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: ACH‐2 from Dr. Thomas Folks,41, 42 were cultured and maintained in RPMI 1640 with 2 mM l‐glutamine (Corning), 10% heat‐inactivated FBS (Sigma–Aldrich), 100 IU penicillin–streptomycin (Corning), and 10 mM HEPES (Corning).
2.2. Cytokines and antibodies
M‐CSF, IL‐3, and IFN‐β were purchased from PeproTech (Rocky Hill, NJ). Anti‐IFNAR2 neutralizing antibody was purchased from PBL Assay Science (Piscataway, NJ). Ultra‐LEAF purified mouse IgG2a, κ isotype control antibody was purchased from BioLegend (San Diego, CA). USP18 [D4E7], p‐STAT1‐Tyr701 [58D6], p‐STAT2‐Tyr690 [D3P2P], and HSP90 [C45G5] monoclonal antibodies, β‐actin and ISG15 polyclonal antibodies, and anti‐mouse IgG, HRP‐linked and anti‐rabbit IgG, HRP‐linked secondary antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti‐HIV‐1 p24 Antibody [39/5.4A] was purchased from Abcam (Cambridge, MA). CD68‐PE [Y1/82A], CD11b‐FITC [ICRF44], CD14‐APC [61D3], and isotype control flow antibodies were purchased from (eBioscience, San Diego, CA).
2.3. HIV‐1 molecular clones
pNL4‐3‐Bal‐IRES‐HSA (HIVHSA), which has been previously described, was a kind gift from Dr. Michel J Tremblay.43 pNL(AD8) (HIVAD) was obtained through NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL(AD8) HIV‐1 AD8 Macrophage‐Tropic R5, 11346 from Fisher BioServices.44
2.4. HIV‐1 virus production
Lenti‐X 293T cells were cultured in a T75 flask and transfected with pNL4‐3‐Bal‐IRES‐HSA using the Viromer® Red transfection reagent (Lipocalyx, Halle, Sachsen‐Anhalt, Germany) or pNL(AD8) using the FuGENE HD transfection reagent (Promega, Madison, WI). The transfection reagent was removed after 4 h and the medium was replenished. After 48 h, the infectious supernatants were collected, clarified, and filtered through 0.45 μM pore size Whatman PES membranes (Fisher Scientific, Suwanee, GA). Heat‐inactivated FBS (Sigma–Aldrich) was added to the virus at a concentration of 10% and stored at −80°C. After 24 h, 1 aliquot of frozen virus was quickly thawed in a 37°C water bath. The virus was titered on TZM‐bl cells for 48 h and a luciferase assay was performed. The titer was determined by the Spearman‐Kärber method45, 46 and expressed as a TCID50/μL.
2.5. TZM‐bl cell luciferase assays
Medium was removed by aspiration and 100 μL of room temperature Bright‐Glo™ Luciferase Assay System (Promega) was added to the cells. After 5 min, the cells were lysed and the lysate was transferred to a black Costar EIA/RIA polystyrene half area 96‐well plate (Corning). Luminescence was measured with the VICTOR™ X4 Multi‐Plate Reader (PerkinElmer, Waltham, MA).
2.6. ELISA
CXCL10 was measured in cell‐free supernatants by Human CXCL10/IP‐10 DuoSet ELISA kit according to the manufacturer's instructions (R&D Systems, Minneapolis, MN). HIV‐1 Gagp24 was measured in cell‐free supernatants lysed with 0.5% Triton X‐100 in PBS by HIV‐1 Gagp24 ELISA kit according to the manufacturer's instructions (Sino Biological, Beijing, China).
2.7. Microarrays
Monocyte‐derived macrophages (MDMs) were infected with 500 TCID50 HIVAD for 7 days. Total RNA was collected with RNeasy Mini Kit (Qiagen, Valencia, CA). Gene expression was assessed with GeneChip™ Human Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA) by the Interdisciplinary Center for Biotechnology Research at the University of Florida. Analysis was performed with Partek Genomics Suite v. 6.6 (Partek Inc., St. Louis, MO). CEL files were imported with GC correction and intensity data were transformed to log base 2. Exons were summarized to genes using the median method and an ANOVA with contrast was performed to determine fold changes between control and treatment groups. Microarray data have been deposited at Gene Expression Omnibus (accession# GSE108897).
2.8. Western blotting
Cells were washed with PBS and lysed in 1× Cell Lysis Buffer (Cell Signaling Technology) with protease Inhibitor Cocktail (Sigma–Aldrich) and 1 mM PMSF (Sigma–Aldrich). Proteins were separated by size by SDS‐PAGE with 4–20% Mini‐PROTEAN® TGX™ precast gels (Bio‐Rad, Hercules, CA) and electroblotted onto Trans‐Blot® Turbo™ Mini PVDF membranes (Bio‐Rad) using the Trans‐Blot® Turbo™ Transfer System (Bio‐Rad) according to the manufacturer's instructions. Membranes were blocked in 5% nonfat milk in TBST. The Super Signal® West Dura Extended Duration Substrate kit was used for chemiluminescence detection of HRP‐linked secondary antibodies. Blots were exposed to CL‐Xposure™ Film (Thermo Scientific, Waltham, MA). Blots were stripped by incubating with Restore™ PLUS Western Blot Stripping Buffer (Thermo Scientific).
2.9. shRNA lentivirus
Human GIPZ lentiviral shRNA gene set for USP18‐specific shRNA were purchased from GE Dharmacon (Lafayette, CO). The shRNA clones were screened in HEK 293 cells and clone V3LHS_645758 (USP18‐5) was determined to most efficiently knockdown USP18 (data not shown) and was used for all subsequent experiments. GIPZ plasmid DNA was cotransfected into Lenti‐X 293T packaging cells cultured in 293T medium with Tet System approved FBS (tetracycline‐free) using the Lenti‐X HTX Packaging System (Clontech) and Xfect Transfection Reagent (Clontech) according to the manufacturer's instructions. The transfection reagent was removed 24 h posttransfection. After 48 h, supernatants containing VSV‐G pseudotyped lentivirus were harvested and clarified by centrifugation at 20,000 x g for 18 h. The supernatant was decanted and the pellet was resuspended in THP‐1 medium. THP‐1 cells were transduced with TransDux (System Biosciences, Mountain View, CA) by spinoculation for 1 hour at 1200 x g at room temperature. Puromycin (1 μg/ml) was used to select for transduced cells 48 h posttransduction.
2.10. siRNA transfections
ON‐TARGETplus Non‐targeting Pool control siRNA (D‐001810‐10‐05) and ON‐TARGETplus USP18 siRNA SMARTpool (L‐004236‐00‐0005) were purchased from GE Dharmacon. siRNA was transfected into cells with Viromer® Blue transfection reagent (Lipocalyx) at a concentration of 25 nM according to the manufacturer's instructions.
2.11. RT‐qPCR
Total RNA was isolated with RNeasy Plus Mini Kit (Qiagen). cDNA synthesis was carried out with High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Predesigned PrimeTime® qPCR Assays for IFIT1, IFIT2, IFIT3, IFITM1, IFITM2, USP18, CXCL9, CXCL10, ISG15, and MX2 were purchased from Integrated DNA Technologies (Coralville, IA). RT‐qPCR reactions were carried out in TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) according to the manufacturer's instructions. RT‐qPCR reactions were performed on the StepOnePlus™ Real‐Time PCR System (Applied Biosystems).
2.12. Reprogramming CD34+ to iPSCs
Donor CD34+ hematopoietic progenitor cells were isolated from peripheral blood utilizing the EasySep Complete Kit for Human Whole Blood CD34+ Cells (Stem Cell Technologies, Vancouver, Canada). Isolated CD34+ cells were expanded in StemSpan SFEM II Medium plus StemSpan CD34+ Expansion supplement for 1 week. Following expansion, cells were infected with Sendai viral vector SeVdp(KOSM)302L encoding 4 reprogramming factors (OCT4, SOX2, KLF4, and c‐MYC).47 Infection was carried out for 2 h at 37°C at an MOI of 2. Cells were reprogrammed on Matrigel‐coated dishes using ReproTeSR (Stem Cell Technologies) following the manufacturer's instructions. Approximately 3 weeks after infection, 6 iPSC colonies were manually isolated to generate iPSC clones. iPSC clones were maintained and expanded on Matrigel‐coated plates in mTeSR1 (Stem Cell Technologies). iPSCs were characterized by assessing morphology, expression of stem cell markers OCT4 and SSEA4 by flow cytometry analysis, and karyotyping. Pluripotency was confirmed by differentiating the cells to ectoderm, endoderm, and mesoderm lineages using the STEMdiff Trilineage Differentiation Kit (Stem Cell Technologies).
