Abstract
Synaptic dysfunction and abnormal processing of amyloid precursor protein (APP) are early pathological features in Alzheimer’s disease (AD). Recently, non-coding RNAs such as microRNAs (miRNAs) and circular RNAs (circRNAs) have been reported to contribute to the pathogenesis of AD. We found an age-dependent elevation of miR-138 in APP/PS1 (presenilin-1) mice. MiR-138 inhibited the expression of ADAM10 [a disintegrin and metalloproteinase domain-containing protein 10], promoted amyloid beta (Aβ) production, and induced synaptic and learning/memory deficits in APP/PS1 mice, while its suppression alleviated the AD-like phenotype in these mice. Overexpression of sirtuin 1 (Sirt1), a target of miR-138, ameliorated the miR-138-induced inhibition of ADAM10 and elevation of Aβ in vitro. The circRNA HDAC9 (circHDAC9) was predicted to contain a miR-138 binding site in several databases. Its expression was inversely correlated with miR-138 in both Aβ-oligomer-treated N2a cells and APP/PS1 mice, and it co-localized with miR-138 in the cytoplasm of N2a cells. CircHDAC9 acted as a miR-138 sponge, decreasing miR-138 expression, and reversing the Sirt1 suppression and excessive Aβ production induced by miR-138 in vitro. Moreover, circHDAC9 was decreased in the serum of both AD patients and individuals with mild cognitive impairment. These results suggest that the circHDAC9/miR-138/Sirt1 pathway mediates synaptic function and APP processing in AD, providing a potential therapeutic target for its treatment.
Keywords: Alzheimer’s disease, Synapse, Memory, Sirtuin-1, microRNA, Circular RNA
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder that is the most frequent cause of dementia, accounting for 60%–80% of all cases with dementia. The prevalence of AD is 10% in people aged 65 and over, and increases to 32% in those aged 85 and over [1]. The symptoms include a gradually worsening ability to remember new information in the initial stage and an inability to communicate in the advanced stage. The mechanism underlying AD remains unclear [2].
Accumulating evidence indicates that non-coding RNAs (ncRNAs) are involved in AD, including microRNAs (miRNAs), long ncRNAs, piwi-interacting RNAs, and circular RNAs (circRNAs) [3–5]. MiRNAs suppress target mRNA translation or induce mRNA decay via binding with seed sequences in the 3′-untranslated region of the target mRNA. Numerous dysregulated miRNAs in AD contribute to the processing of amyloid precursor protein (APP), tau phosphorylation, and synaptic plasticity [6, 7].
CircRNAs are a group of RNAs with covalently closed loop structures, generated during splicing. A number of conserved circRNAs are highly enriched in the mammalian brain, some of which are derived from synaptic genes and regulated depending on the developmental stage [8, 9]. Natural circRNAs function as miRNA sponges in the brain, providing potential novel therapeutic targets for AD treatment [8, 10].
MiR-138 is a conserved miRNA that is enriched in the brain. It is increased in the cerebrospinal fluid (CSF) of AD patients [11], acts as a potential regulator of memory performance in humans, and negatively regulates dendritic spine morphogenesis [12, 13]. Our previous study demonstrated that miR-138 is increased in 12-month-old Tg2576 mice, N2a/APP cells, and HEK293/tau cells. Furthermore, miR-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha [7, 14].
Therefore we set out to determine the role of circHDAC9/miR-138/Sirt1 pathway in the synaptic, learning/memory, and APP processing deficits in AD.
Materials and Methods
Transgenic AD Mice
Male APP/PS1 (APPswe and PSEN1dE9) transgenic mice and control littermates were purchased from the Animal Model Research Center of Nanjing University (Nanjing, Jiangsu, China). This work was approved by the Animal Care and Use Committee of Tongji Hospital, Tongji Medical College of Huazhong University of Science and Technology.
Human Blood Samples
Ten amnestic patients with mild cognitive impairment (MCI) (7 males, 3 females; 70.20 ± 10.40 years old) and seven AD patients (5 males, 2 females; 70.43 ± 5.08 years old) diagnosed as previously described [15], were recruited from Tongji Hospital. A diagnosis of MCI included: (1) memory complaint; (2) clinical dementia rating 0.5; and (3) normal general cognitive function and daily life activities. A diagnosis of AD included: (1) the NINCDS-ADRDA criteria (National Institute of Neurological and Communicative Disorders and Stroke-Alzheimer’s Disease and Related Disorders Association); (2) Mini-Mental State Examination score ≤ 26; and (3) exclusion of brain tumor, vascular dementia, and cerebrovascular diseases. Ten healthy individuals (7 males, 3 females; 64.40 ± 12.69 years old) were recruited as a control group for the MCI patients, and another seven (4 males, 3 females; 70.86 ± 4.11 years old) as controls for the AD patients. No statistically significant differences in age were found between the MCI/AD and corresponding control groups. Serum was collected for RNA extraction. The Ethics Committee of Tongji Hospital approved the study and informed consent was given.
Materials
Anti-APP antibody was from Abcam (Cambridge, UK), anti-ADAM10 from Abcam, anti-BACE1 from Cell Signaling Technology (Danvers, MA, USA), anti-β-actin from Cell Signaling Technology (Danvers, MA, USA), anti-GAPDH from Proteintech (Wuhan, China), anti-sAPPβ from IBL (Fujioka-Shi, Gunma, Japan); and anti-Sirt1 from Cell Signaling Technology (Danvers, MA) or Proteintech (Wuhan, China). The Aβ42 ELISA kit was from Elabscience (Wuhan, China), FISH kit from Bersinbio (Guangzhou, China), RevertAid First Strand cDNA Synthesis Kit from Takara (Kusatsu, Shiga, Japan), miRNA Purification Kit from Cwbiotech (Beijing, China), RIPA lysis buffer from Beyotime (Shanghai, China), and HRP-conjugated secondary antibody from Santa Cruz (Santa Cruz, CA, USA). The miR-138 mimic and agomir/antagomir and the scrambled control were from RiboBio (Guangzhou, China) and the miR-138-overexpressing construct was from GeneCopoeia (San Diego, CA, USA). The Sirt1 and APP plasmids were kindly gifted by Prof. Jianzhi Wang (Tongji Medical College, Huazhong University of Science and Technology). The miR-138 mimic was from RiboBio (Guangzhou, China).