Flow cytometry was used to examine undifferentiated iPSCs stained with OCT4‐PE, SSEA4‐APC, and SSEA1‐PE antibodies (BioLegend). For trilineage differentiated iPSCs, ectoderm was assessed by NESTIN‐PE and PAX6‐Alexa Fluor 647 (BD Biosciences, San Jose, CA). Endoderm was assessed by FOXA2‐PE and SOX17‐Alexa Fluor 647 (BD Biosciences). Mesoderm was assessed by Brachyury‐APC (R&D Systems) and NCAM‐PE (Stem Cell Technologies).
2.13. Differentiation of iPSCs to monocytes
Differentiation was carried out as previously described48 with slight modifications. Briefly, iPSCs were passaged sparsely to 6‐well Matrigel‐coated plates and cultured for 8–10 days in mTeSR1 with daily medium changes until the colonies were approximately 5 mm in diameter. Next, embryoid bodies (EBs) were formed by gently lifting colonies using a cell lifter. Colonies were gently transferred to a 15 mL conical tube using a 10 mL serological pipette. Colonies were allowed to gravity settle for approximately 5 min and supernatant was aspirated. Cells were gently resuspended in mTeSR1 supplemented with 10 μM ROCK inhibitor (Y‐27632) and transferred to Ultra‐Low Attachment Surface 6‐well plates (Corning) containing 4 mL medium per well. EBs were allowed to form for 4 days with a partial (2/3) medium change after 48 h. On day 4, EBs were collected, washed, resuspended in X‐VIVO15 supplemented with GlutaMAX™ (Gibco) and 2‐mercaptoethanol (Gibco), and transferred to adherent, cell culture‐treated plates.
2.14. iPSC USP18 gene knockout using CRISPR/Cas9
iPSCs were transfected using the Human Stem Cell Nucleofector Kit 1 (Lonza, Basal, Switzerland) following manufacturer's recommendations with 3 plasmids: 1 plasmid that encodes S. pyogenes Cas9, 1 plasmid that contains the USP18 gRNA sequence GCAAATCTGTCAGTCCATCC, and a donor plasmid with homology arms that flank the CRISPR site in USP18 exon 2 that delivers a GFP and puromycin resistance gene cassette. Briefly, equal amounts of each of the 3 plasmids (5 μg total) were nucleofected into 8 × 105 cells using Nucleofector II (Amaxa Biosystems, Cologne, Germany) program B‐016. Cells were re‐plated on a Matrigel‐coated dish with mTeSR1 and allowed to recover for 3 days with daily medium changes. GFP fluorescence was observed after 24 h. On day 3, the cells were maintained under puromycin (0.3 μg/ml) selection for 30 days. Ten clones were manually isolated and expanded in mTeSR1. Genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen) for screening. Primer sequences for screening for CRISPR modifications are in Supplemental Table S1.
2.15. DQ‐Ovalbumin and phagocytosis assays
DQ‐Ovalbumin (Invitrogen, Carlsbad, CA) was added to the cells at a concentration of 50 μg/ml. NucBlue® Live ReadyProbes® Reagent (Molecular Probes, Eugene, OR) were also added to the well and incubated at 37°C for 25–30 min. The cells were washed with prewarmed PBS and fixed in 4% paraformaldehyde. pHrodo Red Bioparticles were resuspended in Live Cell Imaging Solution (Gibco) at 1 μg/mL and sonicated for 5 min in a water bath sonicator. The resuspended pHrodo Red‐labeled heat‐killed Escherichia coli or Staphylococcus aureus were added at a final concentration of 1 μg/mL in Live Cell Imaging Solution to macrophages and incubated at 37°C for 1–2 h to allow for uptake and acidification of phagosomes. NucBlue® Live ReadyProbes® Reagent (Molecular Probes) was added before imaging. All fluorescent images were obtained with the EVOS™ FL Cell Imaging System (Invitrogen).
2.16. Macrophage morphology
MDMs or iMacs were washed in PBS and fixed in 4% cold paraformaldehyde for 10 min. The cells were washed again in PBS and permeabilized in 0.1% Triton X‐100. The cells were washed in PBS again and then stained with either ActinGreen™ 488 ReadyProbes® or ActinRed™ 555 ReadyProbes® reagent (Molecular Probes) for 30 min followed by 2 more washes in PBS. The cells were then imaged with the EVOS™ FL Cell Imaging System (Invitrogen).
2.17. Flow cytometry
Fc receptors were blocked with Fc Block (Miltenyi) and stained with antibodies in PBS with 1% human serum. For intracellular staining of CD68, the cells were fixed and permeabilized with the BD Cytofix/Cytoperm™ kit (BD Biosciences). All samples were analyzed with the BD Accuri™ C6 Cytometer (BD Biosciences).
2.18. Integration assay
The integration assay for detection of integrated HIV‐1 proviral DNA was adapted from methods previously described.49, 50 Briefly, genomic DNA was isolated with the DNeasy Blood and Tissue Kit (Qiagen). A preamplification step with 200 nM Alu1, Alu2, and ULF2 primers was performed with 200 ng of genomic DNA in AmpliTaq Gold® 360 Master Mix with 360 GC Enhancer (Applied Biosystems) and incubated in a thermal cycler at 95°C for 10 min followed by 20 cycles of 95°C for 30 sec, 55°C for 30 s, and 72°C for 25 s, and a final extension step at 72°C for 7 min. A second round of PCR was performed with 1/100th of the product from the preamplification step with 200 nM Lambda T2 and UR2 primers and 150 nM Fam‐Int‐HIV‐IABkFQ TaqMan probe in TaqMan® Universal Master Mix (Applied Biosystems) at the thermal cycler conditions recommended by the manufacturer for the master mix. In parallel to the samples, a standard curve also underwent the same preamplification and second amplification steps.
The standard curve was prepared from ACH‐2 cells, which is a T cell line that has exactly 1 integrated HIV‐1 proviral genome. The standard curve was prepared by making 1:10 dilutions of 2000 ng of genomic DNA from ACH‐2 cells. Carrier DNA from uninfected PBMCs was added so that the total genomic DNA in each standard was 2000 ng. Primer sequences are listed in Supplemental Table S1. Using an estimate of ∼6.6 pg of genomic DNA per cell, the number of copies of integrated proviral genomes per 10,000 cells was calculated from the standard curve.
3. RESULTS
3.1. T1 IFNs restrict HIV‐1 replication
It has been demonstrated previously in vitro that T1 IFNs restrict HIV‐1 replication.1, 2, 3, 4, 5, 51 To confirm these previous findings, we pretreated TZM‐bl cells with different doses of IFN‐β for 24 h and infected the cells with HIV‐1. As previously reported, T1 IFN restricted HIV‐1 replication in a dose‐dependent manner (Fig. 1A). We also tested this in MDMs and found that HIV‐1 replication is potently restricted by IFN‐β in a dose‐dependent manner (Fig. 1B) with an IC50 of 504.7 fg/mL (± 142.1 fg/mL).
Figure 1.
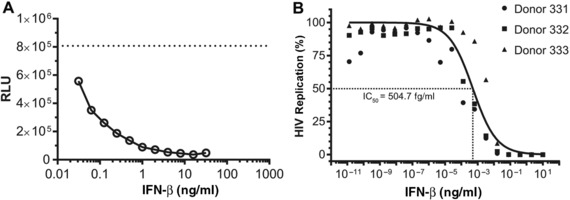
Type I IFN restricts HIV‐1 replication. (A) TZM‐bl cells were plated in 96‐well plates (10,000 cells/well) and allowed to adhere overnight. The following day, the cells were pretreated with IFN‐β for 24 h and then infected with 100 TCID50 HIVHSA. After 48 h, the amount of HIV‐1 replication was measured by luciferase assay. The dashed line indicates the level of luciferase activity in cells that were not pretreated with IFN. (B) Monocytes (100,000 cells/well) were differentiated into MDMs in M‐CSF and then pretreated with IFN‐β for 24 h followed by infection with 150 TCID50 HIVHSA for 7 days. Supernatant Gagp24 was measured by ELISA (n = 3 donors). Data were normalized to Gagp24 levels of infected cells with no IFN‐β treatment (100% replication) and no infection (0% infection) for each donor. A nonlinear regression was plotted of the combined data with GraphPad Prism (v 7.02) using the least squares fit method to determine the IC50
3.2. USP18 is induced by HIV‐1 in MDMs and requires T1 IFN activity
Since HIV‐1 is restricted by T1 IFNs in vitro, we wanted to determine if HIV‐1 was inducing an IFN response in MDMs. To test this, MDMs were infected with HIV‐1 for 7 days and gene expression was analyzed by microarray (Fig. 2A). We observed that all genes up‐regulated at least 2‐fold by HIV‐1 (Table 1) have been reported to be IFN‐inducible in the Interferome (v2.01) database (Fig. 2B). However, this IFN‐like response is not strong enough to inhibit HIV‐1 replication to the same degree as the addition of exogenous IFN (Fig. 1). We were interested in how the IFN response was being attenuated and allowing for HIV‐1 replication to occur despite induction of ISGs. Interestingly, HIV‐1 induces expression of USP18 (Table 1), a negative regulator of T1 IFN signaling.27 Western blot analysis confirmed this at the protein level with 2 different strains of HIV‐1 (Fig. 2C).
Figure 2.
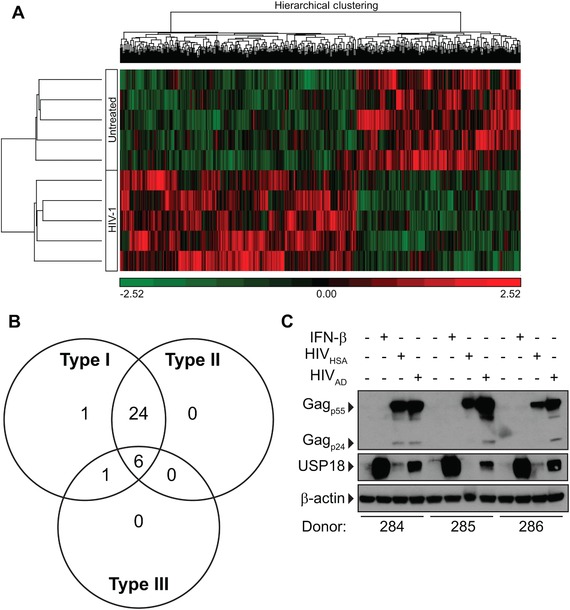
HIV‐1 induces an IFN‐like response in MDMs. MDMs were infected with HIVAD for 7 days and gene expression was assessed by Affymetrix arrays. (A) Heat map showing differential expression of 560 genes that differed significantly (P < 0.05, unadjusted P value) between untreated and HIV‐1 infected samples. (B) Venn diagram showing genes that were induced by HIVAD by 2‐fold or greater are also inducible by type I, type II or type III IFNs. Information based on Interferome (v2.01) database. (C) MDMs were infected with 5000 TCID50 HIVHSA or HIVAD for 7 days. USP18 and Gagp24 expression was assessed by Western blot
Table 1.
HIV‐1 induced gene expression
| Gene symbol | Fold‐change (HIV vs. untreated) |
|---|---|
| CCL7 | 6.06824 |
| IFI44L | 5.89781 |
| IFIT1 | 5.53147 |
| CXCL11 | 5.01958 |
| RSAD2 | 4.75459 |
| IFIT2 | 4.64842 |
| CCL8 | 4.58559 |
| TNFSF10 | 4.42962 |
| CXCL10 | 4.01804 |
| APOBEC3A_B | 3.74453 |
| IFITM1 | 3.33806 |
| USP18 | 2.98445 |
| IFI44 | 2.73129 |
| SIGLEC1 | 2.51668 |
| IFITM2 | 2.48532 |
| OAS3 | 2.4681 |
| GMPR | 2.46577 |
| EPSTI1 | 2.46526 |
| SERPING1 | 2.40793 |
| HERC5 | 2.38334 |
| SP110 | 2.34157 |
| HERC6 | 2.30317 |
| DDX60 | 2.28238 |
| OAS2 | 2.25047 |
| IFI35 | 2.2397 |
| MX1 | 2.22373 |
| MX2 | 2.20131 |
| TLR3 | 2.17625 |
| MFAP5 | 2.16209 |
| NT5C3A | 2.14975 |
| IFIT3 | 2.14181 |
| JUP | 2.13316 |
| IFI27 | 2.04006 |
| FBXO6 | 2.02068 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
It is unknown how HIV‐1 induces USP18 expression in human macrophages. Since USP18 is an ISG, we first pretreated MDMs with α‐IFNAR neutralizing antibodies then infected the cells to see if USP18 was still induced by HIV‐1 in the absence of T1 IFN signaling. The results show that USP18 is not induced by HIV‐1 in the absence of T1 IFN signaling (Fig. 3), providing evidence that HIV‐1 does induce an IFN response in macrophages that has an autocrine effect.
Figure 3.
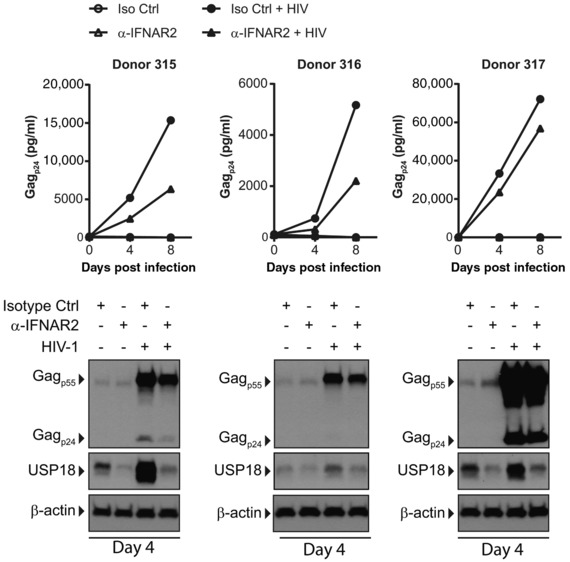
IFNAR blocking inhibits HIV‐1 replication and blocks HIV‐1‐induced USP18 expression in MDMs. MDMs were infected with 1000 TCID50 HIVAD in the presence of α‐IFNAR2 neutralizing antibody or IgG2a isotype control antibody. Supernatants were harvested on days 4 and 8 postinfection and supernatant Gagp24 was measured by ELISA. Protein lysates were collected on day 4 postinfection and Gagp24 and USP18 expression were assessed by Western blot
Surprisingly, HIV‐1 replication was inhibited, not enhanced, in the absence of T1 IFN signaling as measured by Gagp24 ELISA on supernatants (Fig. 3). This finding agrees with previous work where we demonstrated the importance of STAT signaling for HIV‐1 replication in MDMs.35 These data demonstrate that HIV‐1 must strike a balance between some T1 IFN signaling, which is needed for producing transcription factors such as STAT1 and STAT3, and too much IFN signaling, which allows the cells to mount an effective antiviral response. Given that USP18 attenuates the IFN response, we hypothesized that induction of USP18 allowed HIV‐1 to achieve the balance needed to provide the necessary STAT signaling, without too strong an antiviral response.
3.3. USP18 deficiency enhances IFN signaling through JAK‐STAT pathway
USP18 is part of the normal negative feedback response that regulates T1 IFN signaling through IFNAR and JAK‐STAT signaling. In the absence of USP18, signaling through the JAK‐STAT pathway should be enhanced with increased levels of phosphorylated STAT1 and STAT2. We used siRNA to knockdown USP18 expression in MDMs and treated them with IFN‐β for 18 hours and found increased levels of phosphorylated STAT1 and STAT2 (Fig. 4A).
Figure 4.