Stereotaxic Microinfusion
Mice were anesthetized with chloral hydrate (400 mg/kg, intraperitoneal). Holes were drilled above the CA3 field of the hippocampus (anterior/posterior ± 1.9 mm, medial/lateral ± 1.9 mm, dorsal/ventral 2.1 mm). Agomir (350 nmol/L, 1.5 μL) or antagomir (600 nmol/L, 1.5 μL) was bilaterally microinfused into the hippocampus.
Morris Water Maze Test
The Morris water maze (MWM) was a deep pool (diameter: 120 cm, deep: 80 cm) filled with water made opaque with milk. The pool was divided into four quadrants with a platform submerged 1 cm beneath the surface in the target quadrant. Mice were trained for 5 consecutive days to find the platform (n = 12/group). A digital tracking device was used to measure movement. On day 7, the platform was removed, and the crossing time and distance in the target quadrant (%) were recorded.
Golgi Staining
Mice were anesthetized with chloral hydrate (400 mg/kg, intraperitoneal). After perfusion with normal saline and 4% formaldehyde, brain tissue was immersed in 5% potassium dichromate, 4% formaldehyde, and 5% silver nitrate for one month. Slices were then cut on a vibrating microtome (Leica, Wetzlar, Germany).
Fluorescence In situ Hybridization (FISH)
The probes for circHDAC9 (green) and miR-138 (red) were from Bersinbio (Guangzhou, China). N2a cells were harvested and fixed in 4% formaldehyde. The denatured probes were hybridized with N2a cells for 20 h at 42 °C. Nuclei were stained with DAPI. The slices were observed under a fluorescence microscope (LSM800, Carl Zeiss, Darmstadt, Germany).
Real-Time Quantitative PCR
RNA was extracted with TRIzol reagent and reverse-transcribed into cDNA. MiRNAs were isolated with a miRNA purification kit. Real-time quantitative PCR was performed with a SYBR Premix Dimer Eraser kit. Mouse circHDAC9: F, AGCAGTTCCTGGAGAAGCAG, R, GCATCTGTGTCTCGCACTTC; mouse ciRS7: F, ATGTTGGAAGACCTTGGTACTGGC, R, CCAACATCTCCACATCTTCCAGCA; mouse circAGAP1: F, TCCAGCCCAAGTATCTCCAG, R, ATCTGAGACCCAGACGCACT; mouse GAPDH: F, TGTGTCCGTCGTGGATCTGA, R, TTGCTGTTGAAGTCGCAGGAG; human circHDAC9: F, ACCAGCAATTCTTGGAGAAG, R, GCGTCTGCGTCTCACACTTC; human β-actin: F, CCTGCACCACCAACTGCTTA, R, GGCCATCCACAGTCTTCTGG. Primers for miR-138 and U6 were from GeneCopoeia (San Diego, CA). Real-time quantitative PCR was performed with an SYBR Premix Dimer Eraser kit on a Cobas Z480 automatic fluorescence quantitative PCR analyzer (Roche, Basel, Switzerland), and expression of circHDAC9 was normalized to GAPDH or β-actin. MiR-138 expression was normalized to U6.
Western Blotting
Protein was extracted from the hippocampus and cells using RIPA lysis buffer. Equal amounts of protein were separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were incubated with primary antibodies overnight at 4 °C, and further incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at 37 °C. Blots were quantified using ImageJ. Band density values were normalized to GAPDH or β-actin.
Statistics
Data are shown as mean ± SD and analyzed with SPSS version 16.0 (SPSS Inc., Chicago, IL). The difference between two groups was analyzed with unpaired Student’s t-test (two-tailed). The variance among multiple groups was analyzed with one-/two-way analysis of variance with/without repeated measures followed by the Bonferroni post hoc test. All experiments were performed three times, and P < 0.05 was considered statistically significant.
Results
Age-Dependent Elevation of miR-138 in APP/PS1 Mice
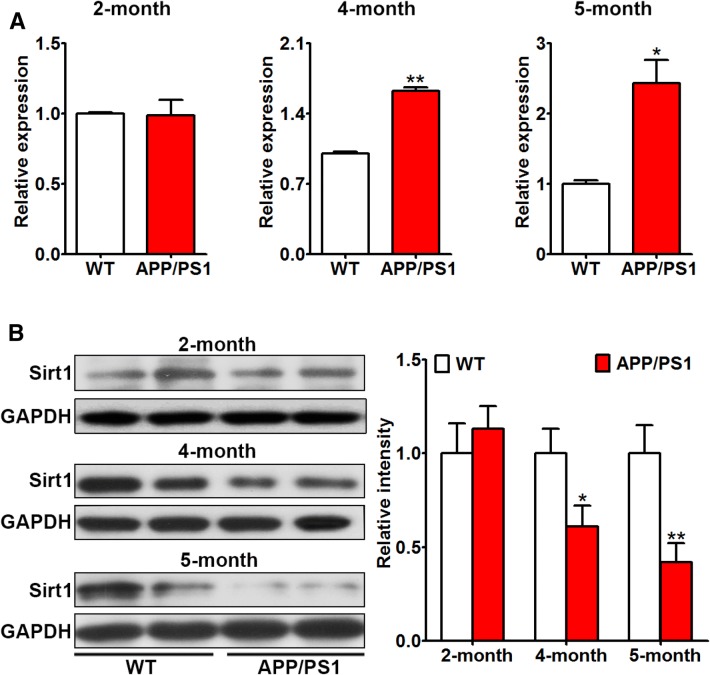
The expression of miR-138 in the hippocampus of APP/PS1 and control mice was assessed in 2-, 4-, and 5-month-old mice. No difference in expression was found in 2-month-old APP/PS1 mice, while the miR-138 level increased to 1.6-fold in in the hippocampus of 4-month-old and 2.4-fold in 5-month-old APP/PS1 mice relative to the control group (Fig. 1A).