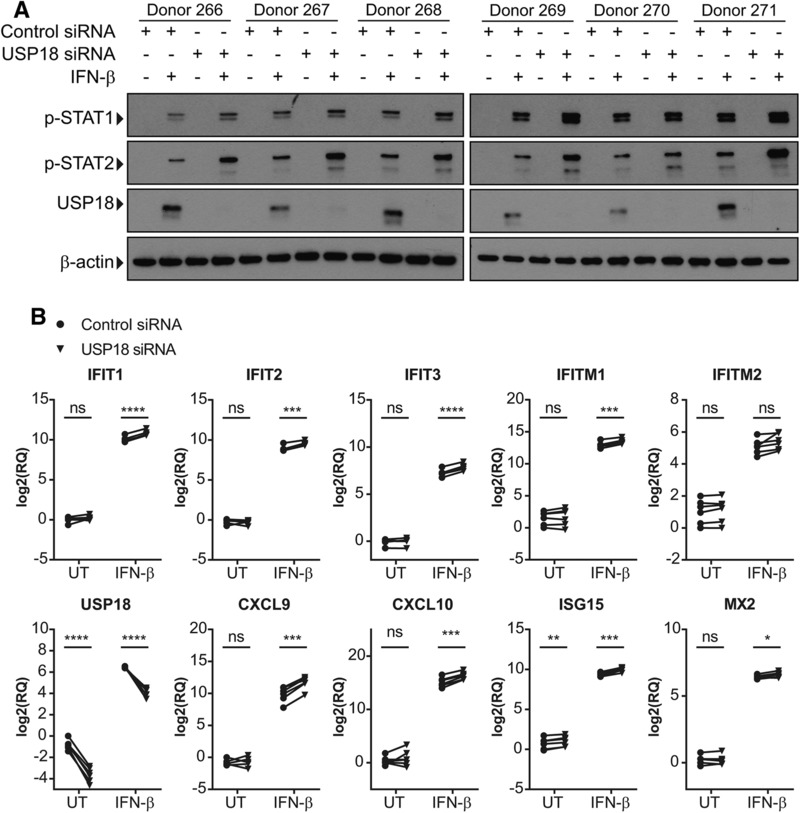
siRNA knockdown of USP18 enhances STAT activation and expression of ISGs in IFN‐β‐treated MDMs. MDMs from 6 donors were transfected with nontargeting (NT) control or USP18 siRNA for 3 h followed by IFN‐β treatment for 18 h. (A) Expression of p‐STAT1, p‐STAT2, and USP18 was measured by Western blot. (B) Expression of ISGs was measured by RT‐qPCR. A paired T‐test was used to compare control siRNA treated and USP18 siRNA treated samples that were untreated or IFN‐β treated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant
Increased signaling through STAT1 and STAT2 should result in enhanced expression of ISGs. To test this, MDMs transfected with USP18 siRNA were treated with IFN‐β for 18 h and gene expression of ISGs was analyzed by RT‐qPCR. In a RT‐qPCR panel looking at IFIT1, IFIT2, IFIT3, IFITM1, IFITM2, CXCL9, CXCL10, ISG15, and MX2, all genes had increased expression when USP18 was knocked down compared with control (Fig. 4B). USP18 expression was also measured to confirm knockdown of USP18 transcripts.
3.4. USP18 knockdown restricts HIV‐1 replication
We next wanted to determine if USP18 expression is necessary for HIV‐1 replication. Although transfecting MDMs with siRNA allowed for efficient knockdown of USP18 (Fig. 4), it also made the cells refractory to infection even with a nontargeting control siRNA (data not shown). Instead, we utilized shRNA knockdown of USP18 in the THP‐1 cell line. THP‐1 cells are suspension cells that can be differentiated into adherent macrophage‐like cells with PMA treatment. They are a well‐established model system for studying HIV‐1 infection in macrophages.52
We generated THP‐1 cell lines that constitutively express nontargeting control or USP18‐specific shRNA (Supplemental Figs. S1A and S1B). After PMA differentiation, THP‐1 cells expressing non‐targeting control or USP18 shRNA were infected by HIV‐1. We found that HIV‐1 replication was significantly restricted in THP‐1 cells with USP18 knockdown. There was significantly less Gagp24 detected in the supernatants and in the protein lysates of USP18 knockdown cells compared with control (Figs. 5A and 5B).
Figure 5.
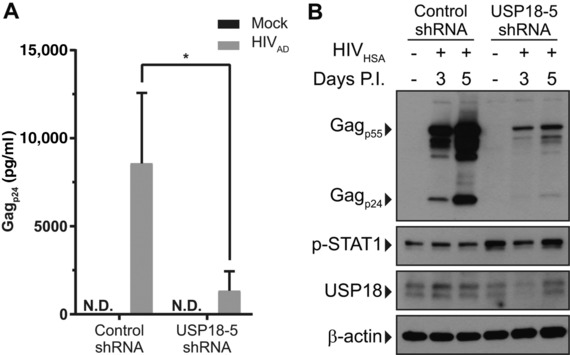
shRNA knockdown of USP18 inhibits HIV‐1 replication in THP‐1 cells. THP‐1 cells expressing control or USP18‐5 shRNA were activated with PMA. (A) THP‐1 cells were infected with 5000 TCID50 HIVAD for 5 days and supernatant Gagp24 was measured by ELISA. (B) THP‐1 cells were infected with 5000 TCID50 HIVHSA for 5 days and intracellular Gagp24 expression was measured by Western blot. A ratio paired T‐test was used to compare supernatant Gagp24 levels between control shRNA and USP18‐5 shRNA expressing cells. N.D., not detected, *P < 0.05. Error bars represent standard deviation between 4 replicates
3.5. iPSC‐derived macrophages
While THP‐1 cells and MDMs with USP18 knockdown provide useful models for investigating the effects of USP18 deficiency on IFN signaling, it may not be the best model. THP‐1 cells are transformed cells with an abnormal karyotype53 and in our hands they do not remain differentiated and adherent in culture for more than 5 days (data not shown). While we could achieve efficient knockdown of USP18 in MDMs, the transfected cells became refractory to infection even when a nontargeting control siRNA was used. To overcome these obstacles, we sought to use an iPSC‐derived macrophage model.
iPSC‐derived macrophages have been previously shown to be a suitable model to study HIV‐1.48 First, we generated iPSCs from CD34+ cells isolated from peripheral blood of healthy adult human donors (Supplemental Fig. S2). The CD34+ cells were expanded in vitro and transduced with a nonintegrating Sendai viral vector (SeVdp(KOSM)302L) encoding OCT4, SOX2, KLF4, and c‐MYC to reprogram them into iPSCs.47, 54 For differentiation, we utilize an EB method in which EBs are formed and cultured in the presence of IL‐3 and M‐CSF to generate monocytes.48 The monocytes are then cultured in M‐CSF alone to produce MDMs from iPSCs (iMacs).
iMacs are CD14+, CD11b+, and CD68+ and have similar morphology to MDMs (Supplemental Figs. S3A and 3B). To confirm that iMacs also function like MDMs, iMacs were treated with DQ‐OVA, which is an ovalbumin that is fluorescently labeled with an intramolecular quencher. If the protein is phagocytosed and processed by acidic proteases, it will be digested freeing the fluorophore and quencher allowing for fluorescence. iMacs fed DQ‐OVA were positive for BODIPY FL dye indicating that ovalbumin was processed by acidic proteases (Supplemental Fig. S3C). To determine if iMacs can phagocytose whole bacteria and process them in acidified phagosomes, heat killed E. coli and S. aureus labeled with a pH sensitive dye were fed to iMacs. After 2 h, the iMacs were RFP positive indicating that they phagocytosed the bacteria and that the phagosomes were acidified (Supplemental Fig. S3C).
3.6. CRISPR knockout of USP18 in iMacs
To test the effects of USP18 deficiency in iMacs, we utilized CRISPR/Cas9 gene editing to knockout USP18 in iPSCs before differentiation to the myeloid lineage. The advantage of knocking out USP18 in iPSCs is that once knockout is achieved, we have a self‐renewing iPSC line that can be a continuous source of new EBs for making iMacs. To knockout USP18, iPSCs were transfected with 3 plasmids that delivered a USP18 targeting single‐guide RNA (sgRNA), Cas9, and a donor plasmid with homology arms to the region where the sgRNA targets USP18 in exon 2 (Fig. 6A). The donor DNA contains a GFP and puromycin resistance gene allowing for selection of successfully modified cells. Puromycin resistant clones were screened by PCR and Western blot for successful knockout of USP18 (Figs. 6B and 6C). Two knockout clones (GFP+ clone 5 and GFP− clone 2) were obtained and used in subsequent experiments. Lack of GFP expression in USP18 knockout clone 2 is likely due to silencing of the EF1‐alpha promoter, a phenomenon that we observe in a portion of modified iPSC lines.
Figure 6.
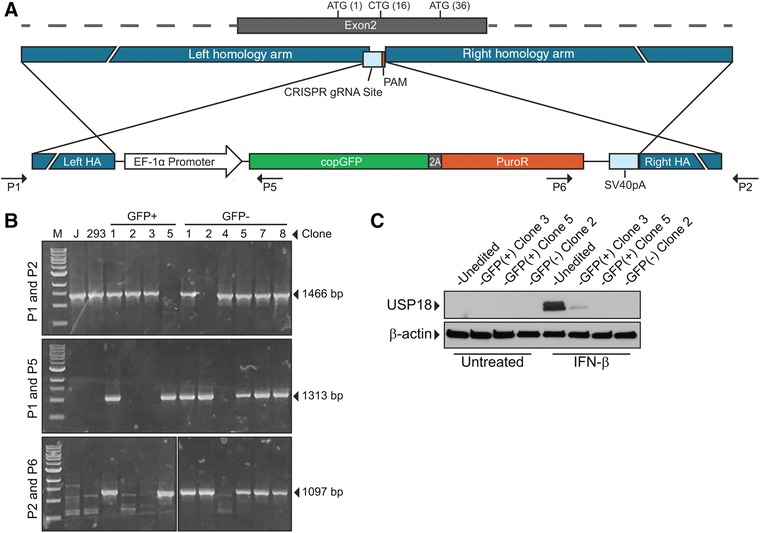
CRISPR/Cas9 knockout of USP18 in iPSCs. (A) Schematic showing the CRISPR gRNA target site in Exon 2 of USP18. Homology regions flanking the CRISPR site were placed in the donor plasmid with an EF‐1α promoter driving expression of a copGFP‐T2A‐PuroR transcript. After a double‐stranded break, homology‐directed repair in the presence of the donor plasmid will allow for integration of selection cassette at CRISPR site disrupting expression of USP18. (B) Clones were screened by PCR with 3 primer sets. Primer set A (P1 and P2) would amplify a 1466 bp product if there was no modification of the USP18 locus. Primer sets B (P1 and P5) and C (P2 and P6) would amplify 1313 bp and 1097 bp products, respectively, if the resistance cassette was integrated at the USP18 locus. Clones that had integration in only 1 allele had the wild‐type allele sequenced to detect INDELs. One lane that had another marker ladder was omitted from the image in the bottom panel. For control, PCR products from Jurkat T cells (J) and HEK 293 cells (293) were also analyzed by gel electrophoresis. (C) Clones that had INDELs resulting in frameshift or clones that had insertion of selectable cassette in both alleles were treated with IFN‐β for 18 h and USP18 expression was measured by Western blot
To determine if USP18 knockout (USP18−/−) iMacs have a similar phenotype to USP18 knockdown in MDMs, USP18−/− iMacs were treated with IFN‐β and levels of phosphorylated STAT1 and STAT2 were measured by Western blot. USP18−/− iMacs had increased and sustained phosphorylated STAT1 and phosphorylated STAT2 (Fig. 7A). Since USP18−/− iMacs also had increased STAT1 and STAT2 signaling, we wanted to determine if USP18−/− iMacs also had enhanced ISG expression after IFN‐β treatment. Indeed, after treatment with IFN‐β for 18 h, USP18−/− iMacs had increased expression of ISGs compared with USP18 sufficient cells (Fig. 7B). These findings are consistent with the results from siRNA knockdown in MDMs.
Figure 7.
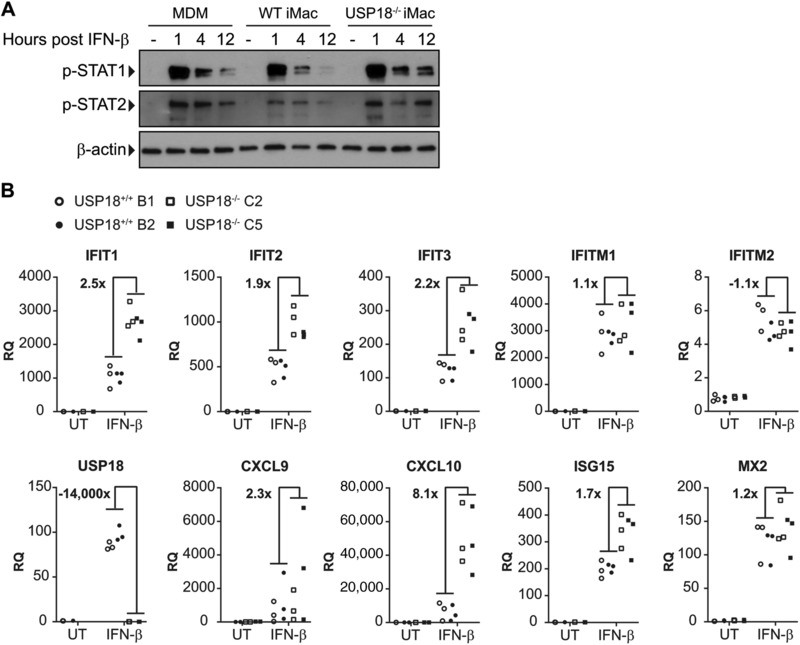
USP18−/− iMacs have enhanced STAT phosphorylation and enhanced ISG expression. (A) iMacs and MDMs were treated with IFN‐β and p‐STAT1 and p‐STAT2 expression was measured by Western blot 1, 4, and 12 h after treatment. (B) iMacs were treated with IFN‐β for 18 h and gene expression of ISGs was measured by RT‐qPCR (n = 3). The average RQ of the USP18−/− clones divided by the average RQ of the USP18+/+ clones are expressed as fold change
3.7. HIV‐1 replication is restricted in USP18 knockout iMacs
We next determined if iMacs can support HIV‐1 replication as previously reported48 and if USP18 is induced by HIV‐1 in iMacs. Indeed, USP18+/+ iMacs support HIV‐1 replication and USP18 is induced by HIV‐1 in these cells (Figs. 8A and 8B). Since USP18 is induced by HIV‐1 in iMacs in a similar manner as MDMs, we concluded that iMacs are a better model for studying USP18 knockout/knockdown than THP‐1 cells.
Figure 8.
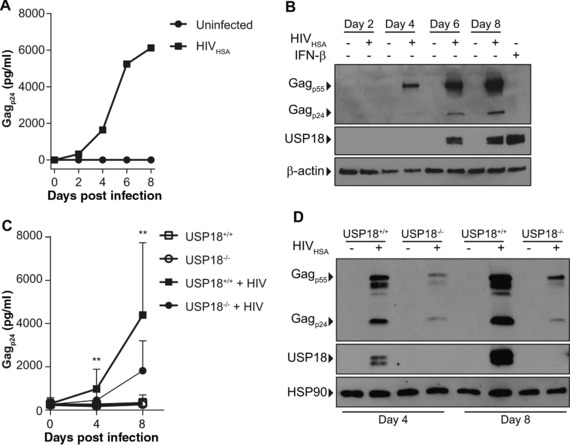
iMacs support HIV‐1 replication. iMacs were infected with 1000 TCID50 HIVHSA for 8 days. (A) Supernatant Gagp24 was measured by ELISA. (B) Intracellular Gagp24 and USP18 expression was measured by Western blot. (C) USP18+/+ and USP18−/− iMacs were infected with 1000 TCID50 HIVHSA for 8 days. Supernatant Gagp24 was measured by ELISA (n = 5). (D) Intracellular Gagp24 and USP18 were measured by Western blot. Representative blot of 3 experiments is shown. A ratio paired T‐test was used to determine statistical significance. **P < 0.01. Error bars represent standard deviation
Next, we wanted to determine if USP18 knockout influenced HIV‐1 replication. USP18 knockout results in an enhanced response to IFN stimulation in macrophages (Fig. 7). Therefore, we hypothesized that HIV‐1 replication would be restricted in USP18−/− iMacs due to enhanced IFN signaling in response to infection. iMacs were infected with HIV‐1 and there was a significant reduction in supernatant Gagp24 and intracellular Gagp24 as determined by ELISA and Western blot in USP18−/− iMacs at days 4 and 8 postinfection (Figs. 8C and 8D).
We also wanted to determine if the restriction in HIV‐1 replication was due to fewer cells becoming infected during the first round of infection or if it was due to decreased spreading. To test this, iMacs were infected and treated with Darunavir, an HIV‐1 protease inhibitor, to prevent subsequent rounds of infection. After 24 h, genomic DNA was collected and a qPCR assay developed to measure integrated proviral genomes showed that there were on average 3800 genomes per 10,000 cells in USP18+/+ iMacs and on average 4900 integrated genomes per 10,000 cells in USP18−/− iMacs treated with Darunavir 24 h postinfection. This suggests that the decrease in viral replication seen over time at day 4 and 8 is due to restricted production and spreading of the virus and not due to decreased or inhibited entry into cells.