Fig. 1.
Expression of miR-138 and Sirt1 in APP/PS1 and control mice. A Expression of miR-138 in 2-, 4-, and 5-month-old mice. B Protein levels of Sirt1 in 2-, 4-, and 5-month-old mice. *P < 0.05, **P < 0.01 vs control.
The expression of Sirt1, a known target of miR-138, was also assessed in 2-, 4-, and 5-month-old mice: it decreased in 4- and 5-month-old APP/PS1 mice, and was negatively correlated with the miR-138 level (Fig. 1B).
MiR-138 Promoted Aβ Production In vivo and In vitro
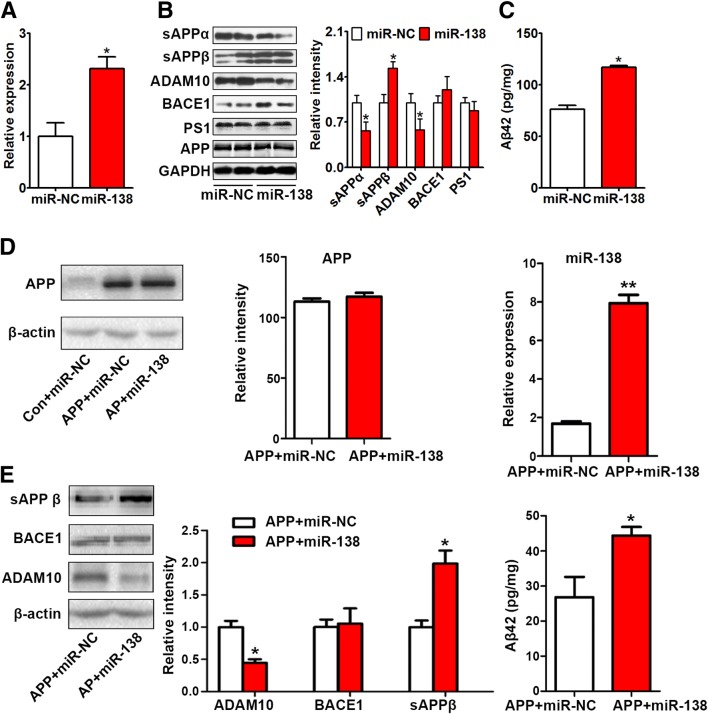
To explore the effect of miR-138 on APP processing, a miR-138 agomir (miR-138) or control agomir (miR-NC) was injected into the hippocampus of 2-month-old APP/PS1 mice. The level of miR-138 was increased after miR-138 agomir injection (Fig. 2A). APP is cleaved by three enzymes, α-, β- and γ-secretases. Non-amyloidogenic cleavage of APP by α-secretase (ADAM10) releases soluble APPα (sAPPα) and the α-carboxy-terminal fragment. Cleavage by β-secretase (beta-site APP cleaving enzyme 1, BACE1) primarily produces soluble APPβ (sAPPβ) and the β-carboxy-terminal fragment, which is further cleaved by γ-secretase (presenilin-1, PS1) to release Aβ. Both ADAM10 and sAPPα were significantly decreased in the miR-138 group, while sAPPβ and Aβ were increased (Fig. 2B, C). The expression of BACE1 and PS1 was unchanged by miR-138 overexpression (Fig. 2B). Overexpression of miR-138 inhibited the α-secretase pathway, thereby promoting the β-secretase pathway.
Fig. 2.
MiR-138 promoted Aβ production in vivo and in vitro. A–C Two-month-old APP/PS1 mice were injected with miR-138 agomir (miR-138) or scrambled control (miR-NC) into the hippocampus and sacrificed 3 days later. The expression of miR-138 (A), APP processing (B), and Aβ (C) were measured by real-time quantitative PCR, Western blotting, and ELISA, respectively. D, E Expression of APP and miR-138 (D), APP processing, and Aβ (E) in N2a cells co-transfected with control/APP and miR-NC/miR-138 and harvested 48 h later. *P < 0.05, **P < 0.01 vs miR-NC or APP + miR-NC group.
Consistent with our in vivo study, overexpression of miR-138 decreased ADAM10 expression but increased sAPPβ and Aβ production in N2a cells (Fig. 2D, E).
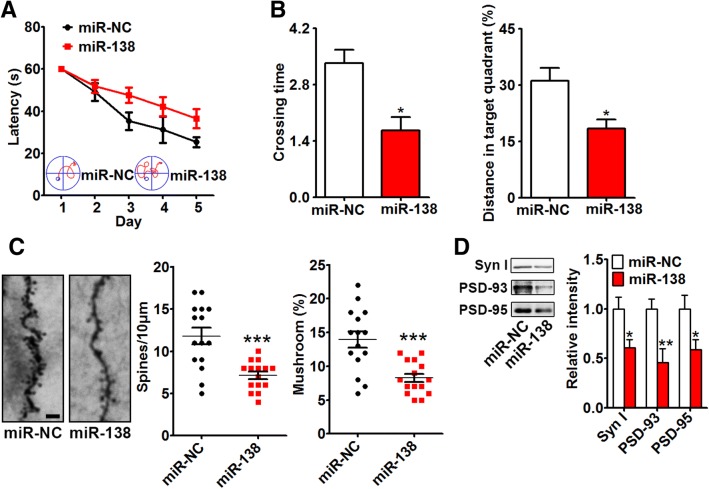
MiR-138 Induced Synaptic and Cognitive Impairments
The effects of miR-138 on synaptic plasticity and spatial learning/memory were assessed in 2-month-old APP/PS1 mice by the Morris water maze test and Golgi staining. Mice injected with miR-138 agomir took longer to find the hidden platform during training. The crossing times and percentage of distance spent in the target quadrant were both significantly lower in the miR-138 group than in the miR-NC group (Fig. 3A, B). Overexpression of miR-138 decreased the spine density, mushroom percentage, and expression of the synapse markers Synapsin I, PSD-93, and PSD-95 (Fig. 3 C, D), leading to synaptic impairment.