4. DISCUSSION
We report here that USP18 deficiency restricts HIV‐1 replication in human macrophages using an iPSC model. The restriction on replication was due to an enhanced IFN response from a lack of feedback inhibition on the JAK/STAT pathway resulting in increased expression of ISGs that have antiviral effects. Previous work done by our group has shown that STAT1 and STAT3 are necessary for efficient HIV‐1 replication. Blocking phosphorylation of either of these 2 molecules resulted in significant reduction in viral replication.35 However, the current study shows that increasing the amount of phosphorylated STAT1 and STAT2 also results in a significant reduction in viral replication. Thus, we propose that USP18 acts as a tuning mechanism which prevents a robust IFN response that would restrict viral replication (Fig. 9).
Figure 9.
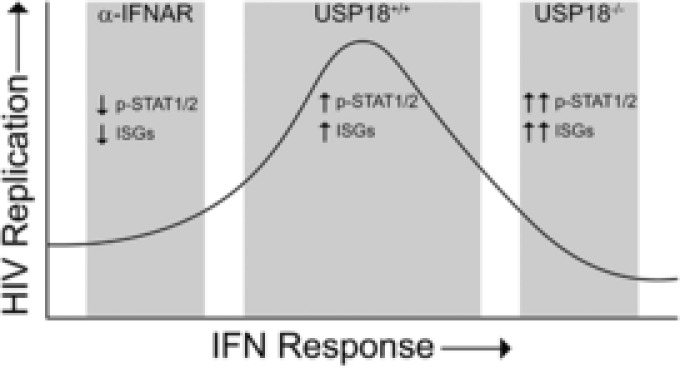
USP18 tunes the IFN response to allow for efficient HIV‐1 replication. A low‐level IFN response in the presence of IFNAR neutralizing antibodies characterized by low levels of phosphorylated STAT1/2 and low expression of ISGs limits HIV‐1 replication. In the absence of USP18, a strong IFN response characterized by high levels of phosphorylated STAT1/2 and high expression of ISGs also limits HIV‐1 replication. In the presence of USP18, the optimal levels of phosphorylated STAT1/2 and ISG expression allows for robust HIV‐1 replication
Previous work has shown that Usp18 −/− mice exhibit protection from intracerebral infection by LCMV and VSV.55 Usp18−/− mice also have an increased ability to restrict Salmonella typhimurium replication and increased survival when lethally challenged with S. typhimurium. The basis of antibacterial protection in USP18 deficiency can be attributed to hypersensitivity to LPS as a consequence of an overactive T1 IFN response.56 Mechanistically, these studies showed that USP18 deficiency resulted in increased ISGylation of proteins and increased phosphorylated STAT1 levels in response to LPS treatment or viral infection. It was based on these properties that we sought to determine if human USP18 regulates HIV‐1 replication.
Differences in biochemical regulation of USP18 in humans compared with mice provide rationale to study USP18 deficiency in human cells. For example, although human USP18 and mouse Usp18 share general functions, human USP18 is ISGylated, which promotes stability of the USP18 protein. Thus, USP18 and ISG15 knockout cells have similarly enhanced ISG responses in humans due to the fact that in ISG15 deficiency, USP18 is destabilized.57, 58 Infants born with naturally occurring USP18 deficiency experience pseudo‐toxoplasmosis, other, rubella, cytomegalovirus, and herpes simplex (pseudo‐TORCH) syndrome as the result of aberrant IFN responses in the absence of congenital infections,59 leading us to believe that the overall hyperactive T1 IFN phenotype observed in mice would be recapitulated in humans.
Similar to mouse studies, our current work shows that USP18 deficient human macrophages also have enhanced levels of phosphorylated STAT1 and phosphorylated STAT2 after treatment with IFN‐β. We expected that if there was enhanced signaling through IFNAR and STAT1/STAT2 that there would be increased expression of ISGs, many of which have known anti‐HIV properties. Indeed, we found that in USP18 deficient cells expression of ISGs was enhanced after treatment with IFN‐β. This is consistent with work done in murine USP18‐deficient macrophages treated with IFN‐β that demonstrated an increase in genes involved in antigen processing and presentation, cytokines and chemokines, as well as genes involved in responses to viral infections.60
In mice, in vivo protection from viral infections is mediated by Usp18 through a mechanism known as enforced viral replication whereby marginal zone macrophages become resistant to the effects of T1 IFN.61 By shielding macrophages from the full antiviral effects of T1 IFN, the cells can express viral proteins and utilize those proteins to prime adaptive T cell‐mediated immunity. HIV‐1 seems to have exploited this property of USP18 as a means of tuning the antiviral response to favor its own replication.
ISG15 and ISGylation of proteins has been previously shown to be important in restricting many different viruses including VSV, HIV‐1, HCV, Chikungunya, Influenza A, Influenza B, LCMV, and Sindbis viruses.62, 63 Previous studies have shown that ISGylation is important for restricting HIV‐1 in 293T, U1.1, HOS‐CD4/CXCR4, HeLa, U2OS, and 293E cell lines.23, 64, 65 However, we are unaware of any studies showing the relevance of ISGylation in primary cells or non‐immortalized cells such as human MDMs or human T cells. Moving forward, a key area of focus should be determining which functional domains of USP18 are required for HIV‐1 replication to dissect the relative roles of deISGylation versus IFNAR blocking in this process.
We have demonstrated in USP18 deficient cells that the first round of HIV‐1 infection is unaffected. The virus enters target cells, reverse transcribes cDNA, and integrates its proviral genome into a host chromosome. This likely results because during the first round of infection, de novo T1 IFN synthesis lags the rapid proviral integration process. Following this initial infection and more prolonged spreading infection in culture, USP18 −/− cells will likely have reduced net proviral integration as the initial T1 IFN response is amplified in these cells. After sensing viral nucleic acids in the infected cells, IFNs are produced and secreted into the surrounding environment. Indeed, we see that T1 IFN‐inducible USP18 protein expression is perturbed when T1 IFN signaling is blocked by anti‐IFNAR.
Autocrine/paracrine signaling is a feed forward antiviral mechanism of T1 IFNs where expression of ISGs establishes a nonpermissive environment for viral replication through expression of hundreds of ISGs and their protein products. Here, we find that attenuation of this antiviral mechanism by USP18 in the context of HIV‐1 infection has a net beneficial effect for the virus by overriding key antiviral signals. The most proximal events of T1 IFN signaling, phosphorylation of JAK/STAT proteins, are augmented in the absence of USP18, and we conclude that this effect is fundamental to the phenotype of restricted HIV‐1 replication in USP18−/− cells.
Uninfected cells that have received paracrine signals from T1 IFNs will up‐regulate ISGs. We demonstrated that USP18 deficient cells have increased expression of IFIT1, IFIT2, IFIT3, IFITM1, IFITM2, CXCL9, CXCL10, ISG15, and MX2 after treatment with exogenous IFN‐β. Many of these genes have been previously shown to restrict HIV‐1 replication.66 IFIT family proteins have been shown to restrict HIV‐1 by shutting down machinery used by the virus for translation of viral proteins.67 The IFITM family members have been shown to inhibit HIV‐1, by interfering with viral fusion or binding.68, 69, 70 This could potentially result in fewer cells being infected. MX2 is another IFN‐inducible factor that has been shown to restrict HIV‐1 replication.71, 72 The effects of these ISGs may simply be enhanced in the absence of USP18. The roles of CXCL9 and CXCL10 in vivo may help to recruit T cells to interact with infected macrophages.
T1 IFN secretion is induced by HIV‐1, but at very low levels not detectable by microarray after 7 days of productive HIV‐1 infection of macrophages. Clearly, the relatively small amounts of IFN produced early in infection have a meaningful effect on virus replication. When IFNAR blocking antibody was used, HIV‐1 infection was reduced rather than enhanced, demonstrating how low levels of IFN can benefit the virus. At the same time, USP18 is not induced by HIV‐1 when anti‐IFNAR is included, demonstrating that low level T1 IFNs being produced are responsible for USP18 induction as well as the IFN‐like transcriptional response. By providing exogenous IFN‐β, we were able to demonstrate that lack of USP18 makes cells more sensitive to the effects of IFN.