Fig. 3.
miR-138 induced synaptic and cognitive impairments. A, B Two-month-old APP/PS1 mice were injected with miR-138 agomir (miR-138) or scrambled control (miR-NC) into the hippocampus and 5 days later were tested in the Morris water maze. The latency to reach the platform from days 1 to 5 (A), and the crossing time and distance in the target quadrant in the test experiment (day 7) were recorded (B), then the mice were sacrificed for Golgi staining. C, D The spine density and mushroom percentage (bar = 5 μm) (C) and the expression of the synapse markers Synapsin I (Syn I), PSD-93, and PSD-95 were assessed (D). *P < 0.05, **P < 0.01, ***P < 0.001 vs miR-NC group.
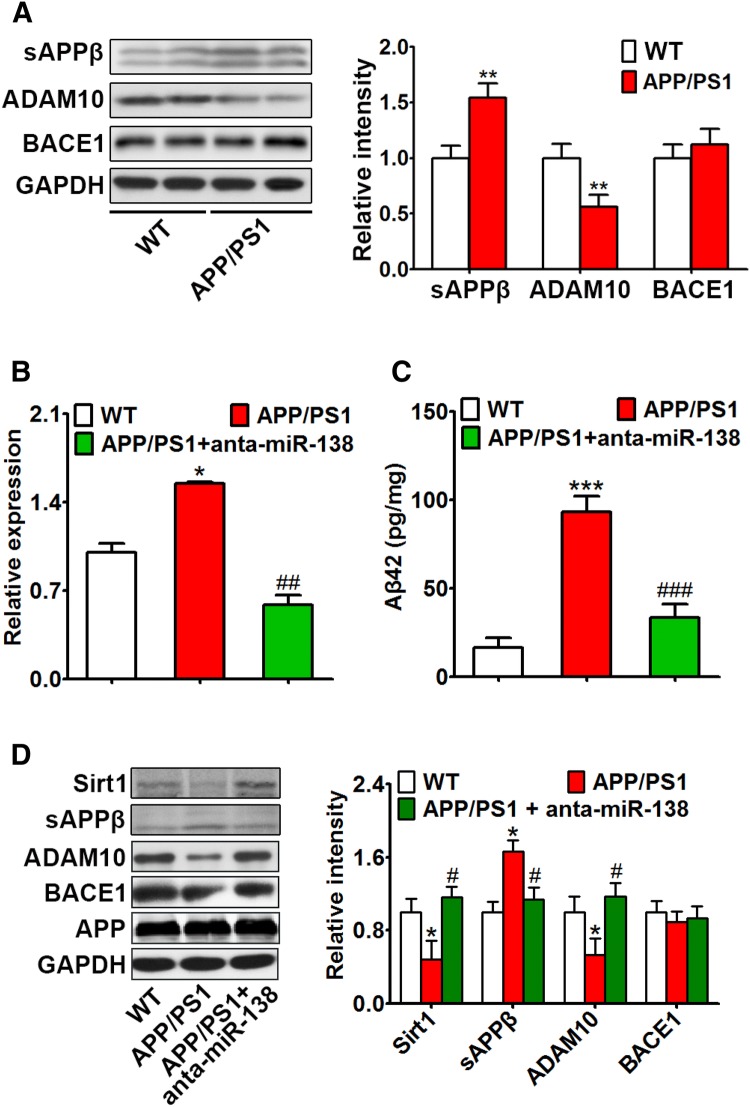
Suppression of miR-138 Alleviated the AD-Like Phenotype
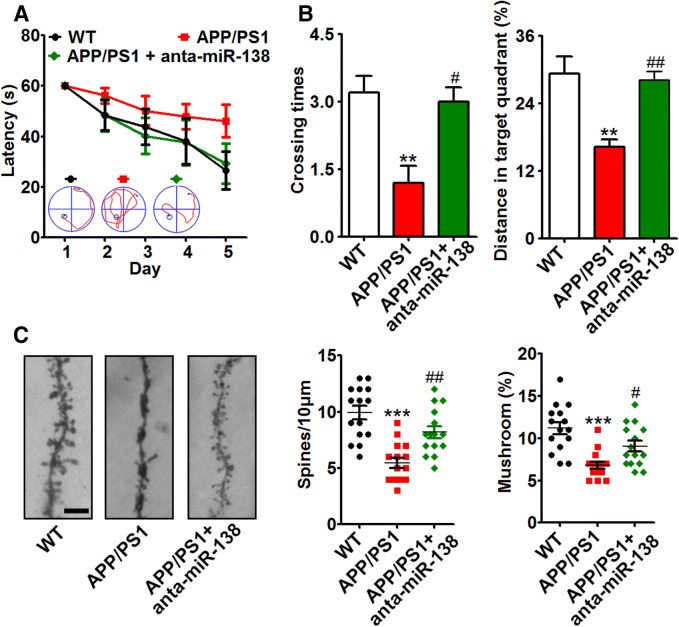
In 4-month-old APP/PS1 mice, the expression of ADAM10 was decreased, while sAPPβ was increased (Fig. 4A). As miR-138 was increased in these mice, miR-138 antagomir (anta-miR-138) was used to inhibit its expression (Fig. 4B). Antagomir-miR-138 rescued the AD-like symptoms, including abnormal APP processing (Fig. 4C, D), spatial learning/memory deficits (Fig. 5A, B), and dendritic spine deterioration (Fig. 5C).
Fig. 4.
Suppression of miR-138 alleviated abnormal APP processing. A Western blots and quantification of the expression of ADAM10, BACE1, and sAPPβ in the hippocampus of APP/PS1 and control mice. B–D Four-month-old APP/PS1 mice were injected with miR-138 antagomir (anta-miR-138) or scrambled control (anta-miR-NC) into the hippocampus, and 5 days later tested in the Morris water maze, then sacrificed for Western blotting and ELISA. The expression of miR-138 (B), Aβ (C), and APP processing (D) were measured by real-time quantitative PCR, ELISA, and Western blotting, respectively. *P < 0.05, **P < 0.01, ***P < 0.001 vs control (WT); #P < 0.05, ##P < 0.01, ###P < 0.001 vs APP/PS1 group.