The strategy used by HIV‐1 for transcription from its LTR promoter utilizes transcription factors such as NFκB, NFAT, and IRFs,73, 74 that would normally be present in a cell that has detected a viral infection and has initiated an antiviral response. By utilizing this transcriptional machinery, HIV‐1 has been able to hijack normal immune processes to improve its own fitness, however in vitro experiments have long demonstrated that inflammatory activators like LPS or IFNs can potently restrict HIV‐1 replication. Thus, the physiologic state that supports HIV‐1 replication is a window of low level immune activation that is due, at least in part, to balanced positive and negative regulators of antiviral immunity (Fig. 9). Infected cells eventually become refractory to T1 IFN signaling due to induction of negative regulators such as USP18. Other negative regulators are also induced by T1 IFNs including SOCS1, SOCS3, and PIAS.75 We focused on USP18 for this study due to its additional function as a deISGylating enzyme and its appearance in gene expression profiling experiments following HIV‐1 infection.
iPSC‐derived cell models have previously been used to study other diseases in a variety of different cell types, including vascular smooth muscle cells, cardiomyocytes, and neurons.76, 77, 78, 79 iPSC‐derived macrophages have previously been shown to be a useful model for HIV‐1 infection48 and we have confirmed those findings in the current study. Our iMac model supports very robust HIV‐1 replication and shows a similar induction of USP18 in response to infection as what occurs in MDMs.
The use of iPSC‐derived cells has several advantages over conventional cell models. First, iPSC technologies can be used to derive cells that may not be obtainable in large numbers from living human donors. Second, iPSCs can be grown indefinitely in culture allowing for iterative mechanistic experiments, thus reducing variability. Third, iPSC‐derived cells can be genetically modified by use of gene editing technologies such as CRISPR/Cas9 and allow for a renewable source of edited cells with knockout and unedited iPSCs derived from the same donor. This complements methods such as siRNA transfection of primary cells such as MDMs.
In conclusion, we have shown in a novel iPSC‐derived macrophage model that USP18 is required for HIV‐1 replication in vitro. We propose that durable and enhanced IFN signaling can restrict HIV‐1 replication in vivo, but only if certain elements of the ISG response are limited. Negative regulators serve an important cellular function by limiting the magnitude of an initial response or helping to resolve a response following early signaling events. Here we show how HIV‐1 capitalized on the biologic effects of 1 negative regulator, USP18, as a means of tuning the T1 IFN response for increased viral fitness.
Many studies have focused on the antiviral effects of ISGs, but understanding how host cells regulate antiviral pathways will unveil more complete biologic mechanisms. Many infections or hereditary diseases are not easily modeled in rodents, so new platforms are required to answer key questions about host gene/proteins including negative regulators of signaling. We have demonstrated that iPSC‐derived cell models, when paired with genome editing technologies, are a powerful tool for studying not only HIV‐1 infection, but can also be used to study other infectious disease models, autoimmunity, and other general biologic problems that lack high quality conventional cell line models.
Supporting information
Supplemental Figure 1.
Supplemental Figure 2.
Supplemental Figure 3.
Supplementary Table 1. Primer Sequences
AUTHORSHIP
J.P.T. helped conceive of the project, designed and performed experiments, and wrote the manuscript. M.N.C. made and titered viral stocks, created knockdown cell lines, and helped create and screen knockout CRISPR clones. K.E.S. and N.T. helped create, maintain, and differentiate iPSCs. M.N. designed and created the Sendai virus used to make iPSC lines. M.A.W. helped conceive of the project, supervised the project, performed experiments, and mentored.
ACKNOWLEDGMENTS
The authors would like to thank Maureen Goodenow and Steve Pomeroy for providing virus stocks and BSL3 facilities. The authors would also like to thank Michael Tremblay fo providing the HIVHSA virus. This work was supported by funding from the National Institute of Allergy and Infectious Diseases grants K22AI09515, R56AI108434, and R56AI122813. J.P.T. was supported by the Ruth L. Kirschstein National Research Service Award Institutional Research Training Grant T32AI007110. The authors would also like to thank Lucas Armitage for his assistance with CRISPR plasmid transfections and Sneha Patel for her assistance with Western blots.
DISCLOSURES
The authors declare no conflicts of interest.
Taylor JP, Cash MN, Santostefano KE, Nakanishi M, Terada N, Wallet MA. CRISPR/Cas9 knockout of USP18 enhances type I IFN responsiveness and restricts HIV‐1 infection in macrophages. J Leukoc Biol. 2018;103:1225–1240. 10.1002/JLB.3MIA0917-352R
REFERENCES
- 1. Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. The role of interferons in the control of HIV replication in macrophages. Clin Immunol Immunopathol. 1990;54:200–219. [DOI] [PubMed] [Google Scholar]
- 2. Gendelman HE, Baca L, Turpin JA, et al. Restriction of HIV replication in infected T cells and monocytes by interferon‐alpha. AIDS Res Hum Retroviruses. 1990;6:1045–1049. [DOI] [PubMed] [Google Scholar]
- 3. Meylan PR, Guatelli JC, Munis JR, Richman DD, Kornbluth RS. Mechanisms for the inhibition of HIV replication by interferons‐alpha, ‐beta, and ‐gamma in primary human macrophages. Virology. 1993;193:138–148. [DOI] [PubMed] [Google Scholar]
- 4. Shirazi Y, Pitha PM. Interferon alpha‐mediated inhibition of human immunodeficiency virus type 1 provirus synthesis in T‐cells. Virology. 1993;193:303–312. [DOI] [PubMed] [Google Scholar]
- 5. Ho DD, Hartshorn KL, Rota TR, et al. Recombinant human interferon alfa‐A suppresses HTLV‐III replication in vitro. Lancet (London, England). 1985;1:602–604. [DOI] [PubMed] [Google Scholar]
- 6. Asmuth DM, Murphy RL, Rosenkranz SL, et al, AIDS Clinical Trials Group A5192 Team . Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon Alfa‐2a in HIV‐1‐monoinfected participants: a phase II clinical trial. J Infect Dis. 2010;201:1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azzoni L, Foulkes AS, Papasavvas E, et al. Pegylated Interferon alfa‐2a monotherapy results in suppression of HIV type 1 replication and decreased cell‐associated HIV DNA integration. J Infect Dis. 2013;207:213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sandler NG, Bosinger SE, Estes JD, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deeks SG, Odorizzi PM, Sekaly R‐P. The interferon paradox: can inhibiting an antiviral mechanism advance an HIV cure. J Clin Invest. 2017;127:103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hardy GAD, Sieg S, Rodriguez B, et al. Interferon‐α is the primary plasma type‐I IFN in HIV‐1 infection and correlates with immune activation and disease markers. PLoS One. 2013;8:e56527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhen A, Rezek V, Youn C, et al. Targeting type I interferon‐mediated activation restores immune function in chronic HIV infection. J Clin Invest. 2017;127:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng L, Yu H, Li G, et al. Type I interferons suppress viral replication but contribute to T cell depletion and dysfunction during chronic HIV‐1 infection. JCI Insight. 2017;2:e94366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilson EB, Yamada DH, Elsaesser H, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teijaro JR, Ng C, Lee AM, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levy DE, Kessler DS, Pine R, Reich N, Darnell JE. Interferon‐induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. [DOI] [PubMed] [Google Scholar]
- 16. Reich N, Evans B, Levy D, Fahey D, Knight E, Darnell JE. Interferon‐induced transcription of a gene encoding a 15‐kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci USA. 1987;84:6394–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE. Interferon‐stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci USA. 1986;83:8929–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang D‐E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. [DOI] [PubMed] [Google Scholar]
- 19. Basters A, Geurink PP, Röcker A, et al. Structural basis of the specificity of USP18 toward ISG15. Nat Struct Mol Biol. 2017;24:270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basters A, Geurink PP, El Oualid F, et al. Molecular characterization of ubiquitin‐specific protease 18 reveals substrate specificity for interferon‐stimulated gene 15. FEBS J. 2014;281:1918–1928. [DOI] [PubMed] [Google Scholar]
- 21. Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell. 2010;38:722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu G, Reinert JT, Pitha‐Rowe I, et al. ISG15 enhances the innate antiviral response by inhibition of IRF‐3 degradation. Cell Mol Biol (Noisy‐le‐grand). 2006;52:29–41. [PubMed] [Google Scholar]