Fig. 5.
Suppression of miR-138 alleviated the synaptic deficit. Four-month-old APP/PS1 mice were injected with miR-138 antagomir (anta-miR-138) or scrambled control (anta-miR-NC) into the hippocampus, and 5 days later, tested in the Morris water maze. A Latency to reach the platform from days 1 to 5. B Crossing time and distance in the target quadrant in the test experiment (day 7). C Spine density and mushroom percentage (bar = 5 μm). **P < 0.01, ***P < 0.001 vs control (WT); #P < 0.05, ##P < 0.01 vs APP/PS1 group.
Overexpression of Sirt1 Ameliorated miR-138-Induced Aβ Elevation In vitro
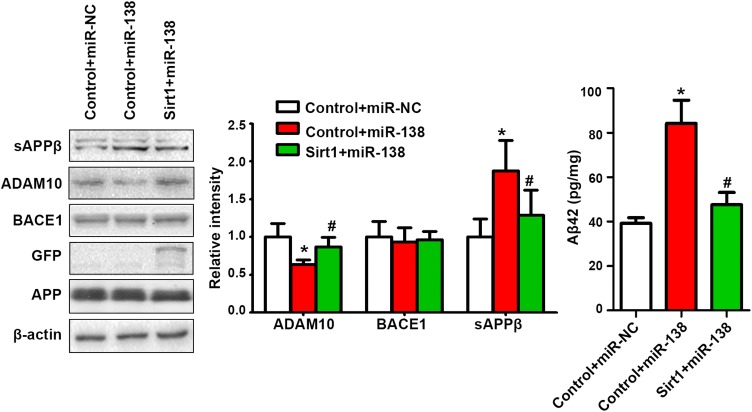
We also investigated the effect of Sirt1 overexpression on miR-138-induced Aβ elevation in vitro. Similar to the effect of antagomir-miR-138 in vivo, delivery of Sirt1 to N2a cells co-transfected with APP and miR-138 significantly reversed the miR-138 induced ADAM10 inhibition and Aβ elevation (Fig. 6).
Fig. 6.
Overexpression of Sirt1 ameliorated miR-138 induced Aβ elevation in vitro. APP processing and Aβ in N2a cells co-transfected with APP, miR-NC/miR-138, and control/Sirt1, and harvested 48 h later. *P < 0.05 vs Control + miR-NC; #P < 0.05 vs Control + miR-138 group.
circHDAC9 Reversed the miR-138-Induced AD-Like Phenotype by Acting as a miR-138 Sponge In vitro
To date, the mechanism of miR-138 elevation in AD is not clear. Accumulating evidence indicates that circRNAs can function as miRNA sponges, regulating their expression and function. Rybak-Wolf et al. have identified 4522 conserved circRNAs in human and mouse brain [9] and Gruner et al. have identified 5528 circRNAs in the mouse hippocampus [16]. We combined the data from the two groups and selected 1795 conserved circRNAs expressed in the mouse hippocampus. Gruner et al. found 25 circRNAs that contained miR-138 binding sites as predicted by TargetScan 7.0 [17]. We further applied miRanda 3.3a (http://www.microrna.org/microrna/) and RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/) to predict the binding sites of miR-138 in 25 selected circRNAs [18]. Only circHDAC9 (mouse, mmu_circ_0003925; human, hsa_circ_0003594) was predicted to contain a miR-138 binding site by all three programs (Table 1).
Table 1.
Prediction of miR-138 binding site in circHDAC9.
| circHDAC9 | TargetScan 7.0 | miRanda 3.3a | RegRNA 2.0 | |||||
|---|---|---|---|---|---|---|---|---|
| Name | Count | Score | Energy | Position | Position | Length | Energy | |
| Mouse | miR-138 | 2 | 157 | − 25.79 | 477 | 475–503 | 29 | − 27.00 |
| Human | 161 | − 26.06 | 480 | 478–506 | 29 | − 23.84 | ||
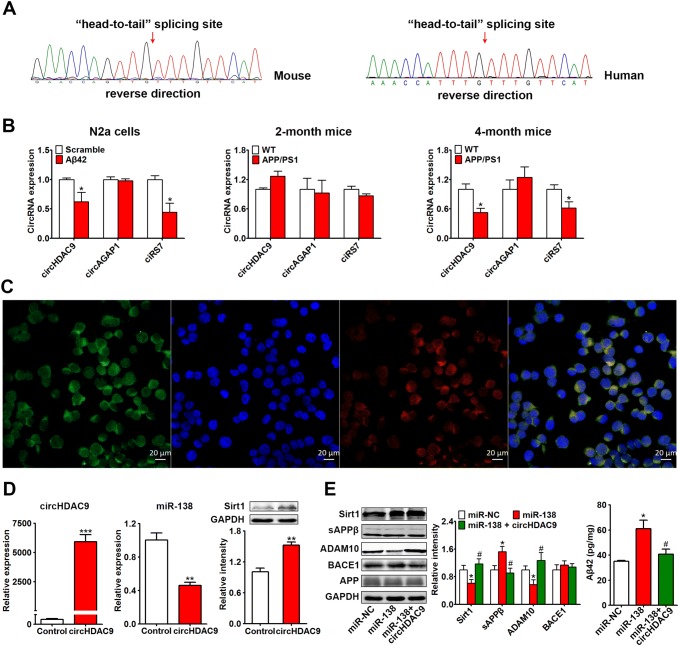
Real-time quantitative PCR primers specific for mouse and human circHDAC9 were designed and validated by Sanger sequencing of the PCR product, which covered the “head to tail” splicing site in circHDAC9 (Fig. 7A). The expression of circHDAC9 in Aβ oligomer-treated N2a cells and in 2-month-old and 4-month-old APP/PS1 mice was found to be reduced in the N2a cells and 4-month-old mice, but not in the two-month-old mice, and was negatively correlated with the miR-138 level (Fig. 7B). CiRS7 is a circRNA that has been reported to be decreased in AD. We used it as a positive control, and found changes similar to circHDAC9. Another brain-enriched circRNA, circAGAP1, had no significant effect in the three models (Fig. 7B).
Fig. 7.
CircHDAC9 acted as a miR-138 sponge. A Sequencing of the “head-to-tail” splicing sites in mouse and human circHDAC9. B Expression of circHDAC9, circAGAP1, and ciRS-7 in N2a cells treated with Aβ oligomers (5 μmol/L, 24 h) and in 2-month-old and 4-month-old APP/PS1 and control mice. C Distribution of circHDAC9 (green) and miR-138 (red) in N2a cells as detected by FISH assay (nuclei stained blue with DAPI). Scale bars, 20 μm. D Expression of circHDAC9, miR-138, and Sirt1, the target of miR-138, assessed by real-time quantitative PCR and Western blotting in N2a cells transfected with circHDAC9 or the control vector for 48 h. E Sirt1, APP processing, and Aβ in N2a cells co-transfected with APP, miR-NC/miR-138, and control/circHDAC9 for 48 h. *P < 0.05 vs scrambled control, miR-NC, or control (WT); #P < 0.05 vs miR-138 group.
Using FISH to investigate the expression and distribution of circHDAC9 and miR-138, we found that both were expressed and co-localized in the cytoplasm (Fig. 7C), indicating a potential interaction between circHDAC9 and miR-138.
To further explore whether circHDAC9 acts as a miR-138 sponge, we generated a construct and transfected it into N2a cells. CircHDAC9 overexpression remarkably decreased the miR-138 level and increased the Sirt1 protein level (Fig. 7D). Moreover, circHDAC9 significantly rescued the miR-138-induced ADAM10 inhibition and Aβ elevation in vitro (Fig. 7E), indicating that circHDAC9 does act as a miR-138 sponge.
CircHDAC9 Was Decreased in Patients with MCI and AD
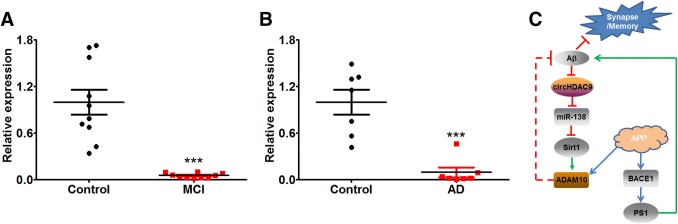
MCI patients have symptoms similar to those with AD and some go on to develop AD. When we assessed the expression of circHDAC9 in the serum of MCI and AD patients, we found that it was remarkably lower in both than in controls (Fig. 8A, B).
Fig. 8.
CircHDAC9 was significantly decreased in both MCI and AD patients. A, B Expression of circHDAC9 in serum from amnestic MCI patients (A) and AD patients (B) along with their healthy controls (***P < 0.001 vs controls). C Schematic of the circHDAC9/miR-138/Sirt1 pathway in AD. APP is cleaved by ADAM10 and BACE1. Aβ decreases circHDAC9 expression and increases miR-138 expression, leading to the down-regulation of Sirt1 and ADAM10, therefore shifting the APP processing from the α-secretase pathway to the β-secretase pathway and increasing the amyloid burden.
Discussion
We have revealed a novel signaling pathway of Aβ-induced pathological events in AD, involving both an upstream regulator and a downstream target of miR-138.
Numerous deregulated miRNAs have been identified in AD patients, transgenic mice, and cell lines. Some of these miRNAs participate in the regulation of Aβ generation, synaptic dysfunction, and tau phosphorylation [19, 20]. Serum miRNA levels have been used as predictive biomarkers in AD [21, 22]; and miR-138 is increased in the CSF of AD patients. In our current and previous studies, we found that miR-138 was increased in AD transgenic mice, including 12-month-old Tg2576 and 4-month-old APP/PS1 mice, as well as cell models including N2a/APP and HEK293/tau cells which stably express human APP and tau, respectively. Taken together, these data imply that miR-138 is widely involved in the pathogenesis of AD. Our previous study demonstrated that miR-138 induces tau phosphorylation by targeting retinoic acid receptor alpha [14]. In the current study, we further demonstrated that miR-138 was higher in APP/PS1 mice than in controls in an age-dependent manner. The expression of miR-138 was increased in 4-month-old APP/PS1 mice by 1.6-fold, and this elevation increased to 2.4-fold in 5-month-old APP/PS1 mice.
Previous studies found no detectable amyloid burden in the hippocampus of 3-month-old APP/PS1 mice, and Aβ plaque deposition starts between 4 and 6 months of age [23, 24]. Liu et al. reported cognitive impairment in 4-month-old APP/PS1 mice [25]. Consistent with previous studies, we found that Aβ was increased in 4-month-old APP/PS1 mice in which synaptic plasticity and learning/memory were impaired. Overexpression of miR-138 resulted in abnormal APP processing and synaptic and learning/memory deficits in 2-month-old APP/PS1 mice, while inhibition of miR-138 alleviated this AD-like phenotype in 4-month-old APP/PS1 mice. Similar results were obtained in cell lines via overexpression of miR-138 or supplement of Sirt1 by plasmid, the target gene of miR-138. In another study, Tian et al. demonstrated that microinjection of miR-138 into the hippocampal CA1 region of male Wistar rats impairs learning/memory in the Morris water maze test [26]. Liu et al. identified miR-138 as a suppressor of axon regeneration via targeting Sirt1 [27], and we demonstrated that overexpression of miR-138 impaired synaptic plasticity. Taken together, these data support the hypothesis that miR-138 is involved in mediating the synaptic and learning/memory deficits and the abnormal APP processing in AD.