- 23. Woods MW, Kelly JN, Hattlmann CJ, et al. Human HERC5 restricts an early stage of HIV‐1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology. 2011;8:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)‐induced ubiquitin‐like ISG15 protein. EMBO J. 2001;20:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mielech AM, Kilianski A, Baez‐Santos YM, Mesecar AD, Baker SC. MERS‐CoV papain‐like protease has deISGylating and deubiquitinating activities. Virology. 2014:450–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ratia K, Kilianski A, Baez‐Santos YM, Baker SC, Mesecar A. Structural Basis for the Ubiquitin‐Linkage Specificity and deISGylating activity of SARS‐CoV papain‐like protease. PLoS Pathog. 2014;10:e1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malakhova OA, Kim KI, Luo J‐K, et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Honke N, Shaabani N, Zhang D‐E, Hardt C, Lang KS. Multiple functions of USP18. Cell Death Dis. 2016;7:e2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arimoto K, Löchte S, Stoner SA, et al. STAT2 is an essential adaptor in USP18‐mediated suppression of type I interferon signaling. Nat Struct Mol Biol. 2017;24:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manini I, Sgorbissa A, Potu H, Tomasella A, Brancolini C. The DeISGylase USP18 limits TRAIL‐induced apoptosis through the regulation of TRAIL levels: cellular levels of TRAIL influences responsiveness to TRAIL‐induced apoptosis. Cancer Biol Ther. 2013;14:1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Potu H, Sgorbissa A, Brancolini C. Identification of USP18 as an important regulator of the susceptibility to IFN‐alpha and drug‐induced apoptosis. Cancer Res. 2010;70:655–665. [DOI] [PubMed] [Google Scholar]
- 32. Sattentau QJ, Stevenson M. Macrophages and HIV‐1: an Unhealthy Constellation. Cell Host Microbe. 2016;19:304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Appay V, Sauce D. Immune activation and inflammation in HIV‐1 infection: causes and consequences. J Pathol. 2008;214:231–241. [DOI] [PubMed] [Google Scholar]
- 34. Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Appelberg KS, Wallet MA, Taylor JP, Cash MN, Sleasman JW, Goodenow MM. HIV‐1 Infection Primes Macrophages through STAT Signaling to Promote Enhanced Inflammation and Viral Replication. AIDS Res Hum Retroviruses. 2017;00. aid.2016.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei X, Decker JM, Liu H, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T‐20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeuchi Y, McClure MO, Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol. 2008;82:12585–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Derdeyn CA, Decker JM, Sfakianos JN, et al. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T‐20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM‐bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol. 2009;83:8289–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Folks TM, Clouse KA, Justement J, et al. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T‐cell clone. Proc Natl Acad Sci USA. 1989;86:2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clouse KA, Powell D, Washington I, et al. Monokine regulation of human immunodeficiency virus‐1 expression in a chronically infected human T cell clone. J Immunol. 1989;142:431–438. [PubMed] [Google Scholar]
- 43. Imbeault M, Lodge R, Ouellet M, Tremblay MJ. Efficient magnetic bead‐based separation of HIV‐1‐infected cells using an improved reporter virus system reveals that p53 up‐regulation occurs exclusively in the virus‐expressing cell population. Virology. 2009;393:160–167. 2009/08/21 ed. [DOI] [PubMed] [Google Scholar]
- 44. Freed EO, Englund G, Martin MA. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spearman C. The method of “right and wrong cases” (“constant stimuli”) without Gauss's formulae. Br J Psychol 1904–1920. 1908;2:227–242. [Google Scholar]
- 46. Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs Arch Exp Pathol Pharmakol. 1931;162:480–483. [Google Scholar]
- 47. Nishimura K, Sano M, Ohtaka M, et al. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming J Biol Chem. 2011:4760–4771. 2010/12/09 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Wilgenburg B, Browne C, Vowles J, Cowley SA. Efficient, long term production of monocyte‐derived macrophages from human pluripotent stem cells under partly‐defined and fully‐defined conditions. PLoS One. 2013;8:e71098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vandergeeten C, Fromentin R, Merlini E, et al. Cross‐clade ultrasensitive PCR‐based assays to measure HIV persistence in large‐cohort studies. J Virol. 2014;88:12385–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77:10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gendelman HE, Baca LM, Turpin J, et al. Regulation of HIV replication in infected monocytes by IFN‐alpha. Mechanisms for viral restriction. J Immunol. 1990;145:2669–2676. [PubMed] [Google Scholar]
- 52. Cassol E, Alfano M, Biswas P, Poli G. Monocyte‐derived macrophages and myeloid cell lines as targets of HIV‐1 replication and persistence. J Leukoc Biol. 2006;80:1018–1030. [DOI] [PubMed] [Google Scholar]
- 53. Odero MD, Zeleznik‐Le NJ, Chinwalla V, Rowley JD. Cytogenetic and molecular analysis of the acute monocytic leukemia cell line THP‐1 with an MLL‐AF9 translocation. Genes Chromosomes Cancer. 2000;29:333–338. [PubMed] [Google Scholar]
- 54. Nakanishi M, Otsu M. Development of Sendai virus vectors and their potential applications in gene therapy and regenerative medicine. Curr Gene Ther. 2012;12:410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ritchie KJ, Hahn CS. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004;10:1374–1378. [DOI] [PubMed] [Google Scholar]
- 56. Kim KI, Malakhova OA, Hoebe K, Yan M, Beutler B, Zhang D‐E. Enhanced antibacterial potential in UBP43‐deficient mice against Salmonella typhimurium infection by up‐regulating type I IFN signaling. J Immunol. 2005;175:847–854. [DOI] [PubMed] [Google Scholar]
- 57. Speer SD, Li Z, Buta S, et al. ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun. 2016;7:11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X, Bogunovic D, Payelle‐Brogard B, et al. Human intracellular ISG15 prevents interferon‐α/β over‐amplification and auto‐inflammation. Nature. 2015;517:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meuwissen MEC, Schot R, Buta S, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo‐TORCH syndrome. J Exp Med. 2016;213:1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zou W, Kim JH, Handidu A, et al. Microarray analysis reveals that Type I interferon strongly increases the expression of immune‐response related genes in Ubp43 (Usp18) deficient macrophages. Biochem Biophys Res Commun. 2007;356:193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Honke N, Shaabani N, Cadeddu G, et al. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol. 2011;13:51–57. [DOI] [PubMed] [Google Scholar]
- 62. Morales DJ, Lenschow DJ. The antiviral activities of ISG15. J Mol Biol. 2013;425:4995–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kunzi MS, Pitha PM, Künzi MS, Pitha PM. Role of interferon‐stimulated gene ISG‐15 in the interferon‐omega‐mediated inhibition of human immunodeficiency virus replication. J Interferon Cytokine Res. 1996;16:919–927. [DOI] [PubMed] [Google Scholar]
- 64. Okumura A, Lu G, Pitha‐Rowe I, Pitha PM. Innate antiviral response targets HIV‐1 release by the induction of ubiquitin‐like protein ISG15. Proc Natl Acad Sci USA. 2006;103:1440–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon‐induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84:4725–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schoggins JW, Wilson SJ, Panis M, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Diamond MS, Farzan M. The broad‐spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2012;13:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu J, Li M, Wilkins J, et al. IFITM proteins restrict HIV‐1 infection by antagonizing the envelope glycoprotein. Cell Rep. 2015;13:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Compton AA, Bruel T, Porrot F, et al. IFITM proteins incorporated into HIV‐1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tartour K, Appourchaux R, Gaillard J, et al. IFITM proteins are incorporated onto HIV‐1 virion particles and negatively imprint their infectivity. Retrovirology. 2014;11:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kane M, Yadav SS, Bitzegeio J, et al. MX2 is an interferon‐induced inhibitor of HIV‐1 infection. Nature. 2013;502:563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goujon C, Moncorgé O, Bauby H, et al. Human MX2 is an interferon‐induced post‐entry inhibitor of HIV‐1 infection. Nature. 2013;502:559–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV‐1 gene expression. Cold Spring Harb Perspect Med. 2012;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Battistini A, Sgarbanti M. HIV‐1 latency: an update of molecular mechanisms and therapeutic strategies. Viruses. 2014;6:1715–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schneider WM, Chevillotte MD, Rice CM. Interferon‐Stimulated Genes: a Complex Web of Host Defenses. Annu Rev Immunol. 2014;32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Biel NM, Santostefano KE, DiVita BB, et al. Vascular smooth muscle cells from hypertensive patient‐derived induced pluripotent stem cells to advance hypertension pharmacogenomics. Stem Cells Transl Med. 2015;4:1380–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gao Y, Guo X, Santostefano K, et al. Genome therapy of myotonic dystrophy type 1 iPS cells for development of autologous stem cell therapy. Mol Ther. 2016;24:1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ferreira RB, Wang M, Law ME, et al. Disulfide bond disrupting agents activate the unfolded protein response in EGFR‐ and HER2‐positive breast tumor cells. Oncotarget. 2017;8:28971–28989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bai F, Ho Lim C, Jia J, et al. Directed differentiation of embryonic stem cells into cardiomyocytes by bacterial injection of defined transcription factors. Sci Rep. 2015;5:15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1.
Supplemental Figure 2.
Supplemental Figure 3.
Supplementary Table 1. Primer Sequences