Sirt1 has been widely reported to be a direct target of miR-138 in several disease models [26, 28, 29]. The decreased expression of Sirt1 in APP/PS1 mice was similar to previous findings. The expression of Sirt1 was reduced in 4- and 5-month-old APP/PS1 mice in our study, and Dong et al. reported decreased expression of Sirt1 in 6- and 10-month-old APP/PS1 mice [30]. A similar reduction has been found in the brain and serum of AD patients [31, 32]. However, recently, cross-platform normalized brain expression profiles of AD brains revealed that Sirt1 mRNA is increased in several brain regions, including the entorhinal cortex, hippocampus, temporal cortex, and frontal cortex [33]. Post-transcriptional analysis of Sirt1 may help evaluate the change in its expression in AD. Network analysis has shown that Sirt1 interacts with MAPT (microtubule-associated protein tau), ADAM10, and MEF2C (myocyte enhancer factor 2) [34]. Sirt1 directly activates the transcription of ADAM10 and therefore suppresses Aβ production [35]. Moreover, Sirt1 regulates the neuro-inflammation and mitochondrial dysfunction in AD [36]. In the current study, we found that the expression of Sirt1 was negatively correlated with miR-138 in APP/PS1 mice, and delivery of Sirt1 rescued the abnormal APP processing induced by miR-138 in cell culture.
Changes in miR-138 occurred in 4-month-old APP/PS1 mice with remarkably increased Aβ, but not in 2-month-old APP/PS1 mice with Aβ production similar to controls. These results suggest that alteration of miR-138 might be triggered by Aβ over-production, and in turn, the increased miR-138 promotes the production of Aβ and its deleterious effects on learning/memory and synaptic plasticity. Intervention in the miR-138 pathways led to alleviation in the AD phenotype and pathology in this study. These improvements might result from reduced Aβ production after miR-138 intervention; however, whether other parallel effects on the different pathways of AD pathogenesis by miR-138 cannot be excluded. Tian et al. revealed that miR-138 improves learning/memory via promoting autophagy in rats with cerebral ischemia/reperfusion injury [26]. MiR-138 is locally enriched at synaptic sites; Siegel et al. demonstrated that miR-138 is localized within dendrites and negatively regulates the morphogenesis of dendritic spines [13], indicating direct functional activity of miR-138 at the synapse.
CircRNAs have been considered to be upstream effectors of miRNAs via functioning as a miRNA sponge, and many circRNAs are enriched in the mammalian brain. Brain-enriched circRNAs are often derived from genes involved in synaptic and neuronal function [10, 16, 37]. ciRS-7 was the first circRNA identified in the brain, functioning as a miR-7 sponge. Loss of ciRS-7 also causes miRNA deregulation and affects brain function [10, 37]. CiRS-7 is decreased in the hippocampus of AD patients [38]. In our study, both circHDAC9 and ciRS-7 were reduced in Aβ-treated N2a cells and 4-month-old APP/PS1 mice but were unchanged in 2-month-old APP/PS1 mice. We also assessed the expression of another brain-enriched circRNA, circAGAP1, and found that it was unchanged both in vivo and in vitro. HDAC9 is known to be decreased in several regions of the AD brain [33]. Moreover, HDAC9 has been reported to regulate dendritic growth in cultured mouse cortical neurons, indicating its involvement in synaptic function [39]. These data suggest that circHDAC9 is specifically decreased in animal and cell models of AD. CircHDAC9 co-localized with miR-138 in the cytoplasm of N2a cells, and was negatively correlated with miR-138 in Aβ-treated N2a cells and 4-month-old APP/PS1 mice. CircHDAC9 acted as a miR-138 sponge, inhibiting miR-138 expression and reversing the abnormal APP processing induced by miR-138 in vitro. Our results imply that circHDAC9 serves as an upstream effector of miR-138 in the Aβ-induced pathogenic changes in AD.
In addition, circRNAs have been shown to serve as non-invasive diagnostic markers for several disorders, including atherosclerosis, degenerative diseases, and cancers. Finding biomarkers at an early stage is essential for the treatment of many complex diseases. [40]. MCI is a prodromal stage of AD; 10%–15% of patients with MCI tend to progress to AD per year [41]. CircHDAC9 was significantly lower in the serum of both MCI and AD patients than in their corresponding controls, indicating its potential for serving as an early marker for AD. However, studies with larger sample sizes are needed to test this.
In conclusion, our results demonstrate an essential role of the Aβ-circHDAC9-miR-138-Sirt1 signal pathway in the synaptic dysfunction, cognitive deficit, and abnormal APP processing in AD (Fig. 8C). Our study provides a novel early diagnostic marker and therapeutic target for AD.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81500925).
Conflict of interest
The authors have no conflict of interest to report.
References
- 1.Alzheimer’s A. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 2016;12:459–509. doi: 10.1016/j.jalz.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Sun BL, Li WW, Zhu C, Jin WS, Zeng F, Liu YH, et al. Clinical research on Alzheimer’s disease: progress and perspectives. Neurosci Bull. 2018;34:1111–1118. doi: 10.1007/s12264-018-0249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun W, Samimi H, Gamez M, Zare H, Frost B. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci. 2018;21:1038–1048. doi: 10.1038/s41593-018-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zusso M, Barbierato M, Facci L, Skaper SD, Giusti P. Neuroepigenetics and Alzheimer’s disease: an update. J Alzheimers Dis. 2018;64:671–688. doi: 10.3233/JAD-180259. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, et al. Identifying circRNA-associated-ceRNA networks in the hippocampus of Abeta1-42-induced Alzheimer’s disease-like rats using microarray analysis. Aging (Albany NY) 2018;10:775–788. doi: 10.18632/aging.101427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millan MJ. Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: An integrative review. Prog Neurobiol. 2017;156:1–68. doi: 10.1016/j.pneurobio.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Liu D, Huang HZ, Wang ZH, Hou TY, Yang X, et al. A Novel microRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in Alzheimer’s disease. Biol Psychiatry. 2018;83:395–405. doi: 10.1016/j.biopsych.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 8.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/JAD-2008-14103. [DOI] [PubMed] [Google Scholar]
- 12.Schroder J, Ansaloni S, Schilling M, Liu T, Radke J, Jaedicke M, et al. MicroRNA-138 is a potential regulator of memory performance in humans. Front Hum Neurosci. 2014;8:501. doi: 10.3389/fnhum.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Tan L, Lu Y, Peng J, Zhu Y, Zhang Y, et al. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 2015;589:726–729. doi: 10.1016/j.febslet.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Lu R, Wang J, Tao R, Wang J, Zhu T, Guo W, et al. Reduced TRPC6 mRNA levels in the blood cells of patients with Alzheimer’s disease and mild cognitive impairment. Mol Psychiatry. 2018;23:767–776. doi: 10.1038/mp.2017.136. [DOI] [PubMed] [Google Scholar]
- 16.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [DOI] [PMC free article] [PubMed]
- 18.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salta E, De Strooper B. microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer’s disease. FASEB J. 2017;31:424–433. doi: 10.1096/fj.201601308. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Veremeyko T, Wong AH, El Fatimy R, Wei Z, Cai W, et al. Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol Aging. 2017;51:156–166. doi: 10.1016/j.neurobiolaging.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Xie B, Liu Z, Jiang L, Liu W, Song M, Zhang Q, et al. Increased serum miR-206 level predicts conversion from amnestic mild cognitive impairment to Alzheimer’s disease: a 5-year follow-up study. J Alzheimers Dis. 2017;55:509–520. doi: 10.3233/JAD-160468. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Vijayan M, Reddy PH. MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer’s disease. Hum Mol Genet. 2017;26:3808–3822. doi: 10.1093/hmg/ddx267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon MN, Holcomb LA, Jantzen PT, DiCarlo G, Wilcock D, Boyett KW, et al. Time course of the development of Alzheimer-like pathology in the doubly transgenic PS1 + APP mouse. Exp Neurol. 2002;173:183–195. doi: 10.1006/exnr.2001.7754. [DOI] [PubMed] [Google Scholar]
- 24.Chen SQ, Cai Q, Shen YY, Wang PJ, Teng GJ, Zhang W, et al. Age-related changes in brain metabolites and cognitive function in APP/PS1 transgenic mice. Behav Brain Res. 2012;235:1–6. doi: 10.1016/j.bbr.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Xu YF, Zhang L, Huang L, Yu P, Zhu H, et al. Effective expression of Drebrin in hippocampus improves cognitive function and alleviates lesions of Alzheimer’s disease in APP (swe)/PS1 (DeltaE9) mice. CNS Neurosci Ther. 2017;23:590–604. doi: 10.1111/cns.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian F, Yuan C, Yue H. MiR-138/SIRT1 axis is implicated in impaired learning and memory abilities of cerebral ischemia/reperfusion injured rats. Exp Cell Res. 2018;367:232–240. doi: 10.1016/j.yexcr.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 27.Liu CM, Wang RY, Saijilafu, Jiao ZX, Zhang BY, Zhou FQ. MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 2013;27:1473–1483. doi: 10.1101/gad.209619.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J, Chen P, Xie W, Wu F. MicroRNA-138 inhibits cell proliferation in hepatocellular carcinoma by targeting Sirt1. Oncol Rep. 2017;38:1067–1074. doi: 10.3892/or.2017.5782. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z, Mo H, Mo L, He J, Wu Z, Lin X. Suppressive effect of microRNA-138 on the proliferation and invasion of osteosarcoma cells via targeting SIRT1. Exp Ther Med. 2017;13:3417–3423. doi: 10.3892/etm.2017.4426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Dong YT, Cao K, Tan LC, Wang XL, Qi XL, Xiao Y, et al. Stimulation of SIRT1 attenuates the level of oxidative stress in the brains of APP/PS1 double transgenic mice and in primary neurons exposed to oligomers of the amyloid-beta peptide. J Alzheimers Dis. 2018;63:283–301. doi: 10.3233/JAD-171020. [DOI] [PubMed] [Google Scholar]
- 31.Lutz MI, Milenkovic I, Regelsberger G, Kovacs GG. Distinct patterns of sirtuin expression during progression of Alzheimer’s disease. Neuromolecular Med. 2014;16:405–414. doi: 10.1007/s12017-014-8288-8. [DOI] [PubMed] [Google Scholar]
- 32.Kumar R, Chaterjee P, Sharma PK, Singh AK, Gupta A, Gill K, et al. Sirtuin1: a promising serum protein marker for early detection of Alzheimer’s disease. PLoS One. 2013;8:e61560. doi: 10.1371/journal.pone.0061560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Zhang DF, Luo R, Wu Y, Zhou H, Kong LL, et al. A systematic integrated analysis of brain expression profiles reveals YAP1 and other prioritized hub genes as important upstream regulators in Alzheimer’s disease. Alzheimers Dement. 2018;14:215–229. doi: 10.1016/j.jalz.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Ni H, Xu M, Zhan GL, Fan Y, Zhou H, Jiang HY, et al. The GWAS Risk Genes for Depression May Be Actively Involved in Alzheimer’s Disease. J Alzheimers Dis. 2018;64:1149–1161. doi: 10.3233/JAD-180276. [DOI] [PubMed] [Google Scholar]
- 35.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Rizzi L, Roriz-Cruz M. Sirtuin 1 and Alzheimer’s disease: An up-to-date review. Neuropeptides 2018. [DOI] [PubMed]
- 37.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357. [DOI] [PubMed]
- 38.Lukiw WJ, Circular RNA. circRNA) in Alzheimer’s disease (AD. Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugo N, Oshiro H, Takemura M, Kobayashi T, Kohno Y, Uesaka N, et al. Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur J Neurosci. 2010;31:1521–1532. doi: 10.1111/j.1460-9568.2010.07218.x. [DOI] [PubMed] [Google Scholar]
- 40.Lu D, Xu AD. Mini Review: Circular RNAs as Potential Clinical Biomarkers for Disorders in the Central Nervous System. Front Genet. 2016;7:53. doi: 10.3389/fgene.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grundman M, Petersen RC, Ferris SH, Thomas RG, Aisen PS, Bennett DA, et al. Mild cognitive impairment can be distinguished from Alzheimer disease and normal aging for clinical trials. Arch Neurol. 2004;61:59–66. doi: 10.1001/archneur.61.1.59. [DOI] [PubMed] [Google Scholar]